Summary
POLR3B encodes the second-largest catalytic subunit of RNA polymerase III, an enzyme involved in transcription. Bi-allelic pathogenic variants in POLR3B are a well-established cause of hypomyelinating leukodystrophy. We describe six unrelated individuals with de novo missense variants in POLR3B and a clinical presentation substantially different from POLR3-related leukodystrophy. These individuals had afferent ataxia, spasticity, variable intellectual disability and epilepsy, and predominantly demyelinating sensory motor peripheral neuropathy. Protein modeling and proteomic analysis revealed a distinct mechanism of pathogenicity; the de novo POLR3B variants caused aberrant association of individual enzyme subunits rather than affecting overall enzyme assembly or stability. We expand the spectrum of disorders associated with pathogenic variants in POLR3B to include a de novo heterozygous POLR3B-related disorder.
Keywords: ataxia, spasticity, neuropathy, intellectual disability, POLR3B, RNA polymerase III assembly
Main text
RNA polymerase III (pol III) is a 17-subunit enzyme involved in the transcription of small non-coding RNAs, which regulate transcription in eukaryotes.1 POLR3B (MIM: 614366) encodes the second-largest catalytic subunit of pol III, POLR3B. Bi-allelic mutations in POLR3B and genes encoding other POLR3 subunits cause hypomyelinating leukodystrophy type 8 (MIM: 614381) with a wide clinical spectrum of associated symptoms. These include endocrine dysfunction (hypogonadism, reproductive failure, and delayed puberty), ocular abnormalities, and abnormal dentition (hypodontia and oligodontia).1,2
The original descriptions of POLR3-related leukodystrophy were of individuals with hypodontia, ataxia, and hypomyelination,3,4 followed by individuals with additional endocrine abnormalities of hypogonadotropic hypogonadism.2 The initial identification of the genetic basis of POLR3-related leukodystrophy enabled definitive association of this constellation of clinical features with recessive mutations in POLR3A (MIM: 614258), POLR3B, POLR1C (MIM: 610060), and POLR3K (MIM: 606007).1,5, 6, 7, 8, 9 In total, five previously distinct clinical phenotypes were described prior to the identification of their shared molecular basis. These include hypomyelination, hypodontia, hypogonadotropic hypogonadism (4H syndrome); ataxia, delayed dentition, and hypomyelination (ADDH); tremor-ataxia with central hypomyelination (TACH); leukodystrophy with oligodontia (LO); and hypomyelination with cerebellar atrophy and hypoplasia of the corpus callosum (HCAHC). These previously distinct clinical phenotypes were subsequently re-classified as 4H leukodystrophy after the discovery of the shared phenotypic spectrum and genetics10 and are now collectively referred to as POLR3-related leukodystrophy.
The classic neurological features of individuals with POLR3B-related leukodystrophy consist primarily of central nervous system involvement, and abnormal electromyogram and nerve conduction studies (EMG/NCSs) have not been reported in these individuals.1,2 To our knowledge, no prior publications have reported sensory or peripheral motor abnormalities of clinical significance in individuals with bi-allelic POLR3B mutations.
Six unrelated individuals were enrolled in the study, which was approved by the research ethics board of the Hospital for Sick Children (REB #1000009004) and the McGill University Health Center (11-105-PED and 2019-4972). Five individuals (subjects 1–4 and 6) were found to have pathogenic variants in POLR3B from exome sequencing completed at GeneDx (Gaithersburg, MD). One additional individual (subject 5) was diagnosed at the University Medical Center Utrecht. All parents/legal guardians provided written informed consent for their children to participate in the study and for publication of clinical information.
We identified six unrelated individuals, from non-consanguineous families, who harbor de novo heterozygous variants in POLR3B (Figure S1) with consistent clinical features that differ from those associated with previously reported bi-allelic mutations1,2 (Table 1).
Table 1.
