Summary
Holobiont phenotype results from a combination of host and symbiont genotypes as well as from prevailing environmental conditions that alter the relationships among symbiotic members. Corals exemplify this concept, where shifts in the algal symbiont community can lead to some corals becoming more or less thermally tolerant. Despite linkage between coral bleaching and disease, the roles of symbiotic bacteria in holobiont resistance and susceptibility to disease remains less well understood. This study thus characterizes the microbiome of disease‐resistant and ‐susceptible Acropora cervicornis coral genotypes (hereafter referred to simply as ‘genotypes’) before and after high temperature‐mediated bleaching. We found that the intracellular bacterial parasite ‘Ca. Aquarickettsia rohweri’ was strikingly abundant in disease‐susceptible genotypes. Disease‐resistant genotypes, however, had notably more diverse and even communities, with correspondingly low abundances of ‘Ca. Aquarickettsia’. Bleaching caused a dramatic reduction of ‘Ca. Aquarickettsia’ within disease‐susceptible corals and led to an increase in bacterial community dispersion, as well as the proliferation of opportunists. Our data support the hypothesis that ‘Ca. Aquarickettsia’ species increase coral disease risk through two mechanisms: (i) the creation of host nutritional deficiencies leading to a compromised host‐symbiont state and (ii) the opening of niche space for potential pathogens during thermal stress.
Introduction
Role of the microbiome in resistance and resilience to disturbance
New studies in diverse host systems reveal the profound influence of the microbiome on animal host fitness and survival after disturbance. Beneficial microbes contribute to increased or expanded host metabolism and pathogen defence, while also occluding niche space otherwise available to opportunists (Mao‐Jones et al., 2010; Ritchie, 2011). Therefore, a diverse microbiome containing beneficial organisms may allow for optimal host response to changing environmental conditions (Zilber‐Rosenberg and Rosenberg, 2008; West et al., 2019). Rapid changes in microbial community composition in response to stressors may reflect the flexibility of the microbiome, allowing rapid adaptation to stress (Ziegler et al., 2019). In contrast, imbalances in microbiomes characterized by the loss of commensal species and an increase in opportunists are often associated with negative health consequences across host systems and are referred to as dysbiosis (Gilbert et al., 2016; Apprill, 2017). These changes in microbiome ecological state (e.g, community composition or variability) may be driven by and exacerbate stress from environmental change (Lewis et al., 2015; Zaneveld et al., 2017). It is suggested, therefore, that microbial dysbiosis may contribute to problems plaguing threatened species such as scleractinian corals and should be considered when developing strategies to preserve these foundational species (Lesser et al., 2007; West et al., 2019; Ware et al., 2020).
Disease is a major disturbance within Caribbean Acroporid corals
Infectious disease is a significant disturbance for corals in general, but especially for species within the Caribbean region including the staghorn coral Acropora cervicornis. Indeed, regional losses of Caribbean coral cover by up to 80% (Gardiner et al., 2003) are primarily attributed to the loss of these important reef‐building species because of white band disease (Aronson and Precht, 2006). Acroporids are now listed as protected under the US Endangered Species Act and critically endangered under the International Union of Conservation of Nature's (IUCN) Red List (Aronson and Precht, 2001; Acropora Biological Review Team, 2005). The continued occurrence of disease, which may be limiting population recovery (Rogers and Muller, 2012; Miller et al., 2014), has led to significant focus on disease dynamics with Caribbean Acropora spp. and the mechanisms that may lead to disease resistance.
Influence of coral genotype on disease susceptibility
The severity of stress responses that lead to disease differs with coral genotype (Drury et al., 2017; Muller et al., 2018) suggesting that the genetic composition of the host may influence disease susceptibility and resistance. Naturally disease‐resistant genotypes, observed throughout the Caribbean, may harbour potential pathogens yet do not succumb to disease‐related health reductions or mortality. This trait seems to be heritable as increases in local levels of Acropora coral population recovery have been documented (Rogers and Muller, 2012; Edmunds, 2014; Croquer et al., 2016). Potential for population recovery, combined with the continued outbreaks of disease events in these species, should theoretically lead to additional increases in community level disease resistance through repeated natural selection events. In fact, prevalence of disease‐resistant Acropora genotypes within the Western Atlantic (e.g. Florida) is notably higher (26% resistant to white band disease; Muller et al., 2018) compared with those in other parts of the Caribbean (5%–8% resistant to white band disease; Vollmer and Kline, 2008; Libro and Vollmer, 2016). Increasing frequency of thermal stress events, however, may reduce disease resistance: only two genotypes, or 13% of those tested by Muller et al. (2018), maintained a disease‐resistant phenotype after bleaching.
