Summary
Plant Phosphate Transporter 1 (PHT1) proteins, probably the only influx transporters for phosphate (Pi) uptake, are partially degraded on sufficient Pi levels to prevent excessive Pi accumulation. Therefore, the basal/constitutive expression level of PHT1 genes is vital for maintaining Pi uptake under Pi‐replete conditions.
Rice (Oryza sativa) OsPHT1;1 is a unique gene as it is highly expressed and not responsive to Pi, however the mechanism for maintaining its basal/constitutive expression remains unknown. Using biochemical and genetic approaches, we identified and functionally characterised the transcription factors maintaining the basal/constitutive expression of OsPHT1;1.
OsWRKY21 and OsWRKY108 interact within the nucleus and both bind to the W‐box in the OsPHT1;1 promoter. Overexpression of OsWRKY21 or OsWRKY108 led to increased Pi accumulation, resulting from elevated expression of OsPHT1;1. By contrast, oswrky21 oswrky108 double mutants showed decreased Pi accumulation and OsPHT1;1 expression in a Pi‐dependent manner. Moreover, similar to ospht1;1 mutants, plants expressing the OsWRKY21–SRDX fusion protein (a chimeric dominant suppressor) were impaired in Pi accumulation in Pi‐replete roots, accompanied by downregulation of OsPHT1;1 expression.
Our findings demonstrated that rice WRKY transcription factors function redundantly to promote Pi uptake by activating OsPHT1;1 expression under Pi‐replete conditions, and represent a novel pathway independent of the central Pi signalling system.
Keywords: phosphate, phosphate transporter, phosphorus, redundancy, rice, signalling, transcription factor, WRKY
Introduction
Phosphorus (P) is one of the macronutrients essential for plant growth and development. In soil, inorganic orthophosphate (Pi) is the major form of P for plant uptake (Raghothama, 1999; Lambers & Plaxton, 2015). The bioavailability of soil P is limited in natural and agricultural ecosystems due to precipitation and fixation, as well as the low mobility of this nutrient. However, excessive P is released into soil by excessive fertilisation, leading to detrimental environmental issues such as eutrophication. Thus, plants during their life cycle have to cope with the heterogeneity and fluctuation of soil Pi (Shen et al., 2011). To maintain in planta P at physiologically optimal levels, the evolution of plants has been accompanied by a suite of adaptive responses for the uptake, distribution and metabolism of P, all of which are achieved by the coordination of various genes (Gu et al., 2016; Oldroyd & Leyser, 2020).
Plant Pi transporters of the Phosphate Transporter 1 (PHT1) family are probably the only influx transporters for Pi uptake (Ayadi et al., 2015). PHT1 members have long been designated as high‐affinity Pi transporters based on the induction of their expression by Pi starvation and kinetic analysis of Pi uptake in heterologous systems (e.g. yeast and Xenopus laevis oocyte). However, it has been realised that many PHT1 proteins are dual‐affinity Pi transporters and that their genuine affinity for Pi may not be correctly reflected by heterologous data (Glass & Kotur, 2013; Ayadi et al., 2015; Chang et al., 2019). In total, nine and 13 PHT1 genes are present in Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa), respectively. In Arabidopsis, eight out of nine PHT1 homologues are induced in Pi‐deficient roots, among which AtPHT1;1 and AtPHT1;4 are two major members for Pi uptake (Muchhal et al., 1996; Mudge et al., 2002; Misson et al., 2004; Aung et al., 2006; Sun et al., 2016). These two PHT1s are responsible for 45–56% of Pi uptake under low‐Pi conditions. AtPHT1;1 and AtPHT1;4 also account for 57–75% of Pi uptake from high‐Pi environments, consistent with their higher transcript abundance compared with other paralogues under Pi‐sufficient conditions (Shin et al., 2004; Aung et al., 2006; Ayadi et al., 2015; Sun et al., 2016). In rice, four PHT1 genes, namely OsPHT1;1, OsPHT1;2, OsPHT1;4 and OsPHT1;8, showed higher expression levels in Pi‐sufficient root (Secco et al., 2013), suggesting that they are involved in Pi uptake/accumulation under Pi‐replete conditions. Loss‐of‐function mutation of OsPHT1;4 or OsPHT1;8 led to decreased Pi accumulation in Pi‐replete plants (Jia et al., 2011; Chen et al., 2015; Zhang et al., 2015). The knockdown plants of OsPHT1;2 derived from RNA interference (RNAi) were found to have decreased total P concentration and root‐to‐shoot translocation of P under low‐Pi conditions (Ai et al., 2009), whereas another OsPHT1;2 knockdown line caused by a T‐DNA insertion in the proximal promoter region showed no alteration in Pi accumulation (Liu et al., 2010). OsPHT1;1 RNAi plants displayed a reduction in Pi accumulation in Pi‐replete shoot but not root (Sun et al., 2012). Despite these important advances, to date, the functional characterisation of OsPHT1;1 and OsPHT1;2 has not been performed with their loss‐of‐function mutants. In addition, the transcriptional regulatory mechanism for maintaining constitutive expression of OsPHT1;1 remains unknown.
The process of maintaining plant P homeostasis is fine tuned by a complex signalling network. Transcriptional regulation of Pi‐starvation‐induced (PSI) genes, including most PHT1s, is largely controlled by various transcription factors (TFs), and represents an early and important loop in this network. A small clade of the MYB TFs classified into the MYB‐coiled coil subfamily Phosphate Starvation Response (PHR) and PHR‐like (PHL) TFs, has been intensively studied and demonstrated to be central regulators of plant Pi signalling (Rubio et al., 2001; Zhou et al., 2008; Bustos et al., 2010; Guo et al., 2015; Sun et al., 2016; Ruan et al., 2017). In addition, maintenance of plant P homeostasis also involves other MYB TFs and TFs from other families, most of which directly or indirectly regulate the transcript abundance of PHT1s (Devaiah et al., 2009; Dai et al., 2012; Baek et al., 2013; Yang et al., 2014; Chen & Schmidt, 2015; Gu et al., 2016 and references therein; Gu et al., 2017; Wang et al., 2018).
WRKY TFs comprise a large family with more than 70 and 100 members in Arabidopsis and rice, respectively. They are implicated in diverse physiological processes and plant tolerance to biotic and abiotic stresses (Ülker & Somssich, 2004; Rushton et al., 2010; Viana et al., 2018). In Arabidopsis, four WRKY TFs have been found to be responsible for Pi signalling. AtWRKY45 positively regulates the expression of AtPH1;1, and AtWRKY75 serves as a transcriptional activator of both AtPHT1;1 and AtPHT1;4, mainly under Pi‐deficient conditions (Devaiah et al., 2007; H. Wang et al., 2014). Under Pi‐sufficient conditions, AtWRKY42 promotes AtPHT1;1 expression, and interacts with AtWRKY6 to co‐ordinately inhibit the expression of PHOSPHATE 1 (AtPHO1), an endomembrane localised Pi efflux transporter for root‐to‐shoot translocation of Pi (Chen et al., 2009; Su et al., 2015; Ye et al., 2018). In rice, only one WRKY TF, namely OsWRKY74, has been reported to regulate P homeostasis by regulating several PSI PHT1 genes (Dai et al., 2016). OsWRKY28 has a positive effect on P accumulation through jasmonic acid‐mediated modulation of root system architecture, independent of PHT1s (Wang et al., 2018). Unlike all other PHT1 counterparts, the expression of OsPHT1;1 is not responsive to Pi, whereas all the reported WRKY TFs, as well as PHR/PHLs, regulate the expression of PSI PHT1s but not OsPHT1;1. In addition, genes with constitutive expression such as OsPHT1;1 may also require TFs (enhancers) to maintain their transcript abundance to a desirable level. In mice, a basic leucine zipper TF, (Nrf2), is responsible for maintaining the constitutive or basal expression of several genes encoding the subunits of glutathione S‐transferase and glutamate cysteine ligase (Chanas et al., 2002).
Despite progress made over the last 2 decades, the functional characterisation of PHT1 genes has long been compromised due to the high genetic redundancy within this gene family and lack of knockout lines (Nussaume et al., 2011; Ayadi et al., 2015). Additionally, the effort on dissecting the plant Pi signalling network with regard to the transcriptional regulation of PHT1s has mainly been made on TFs that render PSI responses. However, insight into the regulatory machinery responsible for maintaining the constitutive expression of PHT1 genes is lacking. Based on these considerations, we focused on a unique PHT1 gene in rice, OsPHT1;1 (PHT1;1 hereafter), which is highly expressed and not responsive to Pi. The physiological role of PHT1;1 was re‐examined using its mutant lines, demonstrating that it is an important PHT1 member functioning mainly under Pi‐replete conditions. Moreover, 28 candidates binding to PHT1;1 promoter were identified, among which a WRKY TF, OsWRKY21 (WRKY21 hereafter), corresponded to the largest number (three) of clones, and was therefore selected for study. WRKY21 and its interacting protein, OsWRKY108 (WRKY108 hereafter), were identified to bind to the W‐box cis‐element(s) in the PHT1;1 promoter. Subsequently, WRKY21 and WRKY108 were demonstrated to regulate P homeostasis redundantly by maintaining constitutive expression of PHT1;1 in a Pi‐dependent manner. Our findings represent a novel pathway in the plant Pi signalling network, and may provide a potential strategy for limiting the excessive absorption of nutrients by crops.
Materials and Methods
Plant materials and growth conditions
The Nipponbare cultivar of rice (Oryza sativa L. ssp. japonica) was used for experiments in this study. Transgenic plants used in experiments were described in vector construction and rice plant transformation. For hydroponic experiments, seeds were soaked in water overnight at 30°C in the dark for 2 d and then transferred to a net floating on 0.5 mM CaCl2 solution. After 3 d, the seedling were transferred to a half‐strength Kimura B solution (pH 5.6) (Yamaji et al., 2013) or full‐strength Yoshida solution (pH 5.6) (Gu et al., 2017), and grown in growth chambers with a 12 h : 12 h, light : dark photoperiod and 30°C : 24°C, day : night temperature, and the relative humidity was controlled at c. 60%.
The wrky21 homozygous allele was crossed with the T0 generation of the wrky108 mutant, and the wrky21 wrky108 double mutant was identified from F2 population via secondary editing by CRISPR‐Ca9 system. The WRKY21 and WRKY108 overexpression lines were crossed with the pht1;1 mutant, respectively, to generate WRKY21‐Ox/PHT1;1 and WRKY108‐Ox/PHT1;1. Their F2 populations were used for the experiments.
RNA extraction, cDNA synthesis and RT‐qPCR
Total RNA was extracted using TRIzol reagent (Invitrogen). First‐strand cDNAs were synthesised from total RNA using the ReverTra Ace™ qPCR RT Master Mix with gDNA Remover (Toyobo). Reverse transcription‐quantitative PCR was performed with the SYBR Premix Ex Taq TM II Kit (TaKaRa Biotechnology, Dalian, China) on the QuantStudio 6 Flex Real‐Time PCR System (Applied Biosystems, Thermo Fisher Scientific, Waltham, MA, USA). Relative expression levels of each sample were determined by normalising to the amount of OsActin1 (LOC_Os03g50885) detected in the same sample and presented as 2−ΔCT. All primers used for RT‐qPCR are listed in Supporting Information Table S1.
Vector construction and rice plant transformation
For the overexpression of WRKY21 and WRKY108, the double‐cauliflower mosaic virus 35S promoter and NOS terminator were subcloned into the vector pCAMBIA1305.1‐GUSPlus. Then ORFs of WRKY21 and WRKY108 were amplified (Table S2) from the rice cDNA library and then cloned into the modified vector pCAMBIA1305.1‐GUSPlus.
Mutants of pht1;1, wrky21 and wrky108 were generated using the CRISPR‐Cas9 system. Spacers (Table S2) residing in exons of each gene were selected from the library provided by Miao et al. (2013). These spacers were ligated to the intermediate vector pOs‐sgRNA and then introduced into expression vector pH‐Ubi‐cas9‐7 using Gateway recombination technology (Invitrogen).
