Summary
The ubiquitin-proteasome system facilitates the degradation of unstable or damaged proteins. UBR1–7, which are members of hundreds of E3 ubiquitin ligases, recognize and regulate the half-life of specific proteins on the basis of their N-terminal sequences (“N-end rule”). In seven individuals with intellectual disability, epilepsy, ptosis, hypothyroidism, and genital anomalies, we uncovered bi-allelic variants in UBR7. Their phenotype differs significantly from that of Johanson-Blizzard syndrome (JBS), which is caused by bi-allelic variants in UBR1, notably by the presence of epilepsy and the absence of exocrine pancreatic insufficiency and hypoplasia of nasal alae. While the mechanistic etiology of JBS remains uncertain, mutation of both Ubr1 and Ubr2 in the mouse or of the C. elegans UBR5 ortholog results in Notch signaling defects. Consistent with a potential role in Notch signaling, C. elegans ubr-7 expression partially overlaps with that of ubr-5, including in neurons, as well as the distal tip cell that plays a crucial role in signaling to germline stem cells via the Notch signaling pathway. Analysis of ubr-5 and ubr-7 single mutants and double mutants revealed genetic interactions with the Notch receptor gene glp-1 that influenced development and embryo formation. Collectively, our findings further implicate the UBR protein family and the Notch signaling pathway in a neurodevelopmental syndrome with epilepsy, ptosis, and hypothyroidism that differs from JBS. Further studies exploring a potential role in histone regulation are warranted given clinical overlap with KAT6B disorders and the interaction of UBR7 and UBR5 with histones.
Keywords: UBR5, ubr-5, UBR7, ubr-7, epilepsy, ptosis, hypothyroidism, kat6b, Notch, epigenetic
Introduction
Protein homeostasis requires diverse cellular components to assist with the biogenesis, functional maintenance, and elimination of polypeptides.1 Although molecular chaperones are important for the biogenesis and maintenance of proteins, the ubiquitin-proteasome system carries out the bulk of regulated protein degradation.2 The ubiquitin-proteasome system relies on a small protein degradation tag, ubiquitin, that is first modified by a ubiquitin activating enzyme (E1). The activated ubiquitin is passed on to a ubiquitin-conjugating enzyme (E2). The E2 enzyme then interacts with an E3 ubiquitin ligase that specifically recognizes a protein destined for degradation. The E2-E3 protein complex covalently attaches ubiquitin to its substrate with high specificity after which additional ubiquitins are added (polyubiquitination) and the target protein is degraded by the proteasome.
An important branch of the ubiquitin-proteasome degradation pathway confers the so-called “N-end rule.” This pathway, discovered by Alexander Varshavsky,3 acts on different N-terminal residues of proteins—which are exposed by removal of the initiating methionine—to determine their half-life.2,4 In humans, seven different E3 ubiquitin ligases (“N-recognins”), UBR1–7, mediate target protein recognition. The UBR proteins are implicated in several different biological functions, including neurogenesis, cardiovascular development, histone turnover regulation, and spermatogenesis. They are also linked to pathologies such as cancer and neurodegeneration.5, 6, 7, 8 Thus far, UBR1 (MIM: 605981) and potentially UBR4 (MIM: 609890) are associated with a human genetic disease. Variants in UBR1 within multiple families result in Johanson-Blizzard syndrome (JBS [MIM: 243800]), an autosomal recessive disorder characterized by multiple ailments, including intellectual disability, exocrine pancreatic insufficiency, and facial malformations.9 A missense rare variant in UBR4 co-segregates with dominant non-progressive ataxia without intellectual disability in one family.10 However, CAMTA1 (MIM: 611501) and TMEM240 (MIM: 616101) were subsequently found to fall in the candidate locus. Variants in those genes are associated with dominant ataxia and intellectual disability11 and with progressive spinocerebellar ataxia,12 respectively, making the association of UBR4 with ataxia require further validation. The biological targets of UBR1 relevant to JBS remain unclear, although the combined loss of Ubr1 and Ubr2 in mice points to a defect in Notch signaling as a pathological mechanism.13 Notably, the Notch signaling pathway is important for the correct development and functions of the central nervous system, cardiovascular system, pancreatic system, and skeletal system; it is also implicated in tumorigenesis.14 The C. elegans ortholog of another UBR protein, UBR5, is also directly implicated in Notch signaling.15 UBR proteins may play important roles in other signaling pathways, including Hedgehog (UBR3 and UBR5),16 and some phenotypes of JBS (including situs inversus and heart and skeletal anomalies) suggest a potential link to cilia,17 the pervasive sensory-signaling organelles associated with a growing number of ciliopathies.18
Here, we report on the identification of pathogenic bi-allelic variants in UBR7 (MIM: 613816) in individuals with a phenotype partly overlapping JBS. We show that in cell culture, UBR7 appears to negatively regulate the degradation of an N-end rule substrate. Using C. elegans as a model system, we found that ubr-7 encodes a nuclear protein that is expressed in a variety of cells, including ciliated and non-ciliated neurons and distal tip cells (DTCs), and that this expression pattern partially overlaps with that of the ubr-5 gene. Mutations in ubr-5 and ubr-7 influence the developmental and embryogenesis defects exhibited in a Notch receptor (glp-1) mutant background. Together, our findings uncover variants in UBR7 as a cause of a human disease and reveal an expanded network of functional interactions between UBR proteins in an important development signaling pathway.
Material and methods
Human subjects and sequencing studies
Informed consent for all subjects was obtained in accordance with research protocols that were approved by the institutional review boards at local institutions.
Individual 1 had exome sequencing as described previously.19 Individual 2 had clinical exome sequencing performed at Baylor Genetics Laboratories, as described elsewhere.20 For individual 3, trio exome sequencing was performed at the Institute of Human Genetics in Munich as previously described,21 and we performed subsequent genome sequencing to confirm the deletion. Individuals 4 and 5 had exome sequencing performed on a clinical basis at Centogene. Individuals 6 and 7 had exome sequencing as previously described.22
Splicing analyses
To evaluate the effect of the splice variant identified in individual 3, we cultured some of his fibroblasts by using standard conditions. For cDNA analyses, RNA was extracted and reverse transcribed with poly-A primers. Primers were designed in exons 4 and 7, thus flanking the splice variant. Gel electrophoresis after PCR showed a shorter band, and subsequent Sanger sequencing confirmed that the splice variant leads to skipping of exon 6.
Lymphoblastoid cell line immortalization
Whole blood peripheral blood mononuclear cells (PBMCs) were isolated with a Ficoll-Plaque Plus density gradient following the manufacturer instructions (GE Healthcare). Cells were then washed three times with RPMI, 1% FBS, and transduced with the Epstein-Barr virus (gift for Dr. Carolina Alfeiri).
Immunoblotting on fibroblasts and lymphoblastoid cell samples
Fibroblasts or Epstein-Barr virus (EBV)-immortalized lymphoblastoid cell lines (LCLs) from affected and healthy individuals were lysed in RIPA buffer, and equal amounts of proteins were loaded on an SDS-PAGE gel. The antibodies used were anti-UBR7 (1/1000) from Millipore Sigma (HPA000861) and anti-GAPDH (1/200000) from Santa Cruz Biotechnology (SC-47724).
N-degron pathway analyses
HEK293T cells were cultured in 6-well plate (1.2 × 106 per well) and transfected with either negative control siRNA (Bioneer, 4390843) or siUBR7(Thermo Fisher Scientific, s30283) with Lipofectamine RNAiMAX reagent (Invitrogen, 13778150). Final concentrations of siRNAs were 40 nM. 24 h after siRNA transfection, 2 μg of indicated URT plasmids were transfected to cells via lipofectamine 2000 reagent. 48 h after transfection, cells were treated for indicated chemicals or harvested for immunoblotting. The sequences of pre-designed siRNAs are as follows: siUBR7 (sense, 5′-GCAAGAGACCUUAUCCUGA-3′; antisense,5′-UCAGGAUAAGGUCUCUUGC-3′).
C. elegans strains
All C. elegans strains used (Table S2) were cultured and maintained with standard techniques and at 20°C unless indicated otherwise. EL619 and EL34 were obtained from Dr. Eleanor Maine’s lab (Syracuse University) and the mutant ubr-7(gk3772) was obtained from Moerman Lab and outcrossed six times with N2. GC833 was obtained from the Caenorhabditis elegans Genetics Center (CGC). We used standard mating procedures to generate the double mutant, triple mutant, and some extrachromosomal transgenic strains.
