Summary
We have previously described a heart-, eye-, and brain-malformation syndrome caused by homozygous loss-of-function variants in SMG9, which encodes a critical component of the nonsense-mediated decay (NMD) machinery. Here, we describe four consanguineous families with four different likely deleterious homozygous variants in SMG8, encoding a binding partner of SMG9. The observed phenotype greatly resembles that linked to SMG9 and comprises severe global developmental delay, microcephaly, facial dysmorphism, and variable congenital heart and eye malformations. RNA-seq analysis revealed a general increase in mRNA expression levels with significant overrepresentation of core NMD substrates. We also identified increased phosphorylation of UPF1, a key SMG1-dependent step in NMD, which most likely represents the loss of SMG8--mediated inhibition of SMG1 kinase activity. Our data show that SMG8 and SMG9 deficiency results in overlapping developmental disorders that most likely converge mechanistically on impaired NMD.
Keywords: NMD, RNA-seq, SMG1C, cataract, intellectual disability, congenital heart disease, microcephaly
Main Text
The mechanism of maintaining a highly dynamic and functional transcriptome is complex. Key to this process is the controlled degradation of transcripts via several pathways. Nonsense-mediated decay (NMD) is one such pathway that contributes not only to the quantitative control of the transcriptome but also to its fidelity by serving as a quality-control mechanism and, as shown more recently, a surveyor of translation efficacy.1,2 The targeting and degradation of mRNAs that harbor premature termination codons (PTCs) was the original function attributed to NMD before it was recognized that this system contributes more diversely to influence the expression of 5%–10% of the genes in a typical eukaryotic cell.3,4 The detrimental consequences of an NMD that is too stringent (such that transcripts from some pseudogenes and those leading to mutant dominant-negative proteins escape degradation) or too nonspecific (such that the expression of critical genes is reduced) are strong drivers of keeping this process under tight control, including through autoregulation.5
In humans, the known NMD components are 3 UPF (up-frameshift, named for the proteins’ role in ribosomal frameshifting in yeast) and 6 SMG (suppressor with morphogenetic effects on genitalia, named after the abnormal genitalia in roundworm mutants) proteins. After splicing, a multiprotein complex known as exon junction complex (EJC) is deposited upstream of splice junctions. In EJC-coupled NMD, the encounter of an EJC downstream of a termination codon triggers the critical step of NMD, i.e., the phosphorylation of UPF1 (MIM: 601430) by SMG1 (MIM: 607032), which in turn triggers the decay cascade mediated by SMG7 (MIM: 610964) and SMG5/6 (MIM 610962 and 610963). This process is tightly controlled by the action of the SMG1-bound SMG8/SMG9 (MIM 613175 and 613176) complex. Specifically, SMG9 recruits SMG8, which directly regulates the kinase activity of SMG1 on UPF1.1 It should be emphasized that the central role of UPF1 has also been demonstrated for NMD-mediated gene expression regulation beyond EJC-induced NMD.6
The crucial developmental role of NMD components is evident from the lethality observed in animal models; e.g., knockout mice for Smg1, Upf1, Smg6, and Smg9 are embryonic lethal.7, 8, 9, 10 In humans, the first link of NMD machinery to human disease was the identification of individuals with intellectual disability and inactivating variants in the X-linked UPF3B (MIM: 300298), a component of the EJC.11 More recently, we described a multiple-congenital-anomalies syndrome linked to inactivating variants in SMG9 and showed a remarkable overlap between the human and mouse phenotypes.10 Here, we expand on those findings by describing a similar syndrome, which we propose to be caused by SMG8 deficiency and which might have a similar underlying pathogenesis related to impaired NMD.