Influence of the microbiome on disease susceptibility
Along with heritable traits of the host, microbiomes contribute to disease susceptibility in diverse host backgrounds ranging from the human intestine (Honda and Littman, 2012) to marine mammals (Nelson et al., 2015) to shrimp (Holt et al., 2020). In Acroporid corals, the persistence of Endozoicomonas bacteria within disease‐resistant A. cervicornis populations in Panama suggests that the presence of this taxon may increase coral disease resistance (Chu and Vollmer, 2016). Histological samples of A. cervicornis from the Florida reef tract also showed that rates of disease‐associated tissue loss were correlated with the relative abundance of Rickettsiales‐like organisms (Di Lauro, 2015). Members of the Alphaproteobacterial order Rickettsiales are obligate intracellular parasites, often associated with invertebrate vectors and sometimes pathogenic to humans (Gillespie et al., 2012; Montagna et al., 2013). We previously showed that a coral‐associated Rickettsiales species, ‘Ca. Aquarickettsia rohweri’, possesses the genomic capacity to parasitize the coral holobiont for amino acids and ATP (Klinges et al., 2019). We also showed that under nutrient‐enriched conditions, these parasites proliferate and reduce Acroporid coral growth (Shaver et al., 2017) with increasing abundances positively associated with increased disease prevalence and tissue loss in other coral species (Zaneveld et al., 2016). Based on their parasitic lifestyles and capabilities to flourish in various environmental conditions, we hypothesize that the mechanism by which ‘Ca. Aquarickettsia’ may influence disease susceptibility is through the overconsumption of host and symbiont nutritional and energy resources (Klinges et al., 2019). We also suspect that under environmental conditions that are optimal for coral growth, these parasites do not generate disease phenotypes. In certain genotypic backgrounds, however, and under detrimental environmental conditions (e.g., bleaching), the dynamics of these coral parasites may influence a host genotype's disease susceptibility. But how these parasites are distributed across different host genotype backgrounds is unknown.
Influence of the environment on disease susceptibility
Host genotypes and host microbiomes alone cannot always predict disease resistance as the mechanisms that generate disease‐susceptible states are highly varied and often require environmentally mediated changes in host physiology and/or pathogen virulence (Lesser et al., 2007; Mao‐Jones et al., 2010). White band disease has been described as the result of the co‐occurrence of a permissible host genotype, a microbial community containing single or multiple pathogens, and environmental conditions conducive to infection (Bruno, 2015). Microbiome dysbiosis has been repeatedly found to occur in response to known coral stressors, such as thermal stress including bleaching, or nutrient pollution, that result in an increased disease prevalence (Thurber et al., 2009; Zaneveld et al., 2016; Ahmed et al., 2019). Therefore, dysbiosis generated by environmental triggers may shift a genotype's disease susceptibility state. But as different genotypes tend to host distinct microbial communities (Glasl et al., 2019), it has proven difficult to determine causality of this effect. One reason for this is that the propensity for microbial dysbiosis has not been well characterized across different genotypes, but these data are key to quantifying the relative contributions of host genetics and the microbiome to host disease susceptibility, especially in critically endangered species undergoing restoration efforts.
As reefs decline and coral populations dwindle towards extinction, the persistence of stress‐resistant genotypes is critical to fuel population recovery. Identifying mechanisms resulting in genotypic disease resistance, and how increasing water temperatures influence that trait, will provide insight into how corals may acclimatize and adapt to ever‐increasing threats. To uncover microbial signatures of disease resistance, we characterized the bacterial community of Acropora cervicornis genotypes that exhibited different disease susceptibility phenotypes (Muller et al., 2018). In this previous study, different genotypes exhibited variable responses to exposure to white‐band disease homogenate after a natural bleaching event. Using the same samples, we further examined correlations between microbiome community structure and disease resistance prior to and after bleaching, as bleaching and disease susceptibility are positively correlated (Muller et al., 2008; Brandt and McManus, 2009; Muller et al., 2018). The objectives of the present study were to determine (i) whether disease‐resistant genotypes possessed a significantly different microbiome compared with disease‐susceptible genotypes, (ii) how high temperature‐induced bleaching shifted the microbiome of disease‐susceptible or resistant genotypes, and (iii) assess the contribution of microbial community members including ‘Ca. Aquarickettsia’ to signatures of disease susceptibility and resistance.
