Summary
Breeding better crops is a cornerstone of global food security. While efforts in plant genetic improvement show promise, it is increasingly becoming apparent that the plant phenotype should be treated as a function of the holobiont, in which plant and microbial traits are deeply intertwined.
Using a minimal holobiont model, we track ethylene production and plant nutritional value in response to alterations in plant ethylene synthesis (KO mutation in ETO1), which induces 1‐aminocyclopropane‐1‐carboxylic acid (ACC) synthase 5 (ACS5), or microbial degradation of ACC (KO mutation in microbial acdS), preventing the breakdown of the plant ACC pool, the product of ACS5.
We demonstrate that similar plant phenotypes can be generated by either specific mutations of plant‐associated microbes or alterations in the plant genome. Specifically, we could equally increase plant nutritional value by either altering the plant ethylene synthesis gene ETO1, or the microbial gene acdS. Both mutations yielded a similar plant phenotype with increased ethylene production and higher shoot micronutrient concentrations. Restoring bacterial AcdS enzyme activity also rescued the plant wild‐t8yp phenotype in an eto1 background.
Plant and bacterial genes build an integrated plant–microbe regulatory network amenable to genetic improvement from both the plant and microbial sides.
Keywords: ACC deaminase, ethylene signaling, genome editing, holobiont, hologenome, plant–microbe interactions
Introduction
Breeding better crops is essential to feed a growing human population while preserving natural resources. Food security is a complex issue encompassing food quality as well as quantity. For instance, a large proportion of the global population is already affected by chronic deficiency in micronutrients and vitamins despite sufficient calorie intake. Malnutrition is a major public health issue worldwide, affecting more than two billion people (Barrett, 2010), and it is predicted to increase as a consequence of soil depletion and climate change. Deficiencies in micronutrients such as iron (Fe), zinc (Zn) and copper (Cu) are among the most widespread and are particularly problematic in children and pregnant women, causing severe health issues and impaired development (Barrett, 2010).
Several approaches have been proposed to help alleviate the issue of hidden hunger. However, all existing strategies show technical, economic or regulatory limitations preventing their full implementation. Grain fortification, improved fertilization and supplement feeding require complex logistics that may not be available to small stakeholders. Plant genetic improvement may help to generate more nutritious crops, but targeted breeding of improved crops is often hampered by technical limitations and regulatory barriers (Lassoued et al., 2018). As a result, genetic improvement strategies have focused on a few model cultivars, which may not align with the environmental conditions where they are most needed. To make better crops available to farmers, straightforward and flexible strategies need to be developed and made available. We propose that shifting genetic improvement efforts from a plant to a holobiont perspective may offer a new solution to this long‐standing problem. Both on the inside and the outside, plants are colonized by myriad bacteria, fungi and protists, profoundly impacting plant growth and physiology (Berendsen et al., 2012). The plant‐associated microbiome has been shown to provide an important degree of phenotypic variability to the plant, affecting plant traits associated with resource allocation, flowering time or stress resistance (Panke‐Buisse et al., 2015; Bartoli et al., 2018). Engineering the microbiome may therefore offer a promising approach to modify plant traits, complementing plant breeding (Wei & Jousset, 2017). However, targeted microbiome modification has remained an arduous process. We ascribe these difficulties to a limited integration of knowledge in the different fields of plant physiology and plant–microbe interactions.
Plant breeding has long been approached from a strict plant‐centered perspective, seeking to alter plant‐specific traits by targeting the plant genome. Plant hormonal signaling networks are a promising target for such breeding strategies (Wilkinson et al., 2012), as slight modifications in their regulation can have profound effects on plant life‐history strategies at all growth stages (Dubois et al., 2018). However, there is a growing awareness that plant hormonal signaling is not an encapsulated process, but instead is the result of traits encoded both in the genome of the plants as well as their associated microorganisms (Fig. 1b) (Ravanbakhsh et al., 2018). In this way, the regulation of many plant hormones is highly permeable, with hormones and their precursors being produced, exchanged and degraded by both plants and their associated microorganisms. As a result, plant hormonal networks can best be approached at a holobiont level, as a joint venture between plant‐ and microbiome‐encoded traits (Ravanbakhsh et al., 2018). Here, we propose that approaching plant–microbe interactions from a regulatory network perspective may offer a novel way to engineer the plant phenotype by targeting naturally occurring microorganisms. Such an approach opens new possibilities to manage plant–microbe interactions by targeting plant traits via targeted microbial genomic selection. Microorganisms were long ago reported to modulate plant development (Puga‐Freitas & Blouin, 2015) and have been increasingly investigated as a replacement for chemical plant protection products during recent decades (Lugtenberg & Kamilova, 2009). However, microbial applications often deliver inconsistent results under field conditions. We ascribe this effect to the traditional classification of microorganisms as standalone ‘beneficial’ or ‘deleterious’ species, often ignoring their strong host‐ and condition‐dependent impact on plant development (Veiga et al., 2013).
Fig. 1.
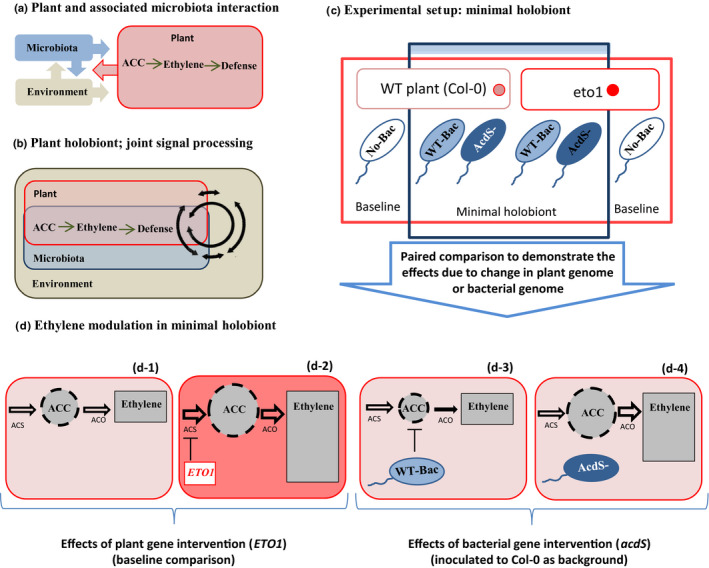
Conceptual figure explaining the hypothesis and experimental approach. Plant 1‐aminocyclopropane‐1‐carboxylic acid synthases (ACS) create a pool of 1‐aminocyclopropane‐1‐carboxylic acid (ACC) in response to environmental inputs. ACC is then transformed into ethylene by the plant enzyme ACC oxidase, triggering a range of traits in a dose‐dependent way, thereby modulating responses to environmental stimuli and plant stress (e.g. pathogen or drought). (a) Ethylene hormone regulation has typically been examined in a disjointed fashion with separate study of microbial vs plant components. (b) Ethylene signaling can also be seen as a coregulation by intertwined activities of plants and their associated microorganisms in an integrated system; plants and associated microbiota together receive the environmental signals, process them, and jointly trigger an integrated response. (c) Our minimal holobiont designed by growing Arabidopsis thaliana plants colonized by the model, bacterium Pseudomonas putida UW4, a naturally occurring root‐colonizing bacterium representative of plant‐associated microbial species that degrade the plant ACC pool (wild‐type ACC deaminase producing bacteria (WT‐Bac)) (Supporting Information Fig. S1). For each plant genetic background, we used an additional sterile treatment to assess plant traits in the absence of microorganisms. (d) Ethylene coregulation by the plant and microbes in a joint network. Knocking out the acdS gene in plant‐associated wild‐type ACC deaminase bacteria or ETO1 gene in the plant should both increase overall ethylene production in the plants (d‐2, d‐4). General analysis of the data was based on a full factorial design (Table S2), with pairwise comparisons used to show the effects resulting from changes in plant genotype (Col‐0 vs eto1) or from changes in bacterial genotype (wild‐type bacteria (WT‐Bac) vs AcdS‐). The area of the rectangles and thickness of the arrows (in panel d) depict relative concentrations and enzyme activities.
