Abstract
Background
Despite advances in treatments, 30% to 50% of stage III‐IV head and neck squamous cell carcinoma (HNSCC) patients relapse within 2 years after treatment. The Big Data to Decide (BD2Decide) project aimed to build a database for prognostic prediction modeling.
Methods
Stage III‐IV HNSCC patients with locoregionally advanced HNSCC treated with curative intent (1537) were included. Whole transcriptomics and radiomics analyses were performed using pretreatment tumor samples and computed tomography/magnetic resonance imaging scans, respectively.
Results
The entire cohort was composed of 71% male (1097)and 29% female (440): oral cavity (429, 28%), oropharynx (624, 41%), larynx (314, 20%), and hypopharynx (170, 11%); median follow‐up 50.5 months. Transcriptomics and imaging data were available for 1284 (83%) and 1239 (80%) cases, respectively; 1047 (68%) patients shared both.
Conclusions
This annotated database represents the HNSCC largest available repository and will enable to develop/validate a decision support system integrating multiscale data to explore through classical and machine learning models their prognostic role.
Keywords: big data, head and neck cancer, prognostic models, radiomics, transcriptomics
1. INTRODUCTION
Head and neck cancers (HNCs) are a group of highly heterogeneous diseases. Worldwide HNCs are the sixth most deadly tumors, accounting more than 700 000 newly detected cases (150 000 in Europe), leading to approximately 350 000 deaths annually 1 (70 000 in Europe). The majority of cases are reported as head and neck squamous cell carcinomas (HNSCCs), arising from the epithelial cells of the mucosal lining of the upper aerodigestive tract. In clinical practice, HNSCCs are classified according to their anatomic site of origin in oral cavity carcinoma (OCC), oropharyngeal carcinoma (OPC), further subclassified according to its relation with human papillomavirus (HPV), 2 hypopharyngeal (HPC), and laryngeal (LC) cancers. To date, the most relevant risk factors for HNSCC are excessive alcohol consumption, smoking, and for OPC, infection with a high‐risk HPV.
Approximately only one‐third of patients with HNSCCs are detected at early stages of the disease 3 (stages I and II according to the Seventh Edition of the Tumor‐lymph Node‐Metastasis classification [TNM7] of the American Joint Committee on Cancer [AJCC]/Union for International Cancer Control [UICC] staging system 4 ). These patients are treated with surgery or radiotherapy, depending on the tumor site and extension, with a 70% to 90% cure rate. However, the majority of patients present with locoregionally advanced stages (III and IV) and require multimodal interventions. 2 For HNSCC patients, the treatment can be extremely impairing, having huge impacts on functionalities and quality of life (QoL). Despite impressive advances in surgery and radiotherapy techniques, 30% to 50% of stage III‐IV HNSCC patients relapse within 2 years after treatment. Notably, salvage treatments (eg, surgery, re‐irradiation, systemic treatments) are ineffective in most cases. 5 , 6
Treatment choice depends on primary tumor site and stage, and the heterogeneity in biological behaviors hampers the development of general or site‐specific prognostic models to guide clinical management for all the different tumors included in HNSCC. In this complex and challenging scenario, there is a substantial need to select the optimal treatment that maximizes the probability of cure, while minimizing the side effects and impact on the QoL of patients. A more personalized strategy would overcome the current approach based on the TNM staging classification. 4 , 7
Starting from the hypothesis that the use of multiparametric variables applied on tumor site‐ and treatment‐specific populations may facilitate prognostic prediction and guidance of the optimal treatment choice, we established an international European consortium, which allowed to build the Big Data to Decide (BD2Decide) project. To achieve this ambitious aim, we developed a new database, where clinical and omics information could be acquired, stored, and analyzed through different approaches. In this article, we present in details the characteristics of the BD2Decide database, the methodology for its creation and curation and the major information retrievable from it. This new database could enable linking together rigorously annotated patient‐specific multiparameter clinical, pathologic, demographic, transcriptomics and radiomics data from the currently largest cohort of patients with locoregionally advanced HNSCC.
2. MATERIALS AND METHODS
The study population was composed of loco‐regionally advanced (stage III‐IVA/B according to TNM7) HNSCC patients receiving treatments with curative intent between 2008 and 2017. A European Consortium was formed in 2015 and in 2016 the project “Big Data and Models for Personalized Head and Neck Cancer Decision Support (BD2Decide)” (ClinicalTrial.gov Identifier NCT02832102) 8 started with the financial support of the EU commission. 9 The Ethical Committee of each participating center approved the protocol. Details about the participating Institutions are reported in Supplementary materials. The follow‐up was closed at September 2019. For data use and retrieval of tumor samples, when available, patients still alive provided informed consent; a waiver for informed consent was obtained according to national regulations. For deceased patients in the retrospective cohort, a waiver was provided by the Institutional Review Board of each center.
Main inclusion criteria were histological confirmation of squamous cell OCC, OPC, LC orHPC, aged ≥18 years, and III and IVA or IV as clinical stage (TNM7). The administration of treatment had to be with curative intent, including any combination of surgery, radiotherapy (three‐dimensional or intensity‐modulated radiotherapy [IMRT]) and chemotherapy. Other requirements were the availability of both adequate archival pretreatment tumor specimen and pretreatment contrast‐enhanced CT scan of the head and neck region, performed with contiguous cuts of ≤2 to 3 mm in slice thickness or MRI scans with T1 (non or pre‐contrast) and T2‐weighted acquisitions (slice thickness < 3 mm). For OPC cases, assessment of p16/HPV status was mandatory. HPV testing was performed with p16 immunohistochemistry (IHC)‐staining (positive/negative) and HPV DNA presence by either in situ hybridization (ISH) or polymerase chain reaction (PCR) (details in Supplementary text, subheading “HPV status analysis”).
Patients with clinical stage IVC, malignancies in the previous 5 years prior to the treatment of HNSCC, or any previous malignancy in the head and neck that was treated with surgery and/or radiation were excluded.
