Summary
Signal peptide-CUB-EGF domain-containing protein 3 (SCUBE3) is a member of a small family of multifunctional cell surface-anchored glycoproteins functioning as co-receptors for a variety of growth factors. Here we report that bi-allelic inactivating variants in SCUBE3 have pleiotropic consequences on development and cause a previously unrecognized syndromic disorder. Eighteen affected individuals from nine unrelated families showed a consistent phenotype characterized by reduced growth, skeletal features, distinctive craniofacial appearance, and dental anomalies. In vitro functional validation studies demonstrated a variable impact of disease-causing variants on transcript processing, protein secretion and function, and their dysregulating effect on bone morphogenetic protein (BMP) signaling. We show that SCUBE3 acts as a BMP2/BMP4 co-receptor, recruits the BMP receptor complexes into raft microdomains, and positively modulates signaling possibly by augmenting the specific interactions between BMPs and BMP type I receptors. Scube3−/− mice showed craniofacial and dental defects, reduced body size, and defective endochondral bone growth due to impaired BMP-mediated chondrogenesis and osteogenesis, recapitulating the human disorder. Our findings identify a human disease caused by defective function of a member of the SCUBE family, and link SCUBE3 to processes controlling growth, morphogenesis, and bone and teeth development through modulation of BMP signaling.
Keywords: SCUBE, BMP, BMP receptors, bone morphogenetic protein, skeletal development, morphogenesis, genomic sequencing, mechanism of disease, intracellular signaling
Introduction
The extracellular microenvironment provides the biochemical signals that control cell behavior and coordinate developmental and physiological processes. In this three-dimensional and dynamic network, a repertoire of secreted and cell surface-anchored modulators is known to contribute to the fine tuning of signal strength by boosting or antagonizing the information transmitted by growth factors, hormones, and cytokines to their cognate receptors.1 Any imbalance of such delicate regulation can considerably affect development and homeostasis.2,3 In this framework, bone morphogenetic protein (BMP) signaling is archetypal. BMPs are secreted extracellular matrix (ECM)-associated proteins of the TGF-β superfamily controlling cell growth, apoptosis, and differentiation, and coordinating a variety of developmental processes, playing an important role in body-plan patterning, morphogenesis, and organogenesis.4,5 Besides the large number of secreted signal molecules and multiple cell-surface receptors, the complexity of BMP signaling regulation is linked to the presence of secreted antagonists that are able to inhibit binding of individual ligands to their cognate receptors, or co-receptors that modulate the activity of the receptor complex.6 Indeed, pathogenic variants in genes encoding these auxiliary proteins can dramatically affect development and physiology.7, 8, 9
The signal peptide-complement protein C1r/C1s, Ugef, and Bmp1 (CUB)-epidermal growth factor (EGF)-like domain-containing (SCUBE) protein family comprises three related multi-functional glycoproteins that act as co-receptors augmenting signaling promoted by a variety of growth factors.10, 11, 12 Their conserved structure is characterized by a signal peptide, nine tandemly arranged EGF-like domains, a spacer region, three cysteine-rich motifs, and a single CUB domain located at the C terminus.11, 12, 13 While the N-terminal and C-terminal regions are involved in protein-protein interactions,12,14 the spacer region and cysteine-rich repeats mediate protein anchoring to the cell surface by at least two different membrane-associating mechanisms involving electrostatic and lectin-glycan interactions.15 Proteolytic cleavage within the spacer region is believed to control the modulatory function mediated by the CUB domain and secretion of the N-terminal portion of the protein in the extracellular microenvironment.16 SCUBE proteins modulate signaling elicited by a variety of growth factors, including members of the BMP, hedgehog, vascular endothelial growth factor and fibroblast growth factor families.17, 18, 19, 20 These proteins are prominently expressed in various developing tissues, which implies their potential role during development, but have distinct expression patterns. While SCUBE1 (MIM: 611746) is mainly expressed in platelets and endothelial cells10 and SCUBE2 (MIM: 611747) is more widely expressed,12 the most recently discovered member of this family, SCUBE3 (MIM: 614708), is expressed in a more restricted fashion, with highest levels in osteoblasts, craniofacial regions, and limb buds.11,21
Here we report that bi-allelic, largely inactivating variants in SCUBE3 underlie a developmental disorder characterized by reduced growth, skeletal features, dental anomalies, and a distinctive craniofacial appearance. We demonstrate that the disease-causing variants affect protein synthesis, secretion, and/or function, with a subset of them directly perturbing BMP2 signaling. We also show that SCUBE3 functions as a BMP co-receptor and that Scube3−/− mice have defective endochondral bone growth due to impaired BMP-mediated chondrogenesis and ossification.
Subjects and methods
Study approval
Affected subjects were analyzed in the context of research projects dedicated to “undiagnosed patients,” or referred for diagnostic genetic testing. All clinical investigations were conducted according to Declaration of Helsinki principles. This project was approved by the local Institutional Ethical Committee of the Ospedale Pediatrico Bambino Gesù (1702_OPBG_2018), Rome. Clinical data, photographs, DNA specimens, and other biological material were collected, used for genetic analyses, and stored after signed informed consents from the participating subjects/families were obtained. Permission was obtained to publish the photographs of all subjects shown in Figure 1.
Figure 1.
Pedigrees of the families included in the study, domain structure of SCUBE3, and clinical features of subjects with bi-allelic pathogenic SCUBE3 variants
(A) Pedigrees and segregation data for the nine families included in the study. Affected and unaffected subjects are indicated by filled and open squares/circles, respectively.
(B) Cartoons showing the genomic organization of SCUBE3 and functional domains of its encoded protein. Location of the disease-causing variants is shown. c.2599+2T>C is predicted to result in multiple aberrantly processed transcripts, with p.Asn801Thrfs∗127 representing the prevalent out-of-frame product (see Figure S3).
(C) Clinical features of affected individuals. (1a) Whole body appearance of subject F1S1; (1b, 1c) facies of subjects F1S1 and F1S2; (1d) dental anomalies in F1S2; (2a–2c) facies of subjects F2S1 and F2S2; (2d) dental anomalies in F2S1; (3a) facies of subject F3S3; (3b) finger joint swelling in F3S3; (3c) dental anomalies in F3S3; (4a, 4b) facies of subject F4S1; (4c) 5th finger camptodactyly in F4S1; (4d) dental anomalies in F4S1; (5a–5d) facies of subjects F5S1 and F5S2; (6a) facies of subject F6S1; (7a) facies of subject F7S1; (8a, 8b) facies of subject F8S1; (8c) bilateral shortening of 4th and 5th toes in F8S1; (9a–9c) facies of subjects F9S1 and F9S2.
(D) Radiological findings in affected subjects. (1) dental anomalies in F1S2 shown by panoramic dental X-ray; (2) bowed radius in F2S1; (3) reduced epiphyses including the metacarpal heads, capitate-3rd-metacarpal and trapezoid-2nd-metacarpal coalitions and camptodactyly in F3S1; (4) trapezoid-2nd metacarpal coalition and cuneiform bones-2nd and 3rd metatarsals coalitions in F8S1; (5) absent 12th rib pair in F3S1; (6–7) partial fusion of C5-C6 and failure of the posterior arch fusion in C7-T1 in F8S1; (8) narrow iliac wings in F3S1; (9) squared vertebral bodies in F2S1; (10) tarsal-metatarsal coalition in both feet in F2S1.
All experimental in vivo procedures were performed in accordance with national and institutional guidelines for animal care and experiments. Animal handling protocols and experiments were reviewed and approved by the Institute Animal Care and Utilization Committee of Academia Sinica (Protocol ID: 19-11-1358).
Genomic analyses
Genomic analyses were performed using DNA samples obtained from leukocytes. All families except families 4 and 6 were analyzed by WES. Families 4 and 6 were investigated by WGS, the former as part of the 100KGP, a national genome sequencing effort delivered jointly by the UK’s National Health Service and Genomics England. Target enrichment kits, sequencing platforms, and WES/WGS statistics are reported in Table S1. Data processing, sequence alignment to GRCh37/GRCh38, and variant filtering and prioritization by allele frequency, predicted functional impact, and inheritance models were performed as previously reported.22, 23, 24, 25, 26, 27, 28 WES data output is summarized in Table S1. Co-segregation analysis was performed by Sanger sequencing in all but family 7. Primer sequences available on request.
Minigene splicing analysis
The SCUBE3 genomic portion encompassing exons 18 to 21 was amplified by PCR from the MCF-7 cell line genomic DNA (ATCC) (primers available on request). The amplified fragments were cloned into the pGEM-T easy vector (Promega), and clones were randomly selected, and sequenced. The minigene construct was generated by subcloning the SCUBE3 genomic region of interest into the pEGFP-C2 vector (Clontech). The minigene plasmids were introduced into human HEK293T and MG-63 cells using Lipofectamine 3000. At 48 h after transfection, total RNA was extracted by the TRIzol method (Life Technologies), first-strand cDNA was synthesized with SuperScript II reverse transcriptase (Life Technologies), and PCR was performed using a primer pair designed to anneal to exons 18 and 21. The amplified fragments were cloned into the pGEM-T easy vector (Promega). After cloning, at least 10 clones of each transfection were randomly selected, and their sequences were determined by Sanger sequencing.
Plasmids and transfection
SCUBE3 constructs were generated to include a FLAG tag immediately after the signal peptide cleavage site (pFLAG-CMV-1, Sigma) and a Myc tag at the C-terminus (pSecTag2, Invitrogen). HEK293T, HepG2, and SAOS-2 cells were transfected by using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions.
Cell cultures
C3H/10T1/2, HEK293T, HepG2, and SAOS-2 cells were from the American Type Culture Collection and cultured according to the supplier’s recommendations. Cells were maintained in Eagle’s basal medium supplemented with 10% heat-inactivated fetal bovine serum, 2 mM L-glutamine, 100 units/mL penicillin, and 100 μg/mL streptomycin at 37°C, 5% CO2. Primary chondrocyte culture was performed as previously described,29 and primary BMSCs were obtained from 4-week-old Scube3−/− and control littermates as described.30
FACS analysis
Transfected cells were collected and resuspended in FACS buffer (2% FBS PBS). Cell suspensions were incubated with fluorescein-labeled FLAG antibody (Sigma) (4°C, 1 h). After washing with ice-cold PBS three times, cells were resuspended in FACS buffer. FACS analyses were performed using a FACSCalibur (BD Biosciences).
Co-immunoprecipitation assays and western blot analyses
For western blot analyses, transfected cells were lysed on ice in lysis buffer (25 mM HEPES [pH 7.6], 150 mM NaCl, 5 mM EDTA, 10 μg/mL aprotinin, 5 μg/mL leupeptin, 10% glycerol, and 1% Triton X-100). Cell lysates (20 μg) or samples from conditioned medium (20 μL) were separated by SDS-polyacrylamide gel and transferred to PVDF membranes. For immunoprecipitation assays, cell lysates (500 μg) were incubated with 1 μg of the indicated antibody and 20 μL of 50% (v/v) protein A-agarose (GE) (2 h with gentle rocking). The precipitated complexes were then separated by SDS-polyacrylamide gel and transferred to PVDF membranes. After blocking, membranes were incubated with-anti-FLAG (Sigma), anti-HA (Cell Signaling Technology), or anti-Myc antibodies (Cell Signaling Technology) for 1 h. After 3 washes, the blots were incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch Laboratories) (1 h). The reactive bands were visualized by use of the VisGlow chemiluminescent substrate, HRP system (Visual Protein).
Luciferase reporter assay
HepG2 cells were transfected with the BMP-responsive luciferase reporter, BRE-luc, together with the individual SCUBE3 constructs (or a mock vector) and a Renilla luciferase reporter as internal control, by using Lipofectamine 3000 (Invitrogen). Cells were cultured for an additional 2 days, harvested, and prepared for reporter assay with the Dual-Luciferase reporter assay system (Promega) according to the manufacturer’s instructions.
