Summary
Chiari I malformation (CM1), the displacement of the cerebellum through the foramen magnum into the spinal canal, is one of the most common pediatric neurological conditions. Individuals with CM1 can present with neurological symptoms, including severe headaches and sensory or motor deficits, often as a consequence of brainstem compression or syringomyelia (SM). We conducted whole-exome sequencing (WES) on 668 CM1 probands and 232 family members and performed gene-burden and de novo enrichment analyses. A significant enrichment of rare and de novo non-synonymous variants in chromodomain (CHD) genes was observed among individuals with CM1 (combined p = 2.4 × 10−10), including 3 de novo loss-of-function variants in CHD8 (LOF enrichment p = 1.9 × 10−10) and a significant burden of rare transmitted variants in CHD3 (p = 1.8 × 10−6). Overall, individuals with CM1 were found to have significantly increased head circumference (p = 2.6 × 10−9), with many harboring CHD rare variants having macrocephaly. Finally, haploinsufficiency for chd8 in zebrafish led to macrocephaly and posterior hindbrain displacement reminiscent of CM1. These results implicate chromodomain genes and excessive brain growth in CM1 pathogenesis.
Keywords: de novo mutations, Chiari I malformation, chromodomain genes, zebrafish disease model, gene burden, macrocephaly, rare variants
Introduction
Chiari I malformation (CM1) is characterized by displacement of the hindbrain through the foramen magnum. CM1 is one of the most common pediatric neurological conditions affecting 1 in 1,000 individuals symptomatically with as many as 1 in 100 meeting radiographic criteria.1,2 Individuals with CM1 present with a wide array of symptoms stemming from compression of neural tissue and may suffer from syringomyelia (SM) or hydrocephalus.3 Approximately 25% of individuals with CM1 develop SM, which can lead to serious sensorimotor symptoms and about 20%–60% of them develop scoliosis, depending on the presence of syringomyelia.4, 5, 6, 7, 8 Whereas CM1 can be associated with other conditions or syndromes, such as hydrocephalus, craniosynostosis, or segmental bony anomalies, the majority are non-syndromic and idiopathic.9
Suboccipital decompression surgery, the primary treatment option for CM1, is costly, invasive, and does not always improve symptoms.10 Without surgery, however, individuals with CM1 may develop progressive headaches, visual disturbance, vertigo, paralysis, paresthesia, dysphagia, bowel and bladder incontinence, and a variety of nonspecific symptoms that impair quality of life.11,12 Therefore, understanding the underlying etiology of CM1 is essential to improve early diagnosis and develop tailored treatment strategies.
A genetic predisposition to CM1 is evidenced by high rates of concordance in twin studies2,13,14 and increased risk to first-degree relatives of individuals with CM1.11,15 One study estimated heritability at 50%, with a range of 30%–70%.16 A limited number of genetics studies have identified candidate genes, but no confirmed genetic etiology for idiopathic CM1 has been identified.1,3,17, 18, 19
In the majority of CM1 cases, no family history is present; this has limited traditional gene mapping efforts before the advent of high-throughput sequencing. The lack of known inheritance of CM1 within a family is also partially attributable to the need for a radiographic study for diagnoses which has only been possible for the past few decades and is not readily available in many parts of the world or for asymptomatic individuals. This apparent complex genetic architecture of CM1 led us to take an approach utilized for other severe pediatric disorders, namely searching for genes with more variants in affected subjects than expected by chance. Others have successfully used this approach for other neurodevelopmental disorders including epilepsy,20, 21, 22, 23, 24 craniosynostosis,25, 26, 27 and autism.28, 29, 30, 31, 32, 33, 34, 35, 36 We hypothesized that like these severe pediatric developmental disorders, CM1 can be explained, at least in part, by damaging de novo mutations and rare transmitted variants with incomplete penetrance. Here we performed exome sequencing a large cohort of individuals with idiopathic CM1 to identify genes and genetic variants associated with CM1 risk.
Subjects and Methods
Subjects and samples
Informed consent was obtained for all participating individuals and genetic studies were performed as approved by the Institutional Review Boards of the relevant institutions. Individuals presenting with Chiari I malformation were recruited from St. Louis Children’s Hospital, University of Utah, Vanderbilt University Medical Center (BioVU), Duke University, and University Vall d’Hebron, Spain. We recruited a cohort of 900 individuals with CM1 and family members, including 668 probands with Chiari I malformation with >5 mm cerebellar tonsil herniation, 76 affected relatives, and 156 unaffected parents and relatives. The CM1 cohort included 67 parent-offspring uniplex CM1 trios, 584 singleton cases, and 55 multiplex kindreds. Individuals with known etiologies or secondary causes of CM1 (i.e., brain tumors, hydrocephalus, craniosynostosis, known genetic syndromes) were excluded from the study. All affected individuals and/or parents provided informed consent. Only individuals with isolated, non-syndromic Chiari I malformation were included. Individuals with rare non-synonymous variants in CHD genes were retrospectively screened for diagnoses of neurodevelopmental disorders and none were found at the time of this study. All individuals had diagnoses of MRI-confirmed CM1 with >5 mm herniation of cerebellar tonsils. In-house control subjects consisted of unrelated healthy individuals or individuals ascertained for conditions other than CM1 (i.e., Alzheimer disease [C.C.], amyotrophic lateral sclerosis [M.H.], and male infertility [D.C.]). All in-house control subjects were of European ancestry. Control trios (n = 1,911) were composed of siblings and parents of probands who are part of the Simon’s Simplex collection.29,37,38 Occipitofrontal head circumferences (OFCs) were measured for the majority of CM1-affected subjects recruited at St. Louis Children’s hospital. In-house control OFCs were for age-matched individuals with adolescent idiopathic scoliosis.
Sequencing analysis and validation
DNA was isolated, and WES was performed using IDT or Agilent exome capture and sequenced on Illumina HiSeq sequencers at the McDonnell Genome Institute at Washington University. Analysis of exome-sequencing data was performed in-house using our previously described methods.39,40 Briefly, FASTQ formatted sequences were aligned to the hg37 human reference sequence (NCBI GRCh37) using BWA.41 Mapped reads were filtered to remove duplicate reads with the same paired start sites. Median depth per sample at captured bases for CM1-affected subjects was 71× (range 21–121) and was 76× (range 17–202) for control subjects. All affected subjects and control subjects had >97% of captured bases covered with >10 reads (Table S1). The Binary sequence Alignment/Map (BAM)42 formatted alignments were then processed using the Genome Analysis Toolkit (GATK) Haplotype Caller43,44 and genotypes jointly called together with all in-house control exome-sequenced individuals using an Agilent/IDT intersection bedfile. Genotypes were filtered for read-depth (>10×), genotype quality (GQ > 20), and allele balance (AB) > 0.3 and < 0.7. Variants were filtered for GATK-calculated variant quality score recalibration (VQSR) and genotype call-rate > 90%. We summarize all sample- and cohort-level sequencing metrics in Table S1. Two exome library kits were used for the case/control cohorts with different target sizes and, therefore, varying coverage of RegSeq hg19 coding regions. To control for these factors, we determined the number of “callable” bp, or the number of bp that have coverage >10×. We then intersected these coordinates with RefSeq hg19 coding exons to determine the “total callable exome,” or the number of bp within RefSeq coding exons that had sufficient coverage for genotype calling. SSC trios were captured using the Nimblegen EZ Exome V2 capture kit and were processed separately using the same calling pipeline but using a Nimblegen EZ Exome V2 specific bedfile. Allele frequencies were annotated using the gnomAD database.45 To reduce the risk of population stratification, only affected subjects and control subjects with principal components confirmed European ancestry were included. Principal components were calculated using EIGENSTRAT46 from whole-exome SNP data using all common (MAF > 5%) SNPs.
De novo variant calling
De novo SNVs (single-nucleotide variants) and indels (insertions-deletions) were called using a custom pipeline using family-level VCFs. Potential de novo sites were called as those where the father’s genotype was 0/0, the mother’s genotype was 0/0, and the child’s genotype was either 0/1 or 1/1. We then applied allele count, read-depth, and allele balance filters: the father alternate allele count = 0, mother alternate allele count = 0, child allele balance > 0.25, father depth > 9, mother depth > 9, child depth > 9, and child genotype quality (GQ) > 20. DNMs were also filtered on the basis of population frequency, using only rare (MAF < 0.1% across all of gnomAD v2.1.1) coding DNMs. For all nonsynonymous coding variants, MetaSVM was used to infer the impact of missense mutations.47 Sanger sequencing of PCR amplicons containing the mutation verified variants in genes of interest and all de novo variants. Quality control and sequencing metrics for DNMs called in CM1 trios or SSC sibling control trios can be found in Table S2. A full list of DNMs found in CM1 trios or SSC sibling control trios can be found in Table S3.
De novo mutation expectation model
We applied a sequence context-based method to calculate the probability of observing a DNM for each base in the coding region adjusting for the sequencing depth in each gene as described previously.48 All protein-coding RefSeq transcripts intersecting with target regions present on the on the IDT xGEN Exome Research panel (v1.0) for CM1 trios or Nimblegen EZ Exome V2 for SSC sibling trios and with annotations in the ANNOVAR RefSeq database were considered.49 Briefly, for each base in the exome, the probability of observing every tri-nucleotide mutating to other tri-nucleotides was determined. ANNOVAR (v2019Oct24) was used to annotate the consequence of each possible substitution. RefSeq was used to annotate variants (based on the file “hg19_refGene.txt” provided by ANNOVAR). For each gene, the coding consequence of each potential substitution was summed for each functional class (synonymous, missense, canonical splice site, stop-gain, stop-loss, start-lost) to determine the gene-specific mutation probabilities.48 Insertions or deletions were excluded from the model and were not considered in the downstream statistical analyses due the high false positive rate of these variants. Each probability was adjusted to control for variable sequencing coverage as previously described: the raw probability was multiplied by a factor in the range 0.9–1, according to the percentage of trios covering that base with at least 10× depth. Positions with a coverage of zero resulted in a probability of zero for that base. The sequencing coverage adjustment was calculated separately for affected subjects and control subjects, which were sequenced in separate batches, thus separate de novo probability tables were generated for affected subjects and control subjects, respectively. To align with ANNOVAR annotations, analysis was limited to variants that were located in the exonic or canonical splice site regions and were not annotated as “unknown” by ANNOVAR. Following the inclusion criteria, we identified potential coding mutations and generated gene-specific mutation probabilities for 18,272 unique genes, covering 31.68 Mb of protein-coding sequence for CM1 trios (Table S4) and 18,515 unique genes covering 33.2 Mb of protein-coding sequencing in SSC sibling control trios (Table S5).