Clinical, pathological, and demographical data (Supplementary Tables S1‐S4) were collected using a web‐based electronic case report form (eCRF) named OpenClinica platform (OpenClinica, LLC; Waltham, Massachusetts),. Since the BD2Decide was aimed at defining data‐driven prognostic factors for locoregionally advanced HNSCC patients, all pretreatment clinical data that could be collected through medical charts were recorded. Moreover, details about treatments, their toxicities, pathologic features, and follow‐up were deemed fundamental to build the largest data set of clinicopathological data about HNSCC patients.
For patients enrolled prospectively, the following quality of life questionnaires (QLQ) were collected: EORTC QLQ‐C30, 10 EORTC HN35, 11 and EQ‐5D‐5L. 12
The selection of these questionnaires was related to their specific independent role in assessing patient‐reported outcome measurements (PROMs): EORTC QLQ‐C30 is valid for all cancer patients; EORTC HN35 is a PROM specifically designed for HNC patients, and it was the most updated version available at the time of study approval; and EQ‐5D‐5L is a questionnaire used in health‐technology analysis (HTA).
Disease‐free survival (DFS) was defined as the time between primary diagnosis and the occurrence of any event (first disease recurrence; relapse; death of any cause) or the last follow‐up in case of patients without evidence of disease. Overall survival (OS) was defined as the time between primary diagnosis and death or last follow‐up. Survival curves were estimated with the Kaplan‐Meier method.
Further details about the following sections are reported in Supplementary materials: participating centers and roles; ethics and privacy; sample size calculation; HPV status analysis; data quality assessment; quality control of clinical and pathological data; ontology, knowledge management system; whole transcriptome analysis; transcriptomics and prognostic clinical models survey; radiomics analysis; population‐related data from external sources; development of the big data environment and the interfaces for Clinical Decision Support System (CDSS) and Visual Analytics Tool (VAT); CDSS; and VAT.
3. RESULTS
A total of 1537 stage III‐IV HNSCC patients were included in the BD2Decide database, 1086 cases (70%) were collected retrospectively (2008‐2014) and the remaining 451 prospectively (2015‐2017). Details on the country of origin (47%—716 from Italian, 43%—670 from Dutch and 10%—151 from German Cancer Centers) are reported in Supplementary Table S5.
The majority of patients (71%, 1097) were male, and the study population comprised 28% OCC (429), 41% OPC (624), 11% HPC (170), and 20% LC (314) cases (Table 1). The clinical TNM7 stage at diagnosis was III and IV in 26% (393) and 74% (1144) of cases, respectively. The median age was 62 years (range 20‐93) and median follow‐up, estimated using the reverse Kaplan‐Meier method, was 50.5 months (95% confidence interval [CI]: 47.9‐54.2). IHC p16 testing was assessed in all 624 OPC cases: 377 (60%) resulted as p16‐positive and as 247 (40%) as p16‐negative; of the 377 p16‐positive cases, 374 were tested for HPV DNA as well and a 4% (16 cases) were HPV‐DNA negative.
TABLE 1.
Patient characteristics
| No. of patients | No. of patients (%) | |
|---|---|---|
| Tumor site | ||
| Oral cavity | 429 | 28% |
| Oropharynx a | 624 | 41% |
| Hypopharynx | 170 | 11% |
| Larynx | 314 | 20% |
| Clinical stage (TNM7) | ||
| III | 393 | 26% |
| IVA | 1001 | 65% |
| IVB | 143 | 9% |
| Gender | ||
| Male | 1097 | 71% |
| Female | 440 | 29% |
| Age at diagnosis | Median 62 years (range 20‐93) | |
| Median follow‐up | 50.5 months (95% CI 47.9‐54.2) | |
377 were p16 positive (60%) and 247 (40%) were p16 negative.
The major risk factors (smoke, alcohol, oral hygiene and familiar history of malignancies) are reported in Table 2:76% of the patients (1170) were current or former smokers and 58% (900) alcohol consumers. Further details on other clinical characteristics and risk factors are reported in Supplementary Table S6.
TABLE 2.
Major risk factors
| No. of patients | No. of patients (%) | |
|---|---|---|
| Smoking | ||
| Current or former | 1170 | 76% |
| Never | 300 | 20% |
| Unknown | 67 | 4% |
| Alcohol | ||
| Current or former | 900 | 58% |
| Never | 551 | 36% |
| Unknown | 86 | 6% |
| Oral hygiene | ||
| Good | 420 | 28% |
| Intermediate | 445 | 29% |
| Poor | 176 | 11% |
| Unknown/not available | 496 | 32% |
| Familiar history of malignancies | ||
| Yes | 381 | 25% |
| No | 547 | 36% |
| Unknown | 609 | 39% |
After the update of the staging system to the TNM eighth edition, there were no changes observed in the 1160 HPV‐negative HNSCCs (all tumor sites) patients with stage III and IVB disease. An upstaging to IVB was observed in 120 (17%) of patients with IVA disease (Table 3). This was mostly attributed to the introduction of N3b in patients having regional lymph node metastases with extranodal extension.
TABLE 3.
Staging changes from TNM7 to TNM8 for p16‐negative HNSCC patients a
| TNM7 | TNM8 | Staging variation | ||
|---|---|---|---|---|
| Stage | No. of patients | Stage | No. of patients | Staging variation |
| III | 337 | III | 337 | No change |
| IVa | 722 | III | 8 | 1% downstaging |
| IVb | 594 | 82% no change | ||
IVb
|
120 | 17% upstaging | ||
| IVb | 101 | IVb | 1 | <1% downstaging |
| IVb | 100 | >99% no change |
All tumor sites.
Details about each treatment modality (Supplementary Table S7), the related adverse events (Supplementary Tables S8‐S9), and the quota of QLQ collected for the patients prospectively enrolled (Supplementary Table S10) are reported in supplementary material.
During follow‐up, 257 patients (17%) experienced locoregional recurrence only, 151 distant relapse only (10%), and 49 both (3%). A second primary malignancy was observed in 59 cases (4%), with concomitant primary disease failure in 7 cases (<1%). Among cases with recurrence/relapse/second primary cancer, 178 and 213 were treated with curative and palliative intent, respectively, while the remaining did not receive any specific therapy. In the overall cohort, 599 patients (39%) expired (Table 4), 938 patients (61%) were alive at last follow‐up, and 782 of them (83%) were disease‐free as well.
