Abstract
Nicotinic acetylcholine receptor (nAChR) participates in diverse biological processes, e.g., mood, learning, and addiction. Glycosylphosphatidylinositol (GPI)-linked lynx1 is an allosteric modulator of nAChR function, including shifts in agonist sensitivity, reduced desensitization, and slower recovery from desensitization. This modulation is thought to be achieved by lynx1’s interaction with nAChR subunits, particularly at the α4:α4 interface. In this study, we used molecular modeling and simulation to study the structure, dynamics, and interactions of lynx1 when bound to nAChRs, as well as unbound, monomeric lynx1, when embedded in membranes. Though lynx1 structures are similar in both states, lynx1 dynamics are more restricted in the bound state than in the unbound one. When bound, interactions between lynx1 and nAChR are observed to be maintained throughout the simulations. Of particular note, lynx1 demonstrates prolonged interactions with the receptor C-loop in one of the nAChR α4 subunits, a region important for agonist binding and possibly the transition between open/close states. During interactions with lynx1, an α4 C-loop tends to be restricted in either close or open state, whereas the C-loop state transitions are more evident in when the nAChR is unbound by lynx1. Interestingly, the conformational change of the C-loop is stochastic, suggesting that lynx1 can influence nAChR (critical for its multimodal action), for instance by shifting its agonist sensitivity and recovery from desensitization.
Graphical Abstract
Introduction
The cholinergic system is involved in diverse functions including mood, anxiety, learning and memory, and addiction 1,2. The nicotinic acetylcholine receptors (nAChRs) of the cholinergic system are comprised of 15 different subunits (α1-10 and β1-4) that assemble into both homopentameric receptors such as (α7)5 3-5 or (α9)5 6 and heteropentameric receptors such as α4β2 7-11. Subunit composition and even the position of specific subunits within pentamers can influence the biophysical, pharmacological, and cell biological properties of the receptor, with profound effects on information processing in neurons that express these nAChRs.
The basic conformational states of nAChRs are the closed state, the open state, and the desensitized state. There is considerable evidence that the movement of the C-loop is important in the transition from the closed (agonist unbound) state to the open (agonist bound) state 12,13. Agonist binding is thought to accompany movement of the C-loop in closer approximation to the agonist binding site (i.e., the so-called capping of the agonist binding site) along with subsequent changes in conformation of the pore lining transmembrane domain 14. Multiple factors can influence these transition states, including subunit composition, nicotine exposure, calcium, etc.
Subunit composition influences the properties of the two possible pentameric stoichiometries of α4β2 nAChRs, the most abundant heteromeric nAChR subtype in the brain. The lower sensitivity subtype (LS) receptors adopt the (α4)3(β2)2 stoichiometry, while the higher sensitivity (HS) receptors adopt the (α4)2(β2)3 stoichiometry. These two stoichiometries exhibit different desensitization kinetics and respond differently to chronic nicotine treatment. HS receptors are upregulated in response to nicotine, while LS receptors are unaffected. This is thought to play an important role in mechanisms of nicotine addiction. Additionally, subunit composition could possibly influence sub-cellular localization of receptors on the plasma membrane 15.
The lynx family proteins are a subset of the Ly6/uPAR superfamily, coding for cysteine-rich proteins adopting a three-fingered toxin fold 16-21. A superfamily is related to the elapid snake venom toxin genes, such as α-bungarotoxin, consisting of multiple internal disulfide bonds between the conserved cysteine residues 22-25. α-bungarotoxin and other such toxins exemplify a highly-conserved receptor binding motif that apparently evolved from lynx proteins 22. α-neurotoxins, such as α-bungarotoxin, have become widely used probes for the investigation of the properties of nAChRs.
Studies have indicated preferential interactions of lynx1 to the LS stoichiometry, particularly the α4:α4 nAChR subunit interface, which only exists with the (α4)3(β2)2 stoichiometry of α4β2 nAChRs 24. Specifically, hydrogen bonds and cation-π interactions were observed between Arg38 of lynx1 and Trp156 / Tyr204 on the α4:α4 interface 26. Computational modeling of lynx1 complexes with (α4)3(β2)2 suggests steric hindrance of lynx1 at the α4:β2 interface that would not exist at α4:α4 interfaces, proving a possible explanation for lynx1’s preferential binding to the LS stoichiometry over the HS one 26.
A glycosylphosphatidylinositol (GPI)-linked prototoxin molecule such as lynx1 has been demonstrated to alter the stoichiometry and assembly of nAChRs 24. Further, the presence or absence of a GPI-anchor has been shown in some cases to influence neuronal signaling 27. Therefore, the existence of the GPI-anchor on lynx1 proteins could contribute to the structure, composition, or function of nascent nAChRs as well. GPI-anchored neurotoxin-like receptor binding proteins are confined to a volume above the extracellular face of the membrane. This topology positions then exert effects by binding to the extracellular portion of receptors.
Lyukmanova et al. 19 determined the first three-dimensional (3D) structure of the water-soluble domain lynx1 (i.e., without the GPI-anchor) by NMR. This structure becomes an important model for the lynx1-mediated nAChR structure-function studies. For example, based on this water-soluble model, a series of lynx1 mutations were also studied 28 for understanding the binding and functional properties of lynx1 protein. However, there is little detailed biophysical information on how lynx1 interacts with nAChR or how these interactions lead to changes in nAChR function. The most abundant nAChRs in central neurons are α4β2 heteropentamers and α7 homopentamers 16,29-31. Due to the lack of 3D structures for α4β2 and α7 nAChRs, most studies used acetylcholine-binding proteins (AChBPs) and Torpedo nAChR to elucidate the nAChR functions. Recently, Walsh et al. 32 determined the 3D structure of human α4β2 nAChR and Nissen et al. 26 modeled a first complex structure of lynx1 with α4β2 nAChR. Preferable binding of lynx1 at the α4:α4 interface of nAChR was obtained through all-atom molecular modeling and molecular dynamics (MD) simulations of lynx1 at the α4:α4 interface (i.e., without the GPI-anchor and with only two α4 interface).
A GPI-anchor is presumably important in the function of lynx1 on nAChR function, but none of the previously published simulations have been conducted with GPI-anchored lynx1. Here, we performed all-atom molecular modeling and MD simulations of GPI-anchored lynx1 with the α4:α4 nAChR interface in the context of a membrane-embedded (α4)3(β2)2 nAChR. We also performed all-atom MD simulations of GPI-anchored lynx1 in a bilayer without nAChR. These simulations provide insights into the structure, orientation, and dynamics of GPI-anchored lynx1 on a bilayer surface in the presence or absence of nAChR, as well as detailed interactions between GPI-anchored lynx1 and nAChR in a bilayer.