Preparation of C. elegans transgenic constructs and imaging
We generated transcriptional GFP reporter constructs for ubr-5 (F36A2.13) and ubr-7 (T22C1.1) by fusing the ubr-5 promoter (2,038 bp) and ubr-7 promoter (1,722 bp) to the coding sequence of NLS-GFP, respectively. To generate the ubr-7 translational GFP fusion construct, the entire exonic and intronic sequence of ubr-7, along with its native promoter (1,722 bp), was fused in-frame to EGFP. Transgenic lines were generated as reported previously,23 and all images were obtained by spinning-disc confocal microscopy. For DAPI staining, the strain expressing UBR-7::GFP was incubated in 5 μg/mL DAPI solution for 30 min and imaged with a Zeiss LSM 880 Airyscan.
C. elegans body length measurements
Strains including N2, ubr-5(om2), ubr-7(gk3772) and ubr-5(om2); ubr-7(gk3772) were measured for body length. Gravid adult worms were allowed to lay eggs for 60 min and these egg progeny were grown at 20°C for 4 days. Adult worms were then imaged with a Zeiss Axioskop 2+ compound microscope. The length of the worms was measured and plotted with dot plots and boxplots in R software. The distribution of each dataset was determined by the Shapiro-Wilk test. The statistical significance (p value) was calculated with Tukey’s honestly significant difference test.
C. elegans developmental profiling
Developmental profiling assays were performed on the following strains: wild type (WT) (N2), ubr-5(om2), ubr-7(gk3772), ubr-5(om2); ubr-7(gk3772), glp-1(ar202), ubr-5(om2); glp-1(ar202), ubr-7(gk3772); glp-1(ar202) and ubr-5(om2);ubr-7(gk3772);glp-1(ar202). Gravid adult worms were allowed to lay eggs for 60 min and these progeny were cultivated at 20°C for 68 h. We counted younger L3 to L4 larvae on each plate and imaged the remaining adults with a Zeiss Axioskop 2+ compound microscope to score their developmental stage as young adult/vulval eversion,24 mature adult lacking eggs, or gravid adult.
C. elegans egg-hatching assay
Egg-hatching assays were performed on the following strains: glp-1(ar202), ubr-5(om2); glp-1(ar202), ubr-7(gk3772); glp-1(ar202) and ubr-5(om2);ubr-7(gk3772);glp-1(ar202). First, L4 larvae were moved to a 22°C incubator. 24 h later, these P0 worms were killed and the eggs on each plate were counted. Then, the plates were placed back in the 22°C incubator, and after 48 h, the larvae on each plate were counted. The percentage of hatched larvae was calculated and plotted with dot plots and boxplots in R software. The distribution of each dataset was determined by the Shapiro-Wilk test. The statistical significance (p value) was calculated by the Dunn’s Kruskal-Wallis multiple comparisons with Holm-Sidak adjustment.
C. elegans sterility assay
Sterility assays were carried out at the semi-permissive temperature of 22°C15 over a period of 7 days. The strains evaluated were the same as the egg-hatching assay. On day 1, L4 larvae (P0) of each strain were moved to the 22°C incubator. On day 2, these P0 worms were killed. On day 4, the P1 progeny were placed onto individual plates. On day 7, the plates that had hatched P2 larvae were counted. The percentage of plates that had hatched P2 larvae was calculated and plotted with dot plots and boxplots in R software. The distribution of each dataset was determined by the Shapiro-Wilk test. The statistical significance (p value) was calculated with Tukey’s honestly significant difference test.
C. elegans ciliary assays
Fluorescent dye-filling assay was performed as described previously.25 C. elegans worms, WT (N2), ubr-5 single mutant, ubr-7 single mutant, and ubr-5;ubr-7 double mutant were grown at 20°C. L4 larvae were incubated in the lipophilic dye Vybrant DiI (Invitrogen; 1:1,000-fold dilution of 1 mM stock in M9 buffer) for 30 min. Animals were then washed twice with M9 and allowed to roam on regular NGM plates for 1 h to clear intestinal dye. Finally, the stained worms were imaged with a Zeiss Axioskop 2+ compound microscope.
We performed chemotaxis assays by using isoamyl alcohol (1:100 dilution) as an attractant as previously described.26 All strains were tested on 2 separate days (minimum n = 4 assays except for the control che-3 mutant). The chemotaxis index was calculated at 60 min.
Osmotic avoidance assays were performed as previously described.27 The percentage of worms that responded by reversing backward when encountering a high-osmotic barrier ring (60% glycerol with Bromphenol blue) was observed over 10 min. All strains were tested on at least 3 separate days (n > 50 animals).
Results
Variants in UBR7 cause a syndrome overlapping with JBS
We describe seven individuals, aged from 2 to 10 years, from six unrelated families (Table 1, Figure 1, and Supplemental Notes for detailed clinical information). They all demonstrated developmental delay, and all males had urogenital anomalies, namely cryptorchidism in 5/6 and small penis in 1/6. Six individuals had seizures and hypotonia. Hypothyroidism was present in 4/7 individuals, and ptosis was noted in 6/7 individuals. Five individuals exhibited cardiac abnormalities: two had ventricular septal defect, one had atrial septal defect, one had a patent ductus arteriosus requiring surgery, and the other had a patent ductus arteriosus and a patent foramen ovale that both closed spontaneously. Five individuals had short stature (height < 3rd percentile). Physical examination revealed various dysmorphic features, including prominent forehead (3/7), hypertelorism (4/7), telecanthus (1/7), epicanthus(1/7), downslanting palpebral fissures (3/7), thick eyebrow (1/7), low-set ears (3/7), long philtrum (2/7), unilateral single transverse palmar crease (1/7), and hypertrichosis (1/7). Individual 1 was found with hypoplastic patellae and also had gastrointestinal dysmotility. A KAT6B (MIM: 605880) disorder was the initial diagnosis considered in this individual given ptosis, hypertelorism, hypothyroidism, patellar hypoplasia, intellectual disability, short stature, and cardiac and genital anomalies, which all overlap; however, KAT6B sequencing was negative (see “GeneReviews” in Web resources). Individual 7 deceased at 2 years of age from sudden unexpected death in epilepsy (SUDEP).
Table 1.
Summary of clinical features of individuals with bi-allelic UBR7 variants
| Individual | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|
| Variant allele 1 | c.37G>T, p.Glu13∗ | c.914G>C, p.Trp305Ser | c.496−2A>G | c.618delT, p.Glu207Argfs∗12 | c.618delT, p.Glu207Argfs∗12 | c.1186−1G>C | c.1186−1G>C |
| Variant allele 2 | c.564_565dup, p.Cys189Phefs∗14 | c.914G>C, p.Trp305Ser | deletion of exons 1–10 | c.618delT, p.Glu207Argfs∗12 | c.618delT, p.Glu207Argfs∗12 | c.1186−1G>C | c.1186−1G>C |
| Gender | M | M | M | M | M | M | F |
| Age at last examination | 9 years | 17 years | 3 years 9 months | 7 years | 3 years 7 months | 5 years | 1 year 10 months (deceased at 2 years) |
| Short stature | + | + | + | − | − | + | + |
| DD/ID | + | + | + | + | + | + | + |
| Epilepsy | − | + | + | + | + | + | + |
| Hypotonia | − | + | + | + | + | + | + |
| Ptosis | + | − | − | + | + | + | + |
| Hypothyroidism | + | − | + | + | + | − | − |
| Genital anomalies | +a | +b | +b | +b | +b | +b | − |
| Cardiac anomalies | +c | +d | +c,e | − | − | +f | +d |
M, male; F, female; DD, developmental delay; ID, intellectual disability.
Small penis.
Cryptorchidism.
Patent ductus arteriosus.
Ventricular septal defect.
Patent foramen ovale.
Atrial septal defect.
