Summary
KDM4B is a lysine-specific demethylase with a preferential activity on H3K9 tri/di-methylation (H3K9me3/2)-modified histones. H3K9 tri/di-demethylation is an important epigenetic mechanism responsible for silencing of gene expression in animal development and cancer. However, the role of KDM4B on human development is still poorly characterized. Through international data sharing, we gathered a cohort of nine individuals with mono-allelic de novo or inherited variants in KDM4B. All individuals presented with dysmorphic features and global developmental delay (GDD) with language and motor skills most affected. Three individuals had a history of seizures, and four had anomalies on brain imaging ranging from agenesis of the corpus callosum with hydrocephalus to cystic formations, abnormal hippocampi, and polymicrogyria. In mice, lysine demethylase 4B is expressed during brain development with high levels in the hippocampus, a region important for learning and memory. To understand how KDM4B variants can lead to GDD in humans, we assessed the effect of KDM4B disruption on brain anatomy and behavior through an in vivo heterozygous mouse model (Kdm4b+/−), focusing on neuroanatomical changes. In mutant mice, the total brain volume was significantly reduced with decreased size of the hippocampal dentate gyrus, partial agenesis of the corpus callosum, and ventriculomegaly. This report demonstrates that variants in KDM4B are associated with GDD/ intellectual disability and neuroanatomical defects. Our findings suggest that KDM4B variation leads to a chromatinopathy, broadening the spectrum of this group of Mendelian disorders caused by alterations in epigenetic machinery.
Keywords: KDM4B, JMJD2B, global developmental delay, intellectual disability, heterozygous variant, dysmorphic hippocampi, agenesis of the corpus callosum, neurodevelopmental disorder
Main Text
Global developmental delay (GDD) affects 1%–3% of children less than 5 years old, and a similar rate of intellectual disability (ID) is present in older children.1 Taken together, GDD and ID are a worldwide pediatric public health problem with a lifetime care cost of up to $1,000,000 per child.2 GDD and ID are often associated with other developmental disorders of childhood and can lead to increased caregiver stress.1,3 The causes of GDD/ID are multifactorial; data suggest that a majority of cases are secondary to a genetic cause.1,4 For example, in severe ID, an estimated 62% of cases are secondary to de novo variants.4,5 A genetic diagnosis can be extraordinarily important for a family to know; it can empower parents to advocate for their child and provide them with prognosis, validation, social support, educational services, and an understanding of reoccurrence risk.1,6 Our understanding of GDD/ID continues to evolve with the advancements in sequencing technologies and functional modeling leading to identification of novel genes and associated pathogenic variants. Using exome sequencing (ES), we have identified a cohort of nine individuals with GDD and heterozygous variants in KDM4B.
KDM4B (MIM: 609765, alterative name JMJD2B) is a lysine-specific histone demethylase with a catalytic activity against different histone modifications, including H3K9me3, H3K9me2, H3K36me3, H3K36me2, H4K20me2, and H1.4K26me3. H3K9me3/2 is the preferred substrate that is enriched at constitutive and facultative heterochromatin, localized at pericentromeric and telomeric regions as well as at nuclear lamina in mammalian cells.7 Accumulation of H3K9 methylation is associated with chromatin compaction and gene repression.8 Previous studies of KDM4B have primarily been in cancer models and have shown that KDM4B is strongly regulated by multiple cellular stimuli, including oxygen concentration and HiF1 alpha, androgens, estrogen, and DNA damage.9, 10, 11, 12, 13, 14
Recent data suggest that KDM4B is expressed during brain development with high expression in the hippocampus, a region important for learning and memory.15 In a neuron-specific KDM4B (JMJD2B)-deficient mouse model, its depletion was associated with an increase in total dendritic spine number within the hippocampus and a notable decrease in maturity of the dendritic spines. These mice also exhibited hyperactive behavior, had deficits in their working memory, and were prone to seizures.15 This study suggests that KDM4B depletion alters connectivity, particularly within the hippocampus. Several other histone lysine demethylases have previously been linked to neurodevelopmental disorders, including KDM5C, which causes an X-linked intellectual disability syndrome, and KDM6A, which has been implicated in Kabuki syndrome.16,17 In the case of KDM5C, its depletion is thought to lead to altered dendritic development in the rodent cerebellar cortex, while haploinsufficiency of KDM6A in Kabuki syndrome is thought to affect neural differentiation.16,18 However, the role of KDM4B in human development has not been investigated.