Methods
To gain insights into how the GPI-anchored lynx1 protein behaves in a membrane bilayer and whether the lynx1-nAChR interactions influence lynx1’s structure, orientation, dynamics, and nAChR C-loop motions, we performed all-atom MD simulations of GPI-anchored lynx1 (lynx1-only system), a GPI-anchored lynx1-nAChR complex system, as well as an apo-nAChR system with three independent replicas of each system. All simulations were performed with NAMD 33. We used the CHARMM36(m) force field for protein 34, carbohydrates 35-37, and lipids 38,39. The NMR structure of lynx1 (PDB ID: 2L03) 19 and structure of nAChR (PDB ID: 6CNK) 32 were taken from the Protein Data Bank 40. We used the TIP3P water model 41 and counter ions of K+ and Cl− with a concentration of 0.15 M to neutralize the system.
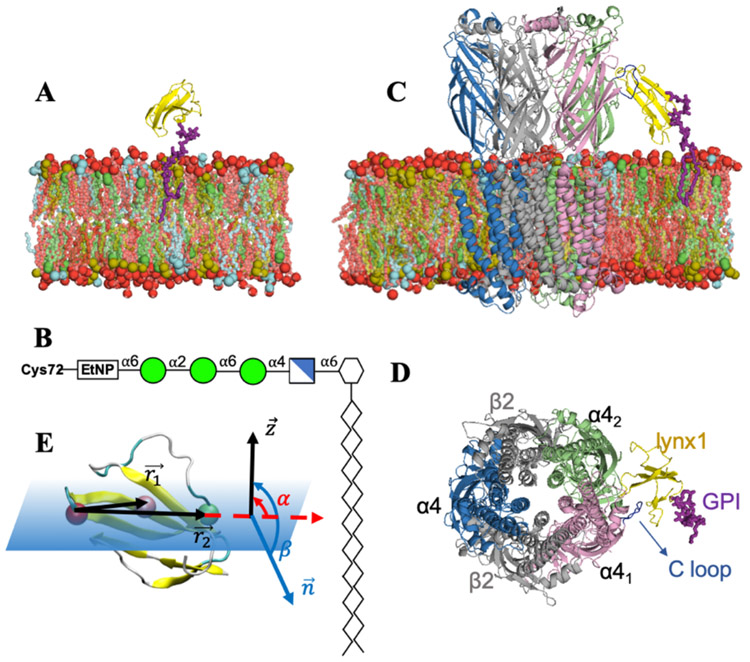
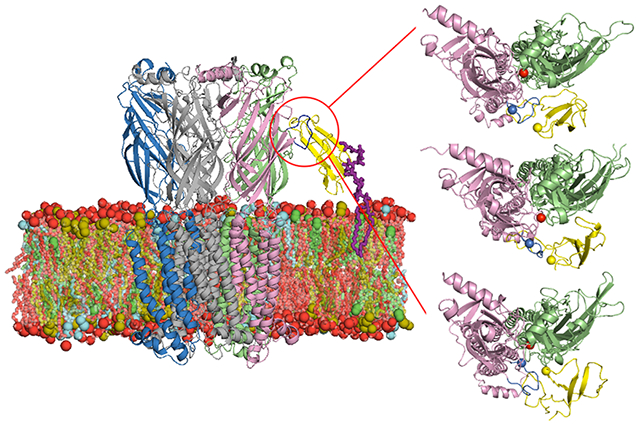
Cholesterol (Chol), phosphatidyl-ethanolamine (PE), and phosphatidyl-choline (PC) are reported to be enriched in synaptic plasma membranes 42. Chol and sphingomyelin (SM) are postulated as important lipid types in membrane trafficking 43-45. We chose to use the same lipid composition as in Sunshine et at. 46 (DOPE:DOPC:PSM:Chol = 2:1:5:2) for all systems (Figs. 1A, C) to represent a neuronal plasma membrane using dioleoyl-phosphatidyl-ethanolamine (DOPE), dioleoyl-phosphatidyl-choline (DOPC), and palmitoyl-sphingomyelin (PSM). Lynx1 was anchored to the membrane with a GPI-anchor linked to the C-terminus of the protein. The GPI structure is shown in Figure 1B. The membrane and GPI-anchor structure were generated through CHARMM-GUIMembrane Builder 47,48 and Glycolipid Modeler 49,50. The initial system size was 100 Å × 100 Å × 115 Å for the lynx1-only and apo-nAChR system and 150 Å × 150 Å × 160 Å for the lynx1-nAChR system. All simulation systems and simulation inputs were generated through CHARMM-GUI 51,52. Visualization was done via VMD and PyMOL 53,54, and all analyses were done by CHARMM 55.
Figure 1.
(A) Snapshot of the lynx1-only system: lynx1 in yellow, GPI-anchor in magenta sticks, and membrane bilayers colored with DOPE in brown, DOPC in cyan, PSM in red, and Chol in green. Lipid head groups are shown as van der Waals spheres and tails as sticks with each lipid color. (B) A GPI-anchored structure used in this study: mannose (green circles), glucosamine (blue cross square), phosphoethanolamine (EtNP), and myo-inositol 1-phosphate (white hexagon). (C and D) Snapshots of the lynx1-nAChR system: (α4)3(β2)2 structure of nAChR was employed here with α4:α4 facing lynx1. β2 colored in light gray and α4 in pink, green, and blue, respectively. The C-loop of α41 in dark blue as critical interaction site with lynx1 protein. The lipid composition is the same in the two systems: DOPE:DOPC:PSM:Chol = 2:1:5:2. In (A) and (C), water molecules, ions, and lipids located in front of proteins are not shown for clarity. (E) Schematic representation of lynx1 tilt (α) and rotation (β) angles with respect to the membrane whose normal is the Z axis. The three selected residues used to define α and β are shown as van der Waals spheres colored red (Arg38), pink (Thr30), and green (Pro47).
In this work, the van der Waals interactions were smoothly switched off over 10-12 Å by a force-based switching function 56 and the long-range electrostatic interactions were calculated by the particle-mesh Ewald method 57 with a mesh size of ~1 Å. The timestep was set to 2 fs and we constrained bonds containing hydrogen atoms with the SHAKE algorithm 58. Temperature was held at 310 K, the constant temperature was controlled by Langevin dynamics with a damping frequency of 50 fs−1 59. We first relaxed the system in a canonical ensemble (NVT) with all the solute atoms subjected to harmonic restraints. A 100-120 ps isothermal–isobaric ensemble (NPT) was then applied to adjust the solvent density. The Langevin piston method was used to control the constant pressure 60,61. A dihedral restraint force constant was set to 1 kcal/(mol·rad2) to maintain the carbohydrate chair conformation during these equilibration steps. Simulations were performed using NAMD 33 (lynx1-only and lynx1-nAChR) and OpenMM 62 (apo-nAChR). For the production run, each lynx1-only replica was simulated for 1.2 μs, each lynx1-nAChR replica for 500 ns, and each apo-nAChR replica for 300 ns. Trajectories were saved every 25 ps. All the results shown in this work were based on the analysis for production simulations.
