Abstract
The full potential of polyketide discovery has yet to be reached due to a lack of suitable technologies and knowledge required to advance engineering of polyketide biosynthesis. Recent investigations on the discovery, enhancement, and non-natural utilization of these biosynthetic gene clusters via computational biology, metabolic engineering, structural biology, and enzymology-guided approaches have facilitated improved access to designer polyketides. Here, we discuss recent successes in gene cluster discovery, host strain engineering, precursor-directed biosynthesis, combinatorial biosynthesis, polyketide tailoring, and high-throughput synthetic biology, as well as challenges and outlooks for rapidly generating useful target polyketides.
Graphical Abstract
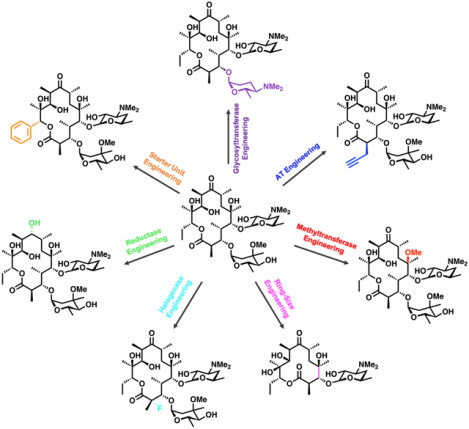
Introduction
Nature has evolved diverse enzymatic machinery for the assembly of highly complex small molecule natural products. Biosynthesized by polyketide synthases (PKSs), polyketides are a large class of natural products and represent a significant source of new drugs, molecular probes, and bioactive small molecules [1,2]. Many polyketides are blockbuster drugs such as erythromycin (antibiotic), epothilone B (antitumor), and lovastatin (anticholesterol), while others, such as solithromycin, are currently in clinical development [3].
Within canonical type I PKS assembly lines, each module is responsible for the incorporation of a single malonyl-derived extender unit. Minimally, modules are comprised of an acyltransferase (AT), acyl carrier protein (ACP), and ketosynthase (KS) catalytic domains, which enable extender unit selection and subsequent decarboxylative Claisen condensation between the extender unit and the growing chain. Additional in-line tailoring domains, including the ketoreductase (KR), dehydratase (DH), enoylreductase (ER), or more rarely, methyltransferase (MT), can also further site-selectively modify the alkylation and oxidation patterns within the growing polyketide chain. The final elongated chain is then cleaved from the PKS and cyclized by a thioesterase (TE) domain to yield a core macrolactone, which can be further decorated by post-PKS enzymes [4].
Despite their broad chemical and structural diversity, naturally occurring polyketides often require optimization for a given application [5]. For example, the macrolide antibiotic erythromycin was synthetically modified to prevent cyclization in the acidic gastric environment, for increased bioavailability, and for additional molecular interactions with the ribosome, spawning multiple generations of improved antibiotics. While synthetic methods have successfully modified polyketide structures, biosynthetic modifications offer a scalable and potentially facile approach for regio- and stereoselective scaffold diversification. Herein, this review describes the current state-of-the-art in synthetic biology to enable access to designer polyketides, leveraging the modularity of PKS machinery. We highlight recent works that have expanded the polyketide chemical space through pathway discovery, refactoring, and engineering and reflect on the future outlook of synthetic biology approaches to polyketide analoging (Figure 1).
Figure 1.
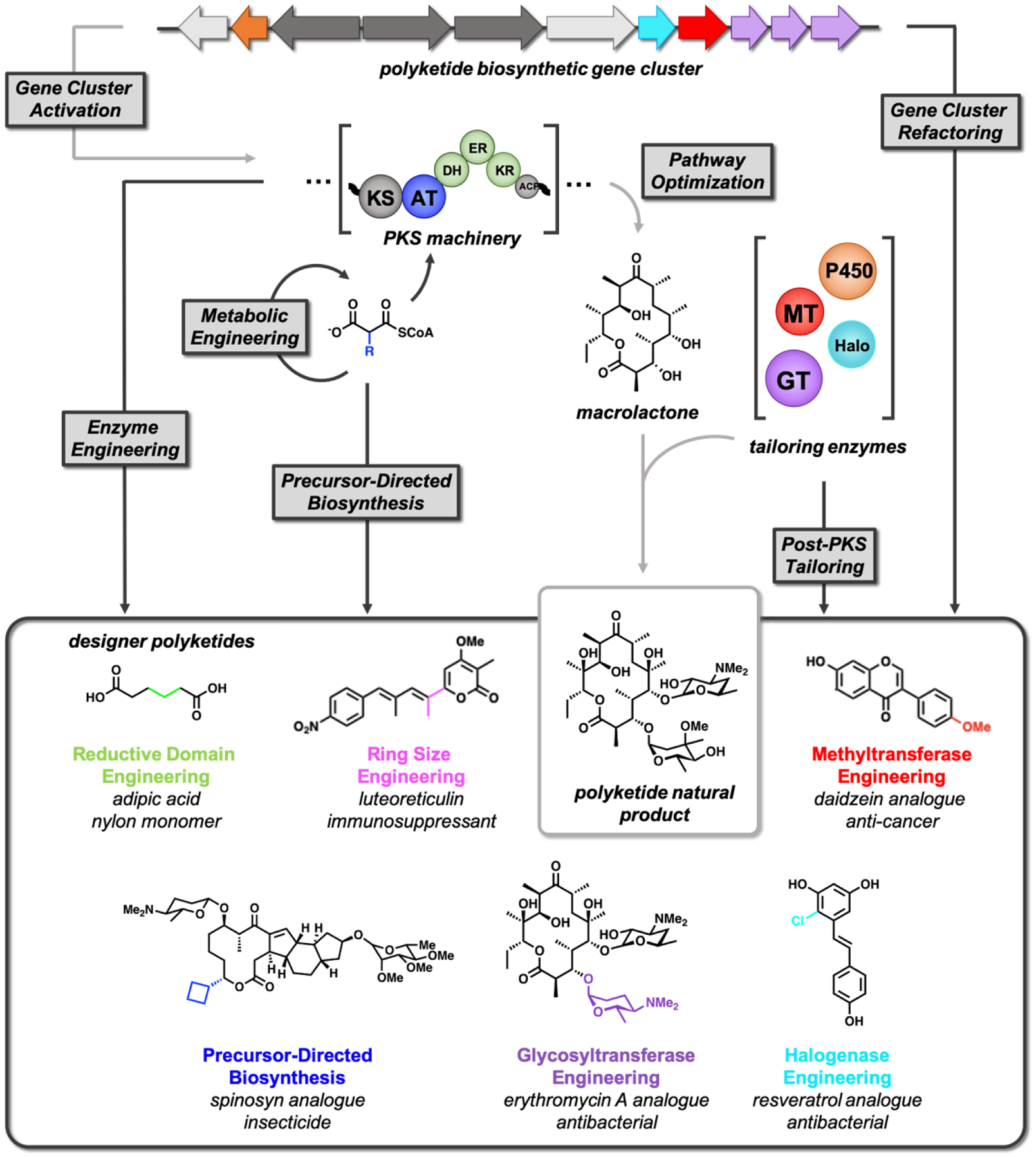
Accessing designer polyketides leveraging diverse engineering strategies. Native pathways are shown in light grey, while engineered and non-natural pathways are shown in dark grey. Modifications to the PKS and post-PKS tailoring enzymes are highlighted in their corresponding colors.