Clinical and genetic characteristics of individuals with de novo POLR3B variants
| Subject | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| POLR3B variant (NM_018082.5) | c.1124A>T (de novo) | c.1277T>C (de novo) | c.3137G>A (de novo) | c.1094C>T (de novo) | c.1087G>A (de novo) | c. 1385C>G (de novo) |
| Predicted effect on protein | p.Asp375Val | p. Leu426Ser | p. Arg1046His | p.Ala365Val | p.Glu363Lys | p.Thr462Arg |
| Age at last review | 16 years | 4 years | 11 years | 22 years | 8 years | 14 years |
| Gender | female | male | male | male | female | female |
| Current height (centile) | 166 cm (50%) | 96 cm (1%) | 152 cm (95%) | 165 cm (5%) | 116 cm (10%) | 157 cm (30%) |
| Current weight (centile) | 74.3 kg (85%) | 13.5 kg (1%) | 51.9 kg (95%) | 95.45 kg (95%) | 19.9 kg (20%) | 55.4 kg (63%) |
| Current OFC (centile) | 51 cm (<3%) | 48 cm (5%) | 55 cm (85%) | 58 cm (~98%) | 47 cm (~3%) | 51 cm (<2%) |
| Age achieved walking | 3.5 years | 22 months | 16 months | 18 months | 2.5 years | 16 months |
| Global developmental delay/intellectual disability | severe; on formal testing verbal reasoning and adaptive skills <1% (ABAS-II, Leiter-R, PPVT-4) | language DQ 51%, visual motor DQ 51% | normal intellectual functioning; grade-level academic performance | IQ at age 21 years: FSIQ 46 (moderate ID) | IQ at age 7 years: £55 (mild to moderate ID) | severe; reading, writing, and adaptive function at kindergarten level at age 14 |
| Communication/speech | first words at 5 years, sentences at 7 years, mainly uses single words and short phrases at 16 years, dysarthria | first words at 2 years, 2–3-word phrases, dysarthria | normal speech | first word at 13 months, put 2 words together at 2.5 years, dysarthria | language development initially normal, can use simple sentences, dysarthria | significant early delay, phrases at 3 years but limited vocabulary, now short phrases, dysarthria |
| Gross motor | ambulatory, spastic gait, ataxia, occasional dystonic posturing | ambulatory, in-toeing L > R | mild gross motor delay as an infant | ambulatory, ataxia, falls monthly, spasticity and increasing unsteadiness | ambulatory, spastic gait | gross motor delay, ambulatory, spastic gait, mild ataxia |
| Fine motor | needs assistance to feed herself, dress, and bathe, can print her name | feeds self with spoon and fork | normal | can use utensils but prefers finger feeding, does not color within lines, needs assistance with most ADLs | needs assistance with all activities of daily living | cuts own food but needs help with buttons and zippers, feeds self, needs assistance to bathe and toilet |
| Appendicular hypertonia/spasticity | severe, progressive | none | none | severe, progressive, LEs≫UEs | clearly present, progressive | moderate, all 4 limbs, LE > UE |
| Deep tendon reflexes | 4+ with sustained ankle clonus, +Hoffman sign | normal and symmetric | 1+ in biceps, triceps, absent in patella and ankles bilaterally | 3+ throughout, except 1+ at the ankles | arms, 3+; legs, 4+ (crossed adductor reflex, sustained ankle clonus) | 3+ bilaterally |
| Extensor plantar responses | yes | no | no | yes | yes | yes |
| Muscle weakness | MRC grade 5/5 UEs, tibialis anterior and extensor hallucis longus 4/5 | normal strength | prominent distal leg weakness | not tested formally but grossly normal, no asymmetry | not tested formally, no obvious weakness | MRC grade 5/5 throughout |
| Sensory examination | not able to cooperate with formal exam | grossly normal | decreased vibration and proprioception in distal limbs | uncooperative for detailed exam, hypersensitivity in the feet bilaterally | detailed sensory exam not possible, light touch preserved | unable to cooperate with formal exam |
| Cerebellar testing | mirror movements, intention tremor, mild dysmetria, gait ataxia, unable to perform tandem gait | normal for age | slightly wide-based gait | slow finger to nose movements, clearly ataxic gait with bilateral circumduction, unable to perform tandem gait or stance | mild intention tremor and dysmetria, unable to balance on one foot or perform tandem gait | no overt appendicular ataxia but mild gait ataxia and unable to perform tandem gait |
| Seizures | no | onset 3 years, head and bilateral arm drop attacks, also generalized seizures | no | onset 6 months, refractory generalized epilepsy, myoclonic, rarely atonic and absence | onset 12 months, head drop attacks, refractory to medications and ketogenic diet | no |