Results
In the present study, we evaluated differences in the microbiomes of disease‐susceptible and disease‐resistant A. cervicornis genotypes before and after bleaching. Disease susceptibility was previously assessed for apparently healthy and bleached A. cervicornis genotypes in the study by Muller et al. (2018) using experimental exposure to homogenate of white band diseased tissue. For this study, only genotypes that had zero probability of disease after bleaching were classified as disease‐resistant, while all other genotypes were classified as susceptible (see Experimental Details).
Disease‐susceptible genotypes contained more ‘Ca. Aquarickettsia’ than disease‐resistant genotypes
The microbiomes of apparently healthy A. cervicornis disease‐susceptible and ‐resistant genotypes were differentiated by the abundance of the bacterial parasite ‘Ca. Aquarickettsia’ (Fig. 1). The genus ‘Ca. Aquarickettsia’ dominated the disease‐susceptible genotypes, with a mean relative abundance of 89.7% ± 2.1% (Supporting Information Figures S1 and S2). In contrast, the mean relative abundance of ‘Ca. Aquarickettsia’ within resistant genotypes was only 2.5% ± 0.8%. In fact, differential abundance analysis showed that ‘Ca. Aquarickettsia’ was significantly more abundant in susceptible genotypes (ANCOM, W = 25; Fig. 2A, Supporting Information Table S1). The dominance of ‘Ca. Aquarickettsia’ in the disease‐susceptible genotypes led to an uneven community that contained only a few additional rare taxa including Corynebacterium (1.0% ± 0.5%), Alteromonas (0.7% ± 0.3%), and Cellulomonas (0.7% ± 0.5%). Microbiomes of resistant genotypes were more even, with the most abundant genera, Corynebacterium, Exiguobacterium, and Staphylococcus, having mean relative abundances of 10.6% ± 7.3%, 10.3% ± 3.0%, and 9.0% ± 4.1%, respectively (Fig. 1, Supporting Information Fig. S1). Notably, Corynebacterium has been detected in sequenced negative controls (Salter et al., 2014; Rosales et al., 2019), but this genus was not detectable in our PCR negatives.
Fig. 1.
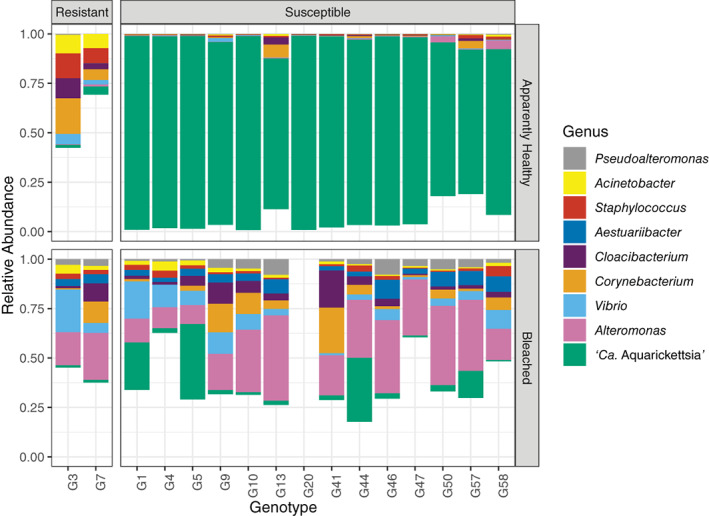
Relative abundance of the most abundant genera across different resistant and susceptible genotypes in pre‐bleached and bleached samples. Taxa are included in the plot if they had a relative abundance greater than 1% across the entire dataset. Each bar represents two replicates. Genotype 20 was not sampled during bleaching.
Fig. 2.