Polyketide Biosynthetic Gene Cluster Discovery
Though there are many elucidated polyketide synthases (PKSs), these pathways likely represent a portion of the genetic machinery available in Nature [6]. Moreover, high rediscovery rates continue to hinder the identification of novel natural compounds. Therefore, several “omics-guided” methods such as transcriptomics [7], metabolomics [8], and genome mining (metagenomics) [9,10] have been utilized in polyketide biosynthetic gene cluster (BGC) discovery. These approaches expand the polyketide engineering toolbox, but more significant barriers for accessing non-natural polyketide derivatives including host selection, starter and extender unit availability, and pathway design, must be overcome.
Accessing Polyketides via Host Strain Selection and Engineering
Exploring and optimizing polyketide biosynthesis can be completed within native production strains given that the BGC and necessary precursors are often already functionally expressed and produced. Typically, enhancing polyketide production focuses on the redirection and optimization of carbon flux via enhanced precursor availability [11,12], promoter and ribosome engineering [13], removal or overexpression of regulatory elements [14,15], deletion of competing biosynthetic pathways, and combinations thereof to significantly enhance titers above that of the wild-type pathway. Strategies including cooperative induction [9], co-culturing [16], and transcription factor decoys [17] have been explored for the activation of cryptic BGCs (cBGCs). The expression of cBGCs has also been enabled through the deletion of “primary” PKS pathways [18,19]. While these methods have been useful in specific well-studied, genetically tractable hosts, the full potential of polyketide cBGCs will not be fully realized until there is a global strategy for pathway activation under traditional and industrially relevant fermentation conditions.
Heterologous pathway expression in well-characterized chassis has addressed challenges posed by native host expression. Heterologous platforms for the rapid and robust biosynthesis of polyketides derived from type I [20,21], II [22,23], and III [24] synthases, offer key opportunities for synthetic biology including the expression of cBGC, development of chimeric PKSs through “plug and play” modifications, and precursor-directed engineering [22]. Usually, the goal of heterologous expression is to decouple secondary and primary metabolism to enhance polyketide production. A recent omics-guided approach in Streptomyces revealed that primary metabolism derived triacylglycerols can limit polyketide biosynthesis. This was overcome by dynamically degrading triacylglycerol and led to enhanced titers of actinorhodin, jadomycin B, oxytetracycline and avermectin B1a in various Streptomyces strains [25]. However, direct BGC transfer [13] and their expression [22] present numerous obstacles including significant cellular burden, resulting in poor expression or inefficient cellular maintenance of the BGC [20]. Advances in DNA manipulation and genomic integration technologies, including transformation-associated recombination (TAR) [26] and direct pathway cloning (DiPaC) [27], continue to make these BGC transfers more feasible.
Altered Macrolactone Sidechains through Precursor-Directed Biosynthesis
The AT is often a target of engineering given that it dictates large portions of the macrolactone structure. Extender unit specificity of AT domains has been altered through domain and motif swaps while targeted mutagenesis strategies have enabled site-selective integration of non-natural building blocks. Notably, AT mutagenesis of the final two modules of the pikromycin PKS revealed the unprecedented capability to introduce consecutive non-natural extender units into a macrolactone [28]. In another recent example, mutation of a conserved tryptophan switched specificity of an AT from ACP- to coenzyme A (CoA)-linked extenders [29], providing an additional potential route to polyketide diversification.
While many of these efforts have had limited utility to in-line, or cis-ATs, opportunities for expanding polyketide chemical space also exist via complex free-standing trans-AT pathways [30]. Though there are examples of trans-ATs like the orthogonal and promiscuous ZmaF whose activities have been probed, their non-natural utility and ability to be leveraged by enzyme engineering in general has not been well explored [31]. Non-canonical modular junctions within these systems, often between a KS and DH, make their products difficult to predict through traditional genome mining [32]. However, programs such as TransATor, which predicts trans-AT containing PKS products by comparing a PKS to KS domains in BGCs and their associated downstream modular enzymes, are powerful tools for discovery [33].
While native and engineered AT promiscuity facilitates the biosynthesis of novel polyketides, the modest diversity of extender units [34] beyond malonyl-CoA (mCoA) and methylmalonyl-CoA (mmCoA) has restricted in vivo efforts. To expand the existing in vivo repertoire of non-natural extender units, mCoA synthetases have been used for the activation of diverse C2-substituted malonates. A native mCoA synthetase from Streptomyces cinnamonensis was used to synthesize allyl-, propargyl-, and propyl-CoAs for the production of monensin analogues [35]. The native promiscuity of enoyl-thioester carboxylase/reductases (ECRs) [36,37] have also been leveraged to produce non-natural extender units in vitro. Moreover, halogenases such as SalL can diversify precursors to produce chlorinated and fluorinated mCoA analogues (Figure 2) [38,39]. Notably, once incorporated, these halogenated analogues can be further leveraged as chemical handles for downstream cross-coupling reactions for further derivatization, as has been done with other classes of natural products [40].
Figure 2.