We identified a cohort of nine individuals with extremely rare variants in KDM4B and presenting with syndromic GDD/ID and neuroanatomical findings. Using a heterozygous mouse model (Kdm4b+/−), we performed a systematic assessment of brain regions to analyze neuroanatomical defects in this model and identified structural anomalies compatible with those identified in our cohort. Overall, this study highlights the importance of KDM4B in human brain development and cognitive function.
We performed trio ES on an individual (I:2) who presented with GDD, agenesis of the corpus callosum, intracranial cysts leading to obstructive hydrocephalus, seizures, abnormal hippocampi, and dysmorphic facial features (Tables 1, S1, and S2) and identified a de novo missense variant in KDM4B. International data sharing through MatchMaker Exchange allowed us to identify eight additional individuals carrying rare missense variants in this gene with an overlapping clinical spectrum (Figure 1A; Table 1). In order to better delineate the symptomatology associated with KDM4B variation, we first compared the clinical features and MRIs of each of the individuals identified. All were diagnosed with GDD and/or ID and language and gross motor skills were found to be the most affected; language delay was present in eight out of nine and gross motor delay was present in six out of nine of the individuals. Three of the individuals (I:2, I:4, and I:7) had a history of seizures, and four of them (I:1, I:2, I:5, and I:7) had structural anomalies appreciated on their brain MRIs ranging from agenesis of the corpus callosum with hydrocephalus to cystic formations, polymicrogyria, and abnormal formation of the hippocampus (Figure 2). Only one of the individuals (I:6) was microcephalic. Dysmorphic features were present in this cohort as well, including abnormal palpebral fissures, short philtrum, sparse hair, and clinodactyly (Table S1).
Table 1.
Clinical and Genetic Findings of Individuals with KDM4B Variants
| Affected Individual (I)/ Country | I:1 (Italy) | I:2 (USA) | I:3 (Netherlands) | I:4 (Netherlands) | I:5 (France) | I:6 (France) | I:7 (Canada) | I:8 (France) | I:9 (France) |
|---|---|---|---|---|---|---|---|---|---|
| Variant from reference sequence: NM_015015.2 | c.3284C>T (p.Pro1095Leu) | c.2303A>G (p.His768Arg) | c.659T>C (p.Leu220Pro) | c.288C>T (r.287_317del; p.Glu97Thrfs∗66); synonymous variant leading to new donor splice site and frameshift | c.2221dup (p.Glu741Glyfs∗41) | c.1778_1779delAG (p.Glu593Glyfs∗41) | c.664C>T (p.Arg222Trp) | c.371_374del (p.Lys124Thrfs∗48) | c.1907−1G>C (p.?) |
| gnomAD frequency | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| CADD score | 25.4 | 25.6 | 32 | − | − | − | 32 | − | − |
| Inheritance | de novo | de novo | de novo | de novo | de novo | paternally inherited | de novo | de novo | maternally inherited |
| Sex | male | male | female | male | male | female | female | male | male |
| Family history | NC | parents from Middle East and consanguineous | NC | NC | NC | father with learning disabilities | NC | mother and father with mild learning disabilities | mother with a similar phenotype |
| Gross motor delay | + | + | + | + | + | − | + | − | − |
| Fine motor delay | + | + | − | − | − | − | + | + | − |
| Language delay | + | + | + | + | + | + | + | + | − |
| GDD or ID | +, GDD | +, GDD | +, GDD | +, GDD | +, GDD and mild ID | +, GDD and severe ID, IQ = 50 | +, GDD | +, GDD and mild ID, IQ 60–72 | +, mild ID |
| Tone | hypotonia | ND | ND | ND | ND | ND | central hypotonia, peripheral hypertonia | facial hypotonia | ND |
| Seizures | − | +, neonatal seizures, none since then | − | +, seizures starting at 2.5 years | − | − | +, history of infantile spasms starting at 3 weeks | − | − |
| Behavioral concerns | ND | ADHD | behavior concerns, autistic features | ND | history of intermittent aggression with other children | eating disorder; compulsive behavior | ND | aggression, ADHD | − |
| Other neurological abnormalities | − | +, obstructive hydrocephalus s/p shunt placement | +, ataxia, autistic features | − | +, general clumsiness | − | − | − | − |
Abbreviations: +, features are present; −, features are absent; NC, noncontributory; ND, not documented.