| Brain MRI | mild cerebellar atrophy, non-specific T2 and FLAIR hyperintensities | normal | normal (except for Chiari I malformation) | non-specific white matter signal abnormalities, stable over time | normal (age 5 years) | initially delayed myelination that normalized by 14 years |
| EMG/NCS (age when performed) | length-dependent primary axonal neuropathy with secondary demyelination, markedly decreased velocities in motor fibers of tibial and deep peroneal nerves (13 years) | normal (4 years) | severe demyelinating sensory motor polyneuropathy, normal amplitudes, mild-moderate prolonged distal latencies, significant slowing of conduction velocities (10 years) | predominantly demyelinating sensory motor neuropathy, slow motor conduction velocities, markedly decreased amplitudes in tibial nerve, absent sural responses (12 years) | combined axonal and demyelinating polyneuropathy of both motor and sensory nerves (7 years) | predominantly demyelinating polyneuropathy with slow conduction velocities (6 and 8 years) |
Abbreviations are as follows: OFC, occipitofrontal circumference; DQ, development quotient; ID, intellectual disability; L, left; R, right; ADLs, activities of daily living; LE, lower extremity; UE, upper extremity; MRC, Medical Research Council
The six participants each had some degree of gait dysfunction ranging from mild instability to more severe gait ataxia. Five participants (subjects 1, 3, 4, 5, and 6) had truncal and/or appendicular ataxia with wide-based ataxic gait, inability to perform tandem gait, or gait instability (Table 1, Supplemental notes). Three participants had hyperreflexia, while one had normal deep tendon reflexes, one had diminished reflexes, and one had a mixed picture of proximal hyperreflexia with distal hyporeflexia.
The majority of individuals also had some degree of delay in other developmental domains; intellectual disability ranging from mild to moderate severity was diagnosed in 5/6 participants. Only one individual (subject 3) had normal academic performance and development apart from early mild motor delay. This individual also had normal language, while the other 5/6 participants had dysarthria and/or varying degrees of delayed early speech development ranging from normal to markedly delayed (the most severely affected, subject 1, achieved first words at 5 years of age). Four individuals (subjects 1, 4, 5, and 6) required assistance with basic activities of daily living, suggestive of significant degrees of intellectual disability and/or coordination and movement difficulties, however no subject had developmental regression. Motor delay also varied from mild to severe; independent ambulation was achieved by all participants between 16 months and 3.5 years of age.
Other clinical features were less consistently present. Seizures appeared to be variably present: 3/6 participants (subjects 2, 4, and 5) had seizures of diverse semiologies. Two are refractory to medical management with seizure onset at 6–12 months of age, while subject 2 responded well to anti-epileptic medications and had seizure onset later at 3 years of age. There did not appear to be an association between presence of seizures and other features, including brain MRI abnormalities, severity of clinical phenotype, or developmental history. Although there was no clear pattern of bulbar involvement, subject 4 had dysphagia and oromotor dyspraxia. Feeding was otherwise normal; only one other subject had early food aversion, which resolved.
Although various additional clinical features were noted (Table 1, Supplemental notes), there were no consistent abnormalities found with respect to growth, endocrinopathies, dentition, vision, or cardiovascular or skeletal systems. All participants were born at term, had normal birth weights, and were born to non-consanguineous parents of varied ethnic origins.
EMG/NCSs for the majority of individuals (5/6) revealed predominantly demyelinating sensory and motor neuropathy (Table 1, Table S2). Subject 4 had a muscle biopsy that showed neuropathic changes. Subject 2 had NCSs at a young age (4 years) that was reported as normal. Brain MRI did not reveal specific abnormalities; three participants (subjects 2, 3, and 5) had normal brain imaging, and two participants (subjects 1 and 4) had non-specific white matter signal abnormalities, one of whom (subject 1) also had mild cerebellar atrophy (Figure S2). Other routine clinical investigations (microarray and metabolic testing) were noncontributory (Supplemental notes).