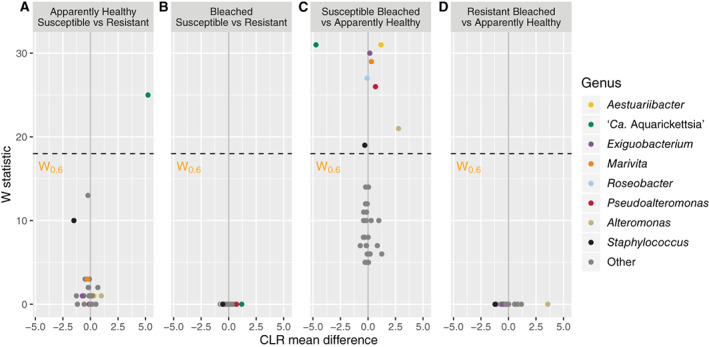
Volcano plot of results from differential abundance analysis using ANCOM. The genus‐level ASV table filtered to include taxa with a total count of 10 in at least 20% of samples (# taxa = 32) was used. ANCOM tests the null hypothesis that the average abundance of a given species in a group is equal to that in the other group. The W statistic represents the strength of the test for the 32 tested species and is the number of times the null‐hypothesis was rejected for a given species. Taxa above the dashed line are significant with the null‐hypothesis rejected 60% of the time. Non‐significant taxa in any contrast are coloured grey. The x‐axis value presents the effect size as the clr (centered log ratio) transformed mean difference in abundance of a given species between the two groups being compared. For the first panel, a positive x‐axis value means the genus is abundant in apparently healthy susceptible samples compared to apparently healthy resistant samples or vice versa for a negative x‐axis value.
‘Ca. Aquarickettsia’ was lost and opportunists increased during bleaching
In contrast to apparently healthy corals, bleached corals showed a marked reduction in the relative abundance of ‘Ca. Aquarickettsia’ regardless of disease resistance. Overall the mean relative abundance of ‘Ca. Aquarickettsia’ was only 7.0% ± 2.3% in the bleached corals. However, as with apparently healthy corals, the relative abundance of ‘Ca. Aquarickettsia’ during bleaching remained higher for disease‐susceptible genotypes (7.9% ± 2.6%) than disease‐resistant genotypes (1.0% ± 0.05%) although the difference was not significant. Instead, the dominant taxon in bleached samples was Alteromonas for both susceptible and resistant genotypes (19.0% ± 2.1% and 15.2% ± 4.9% respectively) (Fig. 1). As this taxon was only 0.7% ± 0.2% in apparently healthy individuals, bleaching thus resulted in a 26‐fold increase in this opportunist compared to apparently healthy corals in both disease categories (18.5% ± 1.9%). Other dominant taxa in bleached samples included several potential opportunists such as: Vibrio (6.1% ± 1.6%), Corynebacterium (4.4% ± 1.1%), Cloacibacterium (3.7% ± 0.9%), and Austauriibacter (3.6% ± 0.4%) (Fig. 1).
Interestingly, neither ‘Ca. Aquarickettsia’ nor any other taxa were significantly different in abundance between bleached‐susceptible and ‐resistant genotypes based on differential abundance analysis with ANCOM (Fig. 2B, Supporting Information Table S1). Instead, opportunistic taxa were most variable between apparently healthy susceptible and bleached susceptible genotypes. Of the 32 observed genera included in the ANCOM analysis, ‘Ca. Aquarickettsia’ (W = 31), Aestuariibacter (W = 31), Exiguobacterium (W = 30), Marivita (W = 29), Roseobacter (W = 27), Pseudoalteromonas (W = 26), Alteromonas (W = 21), and Staphylococcus (W = 19) showed a significant difference (P < 0.05) in abundance between apparently healthy susceptible and bleached susceptible genotypes (Fig. 2C). In contrast, bleaching did not significantly affect the abundance of bacterial taxa in microbiomes from resistant genotypes (Fig. 2D) indicating high stability and resistance to disturbance within the disease resistant genotypes. ANCOM results for all taxa included in the analysis (n = 32) can be found in the Supporting Information Figure S3.
