Abstract
As coronavirus disease 2019 (COVID-19) spreads across the world, the transfusion of efficient convalescent plasma (CP) to the most critical patients can be the primary approach to preventing the virus spread and treating the disease, and this strategy is considered as an intelligent computing concern. In providing an automated intelligent computing solution to select the appropriate CP for the most critical patients with COVID-19, two challenges aspects are bound to be faced: (1) distributed hospital management aspects (including scalability and management issues for prioritising COVID-19 patients and donors simultaneously), and (2) technical aspects (including the lack of COVID-19 dataset availability of patients and donors and an accurate matching process amongst them considering all blood types). Based on previous reports, no study has provided a solution for CP-transfusion-rescue intelligent framework during this pandemic that has addressed said challenges and issues. This study aimed to propose a novel CP-transfusion intelligent framework for rescuing COVID-19 patients across centralised/decentralised telemedicine hospitals based on the matching component process to provide an efficient CP from eligible donors to the most critical patients using multicriteria decision-making (MCDM) methods. A dataset, including COVID-19 patients/donors that have met the important criteria in the virology field, must be augmented to improve the developed framework. Four consecutive phases conclude the methodology. In the first phase, a new COVID-19 dataset is generated on the basis of medical-reference ranges by specialised experts in the virology field. The simulation data are classified into 80 patients and 80 donors on the basis of the five biomarker criteria with four blood types (i.e., A, B, AB, and O) and produced for COVID-19 case study. In the second phase, the identification scenario of patient/donor distributions across four centralised/decentralised telemedicine hospitals is identified ‘as a proof of concept’. In the third phase, three stages are conducted to develop a CP-transfusion-rescue framework. In the first stage, two decision matrices are adopted and developed on the basis of the five ‘serological/protein biomarker’ criteria for the prioritisation of patient/donor lists. In the second stage, MCDM techniques are analysed to adopt individual and group decision making based on integrated AHP-TOPSIS as suitable methods. In the third stage, the intelligent matching components amongst patients/donors are developed on the basis of four distinct rules. In the final phase, the guideline of the objective validation steps is reported. The intelligent framework implies the benefits and strength weights of biomarker criteria to the priority configuration results and can obtain efficient CPs for the most critical patients. The execution of matching components possesses the scalability and balancing presentation within centralised/decentralised hospitals. The objective validation results indicate that the ranking is valid.
Keywords: COVID-19, Serological Biomarkers/Protein, Convalescent Plasma, Prioritisation, Multi-criteria decision making, TOPSIS, AHP
Introduction
In December 2019, a cluster of patients with a novel coronavirus was identified in Wuhan, China. Initially named as 2019 novel coronavirus, the virus has now been named as SARS-CoV-2 by the International Committee of Taxonomy of Viruses [1–3]. This virus can cause the disease known as coronavirus disease 2019 (COVID-19) [4–7]. The COVID-19 pandemic has shocked the world for the first time in decades, resulting in an extraordinary impact on human life [8]. The number of patients worldwide increases consistently, and the number of patients closely infected follows an exponential trend [9]. Researchers from different countries have recently contributed to the application of different technologies that can help medical and healthcare providers stop this pandemic, such as the transfusion framework of convalescent plasma (CP) [10]. CP transfusion to COVID-19 patients, which is considered as one of the most successful protocols, is used in hospitals to treat this disease [11, 12]. Moreover, integration amongst hospitals in terms of the intelligent transfusion of CP across centralised/decentralised telemedicine architecture is necessary to help doctors in the rapid delivery of COVID-19 treatment [10]. For a clear view on how to support the hospital community in managing a CP-transfusion-rescue intelligent framework across the centralised/decentralised telemedicine architecture for the COVID-19 pandemic, five sequential questions are raised and answered as follows.
First question: ‘What is the importance of CP transfusion to COVID-19 patients’?
People who have recently recovered from the threat of deteriorating COVID-19 have antibodies to the coronavirus circulating in their blood [10]. Studies have reported that the virus can be eliminated by managing the healthcare quality of patients and providing them with protective antibodies from the blood of recovered patients via strong practice [13–15]. Thus, the transfusion of these antibodies to deteriorating patients can theoretically boost their immune system. Convalescent blood products (CBPs) are obtained by collecting plasma from a patient who has recovered from a viral or bacterial infection and has developed immunity against the pathogen causing the disease [16]. When transfused, CBPs can neutralise viruses and bacteria, thereby suppressing them in the blood [17]. Furthermore, the transfusion of CBPs from patients who recovered from COVID-19 can be the primary approach for preventing rapid virus spread and treating the disease [18]. For plasma=protein therapies, general safety measures have been established regarding plasma collection from donors. Patients treated with CP (donors) demonstrate shorter hospital stay and lower mortality than those not treated with CP; work is ongoing to test this theory on patients with COVID-19 [19]. Thus, the evaluation of suitability and efficacy of CP towards transfusion is important at this stage. Biologically, convalescent subjects must meet the donor-selection plasma criteria and must comply with the national health requirements and known standard routine procedures [10].
Second question: ‘How can suitability and efficacy of CP towards transfusion be evaluated and what is the key direction’?
Pooled plasma from recovered COVID-19 donors for anti-COVID-19 antibody therapy may undergo several general tests in two stages [10]. The first stage involves general plasma requirements. The second stage is considered as an evaluation of plasma suitability/efficacy by using protein biomarkers that indicate plasma safety/suitability. These biomarkers include PAO2/FIO2, C-reactive protein (CRP; mg/L), IL-6 (pg/mL; cytokines), albumin (g/L), and IgM (enzyme-linked immunosorbent assay [ELISA] titre). In these contexts, the mentioned biomarkers are the suitable CP criteria that can be utilised for transfusion from infected patients to recovered ones (donors). The procedure can ideally help strengthen the immunity of infected patients [20].
The key direction of the above-mentioned points is to select the best CP for the most critical patients with COVID-19 whilst considering the blood types. This process is considered as a problem of multicriteria decision-making (MCDM) and as an intelligent computing concern, which complies with the national health requirements and known standard routine procedures. Thus, an automated intelligent computing framework for selecting the suitable CP for the most critical patients with COVID-19 is proposed [10]. However, at present, many points have not been achieved yet.
Third question: ‘What is the criticism and gap analysis for academic literature that attempt to provide an automated intelligent computing solution to select the best CP for the most critical patients with COVID-19’?
Based on literature, one study has attempted to provide an automated intelligent computing solution as a rescue intelligent framework to select the best CP for the most critical patients with COVID-19 on the basis of the biological requirements using MCDM methods [10]. Two challenge aspects are considered. The first is related to distributed hospital-management issues, and the second is related to technical issues.
Regarding the first challenge aspect related to distributed hospital-management issues, hospitals’ capability may lack an accurate plan for transfusion management care particularly when this pandemic has affected a large scale of patients in different countries [21]. Moreover, major challenges face the health sector when hospitals lack CPs for critical patients, thereby increasing the complexity related to the entire transfusion process in the hospital’s community [22, 23]. Meanwhile, identifying an adequate number of blood donors for COVID-19 patients is difficult particularly because some blood types are almost rare [24]. The health providers during this pandemic still face serious aspects regarding distributed hospital management; for example, patients may be increased in a particular hospital but not in others [25]. This scenario is becoming common for hospital workflow when the demand for CPs increases as in COVID-19. Furthermore, the dataset for COVID-19 in literature either presents with limited number of patients/donors or lacks the use of sufficient biomarker criteria that affect the prioritisation process [26, 27]. COVID-19 samples are difficult to collect because of protection of patient privacy. Finally, the issue of fair management and efficient distribution of CPs amongst patients and donors regarding distributed hospitals simultaneously has not been considered [10]. Accordingly, the full picture of intelligent managing patients/donors with COVID-19 in terms of prioritisation with regard to connected hospitals simultaneously is not presented yet, and this aspect is considered as the primary distributed hospital-management issue.
In the shade of the second challenge linked with technical issues, two decision matrices (DMs) are proposed for the prioritisation of patients or donors based on five serological/protein biomarker criteria in a unique hospital. No results are produced because existing published works are insufficient to produce a satisfied patient/donor dataset considering the serological/protein biomarker criteria for dealing with this subject [28]. This aspect is considered as the first technical issue. Accordingly, the validation phase of the prioritisation results are not discussed in the presented methodology. Similar to the above-mentioned unavailability datasets, an intelligent matching process amongst critical patients has not presented suitable donors, and this aspect is considered as the second technical issue. Thus, providing a full solution to address the two above-mentioned challenge aspects and their issues is necessary.
Fourth question ‘What are the recommended solution for such challenge aspects and their issues’?
According to the first challenge of the distributed hospital-management issues, the use of telemedicine architecture is proposed to provide integration within hospitals to fight the COVID-19 pandemic. It is incorporated to optimise care whilst minimising exposures and viral transmission. The architecture of telemedicine is categorised into three tiers: Tier 1, Tier 2, and Tier 3 [29]. Tiers 1 and 2 are responsible for clients’ side. This architecture is a medical centre connected to distributed hospital servers. This architecture is also called centralised connected hospitals and is considered as the first recommended direction when no shared medical data resources are found amongst the countries [30–32], which can benefit countries during the COVID-19 pandemic by establishing a medical centre. For example, the ministry of health, which controls all hospitals either private or public, can customise the proposed framework [10] and share hospital-data resources to COVID-19 patients and donors. However, the second recommended direction is to determine whether shared medical-data resources can be found amongst the countries. This process can benefit countries through the blockchain technology. This proposed technology can eliminate the third party of centralised phenomena with regard to authentication and adapt to the telemedicine architecture, namely, decentralised connected hospitals [33]. Blockchain technology maintains a continuous update of all transactions occurring across distributed hospital networks in COVID-19 patients and donors.
Based on the above-mentioned discussion contexts on centralised or decentralised telemedicine, a new intelligent healthcare framework must be connected with several hospitals to boost the availability of service, share medical resources, and evade acute shortage of CPs between patients and donors to help doctors hasten COVID-19 treatment. Therefore, the distributed hospital-management issues can be addressed. The current scenario of the hospital interoperability for both architectures with regard to the status of the current pandemic is presented in Fig. 1.
Fig. 1.
Conceptual diagram for hospital interoperability in centralised/decentralised telemedicine
As shown in Fig. 1, the management system within each hospital admits patients with COVID-19 whose health severity differs amongst one another. Three levels are considered for the infected patients: mild, severe, and critical [34]. Moreover, the donors are admitted to the hospitals for the donation process where convalescent subjects must meet donor selection plasma criteria and comply with the national health requirements and known standard routine procedures. Thus, the transfusion of the best CP to the most critical patients with COVID-19 based on serological/protein biomarker measures for all blood types is required, considering that this scenario must be accomplished amongst the connected hospitals to avoid acute plasma shortages or an increase in the number of patients in a particular hospital.
Regarding the second challenge aspect that inlinks with technical issues, a simulation data of 80 patients and 80 donors based on the five biomarker criteria with four blood types (i.e., A, B, AB and O) are produced for the first time for COVID-19 case study. The new dataset is generated on the basis of reliable reference ranges and expert-validated occurrence records in the respiratory field with more than 10 years of experience to include different health conditions. Based on these new datasets, the outcome of prioritisation configuration results is used in the transfusion of CPs through a new matching component guideline between patients and donors’ CPs considering the four blood types. Thus, the technical issues can be addressed.
From the above-mentioned points, the development of a rescue interoperability intelligent framework across telemedicine architecture in centralised or decentralised hospital connections for prioritisation of patients and donors based on the generation datasets can provide a complete solution. A scalable management framework can withstand the CP load amongst connected hospitals, and the proper donor can be matched with compatible patients to improve balance control between patients and donors. In these contexts, balancing a huge number of patients/donors to avoid an acute shortage of CPs can be accomplished. If this framework is appropriately developed, then it would exhibit the potential to save more lives. One way of achieving this aim is to develop a rescue framework that achieves the transfusion approach of CPs. Moreover, the matching process must be considered to enable balance across distributed hospitals. Thus, a prioritisation methodology is often conducted to ensure that CP is given in an appropriate and timely manner [35]. Therefore, for a sustainable health system and best care, improvements must be made to satisfy current requirements, particularly the need to present an interoperability rescue intelligent framework to manage the transfusion of best CPs between patients and donors with COVID-19 across centralised and decentralised connected hospitals. This intelligent framework must be able to integrate the work process of the prioritisation of patients and donors amongst these hospitals simultaneously.
Fifth question: ‘What is the contribution, novelty, and implication of the present study’?
This study has proposed a novel CP-transfusion-rescue intelligent framework across centralised/decentralised telemedicine hospitals on the basis of the matching component process to provide an efficient CP from eligible donors to the most critical patients by using the integrated AHP-TOPSIS methods. A dataset of COVID-19 patients/donors that met the important criteria in the virology field must be augmented to improve the developed intelligent framework. The proposed intelligent framework can improve balancing and scalability across telemedicine hospitals between patients and donors simultaneously.
Methodology
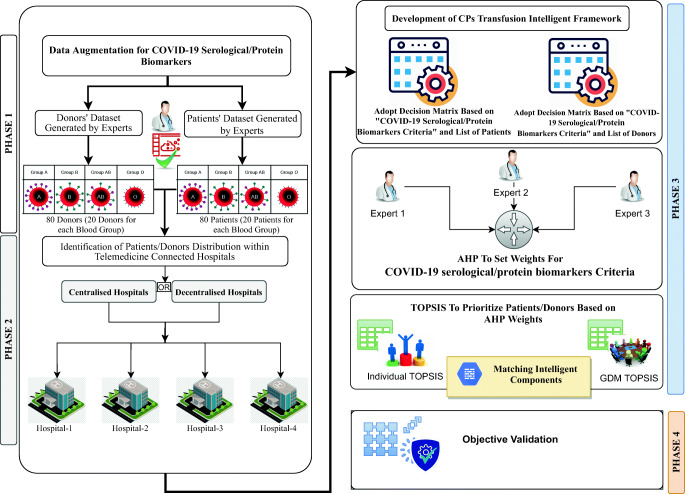
The development methodology of the proposed CP-transfusion-rescue intelligent framework is divided into four sequence phases (i.e., data augmentation [DA] for patients/donors; identification of patients/donors distribution within telemedicine hospitals; development and presentation of CP-transfusion-rescue intelligent framework for COVID-19, including three stages; and objective validation of the constructed results). Figure 2 shows the structure of the research-methodology phases.
Fig. 2.
Methodology phases for the CP-transfusion-rescue intelligent framework
Phase 1: DA
The augmentation of COVID-19 patient/donor datasets based on serological/protein biomarkers is accomplished in this section. Experts are needed to generate reliable clinical datasets to annotate labels. Given the complexity of the biomarker medical data, a COVID-19 medical dataset, whose labels are completely reliable, is unavailable [36]. For these challenges, DA can be used to generate dummy data to help prioritise patients/donors with COVID-19. An expert in the virology field with more than 10 years of experience provides a subjective judgment and generates an augmented dataset on the basis of medical-reference ranges (Tables 10 and 11 in the Appendix) to reduce this gap. These tables also present the reference ranges that serve as an indicator to identify the emergency health levels for the patients. A sample of first patient and donor based on each blood type from the augmented dataset is presented in Table 1. The specifications of the dataset are as follows.
The dataset includes 80 patients and 80 donors, as well as four blood types (i.e., A, B, AB, and O).
Patient/donor clinical data measurements are generated according to five biomarker measurements (i.e., PAO2/FIO2, CRP (mg/L), IL-6 (pg/mL; cytokines), albumin (g/L), and IgM (ELISA titre).
Biomarker measurements for the generated data are varied with regard to the health emergency level (mild, moderate, or severe) based on medical perspective and depend on reliable biomarker-reference ranges.
Table 10.