Burden of de novo mutations
The burden of de novo mutations in CM1-affected subjects was determined using denovolyzeR v.0.2.048 using a coverage adjustment factor based on 67 case trios. The expected number of DNMs was calculated by taking the sum of each functional class specific probability multiplied by the number of probands in the study (67 for CM1 trios or 1,911 for SSC sibling control trios), multiplied by two (diploid genomes). The Poisson test was then used to test for enrichment of observed DNMs versus expected. As separate tests were performed for protein-altering DNMs and LoF DNMs, the Bonferroni multiple-testing threshold is, therefore, equal to 1.36 × 10−6 (0.05/(18,272 genes × 2 tests)). The most significant p value of the two tests was reported. For gene set enrichment, the expected probability was calculated from the probabilities corresponding to the gene set only (i.e., brain-expressed, AD disease genes, CHD genes). Brain-expressed genes were defined as those detected in the brain as part of the Human Protein Atlas.50 Autosomal-dominant (AD) disease genes were those annotated to cause an AD disease in OMIM.51
Recessive and X-linked analyses
We filtered recessive and X-linked genotypes for rare (max MAC in all populations < 10 in gnomAD exomes) homozygous and compound heterozygous variants with high quality: pass GATK Variant Score Quality Recalibration (VSQR), have >10 reads, and have a genotype quality (GQ) ≥ 20, demonstrate allele balance (AB) > 0.3 and < 0.7, and have overall genotype call-rate > 90%. All putative LoF variants (nonsense, canonical splice-site, and start loss), MetaSVM D-Mis missense variants and variants listed in Clinvar as “pathogenic” or “likely pathogenic” were considered. Synonymous variants were also filtered using the same criteria and analyzed separately to determine whether there is an inflation of background rate.
Gene burden and gene-set burden analysis
To quantify the enrichment of rare, non-synonymous/splice-site variants in CM1-affected subjects compared to control subjects for each gene, we compared the collapsed minor allele frequency of variants in affected subjects and control subjects using a Fisher’s exact test utilizing only filtered variants. Variants were filtered by rareness and quality: (1) max minor allele count in all populations < 10 in gnomAD exomes, (2) GQ > 20 and alternate allele ratio 40%, (3) GATK VQSR of “PASS,” and (4) minimum sequencing depth of 8 reads in each participant. For statistical analysis of exome-sequencing data, affected individuals were compared to in-house exome controls. For individual gene association analyses, Fisher’s exact tests were performed using PLINK to compare the collapsed minor allele frequency of rare variants in affected subjects and control subjects. t tests were performed using R to compare the quantitative traits. For gene-set burden analyses, non-synonymous/splice-site variants within a gene were collapsed to obtain the number of rare (gnomAD maxMAC < 10) variants per gene. The numbers of variants within groups of genes were then summed based on membership within a given gene-set and used as the dependent variable in a linear regression. As there were 15,970 RefSeq genes with at least one rare protein-altering variants in either CM1-affected subjects or in-house control subjects, the Bonferroni multiple-testing threshold is, therefore, equal to 3.1 × 10−6 (0.05/(15,970 genes)).
Additional statistical analyses
Occipitofrontal head circumferences (OFCs) were compared to control subjects (n = 50, individuals without CM1 and no known brain disorder) using age and sex corrected values using Wilcoxon rank-sum tests. Expected OFC values for all ages up to 20 years old were calculated using data presented previously.52 Ages for individuals above 20 years old were windsorized at 20. Differences in OFC from expected for age and sex were calculated and the distribution of differences were compared to the expected average difference of 0 using Wilcoxon rank-sum tests. Meta-analysis of DNM enrichment per gene and gene burden association analyses were performed using Fisher’s method using the R package metaP.
Production and characterization of CRISPR-mediated chd8 disruption in zebrafish
Briefly, a single gRNA designed to target exon 2 (gRNA sequence: 5′-GGAAGCAAAGAGGATCACTC-3′) of the chd8 gene were synthesized in vitro and microinjected with codon-optimized Cas9 mRNA into one-cell-stage embryos of the AB∗ zebrafish strain. F0 founders with germline mutations were identified with Illumina sequencing of F0 sperm. F0 fish found to harbor a high rate of germline chimerism for protein-truncating mutations in chd8 mutants were then bred to HUC-GFP zebrafish (express GFP in adult neuronal populations) to produce mutant zebrafish fully heterozygous for both chd8 disruption and HUC-GFP. Animals were raised in in 0.003% phenylthiourea (PTU) to prevent melanin synthesis and allow for unobscured imaging of the brain. At 5 days post-fertilization (dpf), larvae were imaged using a Vertebrate Automated Screening Technology (VAST) system linked to a Zeiss fluorescent confocal microscope which enables rapid, automated confocal fluorescent imaging of larval zebrafish (up to 7 days post fertilization). Multiple founder F0 zebrafish exhibiting high rate of germline chimerism for protein-truncating mutations in chd8 were imaged separately to ensure reproducibility of observed mutant phenotypes. For each group of imaged F1 zebrafish, individual fish were genotyped by Sanger sequencing of the CRISPR-edited exon. ImageJ was used to calculate the average area of Z-position aligned images of mutant or wild-type zebrafish. Brain images were subdivided into forebrain, midbrain, and hindbrain. All volumes were defined and measured by blinded reviewers. All experimental in vivo procedures were performed in accordance with national and institutional guidelines for animal care and experiments. Animal handling protocols and experiments were reviewed and approved by the Institute Animal Care and Utilization Committee of Washington University.
Results
Global burden of de novo mutations in CM1
We observed a total of 81 protein-coding de novo mutations (DNMs) among the 67 trios, which yielded an average of 1.20 de novo coding region mutations per proband, consistent with other studies27,48,53,54 (Table S3). The distribution of DNMs per trio followed the expected Poisson distribution (p = 0.87) (Figure S1 and Table S6). For purposes of analysis, coding DNMs were grouped into three classes: missense, predicted loss-of-function (LoF) (i.e., stop-gains, stop-losses, and variants at essential splice sites), and a combined group, “protein-altering” DNMs, defined as the union of missense and LoF variants. Due to the high false-positive rate of small insertion-deletion variants, these were excluded from the analysis and from per-gene DNM probability calculations. We compared the observed and expected number of de novo mutations and found that protein-altering de novo mutations were overall significantly enriched over expectation (p = 5.3 × 10−4) (Figure 1). Six genes harbored predicted loss-of-function de novo mutations, three of which are predicted to be evolutionary constrained when considering the ratio of observed and expected loss of function variants (oe) in GnomAD: CHD8 (oe = 0.04), CRIM1 (oe = 0.12), ARL8A (oe = 0.07). The average paternal age at birth of the proband for the 67 trios was 32.4 years old. For probands with DNMs, the average paternal age was 32.9 years old and for probands without DNMs, the average paternal age was 31.5 years old (Wilcoxon rank-sum test, p = 0.22) (Table S6).
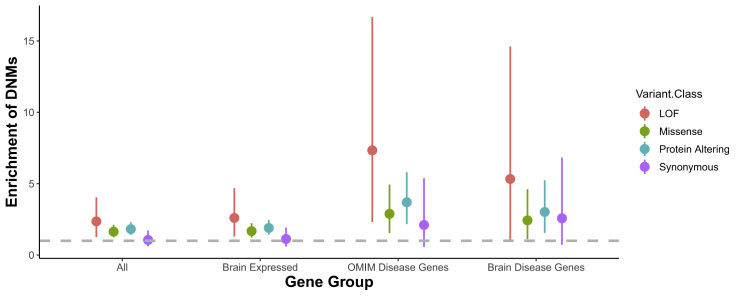
Figure 1.
De novo mutations are enriched in brain genes and in OMIM disease genes
Enrichment of DNMs ±2 SE by variant class for all genes, gene expressed in the brain, genes that are disease causing in OMIM, and the intersection of brain-expressed genes and OMIM disease genes (Brain Disease Genes).
We determined whether CM1 trio probands harbored more coding DNMs than expected based on mutational models.55 Overall, we observed a significant enrichment of protein-altering DNMs (enrichment = 1.67; p = 8.7 × 10−5) and missense DNMs (enrichment = 1.64; p = 2.54 × 10−4) among CM1 trio probands, but there was no difference in the number of synonymous DNMs as expected (enrichment = 1.06; p = 0.44) (Figure 1). We then stratified by gene class to determine whether there was an excess of DNMs in genes that cause autosomal-dominant diseases as reported by OMIM, genes expressed in the human brain as reported by the Human Protein Atlas, or genes expressed in the human brain and that cause an autosomal-dominant disease. We saw strong enrichment in CM1 probands of both protein-altering DNMs and missense DNMs for OMIM dominant disease genes (protein-altering DNM enrichment = 3.28; p = 5.29 × 10−5 and missense enrichment = 2.89; p = 8.12 × 10−4) and genes expressed in the human brain (protein-altering DNM enrichment = 1.75; p = 1.40 × 10−4 and missense enrichment = 1.68; p = 7.16 × 10−4). For the subset of brain expressed dominant disease genes (“brain disease genes”), we observed an enrichment of LoF DNMs (enrichment = 6.32; p = p = 4.1 × 10−2), missense DNMs (enrichment = 2.43; p = 1.36 × 10−2), and all protein-altering DNMs (enrichment = 2.74; p = 2.9 × 10−3), but no enrichment of synonymous DNMs (enrichment = 2.1; p = 0.09). Additionally, we analyzed a cohort of 1,911 Simon’s Simplex Collection sibling trios as a control group. We observed no enrichment of DNMs for any gene functional class (synonymous, missense, protein-altering, or LoF) in any of these gene sets or globally in this dataset (Figure S2). These data indicate that de novo deleterious protein-coding variants, particularly in brain-expressed genes, confer significant risk of CM1.