Alkaline phosphatase activity assay
Cells were washed with PBS and lysed with 0.05% Trixton X-100 solution. Alkaline phosphatase (ALP) activity in lysates was determined by using p-nitrophenol-phosphate as substrate as previously reported.31
Confocal laser scanning microscopy
SAOS-2 cells (2 × 104) were seeded in 24-well cluster plates onto 12-mm cover glasses, transfected with the various FLAG-tagged constructs (48 h), and fixed with 4% paraformaldehyde (30 min, 4°C). Cells were stained with mouse monoclonal IgG anti-FLAG M2 (1:500, SIGMA) (2 h, room temperature), rinsed twice with PBS, and incubated with the secondary goat anti-mouse antibody conjugated with Alexa Fluor 594 (1 h, room temperature) (Molecular Probes). After staining, coverslips were rinsed and mounted on slides by using Vectashield mounting medium (Vector Laboratories) containing 1.5 μg/mL DAPI. Alexa Fluor 488 phalloidin dye was used to stain the cortical actin associated with the plasma membrane. Observations were performed on a Leica TCS-SP8X laser-scanning confocal microscope (Leica Microsystems) equipped with a tunable white light laser source and a 405 nm diode laser. Sequential confocal images were acquired by LAS X software (Leica Microsystems) using a HC PLAPO 63x oil-immersion objective (1.40 numerical aperture).
Isolation of lipid rafts
Lipid rafts were prepared as described.32 In brief, cells were lysed with 1.3 mL ice-cold RIPA buffer. The homogenized lysates were resuspended in a final 40% OptiPrep solution. The mixture was vortexed vigorously and transferred to a 12.5 mL ultracentrifuge tube, and a discontinous OptiPrep gradient (30%, 5%) was formed above the lysate by adding 4 mL 30% OptiPrep and then 4 mL 5% OptiPrep. The gradient was ultracentrifuged (20 h, 39,000 rpm, 4°C). Twelve fractions were collected from top to bottom and designated as fractions 1–12. To verify the separation, a specific caveolin-1 antibody was used.
Mouse phenotyping
Wild-type C57BL/6JNari mice were provided by the National Laboratory Animal Center (NLAC), NARLabs, Taiwan. Scube3−/− mice (C57BL/6JNari) were generated as previously described.21 Adult male mice were used in skeletal phenotype analysis, whereas sex of embryos or neonates was not determined. Radiography, 3-D-reconstructed CT images, and bone mineral density (BMD) analysis were performed as previously described.33 Alcian blue hematoxylin/orange G staining of paraffin sections was performed as described,34 with modification. Briefly, deparaffinized sections were placed in acid-alcohol (0.37% hydrochloric acid in 70% ethanol) for 30 s, alcian blue hematoxylin (1% alcian blue in Mayer’s acid hematoxylin) for 40 min and washed gently in distilled water to remove excess stain from tissue. Sections were placed in acid-alcohol for 3 s, 0.5% ammonium water for 15 s, 95% EtOH for 1 min, and eosin/orange G solution (0.12% eosin, 0.074% phloxin B, and 0.064% orange G in 90% ethanol [pH 4.8]) for 1 min 30 s. After washing with distilled water, sections were dehydrated and mounted in Entellan new (Merck).
Proliferation assays (BrdU) of skeletal tissue sections
To determine the quantity of newly generated cells, the DNA synthesis marker 5-bromo-2′-deoxyuridine (BrdU) was administered intraperitoneally (50 mg/kg body weight) to mice. A single dose of BrdU was injected into mice at E18. All pups were euthanized, fixed, and processed for BrdU immunohistochemistry on P1.
Micromass culture assay
Micromass cultures of whole E12.5 limb buds were prepared as described.35
Quantification and statistical analysis
Data are presented as mean ± SD and were analyzed by Student t test (for 2 groups) or one-way ANOVA (for ≥3 groups) with Tukey multiple comparison analysis by using Prism 7 (GraphPad Software). p < 0.05 was considered statistically significant.
Results
Bi-allelic SCUBE3 variants underlie a disorder affecting development and growth
Nine unrelated consanguineous families with one or more individuals affected by an unrecognized condition severely impairing growth and sharing homozygous putative pathogenic variants in SCUBE3 (GenBank: NM_152753.4) were identified by GeneMatcher,36 Decipher,37 and networking (Table 1, Figures 1A and 1B). All subjects showed a consistent phenotype characterized by peculiar craniofacial features, short stature, and dental and skeletal abnormalities (see below). In all tested individuals, exome/genome sequencing analysis excluded the presence of other functionally relevant variants compatible with known Mendelian diseases based on the expected inheritance model and clinical presentation (Table S1). The identified variants were distributed along the entire SCUBE3 coding sequence and included nonsense, frameshift, missense, and canonical splice site changes as well as a complex intragenic rearrangement. Genomic DNA was available for relatives from all but family 7, and co-segregation of the trait and homozygosity for the SCUBE3 variant was confirmed in all families tested (Figures 1A and S1).
Table 1.
List of the homozygous pathogenic SCUBE3 variants identified in the study.
| Nucleotide change | dbSNP | MAF | Amino Acid Change | M-CAP | REVEL | CADD | ACMG | Domain | Family, subject |
|---|---|---|---|---|---|---|---|---|---|
| c.291C>G | – | N/A | p.Cys97Trp | 0.526729 | 0.911 | 26.1 | VoUS | EGF-like 2 | F1, subjects 1, 2 |
| c.611G>A | – | N/A | p.Gly204Asp | 0.576042 | 0.696 | 26.2 | VoUS | EGF-like 5 | F8, subject 1 |
| c.829+1_952del (in-frame exon 8 deletion) | – | N/A | p.Arg282_Cys322del | N/A | N/A | N/A | VoUS | EGF-like 7 | F6, subjects 1, 2 |
| c.1717C>T | rs1436996181 | 0.4E−5 | p.Arg573∗ | N/A | N/A | 38 | pathogenic | – | F3, subjects 1-4 |
| c.2239+1G>A | – | N/A | p.Val747Aspfs∗46 | N/A | N/A | 34 | pathogenic | – | F4, subject 1 |
| c.2444T>C | rs751478115 | 0.2E−4 | p.Ile815Thr | 0.078026 | 0.776 | 27.9 | VoUS | CUB | F2, subjects 1, 2 F7, subject 1 |
| c.2599+2T>C | – | N/A | p.?a | N/A | N/A | 27.2 | pathogenic | – | F5, subjects 1, 2 |
| c.2785C>T | rs1397172310 | N/A | p.Arg929∗ | N/A | N/A | 45 | pathogenic | – | F9, subjects 1-3 |
VoUS, variant of uncertain significance. Threshold of pathogenicity of the M-CAP, REVEL and CADD predictive tools: M-CAP > 0.025; REVEL > 0.75; CADD_PHRED > 15.
c.2599+2T>C is predicted to result in multiple aberrantly processed transcripts, with p.Asn801Thrfs∗127 representing the prevalent out-of-frame product (see Figure S3).
Two novel variants were predicted to affect transcript processing (c.2239+1G>A [p.Val747Aspfs∗46], family 4; c.2599+2T>C, family 5), and other two were truncating (c.1717C>T [p.Arg573∗], rs1436996181, MAF = 0.000004, family 3; c.2785C>T [p.Arg929∗], not previously reported, family 9). Total RNA was available for the affected individual of family 4, allowing us to confirm the activation of a cryptic donor site of exon 17, retention of four nucleotides of the flanking intronic region, and premature termination of the coding sequence (p.Val747Aspfs∗46) (Figure S2). Biological material from the proband was not available for functional validation of the c.2599+2T>C variant, which was indirectly attained using a minigene splicing assay, confirming aberrant splicing of the genomic portion of SCUBE3 encompassing exons 18–21, with occurrence of multiple mRNA variants all predicted to encode prematurely truncated proteins (Figure S3).
Three different missense changes were identified in four families. These variants affected conserved residues among SCUBE3 orthologs and paralogs (Figure S4) and were predicted to be damaging by multiple in silico predictors (Table 1). The two affected sibs of family 1 (F1S1 [II-1] and F1S2 [II-3]) were homozygous for a novel substitution (c.291C>G [p.Cys97Trp]) within the second EGF-like module, affecting one of the invariant cysteine residues participating to the disulfide bonding network that stabilizes the structure of the EGF-like domains. Substitution of Cys97 implies loss of one of the three conserved disulfide bridges and consequently a structural rearrangement of the motif (Figure S4). Of note, pathogenic variants at equivalent positions in multiple EGF-like modules of the FBN1, FBN2, and LTBP3 proteins have previously been reported (Figure S5),38, 39, 40 further supporting the functional relevance of the variant. Subject F8S1 (family 8, II-1) was homozygous for a previously unreported missense change (c.611G>A [p.Gly204Asp]) affecting a conserved glycine residue within the fifth EGF-like module. The nonconservative substitution was predicted to impair proper module folding locally and impact the interaction with the adjacent EGF-like module (Figure S4). The third missense variant (c.2444T>C [p.Ile815Thr]), identified in families 2 and 7 (F2S1 [V-1], F2S2 [V-3], and F7S1 [II-1]), is a rare substitution (rs751478115, MAF = 0.000024), never reported in the homozygous state, involving a buried apolar residue located in the core of the CUB domain. Its replacement by a polar residue was predicted to considerably destabilize the structure of the hydrophobic cluster involving residues Trp832, Ile896, and Ile911 (Figure S4) and, in turn, perturb the overall folding of the domain and its ability to mediate SCUBE3 binding to interactors.
Finally, homozygosity for a complex intragenic rearrangement in SCUBE3 (deletion plus inverted duplication) was documented in one family (family 6, F6S1 [II-1] and F6S2 [II-2]). The rearrangement was novel and thought to have originated from a two-step fork-switching and template-stalling (FoSTeS) process, causing the complete deletion of exon 8 (c.829_952del [p.Arg282_Cys322del]) with conservation of the coding frame of the SCUBE3 mRNA, resulting in an EGF-like domain lacking the seventh EGF-like module (Figure S6). Equivalent intragenic structural rearrangements resulting in deletions of single EGF-like modules in FBN1 and FBN2 have previously been reported to dramatically affect protein function.41,42
Clinical profiling of subjects with bi-allelic pathogenic SCUBE3 variants
Detailed clinical information was collected for 15 of the 18 individuals carrying the homozygous SCUBE3 variants. These subjects shared a consistent condition characterized by short stature, distinctive facial dysmorphia, dental and skeletal anomalies, in the absence of developmental delay or intellectual disability (Figure 1C, Table 2, Supplemental Note: Case Reports). These subjects showed a reduced birth length (<–2 SDS) (8/11), with or without decrease of other birth parameters, and had severely reduced postnatal height and weight (15/15) (Figure S7) and head circumference (9/13). Facial features included a long/triangular face in older individuals with high/broad forehead (14/14), high nasal bridge (13/15), long nose (13/15), which was short and upturned in the two younger subjects (F6S1 and F7S1), thick lips (7/15), and a short or receding chin at young age (14/14), which may become slightly pointed at older ages (11/13) (Figure 1C). Of note, two individuals (F7S1 and F8S1) had the Pierre-Robin sequence, and one (F1S2) had cleft palate and micrognathia. Peculiar dental defects were frequently observed, including enamel dysplasia and hypodontia/oligodontia (7/15). These signs are quite suggestive of disturbance in odontogenesis and might be considered as major key signs of the condition. The remaining eight affected individuals presented with less specific dental findings, such as dental crowding and misalignment, which however strongly indicate that dental involvement is an invariant feature in subjects with bi-allelic inactivating SCUBE3 variants. Skeletal features occurred in all subjects (15/15), with thin and short long bones (6/11), 11 pairs of ribs (4/13), abnormal vertebrae (6/12), including odontoid hypoplasia and fusions, squared lumbar vertebral bodies (5/13), and scoliosis (6/13) representing recurrent features. Upper and lower limbs presented changes in almost all affected subjects, resulting in short stature with rhizomelia/mesomelia in some instances (Figure 1C). Bowed radius with abnormal radial head occurred in some individuals (F1S1, F1S2, F3S1 [III-1], and F8S1). Shortening of both metacarpal/metatarsal bones and phalanges was documented. Bone age was assessed in four subjects and found delayed in three. Variable joint involvement was also documented (5/8). Other less common features were cardiac anomalies (4/14) and conductive hearing loss (4/12). One subject had ventricular extrasystoles with first degree AV block, while pulmonary hypertension was found in two.