TABLE 4.
Causes of death a
| Cause of death | No. of patients | No. of patients (%) |
|---|---|---|
| Malignant disease under study, or complication due to malignant disease under study | 332 | 55% |
| Adverse event | 47 | 8% |
| Second primary malignant disease, or complication due to second primary malignant disease | 76 | 13% |
| Other / unknown cause (not assessable or insufficient data) | 144 | 24% |
In the cohort of 599 expired patients.
On the entire cohort of 1537 cases, the median OS and DFS were 73.9 months (95% CI: 69.4‐89.2) and 61.4 months (95% CI: 53.9‐68.4), respectively (Figure 1A). According to the TNM8, in the 1160 HPV‐negative HNSCC patients, the median OS was 86.9 months (95% CI: 68.7‐115.9) for patients with stage III disease and 51.3 months (95% CI: 39.8‐60.9) for those with stage IV disease (P < .0001) (Figure 1B).
FIGURE 1.
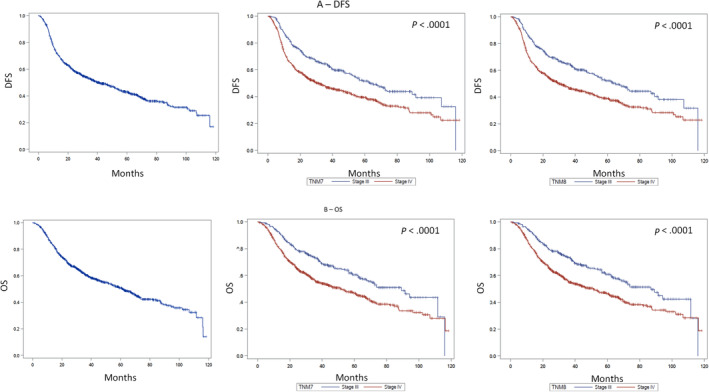
DSFS (A) and OS (B) in p16‐negative HNSCC patients. Left panels: DSFS and OS; central panels: survival according to TNM7; right panels: survival according to TNM8 [Color figure can be viewed at wileyonlinelibrary.com]
Since HNSCC is a heterogeneous group of diseases with different biology, treatments, and outcomes, the survival of the five main HNSCC clinical entities are here reported in detail. We considered the four major primary tumor sites (OCC, OPC, HPC, and LC), but dividing OPC in two different entities according to p16 positivity.
3.1. Oral cavity squamous cell carcinoma
The demographical/clinical and treatment information and the survival data of the 429 OCC patients included in the BD2D database are reported in Supplementary Tables S11 and S12, respectively.
Median age was 63 years (range 20‐93). Almost half of patients (171) were women (40%, the highest percentage between the five HNSCC entities) and 258 were men. Surgery was part of the curative treatment in 374 cases (89%).
Median DFS was 37.83 months (95% CI 27.83‐51.84) and median OS 59.08 months (95% CI 43.49‐84.64). In patients with stage III (TNM8) disease, median DFS and OS were 54.67 months (95% CI 38.36‐91.45) and 89.24 months (95% CI 47.11‐NR), respectively. In those with stage IVA‐B (TNM8) disease, median DFS and OS were 28.98 months (95% CI 21.35‐47.99) and 52.23 months (95% CI 33.06‐69.47), respectively.
3.2. p16‐positive oropharyngeal squamous cell carcinoma
Following oral cavity, the p16‐positive oropharyngeal carcinoma patient cohort was the second most represented of the BD2Decide project. The demographical/clinical and treatment information and the survival data of the 377 p16‐positive OPC patients included in the BD2D database are reported in Supplementary Tables S13 and S14, respectively.
Median age of these patients (60 years, range 29‐86) was the lowest among the five groups. Patients with clinical T4 and/or N3 classification (overall stage III, TNM8) were 105 (29%).
Data about smoking history were available for 98% of cases (370 patients: 252 current or former smokers, 118 never smokers): details about pack‐years were available in 249 out of 252 current/former smokers. According to Ang prognostic classification, 15 64% of patients were at low risk (235 cases: 118 never smokers, 61 with ≤10 pack‐years, 56 with >10 pack‐years, and cN0‐N2a according to TNM7) and 36% at intermediate risk (132 patients with >10 pack‐years and cN2b‐N3 according to TNM7). Radiotherapy was part of multimodal approaches in 86% of subjects (322) and 297 patients (79%) received chemoradiation. Surgery (with or without postoperative radiation) was performed in 107 cases (28%).
In p16‐positive OPC patients, median DFS and OS were not reached (Figure 2). Five‐year DFS and OS of the whole p16‐positive cancer cohort were 76% and 82%, respectively. Five‐year DFS, according to TNM8, was 84% in stage I, 69% in stage II, and 68% in stage III (P = .0008, Figure 2A). Five‐year OS was 89%, 74%, and 77% (P = .004) in stage I, stage II, and stage III disease (TNM8), respectively (Figure 2B). The prognostic accuracy for 5‐year OS was higher for TNM8 (AUC = 0.61, P = .005) than Ang classification (AUC = 0.57, P = .061) and TNM7 (AUC = 0.55, P = .053).
FIGURE 2.
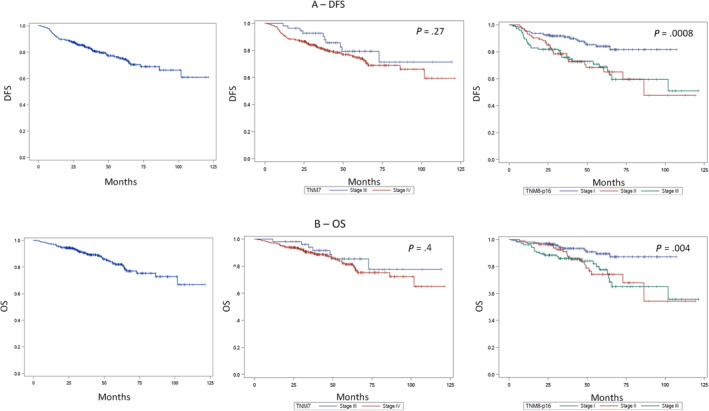
DSFS (A) and OS (B) in p16‐positive OPC patients. Left panels: DSFS and OS; central panels: survival according to TNM7; right panels: survival according to TNM8 [Color figure can be viewed at wileyonlinelibrary.com]
3.3. p16‐negative oropharyngeal squamous cell carcinoma
Sixteen percent of patients included in the BD2D database (247 cases) were affected by p16‐negative OPC. The demographical/clinical and treatment information and the survival data of the p16‐negative OPC patients are reported in Supplementary Tables S15 and S16, respectively.