The (α4)3(β2)2 nAChR stoichiometry selected for this work was based on the most abundant subtypes expressed in the brain and previously reported interactions of lynx1 with the α4:α4 interface 26 (Figs. 1C, D). It was demonstrated previously that the interaction between lynx1 and nAChR occurs preferentially within the α41:α42 interface and mostly with Arg38 of lynx1 protein (Figure 1E). Thus, the initial lynx1-nAChR structure was generated with the α41:α42 interface towards lynx1. After manually opening the C-loop of α41, Arg38 was placed into the α41:α42 interface to generate a starting conformation. This conformation of nAChR was used for the generation of apo-nAChR systems. And this initial conformation was also set as the reference calculating the C-loop motion of nAChR α41 subunit. For the lynx1-nAChR system, distance restraints of 3 Å were applied between Arg38 of lynx1 and Trp156 and Tyr204 on nAChR α41 subunit during the first 100-ns of the MD simulations to relax the system, then all the restrains were released for 500-ns production. Trp156 and Tyr204 are the residues of the α41 subunit located in the α41:α42 interface area that are known to form hydrogen bonds with Arg38 in our previous work 26.
Tilt (α) and rotation (β) angles defined in this study are schematically shown in Figure 1E. Three residues located near the central section of lynx1 were selected for α and β calculation: Arg38, Thr30, and Pro47. A vector from Arg38 to Thr30 () and a vector from Arg38 to Pro 47 () were used to define α as the angle between and (unit vector in the Z direction) and β as the angle between the normal vector formed by and .
The Z-coordinate distribution range of the C-loop in each nAChR subunit was regarded as the potential interaction range for the lynx1-only system in this work. This criterion was made because we observed multi-mode interactions between lynx1 and nAChR C-loop in all complex system replicas (see Results and Discussion). Thus, we regard the C-loop as a critical interaction site for lynx1 protein in this study.
Results and Discussion
Lynx1-nAChR Interactions are Multi-modes
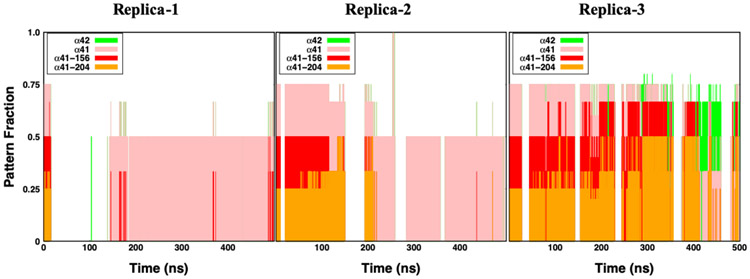
To obtain an insight into which specific regions of the nAChR are involved in lynx1-nAChR interactions, we have produced time series of interaction patterns between lynx1 and its environment in three replicas (Figure 2). Trp156 and Tyr204 are two sites on nAChR α41 subunit that are located within the interaction cut-off distance (4 Å) from lynx1 Arg38 in the initial structure. And, the “α41” label represents residues on nAChR α41 subunit excluding Trp156 and Tyr204. Considering interactions with nAChR α41 / α42 subunits all together (all colored region in Figure 2), Arg38 spent the majority of the time located within the interaction region of nAChR, which provides support of these interactions throughout our simulation time. Even though these interactions can be observed in either replica, the specific residues that Arg38 interacts with are different in each situation.
Figure 2.
Interaction patterns of lynx1 Arg38 with its environment in the lynx1-nAChR complex system. The interaction pattern graph shows the probability of occurrence within 4 Å from each of α42 (green), α41 (pink), α41 Trp156 (red), and α41 Tyr204 (orange) from three individual systems. Note that α41 Trp156 and α41 Tyr204 are initial interaction sites in the lynx1-nAChR complex system. α41 represents residues on α41 excluding Trp156 and Tyr204.
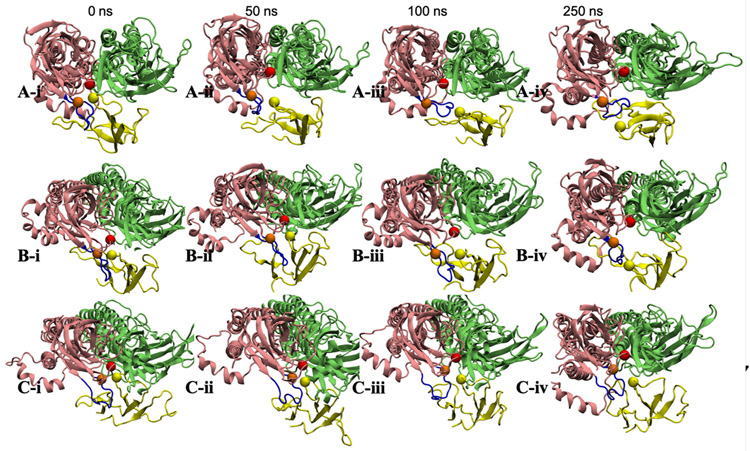
In Figure 3, snapshots of the lynx1 protein and nAChR α41 / α42 subunits were taken at 0-ns, 50-ns, 100-ns, 250-ns production times, respectively. Arg38 is represented in yellow sphere, and Trp156 and Tyr204 in red and orange ones. The C-loop of nAChR α41, colored in dark blue, was initially opened (compared to the structure in PDB 6cnk) and part of lynx1 (including Arg38) was placed within the α41:α42 interface. There are three regions in the C-loop: Ile203-Pro205 (inside facing region), Tyr197-Glu202 (front turn region), and Thr194-Lys196 (outside facing region). As Figure 3A series (A-i to A-iv) represent, Arg38 gradually detached from the α41:α42 interface and the C-loop closed. Interestingly, even though Arg38 left the initial interaction sites, it still kept interacting with other residues on α41 subunit, specifically, with the C-loop front turn residues, preventing the C-loop from opening again. This interaction is clear in Figure 4A that represents the averaged distance between each α41 C-loop residue and Arg38 over the entire simulation. A small distance value indicates a high frequency of Arg38’s presence in close to a specific residue. In replica-1, Cys199-Ala201 located in the front turn region show the closest contacts to Arg38 (Figure 4A), due to the outward movement of Arg38 from initial position as discussed in Figure 3A. Figure 3B series show the positions of α41, α42, and lynx1 when Arg38 maintaining partially detached from the initial interaction interface. In this system, Arg38 did not move far enough for the C-loop to close. Instead, Arg38 actually kept interacting with the front turn on the C-loop, which to some degree stabilizes the movement of the C-loop and lynx1 (Figure 3B-iv). Accordingly, these front turn residues have closer contact to Arg38 than other residues (see replica-2 in Figure 4A). This relative position between Arg38 and the C-loop persists for the rest of the simulation. Though Arg38 interacting mostly with front turn residues, Arg38 in replica-1 detached more complete from initial sites than replica-2 (Figure 2) that differs the C-loop motion dramatically. Unlike the events in Figures 3A and 3B, in Figure 3C, Arg38 was observed to fluctuate only within the initial interaction interface, making Arg38 interact with the inside facing residues in close contact (see replica-3 in Figure 4A and Figure 2). Fluctuating within the α41:α42 interface, Arg38 infrequently moves close to α42 subunit performing some non-specific interactions. In this case, with lynx1 protein kept inside the α41:α42 interface region, the C-loop was stabilized in the open state for the entire simulation time.