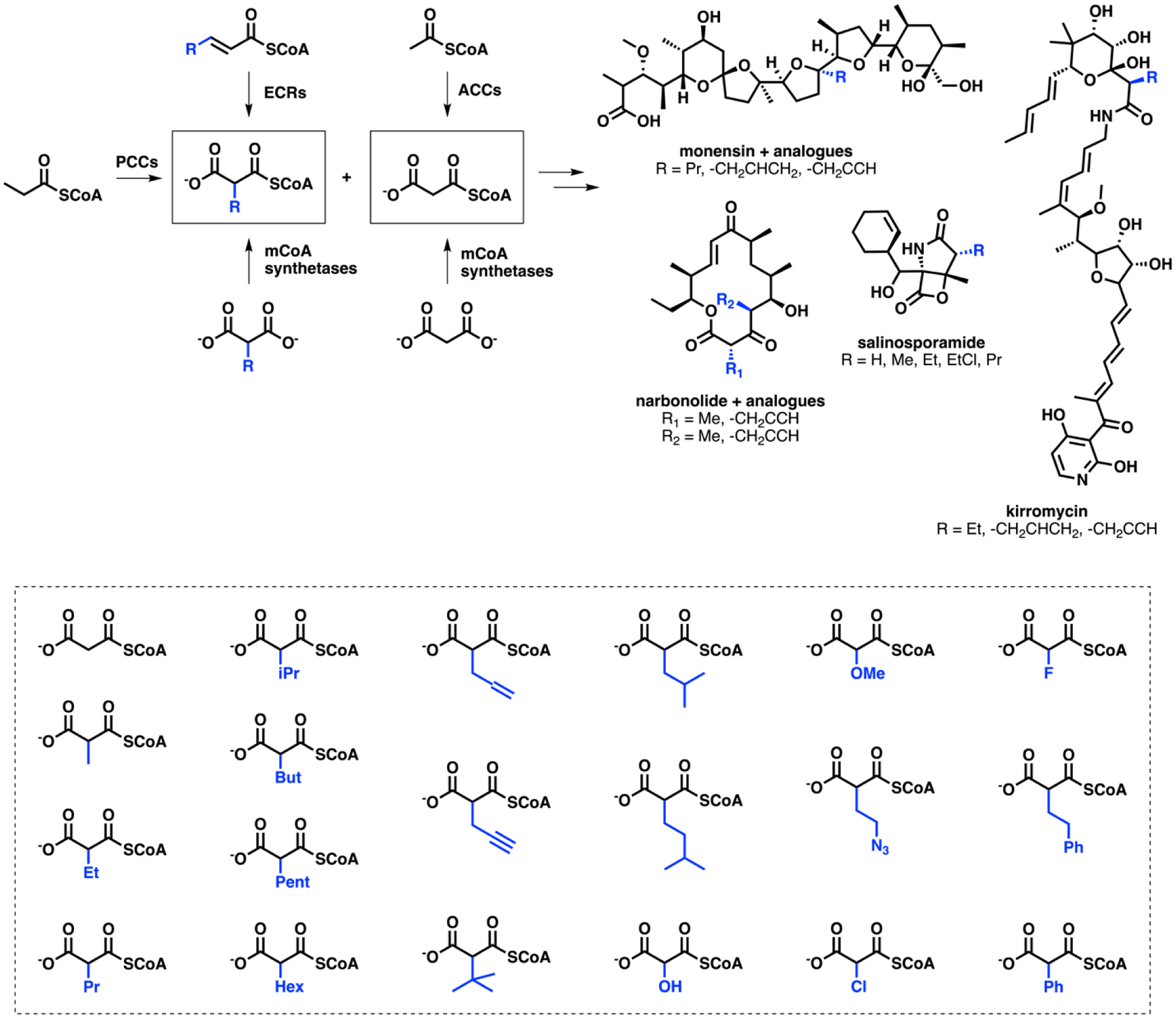
The development of malonyl-CoA analogues for introduction into polyketides. Top: Biosynthesis of malonyl-CoA and various C2-substituted malonyl-CoA analogues. These acyl-CoAs are then utilized by PKSs to produce naturally occurring polyketides and their analogues. Bottom: Panel of previously biosynthesized C2-substituted analogues (See text for references).
Access to Designer Polyketides via Combinatorial Biosynthesis of PKSs
The templated biosynthesis of polyketides by type I PKSs implies the modularity of biosynthetic machinery. Leveraging this paradigm, novel designer molecules are accessible through a “plug-and-play” strategy, wherein modifications to the assembly line, including the insertion, deletion, and exchange of key domains/modules afford predictable scaffold diversification. However, significant changes to protein structure often impair or inactivate the chimeric PKS.
Emerging bioinformatic and evolutionary analyses have challenged the canonical KSn-ATn-ACPn module structure, suggesting that the boundaries be re-defined as ATn-ACPn-KSn+1 [41]. Construction of a hybrid pikromycin-venemycin pathway (Figure 3) with these newly delineated boundaries resulted in higher turnover rates than the traditional boundary swaps and incorporation of the non-natural starter unit 3-hydroxybenzoic into a small combinatorial library of molecules, though non-natural extender selectivity was not explored [42]. Others have also independently identified the KS-AT linker site as a key target for homologous recombination in type I PKSs [43].
Figure 3.
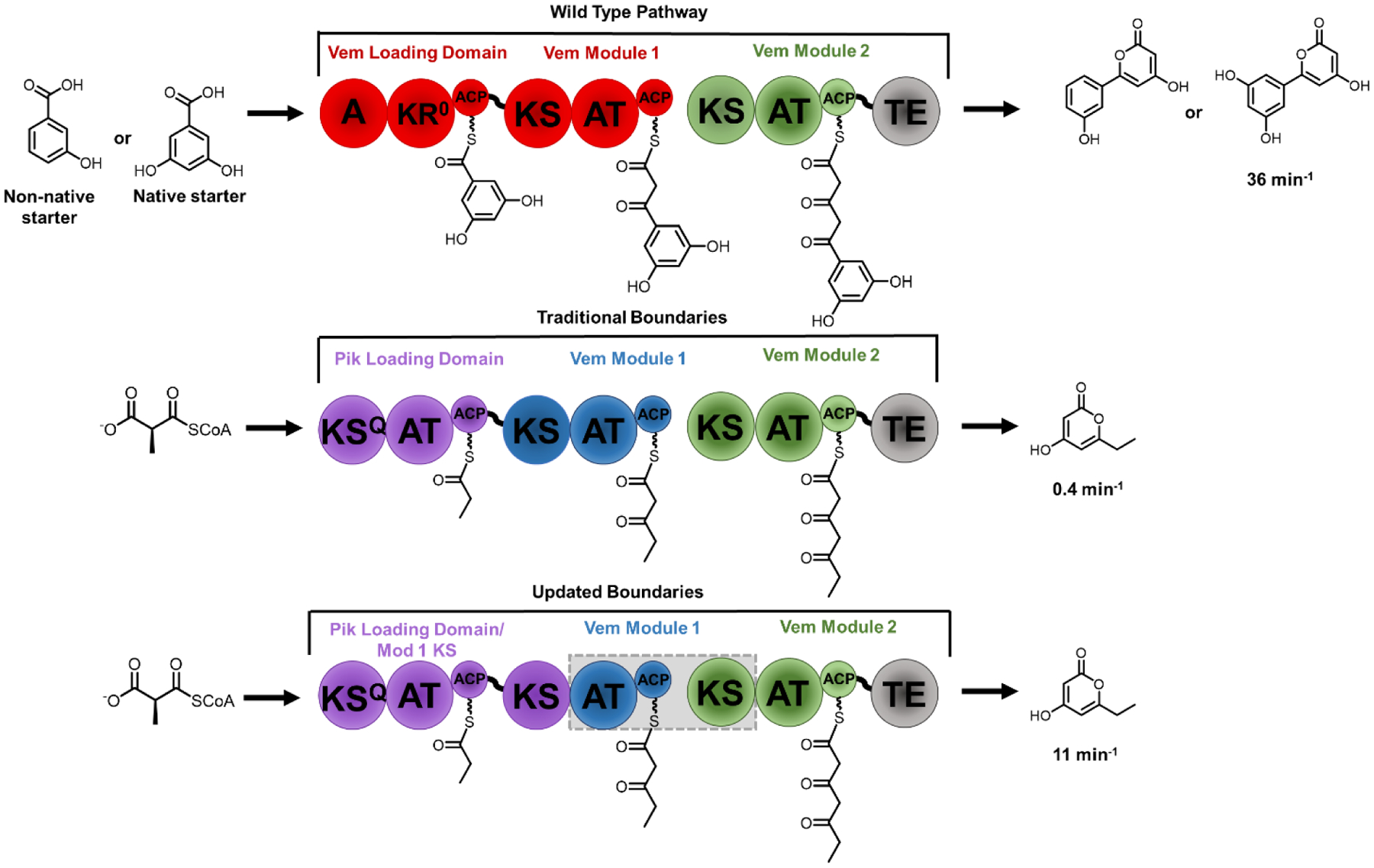
Hybrid synthases of the venemycin PKS (top) were constructed with traditional domain boundaries between the KS and ACP (middle). Module swaps were then constructed using boundaries between the KS and AT (gray box). The new boundary definition resulted in a chimera that was several-fold faster than its traditional boundary counterpart. A (adenylation domain), KR (Ketoreductase), ACP (acyl carrier protein), KS (ketosynthase), AT (acyltransferase), TE (thioesterase), KSQ (ketosynthase-like decarboxylase), KR0 (inactive ketoreductase).
Recent advances aside, there have been varying levels of success in generating chemical diversity in polyketides. Small libraries of compounds with improved antifungal activities have been produced through non-natural incorporation of native extender units by removal of enzymes responsible for extender unit synthesis, inactivation of reductive domains, and knock-outs of tailoring enzymes [44]. Likewise, the fusion of 6-methylsalicylic acid synthase (6MSAS) with a PKS from Pseudallescheria boydii produced a novel compound [45]. Lastly, improvements of a chimeric PKS by point mutations in the KR and host engineering increased production of short-chain ketone fuel additives [46].
Additionally, there have been increased efforts to understand the structure and engineering of specific domains and linkers. This has included reductive loop engineering informed by cheminformatics, which led to higher yields of the expected products [47], phosphopantetheinyltransferase swaps [48], the addition of multiple ACPs on the C-terminus of the final ACP in erythromycin biosynthesis [49], and association of chimeric pathways through docking domain engineering [50–52]. Finally, the recently confirmed “extended conformation” of a module via X-Ray crystallography will better inform future engineering efforts [53].
Non-Natural Chain Lengths