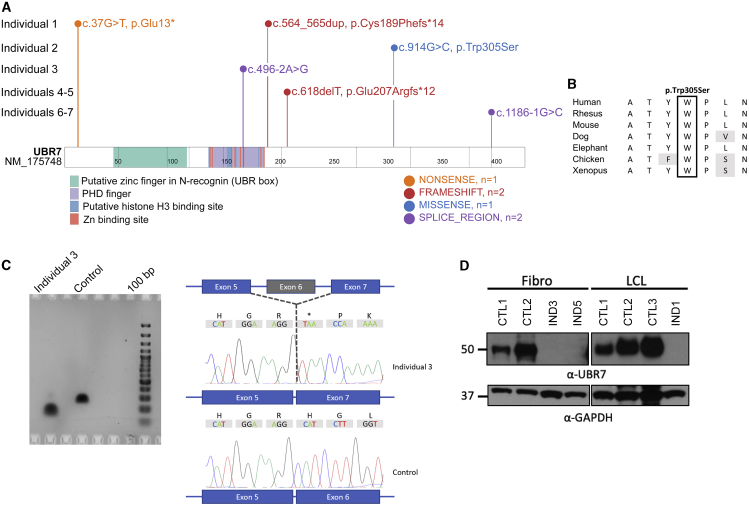
Figure 1.
Pictures and pedigrees of affected individuals
(A) Individual 1 at 5 years of age.
(B) Individual 2 at 10 years of age.
(C) Individual 3 at 9 months of age.
(D) Individual 5 at 3 years of age.
(E) Individual 6 at 5 years of age.
(F) Individual 7 at 2 years of age.
(G) Pedigree of individual 1’s family.
(H) Pedigree of individual 2’s family.
(I) Pedigree of individual 3’s family.
(J) Pedigree of individual 4’s family showing consanguinity loop.
(K) Pedigree of individual 5’s family showing consanguinity loop.
(L) Pedigree of individuals 6 and 7’s family showing consanguinity loop.
Bi-allelic variants in UBR7 were identified in each individual either by exome or genome sequencing. We observed seven distinct variants (Figure 2A). Individual 1 had a nonsense variant (c.37G>T [p.Glu13∗] [GenBank: NM_175748.4]) and a frameshift variant (c.564_565dup [p.Cys189Phefs∗14] [GenBank: NM_175748.4]). Individual 2 was homozygous for a missense variant (c.914G>C [p.Trp305Ser] [GenBank: NM_175748.4]) in a highly conserved region of the protein (Figure 2B). A splice site variant (c.496−2A>G [GenBank: NM_175748.4]) and a deletion encompassing exons 1 to 10 were found in individual 3. Individuals 4 and 5 were homozygous for the same frameshift variant (c.618delT [p.Glu207Argfs∗12] [GenBank: NM_175748.4]) and are both from Saudi Arabian descent, although no relationship could be established between their families. Individual 5 had a cousin also from parents with two loops of consanguinity who displayed brain atrophy. He was not enrolled for the study, so no additional clinical information could be obtained, and he was not tested for a potential familial variant in UBR7. Individuals 6 and 7 were homozygous for a splice site variant (c.1186−1G>C [GenBank: NM_175748.4]).
Figure 2.
Variants identified and their consequences on UBR7 RNA and protein
(A) Schematic representation UBR7 showing the variants identified.
(B) Alignment for residue 305, which is highly conserved among vertebrates.
(C) Fibroblasts from individual 3 (compound heterozygous for a splice variant and a deletion of exons) were cultured, and mRNA was extracted and used to generate complementary DNA. When the cDNA was amplified with primers in exons 4 and 7, exon 6 was shown to be skipped in individual 3’s fibroblasts.
(D) Immunoblotting showed an absence of proteins detected at 55 kDa for individuals 3 and 5 (fibroblasts) and 1 (lymphoblastoid cell line).
The frequency of other loss-of function (LoF) variants in UBR7 in gnomAD is very low, and such variants are never reported at the homozygous state in that database (Table S1).
To provide evidence for the pathogenic potential of the UBR7 variants, we demonstrated that the splicing variant identified in individual 3 led to the skipping of exon 6 (Figure 2C) and that variants in individuals 1, 3, and 5 (for whom cell lines were available) led to nonsense-mediated decay (NMD) and loss of protein (Figure 2). UBR7 immunoblotting on participant cell lines showed an ∼50 kDa-sized band only for the WT cell line (Figure 2D).
UBR7 is a potential negative regulator of the type I/type II N-degron N-end rule pathway
UBR7 is a 48 kDa putative E3 ligase protein with recognizable UBR-box and PHD domains. Previous studies on UBR-box-carrying E3 ligases showed that these act as N-recognins for the degradation of substrates with destabilizing residues. In this N-end rule pathway, arginine (Arg-R) and tyrosine (Tyr-Y) residues represent type I and type II primary destabilizing residues, respectively, whereas methionine (Met-Y) is stabilizing.4 We therefore investigated the possibility that UBR7 participates in the N-end rule pathway.
For these experiments, we used the human embryonic kidney cell line 293T, which expresses UBR7 and allows for efficient manipulation and transfection. To validate the siRNA-mediated UBR7 knockdown in 293T cells, we tested three siRNAs. Of these, siUBR7-1 showed the most efficient knockdown and siUBR7-2 was somewhat less effective (Figure 3A). UBR7 knockdown with siUBR7-1 or siUBR7-2 did not significantly change the level of endogenous RGS4, an N-degron pathway substrate with a cysteine secondary destabilizing residue (Figure 3A). Furthermore, knockdown of UBR7 does not appear to significantly change the cellular ubiquitination of other endogenous proteins as determined by immunoblotting with an FKII antibody that detects monoubiquitinated and polyubiquitinated proteins. These results suggest that UBR7 may not be essential for ubiquitination of N-degron substrates (Figure 3A).
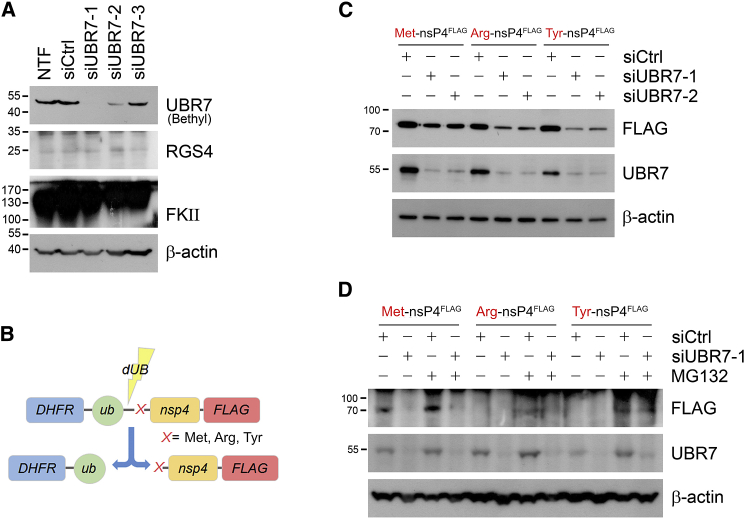
Figure 3.
UBR7 is not a conventional N-recognin
(A) HEK293T cell lines are transfected with Thermo Fisher Scientific pre-designed siRNAs for UBR7 (UBR7-1 targeting exon 4 of NM_175748.3, product ID S30283; UBR7-2 targeting exon 7, S30284; and UBR7-3 targeting exon 9, S30285).
(B) Model N-end rule substrate X-nsp4 processing mechanism involving dubiquitination enzyme.
(C) X-nsp4FLAG(X = Met, Arg, Tyr) constructs are transfected after siRNA-mediated UBR7 silencing.
(D) Same as with (C). but with 10 uM of MG132 treatment for 6 h. Experiments were performed in duplicates with similar results, and representative blots are shown.
We therefore resorted to a ubiquitin reference technique (URT) in which endogenous deubiquitinase (DUB) protein(s) mediate the cleavage of ubiquitin from DHFR-Ub-X-nsp4FLAG to expose either a stabilizing (X = Met; negative control) or a destabilizing type I N-degron (X = Arg) or type II N-degron (X = Tyr) N-terminal residue in X-nsp4FLAG. The experimental approach is shown in Figure 3B. Surprisingly, UBR7 knockdown significantly reduced, rather than increased, Arg- and Tyr-nsP4FLAG levels compared to the negative control (Figure 3C). We confirmed that inhibiting the proteasome with MG132 blocked the degradation of Arg- and Tyr-nsp4FLAG, irrespective of whether UBR7 is silenced or not (Figure 3D).