Table 2.
Phenotypic features of the 15 individuals with biallelic pathogenic SCUBE3 variants included in the study.
| Individual |
F1S1 |
F1S2 |
F2S1 |
F2S2 |
F3S1 |
F3S2 |
F3S3 |
F4S1 |
F5S1 |
F5S2 |
F6S1 |
F7S1 |
F8S1 |
F9S1 |
F9S2 |
Total (%) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SCUBE3 variant/change | p.Cys97Trp | p.Ile815Thr | p.Arg573∗ | p.Val747Asp_fs∗46 | c.2599+2T>C | p.Arg282_ Cys322del | p.Ile815Thr | p.Gly204Asp | p.Arg929∗ | ||||||||
| Age | 19 y | 10 y | 22 y | 15 y | 39 y | 29 y | 25 y | 11.5 y | 11.6 y | 7.3 y | 16 m | 26 m | 12.8 y | 8 y | 22.5 y | ||
| Gender | F | F | M | M | F | F | M | M | M | F | F | M | M | M | F | ||
| Prenatal growth retardation (−2 SDS) | + | − | + | + | + | N/A | N/A | + | N/A | N/A | + | + | + | − | − | 8/11 (82) | |
| Postnatal growth retardation (−2 SDS) | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 15/15 (100) | |
| DD/ID | − | − | − | − | − | − | − | − | − | − | − | + | − | − | + | 1/15 (7) | |
| Speech delay | − | − | − | − | − | − | − | − | − | − | − | + | − | − | + | 3/15 (20) | |
| Hypotonia | − | − | − | − | − | N/A | − | − | − | − | + | + | − | − | − | 2/14 (14) | |
| Craniofacial | |||||||||||||||||
| Microcephaly (−2 SDS) | − | − | + | + | + | N/A | − | + | + | + | + | N/A | + | − | + | 9/13 (69) | |
| Long/triangular face | + | + | + | + | + | + | + | + | + | + | + | N/A | + | + | + | 14/14 (100) | |
| High forehead | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 15/15 (100) | |
| High nasal bridge | + | + | + | + | + | + | + | + | + | + | − | − | + | + | + | 13/15 (73) | |
| Long nose | + | + | + | + | + | + | + | + | + | + | − | − | + | + | + | 13/15 (87) | |
| Thick lips | + | + | − | + | − | − | − | + | − | − | + | + | + | − | − | 7/15 (47) | |
| Short chin | + | + | + | + | + | N/A | + | + | + | + | + | + | + | + | + | 14/14 (100) | |
| Pointed china | + | + | + | + | N/A | N/A | + | + | + | + | − | − | + | + | + | 11/13 (85) | |
| Pierre Robin sequence/cleft palate | − | CP | − | − | − | N/A | − | − | − | − | − | PRS | PRS | − | − | 3/14 (21) | |
| Dental anomalies | H,DC | H,ED, DC | DC | DC | DC | DC | O, DC | MD,ED,DC,ET | ED | ED | DC,MA | MA | DC | DC | O,DC, MA | 15/15 (100) | |
| Skeletal anomalies | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 15/15 (100) | |
| Short hands/brachydactyly | + | + | + | + | + | + | − | + | + | − | − | + | + | + | + | 12/15 (80) | |
| Hip defects | NIW/CV | CV | NIW,CV | NIW, CV | NIW,AD | NIW | N/A | NIW,CV | CV | − | − | − | − | − | − | 9/14 (64) | |
| Thin/short long bones | + | + | N/A | N/A | − | N/A | N/A | + | + | + | − | − | + | − | − | 6/11 (54) | |
| Squared vertebrae | + | + | − | + | + | N/A | N/A | − | − | − | − | − | + | − | − | 5/13 (39) | |
| Abnormal cervical vertebrae | + | + | N/A | N/A | − | N/A | N/A | N/A | − | − | + | − | + | − | − | 4/10 (40) | |
| 11 pairs of ribs | + | + | − | − | + | N/A | N/A | − | − | − | − | − | + | − | − | 4/13 (31) | |
| Scoliosis | + | + | − | + | − | N/A | N/A | − | + | + | − | − | + | − | − | 6/13 (46) | |
| Joint abnormalities | +b | +b | − | − | N/A | N/A | N/A | − | N/A | N/A | +c | N/A | +c | +c | N/A | 5/8 (63) | |
| Hearing loss (conductive) | − | − | + | + | − | N/A | + | − | N/A | N/A | − | − | + | − | − | 4/12 (33) | |
| Cardiac defects | − | − | − | − | − | N/A | − | − | − | CA | ASD | PFO | − | − | ASD | 4/14 (29) | |
| Other defects | − | − | − | − | − | − | − | − | − | − | − | OD,TM, HS, ST | BCC | Ny, SBH | Ny, AST, SBH | ||
AD, acetabular dysplasia; ASD, atrial septal defect; AST, astigmatism; BCC, bilateral coronal synostosis; CA, cardiac arrhythmia (ventricular extrasystoles, atrioventricular block); CP, cleft palate; CV, coxa valga; DC, dental crowding; ED, enamel dysplasia/multiple caries; ET, ectopic teeth; H, hypodontia; HD, hip dislocation (bilateral); HS, hypospadias; MA, misalignment of incisor; MD, macrodontia; N/A, not assessed; NIW, narrow iliac wing; Ny, nystagmus; O, oligodontia; OD, oropharyngeal dysphagia; PFO, patent foramen ovale; PRS, Pierre Robin sequence; SBH, sparse brittle hair; ST, strabismus; TM, tracheo-bronchomalacia.
This feature becomes more evident at older ages.
Joint hypermobility.
Joint stiffness (F6S1); limited flexion of the proximal interphalangeal joints in both hands (F8S1); limited flexions of fingers, and hand grip difficulties (F9S1).
Overall, the consistent phenotype was highly suggestive of a developmental disorder characterized by severe short stature and a distinctive gestalt overlapping, in part, with a recently reported syndromic condition caused by monoallelic loss-of-function variants in BMP2.43 Since the occurrence of a bowed radius and short stature, differential diagnosis may include Lery-weill dyschondrosteosis (MIM: 127300), which however is characterized by the presence of Madelung deformity, whose onset typically occurs mid-to-late childhood and does not present the craniofacial features and enamel dysplasia observed in subjects with bi-allelic inactivating SCUBE3 variants. The disorder differs from other common conditions with short meso-acromelic involvement, such as geleophysis and acromicric dysplasias (MIM: PS231050 and 102370, respectively), by the absence of plump or cone-shaped epiphyses and the distinctive facial features. By reviewing other conditions with short stature and ectodermal defects, the overall phenotype of these subjects may be suggestive of cranioectodermal dysplasia (CED [MIM: PS218330]), a rare ciliopathy characterized by short stature with prevalent rhizomelic shortening, having some facial features reminiscent of the subjects of this series (i.e., frontal bossing, full cheeks, short chin), brachydactyly with mild radiographic changes, and strikingly overlapping dental features. Nonetheless, subjects with bi-allelic SCUBE3 variants lack macrocrania and other skeletal features distinctive of CED.
SCUBE3 binds to BMP2/4 and BMP receptors, is an enhancer for BMP signaling, and is involved in BMP-dependent osteoblast differentiation
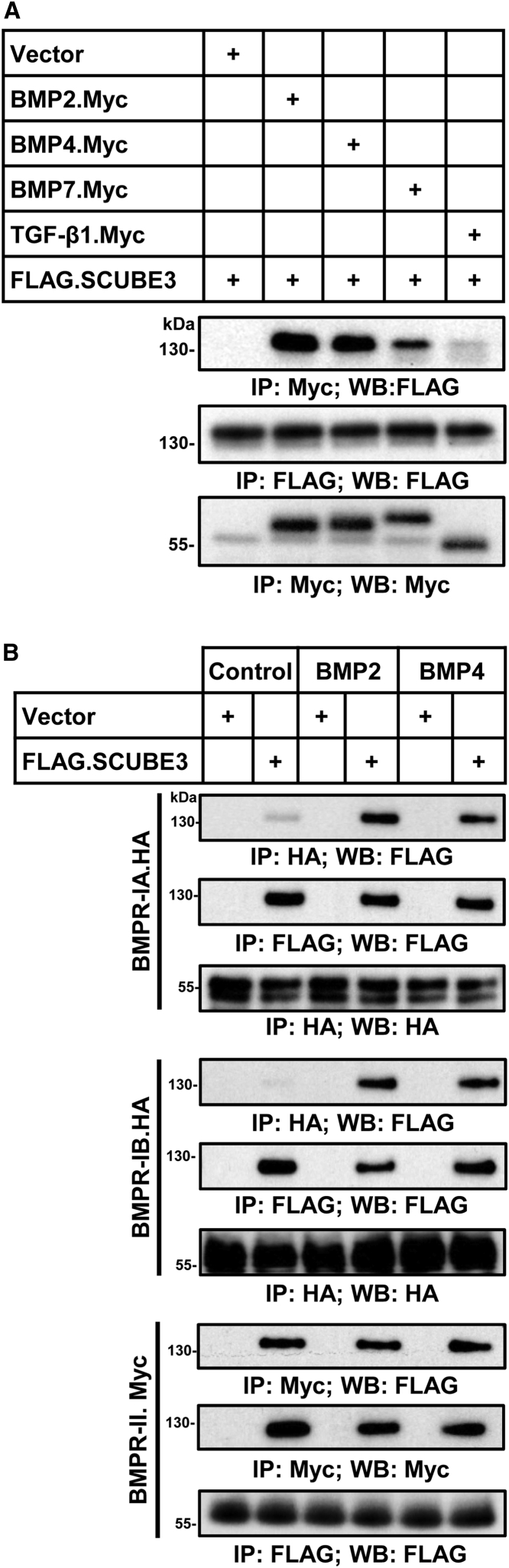
Based on the suggested role of SCUBE3 in BMP/TGF-β signaling,16 the role of BMP2 in skeletogenesis,4 and the remarkable phenotypic overlap between the subjects with pathogenic SCUBE3 variants and those with mutated BMP2, we hypothesized a role of SCUBE3 as an auxiliary modulator of BMP2 function. BMPs bind to a subset of type I or type II serine/threonine kinase receptors (BMPR-I and BMPR-II) forming activated heterotetrameric complexes that phosphorylate SMAD proteins, allowing them to translocate to the nucleus and upregulate transcription of ligand-responsive genes.5 To assess whether SCUBE3 interacts with ligands and receptors of the BMP signaling pathway, co-immunoprecipitation assays were performed using lysates of HEK293T cells transfected to co-overexpress FLAG-tagged SCUBE3 with Myc-tagged BMP2, BMP4, and BMP7, the three major BMP ligands implicated in early limb patterning and skeletogenesis,44 or TGF-β1, as a negative control. Immunoprecipitation with anti-Myc antibody followed by immunoblotting with anti-FLAG antibody documented co-precipitation of SCUBE3 with BMP2 and BMP4 and a slightly reduced co-immunoprecipitation with BMP7 (Figure 2A). The interaction was specific as no significant co-precipitation was attained with TGF-β1. In addition, domain mapping analysis further revealed that the CUB domain of SCUBE3 could specifically interact with the BMP2 ligand protein (Figure S8). The same experiments were performed co-expressing FLAG-SCUBE3 with HA-BMPR-IA, HA-BMPR-IB, or Myc-BMPR-II in cells stimulated with BMP2 or BMP4 (50 ng/mL, 20 min) or left untreated. The BMPR-I or -II protein was immunoprecipitated with its corresponding anti-epitope antibody and individual immunoprecipitates were analyzed by immunoblotting (Figure 2B). SCUBE3 co-immunoprecipitated with BMPR-IA and BMPR-IB in a ligand-dependent manner, while co-immunoprecipitation with BMPRII was ligand independent. Together, these data suggested a potential BMP ligand-inducible association between SCUBE3 and BMPR-IA or -IB but not BMPR-II.