Natural breaks in the colinearity paradigm, such as module skipping, stuttering, or stalling, can similarly enable the biosynthesis of unpredictable macrolactone cores. Although the infidelity of these pathways was thought to be highly unusual, a number of pathways capable of producing multiple or unusual biosynthetic products have been identified. These studies have focused on both single domain inactivation [54] and engineered functionality [55], to dramatically alter the product profile by producing polyketides of variable chain length. Additional mechanisms for non-colinear polyketide biosynthesis have been recently discovered, including the native reversal of selectivity of vatiamides, which allows the biosynthetic pathway to produce multiple products of variable chain lengths and subsequent post-processing modifications [56] as well as the pass-back chain extension mechanism of thalassospiramide biosynthesis [57]. Moreover, some of these processes have been accelerated through engineering, leading to seventeen rapalogs as a result of laboratory evolution that mimicked how PKSs might have evolved in Nature [58].
However, colinearity does not apply to iterative PKSs that catalyze different sets of reactions while maintaining exquisite control to produce ‘cryptic’ templated products. Notably, recent work from Yang et al. identified a possible mechanism in which the KR plays a significant role in chain length determination using intrinsic selectivity across multiple iterative PKSs [59]. The relevance of individual domain processing for the specificity of the chain length as determined by starter unit selection has also been probed [60].
Diversification of Polyketide Scaffolds via Non-Native Post-PKS Tailoring
Modifications to the core scaffolds introduce additional complexity and functionality. By leveraging native and engineered promiscuity of post-PKS tailoring enzymes, new-to-nature polyketides can be generated through the introduction of non-natural or non-native moieties.
Macrolactone glycosylation is critical to the biological functionality of polyketides. Owing to native glycosyltransferase (GT) promiscuity, several groups have leveraged GTs for the biosynthesis of non-native polyketides by leveraging combinatorial libraries derived from wide panels of macrolactone cores and NTP-sugars. Notably, the biological functionality of non-natively glycosylated polyketides can display different activity from their naturally occurring counterparts [61], as demonstrated by a series of erythromycin analogues, some of which have been characterized with potent activity against erythromycin A resistant strains [62]. Although these efforts continue to demonstrate the broad capabilities of these enzymes, the ability to diversify aglycone structures continues to pose a challenge.
Polyketide alkylation via biological and chemical syntheses, has similarly expanded biological functionality, as evidenced by the enhanced bioactivity of the daidzein analogue, 4’-O-methyl daidzein [63]. Often catalyzed by S-adenosylmethionine (SAM) dependent methyltransferases (MTs) [64], post-PKS polyketide alkylation has been diversified using rational reprogramming of promiscuous MTs for use in combinatorial biosynthesis reactions [65]. Interestingly, some natural product MTs have displayed promiscuity for non-native SAM analogues for the transfer of ethyl-, propargyl-, allyl-, and benzyl- moieties [66–68]. Notably though, while MTs have been highly successful for the diversification of non-ribosomal peptides, they have not yet been fully explored with polyketide substrates.
Halogenases also contribute to the diversification of natural products [69]. Flavin-dependent halogenases, such as VemK [70], ChmKN [71], and Rdc2 [72] have been demonstrated to halogenate aromatic polyketides, including resveratrol, to produce natural product analogues with potent bioactivities; however, their mechanisms are not conducive for use on non-aromatic substrates, limiting their potential to diversify other polyketides.
