Introduction
In December 2019, The American Journal of Human Genetics introduced a new annual feature1 designed to chronicle ten key advances in applying genomic information to clinical care in the previous twelve months of published literature. The Genomic Medicine Working Group of the National Advisory Council for Human Genome Research of the National Human Genome Research Institute (NHGRI) has continued this effort by identifying published advances in genomic medicine implementation on a monthly basis and posting them on a searchable website, “Accomplishments in Genomic Medicine” (see Web Resources). From these advances, ten papers viewed as the most significant based on the same criteria used in 2019 (Box 1) were selected for this 2020 Year in Review.
Box 1. Criteria for Inclusion of Papers in Genomic Medicine Year in Review 2019 and 2020.
-
•
Involve use of patients’ individual genomic variant information in clinical decision-making
-
•
Demonstrate impact of direct clinical implementation
-
•
Are likely to be generalizable beyond original setting
-
•
Are likely to have implications for healthcare systems or practice guidelines
-
•
Are of sufficient size to be robust to sampling error
-
•
Are broadly representative of the field beyond NHGRI-sponsored or US-funded programs
Looking back at the ten publications highlighted last year, it is reassuring to see that several received considerable attention in subsequently published papers, as judged by the imperfect metric of PubMed citations. The median number of citations for the 2019 highlighted papers was 16, compared to four for the accomplishments and resources not highlighted in 2019, and only two of the non-highlighted papers received more than 40 citations. In contrast, three of the highlighted papers received three or fewer citations, though there was a clear and understandable trend toward fewer citations of those published later in the year.
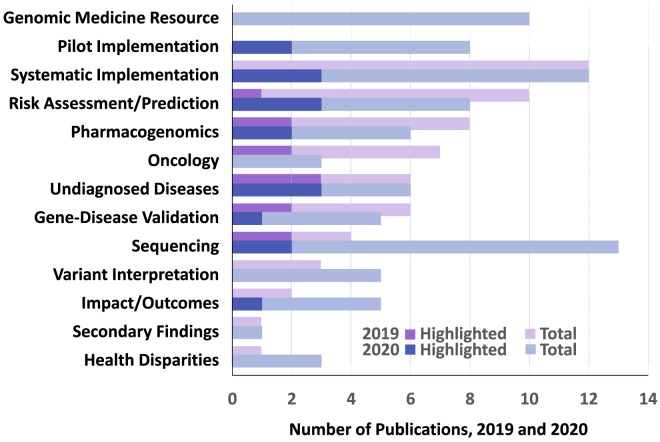
This year the Working Group has again selected its ten most significant advances among the 45 recognized accomplishments and resources published during the 12 months ending August 31, 2020. These papers clustered into 13 broad categories, two of which—“Genomic Medicine Resource” and “Pilot Implementation”—were added for the 2020 review period (Figure 1). The category “Clinical Implementation” was renamed “Systematic Implementation” to distinguish larger, more pragmatic implementation studies from early, single-site efforts. Distributions of these 45 papers across categories were similar to 2019 with the exception of “Sequencing,” which more than tripled in number in 2020, and “Oncology,” which dropped by half.
Figure 1.
Topics of Genomic Medicine Accomplishments and Resources, 2019 and 2020 Years in Review
Topics covered by the 45 genomic medicine resources and “Accomplishments in Genomic Medicine” (see Web Resources) published in the 12 months ending August 30, 2020 (blue bars) and compared to the preceding 12 months (purple bars). Dark blue bars denote the ten papers highlighted in this 2020 review; dark purple denotes papers highlighted in 2019. Papers could be classified as covering more than one topic.
Brief summaries of the ten highlighted papers, ordered by date of publication (except for two trials of the same intervention shown consecutively), are provided below.
Clopidogrel Pharmacogenetics – Genotyping Improves Outcomes, but Is it Convincing?
Common loss-of-function (LOF) CYP2C19 variants reduce bioactivation of the antiplatelet prodrug clopidogrel, but the impact of random assignment to genotype-guided treatment has not been studied. Two randomized trials compared outcomes of rapid genotype-guided versus standard antiplatelet therapy in patients undergoing percutaneous coronary intervention (PCI). Both trended in favor of genotyping but came to different stated conclusions based on prespecified statistical analysis plans. Neither trial was powered to study outcomes in carriers of two LOF alleles, the group unable to bioactivate clopidogrel at all.