This work seeks to develop a more reliable approach to plant–microbe interaction, integrating plants and their microbiota to predictably reach a specific phenotype. To this purpose, we use a simplified model system to adopt a meta‐organism‐level regulation of hormonal balance. Systems biology approaches have long been used in contexts such as pathogenesis (Peyraud et al., 2017). The present work is the culmination of efforts to better bring together plant and microbial coregulation of hormonal balance. This strategy stems from original observations that bacteria reducing plant ethylene concentrations consistently reduced plant fitness under stress conditions (Ravanbakhsh et al., 2017, 2019a). This deleterious impact is in line with the role of ethylene in plant stress tolerance (Kazan, 2015), but in strong contradiction with the general paradigm in plant–microbe interactions, where ethylene‐reducing microorganisms are touted to be ‘beneficial’ (Saleem et al., 2007; Sapre et al., 2019).
We previously compiled existing data in a conceptual holobiont context of plant hormonal regulation (Ravanbakhsh et al., 2018). We then demonstrated that ethylene‐regulating genes encoded in microbial and plant genomes can have comparable effects on plant growth and stress tolerance (Ravanbakhsh et al., 2019a), in contrast to the opposing effects reported in plant–microbe interaction studies (Sapre et al., 2019). According to these results and our conceptual model, plant hormonal balance is the result of an intertwined plant–microbe regulatory network that can be tweaked by altering specific plant or microbial genes. Given the relative ease of microbial manipulations, we anticipate that such a holobiont concept not only has the power to reliably predict plant phenotype, but also opens new opportunities to genetically engineer plants by intervening at the microbiome level rather than at the plant genome level. This present work is the first demonstration that a holobiont breeding concept can be applied to obtain a desired phenotype.
We use ethylene signaling as a model target for genetic intervention and nutritional value as a model multivariate phenotype. Ethylene is one of the core plant hormones and is routinely used as a target in plant breeding programs (Wilkinson et al., 2012). It regulates a wide range of traits linked with plant development, immunity and stress tolerance (Abeles et al., 2012). In particular, ethylene plays a central role in plant nutrition by regulating root cuticula, root architecture and the expression of transporters involved in nutrient translocation to aerial parts (Romera et al., 2016). Ethylene signaling is, furthermore, one of the best described examples of a coregulated cascade controlled by both plants and their associated microorganisms (see Fig. 1b (plant‐associated microbiota joint signal processing) vs Fig. 1a (plant–microbe interaction)).
Ethylene signaling forms a regulatory circuit spanning both plant and associated microorganisms. From a plant perspective, ethylene synthesis starts with the synthesis of the ethylene precursor 1‐aminocyclopropane‐1‐carboxylic acid (ACC) by members of the ACC synthase gene family (ACS1–ACS612 in Arabidopsis thaliana) (Peng et al., 2005). ACC is a mobile molecule that can be transported between plant cells and tissues (Van de Poel & Van Der Straeten, 2014e Poel & Van Der Straeten, 2014), for example from roots to the shoot for conversion to ethylene when plants are subjected to stress conditions (Olson et al., 1995; Shiu et al., 1998). ACC is then transformed into ethylene by the enzyme ACC oxidase, triggering several response traits in a dose‐dependent way (Fig. 2a) (Etheridge et al., 2006). Ethylene signaling is, however, not only regulated by the plant. It is also impacted by several plant‐associated microorganisms that can either synthesize ethylene or alternatively degrade the plant ACC pool (Barnawal et al., 2016; Saleem et al., 2018). In this work, the plant ACC pool will be the focal integrating element that allows for combined interventions on plant and microbial genomes into one predictable plant phenotypic response.
Fig. 2.
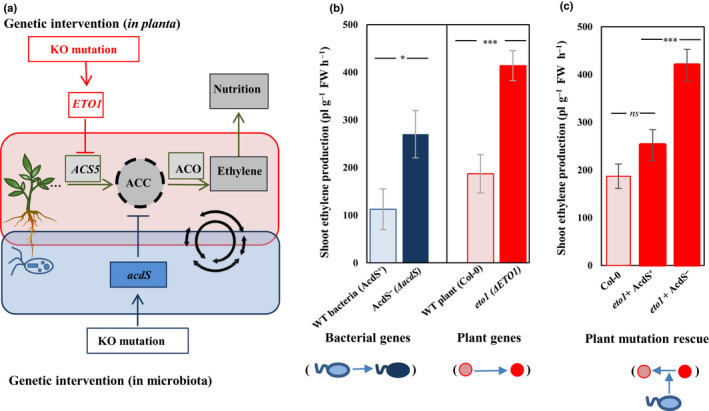
(a) Conceptual overview of the joint coregulation of ethylene signaling by plants and microorganisms. Plant 1‐aminocyclopropane‐1‐carboxylic acid synthases (ACS) create a pool of 1‐aminocyclopropane‐1‐carboxylic acid (ACC) in response to environmental inputs. ACC is later converted into ethylene by the plant enzyme ACC oxidase, triggering a range of traits in a dose‐dependent way. Plant‐associated microorganisms can influence ethylene cycling, for instance by producing the enzyme ACC deaminase (via the bacterial acdS gene) that will degrade ACC. By placing plants and microbes as joint constituents of a common regulatory network, we show that we can intervene in plant phenotype via KO mutations in the bacterial acdS gene or in the plant ETO1 gene, causing an increase in the ACC pool and ultimately ethylene and the traits it regulates. In this study, we selected parallel mutations in the plant (Col‐0 vs eto1 plant mutant) or bacterial genome (wild‐type (WT) vs AcdS‐ mutant bacteria) and assessed their effect on plant ethylene concentrations. The colors show the mutation in the plant (red) and bacterial genome (blue): the darker color shows the mutation that resulted in increased ethylene production; pink, the Col‐0 (WT plant); red, eto1 (ethylene over producer); light blue, Col‐0 associated with WT ACC deaminase‐producing bacteria; dark blue, Col‐0 associated with ACC deaminase‐deficient mutant bacterial. (b) Effect of mutations in bacterial (blue bars) and plant genomes (red bars) on ethylene concentrations in a gnotobiotic system. Both plant‐ and bacterial‐encoded mutations resulted in a similar two‐fold increase in plant ethylene concentrations, demonstrating the parallelism in phenotypic effects between plant‐ and microbiota‐ associated traits. (c) ACC deaminase synthesis by root‐associated bacteria rescues the wild‐type plant phenotype in an eto1 genetic background, validating the homologous nature of plant‐ and microbial‐encoded traits affecting ethylene signaling. WT‐associated bacteria (light blue) rescue the effects of mutation in the plant genome (dark red to pink). Error bars show ± SE. Columns with asterisks are significantly different according to a t‐test (***, P < 0.0001; *, P < 0.05; ns, not significant).