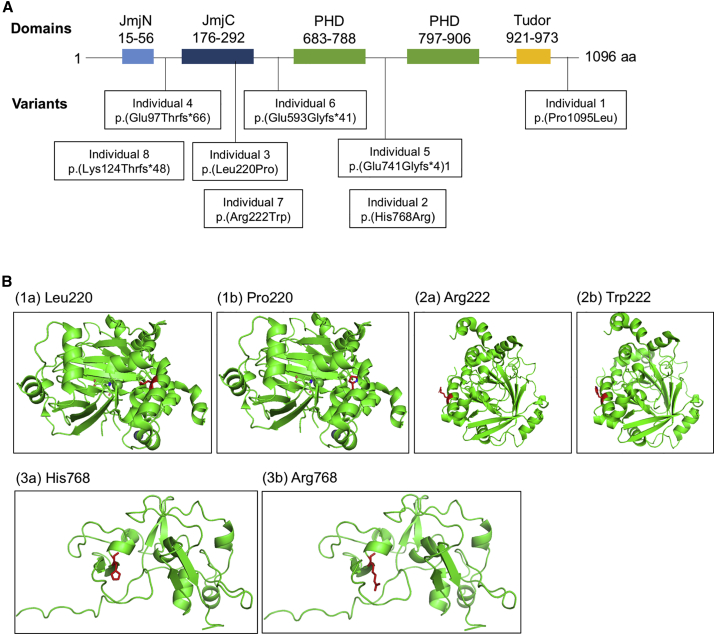
Figure 1.
KDM4B Variant Localization and Molecular Modeling
(A) KDM4B with domains and patient individual variants noted.
(B) Molecular modeling of the missense variants in individuals (I) 2, 3, and 7. (1a) shows the helix formed by the catalytic domain of KDM4B. Residue Leu220 is in red. (2b) shows the local destruction of the helical structure when Pro220 is substituted. Pro220 is shown in red. (2a) shows the residue Arg222 within the catalytic domain of KDM4B; Arg222 is highlighted in red. (2b) shows the substitution of Arg222 for Trp222 in I:7, which creates a bulky hydrophobic side chain most likely altering the folding and stability of the helix formation; Trp222 is shown in red. (3a) shows His768 and the zinc knuckle formation; His768 highlighted in red. (3b) shows the unraveling of the zinc knuckle with the substitution of Arg768 in I:2; Arg768 is shown in red.
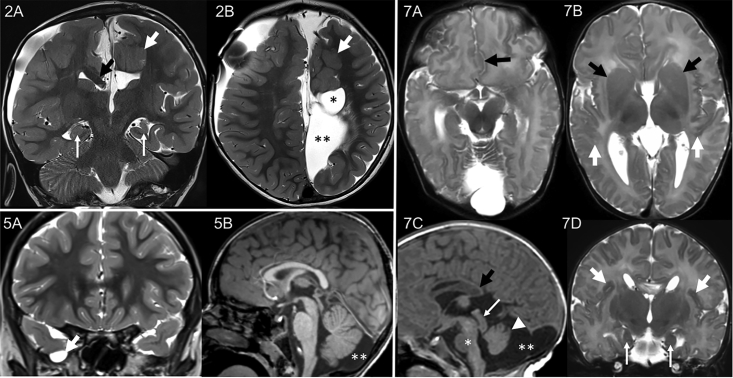
Figure 2.
MRI Findings in Individuals 2, 5, and 7
I:2 had many abnormalities, including agenesis of the corpus callosum with a right bundle of Probst (2A, black arrow), white matter cysts (2B, asterisk), interhemispheric cysts (2B, two asterisks), polymicrogyria (2A and 2B, white arrows), and incompletely rotated dysmorphic hippocampi with the left decreased in volume (2A, thin arrows). The following are not shown: absent left fornix with right fornix fused with the bundle of Probst, subcortical gray matter heterotopia, stenogyria of the right hemisphere (probably due to prior hydrocephalus), and decreased white matter volume. I:5 had a small right middle cranial fossa meningocele (5A, arrow) and mega cisterna magna (5B, two asterisks). I:7 also had many abnormalities, including partial agenesis of the corpus callosum (7C, black arrow), polymicrogyria (7B and 7D, arrows), dysmorphic hippocampi (7D, thin arrows), interdigitation of the frontal lobes (7A, arrow), fused caudate and lentiform nuclei with absent anterior limb internal capsule (7B, arrows), mega cisterna magna (7C, two asterisks), prominent tectum (7C, arrow), incompletely rotated small vermis (7C, arrowhead), and hypoplastic brainstem (7C, asterisk). The following are not shown: dysmorphic partially fused fornices, azygous ACA, small optic nerves, and small cerebellar hemispheres.