Figure 2.
SCUBE3 interacts with ligands and receptors of the BMP signaling pathway
(A) SCUBE3 specifically interacts with BMP proteins but not TGF-β1. The expression plasmids encoding Myc-tagged BMP2, BMP4, BMP7, and TGF-β1 were co-transfected with a FLAG-tagged SCUBE3 construct in HEK293T cells. After 48 h, cell lysates underwent immunoprecipitation (IP), followed by western blot (WB) analysis with indicated antibodies to determine protein-protein interactions. Representative blots from one experiment of three performed are shown.
(B) SCUBE3 interacts with the receptors of the BMP signaling pathway. Plasmids encoding FLAG-tagged SCUBE3 were co-transfected with a HA-tagged BMPRIA, HA-tagged BMPRIB, or Myc-tagged BMPRII construct in HEK293T cells. After 48 h, transfected cells were stimulated with recombinant BMP2 or BMP4 protein for 20 min, and cell lysates were immunoprecipitated. Western blot analysis with the indicated antibodies was performed to determine protein protein interactions. Representative blots from one experiment of three performed are shown.
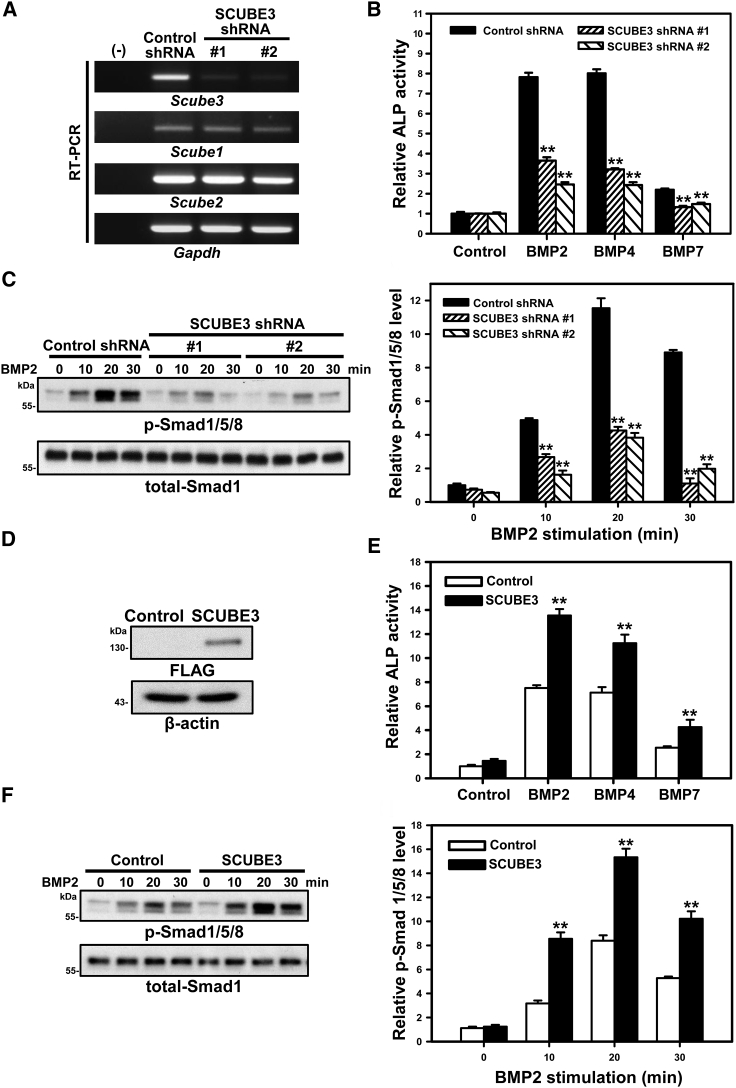
Based on the clinical phenotype of subjects and the observation that impaired BMP2 and BMP4 signaling is associated with defective bone formation and osteoblast differentiation,43,45 we further investigated the effect of SCUBE3 on BMP2/4-stimulated osteoblast differentiation by measuring the induced alkaline phosphatase (ALP) activity and the phosphorylation status of Smad1/5/8 in the mouse mesenchymal progenitor cell line C3H10T1/2. Two different Scube3-targeting short hairpin RNA lentiviruses (SCUBE3-shRNA #1 and #2) were used to knock down endogenous Scube3 expression. The specificity and efficiency of shRNA-mediated knockdown of mRNA expression of Scube3 were validated by RT-PCR analysis (Figure 3A), and a shRNA lentivirus targeting bacterial β-galactosidase (lacZ) was used as negative control. BMP2-, BMP4-, or BMP7-induced ALP activity (Figure 3B) and downstream activation of Smad1/5/8 (Figures 3C and S9A) were significantly and concomitantly suppressed by Scube3 knockdown. Consistently, ectopic SCUBE3 overexpression in C3H10T1/2 cells resulted in a marked induction of ALP activity (Figures 3D and 3E) as well as in an enhanced BMP2/4-induced phosphorylation of Smad1/5/8 (Figures 3F and S9B). These findings provided evidence of a positive modulatory role of SCUBE3 on BMP2/4 signaling exerting osteogenic function by modulating BMP-stimulated osteoblast differentiation, at least in vitro.
Figure 3.
SCUBE3 is an enhancer of BMP signaling and is involved in BMP-induced osteoblast differentiation
(A) Endogenous SCUBE3 expression was suppressed by two different SCUBE3-targeting short hairpin RNA (shRNA) lentiviruses (SCUBE3-shRNA #1 or #2). A luciferase shRNA lentivirus was used as a negative control (Control-shRNA). The efficiency and specificity of Scube3 mRNA knockdown was confirmed by RT-PCR. Gapdh mRNA level was used as internal control. Expression levels from one experiment of three performed are shown.
(B) C3H10T1/2 cells expressing control-shRNA or SCUBE3-targeting shRNAs were cultured without or with BMP (50 ng/mL) for 7 days. Relative ALP activity normalized with control values is shown. The experiments were performed 3 times in triplicate. Data are mean ± SD. ∗∗p < 0.01.
(C) Effect of SCUBE3 knockdown on the BMP2-stimulated phosphorylation of Smad1/5/8 were assessed by western blot analysis, and total-Smad1 expression was used as an internal control (left panel). Quantification by densitometric analysis of BMP2-induced phosphorylation of Smad1/5/8 in control or SCUBE3 knockdown C3H10T1/2 cells. Data are mean ± SD from 3 independent experiments. ∗∗p < 0.01 (right panel).
(D) Exogenous expression of FLAG-tagged SCUBE3 in C3H10T1/2 cells by a recombinant lentivirus. Protein overexpression of SCUBE3 was confirmed by western blot analysis. β-actin was used as internal control. Representative blots from one experiment of three performed are shown.
(E) SCUBE3 overexpression enhances BMP2-stimulated osteoblast differentiation in C3H10T1/2 cells. C3H10T1/2 control (empty lentivirus) or SCUBE3-overexpressing cells were cultured with or without BMP (50 ng/mL) for 7 days. Relative ALP activity was calculated by normalizing with the respective control value. The experiments were performed 3 times in triplicate. Data are mean ± SD. ∗∗p < 0.01.
(F) SCUBE3 overexpression enhances BMP2 downstream signaling in C3H10T1/2 cells. Western blot analysis (left panel) and quantification (right panel) of BMP2-induced phosphorylation of Smad1/5/8 with control and SCUBE3 overexpression in C3H10T1/2 cells. Data are mean ± SD from three independent experiments. ∗∗p < 0.01.
Pathogenic SCUBE3 variants variably affect BMP signaling
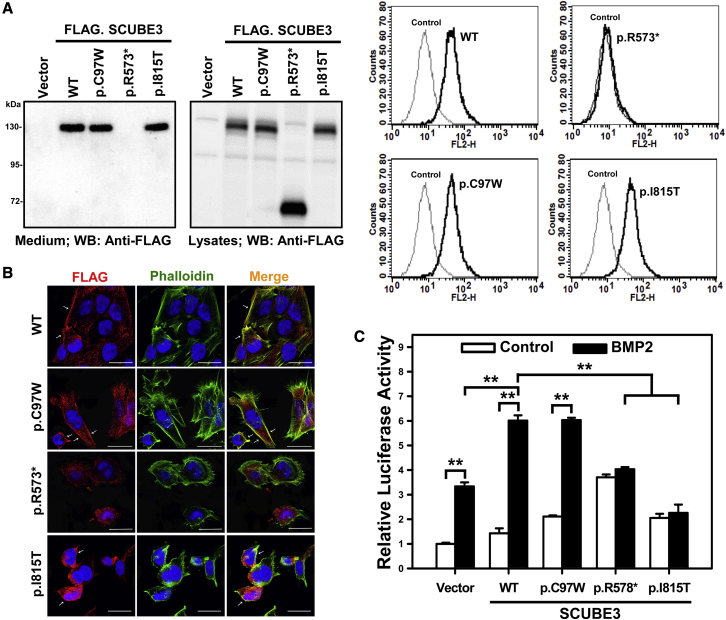
The bi-allelic occurrence and wide spectrum of identified variants pointed to a disruptive impact on SCUBE3 function as the pathogenetic mechanism. To validate this hypothesis, the consequences of a representative panel of variants affecting different domains of the protein (p.Cys97Trp, p.Arg573∗, and p.Ile815Thr) on SCUBE3 stability, secretion, proper membrane anchoring, and functional behavior were investigated by transient transfection experiments. Western blot analyses using whole cell lysates and conditional media, confocal analysis, and flow cytometry documented that the p.Cys97Trp and p.Ile815Thr variant protein levels were comparable to that of the wild-type SCUBE3 and were properly secreted and tethered on the cell surface; in contrast, while the truncated variant protein levels were comparable to the wild-type counterpart, it was neither expressed on the cell surface nor was secreted in the medium, providing evidence of protein mistargeting possibly due to a trafficking defect and endoplasmic reticulum retention (Figures 4A and 4B). For this nonsense variant, an impact on mRNA stability due to nonsense-mediated decay as an alternative loss-of-function effect could not be assessed experimentally and cannot be ruled out.
Figure 4.
Effect of disease-causing SCUBE3 variants on cell-surface protein levels and secretion and on BMP2 signaling
(A) SCUBE3 levels were analyzed in HEK293T cells. Samples from conditioned media and total cell lysates were collected and analyzed by western blot analysis using an anti-FLAG antibody (left panel). Empty vector-transfected cells were used as control. Representative blots from single experiments of three performed each are shown. A set of transfected cells was stained with anti-FLAG antibody and underwent flow cytometry (right panel).