Bleaching does not alter the structure of ‘Ca. Aquarickettsia’ populations
To determine whether different ‘Ca. Aquarickettsia’ variants dominated different disease genotypes we examined associations of ‘Ca. Aquarickettsia’ amplicon sequence variants (ASVs) with different genotypes and assessed the phylogenetic relationship of the variants present. We identified a total of 12 ASVs within the genus ‘Ca. Aquarickettsia’ that shared species‐level homology with ‘Ca. A. rohweri’ (ranging from 98.97% to 99.66% identity). However, strain‐level variation did not demonstrate clear partitioning patterns by host genotype (Fig. 3), though there were clear phylogenetic differences between the most abundant variants (Fig. 3 and Supporting Information Fig. S4). The most abundant of these ASVs (ASV 1, Fig. 3) constituted a mean 19.18% ± 2.00% of all ‘Ca. A. rohweri’ sequences among apparently healthy disease‐susceptible samples, while the second most abundant (ASV 2) averaged 14.61% ± 2.09% in these samples. Although these ASV abundances were considerably lower in disease‐resistant compared to disease‐susceptible genotypes, ASV 1 was still the most abundant of all ‘Ca. Aquarickettsia’ ASVs found in disease‐resistant samples (56.38% ± 37.11%). After bleaching, these two ASVs increased slightly in their relative proportion of the ‘Ca. A. rohweri’ community in disease‐susceptible genotypes, constituting an average of 23.56% ± 12.41% and 17.66% ± 9.44% of ‘Ca. A. rohweri’ reads, respectively, although total relative abundance of ‘Ca. A. rohweri’ declined dramatically in response to bleaching. All other ASVs each averaged less than 10% of the ‘Ca. Aquarickettsia’ community before and after bleaching.
Fig. 3.
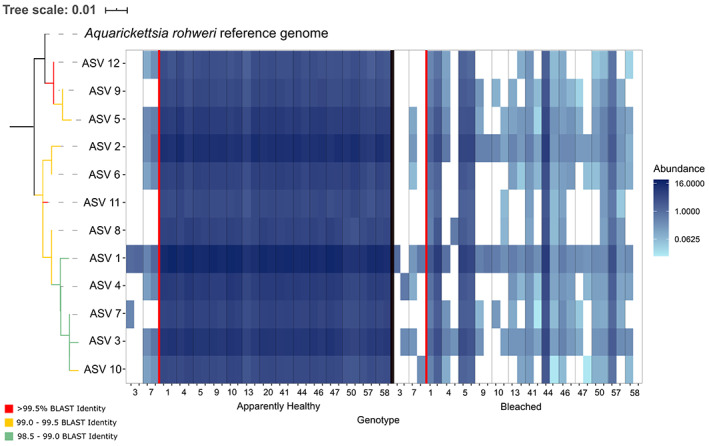
Relative abundance of ASVs from the genus ‘Ca. Aquarickettsia’ across all genotypes in August (apparently healthy state) and September 2015 (bleached state). ASVs are numbered according to their overall abundance in the dataset, and phylogenetic relationships are plotted on the left. Red line separates disease‐resistant and susceptible genotypes. BLASTn percent identity to the 16S rRNA sequence of ‘Ca. Aquarickettsia rohweri’ (NCBI accession number MT544612.1) is plotted as coloured branches on the phylogenetic tree.
Disease susceptibility classification and bleaching state alter patterns in microbiome alpha diversity
Overall, disease‐resistant A. cervicornis genotypes had a significantly greater Simpson's diversity (0.98 ± 0.002) than disease‐susceptible genotypes (0.94 ± 0.005; P < 0.05, χ² = 5.313, Supporting Information Table S2). Bleaching also significantly increased Simpson's diversity (0.98 ± 0.001) compared with apparently healthy corals (0.92 ± 0.005; P < 0.001, χ² = 36.923). The interaction between resistance groupings and bleaching status was also significant (P < 0.001, χ² = 44.271) and pairwise comparisons showed that apparently healthy disease‐susceptible genotypes were significantly less diverse than all other groups (P < 0.001, Fig. 4A).
Fig. 4.
