Abstract
Recently, our understanding of the structural basis of troponin-tropomyosin’s Ca2+-triggered regulation of striated muscle contraction has advanced greatly, particularly via cryo-electron microscopy data. Compelling atomic models of troponin-tropomyosin-actin were published for both apo- and Ca2+-saturated states of the cardiac thin filament. Subsequent electron microscopy and computational analyses have supported and further elaborated the findings. Per cryo-electron microscopy, each troponin is highly extended and contacts both tropomyosin strands, which lie on opposite sides of the actin filament. In the apo-state characteristic of relaxed muscle, troponin and tropomyosin hinder strong myosin-actin binding in several different ways, apparently barricading the actin more substantially than does tropomyosin alone. The troponin core domain, the C-terminal third of TnI, and tropomyosin under the influence of a 64-residue helix of TnT located at the overlap of adjacent tropomyosins are all in positions that would hinder strong myosin binding to actin. In the Ca2+-saturated state, the TnI C-terminus dissociates from actin and binds in part to TnC; the core domain pivots significantly; the N-lobe of TnC binds specifically to actin and tropomyosin; and tropomyosin rotates partially away from myosin’s binding site on actin. At the overlap domain, Ca2+ causes much less tropomyosin movement, so a more inhibitory orientation persists. In the myosin-saturated state of the thin filament, there is a large additional shift in tropomyosin, with molecular interactions now identified between tropomyosin and both actin and myosin. A new era has arrived for investigation of the thin filament and for functional understandings that increasingly accommodate the recent structural results.
Main Text
Striated muscle’s capacity for sharp on-again off-again action, i.e., its twitch, has sparked scientific attention for more than 200 years. An otherwise well-informed scientist might be excused for thinking that this was a long-solved phenomenon. Much is known of course (1), and significant aspects are indeed “solved.” Nevertheless, there has been a huge gap in understanding—the direct structural mechanism of striated muscle inactivation was never explained satisfactorily. Now this gap has narrowed greatly thanks to Yamada et al.’s transformative January 2020 cryo-electron microscopy (cryo-EM) study of cardiac thin filaments (2), aspects of which were promptly supported by excellent independent work (3,4). Substantial further progress is following quickly in Biophysical Journal articles by Lehman and others (5,6) and in preprints (7,8). It is time to take stock of the situation, as this Perspective aims to achieve.
For muscles to relax or, equivalently, remain at rest, the key regulatory factor is the Ca2+-binding thin filament protein troponin. Troponin acts in concert with the coiled-coil protein tropomyosin, to which troponin attaches in a 1:1 stoichiometry (i.e., both are 1:7 relative to actin). Troponin is present universally in striated muscles, where contraction is strictly Ca2+ dependent; troponin and tropomyosin together prevent actin and myosin from producing force and/or movement at a low Ca2+ concentration. Tropomyosin’s direct role in this inhibition was anticipated insightfully decades ago (9,10) and eventually substantiated (11). The 38.5-nm long coiled coil of each tropomyosin molecule tracks along the thin filament’s long pitch helix, where it creates troponin-modulated interference with the myosin-binding site of seven successive actins. Although tropomyosin interferes with the myosin-binding site, tropomyosin alone is insufficient for shutting off muscle contraction. Troponin is required. If troponin is removed from skeletal or cardiac muscle experimentally and tropomyosin remains, muscles contract (12). Correspondingly, the relaxation of smooth muscle, which lacks troponin, requires a robust substitute—an off switch involving dephosphorylation of myosin light chains.
The above discussion then prompts the question that has been lingering for decades—what is the structural mechanism of troponin’s inhibitory action, via tropomyosin or otherwise? Until now, troponin has been an exasperating Second Foundation of thin filament-mediated regulation, known to exist and known to be critical but unknown as to location and methods (13). The recent publications culminate at last with the long worldwide effort to achieve structural understanding of troponin action. Looking back, one cannot but note three high points of the research journey, summits reached by successive generations of scientists working in Japan. In autumn 2001, an international conference at SPring-8 celebrated the 40th anniversary of the discoveries by Ebashi and colleagues of troponin and of muscle contraction as the first calcium ion-carried intracellular signal transduction system (14, 15, 16, 17, 18). There, in a fitting newer achievement, Maeda and associates described the first atomic resolution structure of troponin’s core domain (19). Now, another two decades on, Yamada, Namba, and Fuji (2) of Osaka University have shown the regulated cardiac thin filament structure in action by cryo-EM of actin-tropomyosin-troponin filaments in the absence and presence of Ca2+. Uniquely for reports to date, the study achieved near atomic resolution (4.8–6.6 Å), sufficient to place compelling atomic models of most of troponin into the maps. At last, the structural basis of the regulatory switch emerges more fully into the light, with troponin revealed in situ.
Troponin as barricade
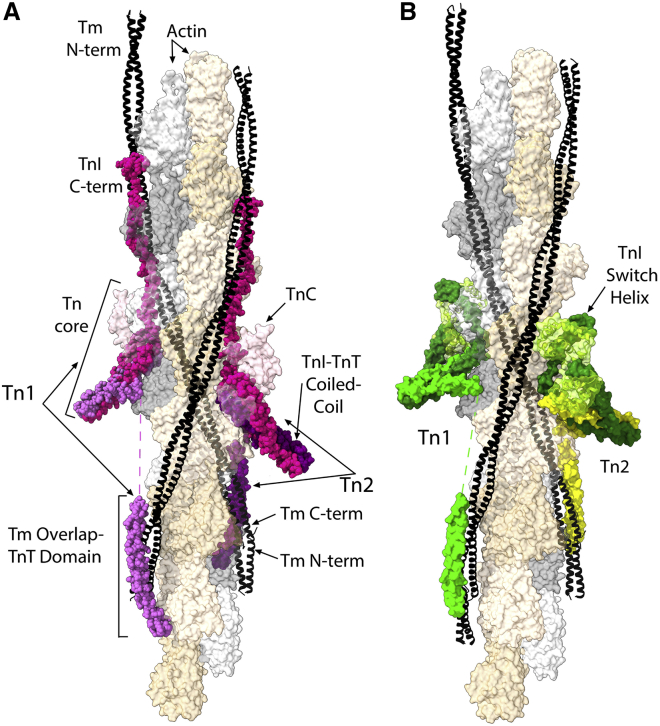
The low Ca2+ concentration structure of the thin filament, characteristic of fully relaxed muscle, is shown in Fig. 1 A. Troponin’s three subunits (20) are colored purple (TnT), magenta (TnI), and light pink (TnC). This is the apo-state of the thin filament; neither Ca2+ nor myosin is bound. To help distinguish the two troponins in the image, the TnT for Tn1, to the left, is colored a brighter purple than the TnT of Tn2, to the right. As the figure shows, the troponin span is large. One troponin or the other directly influences 13 of the 14 actins in the structural unit of two actin strands. No wonder muscles do not produce force under low Ca2+ conditions. Troponin and tropomyosin together form quite a set of barricades against myosin. At bottom, a helix of the troponin “tail” (the N-terminal portion of TnT) overlies the head-to-tail overlap of successive tropomyosins, fortifying tropomyosin’s attachment to an inhibitory position on actin. In the midportion, the core domain of troponin itself barricades two actins from myosin, like a truck parked to protect a national shrine. At the top, the highly extended C-terminal third of TnI stretches along tropomyosin, adding to tropomyosin’s steric hinderance of strong myosin binding to those actins. The size and stoichiometry of the thin filament have always implied a remarkable allostery; there is only one troponin for every seven actins. Underlying this action at a distance is a large wingspan for troponin as well as for tropomyosin.