For tissue localisation, the GUSPlus reporter gene and the NOS terminator were subcloned into the pCAMBIA1300 vector, resulting in a new expression vector designated as pCAMBIA1300‐GN. The 2519‐bp and 2576‐bp DNA fragments upstream of the translation start codons of WRKY21 and WRKY108 were amplified (Table S2) from the rice genomic DNA and fused GUSPlus reporter gene, respectively.
For chromatin immunoprecipitation (ChIP)‐qPCR, full‐length ORFs of WRKY21 and WRKY108 without stop codons were first ligated into an intermediate vector, pSAT6A‐EGFP‐N1. Two new restriction enzyme sites PacI and AscI were introduced into the modified vector pCAMBIA1305.1‐GUSPlus, resulting in the new vector designated as pCAMBIA1305‐PA. The WRKY21‐GFP, WRKY108‐GFP fusion constructs as well as GFP alone were amplified (Table S2) and cloned into the pCAMBIA1305‐PA vector, respectively.
For chimeric repressor expression vectors, both strands (Table S2) of DNA fragments that corresponded to SRDX (LDLDLELRLGFA) were synthesised with a TAA stop codon at 3′ end. The DNA fragments were ligated into PstI and XbaI sites of WRKY21 and WRKY108 overexpression vectors, respectively, using a One Step Cloning Kit (Vazyme Biotech, Nanjing, China).
The above constructs were transformed into mature embryos developed from seeds of wild‐type plants via Agrobacterium tumefaciens‐mediated transformation, as previously described (Jia et al., 2011).
Tissue localisation analysis
Histochemical analysis was performed as described in Ai et al. (2009).
Subcellular localisation and bimolecular fluorescence complementation (BiFC) analysis
The ORFs of WRKY21 and WRKY108 were amplified (Table S3) and subcloned into intermediate vectors, pSAT6A‐EGFP‐N1 and pSAT6‐EGFP‐C1, to generate WRKY21‐eGFP and eGFP‐WRKY21, WRKY108‐eGFP and eGFP‐WRKY108 fusions. These fusion constructs and GFP alone were introduced into pRCS2‐ocs‐nptII. For BiFC, full‐length CDS of WKRY21 and WRKY108 without stop codons were first subcloned into the cloning vector pENTR/D‐TOPO (Invitrogen) (Table S3) to obtain the attL‐containing site and then were introduced into pGTQL1221YN (Plasmid#61704, Addgene) and pGTQL1221YC (Plasmid#61705, Addgene) vectors, respectively. The constructs above were transiently expressed in N. benthamiana leaves by Agrobacterium‐mediated infiltration, as described in Gu et al., 2017. Images were taken using a confocal microscope (Leica SP8X).
Transactivation assay
Full‐length and truncated ORFs of WKYR21 and WRKY108 were amplified (Table S3) and cloned into pBD‐GAL4 Cam to produce fusions with the yeast GAL4 DNA‐binding domain. The constructs were transformed into YRG‐2 cells according to the YeastmakerTM Yeast Transformation System 2 User Manual (Clontech Laboratories, Mountain View, CA, USA). Transformants were selected on synthetic dextrose (SD) medium lacking tryptophan (W). Yeast transformants from SD/−W were then streaked onto SD/−W or SD/−W/−H (histidine) medium for observation.
Yeast one‐hybrid and two‐hybrid assays
A Y1H library screen was performed using the MatchmakerTM Gold Yeast One‐Hybrid Library Screen System kit and the YeastmakerTM Transformation System 2 kit (Clontech Biotechnology). The F0 fragment from the PHT1;1 promoter region −1164 to −112 was amplified (Table S3) and cloned into the pAbAi vector. The construct was linearised and transformed into Y1HGold strain to generate Y1H bait strain, Y1HGold[pAbAi‐F0]. The minimal growth‐inhibitory concentration of aureobasidin A for the Bait‐Reporter yeast strain was determined. RNA was extracted from seedlings grown under Pi‐sufficient conditions; 1.0–2.0 μg total RNA was used to prepare the cDNA library. Screening of interaction clones was carried out according to the manufacturer’s instructions.
The transcriptional activation potential of WRKY21 was examined as previously described. The truncated WRKY21 protein (45 amino acid deletion in C‐terminal) was used as the bait protein. cDNA synthesis and a yeast two‐hybrid screen was performed by Hybrigenics (Paris, France).
The one‐on‐one validation of the library‐scale screening was conducted strictly following the instructions of the kit manual. All primers for vector construction are listed in Table S3.
Electrophoretic mobility shift assay (EMSA)
WRKY21 and WRKY108 CDS were separately cloned into pMal‐c5x (NEB, http://www.neb‐china.com/) and pGST‐21a (GenScript, www.genscript.com.cn). The recombinant plasmids or empty vectors were then transformed into Escherichia coli BL21. Fusion proteins were induced with 0.4 mM β‐d‐1‐thiogalactopyranoside (IPTG) at 16°C for 16–20 h, and were further purified using GST‐Bind™ Resin (GenScript) or Amylose Resin (NEB). Protein concentrations were determined using the bovine serum albumin (BSA) quantitative assay.
Forward and reverse oligonucleotides of probes (Figs 2b, S3a; Table S3) with biotin labelled or unlabelled at the 5′ ends were synthesised by GenScript, and annealed for EMSA and then was conducted using the LightShift Chemiluminescent EMSA kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Biotin‐labelled probes were detected using the ECL substrate Working Solution and imaged using the Odyssey Imaging System (Li‐Cor Biosciences, Lincoln, NE, USA).
Fig. 2.
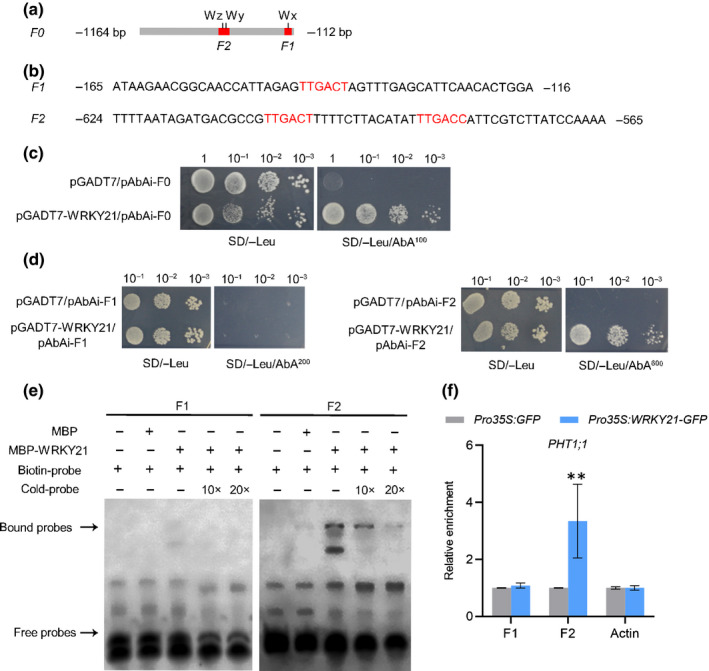
WRKY21 binds to PHT1;1 promoter region in vivo and in vitro. (a, b) Diagram of the 1053‐bp promoter region F0 of PHT1;1 showing the relative positions of the W‐boxes. Three W‐boxes Wx, Wy, Wz (corresponding to the sequences in red (b)) are marked by black vertical bars and relative positions and sizes of two synthesised DNA probes F1, F2 (b) are indicated by red rectangles under the W‐box cis‐elements (a). (c, d) WRKY21 binds to the PHT1;1 promoter region in the Y1H assay. Yeast cells were transformed with a bait vector containing a promoter fragment F0, F1 or F2 fused to AUR1‐C reporter gene, and a prey vector, containing WRKY21 fused to a GAL4 activation domain. Yeast cells were grown in liquid medium to an OD600 of 1.0 and diluted in a 10× dilution series (10−1 to 10−3). From each dilution, 5 μl was spotted onto SD/−Leu medium to select for plasmids, and SD/−Leu supplemented with 100, 200 or 600 ng ml−1 aureobasidin A (AbA) to select for interaction. (e) Electrophoretic mobility shift assay (EMSA) to test the binding of WRKY21 to PHT1;1 promoter fragments. Each biotin‐labelled probe was incubated with MBP‐WRKY21 protein. An excess amount of unlabelled probes (cold probe) only was added to compete with labelled F2 DNA probes. Biotin‐labelled probes alone or biotin‐labelled probes incubated with MBP protein served as negative controls. The WRKY21–DNA complex (bound probes) and free DNA probes are indicated by black arrows, respectively. (f) Chromatin immunoprecipitation (ChIP)‐qPCR analysis of WRKY21 binding to the PHT1;1 promoter region. Rice seeds of Pro35S:GFP and Pro35S:WRKY21‐GFP transgenic plants were germinated in deionised H2O and supplied with sufficient Pi. The whole plants were harvested for ChIP analysis. Enriched DNA fragments (F1 and F2) in the PHT1;1 promoter were quantified using RT‐qPCR. Enrichment was calculated as the ratio of immunoprecipitation to input. Values represent means ± SD (n = 3). Data significantly different from the control are indicated. P‐values were determined using Student’s t‐test. Pro35S:WRKY21‐GFP vs Pro35S:GFP; **, P < 0.01.
ChIP assay
Chromatin immunoprecipitation (ChIP) assay was performed using the EpiQuikTM Plant ChIP Kit (Epigentek) in accordance with the manufacturer's instructions. In brief, c. 1.0 g rice seedlings of Pro35S:GFP and Pro35S:WRKY21‐GFP or Pro35S:WRKY108‐GFP plants were harvested for ChIP experiments. Samples were fixed with 1% formaldehyde and chromatin was isolated and sheared by sonication (Bioruptor Pico, Diagenode) to obtain DNA fragments with an average size of c. 500 bp. Anti‐GFP monoclonal antibodies (Cell Signaling Technology, Danvers, MA, USA) bound to protein A/G‐coated resin fixed at the bottom of strip wells were used to immunoprecipitate genomic DNA fragments. qPCR was performed with immunoprecipitated genomic DNA fragments and enrichment was calculated as the ratio of immunoprecipitation to input.
Pull‐down assay
Fusion proteins of MBP‐WRKY21 and GST‐WRKY108 were purified as described in the EMSA. Purified GST‐WRKY108 or GST was incubated with an equal volume of glutathione resin (GenScript) in phosphate‐buffered saline (PBS) solution for 0.5–1.0 h with rotation, after which equal amounts of purified MBP‐WRKY21 were added to the mixture. After a further incubation for 2 h at 4°C, the resin was washed five times with PBS, and then diluted in 1× SDS loading buffer and boiled for 10 min. The proteins were examined by immunoblotting using anti‐GST and anti‐MBP antibodies.
Measurement of Pi and total P concentration
For determination of Pi and total P concentrations, the methods described by Zhou et al. (2008) and Jia et al. (2011) were strictly followed.
Results
PHT1;1 is involved in Pi accumulation under Pi‐sufficient conditions
To dissect the molecular mechanism underlying Pi uptake from Pi‐replete environments, we focused on PHT1;1, a highly expressed Pi transporter gene not regulated by OsPHR2 (Sun et al., 2012). We generated mutant lines of PHT1;1 by the CRISPR‐Cas9 system and re‐examined its physiological roles (Fig. S1). The pht1;1 mutants and the wild‐type (WT) plants were subjected to a hydroponic culture system supplied with different levels of Pi (Low Pi: LP, 1 µM Pi; Control: Ctrl, 90 µM Pi; High Pi: HP, 300 µM Pi; Fig. 1a). In roots, Pi concentration was significantly decreased in pht1;1 mutants under Ctrl and HP conditions but not LP conditions compared with that in wild‐type plants; in shoots, the decrease in Pi concentration upon PHT1;1 mutation was observed only under HP conditions (Fig. 1b). The same trend was observed for total P concentrations, although the difference was less evident than that for Pi (Fig. S2), probably attributed to the relatively stable organic P levels. These results suggested that PHT1;1 mainly works when external Pi is sufficient, and that it displays functional redundancy with other PSI PHT1 genes under Pi‐deficient conditions.