(B) SAOS-2 cells were transfected with the expression plasmids encoding wild-type (WT) SCUBE3 and disease-associated variants (p.Cys97Trp, p.Arg573∗, and p.Ile815Thr). After 48 h, transfected cells were stained with mouse anti-FLAG antibody, Alexa Fluor 594 goat anti-mouse secondary antibody (red) and were analyzed by confocal microscopy. Alexa Fluor 488 phalloidin dye was used to stain the cortical actin associated with the plasma membrane (green). Nuclei are DAPI stained (blue). Merged images are shown in the right panels. White arrows indicate the distribution of the SCUBE3 proteins on the cellular surface in transfected cells. Red arrows indicate the dispersed localization of the p.Arg573∗ SCUBE3 variant in transfected cells. Scale bars represent 25 μm.
(C) HepG2 cells were transfected with the BMP-responsive luciferase reporter (BRE-luc) and pRL-TK alone or with the indicated expression plasmids. After 24 h, transfected cells were incubated for another 24 h with and without BMP2 (50 ng/mL), then luciferase activity was measured. Relative luciferase activity represents firefly luciferase values normalized to Renilla activity. The experiments were performed 3 times in triplicate. Data are mean ± SD. ∗∗p < 0.01.
SCUBE proteins are known to both homo- and heterodimerize.11 To evaluate a possible impact of mutations on dimerization, HEK293T cells were transfected to express FLAG-tagged wild-type SCUBE3 or individual variants with the respective Myc-tagged SCUBE3 proteins. Co-immunoprecipitation assays documented a reduced homodimerization of the p.Cys97Trp variant protein compared to wild-type SCUBE3, and the p.Arg573∗ and p.Ile815Thr variants (Figure S10A), suggesting a diverse mechanism of functional dysregulation promoted by the variants affecting the EGF-like and CUB domains. Co-transfection experiments using FLAG-tagged SCUBE1, SCUBE2, and SCUBE3 with Myc-tagged wild-type SCUBE3 or the p.Cys97Trp variant documented a less efficient dimerization of the variant with wild-type SCUBE3, while no significant difference was observed in its binding to SCUBE1 and SCUBE2 (Figure S10B).
Based on the positive modulatory role of SCUBE3 on BMP signaling, a BMP2 transactivation assay was finally performed to assess the impact of mutations on proper boosting of BMP signaling. HepG2 cells were transfected with a BMP-responsive luciferase reporter (BRE-Luc) alone or with wild-type SCUBE3 or each variant protein (Figure 4C), and stimulated with BMP2 (50 ng/mL, 24 h) or left unstimulated. While overexpression of the wild-type protein resulted in a 5-fold increase of BMP2 signaling activity, no difference in the transactivation activity of cells expressing the p.Ile815Thr or p.Arg573∗ SCUBE3 protein was observed after BMP2 stimulation, indicating a disruptive effect of both mutants on BMP signaling in vitro. No reduction in the transactivation assay was observed in cells expressing the p.Cys97Trp SCUBE3 protein, supporting further the view of a different perturbing effect of pathogenic variants affecting the EGF-like modules, at least in the used experimental conditions.
Scube3−/− mice have defective skeletogenesis and compromised endochondral bone formation due to defective BMP signaling
We performed a detailed phenotypic and molecular characterization of a previously generated Scube3 knock-out mouse model.21 As previously reported, Scube3−/− mice were viable and did not show any macroscopically visible abnormality at birth. However, Scube3−/− pups (P1, postnatal day 1) were shorter than their control littermates (Figure 5A). Consistent with the clinical features of affected individuals, Scube3−/− animals also showed misaligned upper/lower incisors (Figure 5B) and altered craniofacial development, including a shorter and narrower face, a smaller forehead, and a shorten frontonasal and mandibular region (Figure 5C). Occasionally, a hunchback spine (Figure 5D) and protruding rib cage (Figure 5E) were also observed.
Figure 5.
Impaired endochondral bone formation and chondrogenesis in Scube3-deficient mice
(A) Skeletons of WT Scube3 (+/+) and Scube3 KO (−/−) newborns (P1) stained with Alizarin red (calcified tissue) and Alcian blue (cartilage). Scale bar = 1 cm.
(B) Misaligned incisors observed in −/− 8-week-old mice.
(C) Small lower jaw in −/− 8-week-old mice.
(D) Hunchback spine in −/− 8-week-old mice.
(E) Thoracic cavity of +/+ and −/− animals (P1) stained with Alizarin red and Alcian blue. The asterisk shows the sternum protrusion in −/− mice.
(F) RT-PCT analysis of Scube1/2/3 expression in kidney, testis and femur from +/+ and −/− adult animals (8 week-old). Expression of Gapdh was used as an internal control. The experiments were performed 3 times.
(G) Forelimb of +/+ and −/− animals (P1) stained with Alizarin red and Alcian blue. The dotted line indicates the corresponding length.
(H) Hindlimbs of +/+ and −/− animals (P1) stained with Alizarin red and Alcian blue. fe, femur; t, tibia; fi, fibula. Tibial lengths are marked (upper panel) and quantified (lower panel). Data are mean ± SD (n = 5 in each group). ∗∗p < 0.01.
(I) Photographs of adult (8-week-old) +/+ and −/− mice (nose-to-anus was marked). Scale bar = 1 cm.
(J) Whole-body radiographs. X-ray images of 8-week-old +/+ and −/− male mice.
(K) Body weight from 4- to 8-week-old +/+ (n = 7) and −/− (n = 9) mice. Data are mean ± SD. ∗∗p < 0.01.
(L) Body length of 8-week-old +/+ (n = 7) and −/− (n = 9) mice. Data are mean ± SD. ∗∗p < 0.01.
(M) Femur and tibia length of 8-week-old +/+ (n = 8) and −/− (n = 7) mice. Data are mean ± SD. ∗∗p < 0.01.
(N) Graphic illustration showing the structure of the growth plate, including resting chondrocytes (a), proliferative chondrocytes (b), prehypertrophic chondrocytes (c), hypertrophic chondrocytes (d), and osteoblast/blood vessel (e).
(O) Alcian blue hematoxylin/orange G staining showing a shorter growth plate in E16.5 (n = 5) and P1 (n = 6) −/− mice. Scale bar = 100 μm.
(P) Quantification of different chondrocyte layer lengths in growth plates of P1 +/+ and −/− animals (n = 6).
(Q and R) Chondrocyte micromass cultures of mesenchymal cells from E12.5 +/+ (n = 5) and −/− (n = 5) mouse limbs treated with/without BMP signaling inhibitor (noggin) (50 ng/mL), BMP2 or BMP7 for 9 d (Q). Alcian blue staining was quantified by solubilizing the sample in 6 M guanidine hydrochloride, followed by OD620 measurement by spectrophotometry (R). Data are mean ± SD (n = 5). ∗∗p < 0.01.
Based on the reduced growth and skeletal features associated with bi-allelic SCUBE3 variants, we explored the role of Scube3 on processes linked to skeletal development. RT-PCR and quantitative RT-PCR analyses performed on control newborn pups showed that Scube3 expression is highly expressed in long bone tissues as compared with Scube1 and Scube2 (Figure S11). Of note, loss of Scube3 expression did not induce compensatory upregulation of Scube1 and Scube2 in femoral tissues (Figure 5F). Immunohistochemical staining revealed that Scube3 is specifically expressed in the periosteum and trabecular endosteum, where osteoprogenitor cells and osteoblasts reside (Figure S12). High expression levels were also observed in proliferative/prehypertrophic chondrocytes of the growth plate, where also BMP2 and BMP4 are highly expressed,46,47 suggesting a role of the protein in BMP-mediated chondrogenesis and/or osteoblastogenesis.
In line with the skeletal expression of Scube3, Alcian blue (chondrocyte matrix) and Alizarin red (mineralized tissue) staining documented proper position and number of the skeletal elements in Scube3−/− mice. Yet, skeletogenesis was apparently defective, with all appendicular (forelimbs and hindlimbs) and axial (skull, vertebral column, and rib cage) skeletal elements smaller in Scube3−/− mice compared control littermates (Figures 5A, 5E, 5G, and 5H). The defective skeletal growth persisted up to adulthood, as the adult body size was reduced as compared with control littermates and the general skeletal structure (e.g., skull, axial skeleton, ribs, pelvis, and long bones), as revealed by whole-body radiography, appeared grossly normal but smaller than control animals (Figures 5I and 5J). Quantification of physical parameters revealed reduced body weight (−14%), nose-to-anus body length (−23%), femoral bone length (−14%), and tibial bone length (−9%) (Figures 5K–5M). In line with these findings, micro-CT analysis revealed a marked reduction of trabecular volumetric bone mineral density (−22%), trabecular bone volume (−42%), trabecular thickness (−17%), and trabecular number (−37%) (data not shown). Analysis of the growth plate structure of proximal tibia of Scube3−/− embryos (E16.5) and pups (P1) documented a consistently reduced length of the plate compared to age-matched littermates (Figures 5N and 5O). Histological analysis also revealed a significant reduction in length of the resting, proliferative, and prehypertrophic chondrocyte zone of growth plates in P1 Scube3−/− pups compared to control animals (Figures 5P and S13A). In addition, a significant reduction of the osteoblast/blood vessel invasion zone was observed, indicating reduced endochondral ossification at the growth plates (Figures 5P and S13A). Notably, osteoblasts derived from Scube3−/− embryos (E18.5) cultured in osteogenic differentiation media showed reduced ALP activity compared to the corresponding osteoblast cultures from control embryos (Figure S14). Furthermore, high-density micromass cell cultures of embryonic tibiae mesenchymal cells from Scube3−/− and control animals documented less efficient chondrogenic differentiation in the former (Figures 5Q and 5R). These cells were less responsive to BMP2- or BMP7-stimulated chondrogenic differentiation, suggesting a costimulatory role of Scube3 in BMP-mediated osteo-/chondrogenesis (Figures 5Q and 5R). We further analyzed chondrocyte proliferation by BrdU labeling and showed that chondrocyte proliferation in tibiae was markedly reduced (−70%) in P1 Scube3−/− mice compared to control animals (Figures S13B and S13C). Finally, the expression of a panel of osteogenic markers (i.e., Runx2, Col1a1, Alpl, Bglap, Osx, and Bsp) was markedly reduced in Scube3−/− cultured chondrocytes and isolated long bones (Figure S15).
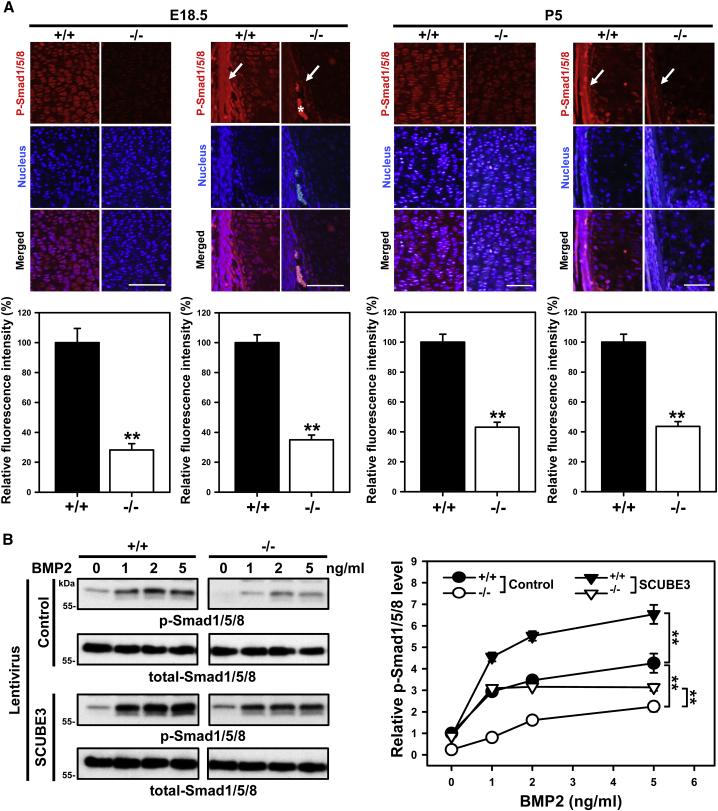
To further confirm the positive co-receptor role of Scube3 in promoting BMP signaling, the phosphorylation status of Smad1/5/8 was examined in proliferative chondrocytes and periosteal osteoprogenitor cells in long-bone sections of embryonic (E18.5) and postnatal stages (P5) Scube3−/− animals by immunofluorescence staining, and in freshly isolated chondrocytes treated with recombinant BMP2 by western blot analysis. In both ex vivo and in vitro conditions, Smad1/5/8 phosphorylation was significantly decreased in Scube3−/− mice-derived tissues/cells (Figures 6A and 6B). Consistently, BMP2 treatment increased Smad1/5/8 phosphorylation in a dose-dependent manner in control chondrocytes, while a significantly less efficient induction was observed in Scube3−/− cultures (Figure 6B). Notably, lentivirus-mediated ectopic expression of Scube3 rescued the defective BMP2 signaling in Scube3−/− chondrocytes and further enhanced BMP2 signaling in control chondrocytes (Figure 6B), confirming the role of the protein in regulating BMP-induced Smad signaling in proliferative chondrocytes and periosteal osteoprogenitor cells.