In a first step, we created a minimal holobiont (Fig. 1c) by growing Arabidopsis thaliana plants colonized by the model bacterium Pseudomonas putida UW4, a naturally occurring root‐colonizing bacterium representative of plant‐associated microbial species that degrade the plant ACC pool (Duan et al., 2013). In this holobiont system, ethylene signaling is a pan‐organism process, encompassing both plant and bacterial traits (Fig. 1b). As ethylene is a known positive regulator of nutrient uptake (Romera et al., 2016), we then sought to reach a more nutritious plant phenotype by increasing plant ethylene concentrations. This was done by either editing the plant genome by KO mutation of ETO1, a repressor of ACC synthases 5, or by editing the bacterial genome with a KO mutation of the enzyme ACC deaminase, which degrades the plant ACC pool (Fig. 2a). For each plant genetic background, we used an additional sterile treatment to assess plant traits in the absence of microorganisms (Fig. 1c).
We hypothesized: that ethylene positively regulates microelement accumulation in aerial parts; that both genetic interventions targeting the genome of the plant or its associated microorganisms can result in a similar increase in ethylene concentration, resulting in similar increases in plant nutritional value (Fig. 1d‐2,d‐4); and that plant‐associated bacteria can rescue the phenotypic effect of mutations in the plant genome.
Materials and Methods
Plant materials
We used Arabidopsis thaliana (L.) Heynh., ecotype Columbia (hereafter referred to as ‘Col‐0’) as a reference accession. We selected the ethylene overproducer mutant1 (eto1) obtained from the Col‐0 background (Wang et al., 2004). This mutant is a KO mutant of ETO1, a negative post‐transcriptional regulator of 1‐aminocyclopropane‐1‐carboxylic acid synthase 5 (ACS5). This mutant is characterized by a constitutively high ethylene biosynthesis resulting from the higher activity of the ACS5 gene (Wang et al., 2004), which catalyzes ACC production, the rate‐limiting step in ethylene biosynthesis (Chae et al., 2003). Seeds were kindly provided by the Plant Ecophysiology Research Group, Utrecht University, the Netherlands.
Bacterial strains
We used the wild isolate P. putida UW4 (later ‘wild‐type’) as a model root‐colonizing bacterium. This bacterium was first reported as Enterobacter cloacae UW4, but later genetic data firmly placed it in the P. putida group (Duan et al., 2013). This strain is a representative of the wide range of root‐associated bacteria producing the enzyme ACC deaminase, degrading the ethylene precursor ACC and using it as a nitrogen source (Glick et al., 1998). ACC deaminase producers are highly prevalent in the rhizosphere (Bouffaud et al., 2018), making plant–microbe coregulation of ethylene signaling a generalizable target for plant improvement. In our holobiont perspective, we conceptually approach this trait as a microbial functional homolog to the plant gene ETO1 (inhibitor of ACS5 gene in A. thaliana) within the plant–microbe hormonal signaling network (Fig. 2b,c). In order to assess the effect of bacterial degradation of ACC on plant phenotype, we used a genetically engineered isogenic mutant (later ‘AcdS‐’) lacking the ACC deaminase gene (acdS) obtained by insertion of a tetracycline resistance gene into the ACC deaminase gene coding region (Li et al., 2000). Both strains were kindly provided by Prof. Bernard Glick, Department of Biology, University of Waterloo, Canada. Bacterial strains were kept as frozen stocks at −80°C. Before experiments, one single colony was grown overnight on 30 g l−1 TSB (wild‐type) or TSB supplemented with 100 μg ml−1 tetracycline (AcdS‐). Tryptic soy broth medium consisted of tryptone (17 g l−1), neutralized soya peptone (3 g l−1), NaCl (5 g l−1), dipotassium phosphate (K2HPO4) (2.5 g l−1), and glucose (2.5 g l−1). Bacteria were harvested by centrifugation (6000 g, 10 min), washed three times with 10 mM MgSO4 and adjusted to a density of 108 cells ml−1 before inoculation.
Experimental setup
Pot experiment
Seeds of Arabidopsis thaliana Col‐0 and its isogenic ethylene overproducer mutant (eto1) were surface‐sterilized by vapor‐phase sterilization in a 2 ml open plastic microcentrifuge tube placed under a 7 l airtight desiccator jar. A 250 ml beaker containing 100 ml bleach and 3 ml concentrated HCl solution was placed in the desiccator jar before sealing for 4 h (Clough & Bent, 1998). After sterilization and stratification for 4 d at 4°C in the dark, plant seeds were sown on 12‐cm‐square Petri dishes containing agar‐solidified Murashige and Skoog (MS) medium supplemented with 0.5% sucrose. Petri dishes were transferred and positioned vertically in a growth chamber (20°C, 70% relative humidity, 9 h photoperiod, 200 µmol m−2 s−1 photosynthetic photon flux density). Ten‐day‐old plant seedlings were selected based on homogeny in size and development and transferred to new pots. Plants (except no‐bacterial control Col‐0 and eto1) (baseline; Fig. 1) were inoculated with 50 µl of a bacterial suspension (root inoculation, 108 cells ml−1; see ‘Bacterial strains’ section), and transferred to pots containing a mixture of fine sand and Perlite (both pasteurized at 90°C for 45 min), saturated with a modified Hoagland nutrient solution (Smeets et al., 2008) and placed in the same walk‐in growth chamber (as described earlier). Growth chambers were continuously ventilated, ensuring the air exchange needed to prevent artefacts as a result of ethylene or volatile organic compound emissions by plants. In this experiment, we combined, in a full factorial design, two plant treatments (Col‐0 and eto1) and three bacterial treatments (wild‐type, AcdS‐ and nonbacterized control), resulting in six treatments (Fig. 1c). We set up eight replicates per treatment. Plants were grown for a period of 35 d after germination, and pots were randomized every 3 d.
Plant harvest, shoot ethylene and nutrient measurements
We measured ethylene production in plant aerial parts, using it as a proxy for ACC biosynthesis. While limited amounts of ethylene can also be synthesized in roots, measuring ethylene production in the shoot is a standard and validated method to estimate ethylene concentrations in plants (Cristescu et al., 2013). For doing so, shoots were carefully separated from the roots with a razor blade and aerial parts placed in serum vials (50 ml). After 2 h, a 1 ml gas sample was taken from each vial and injected into a Chrompack Packard gas chromatograph model 438 A with a Poropack Q column (length 100 cm, packed to 0.34 g cm−3) (Chrompack, Waltham, MA, USA) at 60°C to measure ethylene production. Ethylene concentrations were expressed in pl g−1 FW h−1 (Voesenek et al., 2003). Plant roots were collected to measure bacterial colonization (see later). Plant materials (root and shoot) were collected, dried and digested by standard procedures (dry ashing procedure) (Kalra et al., 1997). The concentration of nutrients in plants was measured by inductively coupled plasma mass spectroscopy (Thermo Scientific ICP 6300; Fischer Scientific, Breda, Netherlands).