Figure 1.
Atomic models of the cardiac thin filament in the apo-state (A) and Ca2+-saturated state (B). Yamada, Namba, and Fuji conducted cryo-EM and determined PDB: 6KN7 and 6KN8, here illustrated (2). Actin (gray and brown) is shown partially transparent, so troponin (Tn; purples (A) and greens (B)) and tropomyosin (Tm; black) can be seen behind as well as in front. (A) The C-terminal third of subunit TnI (magenta) extends adjacent to Tm along two actins toward the pointed end of the filament (at top). (B) This adjacent density is absent, and the smaller, switch helix segment of TnI (dark green) is attached to subunit TnC (lime). The troponin toward the left in (A and B) (Tn1) spans a distance longer by one actin monomer than the span of Tn2 on the right. The model for each Tn has a gap in subunit TnT, where a linker region is detected at low contour. To see this figure in color, go online.
The smallest repeating structural unit of the regulated thin filament includes both of the two actin strands and contains 14 actins, 2 tropomyosins, and 2 troponins. Note that the two opposing troponins are not the same; Tn1 spans a distance along the length of the actin filament that is one actin longer than the span of Tn2. Also, each troponin contacts both actin-tropomyosin strands. Each such strand is a curving, long pitch helix that one can perceive in the figure via the path of the tropomyosin coiled coil (black). The Tn1 troponin core domain at midleft and the Tn1 TnT helix, which lies directly below, are on opposite strands of the actin filament. Very likely, it is correct that these are parts of the same troponin; lower contour electron microscopy images (2) show intervening density. The connecting linker (represented in Fig. 1 by a dashed line) is of an unknown atomic structure and spans a shorter distance for Tn2 than for Tn1. It has low density in the deposited electron microscopy maps, and a different view may yet be offered by others, including those who have examined the TnT connecting linker previously by computational methods (21).
Electron microscopy consistently demonstrates that troponins lie in register on opposite sides of the filament, not only in situ but also in reconstituted thin filaments in which neither actin capping proteins nor Z-disk structures can explain the alignment (2,22,23). Correspondingly, the tropomyosins have been known to lie in register. Now, a new explanation is evident—they must be in register for troponin to attach to both tropomyosins. Furthermore, the resulting structure is more robust than was understood heretofore. Troponin with its dual binding sites gives cross-bracing to the regulated thin filament. Whatever else may result from this feature, it is part of how the regulated thin filament maintains full assembly. Troponin holds onto actin-tropomyosin via both regions, with high affinity, ∼30 nM KD or better, regardless of Ca2+ and myosin (24).
Finally, it is worth noting that the Yamada et al. (2) and Oda et al. (4) cryo-EM studies share several accomplishments, including the placement and orientation of the core domain on the filament (Fig. 1). This is critical for any structural understanding of troponin function. Also critical is the identification of as much of troponin as possible, which was achieved more successfully by the higher resolution Yamada et al. (2) report that is the focus of this article.
The on-off switch Ca2+-mediated change in the troponin core domain
Fig. 1 B shows the thin filament in the same orientation as Fig. 1 A but in the Ca2+-saturated state. The most prominent difference is the disappearance of the extended TnI C-terminal segment. Instead, a much smaller subsegment, called the TnI switch helix (TnI here in dark green), attaches to TnC. The remainder of the TnI C-terminus is no longer seen, but significantly, it no longer lies along tropomyosin in a position that, with tropomyosin, sterically hinders myosin. This change in the TnI C-terminus is the most dramatic alteration within the whole structure, and no doubt, it is pivotal to the ability of Ca2+ to regulate muscle contraction.
Although the atomic structure and interactions of the TnI C-terminus are beyond the present resolution, its identification in the cryo-EM map changes our understanding of regulatory function. First, its steric hindrance of myosin is similar to that of tropomyosin; both must move for strong myosin binding to occur, making both relevant structure and dynamics. Also, the cryo-EM density indicates a predominantly ordered rather than disordered structure in the thin filament apo-state. Human cardiomyopathy genetic data support the view that the C-terminus density reported by Yamada et al. (2) is real, rather than a selection artifact of cryo-EM data processing. At least 15 of TnI’s 40 C-terminal residues have one or more pathogenic missense substitution, according to tabulated data of consortia publications (25,26) applying rigorous clinical genetics criteria. This suggests the TnI C-terminus has an ordered structure, which is dynamic and readily perturbed by single amino acid substitutions.