Seven of the individuals (I:1–I:5, I:7, and I:8) harbored de novo variants, while two (I:6 and I:9) inherited them from affected parents. Four of the variants were missense, three were frameshift, one was a single synonymous substitution predicted to lead to a splicing defect, and one was a canonical splicing variant leading to loss of function (Figures 1A and S1). The combined annotation dependent depletion (CADD) scores of the missense variants ranged from 25.4 to 32, and all identified variants were absent from gnomAD, including the inherited variants from I:6 and I:9, which were identified through this study.19 According to data from gnomAD, KDM4B has a significantly reduced number of truncating and missense variants in controls, indicating a constraint on both types of variants in a control population (probability of being loss-of-function intolerant [pLi] = 1, Z score for missense variants = 3.48).
Molecular modeling was computed to assess how variants in KDM4B would affect the protein’s structure and function. None of the missense variants showed a predicted effect on splicing. KDM4B has five domains, which include two jumonji catalytic demethylase domains, one is the N-terminal portion (JmjN) and one is the C-terminal portion (JmjC) domain; a double tudor domain; and two plant homeodomains (PHDs). The JmjC domain is the catalytic domain of the protein, while the JmjN domain helps to provide essential structure. The tudor domain binds the histones, H3K23me2/3, allowing for demethylation.20 The PHDs contain zinc fingers, which are important for KDM4B’s ability to interact with the chromatin as well. The variant in I:1, p.Pro1095Leu, is located at the tail of KDM4B, which is a flexible region (Figure 1A). This variant may not impair the structure of KDM4B, but it may alter KDM4B’s interactions with binding partners. The variant in I:2, p.His768Arg, is predicted to destabilize the zinc knuckle that connects the two zinc fingers within the PHD. This prediction is based on the structural similarity between the zinc fingers of KDM4B and the BRPF1. His768 has a similar localization in KDM4B to His300 in BRPF1. His300 forms a close contact with a zinc molecule and is important for the zinc knuckle structure. Because His768 plays a similar role in KDM4B, the introduction of arginine will most likely break the contact formed with zinc, destabilizing the zinc knuckle structure as well (Figure 1B). The variants in I:3 and I:7, p.Leu220Pro and p.Arg222Trp, respectively, are both located within the catalytic, JmjC domain. In I:3, the substitution of leucine for proline will destroy the local helical structure of the catalytic domain, most likely impairing KDM4B catalytic function (Figure 1B[1a and 1b]). In I:7, arginine is substituted for tryptophan on the outside of the protein, creating a bulky hydrophobic side chain, which most likely alters the folding and stability of the helix (Figure 1B). The molecular modeling overall suggests that missense variants in KDM4B lead to loss or reduction of KDM4B function.
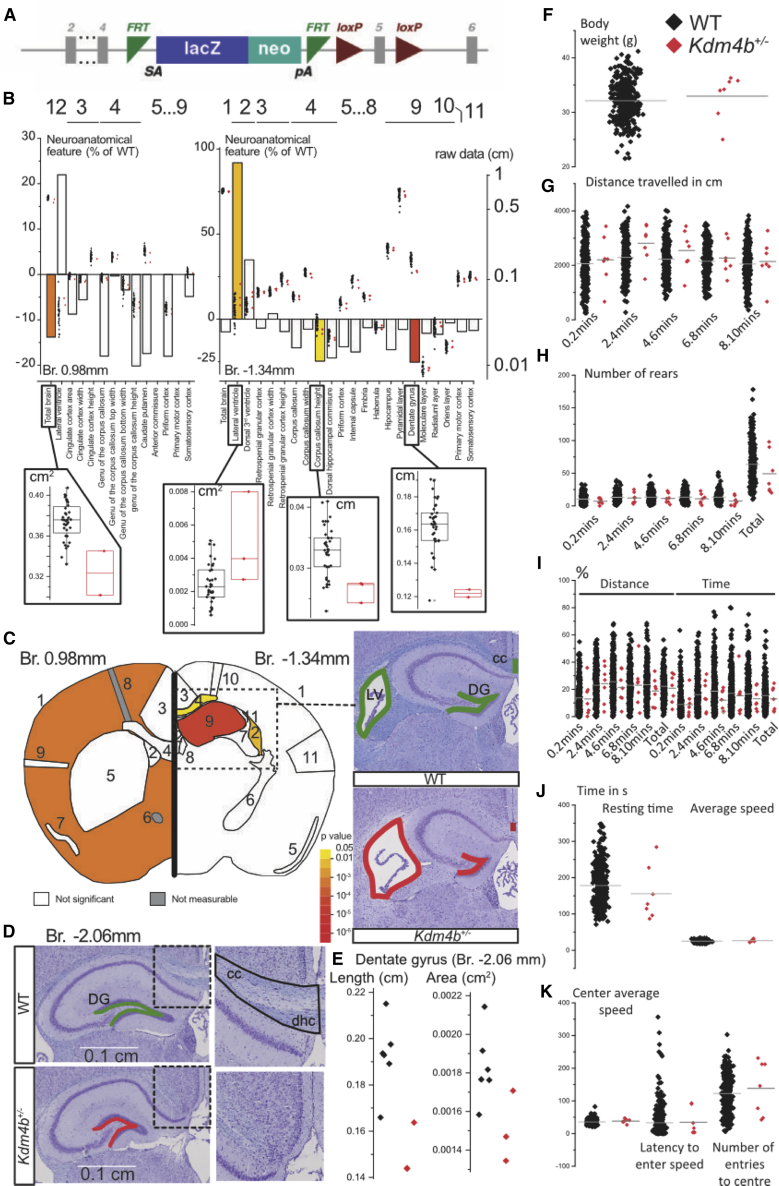