This was the subgroup with the highest frequency (30%, 73) of stage IVB (TNM8) disease. Most patients (217, 96% among those with data availability about smoking history) had a significant tobacco exposure (>10 pack‐years). According to Ang prognostic classification, 15 98% of patients (224) were at high risk, while only 5 were at intermediate risk. Radiotherapy was delivered in 96% of cases (234), and one fourth of patients (60) received surgery.
Median DFS was 33.32 months (95% CI 22.57‐52.27), median OS was 53.22 months (95% CI 38.78‐68.09). In patients with stage III (TNM8) disease, median DFS and OS were 56.71 months (95% CI 22.57‐107.37) and 68.72 months (95% CI 38.78‐111.65), respectively. In those with stage IVA‐B (TNM8) disease, median DFS and OS were 28.45 months (95% CI 21.05‐46.32) and 46.05 months (95% CI 34.84‐66.68), respectively. In this cohort, TNM8 provided a more accurate prognostic prediction than TNM7 (AUC for 5‐year OS: TNM8 0.63, P < .0001 vs TNM7 0.59, P = .0001).
3.4. Hypopharyngeal squamous cell carcinoma
This patient cohort was the less represented in the BD2Decide project. The demographical/clinical and treatment information and the survival data of the 170 HPC patients included in the BD2D database are reported in Supplementary Tables S17 and S18, respectively.
Eighty percent of patients were men and median age was 63 years (range 44‐84). The 95% of patients (161) were treated with radiotherapy and 42% (71) with surgery. Out of these 71 subjects, 38 patients underwent total laryngectomy for primary tumor. Median follow‐up was 62.43 months (95% CI 53.52‐70.72).
Median DFS was 27.79 months (95% CI 17.79‐37.86): median DFS not reached (95% CI 20.09 months‐NR) in stage III and 24.05 months (95% CI 16.68‐34.21) in stage IVA‐B (P = .0072). Median OS was 45.26 months (95% CI 32.99‐62.1): median OS not reached (95% CI 58.62 months‐NR) in stage III and 35.92 months (95% CI 26.12‐54.64) in stage IVA‐B (P = .0043).
3.5. Laryngeal squamous cell carcinoma
One fifth of the patients included in the BD2D database were affected by LC. The demographical/clinical and treatment information and the survival data of the 314 LC patients are reported in Supplementary Tables S19 and S20, respectively.
More than half of them (51%, 161 patients) had a stage III disease. Almost 90% of subjects (270) received radiotherapy within multimodal treatments. Out of the 138 laryngeal cancer patients treated with surgery, 80 received total laryngectomy for primary cancer. Chemoradiation (considering together the following combinations: induction chemotherapy followed by curative radiotherapy; induction chemotherapy followed by surgery and postoperative radiation or chemo‐radiation; definitive concurrent chemoradiation; surgery and postoperative concomitant chemoradiation) was delivered in 97 cases (32%).
Among the five subgroups, the LC patient cohort was the one with the longest survival: median DFS 61.74 months (95% CI 46.74‐70.56), median OS 70.63 months (95% CI 63.09‐87.43). In stage III median DFS and OS were 63.09 (95% CI 51.05‐NR) and 73.72 months (95% CI 62.63‐NR), respectively (Supplementary Table S21). In stage IVA‐B, median DFS was 46.48 months (95% CI 29.18‐67.47) and median OS was 69.44 months (95% CI 51.21‐87.43).
3.6. Omics data
Transcriptomics and radiomics profiles were available for 1284 and 1239 cases, respectively (Figure 3). The distribution of profiles in the different tumor sites and the dropout of tumor specimens or images are reported in Supplementary Table S21.
FIGURE 3.
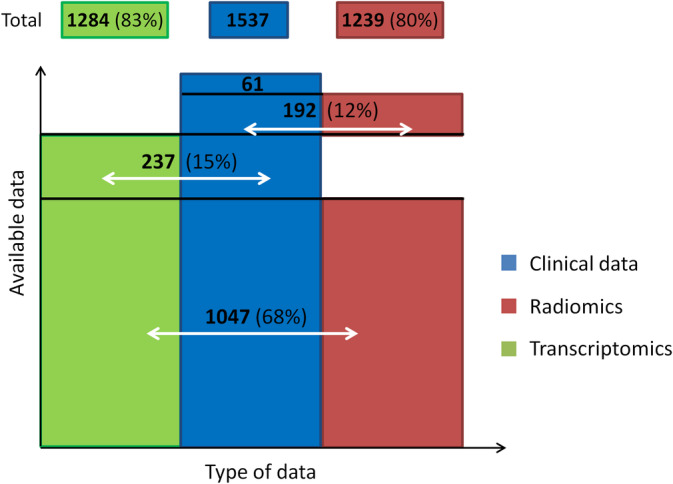
Available data in the BD2Decide final database [Color figure can be viewed at wileyonlinelibrary.com]
Two transcriptomics surveys were conducted during the project to create structured comprehensive transcriptomic databases from public resources for signature identification and testing on the BD2decide data set. 13 , 14 Selected statistical prognostic models 4 , 7 , 15 , 16 , 17 , 18 , 19 , 20 , 21 and transcriptomics signatures 13 , 19 , 22 , 23 , 24 , 25 , 26 , 27 , 28 currently being tested are reported in Supplementary Tables S22 and S23.