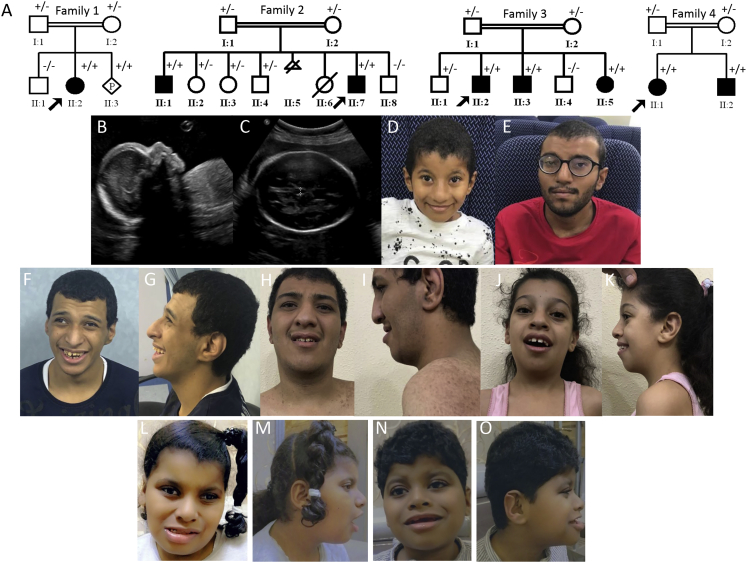
Four consanguineous families presented with overlapping clinical features (Figure 1A). The index individual in family 1 (19DG1424) is a 3-year-old girl with global developmental delay, failure to thrive, microcephaly, facial dysmorphism, and ventricular septal defect (VSD) (see Table 1 and Supplemental Clinical Data). Her Saudi parents are third cousins and have one healthy child but during the course of the study conceived a fetus (MDLREQ2019-6451, Figures 1B and 1C and Supplemental Clinical Data) with features similar to those of the index individual. The index individual in family 2 (19DG1391) is a 7-year-old boy with global developmental delay, intellectual disability, and facial dysmorphism (Figure 1D and Supplemental Clinical Data). His Saudi parents are first cousins and have another affected child (19DG1396, Figure 1E and Supplemental Clinical Data). The index individual in family 3 (19DG0152) and his affected brother (19DG1342) presented with an identical phenotype comprising intellectual disability, history of global developmental delay as younger children, microcephaly, facial dysmorphism, and congenital cataract (Table 1, Figures 1F–1I, and Supplemental Clinical Data). They have a younger sister (19DG0377) with a similar presentation except for the congenital cataract, which she did not have (Table 1, Figures 1J–1K and Supplemental Clinical Data). The index individual in family 4 (10DF10800) is an 8-year-old girl with mild facial dysmorphism, global developmental delay, intellectual disability, microcephaly, congenital cataract, pigmentary retinopathy, hearing loss, short stature, and congenital heart disease (Table 1, Figures 1L–1M, and Supplemental Clinical Data). Her 6-year-old brother was similarly affected with mild facial dysmorphism, global developmental delay, intellectual disability, microcephaly, congenital cataract, pigmentary retinopathy, hearing loss, short stature, and hypospadias (Table 1, Figures 1N–1O, and Supplemental Clinical Data). The consanguineous parents are healthy Omani natives.
Figure 1.
Identification of a Neurodevelopmental Phenotype with Variable Eye and Heart Involvement in Four Families
(A) Pedigrees of the four study families. The genotype of each tested individual is also shown (+/+ homozygous, +/− carrier, −/− wildtype).
(B and C) Ultrasonographic image of individual 1_II:3, showing micrognathia (B) and widened cavum septum pellucidum (C).
(D) Frontal view of individual 2_II:7, showing a prominent nose and nasolabial folds and large ears.
(E) Frontal view of individual 2_II:1, showing a prominent nose.
(F and G) Frontal and profile views of individual 3_II:2, showing microcephaly, a prominent nose and nasolabial folds, large ears, and strabismus.
(H and I) Frontal and profile views of individual 3_II:3, showing an underbite, a prominent nose and nasolabial folds, large ears, and hyperpigmented skin.
(J and K) Frontal and profile views of individual 3_II:5, showing a prominent nose and nasolabial folds and medial flaring of the eyebrows.
(L and M) Frontal and profile views of individual 4_II:1, showing a bulbous nose and upslanting palpebral fissures.
(N and O) Frontal and profile views of individual 4_II:2, showing a depressed nasal bridge and abnormal configuration of the antihelix.
Table 1.