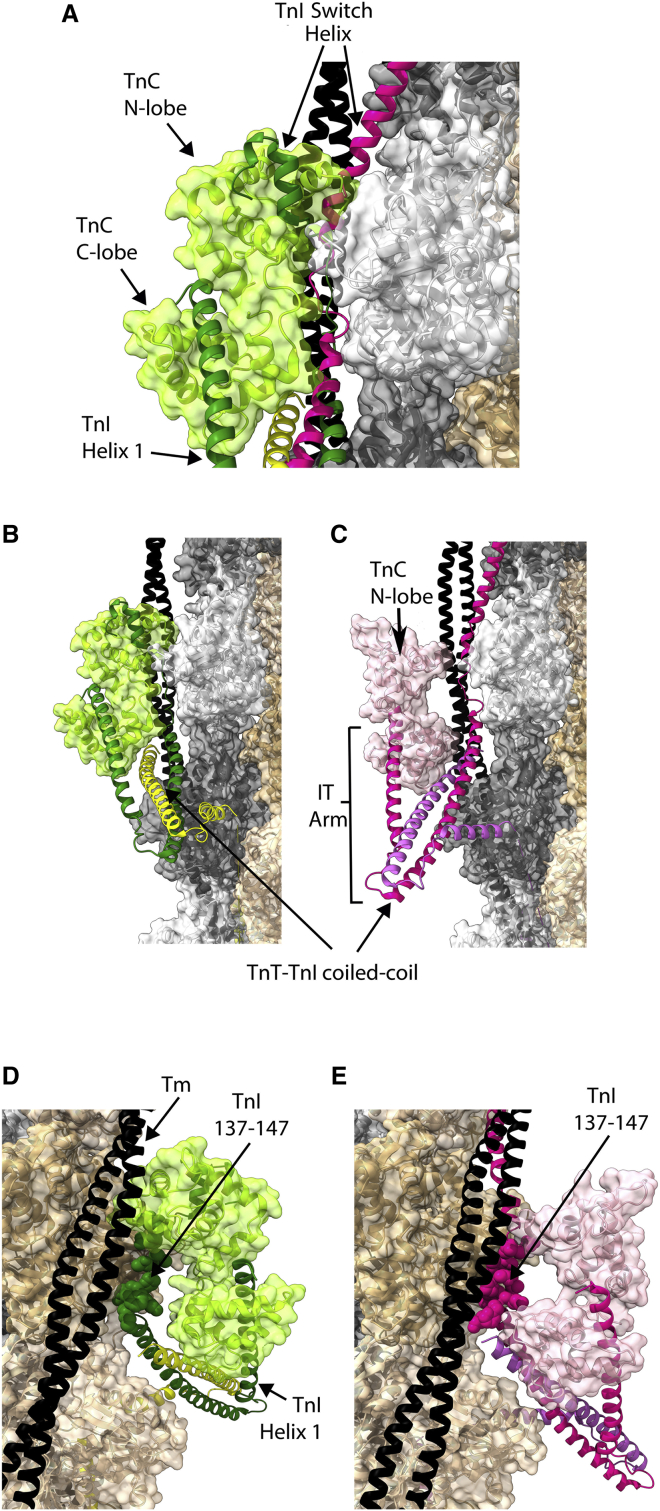
The cryo-EM results indicate that the regulatory mechanism is multifaceted, with several features previously unknown. Fig. 2 A illustrates the primary regulatory switch via superposition. The Ca2+-saturated structure is shown with TnC (light green) in transparent surface format and with TnI (dark green) and TnT (yellow) in cartoon format. Apo-state TnI (magenta) is superimposed. The TnC N-lobe can open upon Ca2+ binding to its EF-hand motif to produce a binding surface for the TnI switch helix (27,28), and TnI switches from one position to the other. Perhaps the TnI switch segment oscillates on and off the thin filament, and if the N-lobe is primed by Ca2+, the switch helix attaches, pulling the rest of the TnI C-terminus off actin. From the cryo-EM findings, however, a different possibility occurs—that a mobile TnC N-lobe effects regulation directly—nudging TnI off the actin surface while also nudging tropomyosin to pivot away from its apo-state orientation. In either case, the switch occurs right at the thin filament surface, where the TnI shift from actin to TnC is a relatively modest distance, as would seem facilitating for rapid, efficient activation.
Figure 2.
Troponin core domain. (A) Shown is the Ca2+-saturated structure plus superimposed TnI as it is found in the apo-state. The regulatory mechanism involves, prominently, a helical segment of TnI’s C-terminus switching its attachment from actin to the TnC N-lobe. Note that the position on actin and tropomyosin of the Ca2+-bound TnC N-lobe is incompatible with TnI retaining its magenta, apo-state location. (B and C) Shown is an identical view comparison of Ca2+- and apo-states. Note changes in tropomyosin, the TnC N-lobe, and the IT arm that includes the TnT-TnI coiled coil. (D and E) Shown is the core domain, as viewed from the opposite side of the filament. Note TnI 137–147 (atoms format) has inhibitory effects as an isolated peptide. Unexpectedly, it has a close proximity to actin, regardless of Ca2+. Color scheme is the same as in Fig. 1. To see this figure in color, go online.
The TnC N-lobe is not fixed within the core domain and is capable of independent rotation (19,29). In a potentially critical finding, the N-lobe nods very closely onto the thin filament upon Ca2+ binding into a position that displaces TnI from the actin surface (Fig. 2 A). Here, the Ca2+-saturated N-lobe forms extensive interactions with actin and with both helices of tropomyosin. Based on the cryo-EM map, the appositions are extensive, specific, and likely fundamental. The N-lobe appears to serve as an active controller of contraction, not just as a mechanism for relieving inhibition by TnI. Finally, comparison between troponin in the apo-state and the Ca2+-saturated state indicates that, although the TnI C-terminus dissociates from actin upon Ca2+ binding, the overall pattern is not one of dissociation. Rather, one set of troponin interactions is replaced by another (30).
Fig. 2, B and C appear quite different from each other, although they are images from the same viewpoint. The difference is that troponin moves considerably between the Ca2+-saturated state (Fig. 2 B) and the apo-state (Fig. 2 C). The TnC N-lobe pivot toward actin, and the TnI dissociation from actin, discussed above, is easily seen by comparison of the two panels. Very prominent is a ∼30° rotation in what is called the “IT arm” of the core domain, which is comprised of a 46 residue TnI-TnT coiled coil and the TnC C-lobe to which long TnI helix 1 is closely attached. These elements shift together as a unit. This finding was anticipated by a 2012 study employing in situ polarized fluorescence in skeletal muscle (31). The spectroscopic report identified the IT arm orientation on the actin filament correctly and showed rotation in the presence of Ca2+. Although the magnitude of the cryo-EM rotation is larger than that detected by in situ fluorescence, this can be explained by geometric constraints; the studies appear to agree.
Something causes the IT arm to rotate when Ca2+ binds to the TnC N-lobe, but the mechanism is unclear. Candidate possibilities can be delineated from paired Fig. 2, D and E, showing a view from the opposite side of the filament. Tropomyosin is not the same in the two panels, so tropomyosin movement is a potential cause of the IT arm rotation. Also, TnI helix 1 (running vertically at right) reaches and contacts the TnC N-lobe in both panels. Finally, note the segment of TnI that immediately precedes the switch helix and is shown in atoms format. This is the classical inhibitory region, TnI 137–147 (32), which has inhibitory effects as an isolated peptide, is a cardiomyopathy site, and has been studied by many ((33,34) and references therein). Surprisingly, it remains anchored to actin; regardless of Ca2+, it does not detach. Near both tropomyosin and other portions of the core domain, it may influence more than one thin filament state, consistent with prior observations.