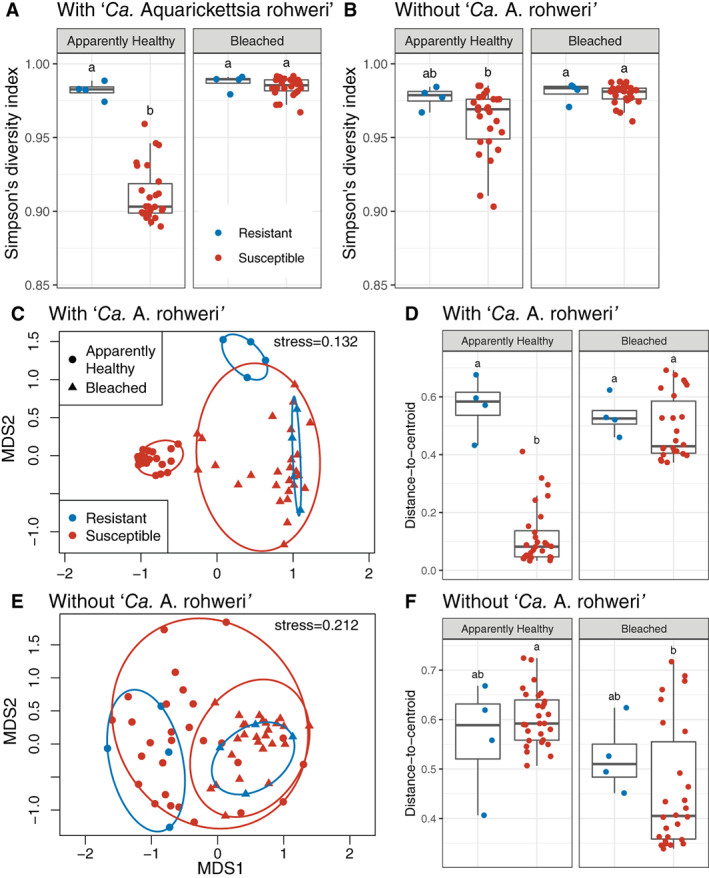
Community diversity metrics by genotype (resistant vs susceptible) and health state (apparently healthy vs bleaching) for microbiomes with and without the dominant taxon ‘Ca. Aquarickettsia rohweri’.
A and B. Shannon's diversity index.
C and E. NMDS ordinations of samples using Bray–Curtis distance.
D and F. Distance‐to‐centroid measures. Note that detection of significant differences between disease‐susceptible and disease‐resistant genotypes may be influenced by the difference in replication (n = 4 versus n = 26, respectively) causing disparate dispersion between the two groups (Anderson and Walsh, 2013). See Supporting Information Tables S3 and S4. Boxes sharing a letter are not significantly different from each other using an FDR corrected significance level of P < 0.05.
To evaluate the effect of the dominant taxon ‘Ca. Aquarickettsia’ on alpha diversity dynamics, we repeated the analyses with ASVs from this taxon removed from the community. After removal, the Simpson's diversity index of disease‐resistant genotypes was not significantly different from that of disease‐susceptible genotypes (P = 0.171, χ² = 1.877), although bleaching still significantly increased Simpson's diversity (P < 0.001, χ² = 15.292, Supporting Information Table S2). The interaction between resistance groupings and bleaching status remained significant (P < 0.001, χ² = 17.582), however, apparently healthy susceptible genotypes were only different from bleached susceptible genotypes (P < 0.001). After in silico removal of ‘Ca. Aquarickettsia’, the Simpson's diversity index of apparently healthy susceptible genotypes was no longer different from apparently healthy resistant genotypes (Fig. 4B).
Bleaching homogenized resistant and susceptible microbiomes
Apparently healthy disease‐susceptible microbiomes were less variable than, and compositionally distinct from, all other microbiomes. Beta diversity analysis showed that disease‐susceptibility classification (PERMANOVA; P < 0.001, R 2 = 0.079), bleaching condition (P < 0.001, R 2 = 0.391), and their interaction (P < 0.001, R 2 = 0.055) produced distinct communities of microbes (Fig. 4C, Supporting Information Table S3). Pairwise comparisons showed that all combinations were significantly different from each other (P < 0.05), except for bleached disease‐susceptible and bleached disease‐resistant genotypes (P = 0.628).
Tests for differences in group dispersion or sample‐to‐sample variation showed that while disease susceptibility did not affect dispersion (PERMDISP; P = 0.287, F = 1.153), dispersion significantly increased when corals were bleached (P < 0.001, F = 22.143, Supporting Information Table S4). The interaction between disease susceptibility groups and bleaching status was a significant predictor of group dispersions (P < 0.001, F = 73.187) with pairwise comparisons showing that apparently healthy disease‐susceptible genotypes were less variable than all other genotypes (P < 0.01) (Fig. 4D). As unbalanced designs are known to bias PERMANOVA results in the presence of heterogeneity of dispersion (Anderson and Walsh 2013), we verified these results with 1000 permutations of down‐sampling to a balanced design (Supporting Information Tables S3 and S4).