Enumeration of rhizosphere bacteria
For plants treated with bacteria, we assessed the density of the introduced wild‐type P. putida UW4 and its isogenic AcdS‐ mutant strain on plant roots immediately after harvest. Roots were gently shaken to remove adhering soil, after which bacteria were recovered by shaking the whole root system roots in 10 mM MgSO4 for 30 min at 200 min−1, sonicating them for 20 s and vortexing for 30 s before serial dilution on appropriate culture medium: The wild‐type strain was enumerated on DF agar (Table S1, composition of medium) (Dworkin & Foster, 1958) supplemented with ACC as sole nitrogen source (Penrose & Glick, 2003). The AcdS‐ mutant strain was enumerated on DF agar supplemented with (NH4)2SO4 as nitrogen source and 100 μg ml−1 tetracycline as selective agent.
Statistical analyses
We used two‐way ANOVAs to evaluate the interactive effects of plant genotype (Col‐0 and eto1) and bacterial treatment (three levels: nonbacterized control, wild‐type and AcdS‐ mutant bacterial treatment) on shoot ethylene concentration, shoot and root DW, and shoot nutrient concentration.
We set up a two‐sided t‐test to evaluate the effects of single‐gene (acdS) mutation in the bacteria (wild‐type vs AcdS‐), in the plant genotype (ETO1) (Col‐0 vs eto1), and also microbial complementation of plant mutations (Col‐0 plant vs eto1 plant associated with wild‐type ACC deaminase‐producing bacteria) on the shoot ethylene concentration and nutrient concentration in the shoot.
We used principal coordinate analysis to compare the effects of gene mutation in plant and bacteria on shoot micronutrient concentration . Two‐way ANOVAs were performed to assess the effect of place of mutation (bacterial vs plant genome) and presence of mutation (mutation connected to ethylene concentrations in either bacterial or plant genome) on ethylene concentration and the concentration of plant nutrient elements. We used a linear regression model to assess the relationship between ethylene production and nutrient concentration in the shoot . The effects of mutation in bacteria and bacterial inoculation on micronutrients in shoots were separated using two‐way ANOVA. All analyses were performed in Spss v.22 (IBM Corp., Armonk, NY, USA) and R (package poa (Haffenden, 2019).
Results
In the current study, we investigated a holobiont breeding approach aiming at increasing desired plant phenotypic traits (nutritional value) by targeting ethylene signaling. As the synthesis of the ethylene precursor ACC is the rate‐limiting step in ethylene production (Chae et al., 2003), we selected genetic interventions targeting the microbial and plant genes involved in controlling the plant ACC pool. We did this by using plants deficient in ETO1 (ethylene overproducer eto1 vs Col‐0 wild‐type plant) or by using root‐associated bacterial mutants deficient in ACC deaminase (AcdS‐ mutant bacteria vs wild‐type ACC deaminase‐producing bacteria) (Fig. 2a).
Mutation in plant (ethylene overproducer eto1 vs Col‐0 wild‐type plant), and bacterial genomes (wild‐type vs mutant (acdS‐)) both led to a significant increase in ethylene and micronutrient concentrations in plant aerial parts. A key additional finding is the absence of significant interactive effects for the plant and bacteria treatments (Table S2), indicating an additive effect of bacterial and plant mutations (Fig. S1). By placing plant and microbes as joint constituents of a common regulatory network, we show that the effects of gene mutation in the associated bacteria can have the same effects as changing in the plant genome. For this purpose, we hereafter use a set of two‐sided t‐test analyses to demonstrate the effects of single‐gene (acdS) mutation in the bacteria, and compare it with the effects of single‐gene mutation in the plant genotype (ETO1 gene) (Fig. 1d) as a focal point.
Effects of mutations in the plant and bacterial genomes on shoot ethylene concentration
Ethylene concentrations varied significantly in different plant and bacterial treatments (Table S2). Inhibiting the ETO1 gene (in eto1 mutant plant phenotype) resulted in a 2.2‐fold increase in the concentrations of shoot ethylene, from 180 to 390 pl g−1 FW h−1, compared with the wild‐type A. thaliana Col‐0 (Figs 2b, S1). Similarly, in the plants inoculated with bacteria, deactivation of ACC deaminase in bacteria resulted in a 2.45‐fold higher ethylene concentration compared with the wild‐type, from 110 to 270 pl g−1 FW h−1 (Figs 1b, S1). The presence of WT, ACC deaminase‐producing bacteria (AcdS+) on roots rescued the effects of the ETO1 mutation, decreasing ethylene concentrations by 1.7‐fold compared with inoculation with AcdS‐ mutant, from 410 to 250 pl g−1 FW h−1. This concentration is comparable to that found in noninoculated Col‐0 plants (Fig. 1c).
Effects of mutations in plant and bacterial genomes on shoot micronutrient concentrations
Deactivation of both the ETO1 gene in the plant or the acdS gene in the plant‐associated bacterium impacted the concentration of several micro‐ and macronutrients in the aerial parts (Fig. 3a). Plant nutrient content was structured along a mutation axis (mutations leading to a change in ethylene concentration, regardless of whether it takes place in plant or bacteria) and a second axis reflecting the place of the mutation (plant vs bacteria effects). Both plant ETO1 gene overexpression and bacterial acdS deactivation resulted in a significant change in the concentration of several shoot micronutrients, including increases in shoot Fe, Zn and Cu concentrations (Fig. 3; Table S2). In addition, the effects as a result of ethylene and as a result of the target of the genetic intervention (plant vs bacteria) were equal: changes in ethylene production explained most of the variation across the treatments, regardless of whether this alteration was a result of a mutation in the plant or the microbial genome (nonsignificant effect of the place of mutation when sequentially fitted after shoot ethylene production) (Table 1). Increasing the ethylene concentration also led to a higher concentration of the macroelement P, which increased both in eto1 and in plants colonized by AcdS‐ bacteria (Fig. S2; Table S2) in an ethylene‐dependent way (Table 1). It is noteworthy that the effect of ethylene on some nutrient concentrations was slightly stronger when ethylene signaling was manipulated by mutations in bacteria rather than in plants (Table 1; ethylene × place interaction) (Fig. 4). This effect was, however, moderate and changes in nutrient concentrations induced by bacteria and plant mutations were generally comparable. The concentrations of micronutrients in the plant shoots were positively correlated with shoot ethylene production in the treatments involving a plant mutation (Col‐0 vs eto1) (Fig. 4a) and those involving a bacterial mutation (wild‐type bacteria vs AcdS‐ mutant bacteria) (Fig. 4b). The concentrations of Fe, Zn and Cu showed strong positive correlations with the concentration of ethylene in the shoot of Col‐0 and eto1 plant genotypes (Fig. 4c–e, red markers), or Col‐0 inoculated with wild‐type ACC deaminase bacteria or ACC deaminase‐deficient mutant (Fig. 4c–e, blue markers). In general, these results suggest that increasing the plant ethylene concentration may serve as a target to selectively increase a range of important nutrients in plant shoots.
Fig. 3.