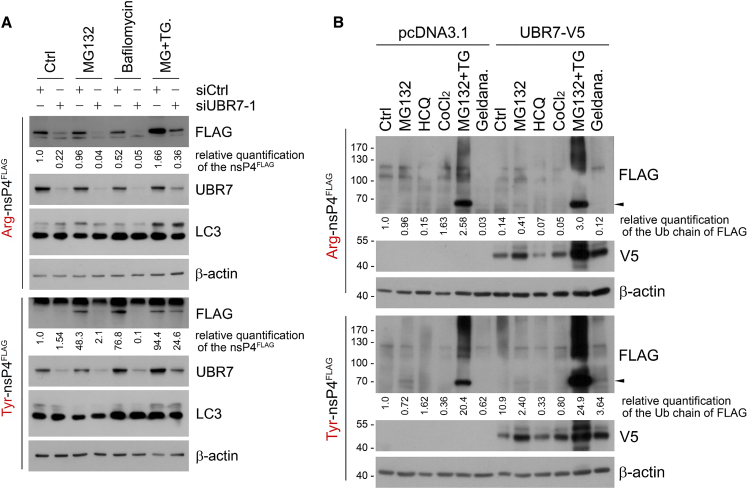
Along with proteasomal inhibition, we induced various proteotoxic stresses on UBR7-silenced cells to query for potential effects on the degradation of the X-nsp4FLAG reporter proteins. We used bafilomycin A1 to inhibit autophagic flux and MG132 + thapsigargin to induce ER stress. Knockdown of UBR7 resulted in reduced levels of Arg- and Tyr-nsp4FLAG, both in control lysates and in lysates from cells treated with the above stressors (Figure 4A). Because UBR1 and UBR2, the major N-recognins for type I and type II N-degrons, are known to compensate for each other, reduced UBR7 levels may have led to over-compensation by other UBR family members. To determine the role of UBR7 as an N-recognin E3 ligase, we overexpressed V5-tagged UBR7 by using plasmid pcDNA6.2-UBR7-V5 in control cells or cells subjected to proteotoxic stresses such as proteasomal inhibition (MG132), HSP90 chaperone inhibition (geldanamycin), oxidative stress (CoCl2), and ER stress (MG132 + thapsigargin). UBR7 was upregulated by proteasomal inhibition and ER stress. Also, both Arg- and Tyr-nsp4 proteins were moderately more poly-ubiquitinated in UBR7-overexpressed ER stress condition, with Tyr-nsp4FLAG levels increased. Therefore, unlike UBR1 and UBR2, UBR7 could be a novel N-recognin and E3 ligase whose function is linked to proteotoxic stress. However, existence of another upstream regulatory mechanism for UBR7 under proteotoxic stress such as misfolded protein stress is also possible because the level of UBR7 itself responds to proteotoxic stress.
Figure 4.
UBR7 could be a proteotoxic stress-specific N-recognin E3 ligase
(A) 200 nM bafilomycin A1, which blocks autophagic flux, is applied for 6 h, and 1 uM MG132 and 200 nM thapsigargin co-treatment, which induces misfolded protein frequency, is applied for 20 h.
(B) UBR7-V5 constructs are transiently expressed and, following stressors, are applied for 24 h except 10 uM of MG132; 25 nM hydroxy chloroquine (HCQ) or 200 nM Bafilomycin A1 for autophagic flux inhibition; 200 μM CoCl2 for oxidative stress induction; 1 μM MG132 and 200 nM thapsigargin co-treatment; and 1.5 μM geldanamycin treatment for misfolded protein inducing stress. Arrowhead indicates X-nsp4FLAG. Experiments were performed in duplicates with similar results, and representative blots are shown.
C. elegans ubr-5 and ubr-7 have partially overlapping expression patterns that include the DTC, involved in Notch signaling
Previous findings revealed a partial functional redundancy between UBR proteins (UBR1 and UBR2) and Notch signaling in the mouse13 and a role for C. elegans UBR-5 in Notch signaling,15 suggesting that multiple UBR proteins may regulate, collectively, this developmental signaling pathway. To provide evidence for this hypothesis, we used C. elegans as a model system to study the potential combined role of UBR-5 and the ortholog of human UBR7 (UBR-7; see Figure S1 for alignment), which is yet to be characterized in this organism, in Notch signaling. In C. elegans, the Notch signaling pathway, which includes the GLP-1 Notch receptor (ortholog of human NOTCH1/2/3 receptor), plays important roles in germline proliferation, cell fate determination, and development.28 We therefore investigated the expression patterns of the ubr-5 and ubr-7 by creating transgenic lines that express GFP from their own respective endogenous promoters (ubr-5p::GFP and ubr-7p::GFP).
The ubr-7 reporter was found to be expressed early in embryonic development, beginning at the gastrulation stage (Figure 5B). In larvae and adults, ubr-7 expression is observed in a variety of cell types, including ciliated and non-ciliated neurons. Notably, strong expression is seen in the DTC (Figure 5B), which is found at the distal ends of the gonad and relies on GLP-1-dependent Notch signaling to regulate the germline stem cell niche.29 The expression pattern for the ubr-5 reporter was found to partially overlap with that of ubr-7. The ubr-5 promoter-GFP construct is similarly expressed in a wide array of non-ciliated and ciliated cell types (Figure 5A). Although weaker than ubr-7, the expression of ubr-5 could be observed in the DTC (Figure 5A).
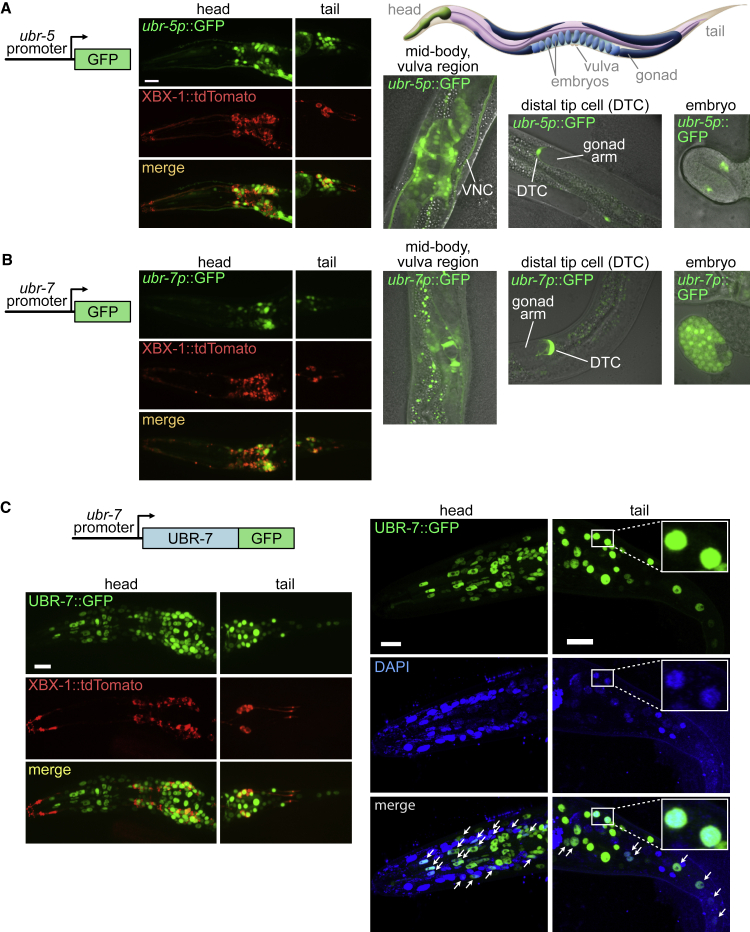
Figure 5.
C. elegans ubr-5 and ubr-7 are broadly expressed in various cell types, including neurons and distal tip cells, and UBR-7 is a nuclear protein
(A and B) Transcriptional (promoter-GFP) reporters of the C. elegans genes ubr-5 (F36A2.13) and ubr-7 (T22C1.1) are expressed in a broad array of cells. ubr-5p::GFP and ubr-7p::GFP reporters (green) are expressed within numerous ciliated and non-ciliated neurons within the head and tail of the animal. The tdTomato-tagged intraflagellar transport (IFT) protein XBX-1 (red) represents a co-marker for amphid (head) and phasmid (tail) ciliated sensory neurons. Expression is observed in the cells around the vulva region and in the ventral nerve cord (VNC) and in the crescent-shaped distal tip cells (DTCs) found at the ends of the gonad arms. Expression of ubr-5p::GFP begins from the 2-fold stage embryo and ubr-7p::GFP from the gastrulation stage embryo onward. Scale bar, 11 μm.