4. DISCUSSION
Although HNSCC arise in the same tissue, the mucosal squamous lining of the upper aerodigestive tract, the disease is remarkably heterogeneous, 29 hampering prognostic modeling. 30 In part this also relates to treatment, for instance OCC are typically surgically treated, whereas OPC can be cured with chemoradiation. 31 Also the role of HPV in OPSCC added a whole new level on this heterogeneity. 31 At present, the majority of studies on HNSCC prognosis encompass heterogeneous and small patient populations, and this hinders the validation of prognostic factors.
Being aware that he heterogeneity of HNSCC demands large efforts, data organization and sample collections at different institutions, we established an international European consortium, which allowed to build the BD2Decide European multicenter project aimed at exploring the potentialities of big data in HNSCC for clinical outcome estimation and research. This work essentially describes the characteristics of a unique large multi‐omics international database useful to analyze both the communalities and the tumor sites and treatment differences in a selected series of locally advanced stage of patients treated with curative intent.
In performing our study and establishing the BD2Decide HNSCC stage III‐IVA/B database, we placed great emphasis on the standardization of the procedures and collection of clinical data, tumor samples, and images from seven different clinical centers. This contributed not only to realize a complete ontological mapping of the HNC domain, in line with existing standard ontologies, but also to produce a formal representation of data dependencies. The “harmonization” efforts empowered us to conduct our systematic analysis. In particular, compared to the literature available data sets, the BD2Decide database contains innovative aspects such as (a) the adequate patient selection; (b) the retrieval and recording of high‐quality data test for up to 170 clinical, pathological, and demographic parameters; (c) the availability of the information in ≥90% of cases for parameters already associated with prognosis; (d) the availability of both transcriptomics and radiomics data for more than 1000 of patients.
To evaluate the representativeness of the study population, patient characteristics and outcomes were compared to literature data. Site distribution in our cohort was in line with the HNC EU incidence reported in Globocan 2018. 1 Men have a 2‐ to 5‐fold higher risk of developing HNSCC than women, and smoking characteristics were in line with those reported in the scientific literature. 32 The proportion of patients currently or formerly exposed to alcohol consumption was consistent with that reported in previous reports (70% in the study population vs 72% in the literature 33 ).
Regarding the attributable fraction of HPV in OPSCC, in the literature the evaluation of p16 positivity without HPV assessment was associated with a risk of false positivity in 12% of cases, indicating the importance of performing additional HPV DNA testing for the prediction of prognosis and when considering treatment de‐intensification. 35 Thus in our OPC cases, 374 of the 377 p16‐positive cases were tested for HPV DNA as well and a mere 4% (16 cases) were HPV‐DNA negative. This result suggests an increment in the accuracy of the tests in the recent years. We observed that in the whole OPC cohort, 60% of patients are p16/HPV DNA positive and data regarding the prospective cohort in this database indicate a proportion of p16‐positive cases higher than the one of the retrospective cohort (70% and 57%, respectively); overall, these percentages are consistent with the incidence data previously reported in the literature. 36
In our study population, 1‐year and 5‐year OS were 88% and 58%, respectively. These outcomes were significantly better than those described in a large epidemiological study (1‐year OS: 68.8%; 5‐year OS: 39.9%), involving European patients with HNSCC. 34 This is not surprising based on the specifics of epidemiological studies that include population‐based results. On the contrary, the BD2Decide population included a selected population (stage III‐IVA/B disease according to TNM7 classification) treated with curative intent in expert centers. In order to clearly identify prognostic factors at diagnosis of locoregionally advanced nonmetastatic HNSCC and to avoid potential negative biases, the study population was highly positively selected. Indeed, patients with either recurrent/metastatic disease or locoregionally advanced stage treated with palliative intent were intentionally excluded. In this setting, as expected, a positive selection bias explains the higher survival rates registered. The observed differences could be attributed to a mixture of the following reasons: (a) the BD2D patients were accrued from three countries of the 20 considered in the Eurocare analysis; (b) the predominance of the OPC and larynx sites could be partially attributed to the referral pattern of the selected cancer centers.
Although the samples and images in our collection derive from different institutions, the application of dedicated standard operating procedures for their collection and management resulted in an acceptable dropout of informative samples/images (12%‐21% for transcriptomics, 7%‐26% for radiomics, depending on the anatomical site of primary tumor). This systematic collection (covering a 10‐year period) demonstrated that in more recent years, the availability of sufficient material for whole transcriptomics analyses, particularly in the case of OPC, tended to be reduced. This could be attributed to the fact that for most OPC cases, where chemoradiation is usually preferred over surgery, the analyzed specimen was an initial small biopsy, and often the quota of tumor cells was scanty. Additionally, we noted that major loss, regarding the quality check of the extracted material, was related to hypopharynx tumors, compared with the other sites. One may speculate that a higher fraction of necrosis could be observed in resection specimens and low‐quantity biopsy material.
The availability of this uniform and large multiomics international database and the high number of cases entering in each of the five clinical entities characterizing HNSCC enabled us to start a series of site and or treatment specific studies. New site‐specific signatures and data‐driven models developed through statistical modeling are yet to be produced and integration of different molecular omics is in progress. In fact, in all cases with available transcriptomics, data regarding the expression of noncoding RNAs have already been acquired and the analysis is ongoing. Moreover, in approximately 450 cases, in the framework of an independently funded project, mutational data are also being analyzed. According to the general frame of the BD2Decide project, all these new omics data will be associated with other clinical and ‐omics data and will become publicly available constituting the core of future studies that will address the potential of big data in HNSCC.
The future use of BD2Decide database will be the incorporation of adaptive learning, allowing updating, improving, and refining the models using routinely collected data. The stepwise classification approach may allow clinicians to reach predictions based on easily obtainable clinical data. The system could be used to inform clinicians regarding the need to include additional sources of information. Furthermore, in case a recursive partitioning procedure is followed, clinicians will be informed regarding the cost‐effectiveness of collecting additional data at the different nodes of the decision tree.