Tropomyosin: E Pur Si Muove
The cryo-EM map resolution reveals individual helices of the tropomyosin coiled coil. For greater detail, proposed atomic models of full-length tropomyosin within the filament cryo-EM envelope are now available from more than one source. In addition to the EGTA and Ca2+ models put forth by Yamada et al. (2) in January 2020, computationally derived alternates were published in July 2020 (5). These latter models have the appeal of greater adherence to canonical coiled-coil measurements, which result in slightly greater length. They remain entirely consistent with the Yamada et al. (2) cryo-EM map, with which they share a great similarity. A significant difference is residue register along the thin filament, which is critical for tropomyosin’s detailed interactions with actin and troponin and with myosin as well.
The overall effect of Ca2+ on tropomyosin is the same in the hands of either group of investigators. Tropomyosin’s center of mass shifts slightly across the thin filament, as found in most past reconstructions. Unexpectedly, this shift is diminished near the overlap domain (see below). Also, now that both of tropomyosin’s two helices are each clearly identified by the cryo-EM data, it’s evident that most of tropomyosin’s Ca2+-induced shift in position is due to rotation rather than translation. Note the tropomyosin difference in Fig. 2, D and E; the apparent two-dimensional crossing point of the coiled-coil changes. As described below, this is because one helix stays relatively fixed at the point of closest actin contact, and its partner helix rocks, as if there were a hinge anchored near the actin surface. Yamada et al. (2) comment that this motion diminishes tropomyosin’s steric hindrance of myosin-actin attachment.
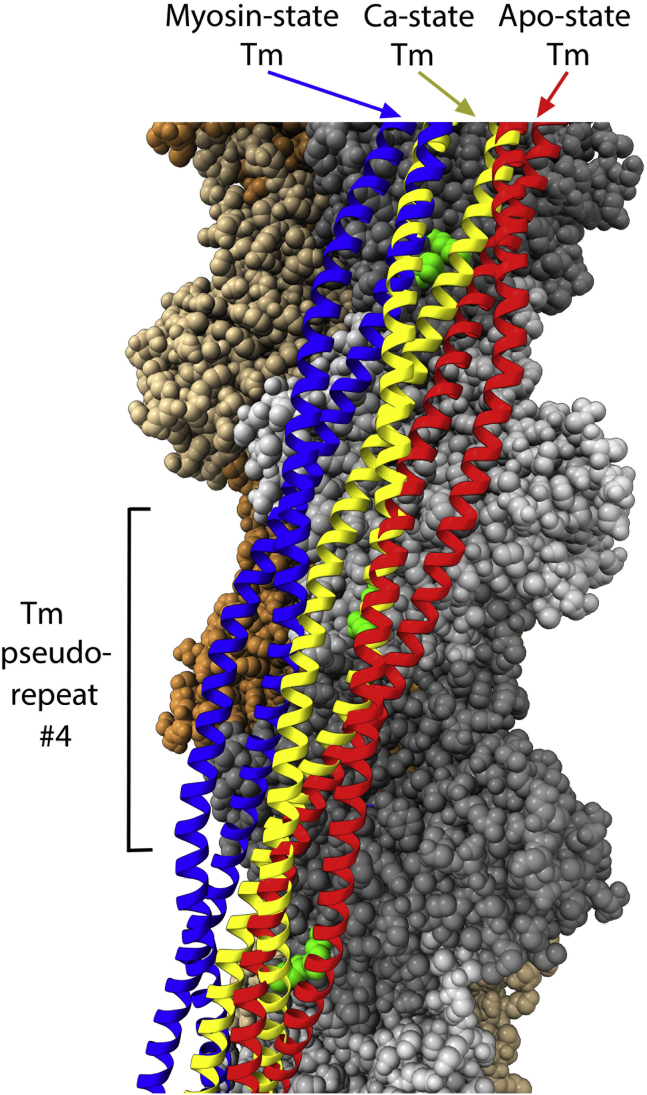
The computational docking study of Pavadai et al. (5) agrees with the above findings, identifying two tropomyosin poses, prepivot, or postpivot. The calculations also identify specific tropomyosin-actin contacts that are consistent along the pseudorepeat structure of tropomyosin. In Fig. 3, tropomyosins in the EGTA and Ca2+ models are shown in red and yellow, respectively. Green-highlighted actin Lys residues 326 and 328 are key tropomyosin interaction sites. On the actin monomer that contacts the fourth of tropomyosin’s seven quasiequivalent regions, these residues are seen to lie near a tropomyosin helix in both yellow and red. The other helix, which is further from this point on the actin surface, pivots from the right side to the left when Ca2+ is added. Tropomyosin consequently clashes less with actin’s myosin-binding site in the Ca2+-saturated filament compared with the apo-state.
Figure 3.
Middle regions of tropomyosin in the three filament end states. Shown are apo-state tropomyosin in red (2), Ca2+-saturated state in yellow (5), and myosin-saturated state in blue (6). Note the proximity between tropomyosin and interacting actin residues Lys 326 and 328 (green). Ca2+ binding to troponin (data not shown) causes tropomyosin to pivot, which exposes more of the site where myosin binds strongly to actin. Best illustrated here for the fourth of tropomyosin’s seven pseudorepeats (but also occurring at repeats 2, 3, and 5), one tropomyosin helix remains largely in place near the actin lysines regardless of Ca2+, whereas the other helix pivots from right to left. When myosin (data not shown) is attached to actin, tropomyosin migrates to a distinct, third location on the filament (blue). To see this figure in color, go online.
Tropomyosin adopts a third position on actin, distinct from those described above, in the presence of strongly bound myosin. To investigate the atomic structure in this circumstance, Doran et al. (6) performed cryo-EM of thin filaments comprised of cardiac tropomyosin, the cardiac myosin isoform, and skeletal muscle actin and also employed computational methods. This August 2020 report (6) showed that the central region of tropomyosin shifts 15° around the filament, a 10 Å movement compared with its position in the presence of troponin and Ca2+. In this myosin-saturated state of the thin filament, tropomyosin (blue in Fig. 3) has translated left, compared with the Ca2+-saturated state (yellow), with little evident rotation. In this position but not the states shown in red or yellow, both myosin (data not shown) and tropomyosin can attach simultaneously to actin without steric conflict. Furthermore, the atomic model identifies specifics of binding. All up and down its length, tropomyosin has close interactions with successive actins and myosins, including consistent contact with actin Lys326 and myosin Arg369. Tropomyosin’s atomic contacts with actin and also myosin (first shown in (35)) are thus identified and can be tested experimentally. The success of the overall approach offers promise for future understanding of tropomyosin’s dynamic, Ca2+-regulated interactions with troponin and with other proteins such as myosin-binding protein C.