We then assessed how KDM4B disruption impacts brain anatomy and behavior through a mouse model, with a focus on the size and shape of the brain structures. Mouse mutants were generated with the knockout-first allele method.21 The strategy relies on the identification of an exon common to all transcript variants (exon 5), upstream of which a LacZ cassette was inserted. Exon 5 of the Kdm4b allele was flanked by loxP sequences bilaterally (Figure 3A). The resulting Kdm4btm1a(EUCOMM)Wtsi mice were then phenotyped.
Figure 3.
Mouse Studies Reveal a Role of KDM4B in the Anatomy of the Brain
(A) Construction of the Kdm4btm1a(EUCOMM)Wtsi allele.
(B) Histograms for three heterozygous Kdm4b mice showing percentage difference relative to 40 WTs. The overlapping dots show actual data points. To present areas and lengths on the same scale, the square root of areas was used. Inserts are full dot and box plots of WT (black) versus Kdm4b+/− mice (red) for each of the four significant parameters (total brain area, lateral ventricle area, corpus callosum height, dentate gyrus length).
(C and D) Left: schematic representation of a section at Bregma +0.98 mm and Bregma −1.34 mm. Colored regions indicate the presence of at least one significant parameter within the brain region at the 0.05 level. White coloring indicates a p value higher than 0.05 and gray shows there is not enough data to calculate a p value. Right: illustrating example of WT and Kdm4b+/− brain images in coronal sections double-stained for Nissl and Luxol at Bregma −1.34 (C) and Bregma −2.06 mm (D).
(E) Dot plot of the coronal section at Bregma −2.06 mm showing further characterization of the hippocampus.
(F) Body weight of seven Kdm4b+/− and 771 baseline control mice.
(G) Distance traveled in cm in the open-field arena shown in sessions of 2 min each.
(H) Number of rears measured in the open-field arena.
(I) Distance and time in the open-field arena expressed in percentages.
(J) Resting time and average speed in the open-field test.
(K) Average speed in the center of the open-field arena, latency to enter the center, and the number of entries to the center.
At weaning age, mouse survival was assessed from 142 successfully genotyped mice originating from several different litters and derived from a heterozygous-by-heterozygous breeding scheme. We obtained the expected number of wild type (WT) and heterozygous Kdm4b+/− mice, but only five (3%) were homozygous and all were female mice, suggesting that the tm1a allele of Kdm4b is not compatible with life in males. To determine the window of death, we carried out a recessive lethality screen at mouse embryonic day 14.5 (E14.5). The rate of homozygosity was, as expected, 22.2% (eight out of 36 embryos assessed).
The reason underlying embryonic lethality in the homozygous knockout mice is unknown and will require further studies; however, these preliminary data suggest death’s occurring late during development between E14.5 and weaning age. Male and female Kdm4b+/− mice were then studied independently. The weights of Kdm4b+/− mice did not significantly differ from their litter-matched controls (Table S3). In line with this, there were no significant differences when comparing to a set of 771 baseline controls accumulated over a 1 year period (Figure 3F; Table S4).