Using genomic DNA from subjects 1, 2, 3, 4, and 6 and their respective parents, we captured the exonic regions and flanking splice junctions of the genome by using the IDT xGen Exome Research Panel v.1.0. Massively parallel next-generation sequencing (NGS) was performed on an Illumina system with 150 bp paired-end reads. Reads were aligned to human genome build GRCh37/UCSC hg19 and analyzed for sequence variants with a custom-developed analysis tool. Additional sequencing technology and variant interpretation protocols have been previously described.11 The general assertion criteria for variant classification are publicly available on the GeneDx ClinVar submission page. For subject 5, trio whole-exome sequencing was performed via the SureSelect XT Human All Exon V5 kit (Agilent) at a mean target depth of 100×. Reads were aligned to Hg19 via BWA (BWA-MEM v.0.7.5a) and variants were called with the GATK haplotype caller (v.2.7-2). Detected variants were annotated, filtered, and prioritized via the Bench lab NGS v.3.1.2 platform (Cartagenia, Leuven, Belgium) and confirmed by Sanger sequencing.
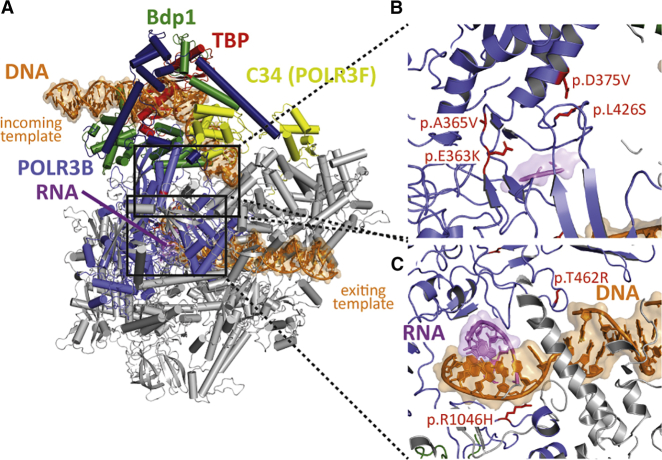
POLR3B variants identified in the participants were localized and correlated on the yeast POLR3 structure. A sequence alignment of the POLR3B subunit was generated and the equivalent positions (Table S1) were then displayed on the yeast model of POLR3 in the transcription initiation state (Figure 1A).12,13 Four of the six de novo variants cluster in a region of POLR3B where transcribed DNA is melted (Figure 1B). No direct contacts with other POLR3 subunits were identified, but the affected region is at the transcription bubble, which may have an impact on transcription itself. The two remaining variants are located in the exiting DNA tunnel and have direct contact with the duplex DNA (Figure 1C) and may thereby affect transcription activity.
Figure 1.
Structural protein modeling
(A) In order to localize the POLR3B variants found in the patients on the yeast POLR3B structure, we used the model of POLR3 in transcription initiation state.13
(B) Of the six variants identified, four of them cluster in a region of POLR3B where transcribed DNA is melted.
(C) No direct contacts with other POLR3 subunits were identified, but the mutated region is at the transcription bubble and may have an impact on transcription itself. Similarly, the two remaining variants are located in the exiting DNA tunnel and have direct contact with the duplex DNA. These variants may affect transcription activity. Of note, none of the identified variants are at the interface with transcription factors or transcription repressors (e.g., Maf1). POLR3B-altered residues found in the six patients and their equivalent yeast positions are as follows: Asp375 = Asp390, Leu426 = Leu444, Arg1046 = Arg1061, Ala365 = Ala380, Glu363 = Glu378, and Thr462 = Ser480.