Figure 3.
Snapshots of α41, α42, and lynx1 in the lynx1-nAChR complex system: α41 in pink, α42 in green, and lynx1 protein in yellow. Arg38 (yellow), α41 Trp156 (red), and α41 Tyr204 (orange) are shown in van der Waals spheres and the C-loop of α41 in dark blue. (i-iv) represents the relative position of the proteins at 0-ns, 50-ns, 100-ns, and 250-ns production times, respectively.
Figure 4.
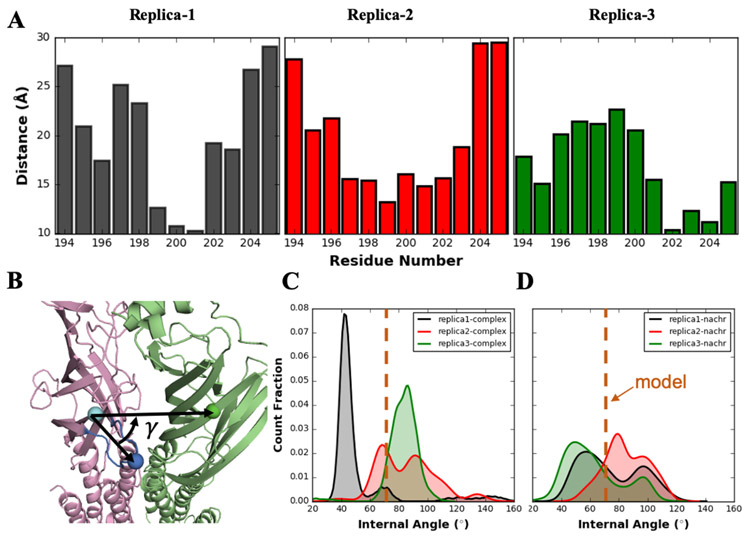
(A) Averaged distance between each α41 C-loop residue and Arg38 over the entire simulation in each replica. (B) Schematic representation of α41 C-loop internal angle (γ) for the representation of C-loop motion. The three selected residues used to define γ are shown as van der Waals spheres colored in cyan (α41 Pro205), blue (α41 Cys199), and green (α42 Phe40). (C) Density distributions of α41 C-loop γ angle in lynx1-nAChR complex and (D) apo-nAChR system. Each replica is colored in same as in (A). The value of initial model structure is represented in orange dashed line
The nAChR C-loop has been reported to be crucial for maintaining functional properties of nAChRs. Lynx1 Arg38 was initially placed inside the α41:α42 interface with the α41 C-loop opened. This is a configuration that is associated with the closed functional state of the nAChR 63,64. As suggested in Figure 3A series, once closed, the C-loop would be hard to reopen due to the interaction between its front turn residues and Arg38 until lynx1 disassociates from the nAChR. Note that the opposite phenomenon was observed in Figure 3C series with maintaining the C-loop in the open state. Figures 4C-D show the α41 C-loop motions in lynx1-nAChR complex and apo-nAChR systems. The internal angle (γ) was employed to represent the open and closed states of C-loop; the smaller the angle is, the C-loop more likely in the closed state. Note that there is no hard threshold to distinguish the open and closed states, so we use the value of the initial structure as a reference. As shown in Figure 3A series in which the C-loop was prevented from re-opening due to the interaction with lynx1, γ values (black curve) are mostly small in Figure 4C. Expectedly, for Figure 3C series, another tight distribution around larger-than-initial γ is shown in Figure 4C (green curve). Opposite to the relatively narrow distribution observed in lynx1-nAChR complex systems, much wider two-peak distributions are shown for apo-nAChR systems (Figure 4D). Without the presence of lynx1, α41 C-loop shows a larger γ fluctuation range mainly with two peaks around 50° and 100°, representing close and open states, respectively. It is clear that due to the interaction with lynx1, α41 C-loop tends to be restricted in either close or open state, and a state transition is harder to be observed than in apo-nAChR. Movement of the C-loop over the ligand to the closed position is thought to transmit important information to the pore lining region of the channel, resulting in channel widening and allowing ion flow. MD simulations of the a7 nAChR alone indicates that a relatively low energetic barrier separates the two C-loop conformations 65. Interactions of lynx1 with an nAChR subdomain (e.g. the C-loop) could provide substantial restrictions on these transitions. This has been supported by our MD results as less transition between the open and closed states are observed in lynx1-nAChR complex than apo-nAChR systems (Figure 4C,D). Although our models involve the C-loop at α4:α4 interfaces, a presumptive non-agonist binding site, presumably interactions with any of the C-loops of the pentamer can impede the tilt required of the pore-lining barrels needed for the enlargement of the pore diameter 66.
Interactions with nAChR Reduce Lynx1’s Activity Dynamics
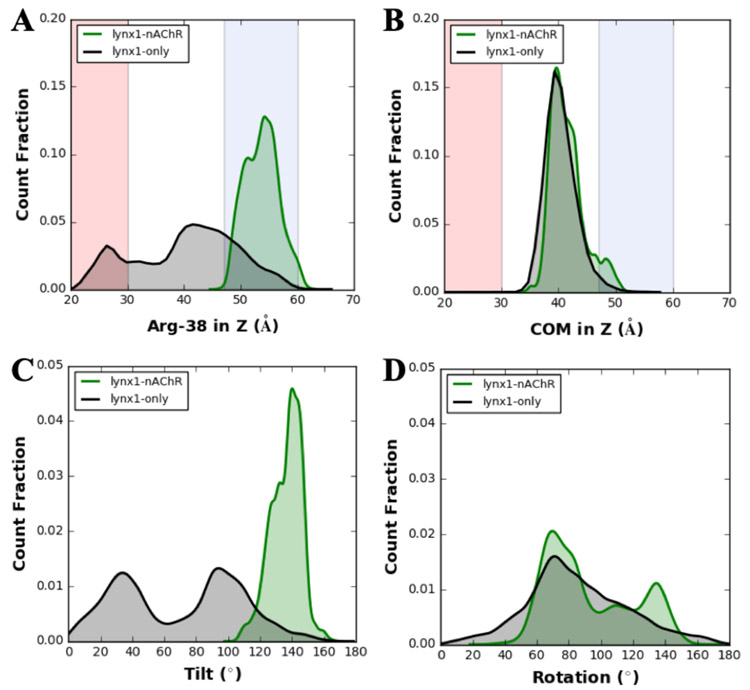
To gain a better understanding of the influence of lynx1-nAChR interactions on lynx1 structure, dynamics, and orientation (with respect to the membrane), we also performed the simulations of the lynx1-only systems. Figure 5A represents the density distributions of the center of mass (COM) of lynx1 Arg38 along the Z axis in either the lynx1-only (black curve) or lynx1-nAChR complex (green curve) system. In lynx1-only, Arg38 fluctuates within a wide Z-coordinate range (25-60 Å) with the possibility of falling into the nAChR interaction range (47-60 Å) and the lipid region (<30 Å). Therefore, some intermittent lynx1-lipids interactions were observed in the lynx1-only system when Arg38’s Z-coordinate is less than 30 Å, which did not happen in the lynx1-nAChR system. It is important to note that because the lynx1-only system mimics the unbound state, the estimation of lynx1-nAChR interactions was extrapolated from the Z-coordinate distribution of the nAChR C-loop of each subunit. Thus, the possibility discussed here is a necessary condition for the actual interaction. On the other hand, Arg38 shows a relatively small Z-coordinate fluctuation (50-60 Å) in the lynx1-nAChR complex system, which manipulates the multi-mode interactions between lynx1 and nAChR C-loop. As for the COM of the entire lynx1 protein backbone (Figure 5B), there is no significant difference observed in these two systems because lynx1 is GPI-anchored to the membrane bilayers. In other words, lynx1 can change its orientation relative to the membrane, but its Z-COM limits are bound.