Fig. 1.
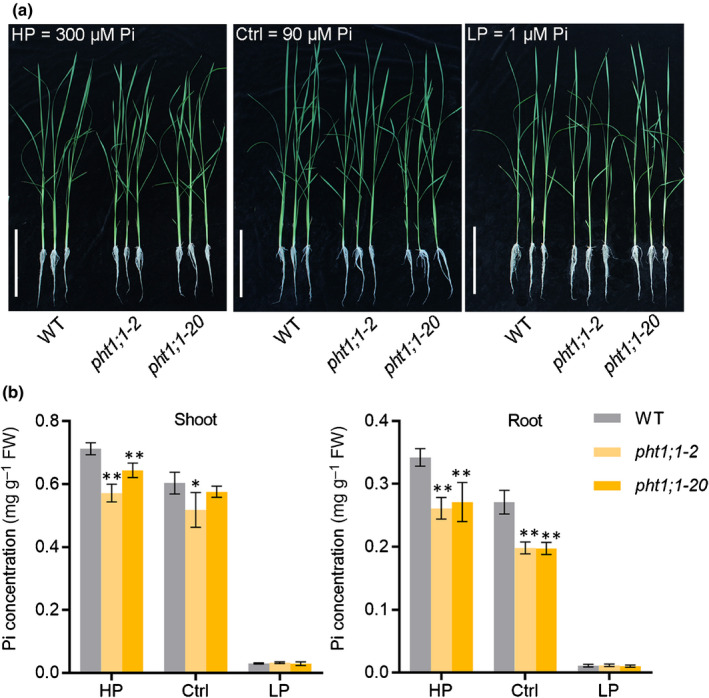
Mutation of PHT1;1 leads to impaired phosphate (Pi) accumulation in rice (Oryza sativa). (a) Phenotype of pht1;1 mutants and wild‐type plants grown under HP (300 μM Pi), Ctrl (90 μM Pi) and LP (1 μM Pi) hydroponic conditions. Bars, 15 cm. (b) Cellular Pi concentration analysis of pht1;1 mutants and wild‐type plants. Four‐leaf‐old seedlings were grown in half‐strength Kimura B nutrient solution supplied with HP (300 μM Pi), Ctrl (90 μM Pi) or LP (1 μM Pi) until the seventh leaf blades were fully expanded. The Pi concentration was measured in shoot (left) and root (right). Error bars indicate ± SD (n = 4). Data significantly different from the corresponding controls are indicated (*, P < 0.05; **, P < 0.01; Student’s t‐test). FW, fresh weight.
Isolation and validation of WRKY21, a transcriptional regulator of PHT1;1
To identify the potential TF(s) regulating the constitutively expressed PHT1;1, we first generated several truncated fragments of its promoter, fused them to a GUS reporter, and produced transgenic rice plants. The results showed that a promoter fragment with a length of 1164 bp displayed the same activity as the full‐length promoter (Fig. S3; Sun et al., 2012). Subsequently, we performed a yeast one‐hybrid (Y1H) screening with part of this fragment (Fragment 0 (F0): −1164 bp to −112 bp) as a bait (Fig. 2a). In total, 28 positive clones were isolated and sequenced (Table S4). Three out of the 28 clones corresponded to the same gene encoding a type‐III WRKY transcription factor, WRKY21.
To validate the interaction between PHT1;1 and WRKY21, a one‐on‐one Y1H assay was performed using the full‐length open reading‐frame (ORF) of WRKY21. The growth of the yeast cells transformed with pGADT7 (the activating domain (AD) of yeast GAL4 TF) and pAbAi‐F0 (the fusion of F0 and the AUR1‐C gene, an antibiotic resistance gene that confers resistance to aureobasidin A (AbA)) was suppressed when 100 ng ml−1 AbA was supplied in the medium, whereas transformation of pGADT7‐WRKY21 (the fusion of GAL4 AD and WRKY21) and pAbAi‐F0 restored yeast growth (Fig. 2c), indicating that WRKY21 directly regulated PHT1;1 expression by binding to the PHT1;1 promoter. To further investigate the potential binding site(s) of WRKY21 in the PHT1;1 promoter, two fragments with W‐boxes (F1 and F2) were used for Y1H. The results showed that WRKY21 could bind to F2 but not to F1 (Fig. 2a,b,d). Moreover, EMSA and ChIP‐qPCR analysis also showed that WRKY21 bound to the corresponding regions of F2 in vitro and in vivo (Fig. 2a,b,e,f). Furthermore, to examine whether WRKY21 could bind to both copies of W‐box within F2, targeted point mutations were introduced into the W‐box motifs and then subjected to Y1H and EMSA analysis. Mutation of the W‐box proximal to the translation start site (Wy) did not affect binding of WRKY21 to F2, whereas mutation of both W‐box motifs or the one located more upstream (Wz) alone abolished binding (Fig S4). These results demonstrated that WRKY21 bound to Wz in the PHT1;1 promoter.
Expression of WRKY21 in response to different Pi regimes and its tissue and subcellular localisation
PHT1;1 expression is not responsive to Pi at the transcriptional level (Sun et al., 2012). To investigate whether WRKY21 displayed a similar expression pattern to PHT1;1, we analysed the expression of WRKY21 in response to different Pi supplies. RT‐qPCR analysis showed that WRKY21 was not responsive to Pi in shoot, similar to that found for PHT1;1; however, WRKY21 was positively regulated by Pi in root (Fig. 3a), suggesting that it exerted its function mainly under Pi‐sufficient conditions.
Fig. 3.
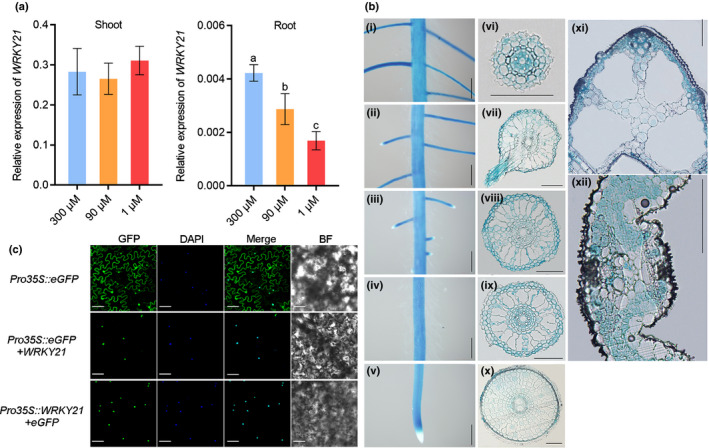
Expression patterns and subcellular localization of WRKY21. (a) Expression of WRKY21 in response to different Pi supplies. Rice seeds were germinated in deionised H2O and supplied with 300 μM, 90 μM or 1 μM Pi. Plant shoots and roots were collected from seedlings. RT‐qPCR analysis was performed using the rice housekeeping gene OsActin1 (LOC_Os03g50885) as an internal control. Values presented are the means ± SD of four biological replicates. Different letters indicate significant differences (P < 0.05, Duncan's test). (b) Histochemical staining for GUS activity in transgenic plants expression a ProWRKY21:GUS fusion. Plants were grown hydroponically and supplied with sufficient Pi. (i–v) Different zones of primary root. (vi) Cross‐section of the basal part of lateral root. (vii) Cross‐section of root segment shown in (ii). (viii) Cross‐section of root segment shown in (iii). (ix) Cross‐section of root segment shown in (iv). (x) Cross‐section of root segment shown in (v). (xi) Cross‐section of leaf sheath. (xii) Cross‐section of leaf blade. Bars in (i–v) and (xi–xii), 100 μm; bars in (vi–x), 500 μm. (c) Subcellular localization of WRKY21. Fusion proteins of WRKY21:eGFP and eGFP:WRKY21 and eGFP were expressed in the Nicotiana benthamiana leaf epidermal cells using Agrobacterium tumefaciens‐mediated infiltration. Green signals indicate GFP, and the blue signals indicate the cell nuclei that were specially stained with 4′,6‐diamidino‐2‐phenylindole (DAPI). Bars, 10 μM. BF, bright field.
To further investigate the expression pattern of WRKY21, ProWRKY21:GUS transgenic rice plants were generated. Seven‐leaf‐old seedlings were sampled for analysis. In primary and adventitious roots, GUS activity was detected in almost all cell types except for root cap, root meristem zone and root hairs (Fig. 3bi–x). In lateral root, the same distribution of GUS activity was observed as that in primary and adventitious roots. In leaf sheath and leaf blade, WRKY21 was expressed throughout the tissues. Overall, the spatial expression pattern of WRKY21 was highly similar to that of PHT1;1 (Figs 3b, S1; Sun et al., 2012), further supporting the physical interaction between WRKY21 and PHT1;1 (Figs 2, S4).
The subcellular localisation of WRKY21 was examined through infiltration of tobacco (Nicotiana benthamiana) epidermal cells. The ORF of WRKY21 was fused with either the 5′ or the 3′ end of the GFP reporter gene. Unlike the GFP control, which was universally distributed to all the compartments of the cell except for the vacuole, the WRKY21:GFP and GFP:WRKY21 fusion proteins were exclusively located in the nucleus (Fig. 3c).
Overexpression of WRKY21 leads to increased Pi accumulation
To evaluate the effect of WRKY21 on maintaining P homeostasis, its overexpression and mutant lines were developed (Fig. S5). All genotypes were grown hydroponically with HP and LP treatments. No difference in growth performance or Pi accumulation was observed in the wrky21 mutant plants. By contrast, overexpression of WRKY21 led to excessive Pi accumulation and impaired growth (Fig. 4). The same trend was observed for total P concentration (Figs S6, S7). These results suggested that WRKY21 is a positive regulator of Pi accumulation and is probably functionally redundant with other TFs.
Fig. 4.
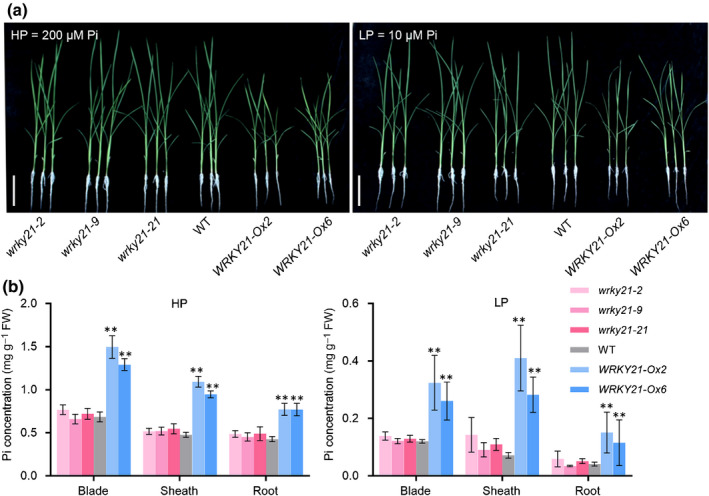
WRKY21 positively affects phosphate (Pi) accumulation in rice (Oryza sativa). (a) Phenotype of WRKY21 transgenic plants and wild‐type plants grown under HP (200 μM Pi) and LP (10 μM Pi) hydroponic conditions. Bars, 10 cm. (b) Pi concentration analysis in leaf tissues and roots of lines (as above). Four‐leaf‐old seedlings were grown under full‐strength Yoshida nutrient solution supplied with HP (200 μM Pi) or LP (1 μM Pi) until the seventh leaf blades were fully expanded. Pi concentration was measured in plants grown under HP (left) and LP (right) conditions. Leaf blades, leaf sheaths and roots were collected for measurement. Error bars indicate ± SD (n = 4). Data significantly different from the corresponding controls are indicated (**, P < 0.01; Student’s t‐test). FW, fresh weight.