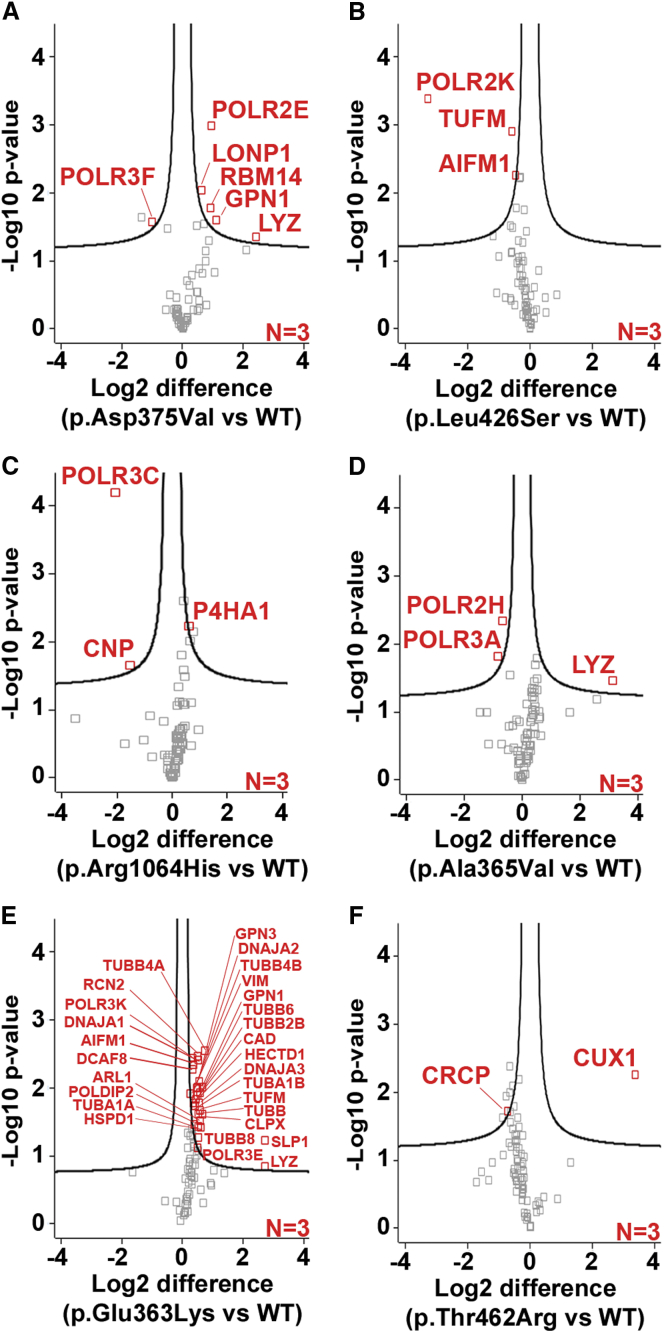
To further define the impact of the de novo variants on POLR3B, we carried out affinity purification coupled with mass spectrometry (AP-MS) to evaluate the assembly of specific RNA pol III subunits in human embryonic kidney cell line 293 (HEK293) cells (Supplemental notes). AP-MS demonstrated that five of the six variants in POLR3B (GenBank: NM_018082.5) caused impairment of the association of at least one individual subunit, either POLR3F (c.1124A>T [p.Asp375Val]), POLR2K (c.1277T>C [p.Leu426Ser]), POLR3C (c.3137G>A [p.Arg1064His]), POLR3A and POLR2H (c.1094C>T [p.Ala365Val]), and CRCP (c. 1385C>G [p.Thr462Arg]), with the enzyme (Figure 2). Although this remains to be directly confirmed, the loss of a pol III subunit is expected to render the enzyme inactive, as has been shown for RNA polymerase II.14, 15, 16 The c.1087G>A (p.Glu363Lys) variant was associated with abnormal POLR3B interactions with multiple proteins (Figure 2, Table S3), but their exact functional role in this context remains to be determined. Expression of POLR3B in fibroblasts from subject 1 was similar to that of control fibroblasts (data not shown), suggesting that the c.1124A>T (p.Asp375Val) variant does not have a direct effect on expression of the protein. Notably, all six de novo POLR3B variants localized in the nucleus as measured by immunoblots of cytoplasmic and nuclear fractions (Figure S3), suggesting that a defect in nucleo-cytoplasmic pol III shuttling is unlikely.
Figure 2.