(C) GFP-tagged UBR-7 (UBR-7::GFP; green) expressed from its own promoter localizes within various cell types in the animal (head and tail regions are shown in the left panels). The tdTomato-tagged IFT protein XBX-1 (red) marks the amphid (head) and phasmid (tail) ciliated neurons. Scale bar, 11 μm. UBR-7::GFP co-localizes with nuclei that are stained with DAPI (right panels; arrows and the inset show examples of co-localization). Scale bar, 10 μm.
The broad expression profiles we witnessed for ubr-5 and ubr-7 are consistent with a recent single-cell, high-resolution transcriptome study of C. elegans embryonic development where the two transcripts are detected in a wide array of different cell types, including neuroblasts and ciliated/non-ciliated neurons.30
Together, these findings show that ubr-5 and ubr-7 are expressed in a partially overlapping set of cells and the DTC is one of the well-established cells relevant to Notch signaling.
C. elegans UBR-7 is a nucleus-localized protein
Having established that UBR-7 functions in a variety of cell types, we sought to determine whether the putative E3 ligase demonstrates a distinct subcellular localization, something which has not yet been shown. We therefore created a transgenic strain that expresses GFP-tagged UBR-7 under its own promoter. We used as a reference point a co-marker, namely tdTomato-tagged XBX-1 (human DYNC2LI1), that functions specifically in head (amphid) and tail (phasmid) sensory neurons that have ciliary organelles at the distal ends of their dendrites. We found that UBR7 is present within a subset of ciliated sensory neurons, as well as in a variety of additional (non-ciliated) neurons and other cell types (Figure 5B). No specific localization of UBR7 could be observed within dendrites, axons, or cilia. Instead, UBR7 is concentrated as puncta that are reminiscent of nuclei (Figure 5C, arrowheads).
To determine whether UBR7 is nucleus localized, we incubated the animals with DAPI stain, which marks many but not all cell nuclei (the procedure is not very efficient in worms). This experiment confirmed that UBR7 overlaps with DAPI in many cells, establishing the primary localization of UBR as nuclear.
C. elegans UBR-5 and UBR-7 regulate Notch signaling-dependent developmental timing
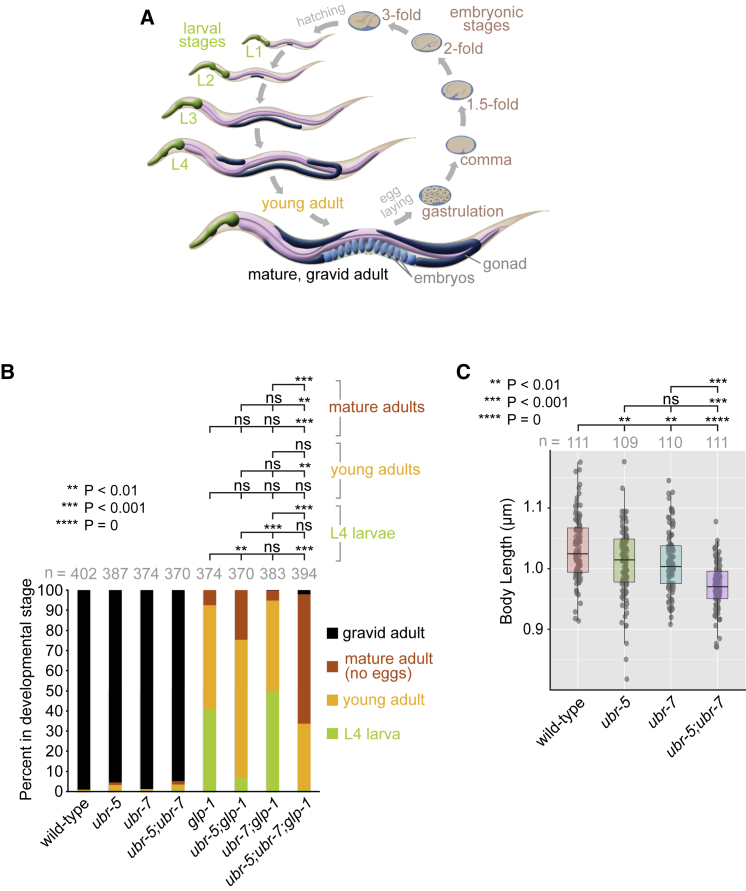
One of the clinical hallmarks of JBS is poor growth. We therefore decided to investigate C. elegans ubr-5 and ubr-7 mutant strains for defects in development. WT animals have a well-characterized developmental progression (Figure 6A). They proceed through embryogenesis, hatch from their eggshell, and progress through four larval stages (L1–L4) interspersed with molting. They then attain a young adult stage and finally a gravid adult stage with a large number of embryos.
Figure 6.
UBR-5 and UBR-7 influence GLP-1 (Notch receptor)-dependent development
(A) Developmental stages of C. elegans hermaphrodite. Embryos continue to develop after being laid, hatch, and proceed through four larval stages (L1–L4) before reaching a young adult stage and then maturing to a gravid adult stage with embryos.
(B) Graph showing the developmental profiles of each indicated strain. Synchronized eggs laid by gravid adults of the indicated strains (WT and mutant) were allowed to develop at 20°C for 68 h. The developmental stages of each strain were then categorized as L4 larva, young adult, mature adult lacking eggs, and gravid adult. The statistical significance (p value) was calculated with Tukey's honestly significant difference test.
(C) Body length measurements of the indicated strains. n, number of animals measured. The statistical significance (p value) was calculated with Tukey's honestly significant difference test. The lower and upper limits of the boxes represent the 25th and 75th percentiles.
To assess the mutants for potential developmental phenotypes, we obtained an available ubr-5 null mutant strain, and using CRISPR-Cas9, we created a ubr-7 mutant that is also likely to be null. A ubr-5;ubr-7 double mutant was also generated to test for potential genetic interactions between ubr-5 and ubr-7. Embryos were staged, and we monitored by microscopy the developmental profiles of the single and double mutants. These mutants did not exhibit any overt developmental delays except for a minor potential delay for the ubr-5 (and ubr-5;ubr-7) mutants in reaching the gravid adult stage (Figure 6B). However, the body lengths of the ubr-5 and ubr-7 mutants are slightly shorter than that of the WT, and the ubr-5;ubr-7 double mutant is significantly different at 0.97 μm versus 1.03 μm in the WT (∼6% shorter; Figure 6C).
Given the functional (genetic) association between UBR-5 and Notch signaling, including the GLP-1 Notch receptor,15 we decided to query for genetic interactions between glp-1 and ubr-5 and/or ubr-7. We confirmed that the glp-1 mutant has developmental defects. Although WT animals have nearly 100% gravid adults after 68 h, glp-1 animals have a high proportion of L4 larvae, young adults, and some mature adults; not one of 374 animals (0%) became gravid (Figure 6B). Further ablation of ubr-7 (ubr-7;glp-1 double mutant) does not alter the glp-1 developmental phenotype. In contrast, the ubr-5;glp-1 double mutant exhibits a statistically significant lower percentage of L4 larvae (Figure 6B). Similar to the glp-1 mutant, none of the ubr-5;glp-1 animals became gravid.
We then tested the ubr-5;ubr-7 mutant in the glp-1 mutant background (ubr-5;ubr-7;glp-1 mutant). Remarkably, this triple mutant strain progressed through development better than the glp-1 and ubr-5;glp-1 mutants: the majority (∼65%) of animals reached a mature stage and the remainder reached a young adult stage (Figure 6B). Furthermore, ∼2% percentage of the animals became gravid. These results are striking; the loss of two conserved UBR proteins in C. elegans does not worsen but rather significantly ameliorates the glp-1 developmental phenotype. Together, our findings reveal a partial rescue of glp-1 developmental phenotypes by the ubr-5 mutant and suggest a potential synergistic role for UBR-5 and UBR-7 in regulating the Notch signaling pathway in C. elegans.
Other glp-1 mutant phenotypes, namely reduced brood size, were not significantly altered when combined with the ubr-5 and/or ubr-7 mutations (Figure S2A). On the other hand, the ubr-7 mutation further reduced the proportion of larvae that hatch in the glp-1 mutant, and the proportion of sterile animals in the ubr-7;glp-1 double mutant was decreased when combined with the ubr-5 mutation (Figures S2A and S2C). Hence, other aspects of GLP-1-dependent functions in Notch signaling may be impacted upon disruption of UBR-5/UBR-7, which may warrant additional studies.