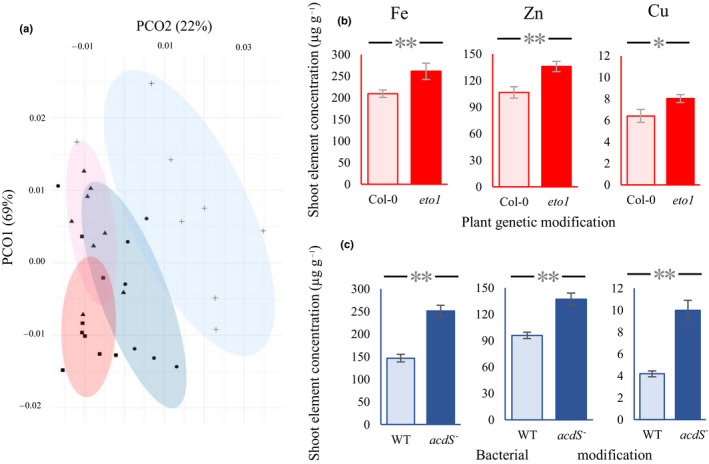
(a) Principal coordinate analysis of the composition of selected essential micronutrients (Fe, Mn, Cu, Zn). (b, c) Effect of mutations in the plant genome (Col‐0 vs eto1 overproducer Arabidopsis thaliana plants grown without bacterial addition) (b) and in the bacterial genome (Pseudomonas putida UW4 vs 1‐aminocyclopropane‐1‐carboxylic acid (ACC) deaminase‐deficient mutant (AcdS‐) growing on Col‐0 plants) (c) on micronutrient concentrations. The original setup of the experiment consisted of six sets of measures (three bacterial inoculations (noninoculated treatment, wild‐type (WT) and acdS‐ mutant of Pseudomonas putida UW4), and two plant genotypes (Arabidopsis thaliana Col‐0 and its ethylene over producer, eto1). However, in this figure, we compare only four measures to show the effect of mutations in the plant genome (Col‐0 vs eto1 overproducer A. thaliana plants grown without bacterial addition) and compare this with the effects resulting from changes in the bacterial genome (P. putida UW4 vs AcdS‐ growing on Col‐0 plants). The effects of mutation in the bacterial and plant genome are shown in blue and red, respectively. Light blue and plus signs (+) show the WT bacteria inoculation (ethylene reducing bacteria); dark blue and black circles show mutant inoculation (high ethylene); light pink and black rectangles show the Col‐0 plant; red and triangles show the eto1 plant genotype. Error bars are ± SE. Columns with asterisks are significantly different according to a t‐test (**, P < 0.001; *, P < 0.05).
Table 1.
ANOVA table summarizing the effects of ethylene production and the background of mutation on the nutrient concentration in Arabidopsis thaliana leaves.
| df | Ethylene concentration | Place of mutation (plants vs bacteria) | Ethylene concentration × place of mutation | ||||
|---|---|---|---|---|---|---|---|
| F | P | F | P | F | P | ||
| Fe | 1 | 26.98 | P < 0.0001 | 0.02 | 0.86 | 9.45 | 0.004 |
| Cu | 1 | 14.65 | P < 0.0001 | 2.31 | 0.11 | 7.56 | 0.008 |
| Zn | 1 | 22.34 | P < 0.0001 | 1.85 | 0.18 | 8.12 | 0.008 |
| B | 1 | 3.36 | 0.12 | 62.8 | P < 0.0001 | 0.07 | 0.75 |
| Mn | 1 | 1.15 | 0.29 | 2.93 | 0.09 | 0.07 | 0.71 |
| P | 1 | 8.75 | P < 0.001 | 0.42 | 0.51 | 11.38 | 0.004 |
| Error | 29 | ||||||
Two‐way ANOVAs compare the effect of place of mutation (bacterial vs plant genome) and presence of mutation (mutation connected to ethylene concentrations either in bacterial or plant genome) on the concentration of plant nutrient elements (Fe, Cu, Zn and P). Changes in ethylene production explained most of the variation across the treatments, regardless of whether this alteration was a result of a mutation in the plant or the microbial genome (nonsignificant effect of the place of mutation when sequentially fitted after shoot ethylene production). Comparison of Col‐0 vs eto1 overproducer Arabidopsis thaliana plants grown without bacterial addition shows the mutations in plant genome, and comparison of the Pseudomonas putida UW4 vs ACC deaminase‐deficient mutant (AcdS‐) growing on Col‐0 plants shows the effects as a result of bacterial genome. The target of intervention (plant or bacterial genome) was sequentially fitted after ethylene concentration. Bold highlights a statistically significant difference.
Fig. 4.
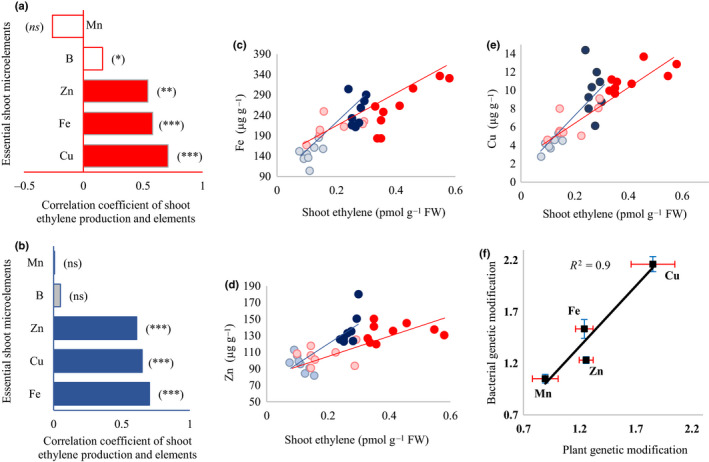
Correlation between ethylene concentrations and microelement concentrations in the shoots of Arabidopsis thaliana. We split the results between mutations in the plant and bacterial genomes. (a) Effect of mutations in the plant genome: nonbacterized Col‐0 and ethylene‐overproducer genotype eto1. (b) Effect of mutations in the bacterial genome: A. thaliana Col‐0 inoculated with Pseudomonas putida UW4 (wild‐type (WT)) or its 1‐aminocyclopropane‐1‐carboxylic acid (ACC) deaminase‐deficient mutant (AcdS‐). (c–e) Correlation between ethylene concentration and Fe (c), Zn (d) and Cu (e) in the shoot of A. thaliana in nonbacterized Col‐0 (light red circles) or the ethylene overproducer eto1 (dark red circles), and Col‐0 plants bacterized with Pseudomonas putida UW4 (light blue circles) or its ACC deaminase‐deficient mutant (AcdS‐) (dark blue circles). Two separate regressions were calculated to highlight the similarity between the effects of mutations in the plant (red line) and bacterial genomes (blue line). (f) Correlation between mutations in the plant and bacterial genomes on selected plant micronutrient. x‐axis: fold‐change in element concentration caused by plant ETO1 gene mutation (eto1/WT); y‐axis: fold‐change in element concentration caused by bacterial acdS deactivation (AcdS‐/WT bacterial treatment). Error bars are ± SE. Columns with asterisks are significantly different according to a t‐test (***, P < 0.0001; **, P < 0.001; *, P < 0.05; ns, not significant).