Augmented dataset for patients
| Patients | Serological/Protein Biomarkers Measurements | ||||
|---|---|---|---|---|---|
| PAO2/FIO2 >300 | C-reactive protein, mg/L (<8) | IL-6, pg/mL (Cytokines) (normal range, 0-7) | Albumin (40–55) g/l | IgM ELISA titer (<200)titers (<200) | |
| Blood group A | |||||
| P1_A | 128 | 93 | 244 | 21 | 94.09 |
| P2_A | 128 | 88 | 294 | 26 | 237.65 |
| P3_A | 146 | 59 | 197 | 25 | 265.78 |
| P4_A | 143 | 124 | 99 | 11 | 181.39 |
| P5_A | 201 | 56 | 277 | 19 | 298.76 |
| P6_A | 267 | 84 | 291 | 19 | 90.21 |
| P7_A | 214 | 110 | 125 | 34 | 227.95 |
| P8_A | 262 | 80 | 344 | 24 | 260.93 |
| P9_A | 183 | 115 | 318 | 11 | 188.18 |
| P10_A | 131 | 138 | 285 | 17 | 246.38 |
| P11_A | 233 | 117 | 292 | 19 | 159.08 |
| P12_A | 189 | 99 | 79 | 18 | 139.68 |
| P13_A | 134 | 135 | 264 | 23 | 257.05 |
| P14_A | 106 | 127 | 78 | 13 | 173.63 |
| P15_A | 215 | 60 | 358 | 22 | 265.78 |
| P16_A | 203 | 55 | 104 | 27 | 297.79 |
| P17_A | 108 | 95 | 332 | 26 | 198.85 |
| P18_A | 121 | 100.98 | 115 | 13 | 142.59 |
| P19_A | 200 | 111.18 | 170 | 16 | 127.07 |
| P20_A | 211 | 154.02 | 280 | 19 | 258.02 |
| Blood group B | |||||
| P1_B | 136 | 83.64 | 168 | 24 | 154 |
| P2_B | 113 | 68.34 | 162 | 22 | 165 |
| P3_B | 103 | 63.24 | 151 | 28 | 264 |
| P4_B | 258 | 133.62 | 270 | 27 | 141 |
| P5_B | 131 | 55.08 | 142 | 29 | 136 |
| P6_B | 115 | 125.46 | 135 | 26 | 212 |
| P7_B | 148 | 117.3 | 139 | 13 | 247 |
| P8_B | 159 | 99.96 | 292 | 28 | 288 |
| P9_B | 111 | 120.36 | 123 | 12 | 270 |
| P10_B | 147 | 125.46 | 231 | 17 | 241 |
| P11_B | 174 | 105.06 | 141 | 14 | 111 |
| P12_B | 263 | 84.66 | 197 | 13 | 236 |
| P13_B | 276 | 160.14 | 211 | 10 | 174 |
| P14_B | 118 | 136.68 | 155 | 13 | 240 |
| P15_B | 201 | 104.04 | 180 | 25 | 222 |
| P16_B | 211 | 56.1 | 242 | 19 | 148 |
| P17_B | 284 | 102 | 299 | 10 | 183 |
| P18_B | 118 | 162.18 | 286 | 29 | 288 |
| P19_B | 139 | 61.2 | 188 | 10 | 153 |
| P20_B | 159 | 143.82 | 260 | 26 | 270 |
| Blood group AB | |||||
| P1_AB | 167 | 69.36 | 216 | 29 | 199 |
| P2_AB | 183 | 100.98 | 267 | 12 | 262 |
| P3_AB | 288 | 98.94 | 72 | 27 | 186 |
| P4_AB | 129 | 63.24 | 186 | 28 | 133 |
| P5_AB | 112 | 124.44 | 173 | 31 | 248 |
| P6_AB | 280 | 116.28 | 167 | 17 | 268 |
| P7_AB | 266 | 122.4 | 158 | 20 | 207 |
| P8_AB | 208 | 112.2 | 79 | 27 | 206 |
| P9_AB | 212 | 74.46 | 166 | 28 | 129 |
| P10_AB | 138 | 102 | 274 | 18 | 145 |
| P11_AB | 234 | 88.74 | 191 | 10 | 240 |
| P12_AB | 259 | 130.56 | 168 | 24 | 163 |
| P13_AB | 225 | 98.94 | 128 | 29 | 255 |
| P14_AB | 109 | 129.54 | 129 | 25 | 269 |
| P15_AB | 183 | 123.42 | 190 | 21 | 174 |
| P16_AB | 141 | 66.3 | 269 | 12 | 199 |
| P17_AB | 143 | 67.32 | 134 | 16 | 246 |
| P18_AB | 280 | 124.44 | 258 | 16 | 90 |
| P19_AB | 261 | 61.2 | 81 | 26 | 137 |
| P20_AB | 207 | 55.08 | 77 | 15 | 198 |
| Blood group O | |||||
| P1_O | 182 | 141.78 | 78 | 31 | 263 |
| P2_O | 158 | 54.06 | 282 | 21 | 94 |
| P3_O | 206 | 55.08 | 233 | 22 | 150 |
| P4_O | 232 | 88.74 | 90 | 23 | 271 |
| P5_O | 188 | 78.54 | 267 | 17 | 285 |
| P6_O | 260 | 158.1 | 214 | 21 | 220 |
| P7_O | 272 | 94.86 | 93 | 25 | 202 |
| P8_O | 196 | 82.62 | 77 | 16 | 100 |
| P9_O | 159 | 142.8 | 245 | 16 | 176 |
| P10_O | 218 | 79.56 | 191 | 22 | 201 |
| P11_O | 158 | 55.08 | 258 | 26 | 188 |
| P12_O | 192 | 144.84 | 194 | 19 | 185 |
| P13_O | 105 | 92.82 | 124 | 22 | 149 |
| P14_O | 282 | 145.86 | 90 | 11 | 259 |
| P15_O | 285 | 82.62 | 151 | 27 | 208 |
| P16_O | 261 | 80.58 | 298 | 26 | 285 |
| P17_O | 250 | 79.56 | 149 | 19 | 268 |
| P18_O | 171 | 169.32 | 243 | 13 | 211 |
| P19_O | 171 | 57.12 | 294 | 28 | 208 |
| P20_O | 268 | 132.6 | 232 | 11 | 228 |
Table 11.
Augmented dataset for donors
| Donors | Serological/Protein Biomarkers Measurements | ||||
|---|---|---|---|---|---|
| PAO2/FIO2 >300 | C-reactive protein, mg/L (<8) | IL-6, pg/mL (Cytokines) (normal range, 0-7) | Albumin (40–55) g/l | IgM ELISA titer (<200) | |
| Blood group A | |||||
| D1_A | 453 | 1.3 | 1.4 | 41.6 | 64.99 |
| D2_A | 524 | 2 | 2.1 | 50.8 | 51.41 |
| D3_A | 301 | 0.54 | 4.1 | 43.8 | 44.62 |
| D4_A | 331 | 5.4 | 5.5 | 38.9 | 43.65 |
| D5_A | 450 | 4.41 | 2 | 54.7 | 34.92 |
| D6_A | 347 | 3.3 | 1.22 | 50 | 66.93 |
| D7_A | 541 | 1.2 | 2.5 | 47.5 | 56.26 |
| D8_A | 380 | 1.3 | 0.8 | 43.9 | 39.77 |
| D9_A | 471 | 4.7 | 3.6 | 45.7 | 44.62 |
| D10_A | 318 | 1.8 | 2.22 | 45.45 | 52.38 |
| D11_A | 304 | 5.88 | 0.9 | 40.4 | 32.98 |
| D12_A | 341 | 2.94 | 3.5 | 49.49 | 33.95 |
| D13_A | 462 | 2.94 | 3.5 | 42.42 | 59.17 |
| D14_A | 326 | 1.96 | 0.9 | 50.5 | 32.01 |
| D15_A | 369 | 5.88 | 2.6 | 48.48 | 51.41 |
| D16_A | 495 | 6.86 | 3.5 | 51.51 | 41.71 |
| D17_A | 456 | 3.92 | 0.9 | 53.53 | 37.83 |
| D18_A | 342 | 6.86 | 4.4 | 54.54 | 57.23 |
| D19_A | 307 | 6.86 | 4.4 | 49.49 | 40.74 |
| D20_A | 335 | 0.98 | 5.2 | 55.55 | 55.29 |
| Blood group B | |||||
| D1_B | 425 | 3.96 | 1.98 | 44.44 | 32.01 |
| D2_B | 301 | 3.96 | 5.94 | 44.44 | 57.23 |
| D3_B | 347 | 6.93 | 3.96 | 45.45 | 33.95 |
| D4_B | 420 | 1.98 | 2.97 | 53.53 | 38.8 |
| D5_B | 318 | 2.97 | 2.97 | 44.44 | 66.93 |
| D6_B | 399 | 3.96 | 4.95 | 49.49 | 54.32 |
| D7_B | 340 | 1.98 | 4.95 | 40.4 | 55.29 |
| D8_B | 358 | 2.97 | 1.98 | 53.53 | 33.95 |
| D9_B | 307 | 6.93 | 2.97 | 43.43 | 44.62 |
| D10_B | 394 | 5.94 | 5.94 | 54.54 | 44.62 |
| D11_B | 326 | 0.99 | 6.93 | 46.46 | 56.26 |
| D12_B | 334 | 3.96 | 3.96 | 43.43 | 32.01 |
| D13_B | 423 | 3.96 | 5.94 | 49.49 | 37.83 |
| D14_B | 485 | 5.94 | 2.97 | 51.51 | 47.53 |
| D15_B | 352 | 0.99 | 4.95 | 43.43 | 56.26 |
| D16_B | 304 | 0.99 | 4.95 | 40.4 | 60.14 |
| D17_B | 396 | 3.96 | 6.93 | 45.45 | 47.53 |
| D18_B | 387 | 3.96 | 0.99 | 54.54 | 32.98 |
| D19_B | 403 | 6.93 | 4.95 | 50.5 | 62.08 |
| D20_B | 358 | 0.99 | 4.95 | 40.4 | 44.62 |
| Blood group AB | |||||
| D1_AB | 449 | 2.97 | 4.95 | 47.47 | 37.83 |
| D2_AB | 462 | 5.94 | 4.95 | 44.44 | 50.44 |
| D3_AB | 478 | 0.99 | 3.96 | 49.49 | 62.08 |
| D4_AB | 375 | 3.96 | 6.93 | 50.5 | 44.62 |
| D5_AB | 418 | 5.94 | 3.96 | 42.42 | 33.95 |
| D6_AB | 403 | 6.93 | 5.94 | 43.43 | 51.41 |
| D7_AB | 457 | 5.94 | 4.95 | 49.49 | 35.89 |
| D8_AB | 325 | 0.99 | 3.96 | 48.48 | 32.01 |
| D9_AB | 486 | 5.94 | 3.96 | 41.41 | 43.65 |
| D10_AB | 391 | 5.94 | 6.93 | 50.5 | 54.32 |
| D11_AB | 314 | 2.97 | 2.97 | 48.48 | 54.32 |
| D12_AB | 458 | 6.93 | 1.98 | 44.44 | 41.71 |
| D13_AB | 366 | 2.97 | 6.93 | 45.45 | 42.68 |
| D14_AB | 346 | 2.97 | 6.93 | 50.5 | 67.9 |
| D15_AB | 368 | 4.95 | 3.96 | 54.54 | 65.96 |
| D16_AB | 318 | 0.99 | 5.94 | 53.53 | 52.38 |
| D17_AB | 382 | 5.94 | 1.98 | 51.51 | 64.99 |
| D18_AB | 407 | 2.97 | 6.93 | 41.41 | 58.2 |
| D19_AB | 370 | 6.93 | 2.97 | 49.49 | 62.08 |
| D20_AB | 417 | 0.99 | 0.99 | 50.5 | 58.2 |
| Blood group O | |||||
| D1_O | 445 | 5.94 | 3.96 | 55.55 | 35.89 |
| D2_O | 419 | 6.93 | 4.95 | 47.47 | 42.68 |
| D3_O | 417 | 1.98 | 2.97 | 40.4 | 44.62 |
| D4_O | 460 | 1.98 | 6.93 | 45.45 | 45.59 |
| D5_O | 330 | 2.97 | 6.93 | 48.48 | 41.71 |
| D6_O | 358 | 2.97 | 0.99 | 54.54 | 45.59 |
| D7_O | 458 | 2.97 | 6.93 | 55.55 | 35.89 |
| D8_O | 483 | 1.98 | 2.97 | 54.54 | 46.56 |
| D9_O | 449 | 1.98 | 2.97 | 47.47 | 64.02 |
| D10_O | 457 | 5.94 | 3.96 | 40.4 | 43.65 |
| D11_O | 486 | 4.95 | 5.94 | 55.55 | 66.93 |
| D12_O | 434 | 1.98 | 6.93 | 41.41 | 36.86 |
| D13_O | 399 | 6.93 | 4.95 | 44.44 | 40.74 |
| D14_O | 382 | 4.95 | 6.93 | 52.52 | 29.1 |
| D15_O | 478 | 3.96 | 1.98 | 45.45 | 61.11 |
| D16_O | 444 | 1.98 | 2.97 | 52.52 | 64.02 |
| D17_O | 398 | 2.97 | 6.93 | 43.43 | 64.02 |
| D18_O | 329 | 4.95 | 6.93 | 46.46 | 59.17 |
| D19_O | 437 | 4.95 | 5.94 | 49.49 | 32.98 |
| D20_O | 471 | 2.97 | 4.95 | 42.42 | 66.93 |
Table 1.
Patient and donor samples from the augmented datasets
| PAO2/FIO2 >300 | C-reactive protein, mg/L (<8) | IL-6, pg/mL (cytokines) (normal range, 0–7) | Albumin (40–55) g/L | IgM ELISA titre (<200) titres (<200) | |
|---|---|---|---|---|---|
| Patients | Serological/Protein Biomarker Measurements | ||||
| P1_A | 128 | 93 | 244 | 21 | 94.09 |
| P1_B | 136 | 83.64 | 168 | 24 | 154 |
| P1_AB | 167 | 69.36 | 216 | 29 | 199 |
| P1_O | 182 | 141.78 | 78 | 31 | 263 |
| Donors | Serological/Protein Biomarker Measurements | ||||
| D1_A | 453 | 1.3 | 1.4 | 41.6 | 64.99 |
| D1_B | 425 | 3.96 | 1.98 | 44.44 | 32.01 |
| D1_AB | 449 | 2.97 | 4.95 | 47.47 | 37.83 |
| D1_O | 445 | 5.94 | 3.96 | 55.55 | 35.89 |
A brief description for each biomarker is illustrated as follows.
PAO2/FIO2 ratio is defined as the ratio of the partial pressure of arterial oxygen to the percentage of inspired oxygen [35], and its reference range must be between 100 and 300.
CRP is a serum amyloid P component belonging to the pentraxin family of calcium-dependent ligand-binding proteins. It serves as a marker of inflammation and ranges between 8 and 250. SARS-CoV-2 seems to increase the CRP levels significantly because of inflammatory reaction, and related tissue destruction was also observed in 2002 in the SARS epidemic. High concentrations indicate a severe disease linked to lung damage and poor prognosis [37].
IL-6 (pg/mL; cytokines) is released by T cells and activated macrophages during the acute-phase response following injury or trauma and may lead to inflammation or infection; it should be between 6 and 300. IL-6 has pro- and anti-inflammatory properties [38].
Albumin is an essential binding and transport protein for various substances in plasma and maintains the osmotic pressure of blood [39]. The reference range is between 5 and 55.
ELISA is used to detect immunoglobulin M (IgM) and IgG antibodies against capsular and O antigens of Haemophilus influenzae. It ranges between 100 and 800.
For further discussion on the augmented data, the P1_A (for example) indicates that this patient is the first augmented one and his blood type is A. Furthermore, the measurements of his biomarker criteria are explained. A total of 20 patients and 20 donors are identified for each blood type.
Phase 2: Identification of patient/donor distribution within telemedicine hospitals
This study adopts four hospitals as ‘a proof of concept’ to represent the managing of patients and donors. Our identification phase proposes that the first hospital has admitted a large scale of patients (40 patients) and a small number of donors (only eight donors) to test the proposed CP-transfusion-rescue intelligent framework. The second hospital has admitted 20 patients, and the number of available donors is 12. The third hospital has admitted 12 patients, and the number of available donors is 20. Finally, the fourth hospital has admitted eight patients and a large number of available donors (40). The scenario of identification of all patients and donors within the four hospitals are shown in Table 2.
Table 2.