WRKY21 physically interacts with WRKY108
Some WRKY TFs have been reported to interact with various proteins (e.g. other WRKY TFs and TFs of other families) to exert their functions redundantly (Xu et al., 2006). Therefore, a yeast two‐hybrid (Y2H) screening was performed to identify potential interacting proteins of WRKY21. Before the library‐scale screening, a series of truncations of WRKY21 was generated and tested for their autoactivation activity. The results showed that the last 45 amino acids at the C terminus were required for the autoactivation activity of WRKY21. Therefore, the truncated version of WRKY21 without the last 45 amino acids at the C terminus, WRKY21‐C‐△45, was used for the screening (Fig. S8a). In total, 100 higher fidelity clones were identified to encode 13 genes. Four clones corresponded to the gene encoding a WRKY transcription factor, WRKY108 (Table S5). To validate the physical interaction between WRKY21 and WRKY108, a one‐on‐one Y2H assay was performed, in which the results of the library‐scale screening was reproduced (Fig. 5a). The interaction between WRKY21 and WRKY108 was further verified by BiFC and pull‐down assays (Fig. 5b,c). All these results suggested that WRKY21 physically interacted with WRKY108 in the nucleus.
Fig. 5.
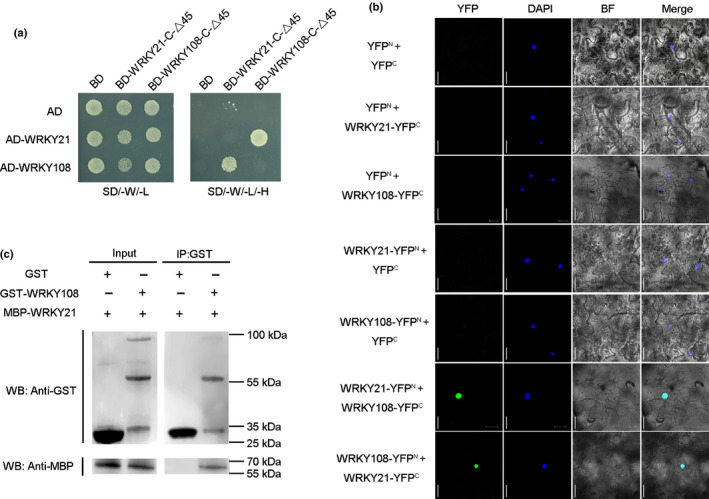
WRKY21 physically interacts with WRKY108. (a) Y2H assay for the interaction of WRKY21 and WRKY108. WRKY21 and WRKY108 were fused to GAL4 activation domain to generate AD‐WRKY21 and AD‐WRKY108. WRKY21‐C‐△45 (described in Supporting Information Fig. S5a) and WRKY108‐C‐△45 (described in Fig. S5b) were fused to GAL4 binding domain to generate BD‐WRKY21‐C‐△45 and BD‐WRKY108‐C‐△45. Yeast cells co‐transformed with AD‐WRKY21/BD‐WRKY21‐C‐△45, AD‐WRKY21/ BD‐WRKY108‐C‐△45, AD‐WRKY108/BD‐WRKY21‐C‐△45 and AD‐WRKY108/BD‐WRKY108‐C‐△45 were grown on selective media SD/−W/−L and SD/−W/−L/−H. Co‐expression of AD/BD, AD/BD‐WRKY21‐C‐△45, AD/BD–WRKY108‐C‐△45, AD‐WRKY21/BD and AD‐WRKY108/BD were used as negative controls. SD/−W/−L, (−Trp−Leu); SD/−W/−L/−H, (−Trp−Leu−His). (b) BiFC analysis for the interaction between WRKY21 and WRKY108. N‐ and C‐terminal fragments of yellow fluorescent protein (YFP) (YFPN and YFPC) were fused to the C terminus of WRKY21 and WRKY108, respectively. Green signals indicate YFP, and the blue signals indicate the cell nucleus that was specifically stained by 4′,6‐diamidino‐2‐phenylindole (DAPI). Bars, 20 μm. (c) GST pull‐down assay for interaction between WRKY21 and WRKY108 in vitro. MBP‐WRKY21 and GST‐WRKY108 were expressed and purified in E. coli and subjected to GST pull‐down assays. GST/GST‐WRKY108 and MBP‐WRKY21 proteins were detected by immunoblotting using anti‐GST and anti‐MBP antibodies, respectively.
The phenotype of WRKY108 overexpression lines mimics that of WRKY21 overexpression plants
To investigate whether WRKY108 was involved in maintaining P homeostasis as its interacting partner WRKY21, we first examined WRKY108 expression in response to Pi. WRKY108 showed exactly the same response to Pi as WRKY21, namely upregulated by Pi in the root but not in the shoot (Figs 3a, S9a). Subsequently, ProWRKY108:GUS transgenic rice plants were generated for tissue localisation analysis. WRKY108 showed the same spatial expression pattern as WRKY21, except that its abundance in the root central cylinder was higher than that in other root cells (Figs 3b, S9b). In addition, the fusions of WRKY108:GFP and GFP:WRKY108 were both detected in the nucleus (Fig. S9c). WRKY108 overexpression and mutant lines were generated (Fig. S10). Similar to that found in WRKY21, WRKY108 overexpression lines but not wrky108 mutants showed excessive Pi/total P accumulation and impaired growth (Figs 4, S6, S7, S11).
WRKY21 and WRKY108 both positively regulates the expression of PHT1;1
Y1H, EMSA and ChIP‐qPCR analyses showed that WRKY108 directly bound to F2 but not F1 in the PHT1;1 promoter. Unlike WRKY21, which only interacted with Wz, WRKY108 bound to both Wy and Wz F2 (Figs 2a, S12). To further investigate the regulation of WRKY21 and WRKY108 on PHT1;1, the expression of PHT1;1 as well as other PHT1 genes was examined in WRKY21/WRKY108 overexpression and mutant plants. Mutation of WRKY21 or WRKY 108 did not alter the expression of PHT1 genes (Fig. S13). By contrast, overexpression of either WRKY21 or WRKY108 led to a significant upregulation of PHT1;1, irrespective of the Pi regimes (Fig. 6a). Under Pi‐sufficient conditions, overexpression of WRKY21 or WRKY108 also resulted in enhanced expression of PHT1;2. In addition, overexpression of WRKY108, but not WRKY21, elevated the expression levels of PHT1;4 and PHT1;8 under Pi‐replete conditions. Moreover, the transcript abundance of PHT1;2 and PHT1;8 was increased in WRKY108 overexpression plants under Pi‐deficient conditions (Fig. 6a). These results indicated that WRKY21 and WRKY108 promoted Pi accumulation by positively regulating the expression of PHT1;1/2/4/8. Notably, PHT1;1/2/4/8 are four rice PHT1 genes with higher basal expression levels under Pi‐replete conditions compared with other PHT1 paralogues (Secco et al., 2013), suggesting that WRKY21 and WRKY108 play crucial roles in Pi accumulation under Pi‐sufficient conditions.
Fig. 6.
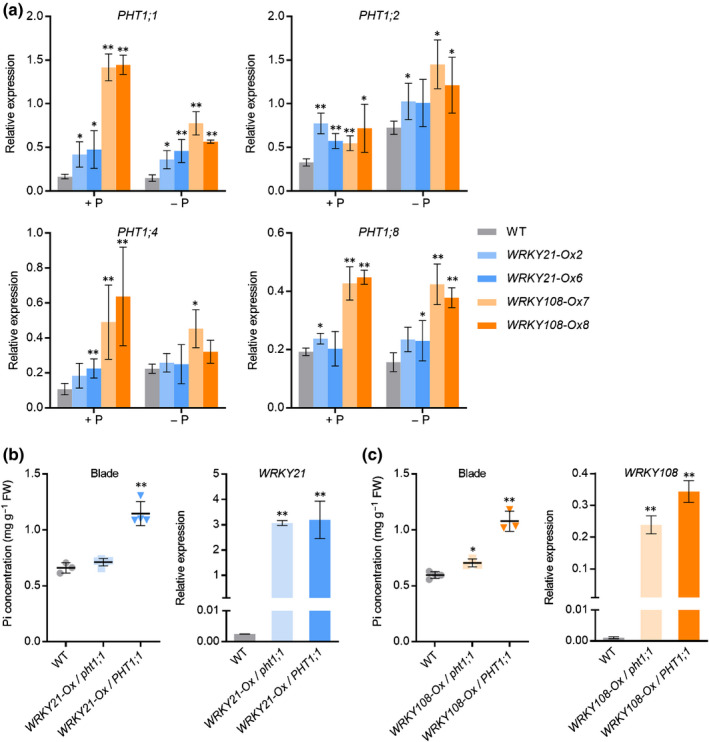
Genetic and molecular analysis of WRKY21/WRKY108 regulating PHT1;1 in rice (Oryza sativa). (a) Effects of WRKY21 and WRKY108 overexpression on transcript levels of four PHT1 genes in rice. Rice seeds of overexpression lines and wild‐type plants were germinated in deionised H2O and cultured hydroponically under Pi‐sufficient (+P, 90 μM) or Pi‐deficient (−P, 0 μM) conditions. Plant roots were collected for RNA extraction and RT‐qPCR analysis. (b, c) Pi concentration in leaf blades and RT‐qPCR analysis of WRKY21(108) expression plants. WT, wild‐type plants; WRKY21‐Ox(WRKY108‐Ox)/pht1;1, plants overexpressing WRKY21(WRKY108) in the pht1;1 mutant background; WRKY21‐Ox(WRKY108‐Ox)/PHT1;1, plants overexpressing WRKY21(WRKY108). Four‐leaf‐old seedlings were grown under nutrient solution supplied with HP (200 μM Pi) until the seventh leaf blades were fully expanded. The fourth and fifth leaf blades were collected for Pi concentration measurement (left panel) and roots were collected for RNA extraction and RT‐qPCR analysis (right panel). The rice housekeeping gene OsActin1 (LOC_Os03g50885) was used as an internal control. Values represent means ± SD of biological replicates (n = 3 or 4). Data significantly different from the corresponding controls are indicated (*, P < 0.05; **, P < 0.01; Student’s t‐test). FW, fresh weight.
PHT1;1 is responsible for WRKY21/108‐mediated accumulation of excessive Pi
Given that WRKY21 and WRKY108 are positive regulators of PHT1;1 expression (Fig. 6a), we reasoned that PHT1;1 might be responsible for the WRKY21/108‐mediated accumulation of excessive Pi. To validate this, WRKY21/108 overexpressors on the pht1;1 background were developed by crossing WRKY21/108 overexpression plants and a pht1;1 mutant line. The resulting segregated filial generations were designated as WRKY21‐Ox/pht1;1 and WRKY108‐Ox/pht1;1. The Pi concentration in WRKY21‐Ox/pht1;1 plants was comparable with that in wild‐type plants (Fig. 6b); the Pi concentration in WRKY108‐Ox/pht1;1 plants was slightly higher than that in wild‐type plants, but significantly decreased compared with that in WRKY108‐Ox/PHT1;1 plants (Fig. 6c). The genetic and molecular evidence demonstrated that WRKY21 and WRKY108 promoted Pi accumulation through activating PHT1;1 expression via binding to the PHT1;1 promoter.
WRKY21 and WRKY108 function redundantly in Pi accumulation in a Pi‐dependent manner
To further investigate the physiological role and molecular mechanism of WRKY21 and WRKY108, wrky21 wrky108 double mutants were generated using a cross between wrky21 and wrky108 single mutants. WRKY21 and WRKY108 reside closely on chromosome 1 (c. 27.5 kb apart), therefore this cross was less likely to produce a wrky21 wrky108 double mutant due to the extremely low chromosomal crossover rate. Nevertheless, we still acquired wrky21 wrky108 double mutants, which was achieved via secondary editing using the CRISPR‐Cas9 system (Fig. S14). Unexpectedly, no alteration in Pi concentration or PHT1;1 expression was observed in the wrky21 wrky108 double mutant compared with that in wild‐type plants under LP, Ctrl and HP conditions (Fig. 7). This indicated that WRKY21 and WRKY108 regulate PHT1;1 expression and Pi accumulation redundantly with other unknown TF(s) and/or play a major role under other conditions. Therefore, we first tested the effect of WRKY21 and WRKY108 on Pi accumulation when even higher concentrations of Pi (1 mM) were supplied. The wrky21 wrky108 double mutants but not the wrky21 and wrky108 single mutants showed a significant decrease in Pi concentration in root and downregulated PHT1;1 expression compared with that of wild‐type plants (Fig. 7), suggesting that the regulation of OsPHT1;1 by WRKY21/108 was dependent on Pi levels.