Density of bacteria recovered on the rhizoplane
The densities of both the wild‐type and ACC deaminase‐deficient mutant (AcdS‐) P. putida UW4 on plant roots were similar (F 2,75 = 1.59, P = 0.21), with a density of 7.51 ± 0.6 log CFU g−1 root FW for ACC deaminase‐producing bacteria, and 7.25 ± 0.3 log CFU g−1 root FW for ACC deaminase‐deficient mutant. This indicates that the lack of ACC deaminase did not affect bacterial fitness under the tested conditions. Shoot ethylene production showed a negative correlation with the density of ACC deaminase‐producing bacteria in Col‐0 (F 1,7 = 4.22, P = 0.06, R 2 = 0.41), and eto1 (F 1,7 = 6.45, P = 0.04, R 2 = 0.39), and a positive correlation with the density of ACC deaminase‐deficient mutant Col‐0 (F 1,7 = 3.88, P = 0.09, R 2 = 0.51) and eto1 (F 1,7 = 5.65, P = 0.05, R 2 = 0.46) plant genotypes.
Discussion
Breeding better crops is a core component of agricultural yield improvement, as it allows for the selection of desired traits contributing to plant yield and quality. Plant breeding is currently undergoing a quantum leap from random mutagenesis to targeted genome editing, with new knowledge showing that plant phenotype can be radically changed with a few mutations in genes of interest (Zsögön et al., 2018). This technological improvement has long left the microbiome in the shadow, at worst an uncontrolled source of phenotypic variation or at best an indirect target for plant genome enhancement. Considering the substantial reduction in the available plant genetic pool that has accompanied the development of modern crop varieties, there is an urgent need to explore new sources of genetic variation to improve the traits needed for enhanced crop quality (Duhamel & Vandenkoornhuyse, 2013). To this effect, in this work, we approach the plant phenotype from a holobiont perspective (Vandenkoornhuyse et al., 2015). The plant holobiont concept was originally developed in the context of evolutionary biology, considering the plant and its microbiota as a cohesive unit of selection. In the present case, plant phenotype is the result of an artificial selection process in which we intervene on either the plant or microbial side of hormonal regulation. Plant‐associated microbial communities harbor a significantly more diverse genetic pool than the host plant (Philippot et al., 2013) and engineering these microorganisms may offer a valuable target to improve plant traits. While several microorganisms are known to impact plant physiology, their effect is still mainly approached from a qualitative and conceptual perspective that hampers their efficient use in agriculture. Here, using plant nutritional value as a model multitrait phenotype, we have shown that we can improve the plant phenotype by editing either the plant genome itself or its associated microbes. We explicitly approached ethylene signaling as a model regulatory cascade influenced by both plants and bacteria. Ethylene signaling is long known to be affected by microbial ACC deaminase (Glick, 2014), yet this research field has become increasingly disconnected from advances on the importance of ethylene signaling in plant physiology (Ravanbakhsh et al., 2018). Ethylene, an important regulator of plant growth and development, controls nutrient uptake via several mechanisms. For instance, ethylene positively regulates the expression of root transporters responsible for micronutrient uptake. It interacts with auxin and phloem signals to alter uptake and transport of Fe (Lucena et al., 2015; Romera et al., 2016) and can modify root architecture in response to P deficiency (Neumann, 2016). Using a combination of minimal holobionts harboring mutations in either the bacterial or plant compartment of their genetic pool, we show that disabling the bacterial enzyme ACC deaminase has the same effect on ethylene production and nutrition as overexpression of ACC synthases in the plant. This moves us to further extend the concept of parallelism between microbiome‐ and plant‐encoded traits (Ravanbakhsh et al., 2019a), and place the plant genome and associated microorganisms in an integrated signaling network to see the collective effects of plant and microbiome parallel genes (Figs 2c, S1). In this context, changing the microbiome and/or the plant genome can collectively define the holobiont phenotype, which can be either highly adaptive or maladaptive, depending on the environmental conditions. Here, both the acdS mutation in bacteria and the ETO1 mutation in plants increased concentrations of macro‐ and micronutrients in plant shoots by altering plant ethylene concentrations. The microbiome can thus serve to rescue or suppress the effects of the plant genome on the plant phenotype, offering an opportunity to shift the holobiont phenotype over time via changes in the microbiome (Theis et al., 2016).
Plant phenotype, characterized here in terms of shoot nutrient concentration, could be well predicted on the basis of shoot ethylene concentration, with little concern for whether the causative mutation was located within the plant or the bacterial genome. The effect of bacteria was so strong that growing plants with wild‐type ACC deaminase bacteria (AcdS+) could inhibit the effects of plant genome alterations on the plant phenotype (shoot ethylene production and DW) of the eto1, confirming the importance of this bacterial trait as an antagonist of the ETO1 gene in the holobiont‐level regulatory network (Figs 2c, S3).
These results have far‐reaching implications for strategies to design better crops. Several microorganisms have been shown to impact plant physiology and to have promising applications in crop improvement. However, their use is currently constrained by the unpredictability of their effects under field conditions. Microbial traits have long been arbitrarily classified as beneficial or negative for plants, without accounting for their interactions with the plant genetic background or the environmental conditions. As a result, observations made under specific conditions cannot be extrapolated to field conditions or even other cultivars of the same plant (Vacheron et al., 2013). By demonstrating that plant‐associated microorganisms are an essential part of the plant hormonal network, we provide a conceptual framework to improve plant–microbe interactions in a predictable way. Plant physiology is already moving from a gene‐for‐gene to a network perspective (Peyraud et al., 2017), presenting the plant phenotype as the result of several interacting cascades that together process environmental information into resource allocation decisions. Including plant‐associated microorganisms in this network will allow for targeted improvement of plant traits by manipulating the microbial genome or metagenome instead of (or in addition to) the plant genome itself (Wei & Jousset, 2017). This work adds an important step in our aim to take microbes out of the context of purely qualitative effects on plant growth towards a predictable framework, connecting microbes to plant phenotype under specific environmental conditions (Ravanbakhsh et al., 2017, 2018, 2019a,b). As plant genetic improvement is a tedious and expensive process from practical and regulatory perspectives, modifying the associated microorganisms may provide a shortcut toward faster and more easily customizable plant improvement strategies.
This study relies on a simplified, minimal holobiont system with one plant and one bacterium, thereby allowing us to disentangle the effects of mutations in plants and bacteria. However, in natural systems and at the full holobiont scale, complex interspecific interaction (Hart et al., 2018) and plant species‐specific responses should be taken into account. From the plant perspective, many A. thaliana genes have homologs in the crops (Gepstein & Horwitz, 1995), so we anticipate that the results can be readily translated to other plant systems. In nature, where plants are associated with a multispecies microbiome, the present work will provide guidelines to selectively improve microbiome‐level properties. For instance, a higher nutritional value may be achieved by shifting microbiome composition to reduce the abundance of ACC deaminase producers or increase the abundance of ethylene producers. By highlighting further the importance of a functioning ethylene signaling in the plant holobiont, this work contributes to the growing number of studies (Ravanbakhsh et al., 2017, 2019a) calling for a re‐evaluation of what constitutes a beneficial plant–microbe interaction. Approaching plant hormonal balance as the result of an intertwined plant–microbe regulatory network opens new perspectives to managing plant phenotype by joint consideration of plant and associated microbial genes. Such an evidence‐based framework will probably contribute to guiding nascent microbiome engineering technologies by predicting the desired trait combinations needed to be expressed to obtain the desired plant phenotype.