Comparison of Clinical Features in SMG8- and SMG9-Mutated Individuals
| Family 1 | Family 2 | Family 3 | Family 4 | SMG9-Mutated Individuals | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Individual | 19DG1424 | MDLREQ20196451 | 19DG1391 | 19DG1396 | 19DG0152 | 19DG1342 | 19DG0377 | 10DF10800_a | 10DF10800_b | |
| Age | 23 months | fetus | 7 y | 19 y | 18 y | 28 y | 10 y | 8 y | 3 y | |
| Gender | f | ? | m | m | m | m | f | f | m | |
| CHD | VSD, PFO, PLSVC, DCS | ?LPAS | − | VSD | − | − | − | ASD, VSD | − | 4/4 |
| Facial dysmorphism | + | + | + | + | + | + | + | + | + | 4/4 |
| GDD | + | NA | + | + | + | + | + | + | + | 4/4 |
| Microcephaly | + | + | − | borderline | + | − | + | + | + | 3/3 |
| Short stature | + | + | − | − | borderline | − | borderline | + | + | 3/3 |
| CNS anomalies | ? | wide CSP and ventricles | AWM | ? | − | ? | ? | AWM, BA | AWM | 4/4 |
| Eye findings | − | ? | − | strabismus | strabismus | cataract | − | cataract | cataract | 3/4 |
| GU | − | ? | − | − | dilated RCS, TUB, hypospadias | hypospadias | − | − | hypospadias | 1/4 |
Age at last assessment is shown. Abbreviations are as follows: f, female; m, male; AWM, abnormal white matter; BA, brain atrophy; CHD, congenital heart disease; CNS, central nervous system; CSP, cavum septum pellucidum; DCS, dilated coronary sinus; GDD, global developmental delay; GU, genitourinary; LPAS, left pulmonary artery sling; PFO, patent foramen ovale; PLSVC, persistent left superior vena cava; RCS, renal collecting system; and VSD, ventricular septal defect.
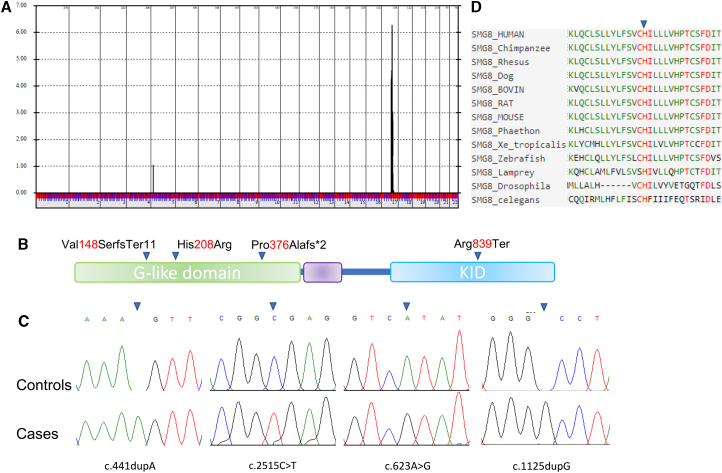
The first three families were initially investigated with clinical exome sequencing, which revealed a different homozygous likely deleterious variant in SMG8 in each of the three families; this revelation led to their subsequent recruitment after providing informed consent under an IRB-approved research protocol (KFSHRC RAC#2080006).12 Genomewide SNP genotyping and autozygome analysis as described previously revealed that all three families map to a single locus (Chr17: 55278337–63977323) with a LOD score of > 6 but with three different haplotypes (Figure 2A). The raw exome files in the three families were reanalyzed in search of novel or very rare (MAF < 0.0001 in gnomAD and a local database of nearly 5,000 ethnically matched exomes) variants within the critical locus. Only the following variants satisfied the search criteria, and all were in SMG8: c.441dupA (p.Val148SerfsTer11) (GenBank: NM_018149.6) in family 1, c.2515C>T (p.Arg839Ter) in family 2, and c.623A>G (p.His208Arg) in family 3 (Figures 2B and 2C). These homozygous variants fully segregated with the phenotype in the respective families.
Figure 2.
A Neurodevelopmental Phenotype Is Linked to SMG8
(A) Genomewide linkage analysis of the four study families combined shows a single linkage peak with LOD score > 6.
(B) Cartoon of SMG8 with the four variants shown.
(C) Sanger tracing of the four SMG8 variants.
(D) Multispecies alignment showing the strong conservation of His208.
The fourth family was independently recruited after providing informed consent under an IRB-approved research protocol (SQU-MREC#1362), and whole-exome sequencing was performed on both affected siblings (Hiseq4000, 150 bp paired-end reads, at 200× coverage). Variant filtration was conducted so that the only remaining variants were those that were novel or rare (<1%) with regard to publicly available variant databases (1000 Genomes, Exome Variant Server, and gnomAD) and an in-house database of 973 ethnically matched exomes. The exome results in the two affected individuals identified two shared homozygous variants within an autozygous region of 20 MB in chromosome 17. Homozygosity for the USH1G variant c.977_986dupTCAGCGTCCC (p.Leu330Glnfs∗109) (GenBank: NM_001282489.3) was consistent with the diagnosis of Usher syndrome, type 1G (MIM: 606943), which explains the bilateral sensorineural hearing impairment and pigmentary retinopathy. The second shared variant detected was SMG8 c.1125dup (p.Pro376Alafs∗2) (GenBank: NM_018149.6), which was considered a variant of unknown significance until our research groups communicated personally and concluded on the basis of the first three families that this is the probable etiology.