Recessive or X-linked disorders in CM1
In order to determine whether a subset of probands in our CM1 cohort were undiagnosed for known recessive or X-linked genetic disorders, we extracted all variants that were rare (max MAF in all populations < 0.1% in gnomAD exomes), putative loss-of-function (LoF) variants, i.e., stop-gain or splice-site variants, Clinvar “pathogenic” or “likely pathogenic” variants or variants predicted to be damaging by MetaSVM. After filtering, a total of 84 rare recessive-autosomal genotypes remained (Table S7) and 16 rare hemizygous X-linked genotypes remained (all in male CM1-affected subjects) (Table S8). Three CM1 probands possessed two nonsense variants in the same gene (LY9, SCNN1D, and TTC40). None of these genes have been previously implicated in a monogenic genetic disorder, however. No individuals with CM1 harbored multiple ClinVar pathogenic variants or both a ClinVar pathogenic variant and a nonsense variant in the same gene. No ClinVar pathogenic variants or rare LoF variants were observed in any known X-linked disease-associated genes in CM1 probands. Two individuals with CM1 harbored rare hemizygous nonsense variants in ADGRG4 (oe = 0.92) and PPP1R3F (oe = 0.73). Neither gene has a high probability of loss intolerance and neither has been associated with human disease, however. The proportion of individuals in both our unrelated in-house control cohort with multiple rare variants in the same gene, i.e., potentially compound heterozygous or harboring two variants on the same allele, was similar to that observed in the CM1 individuals (6% in CM1 versus 8% in controls, p < 0.05).
De novo mutations in CHD8 in CM1
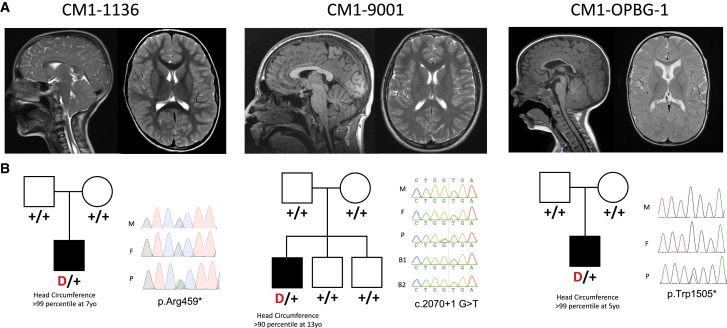
By querying GeneMatcher56 for individuals with DNMs in CHD8, CRIM1, or ARL8A (three genes in which we found one LoF de novo variant each in CM1 trios), we found two additional CM1-affected individuals with predicted loss-of-function de novo mutations in CHD8 (Figure 2). Heterozygous LoF CHD8 mutations were previously reported in individuals with autism spectrum disorder (ASD)/neurodevelopmental disorders, who were described as having macrocephaly and distinctive facial features (MIM: 615032).57,58 However, CM1 was not reported in these cases. The three de novo CHD8 mutations observed in CM1 include two stop-gain mutations (GenBank: NM_001170629.2; c.4414C>T [p.Arg1472∗] and c.4515G>A [p.Trp1505∗]) and a splice donor site mutation (c.2070+1G>T [p.?]). Using denovolyeR, we determined whether any gene harbored more DNMs than expected by chance (Figure S3, Tables S9 and S10). The probability of 3 LoF de novo mutations occurring in CHD8 by chance in a cohort of this size is 1.9 × 10−10, surpassing genome-wide significance (Table 1).48 Notably, all three of these individuals displayed head circumferences above the 90th percentile for age and sex, consistent with the macrocephaly reported with CHD8 mutations. Both GeneMatcher trio probands harboring de novo CHD8 mutations had also been diagnosed with mild to moderate developmental delay, consistent with previous reports.58 The index CM1-affected subject harboring a CHD8 DNM (CM1-1136) was diagnosed with autism spectrum disorder after enrollment in this study. Performing per-gene DNM enrichment analysis in the cohort of control trios identified no genome-wide significant enrichment of DNMs for any gene (Figure S4 and Table S11).
Figure 2.
CHD8 loss-of-function de novo mutations in individuals with CM1
(A) Representative sagittal (left) and axial (right) brain magnetic resonance images of CM1 probands with de novo CHD8 loss-of-function mutations.
(B) Pedigrees with Sanger-verified mutated bases for mothers (M), fathers (F), probands (P), and unaffected brothers (B1 and B2). Positions are with respect to GenBank: NM_001170629.2.
Table 1.
Meta-analysis of protein-altering de novo mutations and rare coding variant gene burden in CHD genes
| Associated disorder | Head size | De novocoding variant | p value | CM1 (n = 668) | Controls (n = 4,964) | OR (CI) | p value | Meta p value | |
|---|---|---|---|---|---|---|---|---|---|
| CHD1 | DD | macro | – | 1 | 8 (1.2%) | 43 (0.87%) | 1.38 (0.56–2.98) | 0.38 | 0.74 |
| CHD2 | DD | – | – | 1 | 15 (2.25%) | 56 (1.13%) | 2.00 (1.04–3.60) | 0.02 | 0.1 |
| CHD3 | DD | macro | – | 1 | 19 (2.84%) | 31 (0.62%) | 4.61 (2.45–8.44) | 1.8 × 10−6 | 2.6 × 10−5 |
| CHD4 | DD | macro | – | 1 | 4 (0.6%) | 37 (0.75%) | 0.80 (0.21–2.24) | 1 | 1 |
| CHD5 | – | – | – | 1 | 16 (2.4%) | 64 (1.29%) | 1.86 (1.00–3.28) | 0.03 | 0.14 |
| CHD6 | – | – | – | 1 | 23 (3.44%) | 107 (2.16%) | 1.61 (0.97–2.55) | 0.05 | 0.2 |
| CHD7 | CHARGE | – | – | 1 | 31 (4.64%) | 165 (3.32%) | 1.41 (0.92–2.08) | 0.09 | 0.31 |
| CHD8 | autism | macro | 3 | 1.6 × 10−7 | 10 (1.5%) | 75 (1.51%) | 1.00 (0.45–1.93) | 0.78 | 9.4 × 10−7 |
| CHD9 | – | – | – | 1 | 11 (1.65%) | 63 (1.27%) | 1.30 (0.61–2.49) | 0.37 | 0.73 |
| Total | – | – | 3 | 7.8 × 10−5 | 137 (20.51%) | 641 (12.91%) | 1.66 (1.35–2.01) | 1.2 × 10−6 | 2.4 × 10−10 |
DNM enrichment p values are for enrichment of protein-altering variants to allow for compatibility with meta-analysis with gene-burden results. Abbreviations: DD, developmental disorder; CHARGE, coloboma of the eye, heart defects, atresia of the choanae, retardation of growth and development, and ear abnormalities and deafness syndrome; macro, macrocephaly; CM1, Chiari I malformation; OR, odds ratio; CI, 95% confidence interval). Total CHD gene DNM enrichment was calculated for the CHD gene set in denovlyzeR using in-house, coverage corrected DNM gene probabilities.
Genome-wide rare variant burden analysis
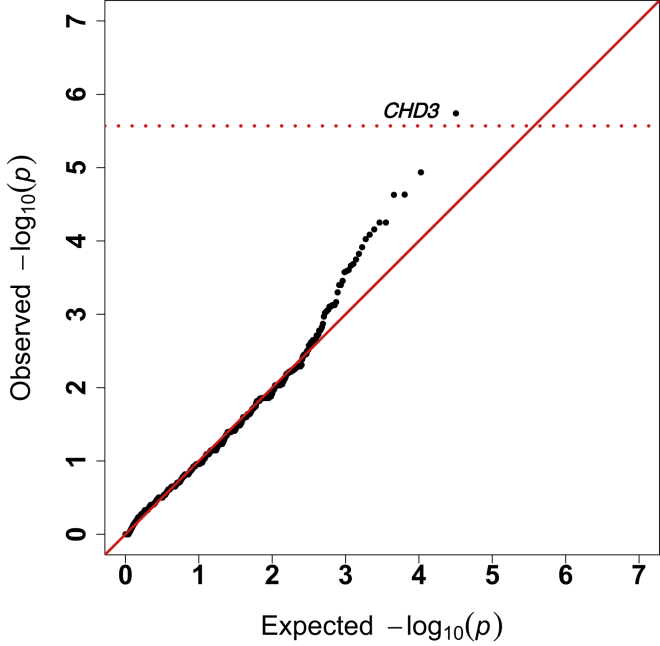
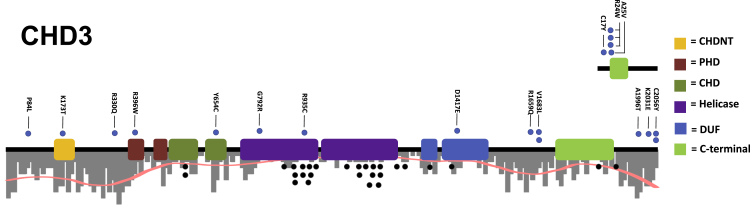
To identify genes enriched for rare coding variants in CM1, exome sequence data from the 668 unrelated CM1 probands of European descent was compared to 4,964 unrelated in-house control subjects of European descent. Gene-burden analysis of rare (<10 minor alleles in the Genome Aggregation Database [gnomAD]) non-synonymous/splice-site variants was performed across all genes (lambda = 0.57) (Table S9). One gene, CHD3, surpassed exome-wide significance (p < 2.5 × 10−6) (Figure 3) with 19 rare variants identified in our cohort of 668 affected. We observed no association between rare synonymous variants in CHD3 and CM1 risk (p < 0.05) and observed no genome-wide significant genes when comparing the burden of synonymous rare variants among CM1-affected individuals and in-house control subjects (lambda = 0.79) (Figure S5 and Table S12). Performing a genome-wide meta-analysis combining per gene protein-altering DNM enrichment and gene burden analysis (after removing all DNMs from the gene burden analysis calculations), the two top genes are CHD8 (p = 9.43 × 10−7) and CHD3 (p = 2.59 × 10−5) (Figure S6 and Table S9). CHD3 is a chromodomain gene with similar function and clinical manifestations to CHD8, and de novo variants in this gene were recently reported to cause Snijders Blok-Campeau syndrome (MIM: 618205).59,60 While the majority of previously reported de novo variants in Snijders Blok-Campeau syndrome occurred within the helicase domains of CHD3,60 most variants identified in CM1 occurred outside these regions (Figure 4). Three variants found in our CM1 cohort (Gencode v24 ENST00000481999.1; c.50G>A [p.Cys17Tyr], c.70C>T [p.Arg24Trp], and c.74C>T [p.Ala25Val]) are present in an alternative coding transcript (ENST00000481999.1) that results from an alternative start site. Notably, CHD3 is known to have neuron-specific alternative splicing patterns61 and both transcripts are expressed at near equal levels in most brain regions in GTEx.62 CM1 was not previously reported with CHD3 DNMs. None of the individuals in our CM1 cohort with CHD3 rare variants had clinical features consistent with Snijders Blok-Campeau syndrome. Additionally, we observed significantly lower mean CADD scores for CHD3 rare variants harbored by CM1-affected subjects compared to the de novo variants reported in Snijders Blok-Campeau syndrome (14.0 versus 34.5, t test p = 1.6 × 10−6).