High-Throughput Approaches to Polyketide Synthetic Biology
Evaluating large numbers of artificial PKS pathways remains a critical engineering bottleneck due to the low-throughput of traditional analytical methods. Emerging high-throughput strategies, including engineered transcription-factor biosensor platforms and colorimetric assays [24], offer promising tools for engineering polyketide biosynthesis. Recently, a newly refactored FapR-based biosensor system for the detection of a variety of C2-substituted CoA- and N-acetylcysteamine (SNAc)-linked extender units [73] was described, both in vivo and in vitro. This biosensor detected the over-production of mmCoA in an engineered E. coli strain and also detected other extender units, paving the way for high-throughput engineering of native and non-natural extender unit supply in producing organisms.
There has also been much effort toward detection of polyketide pathway biosynthesis end-products and intermediates. Significant enhancements to mass spectroscopy-based screening methods, including laser-assisted rapid evaporative ionization mass spectrometry [74], have enabled the high-throughput direct detection of some natural products, including erythromycin A. Moreover, transcriptional regulators such as MphR are currently being leveraged for sensitive and specific screening of novel non-natural polyketides [75]. It is anticipated that such biosensors can be used to improve access to pathway intermediates and novel products through directed evolution by providing the ability to screen millions of pathway variants.
Future Outlook
The discovery of increasingly diverse polyketide enzymatic machinery offers powerful plug-and-play potential for the diversification of complex bioactive compounds. Yet, the ability to effectively engineer these pathways has been limited. However, efforts underpinned by enhanced mechanistic and structural studies of these megasynthases suggest an optimistic outlook for future PKS engineering to yield tailored polyketides. Probing the promiscuity of this machinery utilizing precursor-directed biosynthesis has enabled the mutasynthesis of a variety of critical targets. Moreover, analysis of product profiles derived from single domain and module modifications have elucidated unanticipated breaks in the co-linearity paradigm to enable cryptic and non-natural products. While engineering efforts were previously throttled by low-throughput screens, current and future engineering will be enhanced with the availability of tailored biosensors that enable high-throughput approaches to solve otherwise challenging problems related to polyketide biosynthetic engineering. An exciting future vision for designer polyketides includes augmenting traditional medicinal chemistry approaches with the application of computational methods to identify potential compounds of interest and value [76]. For example, “PKS Enumerator” [77] and the follow-up “SIME: Synthetic Insight-Based Macrolide Enumerator” [78] are cheminformatics tools that generates virtual libraries of macrolactones with multiple user-defined constraints derived from knowledge of polyketide biosynthesis. Such in silico libraries could then be mined using machine learning and/or 3D docking for potential new bioactive compounds. With this in mind, MacrolactoneDB integrates almost 14,000 existing macrolactones and their bioactivity information from various public databases [79]. Machine learning on this data set led to impressive prediction power for activity against several critical targets. Together, our vision for polyketide synthetic biology melds together high-throughput synthetic biology, advances in structural biology, mechanistic studies, and in silico prediction of bioactive compounds to develop new-to-nature polyketides as potential therapeutics and probes.
Acknowledgements
Financial support is provided by the National Institutes of Health (award number GM124112) and the Thomas Lord Distinguished Professorship Endowment.
References and Recommended Reading
- 1.Larsen EM, Wilson MR, Taylor RE: Conformation-activity relationships of polyketide natural products. Nat Prod Rep 2015, 32:1183–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pham JV, Yilma MA, Feliz A, Majid MT, Maffetone N, Walker JR, Kim E, Cho HJ, Reynolds JM, Song MC, et al. : A review of the microbial production of bioactive natural products and biologics. Front Microbiol 2019, 10:1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robertsen HL, Musiol-Kroll EM: Actinomycete-derived polyketides as a source of antibiotics and lead structures for the development of new antimicrobial drugs. Antibiotics 2019, 8:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nivina A, Yuet KP, Hsu J, Khosla C: Evolution and diversity of assembly-line polyketide synthases. Chem Rev 2019, 119:12524–12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yuzawa S, Zargar A, Pang B, Katz L, Keasling JD: Commodity chemicals from engineered modular type I polyketide synthases In Methods in Enzymology. . Academic Press Inc.; 2018:393–415. [DOI] [PubMed] [Google Scholar]
- 6.Wang X, Zhou H, Chen H, Jing X, Zheng W, Li R, Sun T, Liu J, Fu J, Huo L, et al. : Discovery of recombinases enables genome mining of cryptic biosynthetic gene clusters in Burkholderiales species. Proc Natl Acad Sci U S A 2018, 115:E4255–E4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ji ZY, Nie QY, Yin Y, Zhang M, Pan HX, Hou XF, Tang GL: Activation and characterization of cryptic gene cluster: two series of aromatic polyketides biosynthesized by divergent pathways. Angew Chemie - Int Ed 2019, 58:18046–18054. [DOI] [PubMed] [Google Scholar]
- 8.Chiang YM, Szewczyk E, Davidson AD, Keller N, Oakley BR, Wang CCC: A gene cluster containing two fungal polyketide synthases encodes the biosynthetic pathway for a polyketide, asperfuranone, in Aspergillus nidulans. J Am Chem Soc 2009, 131:2965–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Igboeli HA, Marchbank DH, Correa H, Overy D, Kerr RG: Discovery of primarolides A and B from marine fungus Asteromyces cruciatus using osmotic stress and treatment with suberoylanilide hydroxamic acid. Mar Drugs 2019, 17: E435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang JM, Wang HH, Liu X, Hu CH, Zou Y: Heterologous and engineered biosynthesis of nematocidal polyketide-nonribosomal peptide hybrid macrolactone from extreme thermophilic fungi. J Am Chem Soc 2020, 142:1957–1965. [DOI] [PubMed] [Google Scholar]
- 11.You D, Wang M-M, Yin B-C, Ye B-C: Precursor supply for erythromycin biosynthesis: engineering of propionate assimilation pathway based on propionylation modification. ACS Synth Biol 2019, 8:371–380. [DOI] [PubMed] [Google Scholar]
- 12.Yi JS, Kim M, Kim E-J, Kim B-G: Production of pikromycin using branched chain amino acid catabolism in Streptomyces venezuelae ATCC 15439. J Ind Microbiol Biotechnol 2018, 45:293–303. [DOI] [PubMed] [Google Scholar]; **Leveraging branched chain amino acid catabolism, this publication demonstrates the biosynthesis of acyl-CoAs to enhance the biosynthesis of pikromycin by 2.2 fold in Streptomyces venezuelae ATCC 15439.