Figure 5.
(A) Density distributions of lynx1 Arg38 and (B) lynx1 protein center of mass in the Z axis. The background shading is based on the averaged regions of lipids (red), water (white), and tentative nAChR interaction sites (blue) as the nAChR C-loop range in Figure 1. (C and D) Density distributions of (C) tilt and (D) rotation angles of lynx1 (defined in Figure 1). Note that green curves represent the lynx1-nAChR system and black for the lynx1-only system. Each distribution was calculated using all production trajectories of three replicas.
In terms of the tilt/rotation angle, as shown in Figs. 5C and 5D, there is a similar trend, i.e., the tilt/rotation angles show a high fluctuation range in the lynx1-only system and a relatively small one in the lynx1-nAChR complex system. From the schematic shown in Figure 1E, the greater the upward movement of Arg38, the larger tilt angle was observed. Thus, Figure 5C shows a very similar profile as those in Figure 5A. In the lynx1-only system, the tilt angle fluctuates all the way between 0-160° range, with 0° being when Arg38 lies within the lipid area, and 160° referring to the widest possible nAChR interaction range from the lipid. In contrast, in the lynx1-nAChR complex system, the tilt angle exhibits smaller fluctuations (120°-l50°). This is mainly due to the lynx1-nAChR interactions that were elaborated previously. With respect to the rotation angle, lynx1 fully rotates around the protein’s principal axis. Even with Arg38 at the α41:α42 interface and the C-terminal position fixed by the GPI-anchor, there is little limitation for rotating lynx1 as compared to the relatively restricted freedom of tilting. Thus, there is still a relatively large rotation angle change of lynx1 even in the lynx1-nAChR complex system.
We also employed the root mean square deviation (RMSD) and root mean square fluctuation (RMSF) to investigate the structure and dynamics changes of lynx1 in unbound/bound states (Figure S1). In terms of RMSD, both unbound/bound states show similar lyxn1 structure behaviors. In terms of RMSF, residues of β-strands (yellow shaded regions) show a lower fluctuation than other sections of the protein. In bound state, lynx1 has lower fluctuations especially for residues of turn II (residue number: 35-38) and the disordered region (residue number: 53-61). These two regions are located near nAChR and contain the reported interaction residue (Arg38) that is highly involved in the lynx1-nAChR interactions. However, in the back-side regions that are far away from nAChR, there is not much difference observed.
Generally speaking, interactions with lynx1 appear to restrict the conformational change of the C-loop that influences the nAChR function, and on the other hand, these interactions also lock the lynx1 protein (especially the residues facing nAChR) within the interaction region. These reciprocal interactions can only be observed when lynx1 and nAChRs are physically located closely, though it remains to be determined how frequently this happens on the neuron in situ. It is also reasonable to believe that once the interactions form, the influence on either nAChR or lynx1 might last for a significantly long time. We expect the GPI anchor on lynx1 to decrease the search time for nAChR on the membrane surface, by reducing the degree of freedom of lynx1 to the membrane plane. Similarly, this may also prolong the lynx1-nAChR interaction. Note that we only show the influence of lynx1 protein on (α4)3(β2)2 stoichiometry of nAChR in this work, it is possible that lynx1 protein may also have influences on other subtypes of nAChRs (i.e., α7, α3*) that need further investigation.
Conclusions
In this study, we elucidate the interactions between nAChR and lynx1 that either modulate the dynamics fluctuation of lynx1 protein or influence the open/close state change of the C-loop in one of the nAChR subunits that could influence its desensitization related to functional expression. The lynx1-only system was built to represent the unbounded state of lynx1, while the lynx1-nAChR complex systems mimic the bound state. The restrictive influence on lynx1 was inferred from dynamics analysis of systems when it is in bound vs unbound state. When unbound, lynx1 protein (especially Arg38) showed a high dynamic activity. The center of mass along the Z direction of Arg38 show 3 times wider range of fluctuation than in the bound state (30 Å vs. 10 Å). Unbounded lynx1 protein tilt and rotation angles show 5 (160° vs. 30°) and 1.3 (180° vs. 140°) times larger fluctuations, respectively, compared to the bound one. Arg38 intermittently interacts with different lipids when unbound while there are no lipid interactions in the bound one. Lynx1 belongs to the three-finger protein family that has relatively flexible loops. When bound, this loop flexibility would easily be influenced by interactions. The turn and disordered sections in the loops of lynx1 show decreased RMSF values (2 Å) when facing the nAChR. The flexibility of these loose sections is restricted by association with nAChR and which would be extended during long-term interactions.
The simulations described here also provide some insights into how these interactions could influence the activity of the nAChR. In the three replicas performed in this study of the bound state, there were significant and stable interactions between lynx1 (Arg38) and the receptor C-loop. The biological hypothesis of the lynx1 protein as an allosteric modulator on nAChR function with multimodal actions on nAChR function by reducing agonist sensitivity, accelerating the rate of desensitization, and prolonging the recovery from desensitization of nAChRs 23,67. Results presented in this work support these hypotheses. The interactions observed between lynx1 and nAChR show some non-deterministic influences on the C-loop behavior of α41 subunit – preventing a closed C-loop from reopening, maintaining the intermediate state between open and closed, or maintaining the C-loop open state – depending on the position of lynx1 protein. Due to these interactions, nAChR would have a higher barrier between the open to closed transition, thus potentially inhibiting agonist sensitivity or inhibiting the recovery from desensitization.
Supplementary Material
Acknowledgments
Founding Sources
This work was supported in part by grants from NIH R01 DA043567, NSF DBI-1707207, and XSEDE MCB070009.
Footnotes
Supporting Information
Time-series of root mean square deviation (RMSD) and root mean square fluctuation (RMSF) of lynx1 among three individual replicas in lynx1-only system and lynx1-nAChR complex system.