The variants c.441dupA and c.1125dup predict frameshifts that eliminate much of the G-like domain (1–466), which is required for the interaction between SMG8 and both the N-HEAT domain of SMG1 and the G domain of SMG9.13,14 Similarly, the variant c.623A>G replaces a strongly conserved histidine residue (His208) down to C. elegans with arginine in the G-like domain and is predicted to be deleterious by various in silico tools (SIFT = 0; PolyPhen = 0.987; and CADD = 25.8) (Figure 2D). Detailed in silico analysis of the SMG8-H208R suggests that it considerably destabilizes the SMG1-SMG8 complex and makes the structure less active and more prone to conformational distortion. It might also prevent the nearby residues from getting exposed to the solvent and thus influence the overall interaction of the molecule with the environment. Also, because the changes in residual fluctuation and molecule flexibility are clearly observed at the SMG1-SMG8 interface, the mutated SMG8 is predicted to bind with SMG1 with lower binding affinity. As a result, the whole complex becomes less active and more likely to be conformationally distorted, which would most likely alter the dynamic behavior (Supplemental in silico assessment of SMG8-H208R). On the other hand, the nonsense variant c.2515C>T predicts a partial loss of the C terminus KID domain (643–991) of SMG8 (Figure 2). Loss of KID has been shown to permit the formation of the SMG1-SMG8-SMG9 complex (SMG1C) and to influence the SMG8-induced regulatory effect on the SMG1 kinase activity.13
The likely deleterious effect of the candidate SMG8 variants, the remarkable overlap between the phenotypes observed in these four families and the phenotype we described in individuals with deleterious SMG9 variants, and the established interdependence of these two NMD proteins prompted us to investigate a potentially shared mechanism between the two syndromes. First, we wanted to explore the effect of SMG8 and SMG9 deficiency on UPF1 phosphorylation. We performed immunoblot analysis on lymphoblastoid cell lines (LCLs) derived from five individuals with SMG8 (19DG0152, 19DG1391, and 19DG1424) or SMG9 (14DG166110 and 19DG259915) biallelic deleterious variants. As shown in Figure S1A and S1B, we have found by using a specific antibody against p-UPF1 that there is a mild but significant increase in UPF1 phosphorylation. This is in line with the published literature, which suggests that SMG8 and SMG9 act, in addition to serving as a scaffold for the interaction between SMG1 and UPF1, to negatively regulate SMG1 kinase activity and maintain UPF1 in the unphosphorylated (inactive) form within the SURF complex.16, 17, 18, 19
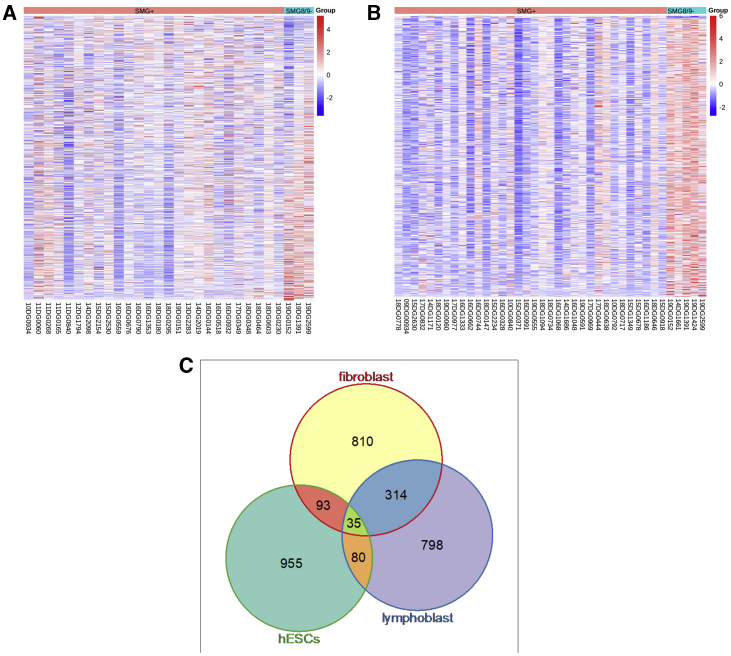
This apparent perturbation of UPF1 phosphorylation regulation encouraged us to further explore a potential NMD dysregulation. Therefore, we performed RNA-seq on fibroblasts derived from a previously described individual (19DG2599)15 with SMG9-related phenotype and from individuals 19DG1391 and 19DG0152 with candidate SMG8 variants in this study. We also performed RNA-seq on LCLs derived from five individuals with SMG8 (19DG0152, 19DG1391, and 19DG1424) or SMG9 (14DG1661 and 19DG2599) biallelic deleterious variants. As corresponding control experiments, we also performed RNA-seq on 26 ethnically matched fibroblast samples and 34 ethnically matched LCL samples with no pathogenic variants in any of the known NMD components, as described before.20 We mapped RNA-seq reads to human reference genome GRCh38 (GENCODE v25) to quantify transcript abundances by using Kallisto.21 We computed gene-level TPM by using the transcript abundances and