Claassens, D.M.F., et al. (2019). A Genotype-Guided Strategy for Oral P2Y12 Inhibitors in Primary PCI. N. Engl. J. Med. 381, 1621–1631
This European trial randomized 2,488 subjects to standard treatment (mainly ticagrelor) or a genotype-guided strategy (clopidogrel for those without LOF variants and standard treatment for LOF variant carriers). Drug assignment was open label: in the genotype-guided group, 61% of patients received clopidogrel, while most in the standard therapy group received ticagrelor, although 7% received clopidogrel. There were two primary outcomes; the first, a combined outcome (death, recurrent myocardial infarction, stroke, or in-stent thrombosis), occurred in 5.9% (standard) versus 5.1% (genotype-guided, p < 0.001 and meeting a preset non-inferiority criterion). The second primary outcome was bleeding (mainly minor), which occurred in 12.5% (standard) versus 9.8% (genotype-guided, p = 0.04). The authors concluded that genotype-guided treatment did not increase the combined endpoint and did reduce bleeding.
Pereira, N.L., et al. (2020). Effect of Genotype-Guided Oral P2Y12 Inhibitor Selection versus Conventional Clopidogrel Therapy on Ischemic Outcomes after Percutaneous Coronary Intervention: The TAILOR-PCI Randomized Clinical Trial. JAMA 324, 761–771
TAILOR-PCI was primarily a US trial that randomized 5,302 patients. The genotype-guided group received ticagrelor if LOF alleles were present and clopidogrel if otherwise, while the standard group received only clopidogrel. The trial had 85% power to detect a hazard ratio (HR) of 0.50 in a composite outcome (similar to that in the European trial) in LOF allele carriers only. The outcome occurred in 35/903 (4.0%) LOF allele carriers in the genotype-guided group and 54/946 (5.9%) in the standard group for an HR of 0.66, p = 0.06. In this trial, genotype-guided therapy did not result in a significant decrease in the primary composite endpoint.
What Happens When You Sequence Prenatal DNA in Nearly Half a Country’s Pregnancies?
van der Meij, K.R.M., et al. (2019). TRIDENT-2: National Implementation of Genome-Wide Non-Invasive Prenatal Testing as a First-Tier Screening Test in the Netherlands. Am. J. Hum. Genet. 105, 1091–1101
Noninvasive prenatal testing (NIPT) based on whole-genome sequencing of maternal blood samples is increasingly used for clinical management of high-risk pregnancies, but its yield as a first-tier test in all pregnant women is not known. In the first year of the TRIDENT-2 study, NIPT was performed in 42% (73,239) of all pregnancies in the Netherlands. Participants could choose to learn about only trisomies 21, 18, and 13 or also other large potential fetal or maternal chromosomal abnormalities; 78% of participants chose to learn about other findings. A total of 343 (0.47%) trisomies 21, 18, and 13 were detected, along with another 207 (0.36%) other abnormalities. Comparing NIPT with invasive diagnostic testing, positive predictive value was high (>95%) for trisomies 21 and 18, moderate for trisomy 13 (54%), and quite variable (6%–64%) for other abnormalities. These findings demonstrate how NIPT can be successfully implemented in a national screening program for trisomies 21 and 18.
Genes Causing LQTS: The Long and Short of It
Adler, A., et al. (2020). An International, Multicentered, Evidence-Based Reappraisal of Genes Reported to Cause Congenital Long QT Syndrome. Circulation 141, 418–428
Increased understanding of the pathogenicity of human genomic variation mandates reappraisal of variants implicated in conditions such as long QT syndrome (LQTS). An international, multicenter, rigorous reappraisal assessed the level of evidence for 17 genes previously reported to cause LQTS. Only three genes (KCNQ1, KCNH2, and SCN5A) met ClinGen criteria as definitively causative for typical LQTS, while four genes (CALM1, CALM2, CALM3, and TRDN) were classified as having strong or definitive evidence for causing LQTS with atypical features. Another nine genes (AKAP9, ANK2, CAV3, KCNE1, KCNE2, KCNJ2, KCNJ5, SCN4B, and SNTA1)—more than half the total investigated—had limited or disputed supporting evidence, and the remaining gene (CACNA1C) had moderate level evidence for causality. These findings call for a rigorous reappraisal of previously reported disease-causing genes to ensure a strong level of evidence for causality to support routine testing in the evaluation and management of patients and families with putative disease-causing variants.