Figure 3.
Gene burden analysis comparing CM1 probands to in-house control subjects
Quantile-quantile plot of observed versus expected p values from exome-wide gene burden association analysis of rare (allele count < 10 in all gnomAD populations).
Figure 4.
Spectrum of CHD3 rare variants in individuals with Chiari I malformation
Protein model of CHD3 with rare nonsynonymous variants indicated. Dots above gene models (blue) were observed in CM1-affected subjects. Dots below the gene models (gray) are de novo missense mutations reported for Snijders Blok-Campeau syndrome. CHDNT domain (yellow), PHD domain (red), chromodomains (dark green), helicase ATP-binding domains (purple), domains of unknown significance (blue), and CHD-C-terminal domain (light green) are indicated as colored blocks. Positions are with reference to sequences ENST00000380358.8 (bottom) and ENST00000481999.1 (top). Two variants (p.Cys2056Tyr and p.Arg24Trp) were observed in multiple unrelated individuals with CM1 of European descent and not in in-house control subjects. Black dots represent the positions of de novo variants observed in individuals with Snijders Blok-Campeau syndrome. A histogram of variants per base-pair observed in the gnomAD database is plotted to depict the relative evolutionary constraint across the protein.
Increased burden of rare variants in CHD gene-set in CM1
Because we found an enrichment of de novo variants in CHD8 and rare variants in CHD3 in our CM1 cohort, we next investigated whether there was an increased burden of rare variants in CHD genes as a group. For this, we performed gene-set burden analysis similar to our prior work with collagen variants in adolescent idiopathic scoliosis.63 In brief, the frequency of individuals carrying at least one rare variant in any CHD gene (CHD1-9) was compared between unrelated CM1 probands and in-house control subjects. We observed a significantly greater number of rare CHD gene missense/nonsense/splice-site variants in individuals with CM1 compared to control subjects (p = 1.2 × 10−6, OR = 1.66) (Tables 1 and S13). We then meta-analyzed the association between the enrichment of protein-altering DNMs in CHD genes compared to expected (p = 7.89 × 10−5, 38-fold enrichment) and gene-set burden analysis across all CHD genes, and we observed a strongly significant association between rare variants in CHD genes and CM1 risk (p = 2.4 × 10−10). Furthermore, even when CHD8 and CHD3 variants were excluded, we find that CHD genes as a group remain significantly associated with CM1 risk with a collapsed minor allele frequency (cMAF) of 27% in CM1-affected subjects compared to 11% in control subjects (p = 1.5 × 10−4, OR = 1.54). Additionally, we observed no significant gene-burden association between rare synonymous variants in any CHD gene or in the set of all CHD genes with CM1 status (see Table S9).
Macrocephaly in CM1 with CHD rare variants
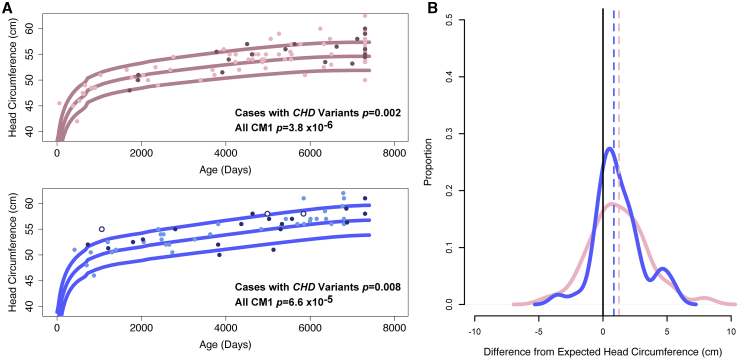
Because macrocephaly is commonly observed in cases with chromodomain gene disruptions,58,60,64 we sought to determine whether the individuals with CM1 in our cohort, particularly those with rare variants in CHD genes, also had larger head circumferences. Importantly, none of the individuals with rare variants in CHD genes in our cohort were diagnosed with the autosomal-dominant disorders previously ascribed to these genes, with the exception of our single CM1 proband with a de novo CHD8 LoF variant who was diagnosed with ASD subsequent to enrolling in this study. Table S14 is a detailed table of phenotypes observed in CM1-affected individuals with CHD rare variants. Individuals with CM1 who harbor rare non-synonymous coding variants in CHD genes (n = 41) displayed head circumferences that were an average of 1.1 cm greater than expected for age and sex compared to in-house control subjects (n = 50) (p = 5 × 10−4), and compared to growth charts available from the U.S. Centers for Disease Control and Prevention (CDC) (p = 3 × 10−4) (Figure 5). Furthermore, when we investigated the head circumference of all CM1-affected individuals in our cohort (n = 240 with available measurements), irrespective of the presence of a rare variant in a CHD gene, those individuals with CM1 had significantly larger head size compared to in-house control subjects (p = 0.003) and CDC normative data (p = 5.6 × 10−13). Individuals with CM1 who harbor a rare CHD variant displayed head circumferences even greater than CM1-affected individuals without CHD variants (+2.15 cm versus +0.94 cm greater than expected for age and sex, Wilcoxon rank-sum test p = 3.0 × 10−4), with 40% (15/38) measured individuals with CM1 and CHD variants displaying head circumferences above the 95th percentile for age and sex. By modeling CM1 risk as a product of age, sex, and head circumference, we estimate that head circumferences >2 standard deviations (SD) above expected result in a 4-fold increased relative risk of CM1. Even after removing individuals with CHD gene variants, this increased average head circumference remained for CM1 compared to in-house control subjects (p = 7.4 × 10−3) and CDC normative data (+1.0 cm, p = 7.4 × 10−11). These data suggest that head circumference alone is a good predictor of CM1 risk in the adolescent population.
Figure 5.
Distribution of head circumference among individuals with CM1
(A) Head circumference in CM1-affected subjects without CHD gene variants (light pink or light blue), CM1-affected subjects with presumed inherited CHD variants (dark pink or dark blue), and individuals with de novo CHD8 LoF variants (dark blue, white fill).
(B) Analysis of standardized head circumference values for individuals with CM1 reveal significantly increased head circumference across ages in both male and female individuals with CM1, with an apparent bimodal distribution or kurtosis toward larger head circumference. p values are for Wilcoxon rank-sum tests comparing the distribution of age corrected observed head circumference to expected.
Modeling of chd8 disruption in zebrafish
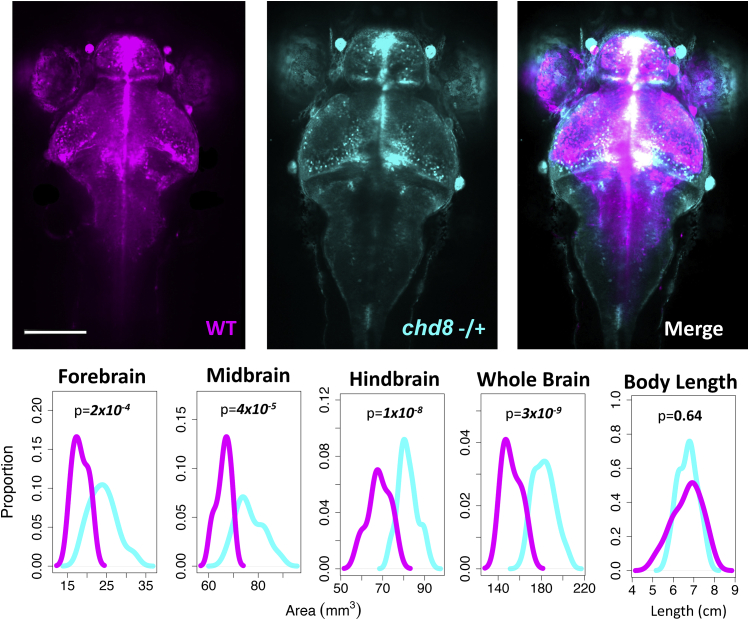
To support the notion that disruption of a single copy of CHD8 is sufficient to cause significantly abnormal brain growth, we generated zebrafish with frameshift protein-truncating mutations in its ortholog, chd8, by CRISPR-Cas9 mutagenesis. We observed significantly larger brain volumes in chd8-/+ zebrafish compared to their wild-type siblings (p = 3 × 10−9). This difference was apparent in forebrain, midbrain, and hindbrain sub-volumes (Figure 6). There was no apparent difference in the overall length of the zebrafish (p = 0.64), suggesting that the difference in brain volume is due to brain-specific overgrowth as opposed to global growth differences or differences in developmental timing.
Figure 6.
chd8 disruption results in enlarged brains early in development
Zebrafish at 6 days post-fertilization (dpf) generated by crossing HuC-GFP AB zebrafish and F0 chd8 germline chimeric knockout zebrafish. After imaging, individual zebrafish were genotyped by Sanger sequencing. Fish genotyped as wild-type (WT) are pseudocolored magenta and fish genotyped as heterozygous for a frameshift mutation are pseudocolored cyan. chd8−/+ n = 40. WT n = 25. Brain region areas (forebrain, midbrain, and hindbrain) were quantified and compared. Differences in mean area for each brain area, total brain area, or body length were compared using a t test. chd8−/+ n = 40. WT n = 25. Scale bar = 160 μm.