Figure 6.
Reduction of Smad1/5/8 phosphorylation in Scube3-deficient proliferative chondrocytes and periosteal osteoprogenitor cells
(A) Immunohistochemical analysis of p-Smad1/5/8 in proliferative chondrocytes (left panel) and periosteal osteoprogenitor cells (right panel) of E18.5 or P5 Scube3 wild-type (+/+) or knock-out (−/−) mouse bone tissues. Sections of tibiae were stained with a rabbit polyclonal antibody for p-Smad1/5/8. Arrows marked p-Smad1/5/8 signal within periosteal osteoprogenitor cells. The asterisk indicates red blood cell autofluorescence. Scale bar = 100 μm. Fluorescence intensity of p-Smad1/5/8 signals was quantified by ImageJ. Data are mean ± SD (n = 5 in each group). ∗∗p < 0.01.
(B) SCUBE3 modulates BMP2 signaling in primary cultured chondrocytes. Chondrocytes were isolated from +/+ and −/− mice (P2). Exogenous SCUBE3 expression was produced by SCUBE3 lentiviruses. Chondrocytes were serum-starved overnight, then stimulated with indicated doses of BMP2 for 20 min. Western blot analysis (left panel) and quantification (right panel) of BMP2-induced phosphorylation of Smad1/5/8 in chondrocytes. Data are mean ± SD from 3 independent experiments. ∗∗p < 0.01.
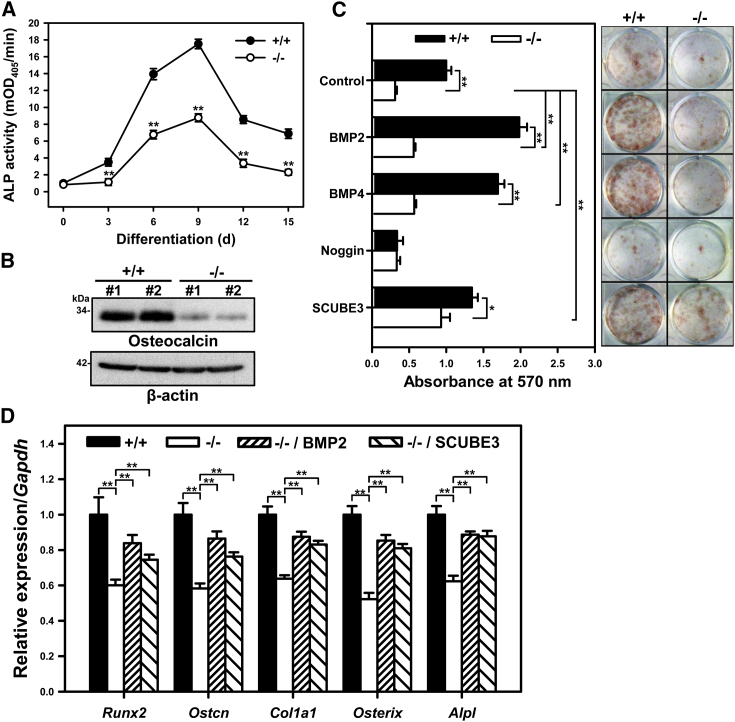
To test the hypothesis that Scube3−/− mice have defective osteoblast differentiation, bone-marrow stromal cells (BMSCs) derived from Scube3−/− and control mice were cultured in osteogenic differentiation media, and the degree of ex vivo osteogenic differentiation was evaluated by measuring ALP activity, osteocalcin expression, and Alizarin red staining for matrix mineralization. ALP activity increased over time and peaked at day 9 in BMSC cultures for both genotypes, with marked reduced level in Scube3−/− cell cultures (by 30% to 50%) (Figure 7A). Likewise, protein levels of osteocalcin were significantly lower in Scube3−/− cell cultures (>90%) (Figure 7B), and matrix mineralization was significantly reduced both basally and following stimulation with BMP2 or BMP4 (Figure 7C). Of note, restoration of SCUBE3 levels by lentiviral ectopic expression rescued, in part, matrix mineralization (Figure 7C). Consistent with these findings, Scube3−/− BMSC osteogenic cultures showed a reduced expression of diverse osteogenic marker genes (e.g., Runx2, Ostcn, Col1a1, Osterix, and Alpl); such defects could be rescued by the exogenous addition of BMP2 or ectopic expression of SCUBE3 (Figure 7D), further demonstrating that BMP2/4 signaling is involved in Scube3-mediated osteoblast differentiation and bone matrix production.
Figure 7.
Ex vivo osteogenic differentiation of Scube3+/+ and Scube3−/− bone-marrow stromal cell (BMSCs) assessed by ALP activity, osteocalcin expression, and Alizarin red staining
(A) ALP activity of Scube3 wild-type (+/+) and knock-out (−/−) BMSCs under osteoblast differentiation conditions at days 3, 6, 9, 12, and 15. The experiments were performed 3 times in triplicate. Data are mean ± SD mOD405, milli-absorbance units at 405 nm. ∗∗p < 0.01.
(B) The protein expression of osteocalcin in +/+ and −/− BMSCs under osteoblast differentiation conditions at day 9. Two independent BMSC cultures (#1 and #2) were used for western blot analysis, and each experiment was performed 3 times.
(C) Bone nodule formation in BMSC cultures under osteoblast differentiation conditions (right panel). Quantification of Alizarin red staining (left panel). The experiments were performed 3 times in triplicate. Data are mean ± SD. ∗p < 0.05; ∗∗p < 0.01.
(D) The mRNA expression of direct targets of BMP signal activity and osteoblast marker genes in +/+ and −/− BMSCs under osteoblast differentiation conditions at day 9. The experiments were performed 3 times in triplicate. Data are mean ± SD. ∗∗p < 0.01.
Overall, the collected data indicate that Scube3−/− mice have craniofacial and dental defects, reduced body size, and defective endochondral bone growth, recapitulating the human disorder, and that defective skeletogenesis and compromised endochondral bone formation results from defective BMP signaling.
Scube3 is required for BMP receptor complex recruitment in lipid rafts
Because BMP receptor association with membrane microdomains (i.e., lipid rafts) is a critical event for BMP signaling48 and overexpressed SCUBE3 is a raft-associated protein (Figures S16A and S16B), we examined whether raft recruitment of BMP receptors is altered in Scube3−/− chondrocytes. Quantitative PCR and western blot analyses showed a comparable level of osteogenic BMP ligands and their receptor subunits in Scube3−/− and control chondrocytes (Figure S17). Chondrocytes were homogenized and fractionated by Opti-Prep density gradient centrifugation. Among the 12 isolated fractions, fraction 5 was identified as the lipid raft fraction, as shown by the distribution of the raft marker protein, caveolin-1.49 BMP receptors (IA, IB, and II) do not localize in the raft fraction under unstimulated conditions (data not shown), but translocated into the lipid raft fraction following BMP stimulation (Figure S16C). Strikingly, this BMP-induced raft association of BMP receptors was completely abolished in Scube3−/− chondrocytes (Figure S16C), indicating a requirement of SCUBE3 for this stimulus-dependent redistribution of receptors. Overall, these data suggest a model in which SCUBE3 facilitates BMP2-induced BMP receptor association with lipid raft microdomains.
Discussion
The three members of the SCUBE gene family encode cell surface-anchored glycoproteins functioning as auxiliary receptors to modulate signaling elicited by a variety of growth factors. Despite their versatile role in signaling, these proteins had not previously been implicated in Mendelian diseases. Here we establish that bi-allelic, mostly inactivating variants in SCUBE3 underlie a recognizable developmental disorder and mechanistically link the defective growth characterizing this novel condition to a compromised endochondral bone formation due to defective BMP signaling.
SCUBE3 has been reported to be highly expressed in primary osteoblasts and long bones, as well as in cartilaginous condensation regions of the axial skeleton, including ribs and vertebrae.11,50 In mice, the protein has been detected in ectodermal, mesodermal, and endodermal derivatives.21 SCUBE3 has been proposed to modulate FGF, hedgehog, and TGF-β signaling,16,20,32,51 even though the precise molecular mechanisms implicated in signal modulation have not been characterized in detail. Conflicting data have apparently been collected regarding its role during development.21,52 Our findings indicate that, despite being dispensable for embryogenesis, Scube3 is required for appropriate skeletal development and growth. Specifically, we show that Scube3 contributes to fine-tuning BMP-dependent endochondral bone formation by regulating both chondrogenic differentiation and chondrocyte proliferation and also osteogenic differentiation. Of note, this osteogenic function is reminiscent of biglycan, which directly binds BMP2 and its receptors to accelerate osteoblast differentiation by positively modulating BMP2/4 activity.53
We show that cell surface-bound SCUBE3 is expressed in osteoblasts and chondrocytes and likely functions as a BMP2/4 co-receptor in a cell-autonomous manner. Mechanistically, our data reveal that SCUBE3 facilitates BMP2/4 ligand binding with BMP type I receptors and promotes BMP-induced BMP receptor complex association with lipid rafts, which are cholesterol- and caveolin-enriched domains serving as signaling platforms.54 These findings suggest a model in which SCUBE3 might form a complex with heterodimerized BMP receptors to facilitate and stabilize the BMP receptor complex in the lipid rafts. The precise nature and kinetics of SCUBE3-assisted recruitment of BMP receptor complexes in lipid rafts remains to be further investigated.
Detailed clinical phenotyping of 15 affected individuals allows us to profile the disorder resulting from impaired SCUBE3 function as a relatively homogeneous recessive trait characterized by short stature, perturbed skeletal and tooth development, and a distinctive facies. Notably, the condition overlaps, in part, with the disorder caused by heterozygous loss-of-function BMP2 mutations, whose cardinal features include short stature, skeletal malformations, dental crowding and enamel defects, palatal anomalies, and congenital heart defects.43 The observation that flawed tooth development and altered bone metabolism are recurrently associated with inactivating mutations in BMP445 further highlights the clinical relatedness and functional link of SCUBE3 with the two BMP ligands. In the context of bone metabolism, subjects harboring BMP4 mutations manifested early-onset osteopenia or osteoporosis. Although we could not perform bone densitometry in any subject of the present cohort, a marked reduction of trabecular volumetric bone mineral density (−22%) in Scube3−/− mice was observed compared to control animals. Remarkably, a similar phenotype had previously been reported in BMP2−/− mice.55 It should be noted that, differently from other human disorders linked to dysfunctional BMP signaling, these clinically related conditions do not show evidence of bone dysplasia.4
Consistent with the clinical features shared by the reported subjects with bi-allelic inactivating SCUBE3 variants, Scube3−/− mice displayed reduced growth associated with smaller forelimbs and hindlimbs. In addition, Scube3−/− mice frequently showed misaligned upper and lower incisors and craniofacial defects, including a shorter and narrower face, smaller forehead, and reduced frontonasal and mandibular regions. Occasionally, a protruding rib cage or hunchback spine was also observed. While no overt phenotype had previously been reported in Scube3−/− mice, features consistent with those we presently observed were reported in homozygous mice carrying a missense change within the seventh EGF-like domain of the protein (p.Asn294Lys), including skeletal defects (reduced upper and lower limbs, vertebral and thoracic malformations), altered bone metabolism, and conductive hearing loss.52 Additional investigations are required to further clarify this issue.