effective lengths of the coding transcripts from chromosomes 1–22 and X. We found that this sample-specific TPM scaling properly normalized our RNA-seq data (see supplemental RNA-seq analysis and Figures S2A and S2B). Genes with the maximum TPM < 1 across all samples were removed, and the constant 1 was added to TPM for each gene in each sample. We performed sample-level quality control of the normalized data with PCA (see supplemental RNA-seq analysis) and found that, although the case samples were clustered together, samples in controls displayed high levels of heterogeneity (Figures S3A and S3B). To alleviate the effects of high variability in the control group, we calculated the z score for each gene and excluded count outliers with |z score| > 2 from the control samples in the differential gene expression (DGE) analysis. With the ANOVA-based DGE analysis (see supplemental RNA-seq analysis), we found that, in the fibroblast dataset (n = 13,619), 1,252 genes were upregulated and 33 were downregulated in the affected individuals, whereas in the LCL dataset (n = 13,727), 1,227 were upregulated and 12 were downregulated, indicating that a large proportion of DE genes were upregulated in affected individuals versus controls, indicating a general tendency toward upregulation, as would be expected with impaired NMD (Figure 3 and Tables S1 and S2). Given the results of this DGE analysis, we performed gene-set enrichment analysis to see whether 34 core NMD substrates (33 expressed in both fibroblast cells and LCLs) compiled in previous studies22,23 were over-represented in these upregulated genes (Figures S4A and S4B). We found that nine of the core NMD substrates (ATF3, DNAJB2, GADD45A, GADD45B, PTPRF, RASSF1, SMG1, TBL2, and UPF2) were upregulated in our fibroblast dataset (3.14-fold enrichment; p < 0.002), whereas seven core NMD substrates (DNAJB2, GADD45B, PCNX2, RASSF1, SMG5, TBL2, and UPF2) were upregulated in our LCL dataset (2.37-fold enrichment; p < 0.025); five core NMD substrates overlapped between these datasets (Figure 3). To further examine the role SMG8 and SMG9 play in NMD, we performed additional enrichment analysis by using 200 high-confidence NMD substrates (187 expressed in fibroblast cells and 177 expressed in LCLs) that were found to be upregulated in the siUPF1-treated hESCs (Table S5 of Lou et al., 201622) (Figures S5A and S5B and Tables S1 and S2).23 Figure S5 depicts the abundance of these high-confidence NMD substrates in both affected individuals and controls. We found that 25 of the high-confidence NMD substrates were upregulated in affected individuals when we based our analysis on the fibroblast dataset (1.45-fold enrichment; p < 0.05), and 32 were upregulated on the basis of the LCL dataset (2.02-fold enrichment; p < 0.0001); there was an overlap of 11 genes (BAG1, DUSP12, C1orf109, TBL2, RASSF1, IRAK4, GADD45B, NFKBIB, ZFAND2A, UPF3B, and DNAJB2) between results from the two datasets. We also attempted to explore the effects of NMD-target PTCs on the abundance of transcripts. However, the weak effect observed in controls did not allow us to make a meaningful comparison (supplemental RNA-seq analysis and Figure S6 in the Supplemental Information). Nonetheless, we performed a follow-up analysis (supplemental RNA-seq analysis) by focusing on differences in relative abundances of transcripts with NMD-target PTCs in affected individuals versus control samples. We found no evidence of PTC-induced NMD impairment (Figure S7). Taken together, these results point to a transcriptional dysregulation pattern that might be related to impaired NMD without affecting specific PTC-induced NMD.