Discussion
The ubiquitin-proteasome system plays pervasive roles in cellular homeostasis by degrading proteins that must be removed in a time-regulated manner, such as cyclins, or are damaged and no longer functional.31,32 It also regulates in eukaryotes the half-life of proteins on the basis of the so-called N-end rule whereby distinct N-terminal residues of proteins confer different levels of instability.4 To achieve a high degree of substrate specificity, the UPS deploys a large number of E3 ubiquitin ligases that interact with specific target proteins destined for degradation (at least 600 are known in humans33). At least seven of these proteins, termed UBR1–7, are believed to act specifically within the N-end rule pathway. Our study shows that variants in UBR7 cause a neurodevelopmental syndrome with epilepsy and hypothyroidism, joining UBR1 as the second UBR to be implicated in a human disorder. Using mammalian cell culture and C. elegans model systems, we provide evidence that UBR7 is a nuclear protein that influences the degradation of an N-end rule substrate and functions together with UBR5 to regulate a Notch signaling-dependent developmental pathway.
A 2011 paper describes homozygosity mapping followed by targeted next-generation sequencing, which revealed variants in 50 novel candidate genes, in 136 consanguineous families.34 In that study, the variant c.371A>G (p.Asn124Ser) (GenBank: NM_175748.4) was identified in three siblings, from first cousin parents, with nonsyndromic autism spectrum disorder and severe intellectual disability. The variant was not homozygous in an unaffected sibling and is absent from gnomAD. This residue is well conserved in vertebrates. No functional studies in cells or model organisms were done for this candidate variant. Although the phenotype appears to be different from that of our cohort, it suggests an important role of UBR7 in neurological development, which is severely affected in the individuals we describe. As there are few details regarding the phenotype of those individuals, it is possible that some features, such as dysmorphisms and thyroid function, were not assessed.
The phenotype of the individuals we describe here presents a minor overlap with JBS, which is an autosomal recessive condition associated with variants in UBR1, encoding for another E3 ubiquitin ligase. Intellectual disability, hypothyroidism, short stature, cardiac anomalies, and genital malformations are common features in both. However, most of our cohort displayed a more severe neurological presentation with epilepsy and hypotonia. Ptosis was noted in four individuals and is not associated with JBS. A single individual also had hypoplastic patellae, which have never been described in JBS. Moreover, exocrine pancreatic insufficiency, which is a striking feature of JBS, was absent in individuals with variants in UBR7. The typical facial dysmorphisms of JBS, namely hypoplastic nasal alae, were not observed either. The pathophysiology underlying hypothyroidism in JBS remains unknown, but its occurrence also in individuals with UBR7 variants suggests that a common pathway is implicated in both conditions.
How different UBR proteins work collectively to regulate the half-lives of cellular proteins displaying different N-terminal residues is not understood because the vast majority of targets are unknown. For example, UBR7 most likely has multiple substrates and therefore influences more than one cellular pathway. Yet the overall effect of a UBR7 loss of function will most likely differ from a complete disruption of any one pathway because the effect will be a less severe extension of protein lifespan. Our work suggests that UBR7 most likely acts on at least some of the same target(s) and cellular pathway(s) as UBR1 because its loss of function causes a disorder with similarities to JBS. Some degree of functional redundancy may exist for mammalian UBR1 and UBR2 where their binding to destabilizing N-terminal residues appears to be very similar.35 Ubr1−/− mice are viable and fertile but are smaller, owing to reduced muscle and lipid content, and exhibit pancreatic dysfunction.36 Ubr2−/− female mice do not survive past embryogenesis, whereas males are viable but infertile because of spermatogenesis defects.35 All Ubr1−/−Ubr2−/− double mutants, however, arrest at midgestation and present with defects in neurological and cardiovascular development. Interestingly, the double mutants show levels of Notch1 that are significantly lower than those of individual knockouts.13 Thus, although these two closely related UBR proteins have different functions, their combined loss has a synergistic effect on the Notch signaling pathway (interestingly, a similar effect is observed on different cyclins).
Given that the C. elegans ortholog of UBR5 was functionally linked to Notch signaling,15 we sought to study the worm counterpart of UBR7 in the context of a potential involvement in this developmental signaling pathway. We found that both C. elegans ubr-5 and ubr-7 are expressed in diverse cell types, including ciliated and non-ciliated neurons, as well as the DTC. The robust expression of ubr-7 in the DTC suggested a role for UBR-7 in Notch signaling. The DTC, present at the ends of gonad arms, is well known to regulate this signaling pathway in a manner that depends on the Notch receptor GLP-1 (orthologous to human NOTCH1/2/3).
The ubr-5 and ubr-7 null mutants exhibit modest body length (developmental) phenotypes, and this developmental defect is more pronounced in the ubr-5;ubr-7 double mutant. Introducing these gene mutations in a sensitized GLP-1 mutant background revealed more prominent functional (genetic) interactions. Compared to the WT, the glp-1 mutant is delayed in its post-embryonic development with a high percentage of L4 larva and young adults compared to mature adults. Furthermore, no embryos are produced in the adults. Addition of the ubr-7 mutant allele to the glp-1 mutant background (ubr-7;glp-1 double mutant) has no effect, but the ubr-5;glp-1 double mutant exhibits an improved developmental profile with more young and mature adults (but still no embryos). Most striking, however, is when mutations in both ubr-5 and ubr-7 are introduced in the glp-1 mutant background (ubr-5;ubr-7;glp-1 triple mutant). Here, more mature adults are observed and a small but significant proportion of adults (2%) reach the gravid stage (i.e., bear embryos), which is not observed in the glp-1 mutant.
Our discovery that UBR-7 (together with UBR-5) is associated with the Notch signaling pathway in C. elegans, similar to that shown for Ubr1 in the mouse, provides further evidence that the developmental anomalies present in the syndrome we described could stem at least in part from disruption of Notch signaling. Consistent with this possibility, Notch signaling plays a major role in development, including neurogenesis, somitogenesis, and vasculogenesis.14,37 The Notch signaling pathway is also important during zebrafish thyroid development,38,39 which provides insights to better understand hypothyroidism in individuals with UBR7 variants.
Interestingly, UBR7 was found to be abundant in human embryonic stem cells (hESCs), whose differentiation depends in part on Notch signaling.40,41 Knockdown of UBR7 in hESCs resulted in changes in the abundance of many different proteins (506), enriched in a multitude of cellular processes. These included biosynthesis of ribonucleosides, energy generation, splicing, ribosome biogenesis, regulation of translation, and DNA repair. However, no definitive links to signaling pathway(s) were found. Nevertheless, the presence of C. elegans UBR-7 within the DTC, which represents the sole stem cell niche in this animal,42 may point to parallel functions in metazoans and nematodes. Given the conservation of UBR7 across non-metazoan species, including plants, slime mold (Dictyostelium discoideum), and some yeast, it will be of interest to determine what other functions this protein may possess that are not related to Notch signaling.
Our analysis of C. elegans UBR-7 revealed that is nuclear localized, suggesting an indirect role in regulating expression rather than a direct influence on the stability of Notch signaling component(s). Consistent with this possibility, two studies identified physical/functional interactions between mammalian UBR7 and histones. Kleiner and colleagues43 uncovered an interaction with Histone 3, the significance of which was not tested. Campos et al.44 found that another histone, H2B, is a target for UBR7-dependent monoubiquitination. In fact, mammalian UBR5, as well as UBR1 and UBR2, is associated with the ubiquitination of histone proteins.45,46 This function may be evolutionarily conserved because the yeast ortholog of UBR1 is implicated in the turnover of an H3-like histone.47 Two additional studies showed the interaction of UBR7 and histone.48,49 The link with histones is especially pertinent given the mentioned clinical overlap with KAT6B disorders (KAT6B is a histone acetyltransferase).
Several manifestations of JBS suggest potential anomalies in the function of primary cilia.17 Primary cilia are implicated in numerous disorders,18 owing to their important roles in several signal transduction pathways, including Hedgehog, receptor tyrosine kinase (RTK), GPCR, and Notch signaling.18,50 Given the expression of ubr-5 and ubr-7 in sensory neurons bearing cilia, we tested these mutants and the double mutant for cilium-associated structural (Figure S3A) and functional defects, including chemotaxis and osmo-avoidance (Figures S3B and S3C). Although we did not uncover obvious phenotypes, more subtle functions for these UBR proteins in cilia and sensory functions may be worth investigating further.