Fig. 7.
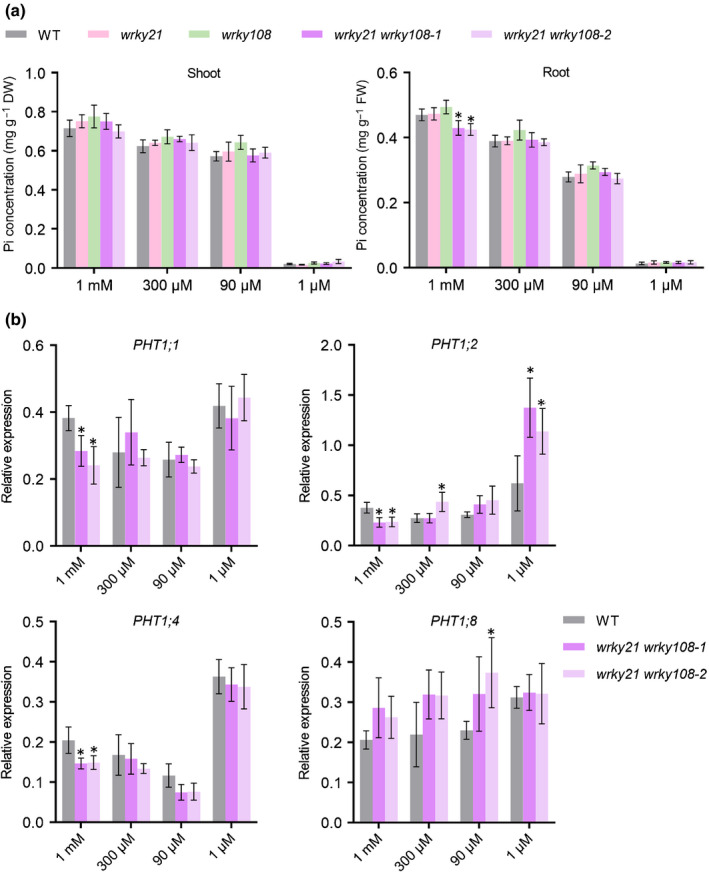
Effect of WRKY21 and WRKY108 mutations on phosphate (Pi) homeostasis in rice (Oryza sativa). (a) Cellular Pi concentration analysis of wrky21 (line wrky21‐2), wrky108 (line wrky108‐17), wrky21 wrky108 mutants and wild‐type plants. Four‐leaf‐old seedlings were grown under half‐strength Kimura B nutrient solution supplied with 1 mM Pi, 300 μM Pi, 90 μM Pi or 1 μM Pi until the seventh leaf blades were fully expanded. Pi concentration was measured in shoot (left) and root (right). (b) RT‐qPCR analysis of the effect of wrky21 wrky108 double mutants on the expression of four PHT1 genes. Plant roots were collected for RNA extraction and RT‐qPCR analysis. The rice housekeeping gene OsActin1 (LOC_Os03g50885) was used as an internal control. Values represent means ± SD of four biological replicates. Data significantly different from that in corresponding controls are indicated (*, P < 0.05; Student’s t‐test). FW, fresh weight.
Expression of a chimeric dominant repressor of WRKY21 leads to a phenotype mimicking that of the pht1;1 mutant
To overcome the potential genetic redundancy, we converted WRKY21 and WRKY108 into suppressors by fusing them with a dominant repressor domain SRDX (Hiratsu et al., 2003), generating rice plants that expressed Pro35S:WRKY21‐SRDX and Pro35S:WRKY108‐SRDX. These transgenic lines and wild‐type plants were grown under Ctrl and HP conditions. No obvious phenotype with regard to Pi accumulation was found in Pro35S:WRKY108‐SRDX plants (Fig. S15a). By contrast, Pro35S:WRKY21‐SRDX plants showed a significant decrease in root Pi concentration, similar to that found in pht1;1 mutants (Figs 1, 8b). A similar trend was observed for total P concentration (Fig. S16). Consistently, PHT1;1 expression was significantly downregulated in Pro35S:WRKY21‐SRDX plants under both Pi regimes (Fig. 8c).
Fig. 8.
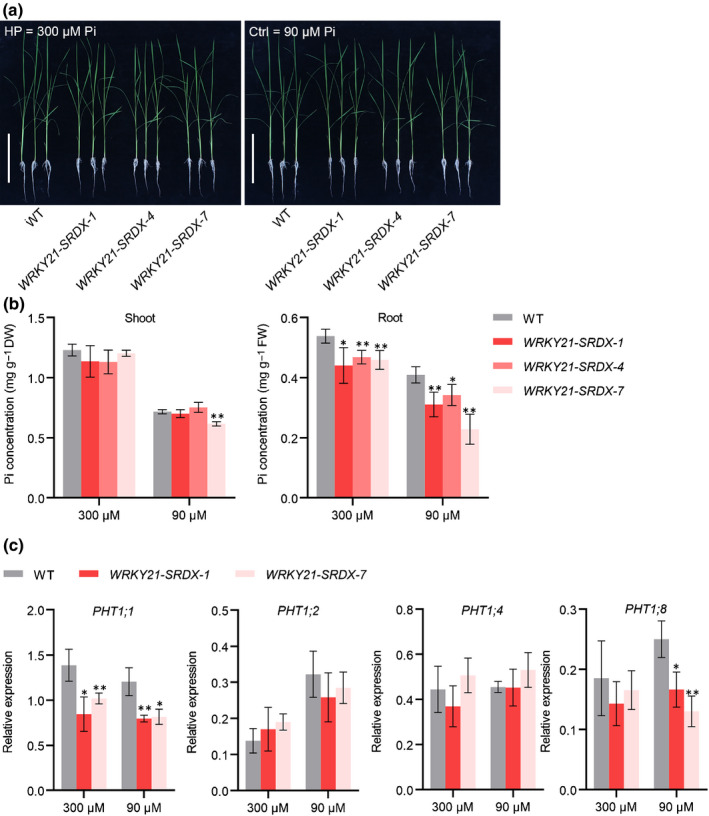
Expression of the chimeric WRKY21 repressor resulted in reduced phosphate (Pi) accumulation in root and suppression of PHT1;1 expression. (a) Phenotype of WRKY21‐SRDX transgenic lines and wild‐type plants grown hydroponically supplied with 300 μM Pi or 90 μM Pi. Bars, 15 cm. (b) Cellular Pi concentration analysis of Pro35S:WRKY21‐SRDX transgenic plants and wild‐type plants. Four‐leaf‐old seedlings were grown under half‐strength Kimura B nutrient solution supplied with 300 μM Pi or 90 μM Pi until the seventh leaf blades were fully expanded. Pi concentration was measured in shoot and root. (c) Effect of chimeric WRKY21 repressor on transcript levels of four PHT1 genes. Plant roots were collected for RNA extraction and RT‐qPCR analysis. The rice housekeeping gene OsActin1 (LOC_Os03g50885) was used as an internal control. Error bars indicate SD (n = 4). Data significantly different from the corresponding controls are indicated (*, P < 0.05; **, P < 0.01; Student’s t‐test). FW, fresh weight.
Discussion
Maintenance of PHT1 transcript level is vital for Pi uptake from Pi‐sufficient environments
Although PHT1 capacities for mediating Pi uptake can be regulated by altering their activities via amino acid substitution (Catarecha et al., 2007; Fontenot et al., 2015), it should be noted that the regulation of PHT1 abundance is a more universal plant innate strategy that occurs at different levels (mainly transcriptional and posttranslational levels). Significant progress has been made in deciphering the mechanisms underlying the posttranslational regulation of PHT1s (Gu et al., 2016 and references therein; Yue et al., 2017; S. Y. Yang et al., 2020; Z. L.Yang et al., 2020), whereas transcriptional regulation is an important determinant of PHT1 abundance, functioning earlier than any posttranslational regulation. In rice, four out of the 13 PHT1 genes (PHT1;1, PHT1;2, PHT1;4 and PHT1;8) showed high basal expression levels in Pi‐replete roots (Figs 6a, 7b, 8c; Secco et al., 2013). Therefore, it could be assumed that these four PHT1 genes, especially PHT1;1 and PHT1;8, which are not very responsive to Pi starvation, play a major role in Pi uptake and accumulation from Pi‐sufficient environments. In support of our assumption, under controlled conditions, mutation of PHT1;1 and PHT1;8 led to a 22–27% and 36–41% reduction, respectively, in root Pi accumulation, and pht1;1 and pht1;8 showed an additive genetic effect as evidenced by a >60% decrease in root Pi accumulation in the pht1;1 pht1;8 double mutant (Figs 1, S17). Under LP conditions, no alteration in Pi accumulation was observed in the pht1;1 or the pht1;8 mutant; by contrast, the pht1;1 pht1;8 double mutant was impaired in Pi accumulation in roots but not in shoots (Figs 1, S17). These results indicated that PHT1;1 and PHT1;8 also played a redundant role in Pi uptake from LP environments, but their contribution was minor and was probably attributed to the induction of PSI PHT1s (i.e. OsPHT1;2, OsPHT1;3, OsPHT1;4, OsPHT1;6, OsPHT1;9 and OsPHT1;10). This differed from that observed in Arabidopsis AtPHT1;1, an orthologue of rice PHT1;1 and PHT1;8 that had high basal expression and was responsible for Pi acquisition from both low‐Pi and high‐Pi environments. Under Pi‐deficient conditions, the mutation of AtPHT1;1 resulted in a c. 20% reduction in Pi accumulation, consistent with its positive response to Pi starvation (Misson et al., 2004; Shin et al., 2004). From this respect, it seems that rice PHT1;4 is more closely related to AtPHT1;1, as the pht1;4 mutant was also impaired in Pi accumulation, irrespective of the Pi supply (Zhang et al., 2015). Additionally, although AtPHT1;1 and AtPHT1;4 are co‐ordinately responsible for a 60–75% Pi uptake from Pi‐sufficient environments, quadruple (atpht1;1 atpht1;2 atpht1;3 atpht1;4) and quintuple (atpht1;1 atpht1;2 atpht1;3 atpht1;4 atphf1) mutants showed a c. 88% and c. 95% reduction, respectively, in Pi uptake. This demonstrated that Arabidopsis PHT1s other than AtPHT1;1 and AtPHT1;4 were also involved in Pi uptake under Pi‐sufficient supply, in support of the lower but nonnegligible expression levels of AtPHT1;3, AtPHT1;5, AtPHT1;8 and AtPHT1;9 under these conditions (Ayadi et al., 2015; Sun et al., 2016). In rice, the expression of PHT1 orthologues, except that of PHT1;1/2/4/8, was barely detectable in Pi‐replete root (Secco et al., 2013). In future work, whether these four rice PHT1s are the only participants for Pi uptake from Pi‐sufficient environments needs to be investigated with multiple mutants.