5. CONCLUSIONS
Overall, we were able to construct a rigorously annotated HNSCC database, which includes clinical data recorded and collected for 1537 patients with stage III‐IVA/B HNSCC treated with curative intent associated transcriptomics and radiomics data for approximately 80% of cases. This represents the largest available database for head and neck cancer in terms of the number of cases, the huge number of available clinical information, and the expected integrated omics. As an example, the BD2D database contains up to 170 well‐annotated clinical variables and almost 3‐fold number of samples than The Cancer Genome Atlas, 37 which includes 528 HNSCC cases. 38 The establishment of our database provides numerous opportunities for conducting different analyses of clinical/radiomics/transcriptomics data to explore their single or combined prognostic role. In agreement with the need of transparency in science, data will be progressively deposited in public repositories and made available to the scientific community. In our view, large studies as BD2Decide will have to be the future for prognostic modeling studies. BD2Decide is a valuable resource, and this type of studies should be extended to even larger cohorts to further decipher the heterogeneity of HNSCC. Large cooperative scientific and clinical efforts and financial resources are needed within the EU or even between continents.
AUTHOR CONTRIBUTIONS
Lisa Licitra, Tito Poli, Ruud H Brakenhoff, Frank J. P. Hoebers, Kathrin Scheckenbach: Conceptualization; Loris De Cecco, Giuseppe Fico, Vasilis Tountopoulos, Ron Shefi, Luca Mainardi: Methodology; Stefano Cavalieri, Mara Serena Serafini, Silvia Rossi, Davide Lanfranco, Frederik WR Wesseling, Simon Keek, Marco Bologna, Irene Nauta, Annalisa Trama, Davide Mattavelli, Thomas Hoffmann: case material selection, recruitment and formal analysis; Stefano Cavalieri, Mara Serena Serafini, Silvia Rossi, Davide Lanfranco, Frederik W. R. Wesseling, Simon Keek, Marco Bologna, Irene Nauta, Silvana Canevari, Laura López Pérez, Thomas Klausch, Johannes Hans Berkhof, Giuseppe Fico, Davide Mattavelli, Thomas Hoffmann: Data Curation; Stefano Cavalieri, Loris De Cecco, Mara Serena Serafini, Silvana Canevari, Laura López Pérez, Marco Bologna, Annalisa Trama, Lisa Licitra: writing‐original draft preparation; all authors: writing‐review & editing; Lisa Licitra, Tito Poli, Ruud H Brakenhoff, Frank J. P. Hoebers, Kathrin Scheckenbach: funding acquisition.
CONFLICT OF INTEREST
Yes Author Lisa Licitra has disclosed funding (to her institution) for clinical studies and research from AstraZeneca, Boehringer Ingelheim, Eisai, Merck Serono, MSD, Novartis, and Roche, has received compensation for service as a consultant/advisor and/or for lectures from AstraZeneca, Bayer, Bristol‐Myers Squibb, Boehringer Ingelheim, Debiopharm, Eisai, Merck Serono, MSD, Novartis, Roche, and Sobi; and has received travel coverage for meetings from Bayer, Bristol‐Myers Squibb, Debiopharm, Merck Serono, MSD, and Sobi. Author Ron Shefi (and the additional person Avner Algom, acknowledged within the “BD2Decide Consortium”) is employed by the company All‐In‐Image Ltd, Israel. Author Vasilis Tountopoulos (and the additional person Thanasis Dalianis, acknowledged within the “BD2Decide Consortium”) is employed by the company Technical Implementation, Innovation Lab, Athens Technology Center, Greece. Author Franco Mercalli (and the additional person Sergio Copelli, acknowledged within the “BD2Decide Consortium”) is employed by the company MultiMed Engineers, Parma, Italy. Stefan Wesarg, acknowledged within the “BD2Decide Consortium”, is employed by the company Fraunhofer, Munich, Germany. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supporting information
Appendix S1. Supporting Information.
Table S1 Clinical characteristics, risk factors and clinical staging
Table S2. Pathologic features
Table S3. Treatments recorded in eCRF
Table S4. Toxicities and follow‐up recorded in eCRF
Table S5. Patients recruited at each clinical participating Institution
Table S6. Clinical characteristics not included in Table 1
Table S7. Treatments
Table S8. Toxicities observed within 30 days from the conclusion of treatments (data available for 1402 cases)
Table S9. Late toxicities (timings measured from the conclusion of treatments)
Table S10. Available of quality of life questionnaires
Table S11. Oral cavity cancer patient characteristics*
Table S12. Median survival of oral cavity cancer patients
Table S13. p16‐positive oropharyngeal cancer patient characteristics*
Table S14. 5‐year survival of p16‐positive oropharyngeal cancer patients
Table S15. p16‐negative oropharyngeal cancer patient characteristics*
Table S16. Median survival of p16‐negative oropharyngeal cancer patients
Table S17. Hypopharyngeal cancer patient characteristics*
Table S18. Median survival of hypopharyngeal cancer patients
Table S19. Laryngeal cancer patient characteristics
Table S20. Median survival of laryngeal cancer patients
Table S21. Dropout
Table S22. Clinical prognostic models
Table S23. Transcriptomics prognostic/predictive models
ACKNOWLEDGMENTS
The authors and the investigators are grateful to Dr. Elena Martinelli, project manager of the BD2Decide project, who lead the Coordination work. Editorial assistance and English language editing (funded by the BD2Decide Consortium) were provided by Charlesworth Author Service.
Cavalieri S, De Cecco L, Brakenhoff RH, et al. Development of a multiomics database for personalized prognostic forecasting in head and neck cancer: The Big Data to Decide EU Project. Head & Neck. 2021;43:601–612. 10.1002/hed.26515
Tito Poli and Lisa Licitra co‐last authors.