In the Yamada et al. (2) structure, Tn1 and Tn2 (Fig. 1) have different spans, but their core domains attach to the same tropomyosin region: pseudorepeat 4 of tropomyosin’s seven quasiequivalent regions (36) (each repeat interacts with a successive actin monomer along the long pitch actin strand). Consider a counterfactual in which both Tn1 and Tn2 have Tn1’s longer longitudinal span. In this scenario, Tn2’s core domain would be attached to a different part of tropomyosin: pseudorepeat 3. Instead, both Tn1 and Tn2 bind to repeat 4, implying specificity. The unknown atomic details of tropomyosin-core domain interactions are an intriguing topic for future investigation. In prior, broader work, activation by Ca2+ is reduced 60% if pseudorepeat 4 is deleted experimentally (37). Furthermore, thin filaments completely resist Ca2+ activation when pseudorepeats 3 and 4 are internally deleted from tropomyosin, but actin-myosin regulation is barely altered if pseudorepeats 2 and 3 are deleted (38,39). Specific interactions of pseudorepeat 4, which binds to the Ca2+-saturated TnC N-lobe, may stabilize the pivoting of tropomyosin’s middle region that is important to regulation.
The overlap domain: an anchor and inhibitor
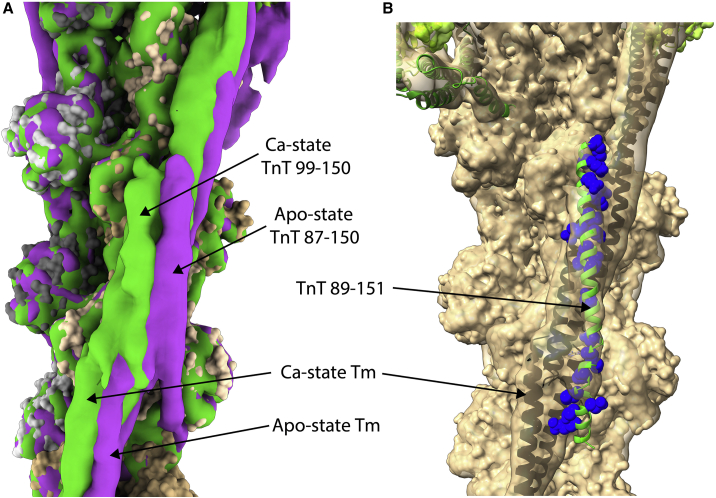
Another denouement should be given its due: the structure of the head-to-tail overlap of tropomyosin N- and C-termini plus an overlying TnT helix. Fig. 4 A is centered on the overlap domain of Tn1 and shows superimposed cryo-EM maps in the apo-state (purple) and Ca2+-saturated state (green). In the presence of Ca2+, both tropomyosin and the overriding TnT density shift slightly. Here, the tropomyosin pivots on the actin, not as much as near the center of tropomyosin but to an extent that clearly alters the position of the TnT density in the figure. Also, note that in the apo-state specifically, the observed TnT density is longer and contacts (at bottom) the actin monomer just beyond the overlap. This apparent difference is not mentioned by Yamada et al. (2). However, the density is clear in the apo-state, and the Ca2+ map, where the density is absent, has slightly better resolution overall. Functional significance to the additional density is supported by the clustering of cardiomyopathic alleles (see below, and by high sequence conservation), which also include 10 residues preceding from Pro77 (40).
Figure 4.
Tropomyosin overlap domain structure on the regulated thin filament. (A) Shown are superimposed EGTA (purple) and Ca2+ (green) cryo-EM maps (2), centered on the region where N- and C-termini of tropomyosins overlap. In both states, there is an overlying density, attributed to subunit TnT of Tn1. In the apo-state, an extension of the TnT density angles off the tropomyosin strand, interacting with the actin monomer at bottom. (B) Shown is a computationally derived atomic model of the overlap domain (5), determined independently (3) but here shown to fit within the Ca2+-state cryo-EM map of the Tn2 overlap domain. TnT residues having multiple interactions with tropomyosin are shown in blue and the remainder of TnT in green. Note the proximity between the top of the TnT helix and TnT of the Tn2 core domain, to which it is connected via unresolved linking residues. To see this figure in color, go online.
A computational study of the overlap domain, conducted independently and published almost simultaneously in January 2020, exhibits excellent agreement with the cryo-EM map (3). Pavadai et al. used docking programs, energy minimization, and molecular dynamics to select among a variety of troponin tail fragments and propose a model of the overlap region on actin. The computational study yielded up the same TnT fragment that was inferred by Yamada et al. (2) in their atomic model and placed it in the same register along the tropomyosin C-terminus and at the same oblique angle. Fig. 4 B shows an iteration (5) of the Pavadai model. Note (in black) the four-helix bundling of the C-terminus of one tropomyosin and the N-terminus of another. As expected (41,42), the termini each are splayed apart and lie at right angles to each other. The overlying, antiparallel TnT tail helix (green) angles slightly across the tropomyosin C-terminus, consistent with data from Oda as well (4). Both N- and C-termini of tropomyosin make multiple contacts with specific TnT residues (blue). Each end of the tropomyosin has close contacts with actin as well. The figure shows the computational model within the Ca2+-saturated Yamada et al. (2) cryo-EM map, illustrating the goodness of fit. This agreement between independently determined structures adds credibility to the specific findings of both studies, including the protein-protein interactions proposed in the simulations.
The cryo-EM and computational studies also agree on another important point. Fuji and co-workers (2) and the Wakabayshi group (4) found by cryo-EM that the overlap region of tropomyosin has less Ca2+-induced movement away from myosin’s attachment site on actin, compared with the effect of Ca2+ on the central part of tropomyosin shown in Fig. 3. Correspondingly, the July 2020 computational study suggests that tropomyosin pivots by 0–15° near the overlap and 50–60° elsewhere (5). The troponin tail domain binds tightly to the thin filament: 1/6th as tightly as whole troponin, which has affinity for actin-tropomyosin of least 108 M−1 (24). Thus, the troponin tail anchors the tropomyosin overlap region in inhibitory positions that interfere with myosin.
A variety of prior functional results correspond to this new structural finding—that the overlap domain may partake in troponin’s functional raison d’être, myosin inhibition. By itself, absent any other part of troponin, troponin tail TnT fragments (43,44) 1) bias tropomyosin toward the blocking position that tropomyosin adopts with troponin and EGTA; 2) greatly inhibit thin filament-myosin S1 ATPase activity, as comprehensively as does whole troponin in the absence of Ca2+; 3) decrease in vitro motility speeds, particularly at low myosin concentrations; and 4) weaken myosin S1-ADP binding to actin-tropomyosin. The skeletal muscle troponin tail also weakens S1-ADP binding to actin-tropomyosin, which is attributed to a different anchoring mechanism that is less blocking to myosin (45).
Continuing this theme of functional importance, TnT residues 79–140 are sites of multiple cardiomyopathic missense mutations that alter TnT attachment, alter apparent Ca2+ affinity, and/or interfere with full relaxation in vivo and in vitro (40,46, 47, 48). This TnT segment is comprised of the overlap domain helix plus the next 10 N-terminal residues, just before where TnT contacts actin in the atomic model.