Identification scenario of patient/donor distribution within the four hospitals
| Hospital-1 Distribution | Hospital-2 Distribution | Hospital-3 Distribution | Hospital-4 Distribution | |||||
|---|---|---|---|---|---|---|---|---|
| Blood Type | Admitted Patients | Available Donors | Admitted Patients | Available Donors | Admitted Patients | Available Donors | Admitted Patients | Available Donors |
| Blood group A | P1_A | D1_A | P11_A | D3_A | P16_A | D6_A | P19_A | D11_A |
| P2_A | D2_A | P12_A | D4_A | P17_A | D7_A | P20_A | D12_A | |
| P3_A | P13_A | D5_A | P18_A | D8_A | D13_A | |||
| P4_A | P14_A | D9_A | D14_A | |||||
| P5_A | P15_A | D10_A | D15_A | |||||
| P6_A | D16_A | |||||||
| P7_A | D17_A | |||||||
| P8_A | D18_A | |||||||
| P9_A | D19_A | |||||||
| P10_A | D20_A | |||||||
| Blood group B | P1_B | D1_B | P11_B | D3_B | P16_B | D6_B | P19_B | D11_B |
| P2_B | D2_B | P12_B | D4_B | P17_B | D7_B | P20_B | D12_B | |
| P3_B | P13_B | D5_B | P18_B | D8_B | D13_B | |||
| P4_B | P14_B | D9_B | D14_B | |||||
| P5_B | P15_B | D10_B | D15_B | |||||
| P6_B | D16_B | |||||||
| P7_B | D17_B | |||||||
| P8_B | D18_B | |||||||
| P9_B | D19_B | |||||||
| P10_B | D20_B | |||||||
| Blood group AB | P1_AB | D1_AB | P11_AB | D3_AB | P16_AB | D6_AB | P19_AB | D11_AB |
| P2_AB | D2_AB | P12_AB | D4_AB | P17_AB | D7_AB | P20_AB | D12_AB | |
| P3_AB | P13_AB | D5_AB | P18_AB | D8_AB | D13_AB | |||
| P4_AB | P14_AB | D9_AB | D14_AB | |||||
| P5_AB | P15_AB | D10_AB | D15_AB | |||||
| P6_AB | D16_AB | |||||||
| P7_AB | D17_AB | |||||||
| P8_AB | D18_AB | |||||||
| P9_AB | D19_AB | |||||||
| P10_AB | D20_AB | |||||||
| Blood group O | P1_O | D1_O | P11_O | D3_O | P16_O | D6_O | P19_O | D11_O |
| P2_O | D2_O | P12_O | D4_O | P17_O | D7_O | P20_O | D12_O | |
| P3_O | P13_O | D5_O | P18_O | D8_O | D13_O | |||
| P4_O | P14_O | D9_O | D14_O | |||||
| P5_O | P15_O | D10_O | D15_O | |||||
| P6_O | D16_O | |||||||
| P7_O | D17_O | |||||||
| P8_O | D18_O | |||||||
| P9_O | D19_O | |||||||
| P10_O | D20_O | |||||||
As shown in Table 2, the number of patients and donors varies across hospitals. This variety is important to test the proposed CP-transfusion-rescue intelligent framework when the hospital has an inverse relationship with regard to distribution between patients and donors either in centralised or decentralised telemedicine connections. In these contexts, any hospital that lacks donors and admits a large number of patients can be tested and vice versa. Thus, the development of the CP-transfusion intelligent framework is needed as presented in the next phase.
Phase 3: Development of the CP-transfusion intelligent framework
This phase includes a three-stage development process as illustrated in Fig. 3. The process can be achieved in either centralised or decentralised telemedicine workflow architecture in the same processes.
Two DMs for the prioritisation of patients and donors are adopted from a previous work [10]. The first DM is for the prioritisation of admitted patients across the four identified hospitals simultaneously in either centralised or decentralised telemedicine workflow architecture. Therefore, any patient in any hospital must be compared and evaluated with all other patients admitted in other hospitals. The second DM can prioritise all donors in the same context.
The best MCDM techniques for the adopted DMs are analysed and selected for handling the prioritisation configurations. In this stage, the evaluation and prioritisation of patients and donors based on the five biomarker criteria are achieved.
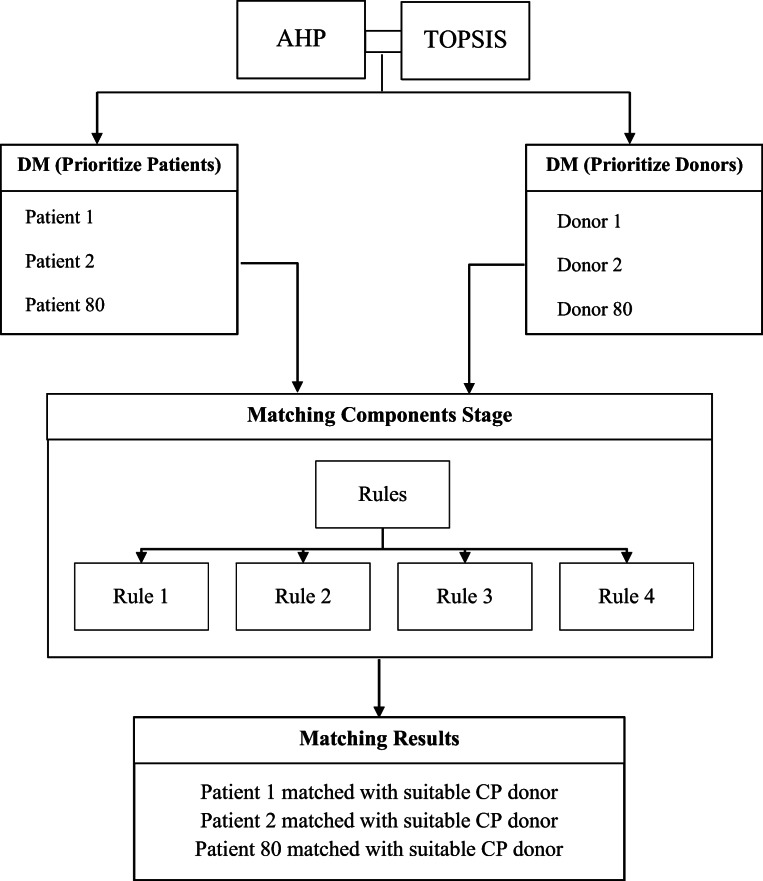
The findings of the prioritisation results from the previous stages are operated with the matching component stage. The developed stage has identified four rules to complete the intelligent-transfusion process between patients and donors.
Fig. 3.
CP-transfusion framework stages
Adopted DMs for the prioritisation of patients/donors
Both DMs are demonstrated in Table 3.
Table 3.
Prioritisation DM for patients and donors
| Serological/Protein Biomarker Criteria | C1 | C2 | C3 | C4 | C5 | |
| Patient Identification Information | ||||||
| Patients | Hospital Number | |||||
| Patient1 | H1 or H2 or H3 or H4 | C1-P1 | C2- P1 | C3-P1 | C4-P1 | C5-P1 |
| Patient2 | H1 or H2 or H3 or H4 | C1-P2 | C2-P2 | C3-P2 | C4-P2 | C5-P2 |
| Patient3 | H1 or H2 or H3 or H4 | C1-P3 | C2-P3 | C3-P3 | C4-P3 | C5-P3 |
| Patient n | H1 or H2 or H3 or H4 | C1-P80 | C2-P80 | C3-P80 | C4-P80 | C5-P80 |
| Serological/Protein Biomarker Criteria | C 1 | C 2 | C 3 | C 4 | C 5 | |
| Donor Identification Information | ||||||
| Donors | Hospital Number | |||||
| Donor1 | H1 or H2 or H3 or H4 | C1-D1 | C2- D1 | C3-D1 | C4-D1 | C5-D1 |
| Donor2 | H1 or H2 or H3 or H4 | C1-D2 | C2-D2 | C3-D2 | C4-D2 | C5-D2 |
| Donor3 | H1 or H2 or H3 or H4 | C1-D3 | C2-D3 | C3-D3 | C4-D3 | C5-D3 |
| Donor n | H1 or H2 or H3 or H4 | C1-D80 | C2-D80 | C3-D80 | C4-D80 | C5-D80 |
C 1= PAO2/FIO2 >300, C 2= C-reactive protein, mg/L (<8), C 3 = IL-6, pg/mL (Cytokines; normal range, 0–7), C4 = Albumin (40–55) g/L, C5 = IgM ELISA titre (<200), P = Patient, D= Donor, H= Hospital
The adopted DM for patients is constructed on the basis of the intersection between ‘serological/protein biomarker criteria’ and ‘COVID-19 infected patient list’. Furthermore, the DM for donors is constructed on the basis of the intersection between ‘serological/protein biomarker criteria’ and ‘COVID-19 donor list’. However, according to the specific problems of the management of COVID-19 patients/donors, prioritisation is achieved through the integration of decision-making methods to considerably reducing the problem complexity.
Adopted MCDM techniques
The recommended solution for our study is to use MCDM that deals with decision problems with regard to the decision criteria. MCDM has the potential to contribute to a fair, transparent, and rational priority-setting process [40–50]. Prioritisation is considered challenging for different kinds of medical perspectives [51–61]. With regard to the adopted DMs, a previous work [10] has suggested the use of the SODOSM method in handling prioritisation. However, the SODOSM method is conducted with regard to the idle solution amongst each criterion within the CP DM. The ideal solution is an alternative for specific criteria [62], and this concept cannot be applied to the COVID-19 case study. The problem in identifying the ideal solution with regard to the reference range for the COVID-19 serological/protein biomarkers has not been detected and recognised [34]. Thus, the use of existing MCDM methods is recommended in the present study.
The newest trend regarding the use of MCDM methods is to combine two or more methods to recoup the weaknesses of a single method [63–73]. AHP and TOPSIS have become a commonly integrated MCDM method [74–76]. One MCDM methodology to address the above-mentioned issues is to apply and require high-level stages of patients’ data.
Integrated AHP-TOPSOS
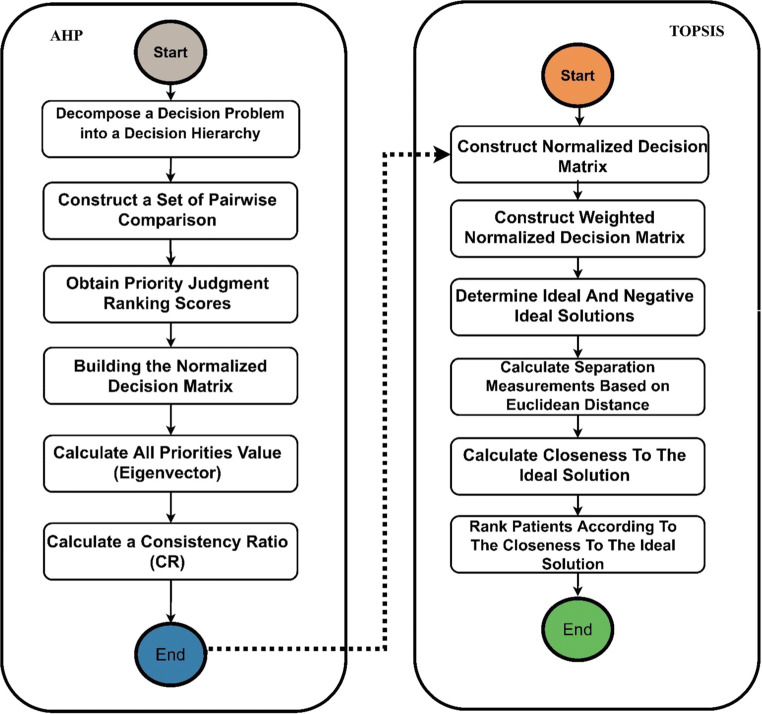
This subsection describes the integration of both methods. Several steps are implemented to assign proper weights to the serological/protein biomarker criteria by using the AHP method together with the TOPSIS method for the prioritisation of patients/donors. The integrated AHP and TOPSIS steps are shown in Fig. 4.
Fig. 4.
Integrated AHP-TOPSIS model for prioritisation using multicriteria decision-making
AHP for setting weights for COVID-19 serological/protein biomarker criteria
This section describes in detail the weighting attributes and proposes a precise approach for setting subjective weights to the COVID-19 serological/protein biomarker criteria for patients and donors on the basis of the AHP method. This section also aims to investigate the effective criteria for such investigation for patients and donors. The procedure of the AHP method is represented by the following steps [77–81].
A. Decomposition of a Decision Problem into a Decision Hierarchy
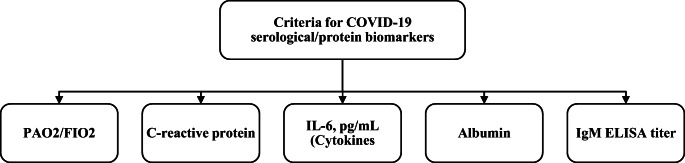
Problem modelling as a hierarchy consists of the decision goal that must be designed for the criteria in AHP. Figure 5 illustrates the hierarchy of the criteria used in the AHP pairwise comparison for serological/protein biomarkers to obtain criterion weights. The top of the hierarchy represents the goal, which is achieved by the eight criteria. Pairwise comparison must be performed amongst all criteria.
B. Construction of Pairwise Comparison Matrix
Fig. 5.
Hierarchy of AHP for the serological/protein biomarker COVID-19 criteria
AHP can build a pairwise comparison matrix to establish a decision:
| 1 |
Elements Xij are obtained from Fig. 5. The comparisons (relative importance) of each criterion are measured according to a numerical scale from 1 to 9 [82, 83]. Table 4 illustrates the relative scales (1–9) used to show each expert’s judgments for each comparison. Experts must critically set these judgments on the basis of their experience and knowledge.
C. Obtaining Priority-Judgment Ranking Scores
Table 4.
Nine scales of pairwise comparisons
| Intensity of Importance | Definition | Explanation |
|---|---|---|
| 1 | Equal importance | Two activities contribute equally to the objective |
| 3 | Weak importance of one over another | Experience and judgment slightly favour one activity over another |
| 5 | Essential or strong importance | Experience and judgment strongly favour one activity over another |
| 7 | Demonstrated importance | Activity is strongly favoured and its dominance is demonstrated in practice |
| 9 | Absolute importance | The evidence favouring one activity over another is of the highest possible order of affirmation |
| 2,4,6,8 | Intermediate values between the two adjacent judgments | When compromise is needed |
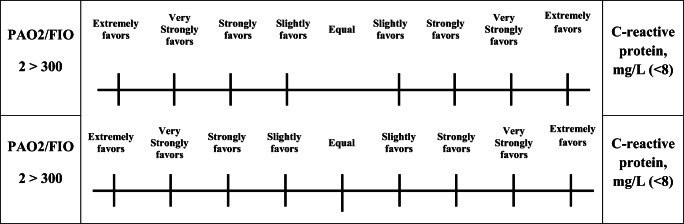
A pairwise comparison questionnaire was designed and distributed to a geographically diverse convenience sample of experts with expertise in respiratory diseases. The experts were asked to show their judgments and the relative importance for all criteria by using the nine scales for comparison. Figure 6 presents a sample of the criteria for pairwise comparisons in the evaluation form distributed amongst the experts.
Fig. 6.
Sample evaluation form
The number of required pairwise comparisons is n × (n − 1)/2, where n is the number of criteria used during evaluation. At this stage, AHP extracts the weight of importance of all serological/protein biomarker COVID-19 criteria from the pairwise comparison by user preferences and judgments from the decision-making team. ‘AHP is technically valid and does not require a large sample size’ [84]. Hence, in this research, three experts with more than 10 years of experience are selected to show their preferences and judgments. Three copies of the evaluation forms are revised by the experts, achieving a total of 10 comparisons by each expert. All comparisons for all criteria are made at this point.
D. Construction of Normalised DM
Every element of matrix A is normalised by dividing each element in a column by the sum of the elements in the same column to create a normalised pairwise comparison matrix Anorm. Anorm is the normalised matrix of A(1), where A(xij) is given by Eq. (2). Anorm (aij) is expressed as follows:
| 2 |
| 3 |
E. Calculation of all Priority Values (Eigenvector)
AHP pairwise comparison uses mathematical calculations to convert judgments to provide weights for all criteria. After obtaining the responses on the pairwise comparisons, a reciprocal matrix is created from the pairwise comparisons. The weights of decision factor i can be calculated as Eq. (4):
| 4 |
where n is the number of the compared elements. The AHP measurement steps must be designed to obtain the weights based on the evaluator’s preference.
F. Calculation of Consistency Ratio (CR)
CR, which expresses the internal consistency of judgments, is calculated. The following terms are defined to develop a quantitative measure of the degree of inconsistency within a pairwise comparison matrix [85]. The consistency index (CI) is calculated with Eq. (5):
| 5 |
The random index (RI) is calculated with Eq. (6):
| 6 |
CI measures the degree of inconsistency. RI is the corresponding measure of the degree of inconsistency of a pairwise comparison matrix. CR is defined in Eq. (7):
| 7 |
CR is the ratio of CI to RI. CR has been previously proposed [86]; it is a quantitative measure of the degree of inconsistency of a pairwise comparison matrix. A pairwise comparison matrix with a corresponding CR must not exceed 10% or 0.1. In this case, the obtained weights are acceptable [87]; otherwise, the obtained weights must be ignored, and decision makers must be asked to answer the designed questionnaires again to reach the acceptable CR ratio.