Using a recently developed robust approach for the assessment of 63 brain parameters across 15 distinct brain regions, we analyzed neuroanatomical defects in adult Kdm4b+/− mice.22 This consisted of a systematic quantification of the same coronal brain regions at Bregma +0.98 mm and Bregma −1.34 mm, as well as an additional coronal section at Bregma −2.06 mm, to further characterize the hippocampus and the corpus callosum. To minimize variation due to sex and age, we only used male mice aged 16 weeks for analysis. In the het mice, a large number of neuroanatomical parameters were reduced in size when compared to WTs with the exception of the ventricles, which were enlarged in size by 92% (p = 0.009) (Figures 3B and 3C). The total brain area parameter was significantly reduced by 13.8% (p = 0.0005) concomitantly with smaller size of the dentate gyrus of the hippocampus (−25.3%, p = 6.9E−5) and decreased height of the soma of the corpus callosum (−24.7%, p = 0.01). At Bregma −2.06 mm, the small size of the dentate gyrus was confirmed and interestingly the corpus callosum was absent (Figures 3D and 3E, Table S5), indicating corpus callosum agenesis. Taken together, these results suggest a contribution of Kdm4b in the regulation of the size of the brain, the hippocampus, and the corpus callosum while contributing to ventriculomegaly.
We also assessed whether haploinsufficiency of Kdm4b could lead to behavioral abnormalities in a similar way to the neuronal conditional homozygous knockout of Jmjd2b (Kdm4b).15 Although open-field findings showed an overall increase of 20% in traveled distance between 2 to 4 min in the het mice, this hyperactive behavior was not statistically significant (Figures 3G–3K).
In this study, we have identified nine individuals with extremely rare variants in KDM4B and syndromic GDD. We have shown an overlap in the neuroanatomical features of the het mice in our murine model with our clinical cohort. In at least two of the three individuals in the study for whom we could obtain MRI images, the MRIs closely mirrored the alterations seen in the het mouse cortex with agenesis of the corpus callosum and dysmorphic hippocampi present. The remaining MRIs were reported as normal, although we were unable to obtain the images for formal review. Molecular modeling of the missense variants suggests that these variants lead to altered KDM4B folding, stability, catalytic activity, and ultimately, loss or reduction of function. Because KDM4B is a histone modifier that acts through lysine demethylation, we hypothesize that the transcription of target genes regulating brain anatomy, in particular the morphology of the hippocampus and the corpus callosum, are suppressed in a dose-dependent manner. The mouse model therefore consistently shows neuroanatomical difference, whereas the human cohort shows a range of severity most likely secondary to the degree of KDM4B haploinsufficiency.
Emerging data suggest that variants in chromatin modifiers are an important cause of neurodevelopmental disability ranging from intellectual disability to autism and schizophrenia.23, 24, 25, 26 Variants in lysine demethylases (KDMs) and lysine methyltransferases (KTMs), in particular, are predicted to cause 22 different developmental delay syndromes.27 These epigenetic modifications can affect the future plasticity of neuronal networks, altering a person’s ability to learn and remember.28 Further understanding of these mechanisms provides important insights into neurogenesis and the broad field of developmental disorders. A previous report on neuron-specific Kdm4b (Jmjd2b) knockout mice by Fukiwara et al. shows that KDM4B (JMJD2B) depletion in mice leads to alteration in dendritic spines within the hippocampi.15 Dendritic spine alteration has previously been reported in other genetic causes of GDD as well, including X-linked ID, Down syndrome, and Fragile X syndrome, highlighting that appropriate synapse formation is critical for neurodevelopment.29 We hypothesize that dendritic spines within the hippocampi may be altered in our human cohort, leading to the developmental delay.
To conclude, this study shows that variants in KDM4B can lead to syndromic GDD in humans, most likely through dysregulation of corticogenesis. These data provide insight into a mechanism of GDD; future studies evaluating the transcriptional targets and their signaling pathways will help to further elucidate the role of KDM4B in neurodevelopment.
Data and Code Availability
The published article includes all datasets generated and analyzed during this study in the Supplemental Information (Tables S3, S4, and S5).
Declaration of Interests
M.J.G.S. and T.S.S. are employees of GeneDx. P.B.A. is on the Scientific Advisory Board of Illumina, Inc. and GeneDx. The other authors declare no competing interests.
Acknowledgments
This work is supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases (R01AR068429-01 to P.B.A.) and by the National Institutes of Health (T32HD098061 to A.R.D.). The mouse neuroanatomical studies were supported by the French National Research Agency (ANR-18-CE12-0009 JCJC to B.Y.). We thank members of the Sanger Institute Mouse Pipelines teams (Ryan Beveridge, Damian Carragher, Jeanne Estabel, Yvette Hooks, Lee Mulderrig, Mark Sanderson, Daniel Sanger, Carl Shannon, and Elizabeth Tuck generated data used in this paper), the Research Support Facility for the provision and management of the Kdm4b mice, and Sylvie Nguyen for histological work. This work was also supported by Wellcome Trust grant WT098051 and Ministero della Salute, Ricerca Corrente 2019 (for E.A. and A.N.).