Overcoming inhibition: new questions revealed
Now that the end states of the thin filament are in view, it becomes important to consider the transitions among them. In this regard, the new structures are hardly simplifying. Instead of trying to appreciate the regulatory significance of dynamic equilibria among three positions or states of tropomyosin (49,50), now one must consider the details of a much more complicated thin filament structure. The “on-off switch” is a metaphor suggesting simplicity, whereas Ca2+ activation of the contractile apparatus is actually quite complex.
From the Yamada et al. (2) results, local structure and local dynamics of Ca2+-saturated troponin and tropomyosin must somehow accommodate initial, strong myosin binding. Absent any movement, troponin-tropomyosin in the Ca2+-saturated state would sterically hinder actin-myosin attachment. The complication is that because troponin is entirely asymmetric, the regulatory protein movements involved in initial myosin binding differ along different actins. This is an important area for future study, mechanistically significant but with information virtually absent. Adding to the complexity, myosin binding is cooperative, and initial binding with activating effects tends to occur via two heads rather than one (7,51, 52, 53).
Apropos of this, in fully active insect muscle, cross-bridges preferentially target those actins farthest from the troponin core domain (54). This is in the isometric state rather than during initial activation, however. Also, the targeting may be driven by thick filament-thin filament geometry that does not apply in vertebrate muscle, or it may be attributable to protein sequence differences (6,40), which could alter or even reverse any such targeting.
In biochemical studies of the apo-state mechanism, investigators have emphasized either the profound kinetic inhibition of attached myosin’s progression toward strong binding and ATP product release under EGTA conditions (55,56) or the success of the three-state model in explaining a wealth of myosin’s other behaviors (45,50,57). Either way, troponin’s extended multipart structure may result in some heterogeneity in the inhibitory mechanism when considering one actin versus another.
The thin filament is a large, cooperative assembly. To elucidate its function, one must understand the transmission of cooperativity, i.e., the changes induced by a bound myosin cross-bridge or calcium ion, that alter binding affinity at additional sites. There is a long-standing conviction that the continuous tropomyosin strand can mediate such cooperativity (57, 58, 59), and the new structures remain consistent with this. However, now one also needs to consider what changes in troponin may be involved. It is unknown how troponin’s multipart structure relates to such cooperativity. Also, one cannot exclude a completely different mechanism: cooperativity between the tropomyosin strands, transmitted via the connecting linker region of TnT. A myosin-induced shift in position of one tropomyosin strand could in theory affect the opposite tropomyosin strand. Interestingly, some cooperativity persists for artificial thin filaments, just one tropomyosin long (60). Finally, Ca2+ binding to three different troponins could plausibly affect each overlap domain, singly or in combination. There is much yet to be learned.
Recent publications have presented to the scientific community, at long last, the structures of the regulated thin filament in its end states: apo, Ca2+ saturated, and myosin saturated. From this better starting point, research can begin again, with new investigations of the dynamics of the filament states, the structures and properties of intermediate states, and troponin structure when myosin is attached to the filament. Also, one looks forward to further data at atomic resolution and to structural discernment of several pieces still missing, each with functional relevance: a PKA-regulated N-terminal strand of TnI (61) and three regions of TnT, the N-terminus (62,63), the C-terminus (64,65), and the linker between the core domain and the overlap region (21). Nature has thrown down the gauntlet to investigators—we have a complicated system to understand. One can hope that now, six decades since Ebashi’s discoveries (14, 15, 16, 17, 18), these are the best days yet for studying thin filament regulation.
Editor: Meyer Jackson.
References
- 1.Gordon A.M., Homsher E., Regnier M. Regulation of contraction in striated muscle. Physiol. Rev. 2000;80:853–924. doi: 10.1152/physrev.2000.80.2.853. [DOI] [PubMed] [Google Scholar]
- 2.Yamada Y., Namba K., Fujii T. Cardiac muscle thin filament structures reveal calcium regulatory mechanism. Nat. Commun. 2020;11:153. doi: 10.1038/s41467-019-14008-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pavadai E., Rynkiewicz M.J., Lehman W. Docking troponin T onto the tropomyosin overlapping domain of thin filaments. Biophys. J. 2020;118:325–336. doi: 10.1016/j.bpj.2019.11.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oda T., Yanagisawa H., Wakabayashi T. Cryo-EM structures of cardiac thin filaments reveal the 3D architecture of troponin. J. Struct. Biol. 2020;209:107450. doi: 10.1016/j.jsb.2020.107450. [DOI] [PubMed] [Google Scholar]
- 5.Pavadai E., Lehman W., Rynkiewicz M.J. Protein-protein docking reveals dynamic interactions of tropomyosin on actin filaments. Biophys. J. 2020;119:75–86. doi: 10.1016/j.bpj.2020.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doran M.H., Pavadai E., Lehman W. Cryo-EM and molecular docking shows myosin loop 4 contacts actin and tropomyosin on thin filaments. Biophys. J. 2020;119:821–830. doi: 10.1016/j.bpj.2020.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Z., Grange M., Raunser S. Molecular plasticity of the native mouse skeletal sarcomere revealed by cryo-ET. bioRxiv. 2020 doi: 10.1101/2020.09.13.295386. [DOI] [Google Scholar]
- 8.Burbaum L., Schneider J., Jasnin M. Molecular-scale visualization of sarcomere contraction within native cardiomyocytes. bioRxiv. 2020 doi: 10.1101/2020.09.09.288977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huxley H.E. Structural changes in the actin and myosin containing filaments during contraction. Cold Spring Harb. Symp. Quant. Biol. 1972;37:361–376. [Google Scholar]
- 10.Parry D.A., Squire J.M. Structural role of tropomyosin in muscle regulation: analysis of the x-ray diffraction patterns from relaxed and contracting muscles. J. Mol. Biol. 1973;75:33–55. doi: 10.1016/0022-2836(73)90527-5. [DOI] [PubMed] [Google Scholar]