Different POLR3B de novo variants distinctively affect the assembly of specific RNA polymerase III subunits
(A–F) FLAG-tagged POLR3B wild type (WT), p.Asp375Val (A), p.Leu426Ser (B), p.Arg1064His (C), p.Ala365Val (D), p.Glu363Lys (E), and p.Thr462Arg (F) were expressed in HEK293 cells for 24 h, purified with an anti-FLAG antibody, and digested with trypsin. The co-purified proteins were identified by liquid chromatography-tandem mass spectrometry (LC-MS/MS). The label-free quantification (LFQ) intensity of each peptide was computed via MaxQuant and Perseus. Volcano plots illustrate the log2-transformed average LFQ-intensity difference between mutant and WT (x axis), and the –log10 p value obtained via a two-tailed t test adjusted with a permutation-based multiple hypothesis testing with 10,000 iterations and an s0 correction factor of 0.1 (y axis). Proteins marked in red are considered significantly different between the conditions.
Recessive mutations in POLR3A, POLR3B, POLR1C, and POLR3K are a well-established cause of hypomyelinating leukodystrophy.1,5, 6, 7,9 The individuals with de novo heterozygous missense variants in POLR3B reported in this study manifest a different set of clinical features, further expanding the phenotypes of POLR3B-related disease to include this distinct disorder. Importantly, brain imaging did not reveal evidence of hypomyelination or leukodystrophy. Participants had pyramidal signs and gait dysfunction of varying severity, as well as some degree of intellectual disability ranging from mild to moderate severity. Most notably, EMG/NCSs for the majority of individuals (5/6) revealed predominantly demyelinating sensory and motor neuropathy. The remaining subject had NCSs at a young age and may not yet have developed this symptom. The shared clinical phenotype of these six individuals provides strong evidence for the pathogenicity of these variants.
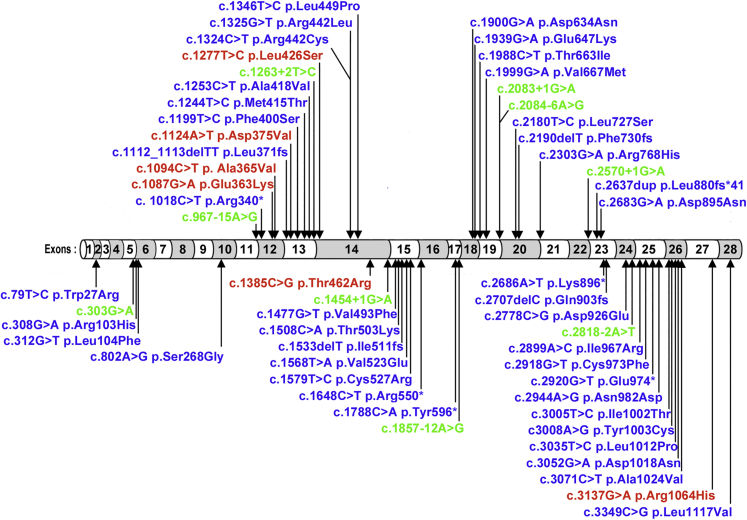
In previous studies, we have shown that the recessive mutations in POLR3B, which generate variant proteins (p.Arg103His and p.Val523Glu) that do not associate with multiple subunits of the 17-subunit RNA pol III and which clearly show impaired ability to assemble into the active enzyme, cause hypomyelinating leukodystrophies.17 In contrast, our studies of these de novo POLR3B variants suggest they cause disruption in the association of one or two enzyme subunits rather than impairment of full enzyme assembly or nuclear transport of the complex. These results are suggestive of a distinct mechanism of pathogenicity for these six de novo heterozygous mutations, most likely a dominant-negative effect. Of note, recessive variants in POLR3B include missense, frameshift, premature truncation, and splicing variants, whereas the de novo POLR3B pathogenic variants we report in this study are missense and do not overlap with the recessive variants (Figure 3). Protein structural modeling suggests that these variants may exert their pathogenicity either via alterations in the local environment or by disruption of the assembly of a functionally active pol III pre-initiation complex,12 however definitive proof will require further studies.
Figure 3.
De novo POLR3B variants differ from known recessive POLR3B pathogenic variants
Recessive variants in POLR3B exons (blue) or introns (green) include missense, frameshift, premature truncation, and splicing variants.18,19 De novo POLR3B pathogenic missense variants reported in this study are marked in red and do not overlap with the recessive variants.