Published: November 23, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.11.001.
Contributor Information
Binnaz Yalcin, Email: binnaz.yalcin@inserm.fr.
Pankaj B. Agrawal, Email: pagrawal@enders.tch.harvard.edu.
Web Resources
Human Splicing Finder, https://www.genomnis.com/access-hsf
Supplemental Information
References
- 1.Moeschler J.B., Shevell M., Committee on Genetics Comprehensive evaluation of the child with intellectual disability or global developmental delays. Pediatrics. 2014;134:e903–e918. doi: 10.1542/peds.2014-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention (CDC) Economic costs associated with mental retardation, cerebral palsy, hearing loss, and vision impairment--United States, 2003. MMWR Morb. Mortal. Wkly. Rep. 2004;53:57–59. [PubMed] [Google Scholar]
- 3.Dikow N., Moog U., Karch S., Sander A., Kilian S., Blank R., Reuner G. What do parents expect from a genetic diagnosis of their child with intellectual disability? J. Appl. Res. Intellect. Disabil. 2019;32:1129–1137. doi: 10.1111/jar.12602. [DOI] [PubMed] [Google Scholar]
- 4.Gilissen C., Hehir-Kwa J.Y., Thung D.T., van de Vorst M., van Bon B.W., Willemsen M.H., Kwint M., Janssen I.M., Hoischen A., Schenck A. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014;511:344–347. doi: 10.1038/nature13394. [DOI] [PubMed] [Google Scholar]
- 5.Vissers L.E., Gilissen C., Veltman J.A. Genetic studies in intellectual disability and related disorders. Nat. Rev. Genet. 2016;17:9–18. doi: 10.1038/nrg3999. [DOI] [PubMed] [Google Scholar]
- 6.Makela N.L., Birch P.H., Friedman J.M., Marra C.A. Parental perceived value of a diagnosis for intellectual disability (ID): a qualitative comparison of families with and without a diagnosis for their child’s ID. Am. J. Med. Genet. A. 2009;149A:2393–2402. doi: 10.1002/ajmg.a.33050. [DOI] [PubMed] [Google Scholar]
- 7.Wilson C., Krieg A.J. KDM4B: A Nail for Every Hammer? Genes (Basel) 2019;10:134. doi: 10.3390/genes10020134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Agger K., Nishimura K., Miyagi S., Messling J.E., Rasmussen K.D., Helin K. The KDM4/JMJD2 histone demethylases are required for hematopoietic stem cell maintenance. Blood. 2019;134:1154–1158. doi: 10.1182/blood.2019000855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beyer S., Kristensen M.M., Jensen K.S., Johansen J.V., Staller P. The histone demethylases JMJD1A and JMJD2B are transcriptional targets of hypoxia-inducible factor HIF. J. Biol. Chem. 2008;283:36542–36552. doi: 10.1074/jbc.M804578200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krieg A.J., Rankin E.B., Chan D., Razorenova O., Fernandez S., Giaccia A.J. Regulation of the histone demethylase JMJD1A by hypoxia-inducible factor 1 alpha enhances hypoxic gene expression and tumor growth. Mol. Cell. Biol. 2010;30:344–353. doi: 10.1128/MCB.00444-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xia X., Lemieux M.E., Li W., Carroll J.S., Brown M., Liu X.S., Kung A.L. Integrative analysis of HIF binding and transactivation reveals its role in maintaining histone methylation homeostasis. Proc. Natl. Acad. Sci. USA. 2009;106:4260–4265. doi: 10.1073/pnas.0810067106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coffey K., Rogerson L., Ryan-Munden C., Alkharaif D., Stockley J., Heer R., Sahadevan K., O’Neill D., Jones D., Darby S. The lysine demethylase, KDM4B, is a key molecule in androgen receptor signalling and turnover. Nucleic Acids Res. 2013;41:4433–4446. doi: 10.1093/nar/gkt106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang J., Jubb A.M., Pike L., Buffa F.M., Turley H., Baban D., Leek R., Gatter K.C., Ragoussis J., Harris A.L. The histone demethylase JMJD2B is regulated by estrogen receptor α and hypoxia, and is a key mediator of estrogen induced growth. Cancer Res. 2010;70:6456–6466. doi: 10.1158/0008-5472.CAN-10-0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mallette F.A., Mattiroli F., Cui G., Young L.C., Hendzel M.J., Mer G., Sixma T.K., Richard S. RNF8- and RNF168-dependent degradation of KDM4A/JMJD2A triggers 53BP1 recruitment to DNA damage sites. EMBO J. 2012;31:1865–1878. doi: 10.1038/emboj.2012.