Apparently healthy disease‐susceptible and disease‐resistant microbiomes were also more similar in sample‐to‐sample variability and composition when ‘Ca. Aquarickettsia’ was removed from the analysis. Overall, disease‐susceptible and disease‐resistant genotypes were no longer different after removal of ‘Ca. Aquarickettsia’ sequences (PERMANOVA; P = 0.314, R 2 = 0.015, Supporting Information Table S3). And although bleaching still produced distinct communities (P < 0.001, R 2 = 0.132), the interaction between bleaching and disease‐susceptibility was no longer significant (P = 0.514, R 2 = 0.014) (Fig. 4E). Pairwise comparisons showed that apparently healthy disease‐susceptible and disease resistant were not different for each other (P = 0.336) in addition to bleached disease‐susceptible and disease‐resistant genotypes (P = 0.550). Likewise, when ‘Ca. Aquarickettsia’ was removed, disease‐susceptibility had no effect on dispersion (PERMDISP; P = 0.627, F = 0.239), while bleaching significantly reduced group dispersions (P < 0.001, F = 32.916, Supporting Information Table S4). The interaction between disease‐susceptible groups and bleaching status was a significant predictor of dispersion (P < 0.001, F = 10.194), and pairwise comparisons showed that for disease‐susceptible genotypes, bleached samples were less variable than apparently healthy samples when ‘Ca. Aquarickettsia’ was removed from the data (Fig. 4F).
Discussion
Coral reef ecosystems continue to be significantly altered by disease epizootics, but why some host populations or even individuals remain resistant while others succumb to outbreaks remains unknown. Previously, Muller et al. (2008) showed that corals susceptible to temperature‐induced bleaching were more likely to suffer from disease‐related mortality. More recently, however, Muller et al. (2018) documented that even after bleaching, some genotypes of A. cervicornis maintained high levels of disease resistance. This variation in disease susceptibility even after bleaching may have resulted from differences in coral host immune responses, holobiont structure, or a combination of the two factors. To explore the effect of holobiont structure, we examined differences in microbial community composition across 15 genotypes of A. cervicornis both before and after a 2015 natural thermal stress event that induced bleaching. To account for unbalanced distribution of samples across disease response phenotypes due to the rarity of disease resistance, we verified statistical analyses by down‐sampling to a balanced design. We found that coral genotypes phenotypically characterized as disease‐susceptible had a distinctive microbial community signature characterized by low diversity, low evenness, and a strikingly high abundance of the genus ‘Ca. Aquarickettsia’. This parasitic genus was roughly 36‐fold more relatively abundant in disease‐susceptible genotypes than in resistant genotypes. After bleaching, however, the microbiomes of these disease‐susceptible genotypes were no longer distinct from resistant genotypes as the microbiomes of all corals became similarly heterogeneous in composition as measured by beta diversity. Importantly, however, in disease‐susceptible genotypes, these shifts were characterized by a decrease in ‘Ca. Aquarickettsia’ and an increase in the relative abundance of a variety of opportunists. Our findings suggest then that ‘Ca. Aquarickettsia’ plays a critical role in the susceptibility of A. cervicornis to white band disease, but that disease resistance is complex and may be influenced both by microbiome history and host factors external to the microbiome, such as genetic capacity to combat disease and the role the host may play on shaping the microbiome.
‘Ca. Aquarickettsia’ sp. dominate the microbiome of disease susceptible A. cervicornis genotypes prior to bleaching
Based on its genomic composition and cosmopolitan distribution, we previously showed that ‘Ca. A. rohweri’ is a ubiquitous parasite of diverse coral hosts that can translocate both amino acids and ATP from the holobiont (Klinges et al., 2019; Fig. 5A). We also found that members of this genus proliferate during nutrient enrichment and their abundance is strongly correlated with reduced coral growth (Shaver et al., 2017) and increased tissue loss and mortality (Zaneveld et al., 2016). Although ‘Ca. A. rohweri’ is highly abundant in disease‐susceptible A. cervicornis genotypes, the presence of this organism does not appear to be an early symptom of disease, as only corals exposed to a white band disease homogenate developed symptoms while controls remained visually healthy (Muller et al., 2018). Furthermore, this species has been found globally in samples of healthy coral spanning diverse genera (Klinges et al., 2019). The considerable dominance of a few ‘Ca. A. rohweri’ variants is apparent as samples of disease‐susceptible genotypes were markedly less diverse than samples of resistant genotypes. There was no apparent association of specific variants of ‘Ca. A. rohweri’ with particular genotypes, but rather a few variants dominated across all samples. The presence of abundant Rickettsiales in apparently healthy A. cervicornis from Florida has been observed in multiple previous studies (Di Lauro, 2015; Rosales et al., 2019, Gignoux‐Wolfsohn et al., 2020), but interestingly, samples of A. cervicornis from Panama are instead dominated by Endozoicomonadaceae (Gignoux‐Wolfsohn et al., 2017). The absence of Endozoicomonadaceae in Floridian individuals in this study and others (Rosales et al., 2019; Gignoux‐Wolfsohn et al., 2020) may be indicative of a compromised health state of these corals compared to Panamanian populations, as Endozoicomonas are proposed to be commensal or beneficial symbionts of corals (Neave et al., 2016).