TOPSIS for the prioritisation of COVID-19 patients/donors
In this stage, TOPSIS is used to prioritise COVID-19 patients/donors based on the weighted criteria from the AHP method to tackle the major weakness of TOPSIS, which is the lack of provision weight for the evaluation criteria [88–91]. In general, the evaluation criteria can be classified into two types: benefit and cost [92, 93]. Benefit criterion indicates that a larger value is more valuable, whereas cost criteria are the opposite. From a medical point of view, all criteria of the serological/protein biomarkers are considered important except for C2 = ‘C-reactive protein’ and C3 = ‘IL-6 (pg/mL, cytokines)’, which are considered as cost criteria. Thus, transferring physicians’ preferences and experiences to an expert system can be proven effective. TOPSIS allocates the scores to each alternative on the basis of their geometric distance from positive and negative ideal solutions. The best alternative is selected, which according to this technique obtains the shortest geometric distance to the positive ideal solution and longest geometric distance to the negative ideal solution. The results of patients’ prioritisation are ranked in descending order, indicating that the patient in order 1 has the poorest critical health condition, and the patient in order 80 has the least critical health condition. In the same context, the results of donors’ prioritisation are also ranked in a descending order, indicating that the donor in order 1 is the least efficient donor, and the donor in order 80 is the most efficient donor. The steps of the TOPSIS method [35] are described as follows.
A. Construction of the Normalised Decision Matrix
This process may transform the various attribute dimensions into non-dimensional attributes. This process enables comparison across the attributes. The matrix (xij)m*n is then normalised from (xij)m*n to the matrix, R = (rij)m*n by using the normalisation method shown in Eq. (8):
| 8 |
This process results in a new matrix R, where R is shown as follows:
| 9 |
B. Construction of the Weighted Normalised Decision Matrix
In this process, a set of weights, w = w1, w2, w3,…, wj,…, wn, from the decision maker is accommodated to the normalised DM. The resulting matrix can be calculated by multiplying each column from the normalised DM (R) with its associated weight wj. Notably, the set of weights is equal to 1, as illustrated in Eq. (10).
| 10 |
This process results in a new matrix V, where V is shown as follows:
| 11 |
C. Determining the Ideal and Negative Ideal Solutions
In this process, two artificial alternatives, A* (the ideal alternative) and A− (the negative ideal alternative), are defined by Eqs. (12) and (13), respectively:
| 12 |
| 13 |
where J is the subset of {i = 1, 2, …, m}, which presents the benefit attribute (i.e., offering an increasing utility with high values), and J− is the complement set of J. The opposite can be added for the cost-type attribute denoted by Jc.
D. Separation-Measurement Calculation Based on Euclidean Distance
In this process, separation measurement is performed by calculating the distance between each alternative in V and the ideal vector A* by using the Euclidean distance, which is given by Eq. (14):
| 14 |
Similarly, the separation measurement for each alternative in ‘V from the negative ideal A−’ is given by Eq. (15):
| 15 |
At the end of step 4, two values, namely, S*i and S−i, for each alternative have been counted, and these two values represent the distance between each alternative and the ideal and negative ideal.
E. Closeness to the Ideal-Solution Calculation
In this process, the closeness of Ai to the ideal solution A* is defined, as shown in Eq. (16):
| 16 |
C*i = 1 if and only if Ai = A−; similarly, C*i = 0 if and only if Ai = A−.
F. Ranking the Alternative According to Closeness to the Ideal Solution
The set of the alternative Ai can be ranked according to the descending order of C*i, indicating that a higher value corresponds with better performance.
G. Group Decision-Making (GDM) Context
GDM is a situation faced when the decision required more than one decision-maker to select the best alternative. GDM methods systematically collect and combine the knowledge and judgment of experts in respiratory diseases. In the group context, each expert gives his/her judgment to the serological/protein biomarker COVID-19 criteria that require subjective judgment. The idea of GDM is to aggregate the result of multiple decisions from the three experts into one unique decision using the arithmetic mean. The academic literature in the area of GDM configurations is applied for several medical domains [94, 95]. In this study, GDM is used to combine the ranking results extracted from each expert preference, and then these ranks are aggregated into one final prioritisation of patients and donors. The use of aggregation can eliminate the variation amongst the obtained results from experts and unify a unique rank.
Matching components
This stage develops a new process for intelligent matching between prioritised patients and prioritised donors across identified hospitals. The rules that enable patient matching with the suitable donors are presented as follows.
Rule1: IF PATIENT GROUP ∈ (A), THEN COMPATIBLE PLASMA DONOR is (A) or (O)
Rule2: IF PATIENT GROUP ∈ (B), THEN COMPATIBLE PLASMA DONOR is (B) or (O)
Rule3: IF PATIENT GROUP ∈ (AB), THEN COMPATIBLE PLASMA DONOR is (AB) or (A) or (B) or (O)
Rule4: IF PATIENT GROUP ∈ (O), THEN COMPATIBLE PLASMA DONOR is (O)
At the end of this step, the transfusion of sufficient CPs from suitable donors to the proper patients can be demonstrated. This transfusion-rescue intelligent process can yield a balancing solution in either centralised or decentralised telemedicine connections and can thus address the lack of CPs.
Phase 4: Objective validation
The results are validated by utilising the objective validation in accordance with previously described methods [66]. The following steps are conducted for each ranking result (patients/donors) to ensure that the results are statistically ranked.
The final prioritisation results are categorised into four equal groups, with each group comprising 20 patients/donors. However, the number of groups or the alternative number within each group does not affect the validation result [80].
The mean ± standard deviation (M ± SD) of each group is obtained on the basis of the normalisation of patient/donor datasets. The first group is statistically proven to be the highest amongst all other groups. The second group must be lower than or equal to those of the first group. The third group must be lower than those of the first and second groups or equal to those of the second group. The fourth group must be lower than those of the first, second, and third groups or equal to those of the third group [96].
Equation (17) indicates the mean () that represents the average of the sum of all the observed results from the sample divided by the total number (n):
| 17 |
Equation (18) presents the measurement of the standard deviation to quantify the variation amount or dispersion of a set of data values.
| 18 |
Results and discussion
This section presents the results of the prioritisation CP-transfusion-rescue intelligent framework. The results of weights of the serological/protein biomarker criteria based on the AHP method for the three experts are presented. Afterwards, individual TOPSIS configurations are applied to provide the ranks of the three experts considering the obtained weights of serological/protein biomarkers. Additionally, the GDM TOPSIS context is applied to eliminate the variation amongst the obtained results from the experts and unify a unique rank. Finally, the results of the intelligent matching component and objective validation are operated in different sections. The sequences of results are illustrated as follows.
AHP weighting results
The AHP results are presented and explained after applying all previously illustrated steps. The results of the weights for the serological/protein biomarker criteria present the importance of each attribute based on the three experts. The weighting results of the three experts are shown in Table 5.
Table 5.
Weights of the serological/protein biomarker criteria for the three experts based on AHP
| Serological/Protein Biomarker Criteria | C1 | C2 | C3 | C4 | C5 |
|---|---|---|---|---|---|
| First Expert Weights | 0.343 | 0.067 | 0.407 | 0.086 | 0.098 |
| Second Expert Weights | 0.054 | 0.118 | 0.283 | 0.054 | 0.491 |
| Third Expert Weights | 0.427 | 0.199 | 0.199 | 0.061 | 0.113 |
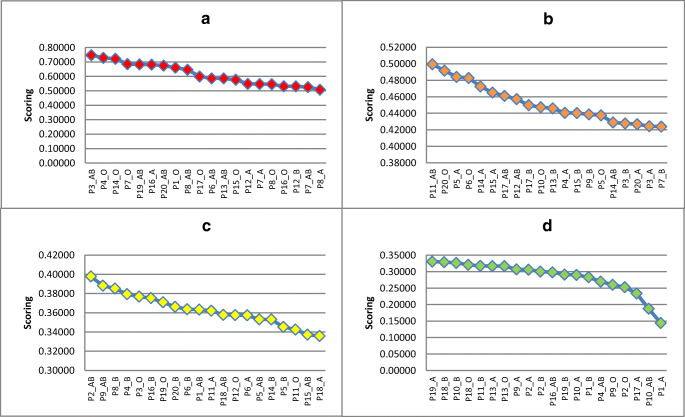
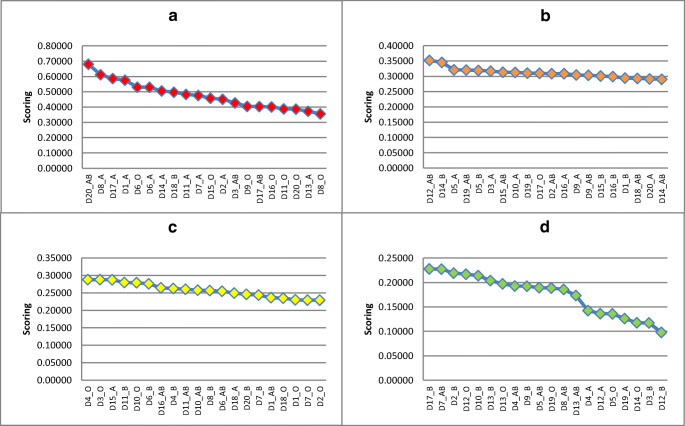
Table 5 shows that the first expert has given ‘C3=IL-6 (pg/mL; cytokines)’ the highest importance (0.407), whereas ‘C2=C-reactive protein, mg/L’ has received the lowest importance (0.067). The second expert has given ‘C5=IgM ELISA titre’ the highest importance (0.491), whereas ‘C1=PAO2/FIO2’ and ‘C4=albumin (g/L)’ have received the lowest importance (0.054). The third expert has given ‘C1= PAO2/FIO2’ the highest importance (0.427), whereas ‘C4= albumin (g/L)’ has received the lowest importance (0.061). The overall CR for the first, second, and third expert is 0.07, 0.09, and 0.06, respectively, which are considered as Additionally, Therefore, the obtained weights from the three experts are acceptable. At this step, the criteria assess the importance of patients and donors according to the best and poorest CP according to the experts through the AHP method.
TOPSIS prioritisation results
In this stage, TOPSIS is used in the prioritisation of COVID-19 patients and donors and can rapidly identify the most suitable option. Furthermore, the AHP method can derive the overall weights. Sample results of individual AHP-TOPSIS for the prioritisation of patients and donors of the three experts are shown in Tables 6 and 7 (the samples include the first 10 orders for each result). Meanwhile, the overall prioritisation results of patients and donors of the three experts based on individual TOPSIS are shown in Tables 12 and 13 (Appendix). The presented results consider the following points.
The set of patients and donors are ranked according to the descending order of C_ (i*), and high values indicate optimal performance for both.
A patient who is near the high record and far from the poorest record (i.e., the patient that gain order 1) is an optimal health condition case and must be given the lowest priority level. Conversely, the patient who is far the high record and near from the poorest record (i.e., the patient who gains order 80) is in the poorest health condition and must be given the highest priority level.
A donor who is near the high record and far from the poorest record (i.e., the donor that gain order 1) is the most highly efficient donor and must be matched with the patients in the poorest health condition.
Table 6.
Samples of the first 10 ranks of patient prioritisation based on individual AHP-TOPSIS
| Patient Rank | First Expert Results | Second Expert Results | Third Expert Results | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Patient Identification Information | C_(i*) Final Score | Patients Identification Information | C_(i*) Final Score | Patient Identification Information | C_(i*) Final Score | ||||
| Patients | Hospital | Patients | Hospital | Patients | Hospital | ||||
| 1 | P3_AB | H1 | 0.886284 | P16_A | H3 | 0.808919 | P19_AB | H4 | 0.784513 |
| 2 | P19_AB | H4 | 0.810943 | P4_O | H1 | 0.795971 | P3_AB | H1 | 0.762962 |
| 3 | P14_O | H2 | 0.750282 | P1_O | H1 | 0.781401 | P7_O | H1 | 0.726115 |
| 4 | P7_O | H1 | 0.747701 | P14_O | H2 | 0.731976 | P15_O | H2 | 0.689345 |
| 5 | P20_AB | H4 | 0.739635 | P9_B | H1 | 0.677319 | P14_O | H2 | 0.682247 |
| 6 | P8_AB | H1 | 0.739048 | P14_AB | H2 | 0.667573 | P4_O | H1 | 0.665293 |
| 7 | P4_O | H1 | 0.72548 | P13_AB | H2 | 0.656204 | P20_AB | H4 | 0.658588 |
| 8 | P1_O | H1 | 0.698919 | P17_O | H3 | 0.655859 | P17_O | H3 | 0.643114 |
| 9 | P8_O | H1 | 0.695913 | P3_B | H1 | 0.650131 | P6_AB | H1 | 0.634803 |
| 10 | P12_A | H2 | 0.683078 | P17_AB | H3 | 0.630744 | P16_A | H3 | 0.627233 |
Table 7.
Samples of the first 10 ranks of donor prioritisation based on individual AHP-TOPSIS
| Donor Rank | First Expert Results | Second Expert Results | Third Expert Results | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Donor Identification Information | C_(i*) Final Score | Donor Identification Information | C_(i*) Final Score | Donor Identification Information | C_(i*) Final Score | ||||
| Donors | Hospital | Donors | Hospital | Donors | Hospital | ||||
| 1 | D17_A | H4 | 0.766737 | D20_AB | H4 | 0.737167 | D20_AB | H4 | 0.584245 |
| 2 | D8_A | H3 | 0.755732 | D6_A | H3 | 0.690513 | D7_A | H3 | 0.548145 |
| 3 | D20_AB | H4 | 0.718893 | D1_A | H1 | 0.673066 | D8_A | H3 | 0.526714 |
| 4 | D14_A | H4 | 0.663588 | D17_AB | H4 | 0.574109 | D1_A | H1 | 0.521456 |
| 5 | D18_B | H4 | 0.662994 | D8_A | H3 | 0.551806 | D17_A | H4 | 0.507695 |
| 6 | D6_O | H3 | 0.651069 | D15_O | H4 | 0.547102 | D2_A | H1 | 0.498713 |
| 7 | D11_A | H4 | 0.640164 | D5_B | H2 | 0.546464 | D3_AB | H2 | 0.483957 |
| 8 | D6_A | H3 | 0.540643 | D6_O | H3 | 0.539688 | D15_O | H4 | 0.427792 |
| 9 | D1_A | H1 | 0.534718 | D16_O | H4 | 0.532662 | D3_A | H2 | 0.425142 |
| 10 | D2_A | H1 | 0.413798 | D9_O | H3 | 0.532389 | D8_O | H3 | 0.422213 |
Table 12.