Discussion
We identified a set of chromodomain genes in which rare coding variants are strongly associated with risk of developing CM1 in a large multi-center exome-sequenced cohort of affected individuals with Chiari I malformation. The enrichment of rare variants in these genes and the presence of three de novo mutations in CHD8 among individuals with CM1 suggests that CHD variants strongly contribute to the pathogenesis of this brain anomaly. Although our data most strongly implicate CHD3 or CHD8 gene variants, our data also suggest that other CHD genes also contribute to CM1 risk. CHD genes share common chromodomain and helicase domains, but structural and functional differences have resulted in three separate subfamilies: subfamily 1 (CHD1 and CHD2), subfamily II (CHD3 and CHD4), and subfamily 3 (CHD5, CHD6, CHD7, CHD8, and CHD9). The main function of the CHD proteins is their ATP-dependent chromatin-remodeling activity, through which they act as either repressors or activators.65 Because chromatin remodeling is crucial during brain development, it is not surprising that the majority of CHD genes have been implicated in neurodevelopmental disorders. De novo variants in or haploinsufficiency of CHD1, CHD2, CHD3, CHD4, CHD7, and CHD8 have been associated with an array of neurodevelopmental phenotypes that include developmental delay, autism spectrum disorder (ASD), facial dysmorphism, macrocephaly, microcephaly, ventriculomegaly, and cleft lip/palate.29,33,57,58,60,64,66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79 Somatic variants in CHD genes have also been implicated in cancer, highlighting their role in cellular proliferation.80 Notably, of the five previously reported individuals with de novo CHD4 mutations, one was diagnosed with CM1, supporting the causal link between CHD variants and CM1. Brain imaging is not standard of care for individuals with isolated autism or neurodevelopmental disorders without concomitant neurological symptoms, however, so CM1 may be present in those previously identified with CHD DNMs.
Because we observed a significant enrichment of protein-altering de novo mutations among individuals with CM1, it is likely that a number of the DNMs that we identified are causative. Among the AD disease genes with DNMs found in CM1 are CHD8, DLX4, GDF5, DSPP, ATXN1, HNRNPK, and HIST1H1E. Mutations in each of these genes are a cause of dominant disease associated with neurodevelopmental disability, facial dysmorphisms, skull abnormalities, or macrocephaly.51 Because only one individual with CM1 in our cohort was diagnosed with these disorders, our data suggest both pleiotropy and phenotypic variability in the phenotypes caused by mutations in these genes. We hypothesize that some of the protein-altering DNMs observed in CM1 (other than those observed in CHD8) may be less damaging and cause less severe disease than has previously been reported, leading to isolated CM1 rather than the full clinical disease. The reduced CADD scores for rare variants in CHD3 in the CM1 subjects in our cohort supports this hypothesis. Our data also suggest the need for widespread genetic testing of cases with early-onset CM1.
The fact that the observed association at CHD3 involved multiple rare missense variants in less constrained regions of CHD3 suggest that they are likely hypomorphic alleles rather than full loss-of-function alleles and likely exhibit incomplete penetrance and variable expressivity. The vast majority of de novo missense variants observed in affected subjects with Snijders Blok-Campeau syndrome were observed within the helicase domains of the protein. Only two variants observed in our CM1 cohort fell within the helicase domains. The majority of variants in CHD3 found among CM1-affected individuals in our cohort were located within the first or last 400–500 bp, regions with no known specific function. Multiple variants in CHD3 (p.Cys17Tyr, p.Arg24Trp, and p.Ala25Val) occur in an alternatively spliced transcript of unknown function that despite its length is expressed as highly as the full-length transcript throughout the brain.81 These variants may display milder phenotypes (i.e., macrocephaly and CM1 rather than Snijders Blok-Campeau syndrome) due to their absence in the canonical full-length transcript. Further work will be required to understand the role of this alternative splice form of CHD3 in protein function and its relationship with disease.
Interestingly, our discovery of the association of CM1 with rare variants in CHD genes, many of which have been associated with macrocephaly, led us to observe that individuals with CM1 who harbor rare variants in CHD genes displayed increased head circumferences. This finding is consistent with previous findings for de novo variants in CHD genes. For example, macrocephaly was also present in 58% of CM1 probands with CHD3 DNMs.60 We previously noted that CM1 is a common feature of syndromes associated with macrocephaly including acromegaly,82 neurofibromatosis type 1,83,84 and overgrowth syndromes.9 Consistent with this hypothesis, 4 out of 65 individuals with chromosome 16p11.2 deletions (MIM: 611913), which is associated with obesity and macrocephaly,85 also had CM1.86 We propose that macrocephaly itself likely predisposes to CM1 in these cases. If the brain grows faster than the skull, the path of least resistance is for the cerebellum to herniate through the foramen magnum, leading to CM1. While some individuals with CM1 have frank macrocephaly, which is defined as a head circumference more than 2 standard deviations above the mean, the head circumferences of most individuals with CM1 are not macrocephalic but are shifted to the right of the bell curve. This work highlights the important potential for variants in known disease genes to contribute to more common, complex inherited human traits like brain volume and CM1.
Our findings with respect to CHD gene variation and CM1 prompted us to test whether macrocephaly was a common feature in our CM1 cohort. We observed an overall increase in head circumference in our entire CM1 cohort, even when we excluded individuals with CHD rare variant, a finding which has not previously been described. Given the prevalence of head circumferences >2 SD above the mean in our cohort, we suggest that increased head circumference is one of the best predictors of CM1 in the general population discovered to date and could be used as a screening tool to potentially identify CM1 before symptom onset, allowing for the possibility of non-surgical or minimally invasive treatment options.
In addition to defining a broader phenotype associated with CHD genes, particularly CHD3 and CHD8, we aimed to characterize the effects of targeted CHD8 disruption by CRISPR in zebrafish. Chd8-null mice die at embryonic stages E8.5–E12.5 and Chd8 heterozygous mice display abnormal corticogenesis and ASD-like behavior.87,88 Previous studies in zebrafish had performed morpholino knockdown experiments and showed increased interorbital distance, suggesting but not conclusively defining the role for the loss of CHD8 as a cause of macrocephaly in zebrafish.58 In particular, morpholinos are notorious for their off-target effects, the study did not thoroughly examine brain morphology, and morpholinos fail to suppress target gene expression for more than the first few days of life. To combat these shortcomings, we created fully heterozygous zebrafish with frameshift insertion/deletion mutations to disrupt protein expression for the life of the fish. Unlike mice, the brains of zebrafish can be monitored in live fish and with much greater detail using fluorescent imaging. Upon high-resolution imaging of chd8+/−, HuC-GFP zebrafish, and wild-type HuC-GFP siblings, we observed stark increases in brain volume in the forebrain, midbrain, hindbrain, and the whole brain of mutant zebrafish without affecting growth of the fish. These data suggest overall brain overgrowth rather than region-specific brain overgrowth is likely the major phenotypic consequence of chd8 disruption in zebrafish and support the role of brain overgrowth as the primary contributing factor to the development of CM1 in individuals with CHD rare variants and for individuals with CM1 who display idiopathic brain overgrowth. This zebrafish model and our ability to detect small differences in brain growth can now be used to further define phenotypic variability and genotype-phenotype associations due to variation in CHD genes.
This study has a number of limitations that we hope to overcome with additional cases, clinical phenotyping, and radiological measurements. First, because we sequenced only 67 CM1 trios, substantial power to detect de novo enrichment will be gained by adding more trios to our cohort. Similarly, additional of a larger CM1 cohort for rare variant association tests will greatly increase our power to detect further associations. Second, although we obtained head circumference measurements on many of the CM1 individuals in this study, some had not been measured. With the evidence from this study demonstrating the role of macrocephaly in CM1 pathophysiology, we hope to better characterize the relationship between brain growth, skull expansion, and CM1 risk, as well as clinical outcomes and best clinical practice for macrocephaly-associated CM1. Next, the true population prevalence of CM1 in the general population is not well characterized. It is possible that some number of individuals within our control cohort have CM1. The prevalence of symptomatic CM1, particularly those requiring surgical treatment, is less that 1/1,000. We estimate, therefore, that there are few symptomatic CM1 individuals within the control subjects used in this study. Our power to detect genetic associations, however, may be decreased due to the presence of CM1 within the control cohort. Lastly, we utilized only individuals of European descent. Individuals of other ethnicities may harbor variants within other genes or pathways that we did not identify due to their exclusion from this study. We hope to expand our cohort to include other underrepresented minority populations in the future to overcome this limitation.
Overall, our results demonstrate an important role for CHD genes in CM1 etiology, an expanded phenotype associated with variants in CHD3, CHD8, and CHD genes as a group, and suggest that excess brain growth contributes more generally to the development of CM1 than previously appreciated. These findings have potential implications for CM1 treatment. Multiple non-neural mechanisms have been proposed to account for CM1, including decreased posterior fossa volume,89, 90, 91, 92 joint laxity near the skull base,93,94 and CSF flow abnormalities.95,96 However, given the genetic data presented here, a primary driver of some forms of CM1 may be overactive neurogenesis. In pediatric CM1-affected subjects where brain overgrowth is observed, but brain and skull expansion is an ongoing process, early intervention may be possible to halt CM1 progression. Further study of these individuals is necessary to understand the natural progression of CM1 caused by brain overgrowth and to understand the best course of treatment for this etiological subset of affected subjects.
Declaration of interests
The authors declare no competing interests.
Published: December 21, 2020; corrected online: January 13, 2021
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.12.001.
Data and code availability
All data supporting the findings of this study are available within the article and its supplemental information files and from the corresponding author upon reasonable request.