Implementing Whole-Genome Sequencing for Diagnosing Rare Diseases on a National Scale
Turro, E., et al. (2020). Whole-Genome Sequencing of Patients with Rare Diseases in a National Health System. Nature 583, 96–102
Whole-genome sequencing (WGS) is increasingly being used for identifying genomic variation associated with rare diseases but has primarily been implemented in specialized centers rather than on a national scale. WGS results were reviewed for 13,037 individuals, 75% of whom had a rare disease or extreme quantitative red blood cell trait, in 57 National Health Service (NHS) hospitals in the UK and 26 hospitals in other countries. Phenotypes were categorized into rare disease domains via the Human Phenotype Ontology. Molecular diagnoses were made in 16% of patients via a manually curated set of genes with established causal roles in disease, leading to specific treatment decisions in several cases. Additional epigenetic studies led to identification of disease-causing variants in regulatory elements. This paper demonstrates the potential value of WGS across a national health system, leading the NHS to plan to increase the availability of WGS-based diagnostics for rare diseases.
Effectiveness of Population-Based Genomic Screening to Identify and Intervene upon Actionable Genetic Conditions
Buchanan, A.H., et al. (2020). Clinical Outcomes of a Genomic Screening Program for Actionable Genetic Conditions. Genet. Med. 22, 1874–1882
The clinical impacts of screening unselected populations for Centers of Disease Control and Prevention (CDC) Tier 1 conditions (hereditary breast and ovarian cancer [HBOC], Lynch syndrome, and familial hypercholesterolemia [FH]) are unknown. This large population-based study of Geisinger MyCode patient participants identified 349 with a pathogenic/likely pathogenic (P/LP) variant in one of nine Tier 1 genes and assessed participants’ awareness of the variant pre-disclosure and risk management and clinical diagnoses post-disclosure. Chart review for relevant medical history prior to testing and subsequent risk management was performed with a median post-disclosure follow-up time of 21.8 months. Eighty-seven percent of participants were unaware they carried a clinically important variant prior to genomic screening. Eighty-six percent were eligible for risk management, and nearly 70% of these completed a risk management procedure. Forty-one participants (13%) had a relevant post-disclosure clinical diagnosis. This is the first large study demonstrating the effectiveness of population-based genomic screening to identify and intervene upon individuals with actionable genetic conditions.
Renown Renowned for Potential Impact on Public Health
Grzymski, J.J., et al. (2020). Population Genetic Screening Efficiently Identifies Carriers of Autosomal Dominant Diseases. Nat. Med. 26, 1235–1239
The value of genetic screening in an unselected population for identifying individuals carrying P/LP genomic variants for HBOC, Lynch syndrome, and FH has not been widely explored. The Healthy Nevada Project at Renown Health performed exome sequencing in 26,906 participants with available electronic medical records and analyzed genomic variants in nine risk genes for these conditions. Roughly 1.3%, 90% of whom had not been previously identified, carried P/LP variants. Among carriers, 22%, 70% of whom were diagnosed before age 65, were diagnosed with clinically relevant disease. Less than 20% of carriers had medical record documentation of inherited genetic disease risk or relevant family history. This suggests that genomic screening for inherited cancer and cardiovascular risk conditions can identify a significant number of at-risk carriers who are not detected by standard medical practice and who may benefit from earlier clinical risk screening.
Predicting Type 1 Diabetes among At-Risk Children
Ferrat, L.A., et al. (2020). A Combined Risk Score Enhances Prediction of Type 1 Diabetes (T1D) Among Susceptible Children. Nat. Med. 26, 1247–1255
Although predicting clinical diabetes onset in children at elevated risk for type 1 diabetes (T1D) is essential to prevent ketoacidosis—which can be particularly dangerous in the very young—and to support development of therapies to preserve pancreatic islet mass, current autoantibody surveillance programs are too costly for use in public health. The Environmental Determinants of Diabetes in the Young (TEDDY) study screened 425,000 children from the US and three European countries and used HLA haplotypes to identify 8,700 high-risk newborns and develop a combined risk score (CRS) incorporating genetic, clinical, and immunological factors. A three-variable CRS model predicting T1D by age 10 significantly improved risk prediction at ages 2–8 years (area under the receiver operator curve ≥ 0.9) compared to autoantibody testing alone. This approach doubles the estimated efficiency of newborn screening to identify children who may benefit from frequent evaluation to predict impending T1D and should be validated in other birth cohorts.