Unlike that found in pht1;1, pht1;4 and pht1;8 mutants, loss‐of‐function mutants of PHT1;2 showed a significant decrease in Pi accumulation under Pi‐deficient, but not Pi‐sufficient, conditions, (Figs 1, S17, S18; Zhang et al., 2015). Given the higher expression levels of PHT1;2 compared with PHT1;1/4/8 under Pi‐sufficient conditions (Secco et al., 2013), the lack of phenotype regarding Pi accumulation in Pi‐replete pht1;2 could be attributed to the posttranscriptional regulation and/or spatial expression pattern of PHT1;2. Indeed, PHT1;2 has been reported to be regulated posttranslationally by Phosphate Transporter Traffic Facilitator 1, CK2 kinase, Nitrogen Limitation Adaptation 1 and Protein Phosphatase 95. PHT1;8 is also under the control of these posttranslational regulators; however, a significant decrease in Pi accumulation was observed under Pi‐sufficient conditions upon mutation of PHT1;8 (Fig. S17; Chen et al., 2011; Chen et al., 2015; Yue et al., 2017; S. Y. Yang et al., 2020; Z. L. Yang et al., 2020). Thus, the potential different extent of posttranslational regulations and/or the varied spatial expression patterns of these PHT1s could be the causes for their diverged effects on Pi accumulation under Pi‐sufficient conditions. It is of interest and significance to test these possibilities. Nevertheless, our results and reported findings suggested that the basal expression levels of PHT1 genes is an important, although not the only, determinant for Pi acquisition from Pi‐sufficient environments.
The WRKY‐PHT1;1 module represents a unique pathway in rice and is independent of the central regulatory system for Pi signalling
The responses of PHT1 genes to the changing Pi availability is an important indicator of in planta demand for Pi that is worth studying. Under Pi‐starvation conditions, the transcription of PSI PHT1s was enhanced, and was largely modulated by the central regulators, PHR(‐like) TFs (Bustos et al., 2010; Guo et al., 2015). TFs of other families, including WRKY TFs, are also found to positively regulate the expression of PHT1 genes in response to Pi deficiency (Devaiah et al., 2007; Dai et al., 2016). AtWRKY75, AtWRKY45, OsWRKY74 are responsible for the PSI expression of at least one PHT1 member (Devaiah et al., 2007; H. Wang et al., 2014; Dai et al., 2016). AtWRKY45 and AtWRKY75 are the two most closely related paralogues, as shown by phylogenetic analysis (Fig. S19; Ross et al., 2007). They both promoted the expression of AtPHT1;1 (the PHT1 member with high transcript level in Pi‐replete Arabidopsis), similar to that found in rice WRKY21 and WRKY108 (Figs 6a, 7b; Devaiah et al., 2007; H. Wang et al., 2014). However, the regulation of PHT1 gene(s) by AtWRKY45/AtWRKY75 mainly occurred under Pi‐starvation conditions, whereas that by WRKY21/WRKY108 mainly took place under Pi‐replete conditions, in accordance with their opposite responses to Pi starvation at the transcriptional level (AtWRKY45/AtWRKY75 and WRKY21/WRKY108 were induced and repressed, respectively, by Pi starvation) (Figs 3a, S9a; Devaiah et al., 2007; H. Wang et al., 2014). Moreover, WRKY21 and WRKY108 were phylogenetically remote from AtWRKY45 and AtWRKY75 (Fig. S19), indicating that WRKY21 and WRKY108 were evolutionarily distinct from AtWRKY45 and AtWRKY75. In addition, AtWRKY42 seemed to be functionally related to WRKY21 and WRKY108, as it was also transcriptionally repressed by Pi starvation and activated AtPH1;1 expression under Pi‐replete conditions (Figs 3a,S9a; Su et al., 2015). Nonetheless, several factors indicated that WRKY21 and WRKY108 were functionally distinct from AtWRKY42 and its interacting homologue AtWRKY6:
(1) AtWRKY42 and AtWRKY6 are classified into Group II WRKY TFs, whereas WRKY21 and WRKY108 belong to the Group III subfamily.
(2) AtWRKY42 has a negative effect on AtPHO1 expression when Pi is ample in the environment, whereas WRKY21 and WRKY108 act as positive regulators of OsPHO1;2 (the functional orthologue of AtPHO1 in rice) when Pi is lacking (Fig. S20; Secco et al., 2010; Jabnoune et al., 2013; Su et al., 2015; Che et al., 2020; Chiou, 2020).
(3) atwrky6 atwrky42 double mutants display elevated Pi accumulation in shoot, whereas wrky21 wrky108 double mutants and Pro35S:WRKY21‐SRDX plants show decreased Pi accumulation only in root (Figs 7a, 8b; Su et al., 2015).
(4) AtWRKY6 is slightly upregulated by Pi starvation, but WRKY21 and WRKY108 are downregulated upon deficiency of Pi (Figs 3a, S9a; Chen et al., 2009; Su et al., 2015).
Our results and reported data indicated that extensive functional divergence regarding WRKY‐modulated P homeostasis has occurred between rice and Arabidopsis. Furthermore, AtWRKY6 and AtWRKY42 are both degraded through the 26S proteasome pathway in response to Pi starvation (Chen et al., 2009; Su et al., 2015; Ye et al., 2018). It would be of interest and significance to investigate whether WRKY21 and WRKY108 are subjected to any posttranslational regulation. Moreover, the effect of the interaction between WRKY21 and WRKY108 (coordination or competition) on PHT1;1 regulation also needs to be investigated in future work.
In addition to transcriptional activation (constitutive expression under Pi‐replete conditions and PSI expression), transcriptional repression is another strategy utilised by plants to monitor the expression of PHT1 genes. Since the first report that negative regulators can bind to the promoter of AtPHT1;4 (Mukatira et al., 2001), several TFs have been demonstrated to act as transcriptional repressors of PHT1 genes under Pi‐replete conditions (S. K. Wang et al., 2014; Gu et al., 2017). In our previous work, two out of the four rice PHT1 genes with high basal expression levels under Pi‐replete conditions, namely PHT1;2 and PHT1;8, were both found to be negatively regulated by OsMYB1. In osmyb1 mutants, the expression levels of PHT1;2 and PHT1;8 were enhanced under Pi‐replete conditions and were comparable to that under Pi‐deficient conditions (Gu et al., 2017), suggesting that their basal expression was maintained by other unknown TF(s), the function of which is/are overridden by the suppressive effect of OsMYB1. Therefore, an intriguing question to be addressed in future work is why plants have evolved both transcriptional suppressors and activators for PHT1 genes with high basal expression levels. TF genes involved in Pi signalling themselves are usually transcriptionally regulated, defining a hierarchical signalling cascade. More than half of the reported WRKY TF genes are upregulated by Pi starvation (Devaiah et al., 2007; Chen et al., 2009; H. Wang et al., 2014; Dai et al., 2016). By contrast, WRKY21 and WRKY108 were both transcriptionally induced by Pi (Figs 3a, S9a), in support of their roles under Pi‐sufficient conditions (Fig. 7). In future work, it would be of interest and significance to investigate the mechanisms underlying the transcriptional responses of WRKY21 and WRKY108 to Pi. This may involve the identification of novel TFs functioning upstream of WRKY21 and WRKY108, although the possibility that WRKY21 and WRKY108 could be autoactivated by their own products cannot be excluded. Altogether, our results demonstrated that WRKY‐PHT1;1 is a unique module in rice that is independent of the central regulatory system for Pi signalling.
Extensive functional redundancy commonly occurs within and among different TF families controlling Pi signalling
The genetic redundancy in plant mineral nutrition is not only observed within downstream components such as transporters, but also in upstream regulatory proteins (e.g. TFs). The single mutants of PHR homologues show no or weak phenotypes regarding Pi accumulation, and double or triple mutants have been found to have additive effects (Bustos et al., 2010; Khan et al., 2014; Guo et al., 2015; Sun et al., 2016; Ruan et al., 2017). Similarly, in this work, no alteration in Pi accumulation in wrky21 and wrky108 single mutants was observed, whereas the wrky21 wrky108 double mutants showed decreased Pi accumulation when 1 mM Pi was supplied (Figs 4b, 7a, S11b), indicating that these two WRKY TFs function redundantly to maintain P homeostasis in a Pi‐dependent manner. To examine the roles of PHT1;1 and WRKY21/108 in the context of soil grown conditions, a soil‐based system was used to examined their effect on P homeostasis. Similar to that found in the hydroponic system, pht1 mutants and WRKY21‐SRDX plants showed significantly decreased total P concentrations in roots supplied with P fertiliser (Figs S6, S7, S21). In contrast with that found in the hydroponic system, both pht1;1 and WRKY21‐SRDX plants were impaired in total P accumulation in shoot also (Figs S6, S7, S21). Despite the difference between hydroponic and soil‐based cultures, both systems demonstrated that WRKY21/108 and other unidentified WRKY(s) function redundantly to promote PHT1;1 expression and therefore P accumulation. It has been reported that, in intensively fertilised farmland, the dissolved reactive P concentration in the soil leachates reaches 10 mg l−1 (>300 μM) (Kalkhajeh et al., 2017). In addition, Pi availability in flooded paddy fields could be higher than that in uplands. A very recent report showed that the soluble Pi in soil solution continuously increased with the duration of rice plant growth (Wang et al., 2020). Therefore, we speculate that, in flooded paddy fields with excessive fertiliser input, the soluble Pi concentration in the soil solution could reach or approach 1 mM. Moreover, transgenic rice plants expressing the WRKY21–SRDX fusion protein were impaired by Pi accumulation under control and HP conditions (Fig. 8b), demonstrating the existence of other WRKY TF(s) that positively regulated PHT1;1 expression and that remain(s) to be identified. Close homologues of WRKY21 and WRKY108, namely OsWRKY79 and OsWRKY60 (Fig. S19), could be potential candidates for further study. Pro35S:WRKY108‐SRDX plants showed no alteration in Pi accumulation as that found in Pro35S:WRKY21‐SRDX plants (Fig. S15). One possible explanation is that WRKY108 but not WRKY21 can form homodimers (data not shown), and the effect of WRKY108‐SRDX on PHT1;1 expression was counteracted by endogenous WRKY108. In addition, genetic redundancy also occurs among TFs from different families. PHR and WRKY TFs are both implicated in regulating the expression of PSI PHT1 genes (Liu et al., 2010; Su et al., 2015; Chang et al., 2019). WRKY and Ethylene Response Factor (ERF) TFs have been reported to synergistically activate the expression of a gene encoding pyruvate decarboxylase (Zhu et al., 2019). Interestingly, in this study, an ERF TF was also detected as a candidate binding to the PHT1;1 promoter (Table S5). Its potential role in maintaining P homeostasis awaits to be investigated.
Based on the results analysed above, we propose a working model for the involvement of the WRKY‐PHT1;1 module in Pi signalling in rice (Fig. 9). PHT1;1 is constitutively and highly expressed irrespective of the Pi regimes. When Pi was deficient, WRKY21 and WRKY108 were expressed at relatively low levels, and the transcription of PHT1;1 was maintained by other unknown TFs. PSI PHT1 genes were positively regulated by the PHR‐P1BS module. When Pi was sufficient, the transcription of WRKY21 and WRKY108 was enhanced. WRKY21 and WRKY108 interacted in the nucleus and bound to the W‐box motif(s) within the PHT1;1 promoter. WRKY21 and WRKY108 together with other unidentified WRKY TF(s) and/or TF(s) from other family/families co‐ordinately promoted the transcription of PHT1;1.
Fig. 9.