In conclusion, our findings provide evidence for an expanded role of UBR proteins, and consequent mis-regulation of Notch signaling, in a neurodevelopmental syndrome with epilepsy, ptosis, and hypothyroidism. Furthermore, we propose that the analysis of other UBR proteins in vertebrates and mammalian model systems may further implicate Notch signaling as a pathomechanism in JBS or other related disorders. Ultimately, understanding the key targets that are affected by the different UBR proteins should unveil additional roles in cell homeostasis that is relevant to human health.
Data and code availability
This study did not generate datasets or code.
Declaration of interests
J.R. is an employee of the Department of Molecular and Human Genetics at Baylor College of Medicine and Baylor Genetics Laboratories. The Department of Molecular and Human Genetics at Baylor College of Medicine receives revenue for clinical genetic testing completed at Baylor Genetics Laboratory. The other authors declare no competing interests.
Acknowledgments
We are grateful to the Caenorhabditis Genetics Center (CGC), knockout consortium, and Donald Moerman (University of British Columbia, Canada) for C. elegans strains. This study was funded by grants from the Canadian Institutes of Health Research (CIHR grant MOP142243 to M.R.L.) and by the Canadian Rare Diseases: Models and Mechanisms Network (funded by CIHR and Genome Canada). M.R.L. acknowledges a senior scholar award from the Michael Smith Foundation for Health Research (MSFHR). P.M.C. is supported by clinician-scientist awards from the CIHR and the Fonds de recherche du Québec–Santé (FRQS). This work was also supported by the National Research Foundation (NRF) grant funded by the MSIP (NRF-2016R1A2B3011389 and NRF-2020R1A5A1019023 to Y.T.K.).
Published: December 18, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.11.018.
Contributor Information
Michel R. Leroux, Email: leroux@sfu.ca.
Philippe M. Campeau, Email: p.campeau@umontreal.ca.
Web resources
GeneReviews, Lemire, G., Campeau, P.M., and Lee, B.H. (1993). KAT6B Disorders, https://www.ncbi.nlm.nih.gov/books/NBK114806/
gnomAD v2.1.1, https://gnomad.broadinstitute.org/
OMIM, https://omim.org/
Supplemental information
References
- 1.Balchin D., Hayer-Hartl M., Hartl F.U. In vivo aspects of protein folding and quality control. Science. 2016;353:aac4354. doi: 10.1126/science.aac4354. [DOI] [PubMed] [Google Scholar]
- 2.Ravid T., Hochstrasser M. Diversity of degradation signals in the ubiquitin-proteasome system. Nat. Rev. Mol. Cell Biol. 2008;9:679–690. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachmair A., Finley D., Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 4.Tasaki T., Kwon Y.T. The mammalian N-end rule pathway: new insights into its components and physiological roles. Trends Biochem. Sci. 2007;32:520–528. doi: 10.1016/j.tibs.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 5.Shearer R.F., Iconomou M., Watts C.K., Saunders D.N. Functional Roles of the E3 Ubiquitin Ligase UBR5 in Cancer. Mol. Cancer Res. 2015;13:1523–1532. doi: 10.1158/1541-7786.MCR-15-0383. [DOI] [PubMed] [Google Scholar]
- 6.Wang D., Ma L., Wang B., Liu J., Wei W. E3 ubiquitin ligases in cancer and implications for therapies. Cancer Metastasis Rev. 2017;36:683–702. doi: 10.1007/s10555-017-9703-z. [DOI] [PubMed] [Google Scholar]
- 7.Rape M. Ubiquitylation at the crossroads of development and disease. Nat. Rev. Mol. Cell Biol. 2018;19:59–70. doi: 10.1038/nrm.2017.83. [DOI] [PubMed] [Google Scholar]
- 8.Harris L.D., Jasem S., Licchesi J.D.F. The Ubiquitin System in Alzheimer’s Disease. Adv. Exp. Med. Biol. 2020;1233:195–221. doi: 10.1007/978-3-030-38266-7_8. [DOI] [PubMed] [Google Scholar]
- 9.Zenker M., Mayerle J., Lerch M.M., Tagariello A., Zerres K., Durie P.R., Beier M., Hülskamp G., Guzman C., Rehder H. Deficiency of UBR1, a ubiquitin ligase of the N-end rule pathway, causes pancreatic dysfunction, malformations and mental retardation (Johanson-Blizzard syndrome) Nat. Genet. 2005;37:1345–1350. doi: 10.1038/ng1681. [DOI] [PubMed] [Google Scholar]
- 10.Conroy J., McGettigan P., Murphy R., Webb D., Murphy S.M., McCoy B., Albertyn C., McCreary D., McDonagh C., Walsh O. A novel locus for episodic ataxia:UBR4 the likely candidate. Eur. J. Hum. Genet. 2014;22:505–510. doi: 10.1038/ejhg.2013.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thevenon J., Lopez E., Keren B., Heron D., Mignot C., Altuzarra C., Béri-Dexheimer M., Bonnet C., Magnin E., Burglen L. Intragenic CAMTA1 rearrangements cause non-progressive congenital ataxia with or without intellectual disability. J. Med. Genet. 2012;49:400–408. doi: 10.1136/jmedgenet-2012-100856. [DOI] [PubMed] [Google Scholar]
- 12.Delplanque J., Devos D., Huin V., Genet A., Sand O., Moreau C., Goizet C., Charles P., Anheim M., Monin M.L. TMEM240 mutations cause spinocerebellar ataxia 21 with mental retardation and severe cognitive impairment. Brain. 2014;137:2657–2663. doi: 10.1093/brain/awu202. [DOI] [PubMed] [Google Scholar]
- 13.An J.Y., Seo J.W., Tasaki T., Lee M.J., Varshavsky A., Kwon Y.T. Impaired neurogenesis and cardiovascular development in mice lacking the E3 ubiquitin ligases UBR1 and UBR2 of the N-end rule pathway. Proc. Natl. Acad. Sci. USA. 2006;103:6212–6217. doi: 10.1073/pnas.0601700103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siebel C., Lendahl U. Notch Signaling in Development, Tissue Homeostasis, and Disease. Physiol. Rev. 2017;97:1235–1294. doi: 10.1152/physrev.00005.2017. [DOI] [PubMed] [Google Scholar]
- 15.Safdar K., Gu A., Xu X., Au V., Taylor J., Flibotte S., Moerman D.G., Maine E.M. UBR-5, a Conserved HECT-Type E3 Ubiquitin Ligase, Negatively Regulates Notch-Type Signaling in Caenorhabditis elegans. G3 (Bethesda) 2016;6:2125–2134. doi: 10.1534/g3.116.027805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kinsella E., Dora N., Mellis D., Lettice L., Deveney P., Hill R., Ditzel M. Use of a Conditional Ubr5 Mutant Allele to Investigate the Role of an N-End Rule Ubiquitin-Protein Ligase in Hedgehog Signalling and Embryonic Limb Development. PLoS ONE. 2016;11:e0157079. doi: 10.1371/journal.pone.0157079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baker K., Beales P.L. Making sense of cilia in disease: the human ciliopathies. Am. J. Med. Genet. C. Semin. Med. Genet. 2009;151C:281–295. doi: 10.1002/ajmg.c.30231. [DOI] [PubMed] [Google Scholar]
- 18.Reiter J.F., Leroux M.R. Genes and molecular pathways underpinning ciliopathies. Nat. Rev. Mol. Cell Biol. 2017;18:533–547. doi: 10.1038/nrm.2017.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kariminejad A., Ajeawung N.F., Bozorgmehr B., Dionne-Laporte A., Molidperee S., Najafi K., Gibbs R.A., Lee B.H., Hennekam R.C., Campeau P.M. Kaufman oculo-cerebro-facial syndrome in a child with small and absent terminal phalanges and absent nails. J. Hum. Genet. 2017;62:465–471. doi: 10.1038/jhg.2016.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Y., Muzny D.M., Xia F., Niu Z., Person R., Ding Y., Ward P., Braxton A., Wang M., Buhay C. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;312:1870–1879. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wagner M., Osborn D.P.S., Gehweiler I., Nagel M., Ulmer U., Bakhtiari S., Amouri R., Boostani R., Hentati F., Hockley M.M. Bi-allelic variants in RNF170 are associated with hereditary spastic paraplegia. Nat. Commun. 2019;10:4790. doi: 10.1038/s41467-019-12620-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Novarino G., Fenstermaker A.G., Zaki M.S., Hofree M., Silhavy J.L., Heiberg A.D., Abdellateef M., Rosti B., Scott E., Mansour L. Exome sequencing links corticospinal motor neuron disease to common neurodegenerative disorders. Science. 2014;343:506–511. doi: 10.1126/science.1247363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inglis P.N., Blacque O.E., Leroux M.R. Functional genomics of intraflagellar transport-associated proteins in C. elegans. Methods Cell Biol. 2009;93:267–304. doi: 10.1016/S0091-679X(08)93014-4. [DOI] [PubMed] [Google Scholar]