Our results contribute to understanding how mutations targeting several different amino acids of the same pol III subunit (i.e., POLR3B) can exhibit distinct pathological effects in human beings. Our study provides a description of the clinical phenotype and functional evidence for a de novo heterozygous POLR3B-related disorder characterized by spasticity, ataxia, and neuropathy. We expand the spectrum of disorders associated with pathogenic variants in POLR3B.
Data and code availability
This study did not generate or analyze any new datasets or code.
Declaration of interests
N.I.W. served as advisor for Orchard and PassageBio and participates in a clinical multicenter trial led by Shire/Takeda. G.B. has no relevant conflict of interest. She has received compensation for advisory boards from Ionis (2019), Shire (2013), Actelion Pharmaceuticals (2011), and Santhera Pharmaceutical (2011) and speaker honoraria from Genzyme (2013) and Actelion Pharmaceutical (2012). She received research grants from Shire/Takeda and Bluebird Bio. B.B. is an employee of GeneDx, Inc.
Acknowledgments
We thank the patients and their families for their participation in the study; this work would not have been possible without their generosity. This study was partly supported by a grant from the Canadian Institutes of Health Research to G.B. and B.C. (CIHR; 201610PJT-377869). G.B. has received the CIHR New Investigator Salary Award (2017–2022). B.C. holds a Bell-Bombardier Research Chair awarded by the IRCM. N.I.W. is a member of the European Reference Network for Rare Neurological Disorders (ERN-RND, project ID 739510).
Published: December 28, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.12.002.
Web resources
Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP), https://evs.gs.washington.edu/EVS/
GeneDx ClinVar submission page, https://www.ncbi.nlm.nih.gov/clinvar/submitters/26957/
OMIM, https://omim.org/
Supplemental information
References
- 1.Tétreault M., Choquet K., Orcesi S., Tonduti D., Balottin U., Teichmann M., Fribourg S., Schiffmann R., Brais B., Vanderver A., Bernard G. Recessive mutations in POLR3B, encoding the second largest subunit of Pol III, cause a rare hypomyelinating leukodystrophy. Am. J. Hum. Genet. 2011;89:652–655. doi: 10.1016/j.ajhg.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolf N.I., Harting I., Innes A.M., Patzer S., Zeitler P., Schneider A., Wolff A., Baier K., Zschocke J., Ebinger F. Ataxia, delayed dentition and hypomyelination: a novel leukoencephalopathy. Neuropediatrics. 2007;38:64–70. doi: 10.1055/s-2007-985137. [DOI] [PubMed] [Google Scholar]
- 3.Wolf N.I., Harting I., Boltshauser E., Wiegand G., Koch M.J., Schmitt-Mechelke T., Martin E., Zschocke J., Uhlenberg B., Hoffmann G.F. Leukoencephalopathy with ataxia, hypodontia, and hypomyelination. Neurology. 2005;64:1461–1464. doi: 10.1212/01.WNL.0000158615.56071.E3. [DOI] [PubMed] [Google Scholar]
- 4.Timmons M., Tsokos M., Asab M.A., Seminara S.B., Zirzow G.C., Kaneski C.R., Heiss J.D., van der Knaap M.S., Vanier M.T., Schiffmann R., Wong K. Peripheral and central hypomyelination with hypogonadotropic hypogonadism and hypodontia. Neurology. 2006;67:2066–2069. doi: 10.1212/01.wnl.0000247666.28904.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernard G., Chouery E., Putorti M.L., Tétreault M., Takanohashi A., Carosso G., Clément I., Boespflug-Tanguy O., Rodriguez D., Delague V. Mutations of POLR3A encoding a catalytic subunit of RNA polymerase Pol III cause a recessive hypomyelinating leukodystrophy. Am. J. Hum. Genet. 2011;89:415–423. doi: 10.1016/j.ajhg.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thiffault I., Wolf N.I., Forget D., Guerrero K., Tran L.T., Choquet K., Lavallée-Adam M., Poitras C., Brais B., Yoon G. Recessive mutations in POLR1C cause a leukodystrophy by impairing biogenesis of RNA polymerase III. Nat. Commun. 