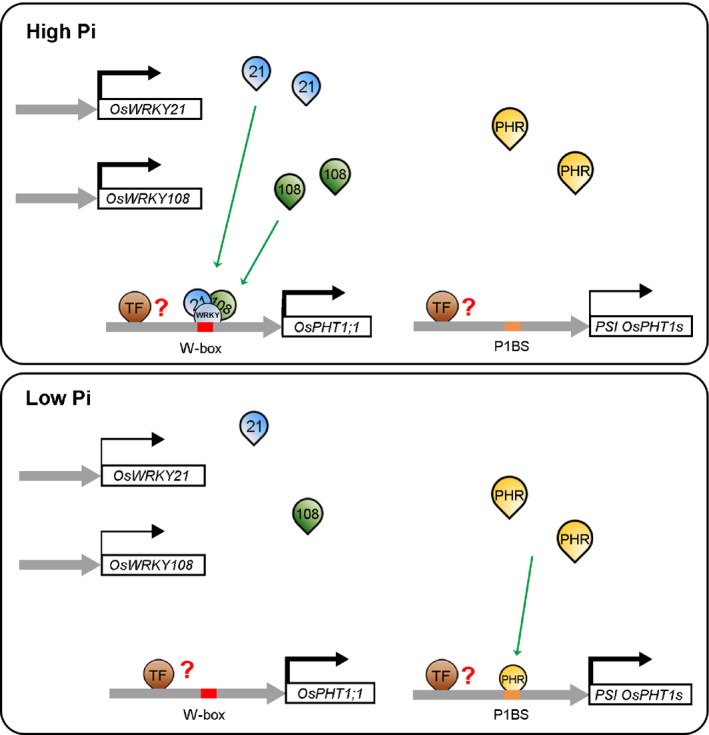
Working model of the WRKY‐PHT1;1 regulatory pathway in maintaining P homeostasis in rice (Oryza sativa). Elbowed arrows in dark indicate the transcriptional expression of OsWRKY21, OsWRKY108, OsPHT1;1 and PSI OsPHT1s genes, and the thickness of the line represents the relative transcription level. Inverted droplet shapes in colours indicate different transcription factors: 21, WRKY21; 108, WRKY108; WRKY, other unknown WRKY members; PHR, Phosphate Starvation Response transcription factors; TF, other unknown transcription factors. Red and orange rectangles indicate W‐box and PHR1 Binding Sequence (P1BS), respectively. Green arrows indicate that TFs can bind directly to the motif(s) on the promoters of OsPHT1 family genes.
Author contributions
MG conceived and designed the experiments. JZ and MG performed most of the experiments. RSHL and XH performed hydroponic culture. XYS and LLC conducted experiments for the soil‐based system. SCW, XLD and HHL constructed partial vectors for generating mutant plants and performed part of the physiological analysis. HYQ performed rice transformation and field management. JZ, MG and GHX analysed the data. JZ and MG wrote the article. GHX gave critical comments and revisions to the paper. JZ and MG contributed equally to this work.
Supporting information
Fig. S1 Identification of pht1;1 mutant lines.
Fig. S2 Total P concentration of pht1;1 mutants and wild‐type plants.
Fig. S3 Histochemical localisation of different truncations of PHT1;1 promoter fused with the GUS reporter gene in transgenic plants.
Fig. S4 WRKY21 binds to F2 fragment of PHT1;1 promoter through the specific W‐box.
Fig. S5 Identification of WRKY21 overexpression and mutant lines.
Fig. S6 Total P concentration of wrky21, wrky108, wrky21 wrky108 mutants and wild‐type plants.
Fig. S7 Total P concentration of WRKY21 and WRKY108 overexpression lines and wild‐type plants.
Fig. S8 Transactivation activity of the WRKY21 and WRKY108 proteins in yeast.
Fig. S9 Expression patterns and subcellular localisation of WRKY108.
Fig. S10 Identification of WRKY108 overexpression and mutant lines.
Fig. S11 WRKY108 positively affects phosphate (Pi) accumulation in rice.
Fig. S12 WRKY108 binds to PHT1;1 promoter region in vivo and in vitro.
Fig. S13 RT‐qPCR analysis of effect of WRKY21/WRKY108 mutation on expression of four PHT1 genes.
Fig. S14 Identification of wrky21 wrky108 double mutants.
Fig. S15 Pi concentration and RT‐qPCR analysis of PHT1;1 in Pro35S:WRKY108‐SRDX transgenic plants and wild‐type plants.
Fig. S16 Total P concentration of Pro35S:WRKY21‐SRDX transgenic lines and wild‐type plants.
Fig. S17 Pi concentration of pht1;8, pht1;1 pht1;8 mutants and wild‐type plants.
Fig. S18 Pi concentration of pht1;2 mutants and wild‐type plants.
Fig. S19 Phylogenetic analysis of the Group II and Group III type WRKY transcription factors from rice and A. thaliana according to Ross et al. (2007).
Fig. S20 Effect of WRKY21 and WRKY108 overexpression or mutation on the transcript level of PHO1;2 in rice.
Fig. S21 Total P concentration of wild‐type plants, pht1;1, wrky21/wrky108 single mutants, wrky21 wrky108 double mutants and WRKY21‐SRDX transgenic lines, at different Pi levels in a soil‐based experiment.
Table S1 Primers used for RT‐qPCR analysis.
Table S2 Primers used for constructs for generating transgenic plants.
Table S3 Primers used for constructs for subcellular location, Y1H, EMSA, Y2H, BiFC and pull‐down assay.
Table S4 Positive interactions from yeast one‐hybrid screening.
Table S5 Candidates from yeast two‐hybrid screening.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
We thank Prof. Chuanzao Mao (Zhejiang University) for providing us the pht1;8 mutant. We are grateful to Professor Lijia Qu (Peking University) and Dr Tzvi Tzfra (University of Michigan) for providing us with vectors for the CRISPR‐Cas9 system and subcellular localisation analysis. We also thank Yuanming Xie for technical assistance in microscopy observation during the COVID‐19 outbreak. This work was supported by the National Key Research and Development Programme of China (2016YFD0100700 and 2017YFD0200204), the Natural Science Foundation of China (31972489, 31930101 and 31872165), and the Innovative Research Team Development Plan of the Ministry of Education of China (IRT_17R56; KYT201802).
References
- Ai PH, Sun SB, Zhao JN, Fan XR, Xin WJ, Guo Q, Yu L, Shen QR, Miller AJ, Xu GH. 2009. Two rice phosphate transporters, OsPht1;2 and OsPht1;6, have different functions and kinetic properties in uptake and translocation. The Plant Journal 57: 798–809. [DOI] [PubMed] [Google Scholar]
- Aung K, Lin SI, Wu CC, Huang YT, Su CL, Chiou TJ. 2006. pho2, a phosphate overaccumulateor, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiology 141: 1000–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayadi A, David P, Arrighi JF, Chiarenza S, Thibaud MC, Nussaume L, Marin E. 2015. Reducing the genetic redundancy of Arabidopsis PHOSPHATE TRANSPORTER1 transporters to study phosphate uptake and signaling. Plant Physiology 167: 1511–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D, Kim MC, Chun HJ, Kang S, Park HC, Shin G, Park J, Shen M, Hong H, Kim W et al 2013. Regulation of miR399f transcription by AtMYB2 affects phosphate starvation responses in Arabidopsis . Plant Physiology 161: 362–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustos R, Castrillo G, Linhares F, Puga MI, Rubio V, Perez‐Perez J, Solano R, Leyva A, Paz‐Ares J. 2010. A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis . PLoS Genetics 6: e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catarecha P, Segura MD, Franco‐Zorrilla JM, García‐Ponce B, Lanza M, Solano R, Paz‐Ares J, Leyva A. 2007. A mutant of the Arabidopsis phosphate transporter PHT1;1 displays enhanced arsenic accumulation. Plant Cell 19: 1123–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanas SA, Jiang Q, McMahon M, McWalter GK, McLellan LI, Elcombe CR, Henderson CJ, Wolf CR, Moffat GJ, Itoh K et al 2002. Loss of the Nrf2 transcription factor causes a marked reduction in constitutive and inducible expression of the glutathione S‐transferase Gsta1, Gsta2, Gstm1, Gstm2, Gstm3 and Gstm4 genes in the livers of male and female mice. Biochemical Journal 365: 405–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang MX, Gu M, Xia YW, Dai XL, Dai CR, Zhang J, Wang SC, Qu HY, Yamaji N, Ma JF et al 2019. OsPHT1;3 mediates uptake, translocation, and remobilization of phosphate under extremely low phosphate regimes. Plant Physiology 179: 656–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che J, Yamaji N, Miyaji T, Mitani‐Ueno N, Kato Y, Shen RF, Ma JF. 2020. Node‐localized transporters of phosphorus essential for seed development in rice. Plant & Cell Physiology 61: 1387–1398. [DOI] [PubMed] [Google Scholar]
- Chen CY, Schmidt W. 2015. The paralogous R3 MYB proteins CAPRICE, TRIPTYCHON and ENHANCER OF TRY AND CPC1 play pleiotropic and partly non‐redundant roles in the phosphate starvation response of Arabidopsis roots. Journal of Experimental Botany 66: 4821–4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JY, Liu Y, Ni J, Wang YF, Bai YH, Shi J, Gan J, Wu ZC, Wu P. 2011. OsPHF1 regulates the plasma membrane localization of low‐ and high‐affinity inorganic phosphate transporters and determines inorganic phosphate uptake and translocation in rice. Plant Physiology 157: 269–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JY, Wang YF, Wang F, Yang J, Gao MX, Li CY, Liu YY, Liu Y, Yamaji N, Ma JF et al 2015. The rice CK2 kinase regulates trafficking of phosphate transporters in response to phosphate levels. Plant Cell 27: 711–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YF, Li LQ, Xu Q, Kong YH, Wang H, Wu WH. 2009. The WRKY6 transcription factor modulates PHOSPHATE1 expression in response to low Pi stress in Arabidopsis . Plant Cell 21: 3554–3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou TJ. 2020. The diverse roles of rice PHO1 in phosphate transport: from root to node to grain. Plant & Cell Physiology 61: 1384–1386. [DOI] [PubMed] [Google Scholar]
- Dai XY, Wang YY, Yang A, Zhang WH. 2012. OsMYB2P‐1, an R2R3 MYB transcription factor, is involved in the regulation of phosphate‐starvation responses and root architecture in rice. Plant Physiology 159: 169–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai XY, Wang YY, Zhang WH. 2016. OsWRKY74, a WRKY transcription factor, modulates tolerance to phosphate starvation in rice. Journal of Experimental Botany 67: 947–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Karithikeyan AS, Raghothama KG. 2007. WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiology 143: 1789–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Madhuvanthi R, Karthikeyan AS, Raghothama KG. 2009. Phosphate starvation responses and gibberellic acid biosynthesis are regulated by the MYB62 transcription factor in Arabidopsis . Molecular Plant 2: 43–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenot EB, DiTusa SF, Kato N, Olivier DM, Dale R, Lin WY, Chiou TJ, Macnaughtan MA, Smith AP. 2015. Increased phosphate transport of Arabidopsis thaliana Pht1;1 by site‐directed mutagenesis of tyrosine 312 may be attributed to the disruption of homomeric interactions. Plant, Cell & Environment 38: 2012–2022. [DOI] [PubMed] [Google Scholar]
- Glass AD, Kotur Z. 2013. A re‐evaluation of the role of Arabidopsis NRT1.1 in high‐affinity nitrate transport. Plant Physiology 163: 1103–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M, Chen AQ, Sun SB, Xu GH. 2016. Complex regulation of plant phosphate transporters and the gap between molecular mechanisms and practical application: what is missing? Molecular Plant 9: 396–416. [DOI] [PubMed] [Google Scholar]
- Gu M, Zhang J, Li HH, Meng DQ, Li R, Dai XL, Wang SC, Liu W, Qu HY, Xu GH. 2017. Maintenance of phosphate homeostasis and root development are coordinately regulated by MYB1, an R2R3‐type MYB transcription factor in rice. Journal of Experimental Botany 68: 3603–3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo MN, Ruan WY, Li CY, Huang FL, Zeng M, Liu YY, Yu YN, Ding XM, Wu YR, Wu ZC et al 2015. Integrative comparison of the role the PHOSPHATE RESPONSE1 subfamily in phosphate signaling and homeostasis in rice. Plant Physiology 168: 1762–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsu K, Matsui K, Koyama T, Ohme‐Takagi M. 2003. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis . The Plant Journal 34: 733–739. [DOI] [PubMed] [Google Scholar]
- Jabnoune M, Secco D, Lecampion C, Robaglia C, Shu QY, Poirier Y. 2013. A rice cis‐natural antisense RNA acts as a translational enhancer for its cognate mRNA and contributes to phosphate homeostasis and plant fitness. Plant Cell 25: 4166–4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia HF, Ren HY, Gu M, Zhao JN, Sun SB, Zhang X, Chen JY, Wu P, Xu GH. 2011. Phosphate transporter gene, OsPht1;8, is involved in phosphate homeostasis in rice. Plant Physiology 156: 1164–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalkhajeh YK, Huang B, Hu W, Holm PE, Bruun Hansen HC. 2017. Phosphorus saturation and mobilization in two typical Chinese greenhouse vegetable soils. Chemosphere 172: 316–324. [DOI] [PubMed] [Google Scholar]