Figure 3.
SMG8 and SMG9 Biallelic Variants Are Associated with Transcriptional Dysregulation Suggestive of Impaired NMD
(A and B) DGE analysis of individuals with SMG8 and SMG9 biallelic variants (SMG8/SMG9⁻) versus individuals who lack such variants (SMG+) by using fibroblasts (A) and LCLs (B).
(C) Summary of the overlap of dysregulated genes between fibroblasts and LCLs in comparison to the list of previously published dysregulated genes in siUPF1-treated hESCs as reported by Lou et al., 2016.21
We have previously described an SMG9-related developmental disorder in humans and mice but had not been able to determine conclusively whether NMD was globally impaired, as would be expected in the setting of severe SMG9 deficiency.10 Here, we describe an overlapping syndrome with a similarly variable frequency of specific malformations but a fully penetrant severe neurodevelopmental phenotype, which we propose to be caused by SMG8 deficiency. In their seminal paper, Yamashita and colleagues showed that SMG8 is part of the SMG1C and that, although SMG8 inhibits SMG1 kinase activity in vitro, its requirement for the recruitment of SMG1 to the surveillance complex means that UPF1 phosphorylation is actually increased when SMG8 is depleted, which is consistent with our results.17 They further showed that SMG8 deficiency reduces the activity of NMD in human cells on both endogenous and engineered target transcripts. Interestingly, although SMG8 depletion in C. elegans mutant that displays paralysis due to NMD activity on a target myosin heavy-chain transcript resulted in suppression of the paralysis phenotype, this was much weaker than the suppression observed with SMG1 depletion.5 Thus, although SMG8 clearly plays an important regulatory role in NMD, its deficiency does not completely eliminate NMD, as shown by others and as suggested by our results.24 We note here that we cannot exclude the possibility that these two proteins might have hitherto unknown functions that are unrelated to their role in NMD and that contribute to the pathogenesis of the SMG9- and SMG8-related syndromes. For instance, these syndromes are clinically distinct from that caused by mutations in UPF3B, which encodes another NMD component. Alternatively, this might be due to specific rather than global NMD perturbation.
In conclusion, we expand the list of NMD-related genes that are linked to human developmental phenotypes. Our findings suggest that deleterious variants in other NMD-related genes should be considered as attractive candidates in individuals with intellectual disability with or without associated syndromic features. We plan to pursue a mouse model to shed more light on the pathogenesis of the SMG8-related phenotype and compare it that of the Smg9 mouse.
Data and Code Availability
The scripts for the DGE analysis, along with the normalized RNA-seq data, have been deposited to Github.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We thank the study participants for their enthusiastic participation. We also thank the Sequencing and Genotyping Core Facilities at KFSRHC for their technical help. This work was supported by King Abdullah University of Science and Technology, Office of Sponsored Research under Awards BAS/1/1624-01, FCC/1/1976-18-01, FCC/1/1976-23-01, FCC/1/1976-25-01, FCC/1/1976-26-01, REI/1/0018-01-01, REI/1/4216-01-01, and URF/1/4098-01-01. This work was also supported by His Majesty Trust Funds at the Sultan Qaboos University, study code SR/MED/GENT/16/01.
Published: November 25, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.11.007.
Contributor Information
Xin Gao, Email: xin.gao@kaust.edu.sa.
Fowzan S. Alkuraya, Email: falkuraya@kfshrc.edu.sa.