Web resources
Genotype Tissue Expression (GTEx) Project, https://www.gtexportal.org/
Online Mendelian Inheritance in Man, https://www.omim.org/
UCSC Genome Browser, http://genome.ucsc.edu/
UniProt database, https://www.uniprot.org/
Supplemental information
References
- 1.Markunas C.A., Soldano K., Dunlap K., Cope H., Asiimwe E., Stajich J., Enterline D., Grant G., Fuchs H., Gregory S.G., Ashley-Koch A.E. Stratified whole genome linkage analysis of Chiari type I malformation implicates known Klippel-Feil syndrome genes as putative disease candidates. PLoS ONE. 2013;8:e61521. doi: 10.1371/journal.pone.0061521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Speer M.C., Enterline D.S., Mehltretter L., Hammock P., Joseph J., Dickerson M., Ellenbogen R.G., Milhorat T.H., Hauser M.A., George T.M. Review Article: Chiari Type I Malformation with or Without Syringomyelia: Prevalence and Genetics. J. Genet. Couns. 2003;12:297–311. doi: 10.1023/A:1023948921381. [DOI] [PubMed] [Google Scholar]
- 3.Urbizu A., Toma C., Poca M.A., Sahuquillo J., Cuenca-León E., Cormand B., Macaya A. Chiari malformation type I: a case-control association study of 58 developmental genes. PLoS ONE. 2013;8:e57241. doi: 10.1371/journal.pone.0057241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelly M.P., Guillaume T.J., Lenke L.G. Spinal Deformity Associated with Chiari Malformation. Neurosurg. Clin. N. Am. 2015;26:579–585. doi: 10.1016/j.nec.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greenberg J.K., Olsen M.A., Yarbrough C.K., Ladner T.R., Shannon C.N., Piccirillo J.F., Anderson R.C., Wellons J.C., 3rd, Smyth M.D., Park T.S., Limbrick D.D., Jr. Chiari malformation Type I surgery in pediatric patients. Part 2: complications and the influence of comorbid disease in California, Florida, and New York. J. Neurosurg. Pediatr. 2016;17:525–532. doi: 10.3171/2015.10.PEDS15369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Godzik J., Kelly M.P., Radmanesh A., Kim D., Holekamp T.F., Smyth M.D., Lenke L.G., Shimony J.S., Park T.S., Leonard J., Limbrick D.D. Relationship of syrinx size and tonsillar descent to spinal deformity in Chiari malformation Type I with associated syringomyelia. J. Neurosurg. Pediatr. 2014;13:368–374. doi: 10.3171/2014.1.PEDS13105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Godzik J., Holekamp T.F., Limbrick D.D., Lenke L.G., Park T.S., Ray W.Z., Bridwell K.H., Kelly M.P. Risks and outcomes of spinal deformity surgery in Chiari malformation, Type 1, with syringomyelia versus adolescent idiopathic scoliosis. Spine J. 2015;15:2002–2008. doi: 10.1016/j.spinee.2015.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Godzik J., Dardas A., Kelly M.P., Holekamp T.F., Lenke L.G., Smyth M.D., Park T.S., Leonard J.R., Limbrick D.D. Comparison of spinal deformity in children with Chiari I malformation with and without syringomyelia: matched cohort study. Eur. Spine J. 2016;25:619–626. doi: 10.1007/s00586-015-4011-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sadler B., Kuensting T., Strahle J., Park T.S., Smyth M., Limbrick D.D., Dobbs M.B., Haller G., Gurnett C.A. Prevalence and Impact of Underlying Diagnosis and Comorbidities on Chiari 1 Malformation. Pediatr. Neurol. 2020;106:32–37. doi: 10.1016/j.pediatrneurol.2019.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Milhorat T.H., Bolognese P.A., Nishikawa M., McDonnell N.B., Francomano C.A. Syndrome of occipitoatlantoaxial hypermobility, cranial settling, and chiari malformation type I in patients with hereditary disorders of connective tissue. J. Neurosurg. Spine. 2007;7:601–609. doi: 10.3171/SPI-07/12/601. [DOI] [PubMed] [Google Scholar]
- 11.Milhorat T.H., Chou M.W., Trinidad E.M., Kula R.W., Mandell M., Wolpert C., Speer M.C. Chiari I malformation redefined: clinical and radiographic findings for 364 symptomatic patients. Neurosurgery. 1999;44:1005–1017. doi: 10.1097/00006123-199905000-00042. [DOI] [PubMed] [Google Scholar]
- 12.Tubbs R.S., Beckman J., Naftel R.P., Chern J.J., Wellons J.C., 3rd, Rozzelle C.J., Blount J.P., Oakes W.J. Institutional experience with 500 cases of surgically treated pediatric Chiari malformation Type I. J. Neurosurg. Pediatr. 2011;7:248–256. doi: 10.3171/2010.12.PEDS10379. [DOI] [PubMed] [Google Scholar]
- 13.Cavender R.K., Schmidt J.H., 3rd Tonsillar ectopia and Chiari malformations: monozygotic triplets. Case report. J. Neurosurg. 1995;82:497–500. doi: 10.3171/jns.1995.82.3.0497. [DOI] [PubMed] [Google Scholar]
- 14.Herman M.D., Cheek W.R., Storrs B.B. Two siblings with the Chiari I malformation. Pediatr. Neurosurg. 1990-1991;16:183–184. doi: 10.1159/000120522. [DOI] [PubMed] [Google Scholar]
- 15.Abbott D., Brockmeyer D., Neklason D.W., Teerlink C., Cannon-Albright L.A. Population-based description of familial clustering of Chiari malformation Type I. J. Neurosurg. 2018;128:460–465. doi: 10.3171/2016.9.JNS161274. [DOI] [PubMed] [Google Scholar]
- 16.Markunas C.A., Enterline D.S., Dunlap K., Soldano K., Cope H., Stajich J., Grant G., Fuchs H., Gregory S.G., Ashley-Koch A.E. Genetic evaluation and application of posterior cranial fossa traits as endophenotypes for Chiari type I malformation. Ann. Hum. Genet. 2014;78:1–12. doi: 10.1111/ahg.12041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lock E.F., Soldano K.L., Garrett M.E., Cope H., Markunas C.A., Fuchs H., Grant G., Dunson D.B., Gregory S.G., Ashley-Koch A.E. Joint eQTL assessment of whole blood and dura mater tissue from individuals with Chiari type I malformation. BMC Genomics. 2015;16:11. doi: 10.1186/s12864-014-1211-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merello E., Tattini L., Magi A., Accogli A., Piatelli G., Pavanello M., Tortora D., Cama A., Kibar Z., Capra V., De Marco P. Exome sequencing of two Italian pedigrees with non-isolated Chiari malformation type I reveals candidate genes for cranio-facial development. Eur. J. Hum. Genet. 2017;25:952–959. doi: 10.1038/ejhg.2017.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duran D., Jin S.C., DeSpenza T., Jr., Nelson-Williams C., Cogal A.G., Abrash E.W., Harris P.C., Lieske J.C., Shimshak S.J., Mane S. Digenic mutations of human OCRL paralogs in Dent’s disease type 2 associated with Chiari I malformation. Hum. Genome Var. 2016;3:16042. doi: 10.1038/hgv.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cushion T.D., Paciorkowski A.R., Pilz D.T., Mullins J.G., Seltzer L.E., Marion R.W., Tuttle E., Ghoneim D., Christian S.L., Chung S.K. De novo mutations in the beta-tubulin gene TUBB2A cause simplified gyral patterning and infantile-onset epilepsy. Am. J. Hum. Genet. 2014;94:634–641. doi: 10.1016/j.ajhg.2014.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guella I., McKenzie M.B., Evans D.M., Buerki S.E., Toyota E.B., Van Allen M.I., Suri M., Elmslie F., Simon M.E.H., van Gassen K.L.I., Epilepsy Genomics Study. Deciphering Developmental Disorders Study De Novo Mutations in YWHAG Cause Early-Onset Epilepsy. Am. J. Hum. Genet. 2017;101:300–310. doi: 10.1016/j.ajhg.2017.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanca O., Andrews J.C., Lee P.T., Patel C., Braddock S.R., Slavotinek A.M., Cohen J.S., Gubbels C.S., Aldinger K.A., Williams J., Undiagnosed Diseases Network De Novo Variants in WDR37 Are Associated with Epilepsy, Colobomas, Dysmorphism, Developmental Delay, Intellectual Disability, and Cerebellar Hypoplasia. Am. J. Hum. Genet. 2019;105:672–674. doi: 10.1016/j.ajhg.2019.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schoch K., Meng L., Szelinger S., Bearden D.R., Stray-Pedersen A., Busk O.L., Stong N., Liston E., Cohn R.D., Scaglia F., UCLA Clinical Genomics Center. Undiagnosed Diseases Network A Recurrent De Novo Variant in NACC1 Causes a Syndrome Characterized by Infantile Epilepsy, Cataracts, and Profound Developmental Delay. Am. J. Hum. Genet. 2017;100:343–351. doi: 10.1016/j.ajhg.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heyne H.O., Singh T., Stamberger H., Abou Jamra R., Caglayan H., Craiu D., De Jonghe P., Guerrini R., Helbig K.L., Koeleman B.P.C., EuroEPINOMICS RES Consortium De novo variants in neurodevelopmental disorders with epilepsy. Nat. Genet. 2018;50:1048–1053. doi: 10.1038/s41588-018-0143-7. [DOI] [PubMed] [Google Scholar]