Overall ranks of 80 patients prioritization based on individual AHP-TOPSIS for three experts
| 1st Expert Results | 2nd Expert Results | 3rd Expert Results | ||||||
|---|---|---|---|---|---|---|---|---|
| Patients Identification Information | C_(i*) Final Score |
Patients Identification Information | C_(i*) Final Score |
Patients Identification Information | C_(i*) Final Score |
|||
| Patients | Hospital | Patients | Hospital | Patients | Hospital | |||
| P3_AB | H1 | 0.886284 | P16_A | H3 | 0.808919 | P19_AB | H4 | 0.784513 |
| P19_AB | H4 | 0.810943 | P4_O | H1 | 0.795971 | P3_AB | H1 | 0.762962 |
| P14_O | H2 | 0.750282 | P1_O | H1 | 0.781401 | P7_O | H1 | 0.726115 |
| P7_O | H1 | 0.747701 | P14_O | H2 | 0.731976 | P15_O | H2 | 0.689345 |
| P20_AB | H4 | 0.739635 | P9_B | H1 | 0.677319 | P14_O | H2 | 0.682247 |
| P8_AB | H1 | 0.739048 | P14_AB | H2 | 0.667573 | P4_O | H1 | 0.665293 |
| P4_O | H1 | 0.72548 | P13_AB | H2 | 0.656204 | P20_AB | H4 | 0.658588 |
| P1_O | H1 | 0.698919 | P17_O | H3 | 0.655859 | P17_O | H3 | 0.643114 |
| P8_O | H1 | 0.695913 | P3_B | H1 | 0.650131 | P6_AB | H1 | 0.634803 |
| P12_A | H2 | 0.683078 | P17_AB | H3 | 0.630744 | P16_A | H3 | 0.627233 |
| P16_A | H3 | 0.606973 | P20_AB | H4 | 0.626686 | P12_B | H2 | 0.618863 |
| P14_A | H2 | 0.560024 | P8_AB | H1 | 0.625991 | P7_AB | H1 | 0.606251 |
| P15_O | H2 | 0.536998 | P5_A | H1 | 0.624698 | P16_O | H3 | 0.602225 |
| P13_AB | H2 | 0.527616 | P6_AB | H1 | 0.623437 | P17_B | H3 | 0.593097 |
| P7_A | H1 | 0.521426 | P3_A | H1 | 0.613166 | P8_A | H1 | 0.592788 |
| P4_A | H1 | 0.501926 | P7_B | H1 | 0.605747 | P12_AB | H2 | 0.575087 |
| P6_AB | H1 | 0.500115 | P5_O | H1 | 0.602552 | P8_AB | H1 | 0.57448 |
| P17_O | H3 | 0.500046 | P16_O | H3 | 0.596653 | P13_AB | H2 | 0.574055 |
| P7_AB | H1 | 0.493374 | P8_B | H1 | 0.595689 | P6_A | H1 | 0.573011 |
| P12_AB | H2 | 0.465017 | P3_AB | H1 | 0.592334 | P20_O | H4 | 0.570277 |
| P12_B | H2 | 0.438512 | P7_A | H1 | 0.58831 | P13_B | H2 | 0.568594 |
| P13_B | H2 | 0.433319 | P18_B | H3 | 0.58774 | P18_AB | H3 | 0.567191 |
| P6_O | H1 | 0.41936 | P7_O | H1 | 0.582611 | P6_O | H1 | 0.553197 |
| P20_O | H4 | 0.414859 | P5_AB | H1 | 0.573807 | P11_AB | H2 | 0.551208 |
| P18_AB | H3 | 0.41028 | P20_B | H4 | 0.570063 | P8_O | H1 | 0.549262 |
| P17_B | H3 | 0.406853 | P14_B | H2 | 0.564311 | P4_B | H1 | 0.535843 |
| P18_A | H3 | 0.400997 | P15_A | H2 | 0.559983 | P7_A | H1 | 0.530552 |
| P9_AB | H1 | 0.399452 | P2_AB | H1 | 0.557422 | P15_A | H2 | 0.523621 |
| P11_AB | H2 | 0.398956 | P11_AB | H2 | 0.54805 | P16_B | H3 | 0.520682 |
| P16_O | H3 | 0.396287 | P8_A | H1 | 0.547855 | P10_O | H1 | 0.520308 |
| P6_A | H1 | 0.384405 | P13_A | H2 | 0.544517 | P5_A | H1 | 0.515585 |
| P4_B | H1 | 0.383991 | P20_A | H4 | 0.540935 | P3_O | H1 | 0.515107 |
| P8_A | H1 | 0.383015 | P12_B | H2 | 0.537488 | P12_A | H2 | 0.514782 |
| P17_AB | H3 | 0.377267 | P14_A | H2 | 0.525715 | P9_AB | H1 | 0.512719 |
| P9_B | H1 | 0.37621 | P10_B | H1 | 0.521287 | P1_O | H1 | 0.499758 |
| P10_O | H1 | 0.375685 | P6_B | H1 | 0.516879 | P11_A | H2 | 0.484766 |
| P11_B | H2 | 0.373836 | P10_A | H1 | 0.51471 | P15_B | H2 | 0.449937 |
| P14_AB | H2 | 0.363693 | P15_B | H2 | 0.5078 | P20_A | H4 | 0.431079 |
| P15_B | H2 | 0.363396 | P2_A | H1 | 0.503824 | P5_O | H1 | 0.42917 |
| P7_B | H1 | 0.359767 | P15_O | H2 | 0.502961 | P19_O | H4 | 0.422135 |
| P13_O | H2 | 0.359723 | P20_O | H4 | 0.489727 | P19_A | H4 | 0.420023 |
| P19_A | H4 | 0.355443 | P4_A | H1 | 0.487784 | P11_O | H2 | 0.399588 |
| P5_B | H1 | 0.341733 | P7_AB | H1 | 0.479225 | P3_A | H1 | 0.387438 |
| P6_B | H1 | 0.34124 | P6_O | H1 | 0.475615 | P1_AB | H1 | 0.385577 |
| P11_A | H2 | 0.33166 | P19_AB | H4 | 0.454427 | P2_O | H1 | 0.383869 |
| P16_B | H3 | 0.319742 | P12_A | H2 | 0.44901 | P12_O | H2 | 0.379677 |
| P3_O | H1 | 0.319142 | P10_O | H1 | 0.446525 | P2_AB | H1 | 0.376916 |
| P12_O | H2 | 0.31455 | P19_O | H4 | 0.445208 | P17_AB | H3 | 0.375411 |
| P5_A | H1 | 0.312335 | P18_O | H3 | 0.434178 | P5_B | H1 | 0.374349 |
| P15_A | H2 | 0.311932 | P1_AB | H1 | 0.433836 | P15_AB | H2 | 0.359753 |
| P3_B | H1 | 0.3108 | P16_AB | H3 | 0.413322 | P11_B | H2 | 0.356557 |
| P20_A | H4 | 0.308699 | P11_O | H2 | 0.398876 | P9_A | H1 | 0.33866 |
| P15_AB | H2 | 0.303549 | P17_A | H3 | 0.392736 | P19_B | H4 | 0.333125 |
| P14_B | H2 | 0.281547 | P8_O | H1 | 0.391768 | P4_A | H1 | 0.332214 |
| P5_O | H1 | 0.281045 | P12_O | H2 | 0.378872 | P14_A | H2 | 0.331591 |
| P3_A | H1 | 0.27223 | P2_B | H1 | 0.358304 | P8_B | H1 | 0.324628 |
| P1_AB | H1 | 0.270697 | P9_A | H1 | 0.356457 | P3_B | H1 | 0.32238 |
| P1_B | H1 | 0.269371 | P17_B | H3 | 0.349989 | P4_AB | H1 | 0.31215 |
| P5_AB | H1 | 0.265328 | P15_AB | H2 | 0.348087 | P16_AB | H3 | 0.310323 |
| P2_B | H1 | 0.263247 | P13_O | H2 | 0.347496 | P7_B | H1 | 0.306191 |
| P2_AB | H1 | 0.259332 | P18_A | H3 | 0.344381 | P18_O | H3 | 0.297537 |
| P19_O | H4 | 0.245389 | P13_B | H2 | 0.336169 | P20_B | H4 | 0.294602 |
| P4_AB | H1 | 0.240941 | P12_AB | H2 | 0.33184 | P2_B | H1 | 0.279074 |
| P8_B | H1 | 0.235501 | P9_O | H1 | 0.328232 | P1_B | H1 | 0.27047 |
| P20_B | H4 | 0.234221 | P5_B | H1 | 0.319709 | P9_B | H1 | 0.262294 |
| P19_B | H4 | 0.231782 | P1_B | H1 | 0.309748 | P18_A | H3 | 0.26198 |
| P18_O | H3 | 0.231169 | P19_B | H4 | 0.307888 | P14_AB | H2 | 0.255414 |
| P11_O | H2 | 0.229241 | P3_O | H1 | 0.296477 | P9_O | H1 | 0.25272 |
| P9_A | H1 | 0.226233 | P16_B | H3 | 0.285519 | P10_B | H1 | 0.251555 |
| P10_B | H1 | 0.207789 | P11_A | H2 | 0.270201 | P2_A | H1 | 0.243476 |
| P2_O | H1 | 0.200036 | P4_AB | H1 | 0.256901 | P13_O | H2 | 0.243129 |
| P9_O | H1 | 0.199245 | P9_AB | H1 | 0.252691 | P6_B | H1 | 0.232698 |
| P18_B | H3 | 0.187665 | P10_AB | H1 | 0.223641 | P13_A | H2 | 0.221635 |
| P13_A | H2 | 0.184238 | P11_B | H2 | 0.22145 | P5_AB | H1 | 0.220233 |
| P2_A | H1 | 0.170126 | P4_B | H1 | 0.218409 | P14_B | H2 | 0.213446 |
| P16_AB | H3 | 0.169731 | P19_A | H4 | 0.216628 | P18_B | H3 | 0.21112 |
| P10_A | H1 | 0.156469 | P2_O | H1 | 0.175417 | P10_AB | H1 | 0.199954 |
| P1_A | H1 | 0.141967 | P6_A | H1 | 0.114848 | P10_A | H1 | 0.199111 |
| P10_AB | H1 | 0.138655 | P1_A | H1 | 0.102404 | P1_A | H1 | 0.188651 |
| P17_A | H3 | 0.12837 | P18_AB | H3 | 0.095855 | P17_A | H3 | 0.180453 |
Table 13.
Overall ranks of 80 donors prioritization based on individual AHP-TOPSIS for three experts
| 1st Expert Results | 2nd Expert Results | 3rd Expert Results | ||||||
|---|---|---|---|---|---|---|---|---|
| Patients Identification Information | C_(i*) Final Score | Patients Identification Information | C_(i*) Final Score | Patients Identification Information | C_(i*) Final Score | |||
| Donors | Hospital | Donors | Hospital | Donors | Hospital | |||
| D17_A | H4 | 0.766737 | D20_AB | H4 | 0.737167 | D20_AB | H4 | 0.584245 |
| D8_A | H3 | 0.755732 | D6_A | H3 | 0.690513 | D7_A | H3 | 0.548145 |
| D20_AB | H4 | 0.718893 | D1_A | H1 | 0.673066 | D8_A | H3 | 0.526714 |
| D14_A | H4 | 0.663588 | D17_AB | H4 | 0.574109 | D1_A | H1 | 0.521456 |
| D18_B | H4 | 0.662994 | D8_A | H3 | 0.551806 | D17_A | H4 | 0.507695 |
| D6_O | H3 | 0.651069 | D15_O | H4 | 0.547102 | D2_A | H1 | 0.498713 |
| D11_A | H4 | 0.640164 | D5_B | H2 | 0.546464 | D3_AB | H2 | 0.483957 |
| D6_A | H3 | 0.540643 | D6_O | H3 | 0.539688 | D15_O | H4 | 0.427792 |
| D1_A | H1 | 0.534718 | D16_O | H4 | 0.532662 | D3_A | H2 | 0.425142 |
| D2_A | H1 | 0.413798 | D9_O | H3 | 0.532389 | D8_O | H3 | 0.422213 |
| D15_O | H4 | 0.400626 | D15_AB | H4 | 0.516875 | D18_B | H4 | 0.411704 |
| D7_A | H3 | 0.391255 | D20_O | H4 | 0.516373 | D14_A | H4 | 0.409338 |
| D12_AB | H4 | 0.374731 | D3_AB | H2 | 0.510694 | D6_O | H3 | 0.400915 |
| D5_A | H2 | 0.366625 | D14_AB | H4 | 0.510371 | D9_O | H3 | 0.391128 |
| D1_B | H1 | 0.347927 | D11_O | H4 | 0.506859 | D11_O | H4 | 0.390001 |
| D17_AB | H4 | 0.341155 | D19_AB | H4 | 0.504252 | D16_A | H4 | 0.389733 |
| D8_B | H3 | 0.310224 | D7_A | H3 | 0.489081 | D14_B | H4 | 0.389617 |
| D8_O | H3 | 0.30895 | D17_A | H4 | 0.486081 | D16_O | H4 | 0.385066 |
| D14_B | H4 | 0.307588 | D17_O | H4 | 0.482839 | D20_O | H4 | 0.384847 |
| D16_A | H4 | 0.292773 | D16_B | H4 | 0.479175 | D13_A | H4 | 0.375746 |
| D9_O | H3 | 0.289059 | D19_B | H4 | 0.474147 | D9_AB | H3 | 0.375283 |
| D16_O | H4 | 0.28668 | D13_A | H4 | 0.471106 | D12_AB | H4 | 0.373154 |
| D3_AB | H2 | 0.285781 | D14_A | H4 | 0.44564 | D5_A | H2 | 0.365358 |
| D13_A | H4 | 0.271212 | D10_A | H3 | 0.444453 | D11_A | H4 | 0.364972 |
| D9_AB | H3 | 0.270683 | D11_A | H4 | 0.443351 | D9_A | H3 | 0.362415 |
| D11_O | H4 | 0.269023 | D2_A | H1 | 0.441505 | D4_O | H2 | 0.359316 |
| D9_A | H3 | 0.266369 | D15_B | H4 | 0.441453 | D6_A | H3 | 0.357396 |
| D10_A | H3 | 0.26265 | D18_O | H4 | 0.43704 | D2_AB | H1 | 0.341342 |
| D20_O | H4 | 0.259857 | D18_A | H4 | 0.432796 | D7_O | H3 | 0.334193 |
| D4_B | H2 | 0.246033 | D18_AB | H4 | 0.431051 | D10_O | H3 | 0.333499 |
| D3_O | H2 | 0.24153 | D20_A | H4 | 0.43014 | D1_B | H1 | 0.326476 |
| D10_O | H3 | 0.240912 | D11_B | H4 | 0.429708 | D1_AB | H1 | 0.325502 |
| D15_A | H4 | 0.237096 | D11_AB | H4 | 0.429375 | D7_AB | H3 | 0.323743 |
| D2_AB | H1 | 0.235943 | D2_B | H1 | 0.42278 | D4_B | H2 | 0.319451 |
| D1_O | H1 | 0.230526 | D18_B | H4 | 0.417339 | D3_O | H2 | 0.316964 |
| D7_AB | H3 | 0.224468 | D7_B | H3 | 0.414701 | D15_B | H4 | 0.314912 |
| D4_O | H4 | 0.221377 | D15_A | H4 | 0.401604 | D1_O | H1 | 0.312265 |
| D19_AB | H4 | 0.220892 | D6_B | H3 | 0.398269 | D12_O | H4 | 0.311765 |
| D7_O | H3 | 0.21809 | D16_AB | H4 | 0.391845 | D20_B | H4 | 0.306247 |
| D1_AB | H1 | 0.216447 | D10_AB | H3 | 0.386365 | D20_A | H4 | 0.302403 |
| D5_B | H2 | 0.201861 | D3_A | H2 | 0.35722 | D17_AB | H4 | 0.297005 |
| D19_O | H2 | 0.193937 | D6_AB | H3 | 0.354627 | D19_O | H4 | 0.289054 |
| D5_AB | H2 | 0.193746 | D2_AB | H1 | 0.349268 | D11_B | H4 | 0.287616 |
| D19_B | H4 | 0.190788 | D14_B | H4 | 0.339489 | D16_B | H4 | 0.285548 |
| D15_AB | H4 | 0.187655 | D8_O | H3 | 0.3356 | D16_AB | H4 | 0.280959 |
| D12_O | H4 | 0.187542 | D12_AB | H4 | 0.308846 | D8_AB | H3 | 0.274243 |
| D2_O | H1 | 0.184113 | D3_O | H2 | 0.304589 | D18_AB | H4 | 0.27346 |
| D11_AB | H4 | 0.182128 | D17_B | H4 | 0.302468 | D13_B | H4 | 0.271593 |
| D13_B | H4 | 0.179719 | D20_B | H4 | 0.297695 | D17_O | H4 | 0.270768 |
| D17_O | H4 | 0.175689 | D9_B | H3 | 0.293812 | D19_B | H4 | 0.267146 |
| D6_B | H3 | 0.175511 | D9_A | H3 | 0.283956 | D2_O | H1 | 0.26436 |
| D18_AB | H4 | 0.175187 | D4_O | H2 | 0.283001 | D5_AB | H2 | 0.262083 |
| D3_A | H2 | 0.167935 | D10_B | H3 | 0.264197 | D6_B | H3 | 0.253384 |
| D6_AB | H3 | 0.164975 | D9_AB | H3 | 0.263613 | D6_AB | H3 | 0.243856 |
| D9_B | H3 | 0.164596 | D10_O | H3 | 0.262724 | D19_AB | H4 | 0.237116 |
| D13_O | H4 | 0.157727 | D4_AB | H2 | 0.26172 | D15_AB | H4 | 0.236321 |
| D10_AB | H3 | 0.156258 | D4_A | H2 | 0.248961 | D8_B | H3 | 0.234667 |
| D10_B | H3 | 0.154847 | D16_A | H4 | 0.243037 | D17_B | H4 | 0.231083 |
| D17_B | H4 | 0.150147 | D2_O | H1 | 0.238695 | D10_A | H3 | 0.230732 |
| D15_B | H4 | 0.147314 | D13_AB | H4 | 0.233873 | D10_AB | H3 | 0.230139 |
| D14_AB | H4 | 0.145891 | D5_A | H2 | 0.231365 | D13_O | H4 | 0.225875 |
| D18_A | H4 | 0.142861 | D8_B | H3 | 0.225048 | D10_B | H3 | 0.223239 |
| D20_A | H4 | 0.142591 | D4_B | H2 | 0.221769 | D15_A | H4 | 0.223182 |
| D12_A | H4 | 0.138953 | D5_O | H2 | 0.219149 | D14_AB | H4 | 0.214246 |
| D16_B | H4 | 0.13349 | D19_A | H4 | 0.211467 | D5_B | H2 | 0.209854 |
| D20_B | H4 | 0.133264 | D1_B | H1 | 0.208165 | D7_B | H3 | 0.196095 |
| D14_O | H4 | 0.126624 | D13_O | H4 | 0.207723 | D4_AB | H2 | 0.190488 |
| D8_AB | H3 | 0.126349 | D1_AB | H1 | 0.167554 | D14_O | H4 | 0.187058 |
| D4_AB | H2 | 0.126309 | D13_B | H4 | 0.160432 | D13_AB | H4 | 0.17699 |
| D11_B | H4 | 0.122043 | D8_AB | H3 | 0.15659 | D18_A | H4 | 0.172942 |
| D16_AB | H4 | 0.121491 | D12_O | H4 | 0.151545 | D11_AB | H4 | 0.169562 |
| D7_B | H3 | 0.120812 | D1_O | H1 | 0.147353 | D18_O | H4 | 0.15621 |
| D3_B | H2 | 0.117283 | D7_O | H3 | 0.134908 | D12_A | H4 | 0.14229 |
| D18_O | H4 | 0.110488 | D7_AB | H3 | 0.134244 | D2_B | H1 | 0.136447 |
| D13_AB | H4 | 0.109836 | D12_A | H4 | 0.127914 | D3_B | H2 | 0.124586 |
| D12_B | H4 | 0.104592 | D5_AB | H2 | 0.112471 | D9_B | H3 | 0.119028 |
| D2_B | H1 | 0.09837 | D3_B | H2 | 0.110035 | D5_O | H2 | 0.116156 |
| D19_A | H4 | 0.088171 | D12_B | H4 | 0.085178 | D12_B | H4 | 0.104485 |
| D4_A | H2 | 0.075218 | D19_O | H4 | 0.083312 | D4_A | H2 | 0.103468 |
| D5_O | H2 | 0.072702 | D14_O | H4 | 0.038946 | D19_A | H4 | 0.079094 |
No unique prioritisation results based on the weights obtained from the three experts are found when the TOPSIS method is applied. Results show variances amongst the ranks obtained from the three experts. Considering the GDM TOPSIS context to provide final and unique prioritisation concerning all decision makers is important to address this challenge. Thus, Table 8 shows the sample ranking results for the first 10 patients and donors based on GDM TOPSIS, whereas the overall prioritisation results are shown in Table 14 (Appendix).