Web Resources
Missense 3D, www.sbg.bio.ic.ac.uk/∼missense3d
OMIM, https://omim.org
Supplemental Information
Mean TPM: mean TPM of all patients, including both control and SMG8/9 patients. Padj: FDR adjusted p value or BH adjusted p value (Benjamini, et al., 1995; reference 18 in Supplemental Information). DE: Whether a gene is differentially expressed or not. Up: upregulated in SMG8/9- patients. Down: downregulated in SMG8/9- patients. High-confidence NMD target: high-confidence NMD substrates upregulated in siUPF1-treated hESCs (Lou et. al, 2016) Core NMD substrate: The 34 core NMD substrates defined by Lou et al.21
Mean TPM: mean TPM of all patients including both control and SMG8/9- patients. Padj: FDR adjust p value or BH adjust p value (Benjamini, et al., 1995; reference 18 in Supplemental Information). DE: Whether a gene is differentially expressed or not. Up: upregulated in SMG8/9 patients. Down: downregulated in SMG8/9- patients. High-confidence NMD target: high-confidence NMD substrates upregulated in siUPF1-treated hESCs21 Core NMD substrate: These 34 core NMD substrates are defined by Lou et, al,21
References
- 1.Kishor A., Fritz S.E., Hogg J.R. Nonsense-mediated mRNA decay: The challenge of telling right from wrong in a complex transcriptome. Wiley Interdiscip. Rev. RNA. 2019;10:e1548. doi: 10.1002/wrna.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karousis E.D., Mühlemann O. Nonsense-mediated mRNA decay begins where translation ends. Cold Spring Harb. Perspect. Biol. 2019;11:a032862. doi: 10.1101/cshperspect.a032862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mühlemann O., Jensen T.H. mRNP quality control goes regulatory. Trends Genet. 2012;28:70–77. doi: 10.1016/j.tig.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Nasif S., Contu L., Mühlemann O. Beyond quality control: The role of nonsense-mediated mRNA decay (NMD) in regulating gene expression. Semin. Cell Dev. Biol. 2018;75:78–87. doi: 10.1016/j.semcdb.2017.08.053. [DOI] [PubMed] [Google Scholar]
- 5.Yepiskoposyan H., Aeschimann F., Nilsson D., Okoniewski M., Mühlemann O. Autoregulation of the nonsense-mediated mRNA decay pathway in human cells. RNA. 2011;17:2108–2118. doi: 10.1261/rna.030247.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lawir D.-F., Sikora K., O’Meara C.P., Schorpp M., Boehm T. Pervasive changes of mRNA splicing in upf1-deficient zebrafish identify rpl10a as a regulator of T cell development. Proc. Natl. Acad. Sci. USA. 2020;117:15799–15808. doi: 10.1073/pnas.1917812117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Medghalchi S.M., Frischmeyer P.A., Mendell J.T., Kelly A.G., Lawler A.M., Dietz H.C. Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum. Mol. Genet. 2001;10:99–105. doi: 10.1093/hmg/10.2.99. [DOI] [PubMed] [Google Scholar]
- 8.McIlwain D.R., Pan Q., Reilly P.T., Elia A.J., McCracken S., Wakeham A.C., Itie-Youten A., Blencowe B.J., Mak T.W. Smg1 is required for embryogenesis and regulates diverse genes via alternative splicing coupled to nonsense-mediated mRNA decay. Proc. Natl. Acad. Sci. USA. 2010;107:12186–12191. doi: 10.1073/pnas.1007336107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li T., Shi Y., Wang P., Guachalla L.M., Sun B., Joerss T., Chen Y.S., Groth M., Krueger A., Platzer M. Smg6/Est1 licenses embryonic stem cell differentiation via nonsense-mediated mRNA decay. EMBO J. 2015;34:1630–1647. doi: 10.15252/embj.201489947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shaheen R., Anazi S., Ben-Omran T., Seidahmed M.Z., Caddle L.B., Palmer K., Ali R., Alshidi T., Hagos S., Goodwin L. Mutations in SMG9, encoding an essential component of nonsense-mediated decay machinery, cause a multiple congenital anomaly syndrome in humans and mice. Am. J. Hum. Genet. 2016;98:643–652. doi: 10.1016/j.ajhg.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tarpey P.S., Raymond F.L., Nguyen L.S., Rodriguez J., Hackett A., Vandeleur L., Smith R., Shoubridge C., Edkins S., Stevens C. Mutations in UPF3B, a member of the nonsense-mediated mRNA decay complex, cause syndromic and nonsyndromic mental retardation. Nat. Genet. 2007;39:1127–1133. doi: 10.1038/ng2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Monies D., Abouelhoda M., Assoum M., Moghrabi N., Rafiullah R., Almontashiri N., Alowain M., Alzaidan H., Alsayed M., Subhani S. Lessons learned from large-scale, first-tier clinical exome sequencing in a highly consanguineous population. Am. J. Hum. Genet. 2019;104:1182–1201. doi: 10.1016/j.ajhg.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu L., Li L., Qi Y., Yu Z., Xu Y. Cryo-EM structure of SMG1-SMG8-SMG9 complex. Cell Res. 2019;29:1027–1034. doi: 10.1038/s41422-019-0255-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gat Y., Schuller J.M., Lingaraju M., Weyher E., Bonneau F., Strauss M., Murray P.J., Conti E. InsP6 binding to PIKK kinases revealed by the cryo-EM structure of an SMG1-SMG8-SMG9 complex. Nat. Struct. Mol. Biol. 2019;26:1089–1093. doi: 10.1038/s41594-019-0342-7. [DOI] [PubMed] [Google Scholar]