- 11.Lehman W., Craig R., Vibert P. Ca(2+)-induced tropomyosin movement in Limulus thin filaments revealed by three-dimensional reconstruction. Nature. 1994;368:65–67. doi: 10.1038/368065a0. [DOI] [PubMed] [Google Scholar]
- 12.Moss R.L., Allen J.D., Greaser M.L. Effects of partial extraction of troponin complex upon the tension-pCa relation in rabbit skeletal muscle. Further evidence that tension development involves cooperative effects within the thin filament. J. Gen. Physiol. 1986;87:761–774. doi: 10.1085/jgp.87.5.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asimov I. Gnome Press; New York: 1951. Foundation. [Google Scholar]
- 14.Ebashi S., Endo M. Calcium ion and muscle contraction. Prog. Biophys. Mol. Biol. 1968;18:123–183. doi: 10.1016/0079-6107(68)90023-0. [DOI] [PubMed] [Google Scholar]
- 15.Ebashi S. Third component participating in the super precipitation of ‘natural actomyosin’. Nature. 1963;200:1010. doi: 10.1038/2001010a0. [DOI] [PubMed] [Google Scholar]
- 16.Ebashi S. Calcium binding activity of vesicular relaxing factor. J. Chir. (Paris) 1961;82:236–244. doi: 10.1093/oxfordjournals.jbchem.a127439. [DOI] [PubMed] [Google Scholar]
- 17.Ebashi S., Ebashi F., Kodama A. Troponin as the Ca++-receptive protein in the contractile system. J. Biochem. 1967;62:137–138. doi: 10.1093/oxfordjournals.jbchem.a128628. [DOI] [PubMed] [Google Scholar]
- 18.Ebashi S., Endo M., Otsuki I. Control of muscle contraction. Q. Rev. Biophys. 1969;2:351–384. doi: 10.1017/s0033583500001190. [DOI] [PubMed] [Google Scholar]
- 19.Takeda S., Yamashita A., Maéda Y. Structure of the core domain of human cardiac troponin in the Ca(2+)-saturated form. Nature. 2003;424:35–41. doi: 10.1038/nature01780. [DOI] [PubMed] [Google Scholar]
- 20.Greaser M.L., Gergely J. Purification and properties of the components from troponin. J. Biol. Chem. 1973;248:2125–2133. [PubMed] [Google Scholar]
- 21.Manning E.P., Tardiff J.C., Schwartz S.D. Molecular effects of familial hypertrophic cardiomyopathy-related mutations in the TNT1 domain of cTnT. J. Mol. Biol. 2012;421:54–66. doi: 10.1016/j.jmb.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Otsuk I. Distribution of troponin components in the thin filament studied by immunoelectron microscopy. J. Biochem. 1975;77:633–639. doi: 10.1093/oxfordjournals.jbchem.a130765. [DOI] [PubMed] [Google Scholar]
- 23.Xu C., Craig R., Lehman W. Tropomyosin positions in regulated thin filaments revealed by cryoelectron microscopy. Biophys. J. 1999;77:985–992. doi: 10.1016/S0006-3495(99)76949-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cassell M., Tobacman L.S. Opposite effects of myosin subfragment 1 on binding of cardiac troponin and tropomyosin to the thin filament. J. Biol. Chem. 1996;271:12867–12872. doi: 10.1074/jbc.271.22.12867. [DOI] [PubMed] [Google Scholar]
- 25.Ho C.Y., Day S.M., Olivotto I. Genotype and lifetime burden of disease in hypertrophic cardiomyopathy: insights from the Sarcomeric Human Cardiomyopathy Registry (SHaRe) Circulation. 2018;138:1387–1398. doi: 10.1161/CIRCULATIONAHA.117.033200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walsh R., Thomson K.L., Watkins H., Exome Aggregation Consortium Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genet. Med. 2017;19:192–203. doi: 10.1038/gim.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vassylyev D.G., Takeda S., Maéda Y. Crystal structure of troponin C in complex with troponin I fragment at 2.3-A resolution. Proc. Natl. Acad. Sci. USA. 1998;95:4847–4852. doi: 10.1073/pnas.95.9.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li M.X., Spyracopoulos L., Sykes B.D. Binding of cardiac troponin-I147-163 induces a structural opening in human cardiac troponin-C. Biochemistry. 1999;38:8289–8298. doi: 10.1021/bi9901679. [DOI] [PubMed] [Google Scholar]
- 29.Vinogradova M.V., Stone D.B., Fletterick R.J. Ca(2+)-regulated structural changes in troponin. Proc. Natl. Acad. Sci. USA. 2005;102:5038–5043. doi: 10.1073/pnas.0408882102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huynh Q., Butters C.A., Tobacman L.S. Effects of cardiac thin filament Ca2+: statistical mechanical analysis of a troponin C site II mutant. Biophys. J. 1996;70:1447–1455. doi: 10.1016/S0006-3495(96)79704-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knowles A.C., Irving M., Sun Y.B. Conformation of the troponin core complex in the thin filaments of skeletal muscle during relaxation and active contraction. J. Mol. Biol. 2012;421:125–137. doi: 10.1016/j.jmb.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 32.Talbot J.A., Hodges R.S. Synthetic studies on the inhibitory region of rabbit skeletal troponin I. Relationship of amino acid sequence to biological activity. J. Biol. Chem. 1981;256:2798–2802. [PubMed] [Google Scholar]
- 33.Kozaili J.M., Leek D., Tobacman L.S. Dual regulatory functions of the thin filament revealed by replacement of the troponin I inhibitory peptide with a linker. J. Biol. Chem. 2010;285:38034–38041. doi: 10.1074/jbc.M110.165753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kobayashi T., Patrick S.E., Kobayashi M. Ala scanning of the inhibitory region of cardiac troponin I. J. Biol. Chem. 2009;284:20052–20060. doi: 10.1074/jbc.M109.001396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Behrmann E., Müller M., Raunser S. Structure of the rigor actin-tropomyosin-myosin complex. Cell. 2012;150:327–338. doi: 10.1016/j.cell.2012.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phillips G.N., Jr., Fillers J.P., Cohen C. Tropomyosin crystal structure and muscle regulation. J. Mol. Biol. 1986;192:111–131. doi: 10.1016/0022-2836(86)90468-7. [DOI] [PubMed] [Google Scholar]
- 37.Hitchcock-DeGregori S.E., Song Y., Greenfield N.J. Functions of tropomyosin’s periodic repeats. Biochemistry. 2002;41:15036–15044. doi: 10.1021/bi026519k. [DOI] [PubMed] [Google Scholar]
- 38.Landis C., Back N., Tobacman L.S. Effects of tropomyosin internal deletions on thin filament function. J. Biol. Chem. 1999;274:31279–31285. doi: 10.1074/jbc.274.44.31279. [DOI] [PubMed] [Google Scholar]
- 39.Lu X., Tobacman L.S., Kawai M. Effects of tropomyosin internal deletion Delta23Tm on isometric tension and the cross-bridge kinetics in bovine myocardium. J. Physiol. 2003;553:457–471. doi: 10.1113/jphysiol.2003.053694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hinkle A., Tobacman L.S. Folding and function of the troponin tail domain. Effects of cardiomyopathic troponin T mutations. J. Biol. Chem. 2003;278:506–513. doi: 10.1074/jbc.M209194200. [DOI] [PubMed] [Google Scholar]