While our functional studies support the idea that disease-causing SCUBE3 variants differentially impact protein synthesis, processing (i.e., secretion), and function (dimerization versus enhancement of BMP2 signaling), the molecular impact of the variants affecting the EGF-like domain on intracellular signaling remains to be characterized. Consistent with our findings, pathogenic variants at equivalent positions within EGF-like domains have been identified in other proteins (e.g., FBN1, FBN2, and LTBP3) and proven to impair TGF-β signaling,38, 39, 40 and a bi-allelic missense variant (p.Asn294Lys) within the seventh EGF-like module in the mouse Scube3 ortholog has been shown to cause morphological abnormalities of the skeleton and alteration of bone metabolism.52 We speculate that this group of mutations may hamper specific processes linked to the extracellular microenvironment, including protein-matrix assembly and protein shedding from the membrane,51,56 which, in turn, control or modulate SCUBE3 function.57
In summary, we identify a developmental disorder caused by defective function of SCUBE3, link this secreted glycoprotein to processes controlling growth, morphogenesis, and teeth development, and provide evidence of its relevant role in BMP signaling-mediated skeletogenesis.
Data and Code Availability
All mutations identified in this work have been submitted to ClinVar (Genbank: NM_152753.4: c.291C>G, SCV001438405; NM_152753.4: c.611G>A, SCV001438406; NM_152753.4: c.829+1_952del, SCV001438407; NM_152753.4: c.1717C>T, SCV001438408; NM_152753.4: c.2239+1G>A, SCV001438409; NM_152753.4: c.2444T>C, SCV001438410; NM_152753.4: c.2599+2T>C, SCV001438411; NM_152753.4: c.2785C>T, SCV001438412). WES datasets have not been deposited in a public repository due to privacy and ethical restrictions but are available from the corresponding authors on request.
Declaration of interests
C.B., P.B., and N.-M.G. declare no additional conflicts of interest beyond their employment affiliation. All the other authors declare no competing interests.
Acknowledgments
The authors wish to thank the participating families and Drs. R.A.J. Nievelstein (University Medical Center Utrecht) and D. Barbuti (Ospedale Pediatrico Bambino Gesù) for radiological advice. The authors would also thank Prof. Kassim Javaid and other members of musculoskeletal GeCIP (Genomics England). This research used data and findings generated by the 100,000 Genomes Project. This work was supported by grants from: Fondazione Bambino Gesù (Vite Coraggiose to M.T.) and Italian Ministry of Health (CCR-2017-23669081 to M.T., Ricerca Corrente 2019 to M.N., and Ricerca Corrente 2019 and 2020 to M.L.D.), Academia Sinica and Ministry of Science and Technology of Taiwan (109-3111-Y-001-001, 109-2320-B-001-012-MY3, 107-3111-Y-001-048, 107-0210-01-19-01, 107-2320-B-001-015-MY3, 106-2320-B-001-017-MY3, 106-0210-01-15-02 to R.-B.Y. and 107-2321-B-001-036-MY3 to Y.-C.L.), São Paulo Research Foundation (FAPESP 2013/03236-5 to A.A.L.J. and 2018/10893-6 to B.L.F.), University of Tübingen Intramural Funding (fortüne 2545-1-0 to B.V.), Ministry of Science, Research and Art Baden-Württemberg (to B.V.), and Department of Science and Technology, Government of India (SB/SO/HS/005/2014 to K.M.G.). The 100,000 Genomes Project is funded by the National Institute for Health Research and NHS England. The 100,000 Genomes Project is managed by Genomics England Limited and uses data provided by patients and collected by the National Health Service as part of their care and support. See supplemental information for consortium details. Additional support was from the Wellcome Trust (203141/Z/16/Z) and the NIHR Biomedical Research Centre Oxford (to A.T.P. and J.C.T.).
Published: December 11, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.11.015.
Contributor Information
Ruey-Bing Yang, Email: rbyang@ibms.sinica.edu.tw.
Marco Tartaglia, Email: marco.tartaglia@opbg.net.
WEB resources
gnomAD Browser, https://gnomad.broadinstitute.org/
InterVar, http://wintervar.wglab.org
OMIM, https://www.omim.org/
Supplemental information
References
- 1.Hynes R.O. The extracellular matrix: not just pretty fibrils. Science. 2009;326:1216–1219. doi: 10.1126/science.1176009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walma D.A.C., Yamada K.M. The extracellular matrix in development. Development. 2020;147:dev175596. doi: 10.1242/dev.175596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonnans C., Chou J., Werb Z. Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Biol. 2014;15:786–801. doi: 10.1038/nrm3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu M., Chen G., Li Y.P. TGF-β and BMP signaling in osteoblast, skeletal development, and bone formation, homeostasis and disease. Bone Res. 2016;4:16009. doi: 10.1038/boneres.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salazar V.S., Gamer L.W., Rosen V. BMP signalling in skeletal development, disease and repair. Nat. Rev. Endocrinol. 2016;12:203–221. doi: 10.1038/nrendo.2016.12. [DOI] [PubMed] [Google Scholar]
- 6.Brazil D.P., Church R.H., Surae S., Godson C., Martin F. BMP signalling: agony and antagony in the family. Trends Cell Biol. 2015;25:249–264. doi: 10.1016/j.tcb.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 7.Gong Y., Krakow D., Marcelino J., Wilkin D., Chitayat D., Babul-Hirji R., Hudgins L., Cremers C.W., Cremers F.P., Brunner H.G. Heterozygous mutations in the gene encoding noggin affect human joint morphogenesis. Nat. Genet. 1999;21:302–304. doi: 10.1038/6821. [DOI] [PubMed] [Google Scholar]
- 8.Michos O., Panman L., Vintersten K., Beier K., Zeller R., Zuniga A. Gremlin-mediated BMP antagonism induces the epithelial-mesenchymal feedback signaling controlling metanephric kidney and limb organogenesis. Development. 2004;131:3401–3410. doi: 10.1242/dev.01251. [DOI] [PubMed] [Google Scholar]
- 9.Bachiller D., Klingensmith J., Shneyder N., Tran U., Anderson R., Rossant J., De Robertis E.M. The role of chordin/Bmp signals in mammalian pharyngeal development and DiGeorge syndrome. Development. 2003;130:3567–3578. doi: 10.1242/dev.00581. [DOI] [PubMed] [Google Scholar]
- 10.Yang R.B., Ng C.K., Wasserman S.M., Colman S.D., Shenoy S., Mehraban F., Komuves L.G., Tomlinson J.E., Topper J.N. Identification of a novel family of cell-surface proteins expressed in human vascular endothelium. J. Biol. Chem. 2002;277:46364–46373. doi: 10.1074/jbc.M207410200. [DOI] [PubMed] [Google Scholar]
- 11.Wu B.T., Su Y.H., Tsai M.T., Wasserman S.M., Topper J.N., Yang R.B. A novel secreted, cell-surface glycoprotein containing multiple epidermal growth factor-like repeats and one CUB domain is highly expressed in primary osteoblasts and bones. J. Biol. Chem. 2004;279:37485–37490. doi: 10.1074/jbc.M405912200. [DOI] [PubMed] [Google Scholar]
- 12.Lin Y.C., Chen C.C., Cheng C.J., Yang R.B. Domain and functional analysis of a novel breast tumor suppressor protein, SCUBE2. J. Biol. Chem. 2011;286:27039–27047. doi: 10.1074/jbc.M111.244418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grimmond S., Larder R., Van Hateren N., Siggers P., Hulsebos T.J., Arkell R., Greenfield A. Cloning, mapping, and expression analysis of a gene encoding a novel mammalian EGF-related protein (SCUBE1) Genomics. 2000;70:74–81. doi: 10.1006/geno.2000.6370. [DOI] [PubMed] [Google Scholar]
- 14.Tu C.F., Su Y.H., Huang Y.N., Tsai M.T., Li L.T., Chen Y.L., Cheng C.J., Dai D.F., Yang R.B. Localization and characterization of a novel secreted protein SCUBE1 in human platelets. Cardiovasc. Res. 2006;71:486–495. doi: 10.1016/j.cardiores.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 15.Tsai M.T., Cheng C.J., Lin Y.C., Chen C.C., Wu A.R., Wu M.T., Hsu C.C., Yang R.B. Isolation and characterization of a secreted, cell-surface glycoprotein SCUBE2 from humans. Biochem. J. 2009;422:119–128. doi: 10.1042/BJ20090341. [DOI] [PubMed] [Google Scholar]
- 16.Wu Y.Y., Peck K., Chang Y.L., Pan S.H., Cheng Y.F., Lin J.C., Yang R.B., Hong T.M., Yang P.C. SCUBE3 is an endogenous TGF-β receptor ligand and regulates the epithelial-mesenchymal transition in lung cancer. Oncogene. 2011;30:3682–3693. doi: 10.1038/onc.2011.85. [DOI] [PubMed] [Google Scholar]
- 17.Tu C.F., Yan Y.T., Wu S.Y., Djoko B., Tsai M.T., Cheng C.J., Yang R.B. Domain and functional analysis of a novel platelet-endothelial cell surface protein, SCUBE1. J. Biol. Chem. 2008;283:12478–12488. doi: 10.1074/jbc.M705872200. [DOI] [PubMed] [Google Scholar]
- 18.Tukachinsky H., Kuzmickas R.P., Jao C.Y., Liu J., Salic A. Dispatched and scube mediate the efficient secretion of the cholesterol-modified hedgehog ligand. Cell Rep. 2012;2:308–320. doi: 10.1016/j.celrep.2012.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin Y.C., Chao T.Y., Yeh C.T., Roffler S.R., Kannagi R., Yang R.B. Endothelial SCUBE2 Interacts With VEGFR2 and Regulates VEGF-Induced Angiogenesis. Arterioscler. Thromb. Vasc. Biol. 2017;37:144–155. doi: 10.1161/ATVBAHA.116.308546. [DOI] [PubMed] [Google Scholar]
- 20.Tu C.F., Tsao K.C., Lee S.J., Yang R.B. SCUBE3 (signal peptide-CUB-EGF domain-containing protein 3) modulates fibroblast growth factor signaling during fast muscle development. J. Biol. Chem. 2014;289:18928–18942. doi: 10.1074/jbc.M114.551929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xavier G.M., Panousopoulos L., Cobourne M.T. Scube3 is expressed in multiple tissues during development but is dispensable for embryonic survival in the mouse. PLoS ONE. 2013;8:e55274. doi: 10.1371/journal.pone.0055274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flex E., Niceta M., Cecchetti S., Thiffault I., Au M.G., Capuano A., Piermarini E., Ivanova A.A., Francis J.W., Chillemi G. Biallelic Mutations in TBCD, Encoding the Tubulin Folding Cofactor D, Perturb Microtubule Dynamics and Cause Early-Onset Encephalopathy. Am. J. Hum. Genet. 2016;99:962–973. doi: 10.1016/j.ajhg.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lam M.T., Coppola S., Krumbach O.H.F., Prencipe G., Insalaco A., Cifaldi C., Brigida I., Zara E., Scala S., Di Cesare S. A novel disorder involving dyshematopoiesis, inflammation, and HLH due to aberrant CDC42 function. J. Exp. Med. 2019;216:2778–2799. doi: 10.1084/jem.20190147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turro E., Astle W.J., Megy K., Gräf S., Greene D., Shamardina O., Allen H.L., Sanchis-Juan A., Frontini M., Thys C., NIHR BioResource for the 100,000 Genomes Project Whole-genome sequencing of patients with rare diseases in a national health system. Nature. 2020;583:96–102. doi: 10.1038/s41586-020-2434-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Homma T.K., Freire B.L., Honjo Kawahira R.S., Dauber A., Funari M.F.A., Lerario A.M., Nishi M.Y., Albuquerque E.V., Vasques G.A., Collett-Solberg P.F. Genetic Disorders in Prenatal Onset Syndromic Short Stature Identified by Exome Sequencing. J. Pediatr. 2019;215:192–198. doi: 10.1016/j.jpeds.2019.08.024. [DOI] [PubMed] [Google Scholar]