- 15.Lemire G., MacDonald S.K., Boycott K.M. SMG9-deficiency syndrome caused by a homozygous missense variant: Expanding the genotypic and phenotypic spectrum of this developmental disorder. Am. J. Med. Genet. A. 2020;182:1829–1831. doi: 10.1002/ajmg.a.61616. [DOI] [PubMed] [Google Scholar]
- 16.Deniaud A., Karuppasamy M., Bock T., Masiulis S., Huard K., Garzoni F., Kerschgens K., Hentze M.W., Kulozik A.E., Beck M. A network of SMG-8, SMG-9 and SMG-1 C-terminal insertion domain regulates UPF1 substrate recruitment and phosphorylation. Nucleic Acids Res. 2015;43:7600–7611. doi: 10.1093/nar/gkv668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamashita A., Izumi N., Kashima I., Ohnishi T., Saari B., Katsuhata Y., Muramatsu R., Morita T., Iwamatsu A., Hachiya T. SMG-8 and SMG-9, two novel subunits of the SMG-1 complex, regulate remodeling of the mRNA surveillance complex during nonsense-mediated mRNA decay. Genes Dev. 2009;23:1091–1105. doi: 10.1101/gad.1767209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arias-Palomo E., Yamashita A., Fernández I.S., Núñez-Ramírez R., Bamba Y., Izumi N., Ohno S., Llorca O. The nonsense-mediated mRNA decay SMG-1 kinase is regulated by large-scale conformational changes controlled by SMG-8. Genes Dev. 2011;25:153–164. doi: 10.1101/gad.606911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Causier B., Li Z., De Smet R., Lloyd J.P.B., Van de Peer Y., Davies B. Conservation of nonsense-mediated mRNA decay complex components throughout eukaryotic evolution. Sci. Rep. 2017;7:16692. doi: 10.1038/s41598-017-16942-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maddirevula S., Kuwahara H., Ewida N., Shamseldin H.E., Patel N., Alzahrani F., AlSheddi T., AlObeid E., Alenazi M., Alsaif H.S. Analysis of transcript-deleterious variants in Mendelian disorders: implications for RNA-based diagnostics. Genome Biol. 2020;21:145. doi: 10.1186/s13059-020-02053-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 22.Kurosaki T., Li W., Hoque M., Popp M.W.-L., Ermolenko D.N., Tian B., Maquat L.E. A post-translational regulatory switch on UPF1 controls targeted mRNA degradation. Genes Dev. 2014;28:1900–1916. doi: 10.1101/gad.245506.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lou C.-H., Dumdie J., Goetz A., Shum E.Y., Brafman D., Liao X., Mora-Castilla S., Ramaiah M., Cook-Andersen H., Laurent L., Wilkinson M.F. Nonsense-mediated RNA decay influences human embryonic stem cell fate. Stem Cell Reports. 2016;6:844–857. doi: 10.1016/j.stemcr.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosains J., Mango S.E. Genetic characterization of smg-8 mutants reveals no role in C. elegans nonsense mediated decay. PLoS ONE. 2012;7:e49490. doi: 10.1371/journal.pone.0049490. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mean TPM: mean TPM of all patients, including both control and SMG8/9 patients. Padj: FDR adjusted p value or BH adjusted p value (Benjamini, et al., 1995; reference 18 in Supplemental Information). DE: Whether a gene is differentially expressed or not. Up: upregulated in SMG8/9- patients. Down: downregulated in SMG8/9- patients. High-confidence NMD target: high-confidence NMD substrates upregulated in siUPF1-treated hESCs (Lou et. al, 2016) Core NMD substrate: The 34 core NMD substrates defined by Lou et al.21
Mean TPM: mean TPM of all patients including both control and SMG8/9- patients. Padj: FDR adjust p value or BH adjust p value (Benjamini, et al., 1995; reference 18 in Supplemental Information). DE: Whether a gene is differentially expressed or not. Up: upregulated in SMG8/9 patients. Down: downregulated in SMG8/9- patients. High-confidence NMD target: high-confidence NMD substrates upregulated in siUPF1-treated hESCs21 Core NMD substrate: These 34 core NMD substrates are defined by Lou et, al,21
Data Availability Statement
The scripts for the DGE analysis, along with the normalized RNA-seq data, have been deposited to Github.