- 41.Li Y., Mui S., Cohen C. The crystal structure of the C-terminal fragment of striated-muscle alpha-tropomyosin reveals a key troponin T recognition site. Proc. Natl. Acad. Sci. USA. 2002;99:7378–7383. doi: 10.1073/pnas.102179999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Greenfield N.J., Huang Y.J., Hitchcock-DeGregori S.E. Solution NMR structure of the junction between tropomyosin molecules: implications for actin binding and regulation. J. Mol. Biol. 2006;364:80–96. doi: 10.1016/j.jmb.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 43.Tobacman L.S., Nihli M., Homsher E. The troponin tail domain promotes a conformational state of the thin filament that suppresses myosin activity. J. Biol. Chem. 2002;277:27636–27642. doi: 10.1074/jbc.M201768200. [DOI] [PubMed] [Google Scholar]
- 44.Madan A., Viswanathan M.C., Cammarato A. TNNT2 mutations in the tropomyosin binding region of TNT1 disrupt its role in contractile inhibition and stimulate cardiac dysfunction. Proc. Natl. Acad. Sci. USA. 2020;117:18822–18831. doi: 10.1073/pnas.2001692117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maytum R., Geeves M.A., Lehrer S.S. A modulatory role for the troponin T tail domain in thin filament regulation. J. Biol. Chem. 2002;277:29774–29780. doi: 10.1074/jbc.M201761200. [DOI] [PubMed] [Google Scholar]
- 46.Cammarato A., Hatch V., Lehman W. Drosophila muscle regulation characterized by electron microscopy and three-dimensional reconstruction of thin filament mutants. Biophys. J. 2004;86:1618–1624. doi: 10.1016/S0006-3495(04)74229-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Palm T., Graboski S., Greenfield N.J. Disease-causing mutations in cardiac troponin T: identification of a critical tropomyosin-binding region. Biophys. J. 2001;81:2827–2837. doi: 10.1016/S0006-3495(01)75924-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gangadharan B., Sunitha M.S., Mercer J.A. Molecular mechanisms and structural features of cardiomyopathy-causing troponin T mutants in the tropomyosin overlap region. Proc. Natl. Acad. Sci. USA. 2017;114:11115–11120. doi: 10.1073/pnas.1710354114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vibert P., Craig R., Lehman W. Steric-model for activation of muscle thin filaments. J. Mol. Biol. 1997;266:8–14. doi: 10.1006/jmbi.1996.0800. [DOI] [PubMed] [Google Scholar]
- 50.McKillop D.F., Geeves M.A. Regulation of the interaction between actin and myosin subfragment 1: evidence for three states of the thin filament. Biophys. J. 1993;65:693–701. doi: 10.1016/S0006-3495(93)81110-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Williams D.L., Jr., Greene L.E. Comparison of the effects of tropomyosin and troponin-tropomyosin on the binding of myosin subfragment 1 to actin. Biochemistry. 1983;22:2770–2774. doi: 10.1021/bi00280a027. [DOI] [PubMed] [Google Scholar]
- 52.Trybus K.M., Taylor E.W. Kinetic studies of the cooperative binding of subfragment 1 to regulated actin. Proc. Natl. Acad. Sci. USA. 1980;77:7209–7213. doi: 10.1073/pnas.77.12.7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kad N.M., Kim S., Baker J.E. Single-myosin crossbridge interactions with actin filaments regulated by troponin-tropomyosin. Proc. Natl. Acad. Sci. USA. 2005;102:16990–16995. doi: 10.1073/pnas.0506326102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tregear R.T., Reedy M.C., Reedy M.K. Cross-bridge number, position, and angle in target zones of cryofixed isometrically active insect flight muscle. Biophys. J. 2004;86:3009–3019. doi: 10.1016/S0006-3495(04)74350-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heeley D.H., White H.D., Taylor E.W. Investigation into the mechanism of thin filament regulation by transient kinetics and equilibrium binding: is there a conflict? J. Gen. Physiol. 2019;151:628–634. doi: 10.1085/jgp.201812198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Houmeida A., Heeley D.H., White H.D. Mechanism of regulation of native cardiac muscle thin filaments by rigor cardiac myosin-S1 and calcium. J. Biol. Chem. 2010;285:32760–32769. doi: 10.1074/jbc.M109.098228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mijailovich S.M., Kayser-Herold O., Geeves M.A. Cooperative regulation of myosin-S1 binding to actin filaments by a continuous flexible Tm-Tn chain. Eur. Biophys. J. 2012;41:1015–1032. doi: 10.1007/s00249-012-0859-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tobacman L.S., Sawyer D. Calcium binds cooperatively to the regulatory sites of the cardiac thin filament. J. Biol. Chem. 1990;265:931–939. [PubMed] [Google Scholar]
- 59.Hill T.L., Eisenberg E., Greene L. Theoretical model for the cooperative equilibrium binding of myosin subfragment 1 to the actin-troponin-tropomyosin complex. Proc. Natl. Acad. Sci. USA. 1980;77:3186–3190. doi: 10.1073/pnas.77.6.3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gong H., Hatch V., Tobacman L.S. Mini-thin filaments regulated by troponin-tropomyosin. Proc. Natl. Acad. Sci. USA. 2005;102:656–661. doi: 10.1073/pnas.0407225102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kobayashi T., Solaro R.J. Calcium, thin filaments, and the integrative biology of cardiac contractility. Annu. Rev. Physiol. 2005;67:39–67. doi: 10.1146/annurev.physiol.67.040403.114025. [DOI] [PubMed] [Google Scholar]
- 62.Jin J.P., Chong S.M. Localization of the two tropomyosin-binding sites of troponin T. Arch. Biochem. Biophys. 2010;500:144–150. doi: 10.1016/j.abb.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tobacman L.S., Lee R. Isolation and functional comparison of bovine cardiac troponin T isoforms. J. Biol. Chem. 1987;262:4059–4064. [PubMed] [Google Scholar]
- 64.Tanokura M., Tawada Y., Ohtsuki I. Chymotryptic subfragments of troponin T from rabbit skeletal muscle. Interaction with tropomyosin, troponin I and troponin C. J. Biochem. 1983;93:331–337. doi: 10.1093/oxfordjournals.jbchem.a134185. [DOI] [PubMed] [Google Scholar]
- 65.Johnson D., Zhu L., Chalovich J.M. Basic residues within the cardiac troponin T C terminus are required for full inhibition of muscle contraction and limit activation by calcium. J. Biol. Chem. 2019;294:19535–19545. doi: 10.1074/jbc.RA119.010966. [DOI] [PMC free article] [PubMed] [Google Scholar]