Table 8.
Samples of the first 10 ranks of patients and donors based on the TOPSIS GDM contexts
| Patient/Donor Rank | Patient Ranking Results | Donor Ranking Results | ||||
|---|---|---|---|---|---|---|
| Patient Identification Information | C_(i*) Final Score | Donor Identification Information | C_(i*) Final Score | |||
| Patients | Hospital | Donors | Hospital | |||
| 1 | P3_AB | H1 | 0.74719 | D20_AB | H4 | 0.68010 |
| 2 | P4_O | H1 | 0.72891 | D8_A | H2 | 0.61142 |
| 3 | P14_O | H2 | 0.72150 | D17_A | H4 | 0.58684 |
| 4 | P7_O | H1 | 0.68548 | D1_A | H1 | 0.57641 |
| 5 | P19_AB | H4 | 0.68329 | D6_O | H3 | 0.53056 |
| 6 | P16_A | H3 | 0.68104 | D6_A | H3 | 0.52952 |
| 7 | P20_AB | H4 | 0.67497 | D14_A | H4 | 0.50619 |
| 8 | P1_O | H1 | 0.66003 | D18_B | H4 | 0.49735 |
| 9 | P8_AB | H1 | 0.64651 | D11_A | H4 | 0.48283 |
| 10 | P17_O | H3 | 0.59967 | D7_A | H3 | 0.47616 |
Table 14.
Overall ranks of 80 patients and 80 donors prioritization based on external TOPSIS GDM contexts for three experts
| Patients/Donors Rank | Patients Ranking Results | Donors Ranking Results | ||||
|---|---|---|---|---|---|---|
| Patients Identification Information | C_(i*) Final Score |
Donors Identification Information | C_(i*) Final Score |
|||
| Patients | Hospital | Donors | Hospital | |||
| 1 | P3_AB | H1 | 0.74719 | D20_AB | H4 | 0.68010 |
| 2 | P4_O | H1 | 0.72891 | D8_A | H2 | 0.61142 |
| 3 | P14_O | H2 | 0.72150 | D17_A | H4 | 0.58684 |
| 4 | P7_O | H1 | 0.68548 | D1_A | H1 | 0.57641 |
| 5 | P19_AB | H4 | 0.68329 | D6_O | H3 | 0.53056 |
| 6 | P16_A | H3 | 0.68104 | D6_A | H3 | 0.52952 |
| 7 | P20_AB | H4 | 0.67497 | D14_A | H4 | 0.50619 |
| 8 | P1_O | H1 | 0.66003 | D18_B | H4 | 0.49735 |
| 9 | P8_AB | H1 | 0.64651 | D11_A | H4 | 0.48283 |
| 10 | P17_O | H3 | 0.59967 | D7_A | H3 | 0.47616 |
| 11 | P6_AB | H1 | 0.58612 | D15_O | H4 | 0.45851 |
| 12 | P13_AB | H2 | 0.58596 | D2_A | H1 | 0.45134 |
| 13 | P15_O | H2 | 0.57643 | D3_AB | H2 | 0.42681 |
| 14 | P12_A | H2 | 0.54896 | D9_O | H3 | 0.40419 |
| 15 | P7_A | H1 | 0.54676 | D17_AB | H4 | 0.40409 |
| 16 | P8_O | H1 | 0.54565 | D16_O | H4 | 0.40147 |
| 17 | P16_O | H3 | 0.53172 | D11_O | H4 | 0.38863 |
| 18 | P12_B | H2 | 0.53162 | D20_O | H4 | 0.38703 |
| 19 | P7_AB | H1 | 0.52628 | D13_A | H4 | 0.37269 |
| 20 | P8_A | H1 | 0.50789 | D8_O | H3 | 0.35559 |
| 21 | P11_AB | H2 | 0.49940 | D12_AB | H4 | 0.35224 |
| 22 | P20_O | H4 | 0.49162 | D14_B | H4 | 0.34556 |
| 23 | P5_A | H1 | 0.48421 | D5_A | H3 | 0.32112 |
| 24 | P6_O | H1 | 0.48272 | D19_AB | H4 | 0.32075 |
| 25 | P14_A | H2 | 0.47244 | D5_B | H2 | 0.31939 |
| 26 | P15_A | H2 | 0.46518 | D3_A | H2 | 0.31677 |
| 27 | P17_AB | H3 | 0.46114 | D15_AB | H4 | 0.31362 |
| 28 | P12_AB | H2 | 0.45731 | D10_A | H3 | 0.31261 |
| 29 | P17_B | H3 | 0.44998 | D19_B | H4 | 0.31069 |
| 30 | P10_O | H1 | 0.44751 | D17_O | H4 | 0.30977 |
| 31 | P13_B | H2 | 0.44603 | D2_AB | H1 | 0.30885 |
| 32 | P4_A | H1 | 0.44064 | D16_A | H4 | 0.30851 |
| 33 | P15_B | H2 | 0.44038 | D9_A | H3 | 0.30425 |
| 34 | P9_B | H1 | 0.43861 | D9_AB | H3 | 0.30319 |
| 35 | P5_O | H1 | 0.43759 | D15_B | H4 | 0.30123 |
| 36 | P14_AB | H2 | 0.42889 | D16_B | H4 | 0.29940 |
| 37 | P3_B | H1 | 0.42777 | D1_B | H1 | 0.29419 |
| 38 | P20_A | H4 | 0.42690 | D18_AB | H4 | 0.29323 |
| 39 | P3_A | H1 | 0.42428 | D20_A | H4 | 0.29171 |
| 40 | P7_B | H1 | 0.42390 | D14_AB | H4 | 0.29017 |
| 41 | P2_AB | H1 | 0.39789 | D4_O | H2 | 0.28790 |
| 42 | P9_AB | H1 | 0.38829 | D3_O | H2 | 0.28769 |
| 43 | P8_B | H1 | 0.38527 | D15_A | H4 | 0.28729 |
| 44 | P4_B | H1 | 0.37941 | D11_B | H4 | 0.27979 |
| 45 | P3_O | H1 | 0.37691 | D10_O | H3 | 0.27905 |
| 46 | P16_B | H3 | 0.37531 | D6_B | H3 | 0.27572 |
| 47 | P19_O | H4 | 0.37091 | D16_AB | H4 | 0.26477 |
| 48 | P20_B | H4 | 0.36630 | D4_B | H2 | 0.26242 |
| 49 | P6_B | H1 | 0.36361 | D11_AB | H4 | 0.26036 |
| 50 | P1_AB | H1 | 0.36337 | D10_AB | H3 | 0.25759 |
| 51 | P11_A | H2 | 0.36221 | D8_B | H3 | 0.25665 |
| 52 | P18_AB | H3 | 0.35778 | D6_AB | H3 | 0.25449 |
| 53 | P12_O | H2 | 0.35770 | D18_A | H4 | 0.24953 |
| 54 | P6_A | H1 | 0.35742 | D20_B | H4 | 0.24574 |
| 55 | P5_AB | H1 | 0.35312 | D7_B | H3 | 0.24387 |
| 56 | P14_B | H2 | 0.35310 | D1_AB | H1 | 0.23650 |
| 57 | P5_B | H1 | 0.34526 | D18_O | H4 | 0.23458 |
| 58 | P11_O | H2 | 0.34257 | D1_O | H1 | 0.23005 |
| 59 | P15_AB | H2 | 0.33713 | D7_O | H3 | 0.22906 |
| 60 | P18_A | H3 | 0.33579 | D2_O | H1 | 0.22906 |
| 61 | P19_A | H4 | 0.33070 | D17_B | H4 | 0.22790 |
| 62 | P18_B | H3 | 0.32884 | D7_AB | H3 | 0.22748 |
| 63 | P10_B | H1 | 0.32688 | D2_B | H1 | 0.21920 |
| 64 | P18_O | H3 | 0.32096 | D12_O | H4 | 0.21695 |
| 65 | P11_B | H2 | 0.31728 | D10_B | H3 | 0.21409 |
| 66 | P13_A | H2 | 0.31680 | D13_B | H4 | 0.20391 |
| 67 | P13_O | H2 | 0.31678 | D13_O | H4 | 0.19711 |
| 68 | P9_A | H1 | 0.30712 | D4_AB | H2 | 0.19284 |
| 69 | P2_A | H1 | 0.30581 | D9_B | H3 | 0.19248 |
| 70 | P2_B | H1 | 0.30021 | D5_AB | H2 | 0.18943 |
| 71 | P16_AB | H3 | 0.29779 | D19_O | H4 | 0.18877 |
| 72 | P19_B | H4 | 0.29093 | D8_AB | H3 | 0.18573 |
| 73 | P10_A | H1 | 0.29010 | D13_AB | H4 | 0.17357 |
| 74 | P1_B | H1 | 0.28320 | D4_A | H2 | 0.14255 |
| 75 | P4_AB | H1 | 0.27000 | D12_A | H4 | 0.13639 |
| 76 | P9_O | H1 | 0.26007 | D5_O | H2 | 0.13600 |
| 77 | P2_O | H1 | 0.25311 | D19_A | H4 | 0.12624 |
| 78 | P17_A | H3 | 0.23385 | D14_O | H4 | 0.11754 |
| 79 | P10_AB | H1 | 0.18742 | D3_B | H2 | 0.11730 |
| 80 | P1_A | H1 | 0.14434 | D12_B | H4 | 0.09808 |
For all ranks, the prioritisation of the 80 patients and 80 donors is stated. The set of patients is ranked in descending order starting from the critical health condition to the mild one. Moreover, donor prioritisation is ranked in descending order from the least efficient donor to the most highly efficient one.
Intelligent matching component results
The result of this section follows the significance of the four rules presented previously to match a proper donor with the suitable patient after achieving prioritisation. A sample of 10 results of the matching component stage is shown in Table 9, whereas the overall results are described in Table 15 (Appendix).
Table 9.
Matching results between patients and donors
| Patient Rank | Patients/admitted hospital | Suitable CP donors/admitted hospital |
|---|---|---|
| 1 | P3_AB/H1 | D13_AB/H4 |
| 2 | P4_O/H1 | D14_O/H4 |
| 3 | P14_O/H2 | D5_O/H2 |
| 4 | P7_O/H1 | D19_O/H4 |
| 5 | P19_AB/H4 | D8_AB/H3 |
| 6 | P16_A/H3 | D19_A/H3 |
| 7 | P20_AB/H4 | D5_AB/H2 |
| 8 | P1_O/H1 | D13_O/H4 |
| 9 | P8_AB/H1 | D4_AB/H2 |
| 10 | P17_O/H3 | D12_O/H4 |
Table 15.
Overall matching results between patients and donors
| Patients Rank | Patients/admitted Hospital | Suitable CP donors/admitted Hospital |
|---|---|---|
| 1 | P3_AB/ H1 | D13_AB/ H4 |
| 2 | P4_O/ H1 | D14_O/ H4 |
| 3 | P14_O/ H2 | D5_O/ H2 |
| 4 | P7_O/ H1 | D19_O/ H4 |
| 5 | P19_AB/ H4 | D8_AB/ H3 |
| 6 | P16_A/ H3 | D19_A/ H3 |
| 7 | P20_AB/ H4 | D5_AB/ H2 |
| 8 | P1_O/ H1 | D13_O/ H4 |
| 9 | P8_AB/H1 | D4_AB/ H2 |
| 10 | P17_O/ H3 | D12_O/ H4 |
| 11 | P6_AB/ H1 | D7_AB/ H3 |
| 12 | P13_AB/ H2 | D1_AB/ H1 |
| 13 | P15_O/ H2 | D2_O/ H1 |
| 14 | P12_A/ H2 | D12_A/ H4 |
| 15 | P7_A/ H1 | D4_A/ H2 |
| 16 | P8_O/ H1 | D7_O/ H3 |
| 17 | P16_O/ H3 | D1_O/ H1 |
| 18 | P12_B/ H2 | D12_B/ H4 |
| 19 | P7_AB/ H1 | D6_AB/ H3 |
| 20 | P8_A/ H1 | D18_A/ H4 |
| 21 | P11_AB/ H2 | D10_AB/ H3 |
| 22 | P20_O/ H4 | D18_O/ H4 |
| 23 | P5_A/ H1 | D15_A/ H4 |
| 24 | P6_O/ H1 | D10_O/ H3 |
| 25 | P14_A/ H2 | D20_A/ H4 |
| 26 | P15_A/ H2 | D9_A/ H3 |
| 27 | P17_AB/ H3 | D11_AB/ H4 |
| 28 | P12_AB/ H2 | D16_AB/H4 |
| 29 | P17_B/ H3 | D3_B/ H2 |
| 30 | P10_O/ H1 | D3_O/ H2 |
| 31 | P13_B/ H2 | D9_B/ H3 |
| 32 | P4_A/ H1 | D16_A/ H4 |
| 33 | P15_B/ H2 | D13_B/ H4 |
| 34 | P9_B/ H1 | D10_B/ H3 |
| 35 | P5_O/ H1 | D4_O/ H2 |
| 36 | P14_AB/ H2 | D14_AB/H4 |
| 37 | P3_B/ H1 | D2_B/ H1 |
| 38 | P20_A/H4 | D10_A/ H3 |
| 39 | P3_A/ H1 | D3_A/ H2 |
| 40 | P7_B/ H1 | D17_B/ H4 |
| 41 | P2_AB/ H1 | D18_AB/ H4 |
| 42 | P9_AB/ H1 | D9_AB/ H3 |
| 43 | P8_B/ H1 | D7_B/ H3 |
| 44 | P4_B/ H1 | D20_B/ H4 |
| 45 | P3_O/ H1 | D17_O/ H4 |
| 46 | P16_B/ H3 | D8_B/ H3 |
| 47 | P19_O/ H4 | D8_O/ H3 |
| 48 | P20_B/ H4 | D4_B/ H2 |
| 49 | P6_B/ H1 | D6_B/ H3 |
| 50 | P1_AB/ H1 | D2_AB/ H1 |
| 51 | P11_A/ H2 | D5_A/ H3 |
| 52 | P18_AB/ H3 | D15_AB/ H4 |
| 53 | P12_O/ H2 | D20_O/ H4 |
| 54 | P6_A/ H1 | D13_A/ H4 |
| 55 | P5_AB/ H1 | D19_AB/ H4 |
| 56 | P14_B/ H2 | D11_B/ H4 |
| 57 | P5_B/ H1 | D1_B/ H1 |
| 58 | P11_O/ H2 | D11_O/ H4 |
| 59 | P15_AB/H2 | D12_AB/ H4 |
| 60 | P18_A/H3 | D2_A/ H1 |
| 61 | P19_A/ H4 | D7_A/ H3 |
| 62 | P18_B/ H3 | D16_B/ H4 |
| 63 | P10_B/ H1 | D15_B/ H4 |
| 64 | P18_O/ H3 | D16_O/ H4 |
| 65 | P11_B/ H2 | D19_B/ H4 |
| 66 | P13_A/ H2 | D11_A/ H4 |
| 67 | P13_O/ H2 | D9_O/ H3 |
| 68 | P9_A/ H1 | D14_A/ H4 |
| 69 | P2_A/ H1 | D6_A/ H3 |
| 70 | P2_B/ H1 | D5_B/ H2 |
| 71 | P16_AB/ H3 | D17_AB/ H4 |
| 72 | P19_B/ H4 | D14_B/ H4 |
| 73 | P10_A/ H1 | D1_A/ H1 |
| 74 | P1_B/ H1 | D18_B/ H4 |
| 75 | P4_AB/ H1 | D3_AB/ H2 |
| 76 | P9_O/ H1 | D15_O/ H4 |
| 77 | P2_O/ H1 | D6_O/ H3 |
| 78 | P17_A/ H3 | D17_A/ H4 |
| 79 | P10_AB/ H1 | D20_AB/ H4 |
| 80 | P1_A/ H1 | D8_A/ H2 |
As shown in the above-mentioned results, each patient is matched with a suitable CP donor on the basis of prioritisation results. Additionally, the matching between patients and donors is operated across four connected hospitals. For example, patient (P1_A) admitted to hospital 1 obtains the suitable CP from donor (D8_A), who is admitted to hospital 2. In these contexts, balancing across hospitals is achieved for all patients/donors and proven within the four hospitals. Moreover, matching amongst patients and donors could be achieved by the inverse relationship between them. For example, the patient (P3_AB) who gains order 1 with score = 0.74719 is considered the most critical condition amongst all patients, and the suitable donor for this patient is the last donor (D12_B) who gains order 80 with a score of 0.09808 according to Rule 3. Therefore, the intelligent matching process of CP transfusion between patients and donors is tested and verified towards the balancing approach across either centralised or decentralised telemedicine hospitals simultaneously.