- 13.Bauman KD, Li J, Murata K, Mantovani SM, Dahesh S, Nizet V, Luhavaya H, Moore BS: Refactoring the cryptic streptophenazine biosynthetic gene cluster unites phenazine, polyketide, and nonribosomal peptide biochemistry. Cell Chem Biol 2019, 26:724–736.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J, Chen Y, Li L, Yang E, Wang Y, Wu H, Zhang L, Wang W, Zhang B: Characterization and engineering of the Lrp/AsnC family regulator SACE_5717 for erythromycin overproduction in Saccharopolyspora erythraea. J Ind Microbiol Biotechnol 2019, 46:1013–1024. [DOI] [PubMed] [Google Scholar]
- 15.Lu Z, Zhang X, Dai J, Wang Y, He W: Engineering of leucine-responsive regulatory protein improves spiramycin and bitespiramycin biosynthesis. Microb Cell Fact 2019, 18: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu P, Marsafari M, Zha J, Koffas M: Microbial coculture for flavonoid synthesis. Trends Biotechnol 2020, doi: 10.1016/j.tibtech.2020.01.008. [DOI] [PubMed] [Google Scholar]
- 17.Wang B, Guo F, Dong S-H, Zhao H: Activation of silent biosynthetic gene clusters using transcription factor decoys. Nat Chem Biol 2019, 15:111–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Culp EJ, Yim G, Waglechner N, Wang W, Pawlowski AC, Wright GD: Hidden antibiotics in actinomycetes can be identified by inactivation of gene clusters for common antibiotics. Nat Biotechnol 2019, 37:1149–1154. [DOI] [PubMed] [Google Scholar]; ** A CRISPR-based method for discovery of new antibiotics in strains already producing well-known antibiotics provides potential for previously-discarded strains.
- 19.Zhou Q, Luo GC, Zhang H, Tang GL: Discovery of 16-demethylrifamycins by removing the predominant polyketide biosynthesis pathway in Micromonospora sp. strain TP-A0468. Appl Environ Microbiol 2019, 85: e02597–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fang L, Guell M, Church GM, Pfeifer BA: Heterologous erythromycin production across strain and plasmid construction. Biotechnol Prog 2017, doi: 10.1002/btpr.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bond CM, Tang Y: Engineering Saccharomyces cerevisiae for production of simvastatin. Metab Eng 2019, 51:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cummings M, Peters AD, Whitehead GFS, Menon BRK, Micklefield J, Webb SJ, Takano E: Assembling a plug-and-play production line for combinatorial biosynthesis of aromatic polyketides in Escherichia coli. PLOS Biol 2019, 17:e3000347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu X, Hua K, Liu D, Wu ZL, Wang Y, Zhang H, Deng Z, Pfeifer BA, Jiang M: Heterologous biosynthesis of type II polyketide products using E. coli. ACS Chem Biol 2019, doi: 10.1021/acschembio.9b00827. [DOI] [PubMed] [Google Scholar]
- 24.Yang D, Kim WJ, Yoo SM, Choi JH, Ha SH, Lee MH, Lee SY: Repurposing type III polyketide synthase as a malonyl- CoA biosensor for metabolic engineering in bacteria. Proc Natl Acad Sci U S A 2018, 115:9835–9844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang W, Li S, Li Z, Zhang J, Fan K, Tan G, Ai G, Lam SM, Shui G, Yang Z, et al. : Harnessing the intracellular triacylglycerols for titer improvement of polyketides in Streptomyces. Nat Biotechnol 2020, 38:76–83. [DOI] [PubMed] [Google Scholar]
- 26.Yamanaka K, Reynolds KA, Kersten RD, Ryan KS, Gonzalez DJ, Nizet V, Dorrestein PC, Moore BS: Direct cloning and refactoring of a silent lipopeptide biosynthetic gene cluster yields the antibiotic taromycin A. Proc Natl Acad Sci U S A 2014, 111:1957–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.D’Agostino PM, Gulder TAM: Direct pathway cloning combined with sequence- and ligation-independent cloning for fast biosynthetic gene cluster refactoring and heterologous expression. ACS Synth Biol 2018, 7:1702–1708. [DOI] [PubMed] [Google Scholar]
- 28.Kalkreuter E, CroweTipton JM, Lowell AN, Sherman DH, Williams GJ: Engineering the substrate specificity of a modular polyketide synthase for installation of consecutive non-natural extender units. J Am Chem Soc 2019, 141:1961–1969. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** This paper demonstrates the first double incorporation of a nonnatural click handle extender unit into a polyketide. It further explores possible bottlenecks between domains.
- 29.Zhang F, Ji H, Ali I, Deng Z, Bai L, Zheng J: Structural and biochemical Insight into the recruitment of acyl carrier protein‐linked extender units in ansamitocin biosynthesis. ChemBioChem 2020, doi: 10.1002/cbic.201900628. [DOI] [PubMed] [Google Scholar]
- 30.Kornfuehrer T, Eustáquio AS: Diversification of polyketide structures via synthase engineering. Medchemcomm 2019, 10:1256–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carpenter SM, Williams GJ: Extender unit promiscuity and orthogonal protein interactions of an aminomalonyl-ACP utilizing trans-acyltransferase from zwittermicin biosynthesis. ACS Chem Biol 2018, 13:3361–3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jenner M, Kosol S, Griffiths D, Prasongpholchai P, Manzi L, Barrow AS, Moses JE, Oldham NJ, Lewandowski JR, Challis GL: Mechanism of intersubunit ketosynthase–dehydratase interaction in polyketide synthases. Nat Chem Biol 2018, 14:270–275. [DOI] [PMC free article] [PubMed] [Google Scholar]; *A docking domain at which trans-ATs associate with a module gives insight into how these enzymes interact, and more importantly,how they can be engineered.