Author contributions
MR and AJ designed the research. MR performed the research and analyzed the data. MR, AJ and GK contributed to interpreting the data and writing the manuscript.
Supporting information
Fig. S1 The concentration of ethylene, Fe, Zn, and Cu in the shoot of Arabidopsis thaliana Col‐0 and eto1 inoculated with the bacterial treatment.
Fig. S2 The effects of mutation in plants or bacterial genome on phosphorus concentration in the shoot of Arabidopsis thaliana.
Fig. S3 Shoot DW of Arabidopsis thaliana Col‐0 and eto1 inoculated with the bacterial treatment.
Table S1 DF minimum medium composition.
Table S2 Two‐way ANOVA table summarizing the interactive effects of plant genotypes and bacterial inoculation on nutrient level and ethylene concentration in plants.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledegments
Laurentius A. C. J. Voesenek and Rashmi Sasidharan from the Plant Ecophysiology Group, Utrecht University are acknowledged for their inputs related to the study design. Fabrice Roux, from the CNRS, France is acknowledged for his inputs on the manuscript. The authors would like to thank Prof. Bernard Glick, Department of Biology, University of Waterloo, Waterloo, ON, Canada for providing the bacterial strains. Gerrit Rouwenhorst from Ecology and Biodiversity Group, Utrecht University, is acknowledged for his valuable advice for plant and soil digestions and doing ICP measurements. Rob Welschen from the Plant Ecophysiology Group, Peter Veenhuizen, and G.P. Verduyn from the Ecology and Biodiversity Group, Utrecht University, are acknowledged for their technical assistance and advice. The authors declare that they have no conflicts of interest.
Contributor Information
Mohammadhossein Ravanbakhsh, Email: M.Ravanbakhsh@uu.nl.
Alexandre Jousset, Email: A.L.C.Jousset@uu.nl.
References
- Abeles FB, Morgan PW, Saltveit M Jr. 2012. Ethylene in plant biology. New York, NY, USA: Academic Press. [Google Scholar]
- Barnawal D, Bharti N, Tripathi A, Pandey SS, Chanotiya CS, Kalra A. 2016. ACC‐deaminase‐producing endophyte Brachybacterium paraconglomeratum strain SMR20 ameliorates chlorophytum salinity stress via altering phytohormone generation. Journal of Plant Growth Regulation 35: 553–564. [Google Scholar]
- Barrett C. 2010. Measuring food insecurity. Science 327: 825–828. [DOI] [PubMed] [Google Scholar]
- Bartoli C, Frachon L, Barret M, Rigal M, Huard‐Chauveau C, Mayjonade B, Zanchetta C, Bouchez O, Roby D, Carrère S et al 2018. In situ relationships between microbiota and potential pathobiota in Arabidopsis thaliana . ISME Journal 12: 2024–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen RL, Pieterse CMJ, Bakker PAHM. 2012. The rhizosphere microbiome and plant health. Trends in Plant Science 17: 478–486. [DOI] [PubMed] [Google Scholar]
- Bouffaud M‐L, Renoud S, Dubost A, Moënne‐Loccoz Y, Muller D. 2018. 1‐Aminocyclopropane‐1‐carboxylate deaminase producers associated to maize and other Poaceae species. Microbiome 6: 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae HS, Faure F, Kieber JJ. 2003. The eto1, eto2, and eto3 mutations and cytokinin treatment increase ethylene biosynthesis in Arabidopsis by increasing the stability of ACS protein. The Plant Cell 15: 545–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . The Plant Journal 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Cristescu SM, Mandon J, Arslanov D, De Pessemier J, Hermans C, Harren FJM. 2013. Current methods for detecting ethylene in plants. Annals of Botany 111: 347–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J, Jiang W, Cheng Z, Heikkila JJ, Glick BR. 2013. The complete genome sequence of the plant growth‐promoting bacterium Pseudomonas sp. UW4. PLoS ONE 8: e58640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois M, den Broeck LV, Inzé D. 2018. The pivotal role of ethylene in plant growth. Trends in Plant Science 23: 311–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duhamel M, Vandenkoornhuyse P. 2013. Sustainable agriculture: possible trajectories from mutualistic symbiosis and plant neodomestication. Trends in Plant Science 18: 597–600. [DOI] [PubMed] [Google Scholar]
- Dworkin M, Foster JW. 1958. Expriment with some micronutrients which utilize ethylene and hydrogen. Journal of Bacteriology 75: 592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etheridge N, Hall BP, Schaller GE. 2006. Progress report: ethylene signaling and responses. Planta 223: 387–391. [DOI] [PubMed] [Google Scholar]
- Gepstein S, Horwitz BA. 1995. The impact of Arabidopsis research on plant biotechnology. Biotechnology Advances 13: 403–414. [DOI] [PubMed] [Google Scholar]
- Glick BR. 2014. Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiological Research 169: 30–39. [DOI] [PubMed] [Google Scholar]
- Glick BR, Penrose DM, Li J. 1998. A model for the lowering of plant ethylene concentrations by plant growth‐promoting bacteria. Journal of Theoretical Biology 190: 63–68. [DOI] [PubMed] [Google Scholar]
- Haffenden H. 2019. PoA: finds the price of anarchy for routing games , R v.1.2.1 [WWW document] URL https://cran.r‐project.org/web/packages/PoA/index.html.