The BD2Decide Consortium includes Giuseppina Calaresoa, Pasquale Quattronea, Ester Orlandia, Federica Perronea, Silvia Franciscib, Danielle Heidemanc, Elisabeth Bloemenac, Marije Vergeerc, Pim de Graafc, Mari van den Houtd, Bernd Kremerd, Elena Schultee, Rene Grässline, Liss Hernandezf, Maria Teresa Arredondof, Thanasis Dalianisg, Avner Algomh, Sergio Copellii, Stefan Wesargj where (a) indicates Fondazione IRCCS Istituto Nazionale dei Tumori di Milano, Milan, Italy; (b) Istituto Superiore di Sanità, Rome, Italy; (c) Amsterdam University Medical Centers, Amsterdam, The Netherlands; (d) Maastricht University Medical Center, Maastricht, the Netherlands; (e) Medical Faculty, Heinrich Heine University Düsseldorf, Düsseldorf, Germany; (f) Universidad Politécnica de Madrid, Madrid, Spain; (g) Technical Implementation, Innovation Lab, Athens Technology Center, Athens, Greece; (h) All‐In‐Image Ltd, Israel; (i) MultiMed Engineers srl, Parma, Italy; (j) Fraunhofer. Munich, Germany.
Funding information European Union Horizon 2020 Framework Programme, Grant/Award Number: 689715
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394‐424. 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2. Cramer JD, Burtness B, Le QT, Ferris RL. The changing therapeutic landscape of head and neck cancer. Nat Rev Clin Oncol. 2019;16(11):669‐683. 10.1038/s41571-019-0227-z. [DOI] [PubMed] [Google Scholar]
- 3. Lo Nigro C, Denaro N, Merlotti A, Merlano M. Head and neck cancer: improving outcomes with a multidisciplinary approach. Cancer Manag Res. 2017;9:363‐371. 10.2147/CMAR.S115761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sobin LH, MK WC Gospodarowicz. TNM classification of malignant tumors. 7th ed Oxford: Wiley‐Blackwell; 2010:349–445. doi: 10.1016/B978-1-4377-0272-9.50014-0 [DOI] [Google Scholar]
- 5. Mehra R, Ang KK, Burtness B. Management of human papillomavirus‐positive and human papillomavirus‐negative head and neck cancer. Semin Radiat Oncol. 2012;22(3):194‐197. 10.1016/j.semradonc.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 6. Pignon J‐P, le Maître A, Maillard E, Bourhis J. MACH‐NC collaborative group. Meta‐analysis of chemotherapy in head and neck cancer (MACH‐NC): an update on 93 randomised trials and 17,346 patients. Radiother Oncol. 2009;92(1):4‐14. 10.1016/j.radonc.2009.04.014. [DOI] [PubMed] [Google Scholar]
- 7. Amin MB, Greene FL, Edge SB, et al. The eighth edition AJCC cancer staging manual: continuing to build a bridge from a population‐based to a more “personalized” approach to cancer staging. CA Cancer J Clin. 2017;67(2):93‐99. 10.3322/caac.21388. [DOI] [PubMed] [Google Scholar]
- 8. Big Data and Models for Personalized Head and Neck Cancer Decision Support (BD2Decide) Full Text View—ClinicalTrials.gov. https://clinicaltrials.gov/ct2/show/NCT02832102. Accessed September 20, 2019.
- 9. Home, BD2DECIDE http://www.bd2decide.eu/. Accessed September 20, 2019.
- 10. Aaronson NK, Ahmedzai S, Bergman B, et al. The European organization for research and treatment of cancer QLQ‐C30: A quality‐of‐life instrument for use in international clinical trials in oncology. J Natl Cancer Inst. 1993;85(5):365‐376. 10.1093/jnci/85.5.365. [DOI] [PubMed] [Google Scholar]
- 11. Bjordal K, Hammerlid E, Ahlner‐Elmqvist M, et al. Quality of life in head and neck cancer patients: validation of the European Organization for Research and Treatment of Cancer Quality of Life Questionnaire‐H&N35. J Clin Oncol. 1999;17(3):1008‐1019. 10.1200/JCO.1999.17.3.1008. [DOI] [PubMed] [Google Scholar]
- 12. Wisløff T, Hagen G, Hamidi V, Movik E, Klemp M, Olsen JA. Estimating qaly gains in applied studies: A review of cost‐utility analyses published in 2010. Pharmacoeconomics. 2014;32(4):367‐375. 10.1007/s40273-014-0136-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Locati, Serafini, Iannò et al. Mining of self‐organizing map gene‐expression portraits reveals prognostic stratification of HPV‐positive head and neck squamous cell carcinoma. Cancers (Basel). 2019;11(8):1057 10.3390/cancers11081057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Serafini MS, Lopez‐Perez L, Fico G, Licitra L, De Cecco L, Resteghini C. Transcriptomics and Epigenomics in head and neck cancer: available repositories and molecular signatures. Cancers Head Neck. 2020;5(1):2 10.1186/s41199-020-0047-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ang KK, Harris J, Wheeler R, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363(1):24‐35. 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rietbergen MM, Brakenhoff RH, Bloemena E, et al. Human papillomavirus detection and comorbidity: critical issues in selection of patients with oropharyngeal cancer for treatment De‐escalation trials. Ann Oncol. 2013;24(11):2740‐2745. 10.1093/annonc/mdt319. [DOI] [PubMed] [Google Scholar]
- 17. Rios Velazquez E, Hoebers F, Aerts HJWL, et al. Externally validated HPV‐based prognostic nomogram for oropharyngeal carcinoma patients yields more accurate predictions than TNM staging. Radiother Oncol. 2014;113(3):324‐330. 10.1016/j.radonc.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 18. Oramod . https://oramod.eu/. Accessed September 30, 2019.