Competing interests: The authors declare no competing interests, except JMM is a founder of Ophidion, Inc, a biotechnology company
References
- (1).Picciotto MR Nicotine as a Modulator of Behavior: Beyond the Inverted U. Trends Pharmacol. Sci 2003, 24 (9), 493–499. 10.1016/S0165-6147(03)00230-X. [DOI] [PubMed] [Google Scholar]
- (2).Zoli MRP and Neuroprotection via NAChRs M: The Role of NAChRs in Neurodegenerative Disorders Such as Alzheimer’s and Parkinson’s Disease. Front. Biosci 2008, 13 (4), 492–504. [DOI] [PubMed] [Google Scholar]
- (3).Papke RL; Bagdas D; Kulkarni AR; Gould T; Alsharari SD; Thakur GA; Damaj MI The Analgesic-like Properties of the Alpha7 NAChR Silent Agonist NS6740 Is Associated with Non-Conducting Conformations of the Receptor. Neuropharmacology 2015, 91, 34–42. 10.1016/j.neuropharm.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Freitas K; Carroll FI; Damaj MI The Antinociceptive Effects of Nicotinic Receptors A7-Positive Allosteric Modulators in Murine Acute and Tonic Pain Models. J. Pharmacol. Exp. Ther 2013, 344 (1), 264–275. 10.1124/jpet.112.197871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Freitas K; Ghosh S; Ivy Carroll F; Lichtman AH; Imad Damaj M Effects of Alpha 7 Positive Allosteric Modulators in Murine Inflammatory and Chronic Neuropathic Pain Models. Neuropharmacology 2013, 65, 156–164. 10.1016/j.neuropharm.2012.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Hone AJ; Servent D; McIntosh JM A9-Containing Nicotinic Acetylcholine Receptors and the Modulation of Pain. Br. J. Pharmacol 2018, 175 (11), 1915–1927. 10.1111/bph.13931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Bitner RS; Nikkel AL; Curzon P; Donnelly-Roberts DL; Puttfarcken PS; Namovic M; Jacobs IC; Meyer MD; Decker MW Reduced Nicotinic Receptor-Mediated Antinociception Following in Vivo Antisense Knock-down in Rat. Brain Res. 2000, 871 (1), 66–74. 10.1016/S0006-8993(00)02442-2. [DOI] [PubMed] [Google Scholar]
- (8).AlSharari SD; Carroll FI; McIntosh JM; Damaj MI The Antinociceptive Effects of Nicotinic Partial Agonists Varenicline and Sazetidine-A in Murine Acute and Tonic Pain Models. J. Pharmacol. Exp. Ther 2012, 342 (3), 742–749. 10.1124/jpet.112.194506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Cucchiaro G; Chaijale N; Commons KG The Dorsal Raphe Nucleus as a Site of Action of the Antinociceptive and Behavioral Effects of the Alpha4 Nicotinic Receptor Agonist Epibatidine. Neuropharmacology 2006, 50 (7), 769–776. 10.1016/j.neuropharm.2005.11.026. [DOI] [PubMed] [Google Scholar]
- (10).Marubio LM; Arroyo-Jimenez MDM; Cordero-Erausquin M; Léna C; Le Novère N; De Kerchove d’Exaerde A; Huchet M; Damaj MI; Changeux JP Reduced Antinociception in Mice Lacking Neuronal Nicotinic Receptor Subunits. Nature 1999, 398 (6730), 805–810. 10.1038/19756. [DOI] [PubMed] [Google Scholar]
- (11).Damaj MI; Fonck C; Marks MJ; Deshpande P; Labarca C; Lester HA; Collins AC; Martin BR Genetic Approaches Identify Differential Roles for α 4β2 Nicotinic Receptors in Acute Models of Antinociception in Mice. J. Pharmacol. Exp. Ther 2007, 321 (3), 1161–1169. 10.1124/jpet.106.112649. [DOI] [PubMed] [Google Scholar]
- (12).Dani JA Neuronal Nicotinic Acetylcholine Receptor Structure and Function and Response to Nicotine. Int. Rev. Neurobiol 2016, 124, 3–19. 10.1016/bs.irn.2015.07.001.Neuronal. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Auerbach A Agonist Activation of a Nicotinic Acetylcholine Receptor. Neuropharmacology 2015, 96 (PB), 150–156. 10.1016/j.neuropharm.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Purohit P; Auerbach A Loop C and the Mechanism of Acetylcholine Receptor-Channel Gating. J. Gen. Physiol 2013, 141 (4), 467–478. 10.1085/jgp.201210946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Srinivasan R; Henderson BJ; Lester HA; Richards CI Pharmacological Chaperoning of NAChRs: A Therapeutic Target for Parkinson’s Disease. Pharmacol Res. 2014, No. 83, 20–29. 10.1016/j.phrs.2014.02.005.Pharmacological. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Wu J; Lukas RJ Naturally-Expressed Nicotinic Acetylcholine Receptor Subtypes. Biochem. Pharmacol. 2011, 82 (8), 800–807. 10.1016/j.bcp.2011.07.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Ochoa V; George AA; Nishi R; Whiteaker P The Prototoxin LYPD6B Modulates Heteromeric A3β4-Containing Nicotinic Acetylcholine Receptors, but Not A7 Homomers. FASEB J. 2016, 30 (3), 1109–1119. 10.1096/fj.15-274548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Lukas RJ; Changeux JP; Le Novère N; Albuquerque EX; Balfour DJK; Berg DK; Bertrand D; Chiappinelli VA; Clarke PBS; Collins AC; et al. International Union of Pharmacology. XX. Current Status of the Nomenclature for Nicotinic Acetylcholine Receptors and Their Subunits. Pharmacol. Rev 1999, 51 (2), 397–401. [PubMed] [Google Scholar]
- (19).Lyukmanova EN; Shenkarev ZO; Shulepko MA; Mineev KS; D’Hoedt D; Kasheverov IE; Filkin SY; Krivolapova AP; Janickova H; Dolezal V; et al. NMR Structure and Action on Nicotinic Acetylcholine Receptors of Water-Soluble Domain of Human LYNX1. J. Biol. Chem 2011, 286 (12), 10618–10627. 10.1074/jbc.M110.189100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Puddifoot CA; Wu M; Sung RJ; Joiner WJ Ly6h Regulates Trafficking of Alpha7 Nicotinic Acetylcholine Receptors and Nicotine-Induced Potentiation of Glutamatergic Signaling. J. Neurosci 2015, 35 (8), 3420–3430. 10.1523/JNEUROSCI.3630-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).George AA; Bloy A; Miwa JM; Lindstrom JM; Lukas RJ; Whiteaker P Isoform-Specific Mechanisms of A3β4*-Nicotinic Acetylcholine Receptor Modulation by the Prototoxin Lynx1. FASEB J. 2017, 31 (4), 1398–1420. 10.1096/fj.201600733R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Miwa JM; Ibaňez-Tallon I; Crabtree GW; Sánchez R; Šali A; Role LW; Heintz N Lynx1, an Endogenous Toxin-like Modulator of Nicotinic Acetylcholine Receptors in the Mammalian CNS. Neuron 1999, 23 (1), 105–114. 10.1016/S0896-6273(00)80757-6. [DOI] [PubMed] [Google Scholar]