21st Century Technologies for 21st Century Babies
Adhikari, A.N., et al. (2020). The Role of Exome Sequencing in Newborn Screening for Inborn Errors of Metabolism. Nat. Med. 26, 1392–1397
The potential role of whole-exome sequencing (WES) in newborn screening (NBS) for inborn errors of metabolism (IEM) has not been evaluated against standard tandem mass spectrometry (MS/MS). WES detected 805 true positives and 385 false positives in blood spots from 1,190 ancestrally diverse newborns who had IEM detected by MS/MS. WES had a sensitivity of 88.0% and specificity of 98.4% (varying by disorder type) versus 99.0% and 99.8%, respectively, previously reported for MS/MS. The authors propose that lack of a putative pathogenic variant in a relevant gene could reduce false positives from MS/MS, although two true positives lacked such variants. WES enabled a more specific diagnosis in several true positives and could thus be useful in follow-up of positive MS/MS testing if WES could be obtained rapidly. WES may also be useful in extending the range of NBS disorders to those not amenable to MS/MS.
Is Anything Truly Monogenic?
Fahed, A.C., et al. (2020). Polygenic Background Modifies Penetrance of Monogenic Variants for Tier 1 Genomic Conditions. Nat. Commun. 11, 3635
Genomic variation influencing disease risk is commonly divided into monogenic variants of large effect and polygenic variants of small effect, but the interplay between monogenic and polygenic risk has not been widely examined. Data from over 80,000 individuals from the UK BioBank and Color Genomics laboratory carrying monogenic risk variants for HBOC, Lynch syndrome, and FH were analyzed to show that polygenic background significantly influences the risk of developing disease by age 75. Odds ratios for coronary disease, for example, in monogenic carriers of FH variants compared to non-carriers ranged from 1.6 to 21.4 across percentiles of polygenic risk. This work demonstrates that risk conferred by monogenic variants can vary substantially by polygenic background and that consideration of polygenic background may improve risk estimation in monogenic variant carriers. This approach should be extended to other monogenic conditions and to cohorts with a broader ancestral diversity.
Conclusion
Genomic medicine implementation research continues its rapid forward pace, and new themes, including randomized trials of pharmacogenetic interventions (Claassens et al. and Pereira et al.), national implementation of genome sequencing for prenatal screening (van der Meij et al.) and rare disease diagnostics (Turro et al.), role of genome sequencing in newborn screening (Adhikari et al.), use of genetics in predicting type 1 diabetes (Ferrat et al.), and polygenic modification of monogenic disease risk (Fahed et al.), were highlighted this year. Themes carried over from the 2019 review, in addition to clopidogrel pharmacogenetics as mentioned above, include population-based screening for monogenic disorders (Buchanan et al. and Grzymski et al.) and re-appraisal and re-analysis of pathogenicity of genomic variants (Adler et al.). Several papers leave readers to form their own conclusions, such as whether the effects of pharmacogenetic testing—while not statistically significant—are still large enough to be convincing (Claassens et al. and Pereira et al.) and whether prenatal genome sequencing for conditions other than the major trisomies is reliable enough to be used in clinical care (van der Meij et al.). Many of the papers point the way to additional research, such as expanding population screening for genetic risk to other cohorts and non-European ancestries (Buchanan et al., Grzymski et al., and Ferrat et al.), implementing genome sequencing for rare diseases in other health systems (van der Meij et al. and Torro et al.), using WES to expand newborn screening beyond conditions detectable only by standard MS/MS (Adhikari et al.), and using polygenic risk for more precise risk stratification in patients with monogenic variants (Fahed et al.). As in 2019, although all of these papers represent significant advances in the application of genomic medicine, none evaluated questions of cost-effectiveness or societies’ willingness to pay that must be answered before clinical use can be widely adopted. Continuing gaps in sociodemographic diversity of populations studied and challenges in genomic variant interpretation need to be addressed. Additional attention is also needed to the development and collection of outcome measures to permit an unbiased assessment of the role of genomics in improving patient care.
Web Resources
Genomic Medicine Working Group Accomplishments in Genomic Medicine, https://www.genome.gov/health/Genomics-and-Medicine/accomplishments
MyCode Community Health Initiative, https://www.geisinger.org/precision-health/mycode
NHGRI definition of genomic medicine, https://www.genome.gov/health/Genomics-and-Medicine
References
- 1.Manolio T.A., Bult C.J., Chisholm R.L., Deverka P.A., Ginsburg G.S., Jarvik G.P., McLeod H.L., Mensah G.A., Relling M.V., Roden D.M. Genomic Medicine Year in Review: 2019. Am. J. Hum. Genet. 2019;105:1072–1075. doi: 10.1016/j.ajhg.2019.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]