- 19. Mes SW, te Beest D, Poli T, et al. Prognostic modeling of oral cancer by gene profiles and clinicopathological co‐variables. Oncotarget. 2017;8(35):59312‐59323. 10.18632/oncotarget.19576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Datema FR, Ferrier MB, van der Schroeff MP, Baatenburg de Jong RJ. Impact of comorbidity on short‐term mortality and overall survival of head and neck cancer patients. Head Neck. 2010;32(6):728‐736. 10.1002/hed.21245. [DOI] [PubMed] [Google Scholar]
- 21. Egelmeer AGTM, Velazquez ER, De Jong JMA, et al. Development and validation of a nomogram for prediction of survival and local control in laryngeal carcinoma patients treated with radiotherapy alone: A cohort study based on 994 patients. Radiother Oncol. 2011;100(1):108‐115. 10.1016/j.radonc.2011.06.023. [DOI] [PubMed] [Google Scholar]
- 22. De Cecco L, Bossi P, Locati L, Canevari S, Licitra L. Comprehensive gene expression meta‐analysis of head and neck squamous cell carcinoma microarray data defines a robust survival predictor. Ann Oncol. 2014;25(8):1628‐1635. 10.1093/annonc/mdu173. [DOI] [PubMed] [Google Scholar]
- 23. Eschrich SA, Pramana J, Zhang H, et al. A gene expression model of intrinsic tumor Radiosensitivity: prediction of response and prognosis after chemoradiation. Int J Radiat Oncol Biol Phys. 2009;75(2):489‐496. 10.1016/j.ijrobp.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Foy JP, Bazire L, Ortiz‐Cuaran S, et al. A 13‐gene expression‐based radioresistance score s the heterogeneity in the response to radiation therapy across HPV‐negative HNSCC molecular subtypes. BMC Med. 2017;15(1):165 10.1186/s12916-017-0929-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hensen EF, De Herdt MJ, Goeman JJ, et al. Gene‐expression of metastasized versus non‐metastasized primary head and neck squamous cell carcinomas: A pathway‐based analysis. BMC Cancer. 2008;8:168 10.1186/1471-2407-8-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lohavanichbutr P, Méndez E, Holsinger FC, et al. A 13‐gene signature prognostic of HPV‐negative OSCC: discovery and external validation. Clin Cancer Res. 2013;19(5):1197‐1203. 10.1158/1078-0432.CCR-12-2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang W, Lim WK, Leong HS, et al. An eleven gene molecular signature for extra‐capsular spread in oral squamous cell carcinoma serves as a prognosticator of outcome in patients without nodal metastases. Oral Oncol. 2015;51(4):355‐362. 10.1016/j.oraloncology.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 28. Zhang Y, Koneva LA, Virani S, et al. Subtypes of HPV‐positive head and neck cancers are associated with HPV characteristics, copy number alterations, PIK3CA mutation, and pathway signatures. Clin Cancer Res. 2016;22(18):4735‐4745. 10.1158/1078-0432.CCR-16-0323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Leemans CR, Snijders PJF, Brakenhoff RH. The molecular landscape of head and neck cancer. Nat Rev Cancer. 2018;18(5):269‐282. 10.1038/nrc.2018.11. [DOI] [PubMed] [Google Scholar]
- 30. Rietbergen MM, Witte BI, Velazquez ER, et al. Different prognostic models for different patient populations: validation of a new prognostic model for patients with oropharyngeal cancer in Western Europe. Br J Cancer. 2015;112(11):1733‐1736. 10.1038/bjc.2015.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.National Comprehensive Cancer Network. Head and Neck Cancers (Version 2.2020 ‐ June 9, 2020). http://www.nccn.org/professionals/physician_gls/pdf/head‐and‐neck.pdf. Accessed October 25, 2020.
- 32. ESMO , Head & Neck Cancers: Essentials for Clinicians. Lugano, Switzerland: ESMO Press; 2017. https://oncologypro.esmo.org/Education‐Library/Essentials‐for‐Clinicians/Head‐Neck‐Cancers. Accessed September 20, 2019. [Google Scholar]
- 33. Freedman ND, Schatzkin A, Leitzmann MF, Hollenbeck AR, Abnet CC. Alcohol and head and neck cancer risk in a prospective study. Br J Cancer. 2007;96(9):1469‐1474. 10.1038/sj.bjc.6603713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gatta G, Botta L, Sánchez MJ, et al. Prognoses and improvement for head and neck cancers diagnosed in Europe in early 2000s: the EUROCARE‐5 population‐based study. Eur J Cancer. 2015;51(15):2130‐2143. 10.1016/j.ejca.2015.07.043. [DOI] [PubMed] [Google Scholar]
- 35. Nauta IH, Rietbergen MM, van Bokhoven AAJD, et al. Evaluation of the eighth TNM classification on p16‐positive oropharyngeal squamous cell carcinomas in The Netherlands and the importance of additional HPV DNA testing. Ann Oncol Off J Eur Soc Med Oncol. 2018;29(5):1273‐1279. 10.1093/annonc/mdy060. [DOI] [PubMed] [Google Scholar]
- 36. Boscolo‐Rizzo P, Zorzi M, Del MA, et al. The evolution of the epidemiological landscape of head and neck cancer in Italy: is there evidence for an increase in the incidence of potentially HPV‐related carcinomas? PLoS One. 2018;13(2):e0192621 10.1371/journal.pone.0192621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lawrence MS, Sougnez C, Lichtenstein L, et al. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517(7536):576‐582. 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. The Cancer Genome Atlas Program National Cancer Institute. https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga. Accessed March 16, 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1. Supporting Information.
Table S1 Clinical characteristics, risk factors and clinical staging
Table S2. Pathologic features
Table S3. Treatments recorded in eCRF
Table S4. Toxicities and follow‐up recorded in eCRF
Table S5. Patients recruited at each clinical participating Institution
Table S6. Clinical characteristics not included in Table 1
Table S7. Treatments
Table S8. Toxicities observed within 30 days from the conclusion of treatments (data available for 1402 cases)
Table S9. Late toxicities (timings measured from the conclusion of treatments)
Table S10. Available of quality of life questionnaires
Table S11. Oral cavity cancer patient characteristics*
Table S12. Median survival of oral cavity cancer patients
Table S13. p16‐positive oropharyngeal cancer patient characteristics*
Table S14. 5‐year survival of p16‐positive oropharyngeal cancer patients
Table S15. p16‐negative oropharyngeal cancer patient characteristics*
Table S16. Median survival of p16‐negative oropharyngeal cancer patients
Table S17. Hypopharyngeal cancer patient characteristics*
Table S18. Median survival of hypopharyngeal cancer patients
Table S19. Laryngeal cancer patient characteristics
Table S20. Median survival of laryngeal cancer patients
Table S21. Dropout
Table S22. Clinical prognostic models
Table S23. Transcriptomics prognostic/predictive models
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