- Hart MM, Antunes PM, Chaudhary VB, Abbott LK. 2018. Fungal inoculants in the field: is the reward greater than the risk? Functional Ecology 32: 126–135. [Google Scholar]
- Kalra L, Perez I, Gupta S, Wittink M. 1997. The Influence of visual neglect on stroke rehabilitation. Stroke 28: 1386–1391. [DOI] [PubMed] [Google Scholar]
- Kazan K. 2015. Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends in Plant Science 20: 219–229. [DOI] [PubMed] [Google Scholar]
- Lassoued R, Smyth SJ, Phillips PWB, Hesseln H. 2018. Regulatory uncertainty around new breeding techniques. Frontiers in Plant Science 9: 1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Ovakim DH, Charles TC, Glick BR. 2000. An ACC deaminase minus mutant of Enterobacter cloacae UW4 no longer promotes root elongation. Current Microbiology 41: 101–105. [DOI] [PubMed] [Google Scholar]
- Lucena C, Romera FJ, García MJ, Alcántara E, Pérez‐Vicente R. 2015. Ethylene participates in the regulation of Fe deficiency responses in strategy I plants and in rice. Frontiers in Plant Science 6: 1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugtenberg B, Kamilova F. 2009. Plant‐growth‐promoting rhizobacteria. Annual Review of Microbiology 63: 541–556. [DOI] [PubMed] [Google Scholar]
- Neumann G. 2016. The role of ethylene in plant adaptations for phosphate acquisition in soils – a review. Frontiers in Plant Science 6: 1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson DC, Oetiker JH, Yang SF. 1995. Analysis of LE‐ACS3, a 1‐Aminocyclopropane‐1‐carboxylic acid synthase gene expressed during flooding in the roots of tomato plants. Journal of Biological Chemistry 270: 14056–14061. [DOI] [PubMed] [Google Scholar]
- Panke‐Buisse K, Poole AC, Goodrich JK, Ley RE, Kao‐Kniffin J. 2015. Selection on soil microbiomes reveals reproducible impacts on plant function. ISME Journal 9: 980–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H‐P, Lin T‐Y, Wang N‐N, Shih M‐C. 2005. Differential expression of genes encoding 1‐aminocyclopropane‐1‐carboxylate synthase in Arabidopsis during hypoxia. Plant Molecular Biology 58: 15–25. [DOI] [PubMed] [Google Scholar]
- Penrose DM, Glick BR. 2003. Methods for isolating and characterizing ACC deaminase‐containing plant growth‐promoting rhizobacteria. Physiologia Plantarum 118: 10–15. [DOI] [PubMed] [Google Scholar]
- Peyraud R, Dubiella U, Barbacci A, Genin S, Raffaele S, Roby D. 2017. Advances on plant–pathogen interactions from molecular toward systems biology perspectives. The Plant Journal 90: 720–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippot L, Raaijmakers JM, Lemanceau P, van der Putten WH. 2013. Going back to the roots: the microbial ecology of the rhizosphere. Nature Reviews Microbiology 11: 789–799. [DOI] [PubMed] [Google Scholar]
- Puga‐Freitas R, Blouin M. 2015. A review of the effects of soil organisms on plant hormone signalling pathways. Environmental and Experimental Botany 114: 104–116. [Google Scholar]
- Ravanbakhsh M, Kowalchuk GA, Jousset A. 2019a. Root‐associated microorganisms reprogram plant life history along the growth–stress resistance tradeoff. ISME Journal 13: 3093–3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravanbakhsh M, Kowalchuk GA, Jousset A. 2019b. Optimization of plant hormonal balance by microorganisms prevents plant heavy metal accumulation. Journal of Hazardous Materials 379: 120787. [DOI] [PubMed] [Google Scholar]
- Ravanbakhsh M, Sasidharan R, Voesenek LACJ, Kowalchuk GA, Jousset A. 2017. ACC deaminase‐producing rhizosphere bacteria modulate plant responses to flooding. Journal of Ecology 105: 979–986. [Google Scholar]
- Ravanbakhsh M, Sasidharan R, Voesenek LACJ, Kowalchuk GA, Jousset A. 2018. Microbial modulation of plant ethylene signaling: ecological and evolutionary consequences. Microbiome 6: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romera FJ, Smith AP, Pérez‐Vicente R. 2016. Ethylene’s role in plant mineral nutrition. Frontiers in Plant Science 6: 911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleem AR, Brunetti C, Khalid A, Rocca GD, Raio A, Emiliani G, Carlo AD, Mahmood T, Centritto M. 2018. Drought response of Mucuna pruriens (L.) DC. inoculated with ACC deaminase and IAA producing rhizobacteria. PLoS ONE 13: e0191218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleem M, Arshad M, Hussain S, Bhatti AS. 2007. Perspective of plant growth promoting rhizobacteria (PGPR) containing ACC deaminase in stress agriculture. Journal of Industrial Microbiology & Biotechnology 34: 635–648. [DOI] [PubMed] [Google Scholar]
- Sapre S, Gontia‐Mishra I, Tiwari S. 2019. ACC Deaminase‐producing bacteria: a key player in alleviating abiotic stresses in plants In: Kumar A, Meena VS, eds. Plant growth promoting rhizobacteria for agricultural sustainability: from theory to practices. Singapore City, Singapore: Springer, 267–291. [Google Scholar]
- Shiu OY, Oetiker JH, Yip WK, Yang SF. 1998. The promoter of LE‐ACS7, an early flooding‐induced 1‐aminocyclopropane‐1‐carboxylate synthase gene of the tomato, is tagged by a Sol3 transposon. Proceedings of the National Academy of Sciences, USA 95: 10334–10339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeets K, Ruytinx J, Semane B, Van Belleghem F, Remans T, Van Sanden S, Vangronsveld J, Cuypers A. 2008. Cadmium‐induced transcriptional and enzymatic alterations related to oxidative stress. Environmental and Experimental Botany 63: 1–8. [Google Scholar]
- Theis KR, Dheilly NM, Klassen JL, Brucker RM, Baines JF, Bosch TCG, Cryan JF, Gilbert SF, Goodnight CJ, Lloyd EA et al 2016. Getting the hologenome concept right: an eco‐evolutionary framework for hosts and their microbiomes. mSystems 1: e00028–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacheron J, Desbrosses G, Bouffaud M‐L, Touraine B, Moënne‐Loccoz Y, Muller D, Legendre L, Wisniewski‐Dyé F, Prigent‐Combaret C. 2013. Plant growth‐promoting rhizobacteria and root system functioning. Frontiers in Plant Science 4: 365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Poel B, Van Der Straeten D. 2014. 1‐aminocyclopropane‐1‐carboxylic acid (ACC) in plants: more than just the precursor of ethylene! Frontiers in Plant Science 5: 640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenkoornhuyse P, Quaiser A, Duhamel M, Van AL, Dufresne A. 2015. The importance of the microbiome of the plant holobiont. New Phytologist 206: 1196–1206. [DOI] [PubMed] [Google Scholar]
- Veiga RSL, Faccio A, Genre A, Pieterse CMJ, Bonfante P, van der Heijden MGA. 2013. Arbuscular mycorrhizal fungi reduce growth and infect roots of the non‐host plant Arabidopsis thaliana . Plant, Cell & Environment 36: 1926–1937. [DOI] [PubMed] [Google Scholar]
- Voesenek LACJ, Jackson MB, Toebes AHW, Huibers W, Vriezen WH, Colmer TD. 2003. De‐submergence‐induced ethylene production inRumex palustris: regulation and ecophysiological significance. The Plant Journal 33: 341–352. [DOI] [PubMed] [Google Scholar]
- Wang KL‐C, Yoshida H, Lurin C, Ecker JR. 2004. Regulation of ethylene gas biosynthesis by the Arabidopsis ETO1 protein. Nature 428: 945. [DOI] [PubMed] [Google Scholar]
- Wei Z, Jousset A. 2017. Plant breeding goes microbial. Trends in Plant Science 22: 555–558. [DOI] [PubMed] [Google Scholar]
- Wilkinson S, Kudoyarova GR, Veselov DS, Arkhipova TN, Davies WJ. 2012. Plant hormone interactions: innovative targets for crop breeding and management. Journal of Experimental Botany 63: 3499–3509. [DOI] [PubMed] [Google Scholar]
- Zsögön A, Čermák T, Naves ER, Notini MM, Edel KH, Weinl S, Freschi L, Voytas DF, Kudla J, Peres LEP. 2018. De novo domestication of wild tomato using genome editing. Nature Biotechnology 36: 1211–1216. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 The concentration of ethylene, Fe, Zn, and Cu in the shoot of Arabidopsis thaliana Col‐0 and eto1 inoculated with the bacterial treatment.
Fig. S2 The effects of mutation in plants or bacterial genome on phosphorus concentration in the shoot of Arabidopsis thaliana.
Fig. S3 Shoot DW of Arabidopsis thaliana Col‐0 and eto1 inoculated with the bacterial treatment.
Table S1 DF minimum medium composition.
Table S2 Two‐way ANOVA table summarizing the interactive effects of plant genotypes and bacterial inoculation on nutrient level and ethylene concentration in plants.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.