- 24.Seydoux G., Savage C., Greenwald I. Isolation and characterization of mutations causing abnormal eversion of the vulva in Caenorhabditis elegans. Dev. Biol. 1993;157:423–436. doi: 10.1006/dbio.1993.1146. [DOI] [PubMed] [Google Scholar]
- 25.Jauregui A.R., Nguyen K.C., Hall D.H., Barr M.M. The Caenorhabditis elegans nephrocystins act as global modifiers of cilium structure. J. Cell Biol. 2008;180:973–988. doi: 10.1083/jcb.200707090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanders A.A., Kennedy J., Blacque O.E. Image analysis of Caenorhabditis elegans ciliary transition zone structure, ultrastructure, molecular composition, and function. Methods Cell Biol. 2015;127:323–347. doi: 10.1016/bs.mcb.2015.01.010. [DOI] [PubMed] [Google Scholar]
- 27.Jensen V.L., Carter S., Sanders A.A., Li C., Kennedy J., Timbers T.A., Cai J., Scheidel N., Kennedy B.N., Morin R.D. Whole-Organism Developmental Expression Profiling Identifies RAB-28 as a Novel Ciliary GTPase Associated with the BBSome and Intraflagellar Transport. PLoS Genet. 2016;12:e1006469. doi: 10.1371/journal.pgen.1006469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greenwald I., Kovall R. Notch signaling: genetics and structure. WormBook. 2013;17:1–28. doi: 10.1895/wormbook.1.10.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Byrd D.T., Kimble J. Scratching the niche that controls Caenorhabditis elegans germline stem cells. Semin. Cell Dev. Biol. 2009;20:1107–1113. doi: 10.1016/j.semcdb.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Packer J.S., Zhu Q., Huynh C., Sivaramakrishnan P., Preston E., Dueck H., Stefanik D., Tan K., Trapnell C., Kim J. A lineage-resolved molecular atlas of C. elegans embryogenesis at single-cell resolution. Science. 2019;365:eaax1971. doi: 10.1126/science.aax1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Glotzer M., Murray A.W., Kirschner M.W. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- 32.Benanti J.A. Coordination of cell growth and division by the ubiquitin-proteasome system. Semin. Cell Dev. Biol. 2012;23:492–498. doi: 10.1016/j.semcdb.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berndsen C.E., Wolberger C. New insights into ubiquitin E3 ligase mechanism. Nat. Struct. Mol. Biol. 2014;21:301–307. doi: 10.1038/nsmb.2780. [DOI] [PubMed] [Google Scholar]
- 34.Najmabadi H., Hu H., Garshasbi M., Zemojtel T., Abedini S.S., Chen W., Hosseini M., Behjati F., Haas S., Jamali P. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature. 2011;478:57–63. doi: 10.1038/nature10423. [DOI] [PubMed] [Google Scholar]
- 35.Kwon Y.T., Xia Z., An J.Y., Tasaki T., Davydov I.V., Seo J.W., Sheng J., Xie Y., Varshavsky A. Female lethality and apoptosis of spermatocytes in mice lacking the UBR2 ubiquitin ligase of the N-end rule pathway. Mol. Cell. Biol. 2003;23:8255–8271. doi: 10.1128/MCB.23.22.8255-8271.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwon Y.T., Xia Z., Davydov I.V., Lecker S.H., Varshavsky A. Construction and analysis of mouse strains lacking the ubiquitin ligase UBR1 (E3alpha) of the N-end rule pathway. Mol. Cell. Biol. 2001;21:8007–8021. doi: 10.1128/MCB.21.23.8007-8021.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lasky J.L., Wu H. Notch signaling, brain development, and human disease. Pediatr. Res. 2005;57:104R–109R. doi: 10.1203/01.PDR.0000159632.70510.3D. [DOI] [PubMed] [Google Scholar]
- 38.Marelli F., Persani L. Role of Jagged1-Notch pathway in thyroid development. J. Endocrinol. Invest. 2018;41:75–81. doi: 10.1007/s40618-017-0715-x. [DOI] [PubMed] [Google Scholar]
- 39.Porazzi P., Marelli F., Benato F., de Filippis T., Calebiro D., Argenton F., Tiso N., Persani L. Disruptions of global and JAGGED1-mediated notch signaling affect thyroid morphogenesis in the zebrafish. Endocrinology. 2012;153:5645–5658. doi: 10.1210/en.2011-1888. [DOI] [PubMed] [Google Scholar]
- 40.Saez I., Koyuncu S., Gutierrez-Garcia R., Dieterich C., Vilchez D. Insights into the ubiquitin-proteasome system of human embryonic stem cells. Sci. Rep. 2018;8:4092. doi: 10.1038/s41598-018-22384-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu J., Sato C., Cerletti M., Wagers A. Notch signaling in the regulation of stem cell self-renewal and differentiation. Curr. Top. Dev. Biol. 2010;92:367–409. doi: 10.1016/S0070-2153(10)92012-7. [DOI] [PubMed] [Google Scholar]
- 42.Kimble J.E., White J.G. On the control of germ cell development in Caenorhabditis elegans. Dev. Biol. 1981;81:208–219. doi: 10.1016/0012-1606(81)90284-0. [DOI] [PubMed] [Google Scholar]
- 43.Kleiner R.E., Hang L.E., Molloy K.R., Chait B.T., Kapoor T.M. A Chemical Proteomics Approach to Reveal Direct Protein-Protein Interactions in Living Cells. Cell Chem. Biol. 2018;25:110–120.e3. doi: 10.1016/j.chembiol.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Campos E.I., Smits A.H., Kang Y.H., Landry S., Escobar T.M., Nayak S., Ueberheide B.M., Durocher D., Vermeulen M., Hurwitz J., Reinberg D. Analysis of the Histone H3.1 Interactome: A Suitable Chaperone for the Right Event. Mol. Cell. 2015;60:697–709. doi: 10.1016/j.molcel.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.An J.Y., Kim E., Zakrzewska A., Yoo Y.D., Jang J.M., Han D.H., Lee M.J., Seo J.W., Lee Y.J., Kim T.Y. UBR2 of the N-end rule pathway is required for chromosome stability via histone ubiquitylation in spermatocytes and somatic cells. PLoS ONE. 2012;7:e37414. doi: 10.1371/journal.pone.0037414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gudjonsson T., Altmeyer M., Savic V., Toledo L., Dinant C., Grøfte M., Bartkova J., Poulsen M., Oka Y., Bekker-Jensen S. TRIP12 and UBR5 suppress spreading of chromatin ubiquitylation at damaged chromosomes. Cell. 2012;150:697–709. doi: 10.1016/j.cell.2012.06.039. [DOI] [PubMed] [Google Scholar]
- 47.Cheng H., Bao X., Gan X., Luo S., Rao H. Multiple E3s promote the degradation of histone H3 variant Cse4. Sci. Rep. 2017;7:8565. doi: 10.1038/s41598-017-08923-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adhikary S., Chakravarti D., Terranova C., Sengupta I., Maitituoheti M., Dasgupta A., Srivastava D.K., Ma J., Raman A.T., Tarco E. Atypical plant homeodomain of UBR7 functions as an H2BK120Ub ligase and breast tumor suppressor. Nat. Commun. 2019;10:1398. doi: 10.1038/s41467-019-08986-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ji X., Dadon D.B., Abraham B.J., Lee T.I., Jaenisch R., Bradner J.E., Young R.A. Chromatin proteomic profiling reveals novel proteins associated with histone-marked genomic regions. Proc. Natl. Acad. Sci. USA. 2015;112:3841–3846. doi: 10.1073/pnas.1502971112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shearer R.F., Frikstad K.M., McKenna J., McCloy R.A., Deng N., Burgess A., Stokke T., Patzke S., Saunders D.N. The E3 ubiquitin ligase UBR5 regulates centriolar satellite stability and primary cilia. Mol. Biol. Cell. 2018;29:1542–1554. doi: 10.1091/mbc.E17-04-0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate datasets or code.