Validation results
In this section, as explained in phase 6, objective validation can be achieved by dividing the prioritisation results for patients and donors into four equal groups. Each group comprises 20 patients. The mean ± SD is calculated for each group on the basis of normalisation scores generated by TOPSIS to ensure that the prioritised patients/donors undergo systematic ranking. The prioritisation results presented in Table 8 are visualised in graphical formats (Fig. 7 for patients and Fig. 8 for donors) after categorising them into four groups based on descending patients’ scores for comparison.
Fig. 7.
Results of the four groups of patients. a First group. b Second group. c Third group. d Fourth group
Fig. 8.
Results of the four groups of donors. a First group. b Second group. c Third group. d Fourth group
The initial observation of the ranking results of the four patients and donors groups show that the groups are systematically distributed as the ranking results of the second group starting from the end of the ranking results of the first group and so on for other groups. Statistical analysis is performed amongst the patient groups, and Eqs. (15) and (16) are applied to obtain the M ± SD. In the first group, the value is M = 0.14538 ± 0.08301. The first group obtains the highest score amongst the four groups. The second group has a value of M = 0.11977 ± 0.07101 and a lower score than the first group but higher scores than the third and fourth groups. The third group has a value of M = 0.10887 ± 0.05795 and a lower score than the first, second, and third groups but a higher score than the fourth groups. The fourth group has a value of M = 0.09705 ± 0.04771 and has the lowest scores amongst the four groups. Furthermore, statistical analysis is performed amongst the donor groups, and Eqs. (15) and (16) are applied to obtain the M ± SD. In the first group, the value is M = 0.12993 ± 0.08581. The first group obtains the highest score amongst the four groups. The second group has a value of M = 0.10759 ± 0.07600 and has a lower score than the first group but higher scores than the third and fourth groups. The third group has a value of M = 0.10041 ± 0.07080 and has a lower score than the first, second, and third groups but higher scores than the fourth groups. The fourth group has a value of M = 0.08921 ± 0.06532 and the lowest scores amongst the four groups. The statistical results for patients and donors indicate that the results have undergone systematic ranking and are valid.
Claim points
The claim points of this study can be summarised as follows.
Serological/Protein Biomarkers and Strength Weights: Even within the area of infectious-disease research, various disciplines in clinical research such as molecular biology, microbiology, mycology, and epidemiology are involved. Despite this multidisciplinary mix, some efforts have been made to prioritise patients based on the criteria that are applicable and defined. According to the scope of the presented study which is COVID-19, the indication of the safety and suitability of CP for patients and donors is demarcated through the constructed weights of biomarker criteria for the first time based on three experts.
New COVID-19 Datasets and Evaluation: The augmented dataset is generated by a specialised expert based on standard medical-reference ranges that are applicable to patients and donors. Thus, we describe new measurement data about COVID-19-related CPs for patients/donors and release such data for public use. In this context, the transparency of the developed intelligent framework and associated processes is confirmed.
Intelligent Matching Component Execution: We We demonstrate the enhanced mechanical priority of two-dimensional patients/donors by using the new components of the four rules. Thus, the prioritisation results make the scoring more transparent for matching each of the critical patients with suitable donors according to their severity and blood types explicitly. The transfusion and balancing approach across the distributed telemedicine hospitals are involved in improving hospital management. In the case of a new patient or donor admitted to any hospital, the DM can repeat the rank across the distributed hospitals in real time.
Scalable Transfusion of CP within Centralised/Decentralised Connected Hospitals: Identifying and selecting eligible donors with a sufficient amount of plasma for efficient utilisation can be challenging within distributed hospitals. The selection of the best CP for critical COVID-19 patients is also challenging because this process is considered as a problem of MCDM, which complies with the national health requirements and known standard routine procedures [97]. The present study addresses these issues and indicates valid results on the basis of a fully automated intelligent computing framework. Thus, the transfusion of the CPs amongst patients and donors in either centralised or decentralised connected hospitals is accomplished, and the shortages of acute plasma in any hospital can be avoided.
Conclusion
The COVID-19 pandemic is critical; it requires rapid scalable load balancing, collaborative management, and decision making. This paper develops a novel interoperability CP-transfusion-rescue intelligent framework across centralised/decentralised telemedicine hospitals on the basis of the matching component process to provide an efficient CP from eligible donors to the most critical patients using the MCDM methods. A dataset, including COVID-19 patients/donors that have met the important criteria in the virology field, is augmented to improve the developed framework and achieve multiface requirements that can address multidimensional problems in the risk management of the COVID-19 pandemic. One main characteristic of the methodology described in this study is that it addresses the prioritisation task on the basis of the same DM for patients and donors. However, the field of COVID-19 infectious diseases has numerous aspects that need to be addressed comprehensively and continuously because this pandemic has shocked clinical organisations for the first time in decades. Therefore, various questions have emerged, and they should be assessed carefully before establishing the prioritisation characteristic intelligent framework. Do the five criteria cover all clinical characteristic aspects for the prioritisation configurations of patients and donors? If other criteria are missing, then how could the newly defined clinical criteria be added to the proposed DMs to be sufficient for the prioritisation process? How large should the group of participating clinical experts be and how should it be composed? If the methodology of prioritisation follows this COVID-19 clinical characteristic approach, then the original purpose for the transfusion of CP would be severely constrained and would result in desirable scalable load balancing amongst patients and donors within telemedicine hospitals. However, this new type of disease often requires more methodological approaches. Therefore, a patient/donor priority list may still be beneficial if decision makers keep in mind how to identify the importance of COVID-19 criteria from clinical prospective studies after collecting baseline information.
Acknowledgment
The authors are grateful to the Universiti Pendidikan Sultan Idris, Malaysia for funding this study under UPSI Rising Star Grant No. 2019–0125–109–01.
Appendix 1
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Albahri O, et al. Systematic review of artificial intelligence techniques in the detection and classification of COVID-19 medical images in terms of evaluation and benchmarking: Taxonomy analysis, challenges, future solutions and methodological aspects. J Infect Pub Health. 2020;13(10):1381–1396. doi: 10.1016/j.jiph.2020.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albahri A, et al. Multi-Biological Laboratory Examination Framework for the Prioritization of Patients with COVID-19 Based on Integrated AHP and Group VIKOR Methods. Int J Inf Technol Decis Mak. 2020;19(05):1247–1269. doi: 10.1142/S0219622020500285. [DOI] [Google Scholar]
- 3.Albahri A, Hamid RA. Role of biological data mining and machine learning techniques in detecting and diagnosing the novel coronavirus (COVID-19): A systematic review. J Med Syst. 2020;44(7):122. doi: 10.1007/s10916-020-01582-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.W. H. Organization (2020) Coronavirus disease 2019 (COVID-19): situation report, 82
- 5.Catelli R, Gargiulo F, Casola V, De Pietro G, Fujita H, Esposito M. Crosslingual named entity recognition for clinical de-identification applied to a COVID-19 Italian data set. Appl Soft Comput. 2020;97:106779. doi: 10.1016/j.asoc.2020.106779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hernandez-Matamoros A, Fujita H, Hayashi T, Perez-Meana H. Forecasting of COVID19 per regions using ARIMA models and polynomial functions. Appl Soft Comput. 2020;96:106610. doi: 10.1016/j.asoc.2020.106610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alamoodi A, Zaidan B, Zaidan A, Albahri O, Mohammed K, Malik R, ... & Hameed H (2020). Sentiment analysis and its applications in fighting COVID-19 and infectious diseases: A systematic review. Expert Syst Appl 114155 [DOI] [PMC free article] [PubMed]
- 8.Richardson S et al. (2020) Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York City area. JAMA [DOI] [PMC free article] [PubMed]
- 9.Remuzzi A, Remuzzi G (2020) COVID-19 and Italy: what next?. Lancet 395 [DOI] [PMC free article] [PubMed]
- 10.Albahri O, et al. Helping doctors hasten COVID-19 treatment: Towards a rescue framework for the transfusion of best convalescent plasma to the most critical patients based on biological requirements via ml and novel MCDM methods. Comput Methods Prog Biomed. 2020;196:105617. doi: 10.1016/j.cmpb.2020.105617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang CJ, Ng CY, Brook RH. Response to COVID-19 in Taiwan: big data analytics, new technology, and proactive testing. Jama. 2020;323(14):1341–1342. doi: 10.1001/jama.2020.3151. [DOI] [PubMed] [Google Scholar]
- 12.Anderson M, McKee M, Mossialos E Covid-19 exposes weaknesses in European response to outbreaks. BMJ 368:m1075. 10.1136/bmj.m1075 [DOI] [PubMed]
- 13.Shah S, et al. Evidence-Based Risk Mitigation and Stratification During COVID-19 for Return to Interventional Pain Practice: American Society of Interventional Pain Physicians (ASIPP) Guidelines. Pain Phys. 2020;23(4S):S161–S182. doi: 10.36076/ppj.2020/23/S161. [DOI] [PubMed] [Google Scholar]
- 14.Tabish M, et al. Approaches for prevention and environmental management of novel COVID-19. Environ Sci Pollut Res. 2020;24(3):1–11. doi: 10.1007/s11356-020-10640-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deeks JJ, et al. Antibody tests for identification of current and past infection with SARS-CoV-2. Cochrane Database Syst Rev. 2020;31(6):CD013652. doi: 10.1002/14651858.CD013652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marano G, et al. Convalescent plasma: new evidence for an old therapeutic tool? Blood Transfus. 2016;14(2):152. doi: 10.2450/2015.0131-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burnouf T, Seghatchian J. Ebola virus convalescent blood products: where we are now and where we may need to go. Transfus Apher Sci. 2014;51(2):120–125. doi: 10.1016/j.transci.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maxmen A. How blood from coronavirus survivors might save lives. Nature. 2020;580(7801):16–17. doi: 10.1038/d41586-020-00895-8. [DOI] [PubMed] [Google Scholar]
- 19.Chen L, Xiong J, Bao L, Shi Y. Convalescent plasma as a potential therapy for COVID-19. Lancet Infect Dis. 2020;20(4):398–400. doi: 10.1016/S1473-3099(20)30141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rajam G, Sampson J, Carlone GM, Ades EW. An augmented passive immune therapy to treat fulminant bacterial infections. Recent Patents Anti-Infect Drug Discov. 2010;5(2):157–167. doi: 10.2174/157489110791233496. [DOI] [PubMed] [Google Scholar]
- 21.Isidori A, de Leval L, Gergis U, Musto P, Porcu P. Management of Patients With Hematologic Malignancies During the COVID-19 Pandemic: Practical Considerations and Lessons to Be Learned. Front Oncol Rev. 2020;10(1439):33–44. doi: 10.3389/fonc.2020.01439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abolghasemi H, et al. Clinical efficacy of convalescent plasma for treatment of COVID-19 infections: Results of a multicenter clinical study. Transfus Apher Sci. 2020;59:102875. doi: 10.1016/j.transci.2020.102875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piechotta V, et al. Convalescent plasma or hyperimmune immunoglobulin for people with COVID-19: a living systematic review. Cochrane Database Syst Rev. 2020;7(7):Cd013600. doi: 10.1002/14651858.CD013600.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vassallo RR, Hilton JF, Bravo MD, Vittinghoff E, Custer B, Kamel H. Recovery of Iron Stores After Adolescents Donate Blood. Pediatrics. 2020;146(1):e20193316. doi: 10.1542/peds.2019-3316. [DOI] [PubMed] [Google Scholar]
- 25.Albahri O, et al. Fault-tolerant mHealth framework in the context of IoT-based real-time wearable health data sensors. IEEE Access. 2019;7:50052–50080. doi: 10.1109/ACCESS.2019.2910411. [DOI] [Google Scholar]
- 26.Hanratty B, Burton JK, Goodman C, Gordon AL, Spilsbury K. Covid-19 and lack of linked datasets for care homes. BMJ. 2020;369:m2463. doi: 10.1136/bmj.m2463. [DOI] [PubMed] [Google Scholar]
- 27.Hall LO, Paul R, Goldgof DB, Goldgof GM (2020) Finding covid-19 from chest x-rays using deep learning on a small dataset. arXiv preprint arXiv:2004.02060 22(3): 333-335
- 28.Loey M, Smarandache F, Khalifa NEM (2020) A deep transfer learning model with classical data augmentation and CGAN to detect COVID-19 from chest CT radiography digital images," ed: Preprints.org [DOI] [PMC free article] [PubMed]
- 29.Albahri AS, Alwan JK, Taha ZK, Ismail SF, Hamid RA, Zaidan AA, ... & Alsalem MA (2021) IoT-based telemedicine for disease prevention and health promotion: State-of-the-Art. J Netw Comput Appl 173: 102873
- 30.(2019) Use of internet of things to provide a new model for remote heart attack prediction. Telemed and e-Health 25(6): 499-510 [DOI] [PubMed]
- 31.Mohammed K, et al. A Uniform Intelligent Prioritisation for Solving Diverse and Big Data Generated From Multiple Chronic Diseases Patients Based on Hybrid Decision-Making and Voting Method. IEEE Access. 2020;8:91521–91530. doi: 10.1109/ACCESS.2020.2994746. [DOI] [Google Scholar]
- 32.Albahri A, et al. Based multiple heterogeneous wearable sensors: A smart real-time health monitoring structured for hospitals distributor. IEEE Access. 2019;7:37269–37323. doi: 10.1109/ACCESS.2019.2898214. [DOI] [Google Scholar]
- 33.Mohsin A, et al. Based blockchain-PSO-AES techniques in finger vein biometrics: A novel verification secure framework for patient authentication. Comput Stand Interfaces. 2019;66:103343. doi: 10.1016/j.csi.2019.04.002. [DOI] [Google Scholar]
- 34.Shen C, et al. Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. JAMA. 2020;323(16):1582–1589. doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seising R, Tabacchi ME (2013) Fuzziness and Medicine: Philosophical Reflections and Application Systems in Health Care: A Companion Volume to Sadegh-Zadeh’s Handbook of Analytical Philosophy of Medicine. Springer pp. XXVI, 447
- 36.Hu R, Ruan G, Xiang S, Huang M, Liang Q, Li J (2020) Automated Diagnosis of COVID-19 Using Deep Learning and Data Augmentation on Chest CT. medRxiv 2020.04.24.20078998