- 33.Helfrich EJN, Ueoka R, Dolev A, Rust M, Meoded RA, Bhushan A, Califano G, Costa R, Gugger M, Steinbeck C, et al. : Automated structure prediction of trans-acyltransferase polyketide synthase products. Nat Chem Biol 2019, 15:813–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Demachi A, Uchida R, Arima S, Nagamitsu T, Hashimoto J, Komatsu M, Kozone I, Shin-ya K, Tomoda H, Ikeda H: An unusual extender unit Is incorporated into the modular polyketide synthase of scopranones biosynthesis. Biochemistry 2019, 58:5066–5073. [DOI] [PubMed] [Google Scholar]
- 35.Grote M, Schulz F: Exploring the promiscuous enzymatic activation of unnatural polyketide extender units in vitro and in vivo for monensin biosynthesis. ChemBioChem 2019, 20:1183–1189. [DOI] [PubMed] [Google Scholar]
- 36.Vögeli B, Geyer K, Gerlinger PD, Benkstein S, Cortina NS, Erb TJ: Combining promiscuous acyl-CoA oxidase and enoyl-CoA carboxylase/reductases for atypical polyketide extender unit biosynthesis. Cell Chem Biol 2018, 25:833–839.e4. [DOI] [PubMed] [Google Scholar]
- 37.Wilson MC, Moore BS: Beyond ethylmalonyl-CoA: The functional role of crotonyl-CoA carboxylase/reductase homologs in expanding polyketide diversity. Nat Prod Rep 2012, 29:72–86. [DOI] [PubMed] [Google Scholar]
- 38.Pereira PRM, Araújo J de O, Silva JRA, Alves CN, Lameira J, Lima AH: Exploring chloride selectivity and halogenase regioselectivity of the SalL enzyme through quantum mechanical/molecular mechanical modeling. J Chem Inf Model 2020, doi: 10.1021/acs.jcim.9b01079. [DOI] [PubMed] [Google Scholar]
- 39.Davis TD, Kunakom S, Burkart MD, Eustaquio AS: Preparation, assay, and application of chlorinase SalL for the chemoenzymatic synthesis of S-adenosyl-L-methionine and analogs In Methods in Enzymology. . Academic Press Inc.; 2018:367–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharma SV, Tong X, Pubill-Ulldemolins C, Cartmell C, Bogosyan EJA, Rackham EJ, Marelli E, Hamed RB, Goss RJM: Living GenoChemetics by hyphenating synthetic biology and synthetic chemistry in vivo. Nat Commun 2017, 8:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang L, Hashimoto T, Qin B, Hashimoto J, Kozone I, Kawahara T, Okada M, Awakawa T, Ito T, Asakawa Y, et al. : Characterization of giant modular PKSs provides insight into genetic mechanism for structural diversification of aminopolyol polyketides. Angew Chemie - Int Ed 2017, 56:1740–1745. [DOI] [PubMed] [Google Scholar]
- 42.Miyazawa T, Hirsch M, Zhang Z, Keatinge-Clay AT: An in vitro platform for engineering and harnessing modular polyketide synthases. Nat Commun 2020, 11:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]; *This paper describes a one pot assay in which optimal domain and module boundaries can be determined relatively quickly.
- 43.Peng H, Ishida K, Sugimoto Y, Jenke-Kodama H, Hertweck C: Emulating evolutionary processes to morph aureothin-type modular polyketide synthases and associated oxygenases. Nat Commun 2019, 10:3918. [DOI] [PMC free article] [PubMed] [Google Scholar]; *With the aim to understand the evolution of two pathways, this paper demonstrates that much can be learned about PKS and post-PKS limitations in engineering from mimicking evolutionary processes.
- 44.Beom JY, Jung JA, Lee KT, Hwangbo A, Song MC, Lee Y, Lee SJ, Oh JH, Ha SJ, Nam SJ, et al. : Biosynthesis of nonimmunosuppressive FK506 analogues with antifungal activity. J Nat Prod 2019, 82:2078–2086. [DOI] [PubMed] [Google Scholar]
- 45.Liao JL, Pang KL, Sun GH, Pai TW, Hsu PH, Lin JS, Sun KH, Hsieh CC, Tang SJ: Chimeric 6-methylsalicylic acid synthase with domains of acyl carrier protein and methyltransferase from Pseudallescheria boydii shows novel biosynthetic activity. Microb Biotechnol 2019, 12:920–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yuzawa S, Mirsiaghi M, Jocic R, Fujii T, Masson F, Benites VT, Baidoo EEK, Sundstrom E, Tanjore D, Pray TR, et al. : Short-chain ketone production by engineered polyketide synthases in Streptomyces albus. Nat Commun 2018, 9:4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alanjary M, Cano-Prieto C, Gross H, Medema MH: Computer-aided re-engineering of nonribosomal peptide and polyketide biosynthetic assembly lines. Nat Prod Rep 2019, 36:1249–1261. [DOI] [PubMed] [Google Scholar]
- 48.Liu T, Mazmouz R, Neilan BA: An in vitro and in vivo study of broad-range phosphopantetheinyl transferases for heterologous expression of cyanobacterial natural products. ACS Synth Biol 2018, 7:1143–1151. [DOI] [PubMed] [Google Scholar]
- 49.Wang Z, Bagde SR, Zavala G, Matsui T, Chen X, Kim CY: De novo design and implementation of a tandem acyl carrier protein domain in a type I modular polyketide synthase. ACS Chem Biol 2018, 13:3072–3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klaus M, D’Souza AD, Nivina A, Khosla C, Grininger M: Engineering of chimeric polyketide synthases using SYNZIP docking domains. ACS Chem Biol 2019, 14:426–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meinke JL, Simon AJ, Wagner DT, Morrow BR, You S, Ellington AD, Keatinge-Clay AT: Employing 25-residue docking motifs from modular polyketide synthases as orthogonal protein connectors. ACS Synth Biol 2019, 8:2017–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li T, Tripathi A, Yu F, Sherman DH, Rao A: DDAP: docking domain affinity and biosynthetic pathway prediction tool for type I polyketide synthases. Bioinformatics 2019, 36:942–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li X, Sevillano N, La Greca F, Deis L, Liu Y-C, Deller MC, Mathews II, Matsui T, Cane DE, Craik CS, et al. : Structure–function analysis of the extended conformation of a polyketide synthase module. J Am Chem Soc 2018, 140:6518–6521. [DOI] [PMC free article] [PubMed] [Google Scholar]; *Structural studies in the paper revealed that modules maintain an open conformation during chain elongation. This knowledge will guide chimeric PKS engineering in the future.