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujiwara K., Fujita Y., Kasai A., Onaka Y., Hashimoto H., Okada H., Yamashita T. Deletion of JMJD2B in neurons leads to defective spine maturation, hyperactive behavior and memory deficits in mouse. Transl. Psychiatry. 2016;6:e766. doi: 10.1038/tp.2016.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwase S., Lan F., Bayliss P., de la Torre-Ubieta L., Huarte M., Qi H.H., Whetstine J.R., Bonni A., Roberts T.M., Shi Y. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128:1077–1088. doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 17.Lederer D., Grisart B., Digilio M.C., Benoit V., Crespin M., Ghariani S.C., Maystadt I., Dallapiccola B., Verellen-Dumoulin C. Deletion of KDM6A, a histone demethylase interacting with MLL2, in three patients with Kabuki syndrome. Am. J. Hum. Genet. 2012;90:119–124. doi: 10.1016/j.ajhg.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dhar S.S., Lee S.H., Kan P.Y., Voigt P., Ma L., Shi X., Reinberg D., Lee M.G. Trans-tail regulation of MLL4-catalyzed H3K4 methylation by H4R3 symmetric dimethylation is mediated by a tandem PHD of MLL4. Genes Dev. 2012;26:2749–2762. doi: 10.1101/gad.203356.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karczewski, K.J., Francioli, L.C., Tiao, G., Cummings, B.B., Alföldi, J., Wang, Q., Collins, R.L., Laricchia, K.M., Ganna, A., Birnbaum, D.P., et al. Genome Aggregation Database (gnomAD) Consortium. bioRxiv. 10.1101/531210. [DOI]
- 20.Su Z., Wang F., Lee J.H., Stephens K.E., Papazyan R., Voronina E., Krautkramer K.A., Raman A., Thorpe J.J., Boersma M.D. Reader domain specificity and lysine demethylase-4 family function. Nat. Commun. 2016;7:13387. doi: 10.1038/ncomms13387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skarnes W.C., Rosen B., West A.P., Koutsourakis M., Bushell W., Iyer V., Mujica A.O., Thomas M., Harrow J., Cox T. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mikhaleva A., Kannan M., Wagner C., Yalcin B. Histomorphological Phenotyping of the Adult Mouse Brain. Curr. Protoc. Mouse Biol. 2016;6:307–332. doi: 10.1002/cpmo.12. [DOI] [PubMed] [Google Scholar]
- 23.Bjornsson H.T. The Mendelian disorders of the epigenetic machinery. Genome Res. 2015;25:1473–1481. doi: 10.1101/gr.190629.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barco A. Neuroepigenetic disorders: progress, promises and challenges. Neuropharmacology. 2014;80:1–2. doi: 10.1016/j.neuropharm.2014.02.013. [DOI] [PubMed] [Google Scholar]
- 25.Cotney J., Muhle R.A., Sanders S.J., Liu L., Willsey A.J., Niu W., Liu W., Klei L., Lei J., Yin J. The autism-associated chromatin modifier CHD8 regulates other autism risk genes during human neurodevelopment. Nat. Commun. 2015;6:6404. doi: 10.1038/ncomms7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weissman J., Naidu S., Bjornsson H.T. Abnormalities of the DNA methylation mark and its machinery: an emerging cause of neurologic dysfunction. Semin. Neurol. 2014;34:249–257. doi: 10.1055/s-0034-1386763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Faundes V., Newman W.G., Bernardini L., Canham N., Clayton-Smith J., Dallapiccola B., Davies S.J., Demos M.K., Goldman A., Gill H., Clinical Assessment of the Utility of Sequencing and Evaluation as a Service (CAUSES) Study. Deciphering Developmental Disorders (DDD) Study Histone Lysine Methylases and Demethylases in the Landscape of Human Developmental Disorders. Am. J. Hum. Genet. 2018;102:175–187. doi: 10.1016/j.ajhg.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vogel-Ciernia A., Wood M.A. Neuron-specific chromatin remodeling: a missing link in epigenetic mechanisms underlying synaptic plasticity, memory, and intellectual disability disorders. Neuropharmacology. 2014;80:18–27. doi: 10.1016/j.neuropharm.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fiala J.C., Spacek J., Harris K.M. Dendritic spine pathology: cause or consequence of neurological disorders? Brain Res. Brain Res. Rev. 2002;39:29–54. doi: 10.1016/s0165-0173(02)00158-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all datasets generated and analyzed during this study in the Supplemental Information (Tables S3, S4, and S5).