- (23).Ibañez-Tallon I; Miwa JM; Wang HL; Adams NC; Crabtree GW; Sine SM; Heintz N Novel Modulation of Neuronal Nicotinic Acetylcholine Receptors by Association with the Endogenous Prototoxin Lynx1. Neuron 2002, 33 (6), 893–903. 10.1016/S0896-6273(02)00632-3. [DOI] [PubMed] [Google Scholar]
- (24).Nichols WA; Henderson BJ; Yu C; Parker RL; Richards CI; Lester HA; Miwa JM Lynx1 Shifts A4β2 Nicotinic Receptor Subunit Stoichiometry by Affecting Assembly in the Endoplasmic Reticulum. J. Biol. Chem 2014, 289 (45), 31423–31432. 10.1074/jbc.M114.573667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Tsetlin VI Three-Finger Snake Neurotoxins and Ly6 Proteins Targeting Nicotinic Acetylcholine Receptors: Pharmacological Tools and Endogenous Modulators. Trends Pharmacol. Sci 2015, 36 (2), 109–123. 10.1016/j.tips.2014.11.003. [DOI] [PubMed] [Google Scholar]
- (26).Nissen NI; Anderson KR; Wang H; Lee HS; Garrison C; Eichelberger SA; Ackerman K; Im W; Miwa JM Augmenting the Antinociceptive Effects of Nicotinic Acetylcholine Receptor Activity through Lynx1 Modulation. PLoS One 2018, 13 (7), 1–23. 10.1371/journal.pone.0199643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Miwa JM; Walz A Enhancement in Motor Learning through Genetic Manipulation of the Lynx1 Gene. PLoS One 2012, 7 (11). 10.1371/journal.pone.0043302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Lyukmanova EN; Shulepko MA; Buldakova SL; Kasheverov IE; Shenkarev ZO; Reshetnikov RV; Filkin SY; Kudryavtsev DS; Ojomoko LO; Kryukova EV; et al. Water-Soluble LYNX1 Residues Important for Interaction with Muscle-Type and/or Neuronal Nicotinic Receptors. J. Biol. Chem 2013, 288 (22), 15888–15899. 10.1074/jbc.M112.436576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Le Novere N; Changeux JP Molecular Evolution of the Nicotinic Acetylcholine Receptor: An Example of Multigene Family in Excitable Cells. J. Mol. Evol 1995, 40 (2), 155–172. 10.1007/BF00167110. [DOI] [PubMed] [Google Scholar]
- (30).Gotti C; Clementi F Neuronal Nicotinic Receptors: From Structure to Pathology. Prog. Neurobiol 2004, 74 (6), 363–396. 10.1016/j.pneurobio.2004.09.006. [DOI] [PubMed] [Google Scholar]
- (31).Hurst R; Rollema H; Bertrand D Nicotinic Acetylcholine Receptors: From Basic Science to Therapeutics. Pharmacol. Ther 2013, 137 (1), 22–54. 10.1016/j.pharmthera.2012.08.012. [DOI] [PubMed] [Google Scholar]
- (32).Walsh RM; Roh SH; Gharpure A; Morales-Perez CL; Teng J; Hibbs RE Structural Principles of Distinct Assemblies of the Human A4β2 Nicotinic Receptor. Nature 2018, 557 (7704), 261–265. 10.1038/s41586-018-0081-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Phillips JC; Braun R; Wang W; Gumbart J; Tajkhorshid E; Villa E; Chipot C; Skeel RD; Kalé L; Schulten K Scalable Molecular Dynamics with NAMD. J. Comput. Chem 2005, 26 (16), 1781–1802. 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Huang J; Rauscher S; Nawrocki G; Ran T; Feig M; De Groot BL; Grubmüller H; MacKerell AD CHARMM36m: An Improved Force Field for Folded and Intrinsically Disordered Proteins. Nat. Methods 2016, 14 (1), 71–73. 10.1038/nmeth.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Guvench Olgun, Greene Shannon N., Kamath Ganesh, Brady John W., Venable Richard M., Pastor Richard W., J. ADM Additive Empirical Force Field for Hexopyranose Monosaccharides. J. Comput. Chem 2008, 29 (15), 2543–2564. 10.1002/jcc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Guvench† Olgun, Hatcher† Elizabeth, Venable‡ Richard M., P. RW and J. ADM CHARMM Additive All-Atom Force Field for Glycosidic Linkages between Hexopyranoses. J. Phys. Chem. B 2009, 5 (9), 2353–2370. 10.1021/jp105758h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Hatcher E; Guvench O; MacKerell AD Charmm Additive All-Atom Force Field for Aldopentofuranoses, Methyl-Aldopentofuranosides, and Fructofuranose. J. Phys. Chem. B 2009, 113 (37), 12466–12476. 10.1021/jp905496e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Klauda JB; Venable RM; Freites JA; O’Connor JW; Tobias DJ; Mondragon-Ramirez C; Vorobyov I; MacKerell AD; Pastor RW Update of the CHARMM All-Atom Additive Force Field for Lipids: Validation on Six Lipid Types. J. Phys. Chem. B 2010, 114 (23), 7830–7843. 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Klauda JB; Monje V; Kim T; Im W Improving the CHARMM Force Field for Polyunsaturated Fatty Acid Chains. J. Phys. Chem. B 2012, 116 (31), 9424–9431. 10.1021/jp304056p. [DOI] [PubMed] [Google Scholar]
- (40).Abola Enrique E., Bernstein Frances C., T. F. K The Protein Data Bank In Neutrons in Biology; Springer: Boston,MA, 1984; p 441. [Google Scholar]
- (41).Jorgensen WL; Chandrasekhar J; Madura JD; Impey RW; Klein ML Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys 1983, 79 (2), 926–935. 10.1063/1.445869. [DOI] [Google Scholar]
- (42).Cotman C; Blank ML; Moehl A; Snyder F Lipid Composition of Synaptic Plasma Membranes Isolated from Rat Brain by Zonal Centrifugation. Biochemistry 1969, 8 (11), 4606–4612. 10.1021/bi00839a056. [DOI] [PubMed] [Google Scholar]
- (43).Brown DA; London E Functions of Lipid Rafts in Biological Membranes. Annu. Rev. Cell Dev. Biol 1998, 14 (1), 111–136. 10.1146/annurev.cellbio.14.1.111. [DOI] [PubMed] [Google Scholar]
- (44).Rauch S; Fackler OT Lipid Rafts and Signal Transduction. Signal Transduct. 2007, 7 (1), 53–63. 10.1002/sita.200600113. [DOI] [Google Scholar]