- 25.Goos J.A.C., Vogel W.K., Mlcochova H., Millard C.J., Esfandiari E., Selman W.H., Calpena E., Koelling N., Carpenter E.L., Swagemakers S.M.A. A de novo substitution in BCL11B leads to loss of interaction with transcriptional complexes and craniosynostosis. Hum. Mol. Genet. 2019;28:2501–2513. doi: 10.1093/hmg/ddz072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ehmke N., Graul-Neumann L., Smorag L., Koenig R., Segebrecht L., Magoulas P., Scaglia F., Kilic E., Hennig A.F., Adolphs N. De Novo Mutations in SLC25A24 Cause a Craniosynostosis Syndrome with Hypertrichosis, Progeroid Appearance, and Mitochondrial Dysfunction. Am. J. Hum. Genet. 2017;101:833–843. doi: 10.1016/j.ajhg.2017.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Timberlake A.T., Furey C.G., Choi J., Nelson-Williams C., Loring E., Galm A., Kahle K.T., Steinbacher D.M., Larysz D., Persing J.A., Lifton R.P., Yale Center for Genome Analysis De novo mutations in inhibitors of Wnt, BMP, and Ras/ERK signaling pathways in non-syndromic midline craniosynostosis. Proc. Natl. Acad. Sci. USA. 2017;114:E7341–E7347. doi: 10.1073/pnas.1709255114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Awadalla P., Gauthier J., Myers R.A., Casals F., Hamdan F.F., Griffing A.R., Côté M., Henrion E., Spiegelman D., Tarabeux J. Direct measure of the de novo mutation rate in autism and schizophrenia cohorts. Am. J. Hum. Genet. 2010;87:316–324. doi: 10.1016/j.ajhg.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Roak B.J., Stessman H.A., Boyle E.A., Witherspoon K.T., Martin B., Lee C., Vives L., Baker C., Hiatt J.B., Nickerson D.A. Recurrent de novo mutations implicate novel genes underlying simplex autism risk. Nat. Commun. 2014;5:5595. doi: 10.1038/ncomms6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Roak B.J., Deriziotis P., Lee C., Vives L., Schwartz J.J., Girirajan S., Karakoc E., Mackenzie A.P., Ng S.B., Baker C. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat. Genet. 2011;43:585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uddin M., Tammimies K., Pellecchia G., Alipanahi B., Hu P., Wang Z., Pinto D., Lau L., Nalpathamkalam T., Marshall C.R. Brain-expressed exons under purifying selection are enriched for de novo mutations in autism spectrum disorder. Nat. Genet. 2014;46:742–747. doi: 10.1038/ng.2980. [DOI] [PubMed] [Google Scholar]
- 32.Iossifov I., O’Roak B.J., Sanders S.J., Ronemus M., Krumm N., Levy D., Stessman H.A., Witherspoon K.T., Vives L., Patterson K.E. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neale B.M., Kou Y., Liu L., Ma’ayan A., Samocha K.E., Sabo A., Lin C.F., Stevens C., Wang L.S., Makarov V. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Roak B.J., Vives L., Girirajan S., Karakoc E., Krumm N., Coe B.P., Levy R., Ko A., Lee C., Smith J.D. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanders S.J., Murtha M.T., Gupta A.R., Murdoch J.D., Raubeson M.J., Willsey A.J., Ercan-Sencicek A.G., DiLullo N.M., Parikshak N.N., Stein J.L. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanders S.J., Ercan-Sencicek A.G., Hus V., Luo R., Murtha M.T., Moreno-De-Luca D., Chu S.H., Moreau M.P., Gupta A.R., Thomson S.A. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krumm N., Turner T.N., Baker C., Vives L., Mohajeri K., Witherspoon K., Raja A., Coe B.P., Stessman H.A., He Z.X. Excess of rare, inherited truncating mutations in autism. Nat. Genet. 2015;47:582–588. doi: 10.1038/ng.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turner T.N., Wilfert A.B., Bakken T.E., Bernier R.A., Pepper M.R., Zhang Z., Torene R.I., Retterer K., Eichler E.E. Sex-Based Analysis of De Novo Variants in Neurodevelopmental Disorders. Am. J. Hum. Genet. 2019;105:1274–1285. doi: 10.1016/j.ajhg.2019.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buchan J.G., Alvarado D.M., Haller G.E., Cruchaga C., Harms M.B., Zhang T., Willing M.C., Grange D.K., Braverman A.C., Miller N.H. Rare variants in FBN1 and FBN2 are associated with severe adolescent idiopathic scoliosis. Hum. Mol. Genet. 2014;23:5271–5282. doi: 10.1093/hmg/ddu224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cruchaga C., Karch C.M., Jin S.C., Benitez B.A., Cai Y., Guerreiro R., Harari O., Norton J., Budde J., Bertelsen S., Alzheimer’s Research UK (ARUK) Consortium Rare coding variants in the phospholipase D3 gene confer risk for Alzheimer’s disease. Nature. 2014;505:550–554. doi: 10.1038/nature12825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krawitz P., Rödelsperger C., Jäger M., Jostins L., Bauer S., Robinson P.N. Microindel detection in short-read sequence data. Bioinformatics. 2010;26:722–729. doi: 10.1093/bioinformatics/btq027. [DOI] [PubMed] [Google Scholar]
- 42.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., Del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics. 2013;43:1–33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 47.Dong C., Wei P., Jian X., Gibbs R., Boerwinkle E., Wang K., Liu X. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum. Mol. Genet. 2015;24:2125–2137. doi: 10.1093/hmg/ddu733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ware J.S., Samocha K.E., Homsy J., Daly M.J. Interpreting de novo Variation in Human Disease Using denovolyzeR. Curr. Protoc. Hum. Genet. 2015;87:1–15. doi: 10.1002/0471142905.hg0725s87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hawrylycz M.J., Lein E.S., Guillozet-Bongaarts A.L., Shen E.H., Ng L., Miller J.A., van de Lagemaat L.N., Smith K.A., Ebbert A., Riley Z.L. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Amberger J.S., Bocchini C.A., Schiettecatte F., Scott A.F., Hamosh A. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43:D789–D798. doi: 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rollins J.D., Collins J.S., Holden K.R. United States head circumference growth reference charts: birth to 21 years. J. Pediatr. 2010;156:907–913.e2. doi: 10.1016/j.jpeds.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 53.Homsy J., Zaidi S., Shen Y., Ware J.S., Samocha K.E., Karczewski K.J., DePalma S.R., McKean D., Wakimoto H., Gorham J. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science. 2015;350:1262–1266. doi: 10.1126/science.aac9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bishop M.R., Diaz Perez K.K., Sun M., Ho S., Chopra P., Mukhopadhyay N., Hetmanski J.B., Taub M.A., Moreno-Uribe L.M., Valencia-Ramirez L.C. Genome-wide Enrichment of De Novo Coding Mutations in Orofacial Cleft Trios. Am. J. Hum. Genet. 2020;107:124–136. doi: 10.1016/j.ajhg.2020.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Samocha K.E., Robinson E.B., Sanders S.J., Stevens C., Sabo A., McGrath L.M., Kosmicki J.A., Rehnström K., Mallick S., Kirby A. A framework for the interpretation of de novo mutation in human disease. Nat. Genet. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Merner N., Forgeot d’Arc B., Bell S.C., Maussion G., Peng H., Gauthier J., Crapper L., Hamdan F.F., Michaud J.L., Mottron L. A de novo frameshift mutation in chromodomain helicase DNA-binding domain 8 (CHD8): A case report and literature review. Am. J. Med. Genet. A. 2016;170A:1225–1235. doi: 10.1002/ajmg.a.37566. [DOI] [PubMed] [Google Scholar]
- 58.Bernier R., Golzio C., Xiong B., Stessman H.A., Coe B.P., Penn O., Witherspoon K., Gerdts J., Baker C., Vulto-van Silfhout A.T. Disruptive CHD8 mutations define a subtype of autism early in development. Cell. 2014;158:263–276. doi: 10.1016/j.cell.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Drivas T.G., Li D., Nair D., Alaimo J.T., Alders M., Altmüller J., Barakat T.S., Bebin E.M., Bertsch N.L., Blackburn P.R. A second cohort of CHD3 patients expands the molecular mechanisms known to cause Snijders Blok-Campeau syndrome. Eur. J. Hum. Genet. 2020;28:1422–1431. doi: 10.1038/s41431-020-0654-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Snijders Blok L., Rousseau J., Twist J., Ehresmann S., Takaku M., Venselaar H., Rodan L.H., Nowak C.B., Douglas J., Swoboda K.J., DDD study CHD3 helicase domain mutations cause a neurodevelopmental syndrome with macrocephaly and impaired speech and language. Nat. Commun. 2018;9:4619. doi: 10.1038/s41467-018-06014-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Porter R.S., Jaamour F., Iwase S. Neuron-specific alternative splicing of transcriptional machineries: Implications for neurodevelopmental disorders. Mol. Cell. Neurosci. 2018;87:35–45. doi: 10.1016/j.mcn.2017.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Battle A., Brown C.D., Engelhardt B.E., Montgomery S.B., GTEx Consortium. Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group. Statistical Methods groups—Analysis Working Group. Enhancing GTEx (eGTEx) groups. NIH Common Fund. NIH/NCI. NIH/NHGRI. NIH/NIMH. NIH/NIDA. Biospecimen Collection Source Site—NDRI. Biospecimen Collection Source Site—RPCI. Biospecimen Core Resource—VARI. Brain Bank Repository—University of Miami Brain Endowment Bank. Leidos Biomedical—Project Management. ELSI Study. Genome Browser Data Integration &Visualization—EBI. Genome Browser Data Integration &Visualization—UCSC Genomics Institute, University of California Santa Cruz. Lead analysts. Laboratory, Data Analysis &Coordinating Center (LDACC) NIH program management. Biospecimen collection. Pathology. eQTL manuscript working group Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. [Google Scholar]
- 63.Haller G., Alvarado D., Mccall K., Yang P., Cruchaga C., Harms M., Goate A., Willing M., Morcuende J.A., Baschal E. A polygenic burden of rare variants across extracellular matrix genes among individuals with adolescent idiopathic scoliosis. Hum. Mol. Genet. 2016;25:202–209. doi: 10.1093/hmg/ddv463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weiss K., Terhal P.A., Cohen L., Bruccoleri M., Irving M., Martinez A.F., Rosenfeld J.A., Machol K., Yang Y., Liu P., DDD Study De Novo Mutations in CHD4, an ATP-Dependent Chromatin Remodeler Gene, Cause an Intellectual Disability Syndrome with Distinctive Dysmorphisms. Am. J. Hum. Genet. 2016;99:934–941. doi: 10.1016/j.ajhg.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hall J.A., Georgel P.T. CHD proteins: a diverse family with strong ties. Biochem. Cell Biol. 2007;85:463–476. doi: 10.1139/O07-063. [DOI] [PubMed] [Google Scholar]