- 26.Vona B., Mazaheri N., Lin S.J., Dunbar L.A., Maroofian R., Azaiez H., Booth K.T., Vitry S., Rad A., Varshney P. Biallelic mutation of CLRN2 causes non-syndromic hearing loss in humans. bioRxiv. 2020 doi: 10.1101/2020.07.29.222828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monies D., Abouelhoda M., Assoum M., Moghrabi N., Rafiullah R., Almontashiri N., Alowain M., Alzaidan H., Alsayed M., Subhani S. Lessons Learned from Large-Scale, First-Tier Clinical Exome Sequencing in a Highly Consanguineous Population. Am. J. Hum. Genet. 2019;104:1182–1201. doi: 10.1016/j.ajhg.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bauer P., Kandaswamy K.K., Weiss M.E.R., Paknia O., Werber M., Bertoli-Avella A.M., Yüksel Z., Bochinska M., Oprea G.E., Kishore S. Development of an evidence-based algorithm that optimizes sensitivity and specificity in ES-based diagnostics of a clinically heterogeneous patient population. Genet. Med. 2019;21:53–61. doi: 10.1038/s41436-018-0016-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mak K.K., Kronenberg H.M., Chuang P.T., Mackem S., Yang Y. Indian hedgehog signals independently of PTHrP to promote chondrocyte hypertrophy. Development. 2008;135:1947–1956. doi: 10.1242/dev.018044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maniatopoulos C., Sodek J., Melcher A.H. Bone formation in vitro by stromal cells obtained from bone marrow of young adult rats. Cell Tissue Res. 1988;254:317–330. doi: 10.1007/BF00225804. [DOI] [PubMed] [Google Scholar]
- 31.Nishimura R., Hata K., Harris S.E., Ikeda F., Yoneda T. Core-binding factor alpha 1 (Cbfa1) induces osteoblastic differentiation of C2C12 cells without interactions with Smad1 and Smad5. Bone. 2002;31:303–312. doi: 10.1016/s8756-3282(02)00826-8. [DOI] [PubMed] [Google Scholar]
- 32.Zhu D., Xiong W.C., Mei L. Lipid rafts serve as a signaling platform for nicotinic acetylcholine receptor clustering. J. Neurosci. 2006;26:4841–4851. doi: 10.1523/JNEUROSCI.2807-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin Y.C., Roffler S.R., Yan Y.T., Yang R.B. Disruption of Scube2 Impairs Endochondral Bone Formation. J. Bone Miner. Res. 2015;30:1255–1267. doi: 10.1002/jbmr.2451. [DOI] [PubMed] [Google Scholar]
- 34.Zhang M., Ho H.C., Sheu T.J., Breyer M.D., Flick L.M., Jonason J.H., Awad H.A., Schwarz E.M., O’Keefe R.-J. EP1(-/-) mice have enhanced osteoblast differentiation and accelerated fracture repair. J. Bone Miner. Res. 2011;26:792–802. doi: 10.1002/jbmr.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoffman L.M., Garcha K., Karamboulas K., Cowan M.F., Drysdale L.M., Horton W.A., Underhill T.M. BMP action in skeletogenesis involves attenuation of retinoid signaling. J. Cell Biol. 2006;174:101–113. doi: 10.1083/jcb.200604150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Firth H.V., Richards S.M., Bevan A.P., Clayton S., Corpas M., Rajan D., Van Vooren S., Moreau Y., Pettett R.M., Carter N.P. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am. J. Hum. Genet. 2009;84:524–533. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Collod-Béroud G., Béroud C., Ades L., Black C., Boxer M., Brock D.J., Holman K.J., de Paepe A., Francke U., Grau U. Marfan Database (third edition): new mutations and new routines for the software. Nucleic Acids Res. 1998;26:229. doi: 10.1093/nar/26.1.229. 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Belleh S., Zhou G., Wang M., Der Kaloustian V.M., Pagon R.A., Godfrey M. Two novel fibrillin-2 mutations in congenital contractural arachnodactyly. Am. J. Med. Genet. 2000;92:7–12. [PubMed] [Google Scholar]
- 40.Intarak N., Theerapanon T., Thaweesapphithak S., Suphapeetiporn K., Porntaveetus T., Shotelersuk V. Genotype-phenotype correlation and expansion of orodental anomalies in LTBP3-related disorders. Mol. Genet. Genomics. 2019;294:773–787. doi: 10.1007/s00438-019-01547-x. [DOI] [PubMed] [Google Scholar]
- 41.Liu W., Qian C., Comeau K., Brenn T., Furthmayr H., Francke U. Mutant fibrillin-1 monomers lacking EGF-like domains disrupt microfibril assembly and cause severe marfan syndrome. Hum. Mol. Genet. 1996;5:1581–1587. doi: 10.1093/hmg/5.10.1581. [DOI] [PubMed] [Google Scholar]
- 42.Xu P., Li R., Huang S., Sun M., Liu J., Niu Y., Zou Y., Li J., Gao M., Li X. A Novel Splicing Mutation in the FBN2 Gene in a Family With Congenital Contractural Arachnodactyly. Front. Genet. 2020;11:143. doi: 10.3389/fgene.2020.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tan T.Y., Gonzaga-Jauregui C., Bhoj E.J., Strauss K.A., Brigatti K., Puffenberger E., Li D., Xie L., Das N., Skubas I. Monoallelic BMP2 Variants Predicted to Result in Haploinsufficiency Cause Craniofacial, Skeletal, and Cardiac Features Overlapping Those of 20p12 Deletions. Am. J. Hum. Genet. 2017;101:985–994. doi: 10.1016/j.ajhg.2017.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bandyopadhyay A., Tsuji K., Cox K., Harfe B.D., Rosen V., Tabin C.J. Genetic analysis of the roles of BMP2, BMP4, and BMP7 in limb patterning and skeletogenesis. PLoS Genet. 2006;2:e216. doi: 10.1371/journal.pgen.0020216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu M., Wang H., Fan Z., Xie C., Liu H., Liu Y., Han D., Wong S.W., Feng H. BMP4 mutations in tooth agenesis and low bone mass. Arch. Oral Biol. 2019;103:40–46. doi: 10.1016/j.archoralbio.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feng J.Q., Xing L., Zhang J.H., Zhao M., Horn D., Chan J., Boyce B.F., Harris S.E., Mundy G.R., Chen D. NF-kappaB specifically activates BMP-2 gene expression in growth plate chondrocytes in vivo and in a chondrocyte cell line in vitro. J. Biol. Chem. 2003;278:29130–29135. doi: 10.1074/jbc.M212296200. [DOI] [PubMed] [Google Scholar]
- 47.Nilsson O., Parker E.A., Hegde A., Chau M., Barnes K.M., Baron J. Gradients in bone morphogenetic protein-related gene expression across the growth plate. J. Endocrinol. 2007;193:75–84. doi: 10.1677/joe.1.07099. [DOI] [PubMed] [Google Scholar]
- 48.Hartung A., Bitton-Worms K., Rechtman M.M., Wenzel V., Boergermann J.H., Hassel S., Henis Y.I., Knaus P. Different routes of bone morphogenic protein (BMP) receptor endocytosis influence BMP signaling. Mol. Cell. Biol. 2006;26:7791–7805. doi: 10.1128/MCB.00022-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Magee T., Pirinen N., Adler J., Pagakis S.N., Parmryd I. Lipid rafts: cell surface platforms for T cell signaling. Biol. Res. 2002;35:127–131. doi: 10.4067/s0716-97602002000200003. [DOI] [PubMed] [Google Scholar]
- 50.Haworth K., Smith F., Zoupa M., Seppala M., Sharpe P.T., Cobourne M.T. Expression of the Scube3 epidermal growth factor-related gene during early embryonic development in the mouse. Gene Expr. Patterns. 2007;7:630–634. doi: 10.1016/j.modgep.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 51.Yang M., Guo M., Hu Y., Jiang Y. Scube regulates synovial angiogenesis-related signaling. Med. Hypotheses. 2013;81:948–953. doi: 10.1016/j.mehy.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 52.Fuchs H., Sabrautzki S., Przemeck G.K., Leuchtenberger S., Lorenz-Depiereux B., Becker L., Rathkolb B., Horsch M., Garrett L., Östereicher M.A. The First Scube3 Mutant Mouse Line with Pleiotropic Phenotypic Alterations. G3 (Bethesda) 2016;6:4035–4046. doi: 10.1534/g3.116.033670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jongwattanapisan P., Terajima M., Miguez P.A., Querido W., Nagaoka H., Sumida N., Gurysh E.G., Ainslie K.M., Pleshko N., Perera L., Yamauchi M. Identification of the effector domain of biglycan that facilitates BMP-2 osteogenic function. Sci. Rep. 2018;8:7022. doi: 10.1038/s41598-018-25279-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lajoie P., Goetz J.G., Dennis J.W., Nabi I.R. Lattices, rafts, and scaffolds: domain regulation of receptor signaling at the plasma membrane. J. Cell Biol. 2009;185:381–385. doi: 10.1083/jcb.200811059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kamiya N., Mishina Y. New insights on the roles of BMP signaling in bone-A review of recent mouse genetic studies. Biofactors. 2011;37:75–82. doi: 10.1002/biof.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jakobs P., Schulz P., Schürmann S., Niland S., Exner S., Rebollido-Rios R., Manikowski D., Hoffmann D., Seidler D.G., Grobe K. Ca2+ coordination controls sonic hedgehog structure and its Scube2-regulated release. J. Cell Sci. 2017;130:3261–3271. doi: 10.1242/jcs.205872. [DOI] [PubMed] [Google Scholar]
- 57.Wouters M.A., Rigoutsos I., Chu C.K., Feng L.L., Sparrow D.B., Dunwoodie S.L. Evolution of distinct EGF domains with specific functions. Protein Sci. 2005;14:1091–1103. doi: 10.1110/ps.041207005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All mutations identified in this work have been submitted to ClinVar (Genbank: NM_152753.4: c.291C>G, SCV001438405; NM_152753.4: c.611G>A, SCV001438406; NM_152753.4: c.829+1_952del, SCV001438407; NM_152753.4: c.1717C>T, SCV001438408; NM_152753.4: c.2239+1G>A, SCV001438409; NM_152753.4: c.2444T>C, SCV001438410; NM_152753.4: c.2599+2T>C, SCV001438411; NM_152753.4: c.2785C>T, SCV001438412). WES datasets have not been deposited in a public repository due to privacy and ethical restrictions but are available from the corresponding authors on request.