- (45).Helms JB; Zurzolo C Lipids as Targeting Signals: Lipid Rafts and Intracellular Trafficking. Traffic 2004, 5 (4), 247–254. 10.1111/j.1600-0854.2004.0181.x. [DOI] [PubMed] [Google Scholar]
- (46).Sunshine C; McNamee MG Lipid Modulation of Nicotinic Acetylcholine Receptor Function: The Role of Membrane Lipid Composition and Fluidity. Biochim. Biophys. Acta - Biomembr 1994, 1191 (1), 59–64. 10.1016/0005-2736(94)90233-X. [DOI] [PubMed] [Google Scholar]
- (47).Jo S; Lim JB; Klauda JB; Im W CHARMM-GUI Membrane Builder for Mixed Bilayers and Its Application to Yeast Membranes. Biophys. J 2009, 97 (1), 50–58. 10.1016/j.bpj.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Wu EL; Cheng X; Jo S; Rui H; Song KC; Dávila-Contreras EM; Qi Y; Lee J; Monje-Galvan V; Venable RM; et al. CHARMM-GUI Membrane Builder toward Realistic Biological Membrane Simulations. J. Comput. Chem 2014, 35 (27), 1997–2004. 10.1002/jcc.23702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Lee J; Patel DS; Ståhle J; Park SJ; Kern NR; Kim S; Lee J; Cheng X; Valvano MA; Holst O; et al. CHARMM-GUI Membrane Builder for Complex Biological Membrane Simulations with Glycolipids and Lipoglycans. J. Chem. Theory Comput 2019, 15 (1), 775–786. 10.1021/acs.jctc.8b01066. [DOI] [PubMed] [Google Scholar]
- (50).Jo S; Song KC; Desaire H; MacKerell AD; Im W Glycan Reader: Automated Sugar Identification and Simulation Preparation for Carbohydrates and Glycoproteins. J. Comput. Chem 2011, 32 (14), 3135–3141. 10.1002/jcc.21886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Jo Sunhwan Taehoon Kim Iyer Vidyashankara G. Im Wonpil. CHARMM-GUI: A Web-Based Graphical UserInterface for CHARMM. J. Comput. Chem 2008, 29 (11), 1859–1865. 10.1002/jcc. [DOI] [PubMed] [Google Scholar]
- (52).Lee J; Cheng X; Swails JM; Yeom MS; Eastman PK; Lemkul JA; Wei S; Buckner J; Jeong JC; Qi Y; et al. CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field. J. Chem. Theory Comput 2016, 12 (1), 405–413. 10.1021/acs.jctc.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Humphrey W; Dalke A; Schulten K VMD: Visual Molecular Dynamics. J. Mol. Graph 1996, 14 (1), 33–38. 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- (54).DeLano WL . PyMOL: An Open-Source Molecular Graphics Tool. Ccp4.Ac.Uk 2002, 1 (40), 82–92. [Google Scholar]
- (55).Brooks BR; Bruccoleri RE; Olafson BD; States DJ; Swaminathan S; Karplus M CHARMM: A Program for Macromolecular Energy, Minimization, and Dynamics Calculations. J. Comput. Chem 1983, 4 (2), 187–217. 10.1002/jcc.540040211. [DOI] [Google Scholar]
- (56).Dion M; Rydberg H; Schröder E; Langreth DC; Lundqvist BI Van Der Waals Density Functional for General Geometries. Phys. Rev. Lett 2004, 92 (24), 22–25. 10.1103/PhysRevLett.92.246401. [DOI] [PubMed] [Google Scholar]
- (57).York DM; Darden TA; Pedersen LG The Effect of Long-Range Electrostatic Interactions in Simulations of Macromolecular Crystals: A Comparison of the Ewald and Truncated List Methods. J. Chem. Phys 1993, 99 (10), 8345–8348. 10.1063/1.465608. [DOI] [Google Scholar]
- (58).Ryckaert JP; Ciccotti G; Berendsen HJC Numerical Integration of the Cartesian Equations of Motion of a System with Constraints: Molecular Dynamics of n-Alkanes. J. Comput. Phys 1977, 23 (3), 327–341. 10.1016/0021-9991(77)90098-5. [DOI] [Google Scholar]
- (59).Papanagnou P; Stivarou T; Tsironi M Unexploited Antineoplastic Effects of Commercially Available Anti-Diabetic Drugs. Pharmaceuticals 2016, 9 (2), 1695–1697. 10.3390/ph9020024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Nosé S; Klein ML A Study of Solid and Liquid Carbon Tetrafluoride Using the Constant Pressure Molecular Dynamics Technique. J. Chem. Phys 1983, 78 (11), 6928–6939. 10.1063/1.444641. [DOI] [Google Scholar]
- (61).Andersen HC Molecular Dynamics Simulations at Constant Pressure and/or Temperature. J. Chem. Phys 1980, 72 (4), 2384–2393. 10.1063/1.439486. [DOI] [Google Scholar]
- (62).Eastman P; Friedrichs MS; Chodera JD; Radmer RJ; Bruns CM; Ku JP; Beauchamp KA; Lane TJ; Wang LP; Shukla D; et al. OpenMM 4: A Reusable, Extensible, Hardware Independent Library for High Performance Molecular Simulation. J. Chem. Theory Comput 2013, 9 (1), 461–469. 10.1021/ct300857j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Yakel JL Nicotinic ACh Receptors in the Hippocampal Circuit; Functional Expression and Role in Synaptic Plasticity. J. Physiol 2014, 592 (19), 4147–4153. 10.1113/jphysiol.2014.273896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Zhang TA; Tang J; Pidoplichko VI; Dani JA Addictive Nicotine Alters Local Circuit Inhibition during the Induction of in Vivo Hippocampal Synaptic Potentiation. J. Neurosci 2010, 30 (18), 6443–6453. 10.1523/JNEUROSCI.0458-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Cheng X; Wang H; Grant B; Sine SM; McCammon JA Targeted Molecular Dynamics Study of C-Loop Closure and Channel Gating in Nicotinic Receptors. PLoS Comput. Biol 2006, 2 (9), 1173–1184. 10.1371/journal.pcbi.0020134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Wang J; Kuryatov A; Sriram A; Jin Z; Kamenecka TM; Kenny PJ; Lindstrom J An Accessory Agonist Binding Site Promotes Activation of A4β2* Nicotinic Acetylcholine Receptors. J. Biol. Chem 2015, 290 (22), 13907–13918. 10.1074/jbc.M115.646786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Miwa JM; Stevens TR; King SL; Caldarone BJ; Ibanez-Tallon I; Xiao C; Fitzsimonds RM; Pavlides C; Lester HA; Picciotto MR; et al. The Prototoxin Lynx1 Acts on Nicotinic Acetylcholine Receptors to Balance Neuronal Activity and Survival In Vivo. Neuron 2006, 51 (5), 587–600. 10.1016/j.neuron.2006.07.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.