2015;6:7623. doi: 10.1038/ncomms8623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dorboz I., Dumay-Odelot H., Boussaid K., Bouyacoub Y., Barreau P., Samaan S., Jmel H., Eymard-Pierre E., Cances C., Bar C. Mutation in POLR3K causes hypomyelinating leukodystrophy and abnormal ribosomal RNA regulation. Neurol. Genet. 2018;4:e289. doi: 10.1212/NXG.0000000000000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saitsu H., Osaka H., Sasaki M., Takanashi J., Hamada K., Yamashita A., Shibayama H., Shiina M., Kondo Y., Nishiyama K. Mutations in POLR3A and POLR3B encoding RNA Polymerase III subunits cause an autosomal-recessive hypomyelinating leukoencephalopathy. Am. J. Hum. Genet. 2011;89:644–651. doi: 10.1016/j.ajhg.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gauquelin L., Cayami F.K., Sztriha L., Yoon G., Tran L.T., Guerrero K., Hocke F., van Spaendonk R.M.L., Fung E.L., D’Arrigo S., DDD Study Clinical spectrum of POLR3-related leukodystrophy caused by biallelic POLR1C pathogenic variants. Neurol. Genet. 2019;5:e369. doi: 10.1212/NXG.0000000000000369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cayami F.K., Bugiani M., Pouwels P.J.W., Bernard G., van der Knaap M.S., Wolf N.I. 4H Leukodystrophy: Lessons from 3T Imaging. Neuropediatrics. 2018;49:112–117. doi: 10.1055/s-0037-1608780. [DOI] [PubMed] [Google Scholar]
- 11.Retterer K., Juusola J., Cho M.T., Vitazka P., Millan F., Gibellini F., Vertino-Bell A., Smaoui N., Neidich J., Monaghan K.G. Clinical application of whole-exome sequencing across clinical indications. Genet. Med. 2016;18:696–704. doi: 10.1038/gim.2015.148. [DOI] [PubMed] [Google Scholar]
- 12.Abascal-Palacios G., Ramsay E.P., Beuron F., Morris E., Vannini A. Structural basis of RNA polymerase III transcription initiation. Nature. 2018;553:301–306. doi: 10.1038/nature25441. [DOI] [PubMed] [Google Scholar]
- 13.Vorländer M.K., Khatter H., Wetzel R., Hagen W.J.H., Müller C.W. Molecular mechanism of promoter opening by RNA polymerase III. Nature. 2018;553:295–300. doi: 10.1038/nature25440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woychik N.A., Young R.A. RNA polymerase II subunit RPB10 is essential for yeast cell viability. J. Biol. Chem. 1990;265:17816–17819. [PubMed] [Google Scholar]
- 15.Archambault J., Schappert K.T., Friesen J.D. A suppressor of an RNA polymerase II mutation of Saccharomyces cerevisiae encodes a subunit common to RNA polymerases I, II, and III. Mol. Cell. Biol. 1990;10:6123–6131. doi: 10.1128/mcb.10.12.6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Runner V.M., Podolny V., Buratowski S. The Rpb4 subunit of RNA polymerase II contributes to cotranscriptional recruitment of 3′ processing factors. Mol. Cell. Biol. 2008;28:1883–1891. doi: 10.1128/MCB.01714-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choquet K., Pinard M., Yang S., Moir R.D., Poitras C., Dicaire M.J., Sgarioto N., Larivière R., Kleinman C.L., Willis I.M. The leukodystrophy mutation Polr3b R103H causes homozygote mouse embryonic lethality and impairs RNA polymerase III biogenesis. Mol. Brain. 2019;12:59. doi: 10.1186/s13041-019-0479-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolf N.I., Vanderver A., van Spaendonk R.M., Schiffmann R., Brais B., Bugiani M., Sistermans E., Catsman-Berrevoets C., Kros J.M., Pinto P.S., 4H Research Group Clinical spectrum of 4H leukodystrophy caused by POLR3A and POLR3B mutations. Neurology. 2014;83:1898–1905. doi: 10.1212/WNL.0000000000001002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richards M.R., Plummer L., Chan Y.M., Lippincott M.F., Quinton R., Kumanov P., Seminara S.B. Phenotypic spectrum of POLR3B mutations: isolated hypogonadotropic hypogonadism without neurological or dental anomalies. J. Med. Genet. 2017;54:19–25. doi: 10.1136/jmedgenet-2016-104064. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate or analyze any new datasets or code.