- 54.Peng H, Ishida K, Hertweck C: Loss of single-domain function in a modular assembly line alters the size and shape of a complex polyketide. Angew Chemie - Int Ed 2019, doi: 10.1002/anie.201911315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sundaram S, Kim HJ, Bauer R, Thongkongkaew T, Heine D, Hertweck C: On-line polyketide cyclization into diverse medium-sized lactones by a specialized ketosynthase domain. Angew Chemie Int Ed 2018, 57:11223–11227. [DOI] [PubMed] [Google Scholar]
- 56.Moss NA, Seiler G, Leão TF, Castro-Falcón G, Gerwick L, Hughes CC, Gerwick WH: Nature’s combinatorial biosynthesis produces vatiamides A-F. Angew Chemie Int Ed 2019, 58:9027–9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang JJ, Tang X, Huan T, Ross AC, Moore BS: Pass-back chain extension expands multimodular assembly line biosynthesis. Nat Chem Biol 2020, 16:42–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wlodek A, Kendrew SG, Coates NJ, Hold A, Pogwizd J, Rudder S, Sheehan LS, Higginbotham SJ, Stanley-Smith AE, Warneck T, et al. : Diversity oriented biosynthesis via accelerated evolution of modular gene clusters. Nat Commun 2017, 8:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang XL, Friedrich S, Yin S, Piech O, Williams K, Simpson TJ, Cox RJ: Molecular basis of methylation and chain-length programming in a fungal iterative highly reducing polyketide synthase. Chem Sci 2019, 10:8478–8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sato K, Katsuyama Y, Yokota K, Awakawa T, Tezuka T, Ohnishi Y: Involvement of β‐alkylation machinery and two sets of ketosynthase‐chain‐length factors in the biosynthesis of fogacin polyketides in Actinoplanes missouriensis. ChemBioChem 2019, 20:1039–1050. [DOI] [PubMed] [Google Scholar]
- 61.Malmierca MG, Pérez-Victori I, Martín J, Reyes F, Méndez C, Salas JA, Olano C: New sipanmycin analogues generated by combinatorial biosynthesis and mutasynthesis approaches relying on the substrate flexibility of key enzymes in the biosynthetic pathway. Appl Environ Microbiol 2020, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fang L, Zhang G, El‐Halfawy O, Simon M, Brown ED, Pfeifer BA: Broadened glycosylation patterning of heterologously produced erythromycin. Biotechnol Bioeng 2018, 115:2771–2777. [DOI] [PMC free article] [PubMed] [Google Scholar]; *Investigation into the glycodiversification of erythromycin for combinatorial biosynthesis for enhanced bioactivity. This paper elucidated the utility of olivose, forosamine, and noviose erythromycin analogues to act against erythromycin resistant B. subtilis.
- 63.Koirala N, Pandey RP, Thuan NH, Ghimire GP, Jung HJ, Oh TJ, Sohng JK: Metabolic engineering of Escherichia coli for the production of isoflavonoid-4′-O-methoxides and their biological activities. Biotechnol Appl Biochem 2019, 66:484–493. [DOI] [PubMed] [Google Scholar]
- 64.Grocholski T, Yamada K, Sinkkonen J, Tirkkonen H, Niemi J, Metsä-Ketelä M: Evolutionary trajectories for the functional diversification of anthracycline methyltransferases. ACS Chem Biol 2019, 14:850–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang X, Wang C, Duan L, Zhang L, Liu H, Xu Y, Liu Q, Mao T, Zhang W, Chen M, et al. : Rational reprogramming of O-methylation regioselectivity for combinatorial biosynthetic tailoring of benzenediol lactone scaffolds. J Am Chem Soc 2019, 141:4355–4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McKean IJW, Sadler JC, Cuetos A, Frese A, Humphreys LD, Grogan G, Hoskisson PA, Burley GA: S‐adenosyl methionine cofactor modifications enhance the biocatalytic repertoire of small molecule C‐alkylation. Angew Chemie 2019, 58:17583–17588. [DOI] [PubMed] [Google Scholar]
- 67.Tengg M, Stecher H, Offner L, Plasch K, Anderl F, Weber H, Schwab H, Gruber-Khadjawi M: Methyltransferases: green catalysts for Friedel-Crafts alkylations. ChemCatChem 2016, 8:1354–1360. [Google Scholar]
- 68.Stecher H, Tengg M, Ueberbacher BJ, Remler P, Schwab H, Griengl H, Gruber-Khadjawi M: Biocatalytic Friedel-Crafts alkylation using non-natural cofactors. Angew Chemie Int Ed 2009, 48:9546–9548. [DOI] [PubMed] [Google Scholar]
- 69.Latham J, Brandenburger E, Shepherd SA, Menon BRK, Micklefield J: Development of halogenase enzymes for use in synthesis. Chem Rev 2018, 118:232–269. [DOI] [PubMed] [Google Scholar]
- 70.Song R, Shi H, Zhu J, Wang H, Shen Y: A single-component flavoenzyme catalyzed regioselective halogenation of pyrone in the biosynthesis of venemycins. ACS Chem Biol 2019, 14:2533–2537. [DOI] [PubMed] [Google Scholar]
- 71.Ugai T, Minami A, Tanaka S, Ozaki T, Liu C, Shigemori H, Hashimoto M, Oikawa H: Biosynthetic machinery of 6‐hydroxymellein eerivatives leading to cyclohelminthols and palmaenones. ChemBioChem 2020, 21:360–367. [DOI] [PubMed] [Google Scholar]
- 72.Wang S, Zhang S, Xiao A, Rasmussen M, Skidmore C, Zhan J: Metabolic engineering of Escherichia coli for the biosynthesis of various phenylpropanoid derivatives. Metab Eng 2015, 29:153–159. [DOI] [PubMed] [Google Scholar]
- 73.Kalkreuter E, Keeler AM, Malico AA, Bingham KS, Gayen AK, Williams GJ: Development of a genetically encoded biosensor for detection of polyketide synthase extender units in Escherichia coli. ACS Synth Biol 2019, 8:1391–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]; **This paper focuses on the development of a FapR biosensor platform both in vivo and in vitro for the detection of malonyl-CoA and its C2 substituted analogues towards the development of non-natural polyketides.
- 74.Gowers G-OF, Cameron SJS, Perdones-Montero A, Bell D, Chee SM, Kern M, Tew D, Ellis T, Takáts Z: Off-colony screening of biosynthetic libraries by rapid laser-enabled mass spectrometry. ACS Synth Biol 2019, 8:2566–2575. [DOI] [PubMed] [Google Scholar]
- 75.Kasey C, Zerrad M, Li Y, Cropp TA, Williams GJ: Development of transcription factor-based designer macrolide biosensors for metabolic engineering and synthetic biology. ACS Synth Biol 2018, 7:227–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Norinder U, Munic Kos V: QSAR models for predicting five levels of cellular accumulation of lysosomotropic macrocycles. Int J Mol Sci 2019, 20:5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zin PPK, Williams G, Fourches D: Cheminformatics-based enumeration and analysis of large libraries of macrolide scaffolds. J Cheminform 2018, 10:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zin PPK, Williams G, Fourches D: SIME: Synthetic insight-based macrolide enumerator to generate the V1B library of 1 billion macrolides. J Cheminform 2020, 12:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zin PPK, Williams GJ, Ekins S: Cheminformatics analysis and modeling with MacrolactoneDB. Sci Rep 2020, 10:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]