- Khan GA, Bouraine S, Wege S, Li YY, Carbonnel MD, Berthomieu P, Poirier Y, Rouached H. 2014. Coordination between zinc and phosphate homeostasis involves the transcription factor PHR1, the phosphate exporter PHO1, and its homologur PHO1;H3 in Arabidopsis. Journal of Experimental Botany 65: 871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambers H, Plaxton WC. 2015. Phosphorus: back to the roots. Annual Plant Reviews 48: 3–22. [Google Scholar]
- Liu F, Wang ZY, Ren HY, Shen C, Li Y, Ling HQ, Wu CY, Lian XM, Wu P. 2010. OsSPX1 suppresses the function of OsPHR2 in the regulation of expression of OsPT2 and phosphate homeostasis in shoots of rice. The Plant Journal 62: 508–517. [DOI] [PubMed] [Google Scholar]
- Misson J, Thibaud MC, Bechtold N, Raghothama K, Nussaume L. 2004. Transcriptional regulation and functional properties of Arabidopsis Pht1;4, a high affinity transporter contributing greatly to phosphate uptake in phosphate deprived plants. Plant Molecular Biology 55: 727–741. [DOI] [PubMed] [Google Scholar]
- Muchhal US, Pardo JM, Raghothama KG. 1996. Phosphate transporters from the higher plant Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 93: 10519–10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudge SR, Rae AL, Diatloff E, Smith FW. 2002. Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. The Plant Journal 31: 341–353. [DOI] [PubMed] [Google Scholar]
- Mukatira UT, Liu CM, Varadarajan DK, Raghothama KG. 2001. Negative regulation of phosphate starvation‐induced genes. Plant Physiology 127: 1854–1862. [PMC free article] [PubMed] [Google Scholar]
- Nussaume L, Kanno S, Javot H, Marin E, Pochon N, Ayadi A, Nakanishi TM, Thibaud MC. 2011. Phosphate import in plants: focus on the PHT1 transporters. Frontier in Plant Science 2: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd GED, Leyser O. 2020. A plant’s diet, surviving in a variable nutrient environment. Science 368: aba0196. [DOI] [PubMed] [Google Scholar]
- Raghothama KG. 1999. Phosphate acquisition. Annual Review of Plant Physiology and Plant Molecular Biology 50: 665–693. [DOI] [PubMed] [Google Scholar]
- Ross CA, Liu Y, Shen QJ. 2007. The WRKY gene family in Rice (Oryza sativa). Journal of Integrative Plant Biology 6: 827–842. [Google Scholar]
- Ruan WY, Guo MN, Wu P, Yi KK. 2017. Phosphate starvation induced OsPHR4 mediates Pi‐signaling and homeostasis in rice. Plant Molecular Biology 93: 327–340. [DOI] [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, Martin AC, Iglesias J, Leyva A, Paz‐Ares J. 2001. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes and Development 15: 2122–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton PJ, Somssich IE, Ringler P, Shen QJ. 2010. WRKY transcription factors. Trends in Plant Science 15: 247–842. [DOI] [PubMed] [Google Scholar]
- Secco D, Baumann A, Poirier Y. 2010. Characterization of the rice PHO1 gene family reveals a key role for OsPHO1;2 in phosphate homeostasis and the evolution of a distinct clade in dicotyledons. Plant Physiology 152: 1693–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secco D, Jabnoune M, Walker H, Shou HX, Wu P, Poirier Y, Whelan J. 2013. Spatio‐temporal transcript profiling of rice roots and shoots in response to phosphate starvation and recovery. Plant Cell 25: 4285–4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen JB, Yuan LX, Zhang JL, Li HG, Bai ZH, Chen XP, Zhang WF, Zhang FS. 2011. Phosphorus dynamics: from soil to plant. Plant Physiology 156: 997–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H, Shin HS, Dewbre GR, Harrison MJ. 2004. Phosphate transporter in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low‐ and high‐phosphate environments. The Plant Journal 39: 629–642. [DOI] [PubMed] [Google Scholar]
- Su T, Xu Q, Zhang FC, Chen Y, Li LQ, Wu WH, Chen YF. 2015. WRKY42 modulates phosphate homeostasis through regulation phosphate translocation and acquisition in Arabidopsis. Plant Physiology 167: 1579–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Song L, Zhang Y, Zheng Z, Liu D. 2016. Arabidopsis PHL2 and PHR1 act redundantly as the key components of the central regulatory system controlling transcriptional responses to phosphate starvation. Plant Physiology 170: 499–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun SB, Gu M, Cao Y, Huang XP, Zhang X, Ai PH, Zhao JN, Fan XR, Xu GH. 2012. A constitutive expressed phosphate transporter, OsPht1;1, modulates phosphate uptake and translocation in phosphate‐replete rice. Plant Physiology 159: 1571–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ülker B, Somssich IE. 2004. WRKY transcription factor: from DNA binding towards biologica function. Current Opinion in Plant Biology 7: 491–498. [DOI] [PubMed] [Google Scholar]
- Viana VE, Marini N, Busanello C, Pegoraro C, Fernando JA, Da Maia LC, de Oliveira AC. 2018. Regulation of rice responses to submergence by WRKY transcription factors. Biologia Plantarum 62: 551–560. [Google Scholar]
- Wang H, Xu Q, Kong YH, Chen Y, Duan JY, Wu WH, Chen YF. 2014. Arabidopsis WRKY45 transcription factor activates PHOSPHATE TRANSPORTER1;1 expression in response to phosphate starvation. Plant Physiology 164: 2020–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang PT, Xu X, Tang Z, Zhang WW, Huang XY, Zhao FJ. 2018. OsWRKY28 regulates phosphate and arsenate accumulation, root system architecture and fertility in rice. Frontier in Plant Science 9: 1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang PT, Yamaji N, Inoue K, Mochida K, Ma JF. 2020. Plastic transport systems of rice for mineral elements in response to diverse soil environmental changes. New Phytologist 226: 159–169. [DOI] [PubMed] [Google Scholar]
- Wang SK, Zhang SN, Sun CD, Xu YX, Chen Y, Yu CL, Qian Q, Jiang DA, Qi YH. 2014. Auxin response factor(OsARF12), a novel regulator for phosphate homeostasis in rice (Oryza sativa). New Phytologist 201: 91–103. [DOI] [PubMed] [Google Scholar]
- Xu XP, Chen CH, Fan BF, Chen ZX. 2006. Physical and Functional interactions between pathogen‐induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18: 1310–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji N, Xia J, Mitani‐Ueno N, Yokosho K, Ma JF. 2013. Preferential delivery of zinc to developing tissues in rice is mediated by P‐type heavy metal ATPase OsHMA2. Plant Physiology 162: 927–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SY, Lu WC, Ko SS, Sun CM, Hung JC, Chiou TJ. 2020. Upstream open reading frame and phosphate‐regulated expression of rice OsNLA1 controls phosphate transport and reproduction. Plant Physiology 182: 393–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WT, Baek D, Yun DJ, Hwang WH, Park DS, Nam MH, Chung ES, Chung YS, Yi YB, Kim DH. 2014. Overexpression of OsMYB4P, an R2R3‐type MYB transcriptional activator, increases phosphate acquisition in rice. Plant Physiology and Biochemistry 80: 259–267. [DOI] [PubMed] [Google Scholar]
- Yang ZL, Yang J, Wang Y, Wang F, Mao WX, He QJ, Xu JM, Wu ZC, Mao CZ. 2020. PROTEIN PHOSPHATASE95 regulates phosphate homeostasis by affecting phosphate transporter trafficking in rice. Plant Cell 32: 740–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q, Wang H, Su T, Wu WH, Chen YF. 2018. The ubiquitin E3 ligase PRU1 regulates WRKY6 degradation to modulate phosphate homeostasis in response to low‐Pi stress in Arabidopsis. Plant Cell 30: 1062–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue WH, Ying YH, Wang C, Zhao Y, Dong CH, Whelan J, Shou HX. 2017. OsNLA1, a RING‐type ubiquitin ligase, maintains phosphate homeostasis in Oryza sative via degradation of phosphate transporters. The Plant Journal 82: 556–569. [DOI] [PubMed] [Google Scholar]
- Zhang F, Sun YF, Pei WX, Jain A, Sun R, Cao Y, Wu XN, Jiang TT, Zhang L, Fan XR et al 2015. Involvement of OsPHT1;4 in phosphate acquisition and mobilization facilitates embryo development in rice. The Plant Journal 82: 556–569. [DOI] [PubMed] [Google Scholar]
- Zhou J, Jiao FC, Wu ZC, Li YY, Wang XM, He XW, Zhong WQ, Wu P. 2008. OsPHR2 is involved in phosphate‐starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiology 146: 1673–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu QG, Gong ZY, Huang JW, Grierson D, Chen KS, Yin XR. 2019. High‐CO2/Hyposia‐responsive transcription factors DkERF24 and DkWRKY1 interact and active DkPDC2 promoter. Plant Physiology 180: 621–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Identification of pht1;1 mutant lines.
Fig. S2 Total P concentration of pht1;1 mutants and wild‐type plants.
Fig. S3 Histochemical localisation of different truncations of PHT1;1 promoter fused with the GUS reporter gene in transgenic plants.
Fig. S4 WRKY21 binds to F2 fragment of PHT1;1 promoter through the specific W‐box.
Fig. S5 Identification of WRKY21 overexpression and mutant lines.
Fig. S6 Total P concentration of wrky21, wrky108, wrky21 wrky108 mutants and wild‐type plants.
Fig. S7 Total P concentration of WRKY21 and WRKY108 overexpression lines and wild‐type plants.
Fig. S8 Transactivation activity of the WRKY21 and WRKY108 proteins in yeast.
Fig. S9 Expression patterns and subcellular localisation of WRKY108.
Fig. S10 Identification of WRKY108 overexpression and mutant lines.
Fig. S11 WRKY108 positively affects phosphate (Pi) accumulation in rice.
Fig. S12 WRKY108 binds to PHT1;1 promoter region in vivo and in vitro.
Fig. S13 RT‐qPCR analysis of effect of WRKY21/WRKY108 mutation on expression of four PHT1 genes.
Fig. S14 Identification of wrky21 wrky108 double mutants.
Fig. S15 Pi concentration and RT‐qPCR analysis of PHT1;1 in Pro35S:WRKY108‐SRDX transgenic plants and wild‐type plants.
Fig. S16 Total P concentration of Pro35S:WRKY21‐SRDX transgenic lines and wild‐type plants.
Fig. S17 Pi concentration of pht1;8, pht1;1 pht1;8 mutants and wild‐type plants.
Fig. S18 Pi concentration of pht1;2 mutants and wild‐type plants.
Fig. S19 Phylogenetic analysis of the Group II and Group III type WRKY transcription factors from rice and A. thaliana according to Ross et al. (2007).
Fig. S20 Effect of WRKY21 and WRKY108 overexpression or mutation on the transcript level of PHO1;2 in rice.
Fig. S21 Total P concentration of wild‐type plants, pht1;1, wrky21/wrky108 single mutants, wrky21 wrky108 double mutants and WRKY21‐SRDX transgenic lines, at different Pi levels in a soil‐based experiment.
Table S1 Primers used for RT‐qPCR analysis.
Table S2 Primers used for constructs for generating transgenic plants.
Table S3 Primers used for constructs for subcellular location, Y1H, EMSA, Y2H, BiFC and pull‐down assay.
Table S4 Positive interactions from yeast one‐hybrid screening.
Table S5 Candidates from yeast two‐hybrid screening.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.