- 66.Pilarowski G.O., Vernon H.J., Applegate C.D., Boukas L., Cho M.T., Gurnett C.A., Benke P.J., Beaver E., Heeley J.M., Medne L. Missense variants in the chromatin remodeler CHD1 are associated with neurodevelopmental disability. J. Med. Genet. 2018;55:561–566. doi: 10.1136/jmedgenet-2017-104759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kulkarni S., Nagarajan P., Wall J., Donovan D.J., Donell R.L., Ligon A.H., Venkatachalam S., Quade B.J. Disruption of chromodomain helicase DNA binding protein 2 (CHD2) causes scoliosis. Am. J. Med. Genet. A. 2008;146A:1117–1127. doi: 10.1002/ajmg.a.32178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Veredice C., Bianco F., Contaldo I., Orteschi D., Stefanini M.C., Battaglia D., Lettori D., Guzzetta F., Zollino M. Early onset myoclonic epilepsy and 15q26 microdeletion: observation of the first case. Epilepsia. 2009;50:1810–1815. doi: 10.1111/j.1528-1167.2009.02078.x. [DOI] [PubMed] [Google Scholar]
- 69.Capelli L.P., Krepischi A.C., Gurgel-Giannetti J., Mendes M.F., Rodrigues T., Varela M.C., Koiffmann C.P., Rosenberg C. Deletion of the RMGA and CHD2 genes in a child with epilepsy and mental deficiency. Eur. J. Med. Genet. 2012;55:132–134. doi: 10.1016/j.ejmg.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 70.Carvill G.L., Heavin S.B., Yendle S.C., McMahon J.M., O’Roak B.J., Cook J., Khan A., Dorschner M.O., Weaver M., Calvert S. Targeted resequencing in epileptic encephalopathies identifies de novo mutations in CHD2 and SYNGAP1. Nat. Genet. 2013;45:825–830. doi: 10.1038/ng.2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Verhoeven W.M., Egger J.I., Knegt A.C., Zuydam J., Kleefstra T. Absence epilepsy and the CHD2 gene: an adolescent male with moderate intellectual disability, short-lasting psychoses, and an interstitial deletion in 15q26.1-q26.2. Neuropsychiatr. Dis. Treat. 2016;12:1135–1139. doi: 10.2147/NDT.S102272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Basson M.A., van Ravenswaaij-Arts C. Functional Insights into Chromatin Remodelling from Studies on CHARGE Syndrome. Trends Genet. 2015;31:600–611. doi: 10.1016/j.tig.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Félix T.M., Hanshaw B.C., Mueller R., Bitoun P., Murray J.C. CHD7 gene and non-syndromic cleft lip and palate. Am. J. Med. Genet. A. 2006;140:2110–2114. doi: 10.1002/ajmg.a.31308. [DOI] [PubMed] [Google Scholar]
- 74.Vissers L.E., van Ravenswaaij C.M., Admiraal R., Hurst J.A., de Vries B.B., Janssen I.M., van der Vliet W.A., Huys E.H., de Jong P.J., Hamel B.C. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat. Genet. 2004;36:955–957. doi: 10.1038/ng1407. [DOI] [PubMed] [Google Scholar]
- 75.Gao X., Gordon D., Zhang D., Browne R., Helms C., Gillum J., Weber S., Devroy S., Swaney S., Dobbs M. CHD7 gene polymorphisms are associated with susceptibility to idiopathic scoliosis. Am. J. Hum. Genet. 2007;80:957–965. doi: 10.1086/513571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.O’Roak B.J., Vives L., Fu W., Egertson J.D., Stanaway I.B., Phelps I.G., Carvill G., Kumar A., Lee C., Ankenman K. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338:1619–1622. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sugathan A., Biagioli M., Golzio C., Erdin S., Blumenthal I., Manavalan P., Ragavendran A., Brand H., Lucente D., Miles J. CHD8 regulates neurodevelopmental pathways associated with autism spectrum disorder in neural progenitors. Proc. Natl. Acad. Sci. USA. 2014;111:E4468–E4477. doi: 10.1073/pnas.1405266111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fujita K., Aida N., Asakura Y., Kurosawa K., Niwa T., Muroya K., Adachi M., Nishimura G., Inoue T. Abnormal basiocciput development in CHARGE syndrome. AJNR Am. J. Neuroradiol. 2009;30:629–634. doi: 10.3174/ajnr.A1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.de Geus C.M., Bergman J.E.H., van Ravenswaaij-Arts C.M.A., Meiners L.C. Imaging of Clival Hypoplasia in CHARGE Syndrome and Hypothesis for Development: A Case-Control Study. AJNR Am. J. Neuroradiol. 2018;39:1938–1942. doi: 10.3174/ajnr.A5810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lamar K.J., Carvill G.L. Chromatin Remodeling Proteins in Epilepsy: Lessons From CHD2-Associated Epilepsy. Front. Mol. Neurosci. 2018;11:208. doi: 10.3389/fnmol.2018.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gamazon E.R., Segrè A.V., van de Bunt M., Wen X., Xi H.S., Hormozdiari F., Ongen H., Konkashbaev A., Derks E.M., Aguet F., GTEx Consortium Using an atlas of gene regulation across 44 human tissues to inform complex disease- and trait-associated variation. Nat. Genet. 2018;50:956–967. doi: 10.1038/s41588-018-0154-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Loukas M., Shayota B.J., Oelhafen K., Miller J.H., Chern J.J., Tubbs R.S., Oakes W.J. Associated disorders of Chiari Type I malformations: a review. Neurosurg. Focus. 2011;31:E3. doi: 10.3171/2011.6.FOCUS11112. [DOI] [PubMed] [Google Scholar]
- 83.Miraglia E., Fabbrini G., Di Biasi C., Iacovino C., Ferrazzano G., Gualdi G., Calvieri S., Giustini S. Chiari type 1 malformation in Neurofibromatosis type 1: experience of a center and review of the literature. Clin. Ter. 2016;167:e6–e10. doi: 10.7417/T.2016.1912. [DOI] [PubMed] [Google Scholar]
- 84.Pozetti M., Belsuzarri T.A., Belsuzarri N.C., Seixas N.B., Araujo J.F. Neurofibromatosis type 1 and Chiari type 1 malformation: A case report and literature review of a rare association. Surg. Neurol. Int. 2016;7(Suppl 16):S469–S472. doi: 10.4103/2152-7806.185778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jacquemont S., Reymond A., Zufferey F., Harewood L., Walters R.G., Kutalik Z., Martinet D., Shen Y., Valsesia A., Beckmann N.D. Mirror extreme BMI phenotypes associated with gene dosage at the chromosome 16p11.2 locus. Nature. 2011;478:97–102. doi: 10.1038/nature10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zufferey F., Sherr E.H., Beckmann N.D., Hanson E., Maillard A.M., Hippolyte L., Macé A., Ferrari C., Kutalik Z., Andrieux J., Simons VIP Consortium. 16p11.2 European Consortium A 600 kb deletion syndrome at 16p11.2 leads to energy imbalance and neuropsychiatric disorders. J. Med. Genet. 2012;49:660–668. doi: 10.1136/jmedgenet-2012-101203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Suetterlin P., Hurley S., Mohan C., Riegman K.L.H., Pagani M., Caruso A., Ellegood J., Galbusera A., Crespo-Enriquez I., Michetti C. Altered Neocortical Gene Expression, Brain Overgrowth and Functional Over-Connectivity in Chd8 Haploinsufficient Mice. Cereb. Cortex. 2018;28:2192–2206. doi: 10.1093/cercor/bhy058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gompers A.L., Su-Feher L., Ellegood J., Copping N.A., Riyadh M.A., Stradleigh T.W., Pride M.C., Schaffler M.D., Wade A.A., Catta-Preta R. Germline Chd8 haploinsufficiency alters brain development in mouse. Nat. Neurosci. 2017;20:1062–1073. doi: 10.1038/nn.4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Musolf A.M., Ho W.S.C., Long K.A., Zhuang Z., Argersinger D.P., Sun H., Moiz B.A., Simpson C.L., Mendelevich E.G., Bogdanov E.I. Small posterior fossa in Chiari I malformation affected families is significantly linked to 1q43-44 and 12q23-24.11 using whole exome sequencing. Eur. J. Hum. Genet. 2019;27:1599–1610. doi: 10.1038/s41431-019-0457-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nwotchouang B.S.T., Eppelheimer M.S., Bishop P., Biswas D., Andronowski J.M., Bapuraj J.R., Frim D., Labuda R., Amini R., Loth F. Three-Dimensional CT Morphometric Image Analysis of the Clivus and Sphenoid Sinus in Chiari Malformation Type I. Ann. Biomed. Eng. 2019;47:2284–2295. doi: 10.1007/s10439-019-02301-5. [DOI] [PubMed] [Google Scholar]
- 91.Neufeld E.A., Menacho S.T., Shah L.M. Craniocervical Junction and Posterior Fossa Dimensions can Affect Need for Decompressive Craniectomy in Posterior Cranial Fossa Hemorrhage. World Neurosurg. 2019;127:e570–e577. doi: 10.1016/j.wneu.2019.03.208. [DOI] [PubMed] [Google Scholar]
- 92.Basaran R., Efendioglu M., Senol M., Ozdogan S., Isik N. Morphometric analysis of posterior fossa and craniovertebral junction in subtypes of Chiari malformation. Clin. Neurol. Neurosurg. 2018;169:1–11. doi: 10.1016/j.clineuro.2018.03.017. [DOI] [PubMed] [Google Scholar]
- 93.Henderson F.C., Sr., Austin C., Benzel E., Bolognese P., Ellenbogen R., Francomano C.A., Ireton C., Klinge P., Koby M., Long D. Neurological and spinal manifestations of the Ehlers-Danlos syndromes. Am. J. Med. Genet. C. Semin. Med. Genet. 2017;175:195–211. doi: 10.1002/ajmg.c.31549. [DOI] [PubMed] [Google Scholar]
- 94.Castori M., Morlino S., Ghibellini G., Celletti C., Camerota F., Grammatico P. Connective tissue, Ehlers-Danlos syndrome(s), and head and cervical pain. Am. J. Med. Genet. C. Semin. Med. Genet. 2015;169C:84–96. doi: 10.1002/ajmg.c.31426. [DOI] [PubMed] [Google Scholar]
- 95.Massimi L., Pennisi G., Frassanito P., Tamburrini G., Di Rocco C., Caldarelli M. Chiari type I and hydrocephalus. Childs Nerv. Syst. 2019;35:1701–1709. doi: 10.1007/s00381-019-04245-6. [DOI] [PubMed] [Google Scholar]
- 96.Piper R.J., Magdum S.A. Chiari 1 malformation and raised intracranial pressure. Childs Nerv. Syst. 2019;35:1719–1725. doi: 10.1007/s00381-019-04232-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within the article and its supplemental information files and from the corresponding author upon reasonable request.