- 37.Wang L. C-reactive protein levels in the early stage of COVID-19. Med Mal Infect. 2020;50(4):332–334. doi: 10.1016/j.medmal.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coperchini F, Chiovato L, Croce L, Magri F, Rotondi M. "The cytokine storm in COVID-19: An overview of the involvement of the chemokine/chemokine-receptor system," (in eng) Cytokine Growth Factor Rev. 2020;53:25–32. doi: 10.1016/j.cytogfr.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aziz M, Fatima R, Lee-Smith W, Assaly R. The association of low serum albumin level with severe COVID-19: a systematic review and meta-analysis. Crit Care. 2020;24(1):1–4. doi: 10.1186/s13054-020-02995-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zaidan A, Zaidan B, Hussain M, Haiqi A, Kiah MM, Abdulnabi M. Multi-criteria analysis for OS-EMR software selection problem: A comparative study. Decis Support Syst. 2015;78(4):15–27. doi: 10.1016/j.dss.2015.07.002. [DOI] [Google Scholar]
- 41.Abdullateef BN, Elias NF, Mohamed H, Zaidan A, Zaidan B. An evaluation and selection problems of OSS-LMS packages. SpringerPlus. 2016;5(1):248–255. doi: 10.1186/s40064-016-1828-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yas QM, Zadain A, Zaidan B, Lakulu M, Rahmatullah B. Towards on develop a framework for the evaluation and benchmarking of skin detectors based on artificial intelligent models using multi-criteria decision-making techniques. Int J Pattern Recognit Artif Intell. 2017;31(03):1759002. doi: 10.1142/S0218001417590029. [DOI] [Google Scholar]
- 43.Zaidan B, Zaidan A, Karim HA, Ahmad NN. A new digital watermarking evaluation and benchmarking methodology using an external group of evaluators and multi-criteria analysis based on ‘large-scale data’. J Softw Pract Exp. 2017;47(10):1365–1392. doi: 10.1002/spe.2465. [DOI] [Google Scholar]
- 44.Zaidan B, Zaidan A. Software and hardware FPGA-based digital watermarking and steganography approaches: Toward new methodology for evaluation and benchmarking using multi-criteria decision-making techniques. J Circ Syst Comput. 2017;26(07):1750116. doi: 10.1142/S021812661750116X. [DOI] [Google Scholar]
- 45.Zaidan BB, Zaidan AA, Karim HA, Ahmad NN A new approach based on multi-dimensional evaluation and benchmarking for data hiding techniques. Int J Inf Technol Dec Mak 0(0): 1-42
- 46.Jumaah F, Zaidan A, Zaidan B, Bahbibi R, Qahtan M, Sali AJTS (2018) Technique for order performance by similarity to ideal solution for solving complex situations in multi-criteria optimization of the tracking channels of GPS baseband telecommunication receivers. 68(3): 425-443
- 47.Rahmatullah B, Zaidan AA, Mohamed F, Sali A (2017) Multi-complex attributes analysis for optimum GPS baseband receiver tracking channels selection. In: 2017 4th International Conference on Control, Decision and Information Technologies (CoDIT), pp. 1084-1088
- 48.Salman OH, Zaidan A, Zaidan B, Naser K. Novel methodology for triage and prioritizing using “big data” patients with chronic heart diseases through telemedicine environmental. Int J Inf Technol Decis Mak. 2017;16(05):1211–1245. doi: 10.1142/S0219622017500225. [DOI] [Google Scholar]
- 49.Yas QM, Zaidan A, Zaidan B, Rahmatullah B, Karim HA. Comprehensive insights into evaluation and benchmarking of real-time skin detectors: Review, open issues & challenges, and recommended solutions. Measurement. 2018;114:243–260. doi: 10.1016/j.measurement.2017.09.027. [DOI] [Google Scholar]
- 50.Zaidan B, Zaidan A. Comparative study on the evaluation and benchmarking information hiding approaches based multi-measurement analysis using TOPSIS method with different normalisation, separation and context techniques. Measurement. 2018;117:277–294. doi: 10.1016/j.measurement.2017.12.019. [DOI] [Google Scholar]
- 51.Tinetti M, et al. Challenges and strategies in patients' health priorities-aligned decision-making for older adults with multiple chronic conditions. PLoS One. 2019;14(6):e0218249. doi: 10.1371/journal.pone.0218249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martínez V, Navarro C, Cano C, Fajardo W, Blanco A. DrugNet: Network-based drug–disease prioritization by integrating heterogeneous data. Artif Intell Med. 2015;63(1):41–49. doi: 10.1016/j.artmed.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 53.Miao F, et al. Continuous blood pressure measurement from one-channel electrocardiogram signal using deep-learning techniques. Artif Intell Med. 2020;108:101919. doi: 10.1016/j.artmed.2020.101919. [DOI] [PubMed] [Google Scholar]
- 54.Kalid N, et al. Based on real time remote health monitoring systems: a new approach for prioritization “large scales data” patients with chronic heart diseases using body sensors and communication technology. J Med Syst. 2018;42(4):69. doi: 10.1007/s10916-018-0916-7. [DOI] [PubMed] [Google Scholar]
- 55.Zaidan A, et al. A review on smartphone skin cancer diagnosis apps in evaluation and benchmarking: coherent taxonomy, open issues and recommendation pathway solution. Heal Technol. 2018;8(4):223–238. doi: 10.1007/s12553-018-0223-9. [DOI] [Google Scholar]
- 56.Albahri O, et al. Systematic review of real-time remote health monitoring system in triage and priority-based sensor technology: Taxonomy, open challenges, motivation and recommendations. J Med Syst. 2018;42(5):80. doi: 10.1007/s10916-018-0943-4. [DOI] [PubMed] [Google Scholar]
- 57.Alsalem M, et al. Systematic review of an automated multiclass detection and classification system for acute Leukaemia in terms of evaluation and benchmarking, open challenges, issues and methodological aspects. J Med Syst. 2018;42(11):204. doi: 10.1007/s10916-018-1064-9. [DOI] [PubMed] [Google Scholar]
- 58.Kalid N, Zaidan A, Zaidan B, Salman OH, Hashim M, Muzammil H. Based real time remote health monitoring systems: A review on patients prioritization and related" big data" using body sensors information and communication technology. J Med Syst. 2018;42(2):30. doi: 10.1007/s10916-017-0883-4. [DOI] [PubMed] [Google Scholar]
- 59.Albahri A, Zaidan A, Albahri O, Zaidan B, Alsalem M. Real-time fault-tolerant mHealth system: Comprehensive review of healthcare services, opens issues, challenges and methodological aspects. J Med Syst. 2018;42(8):137. doi: 10.1007/s10916-018-0983-9. [DOI] [PubMed] [Google Scholar]
- 60.Albahri O, Zaidan A, Zaidan B, Hashim M, Albahri A, Alsalem M. Real-time remote health-monitoring Systems in a Medical Centre: A review of the provision of healthcare services-based body sensor information, open challenges and methodological aspects. J Med Syst. 2018;42(9):164. doi: 10.1007/s10916-018-1006-6. [DOI] [PubMed] [Google Scholar]
- 61.Zughoul O, et al. Comprehensive insights into the criteria of student performance in various educational domains. IEEE Access. 2018;6(4):73245–73264. doi: 10.1109/ACCESS.2018.2881282. [DOI] [Google Scholar]
- 62.Badi I, Pamucar D. Supplier selection for steelmaking company by using combined Grey-MARCOS methods. Decis Mak Appl Manag Eng. 2020;3(2):37–48. doi: 10.31181/dmame2003037b. [DOI] [Google Scholar]
- 63.Alao MA, Ayodele TR, Ogunjuyigbe ASO, Popoola OM. Multi-criteria decision based waste to energy technology selection using entropy-weighted TOPSIS technique: The case study of Lagos, Nigeria. Energy. 2020;201:117675. doi: 10.1016/j.energy.2020.117675. [DOI] [Google Scholar]
- 64.Almahdi E, Zaidan A, Zaidan B, Alsalem M, Albahri O, Albahri A. Mobile patient monitoring systems from a benchmarking aspect: Challenges, open issues and recommended solutions. J Med Syst. 2019;43(7):207. doi: 10.1007/s10916-019-1336-z. [DOI] [PubMed] [Google Scholar]
- 65.Alsalem M, et al. Multiclass benchmarking framework for automated acute Leukaemia detection and classification based on BWM and group-VIKOR. J Med Syst. 2019;43(7):212. doi: 10.1007/s10916-019-1338-x. [DOI] [PubMed] [Google Scholar]
- 66.Almahdi E, Zaidan A, Zaidan B, Alsalem M, Albahri O, Albahri A. Mobile-based patient monitoring systems: A prioritisation framework using multi-criteria decision-making techniques. J Med Syst. 2019;43(7):219. doi: 10.1007/s10916-019-1339-9. [DOI] [PubMed] [Google Scholar]
- 67.Mohammed K, et al. Real-time remote-health monitoring systems: a review on patients prioritisation for multiple-chronic diseases, taxonomy analysis, concerns and solution procedure. J Med Syst. 2019;43(7):223. doi: 10.1007/s10916-019-1362-x. [DOI] [PubMed] [Google Scholar]
- 68.Khatari M, Zaidan A, Zaidan B, Albahri O, Alsalem M. Multi-criteria evaluation and benchmarking for active queue management methods: Open issues challenges and recommended pathway solutions. Int J Inf Technol Decis Mak. 2019;18(4):1187–1242. doi: 10.1142/S0219622019300039. [DOI] [Google Scholar]
- 69.Ibrahim N, et al. Multi-criteria evaluation and benchmarking for young learners’ English language mobile applications in terms of LSRW skills. IEEE Access. 2019;7(7):146620–146651. doi: 10.1109/ACCESS.2019.2941640. [DOI] [Google Scholar]
- 70.Talal M, et al. Comprehensive review and analysis of anti-malware apps for smartphones. Telecommun Syst. 2019;72(2):285–337. doi: 10.1007/s11235-019-00575-7. [DOI] [Google Scholar]
- 71.Napi NM, Zaidan AA, Zaidan BB, Albahri OS, Alsalem MA, Albahri AS. Medical emergency triage and patient prioritisation in a telemedicine environment: a systematic review. Heal Technol. 2019;9(5):679–700. doi: 10.1007/s12553-019-00357-w. [DOI] [Google Scholar]
- 72.Enaizan O, et al. Electronic medical record systems: Decision support examination framework for individual, security and privacy concerns using multi-perspective analysis. Heal Technol. 2020;10(3):795–822. doi: 10.1007/s12553-018-0278-7. [DOI] [Google Scholar]
- 73.Zaidan A, Zaidan B, Alsalem M, Albahri O, Albahri A, Qahtan M. Multi-agent learning neural network and Bayesian model for real-time IoT skin detectors: a new evaluation and benchmarking methodology. Neural Comput Applic. 2020;32(12):8315–8366. doi: 10.1007/s00521-019-04325-3. [DOI] [Google Scholar]
- 74.Kaur R, Singh S, Kumar H. AuthCom: Authorship verification and compromised account detection in online social networks using AHP-TOPSIS embedded profiling based technique. Expert Syst Appl. 2018;113:397–414. doi: 10.1016/j.eswa.2018.07.011. [DOI] [Google Scholar]
- 75.Zaidan AA, Zaidan BB, Al-Haiqi A, Kiah MLM, Hussain M, Abdulnabi M. Evaluation and selection of open-source EMR software packages based on integrated AHP and TOPSIS. J Biomed Inform. 2015;53(8):390–404. doi: 10.1016/j.jbi.2014.11.012. [DOI] [PubMed] [Google Scholar]
- 76.Rajak M, Shaw K. Evaluation and selection of mobile health (mHealth) applications using AHP and fuzzy TOPSIS. Technol Soc. 2019;59:101186. doi: 10.1016/j.techsoc.2019.101186. [DOI] [Google Scholar]
- 77.Lahby M, Cherkaoui L, Adib A. A Novel Ranking Algorithm Based Network Selection For Heterogeneous Wireless Access. J Netw. 2013;8:263–272. [Google Scholar]
- 78.Saksrisathaporn K, Bouras A, Reeveerakul N, Charles A. Application of a Decision Model by Using an Integration of AHP and TOPSIS Approaches within Humanitarian Operation Life Cycle. Int J Inf Technol Decis Mak. 2016;15(04):887–918. doi: 10.1142/S0219622015500261. [DOI] [Google Scholar]
- 79.Tariq I, et al. MOGSABAT: a metaheuristic hybrid algorithm for solving multi-objective optimisation problems. Neural Comput Applic. 2020;32(8):3101–3115. doi: 10.1007/s00521-018-3808-3. [DOI] [Google Scholar]
- 80.Abdulkareem KH et al. (2020) A new standardisation and selection framework for real-time image dehazing algorithms from multi-foggy scenes based on fuzzy Delphi and hybrid multi-criteria decision analysis methods. Neural Comput Applic
- 81.Mohammed K, et al. Novel technique for reorganisation of opinion order to interval levels for solving several instances representing prioritisation in patients with multiple chronic diseases. Comput Methods Prog Biomed. 2020;185:105151. doi: 10.1016/j.cmpb.2019.105151. [DOI] [PubMed] [Google Scholar]
- 82.Saaty TL. A scaling method for priorities in hierarchical structures. J Math Psychol. 1977;15(3):234–281. doi: 10.1016/0022-2496(77)90033-5. [DOI] [Google Scholar]
- 83.Wind Y, Saaty TL. Marketing Applications of the Analytic Hierarchy Process. Manag Sci. 1980;26(7):641–658. doi: 10.1287/mnsc.26.7.641. [DOI] [Google Scholar]
- 84.Ahmadi H, Nilashi M, Ibrahim O. Organizational decision to adopt hospital information system: An empirical investigation in the case of Malaysian public hospitals. Int J Med Inform. 2015;84(3):166–188. doi: 10.1016/j.ijmedinf.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 85.Pecchia L, Bath PA, Pendleton N, Bracale M. Analytic Hierarchy Process (AHP) for examining healthcare professionals' assessments of risk factors. The relative importance of risk factors for falls in community-dwelling older people. Methods Inf Med. 2011;50(5):435–444. doi: 10.3414/ME10-01-0028. [DOI] [PubMed] [Google Scholar]
- 86.Saaty TL, Ozdemir MS. Why the magic number seven plus or minus two. Math Comput Model. 2003;38(3):233–244. doi: 10.1016/S0895-7177(03)90083-5. [DOI] [Google Scholar]
- 87.Sherekar V, Tatikonda M, Student M. Impact of factor affecting on labour productivity in construction projects by AHP method. Int J Eng Sci Comput. 2016;6(6):6771–6775. [Google Scholar]
- 88.Zaidan A, Zaidan B, Alsalem M, Momani F, Zughoul O. Novel Multiperspective Hiring Framework for the Selection of Software Programmer Applicants Based on AHP and Group TOPSIS Techniques. Int J Inf Technol Decis Mak. 2020;18(4):1–73. [Google Scholar]
- 89.Abdulkareem KH, et al. A Novel Multi-Perspective Benchmarking Framework for Selecting Image Dehazing Intelligent Algorithms Based on BWM and Group VIKOR Techniques. Int J Inf Technol Decis Mak. 2020;19(3):909–957. doi: 10.1142/S0219622020500169. [DOI] [Google Scholar]
- 90.Salih MM, Zaidan B, Zaidan A. Fuzzy decision by opinion score method. Appl Soft Comput. 2020;96(4):106595. doi: 10.1016/j.asoc.2020.106595. [DOI] [Google Scholar]
- 91.R. T. Mohammed et al. Review of the Research Landscape of Multi-criteria Evaluation and Benchmarking Processes for Many-objective Optimisation Methods: Coherent Taxonomy, Challenges and Recommended Solution. Int J Inf Technol Decis Mak 0, no. ja: p. null
- 92.Albahri AS, Hamid RA (2020) Detection-based Prioritisation: Framework of Multi-laboratory Characteristics for Asymptomatic COVID-19 Carriers Based on Integrated Entropy–TOPSIS Methods. Artif Intell Med 101983 [DOI] [PMC free article] [PubMed]
- 93.Albahri OS, Zaidan AA, Salih MM, Zaidan BB, Khatari MA, Ahmed MA, ... & Alazab M (2020) Multidimensional benchmarking of the active queue management methods of network congestion control based on extension of fuzzy decision by opinion score method. Int J Intell Syst
- 94.Hatami-Marbini A, Kangi F. An extension of fuzzy TOPSIS for a group decision making with an application to Tehran stock exchange. Appl Soft Comput. 2017;52:1084–1097. doi: 10.1016/j.asoc.2016.09.021. [DOI] [Google Scholar]
- 95.Grigoroudis E, Orfanoudaki E, Zopounidis C. Strategic performance measurement in a healthcare organisation: A multiple criteria approach based on balanced scorecard. Omega. 2012;40(1):104–119. doi: 10.1016/j.omega.2011.04.001. [DOI] [Google Scholar]
- 96.Qader M, Zaidan B, Zaidan A, Ali S, Kamaluddin M. A methodology for football players selection problem based on multi-measurements criteria analysis. Measurement. 2017;111:38–50. doi: 10.1016/j.measurement.2017.07.024. [DOI] [Google Scholar]
- 97.Lai KY, Ng WYG, Cheng FF. Human Ebola virus infection in West Africa: a review of available therapeutic agents that target different steps of the life cycle of Ebola virus. Infect Dis Poverty. 2014;3:43–43. doi: 10.1186/2049-9957-3-43. [DOI] [PMC free article] [PubMed] [Google Scholar]