Fig. 5.
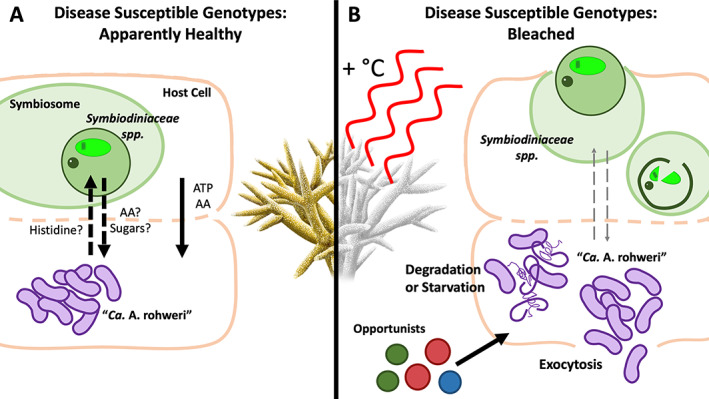
Model of predicted impacts of heat stress on Symbiodiniaceae and ‘Ca. Aquarickettsia rohweri’ interactions in Acropora cervicornis. Intracellular ‘Ca. Aquarickettsia’ sp. acquire amino acids (AA) and ATP either from Symbiodiniaceae or from the coral host, or both. Decreased metabolic activity of the coral and algal symbiont during bleaching could lead to the starvation of ‘Ca. Aquarickettsia’ populations, as there are fewer available resources to parasitize and the bacterium possesses sparse metabolic capabilities of its own. Due to the close colocalization of ‘Ca. Aquarickettsia’ with Symbiodiniaceae, these two organisms could also be lost simultaneously via host mechanisms targeted at the symbiont, inducing expulsion and degradation. AA amino acids, ATP adenosine triphosphate.
‘Ca. Aquarickettsia’ populations decline in response to bleaching but predispose corals to disease
As ‘Ca. Aquarickettsia’ species lack the ability to produce most essential amino acids and thus rely heavily on the coral host, Symbiodiniaceae, or both, we predicted that ‘Ca. Aquarickettsia’ would be negatively affected by elevated ocean temperatures as host and symbiont metabolic activity are dramatically reduced during bleaching (Weis, 2008, Fig. 5). ‘Ca. Aquarickettsia’ mean abundances indeed declined dramatically in disease‐susceptible genotypes during bleaching to the extent that they no longer differed in relative abundance compared to the disease‐resistant genotypes (Fig. 1). The loss of microbiome distinctiveness between disease‐susceptibility groups after bleaching suggests that bleaching eliminates the microbiome signatures of disease‐resistance by homogenizing the microbiomes of these two groups. Bleaching did, however, increase community dispersion, upholding results from previous studies which suggested that stressors induce microbiome instability (Zaneveld et al., 2017; Maher et al., 2019). However, this effect was reversed with the removal of ‘Ca. Aquarickettsia’ from the dataset (Fig. 4). These results suggest that the variation in ‘Ca. Aquarickettsia’ abundance among disease‐susceptible genotypes, rather than variation in minor taxa, affected dispersion. Furthermore, the homogenization of the communities after bleaching suggests that in this scenario, ‘Ca. Aquarickettsia’ is likely not a primary pathogen because it was no longer in higher abundances after bleaching when disease emerged in these susceptible genotypes. Nevertheless, the rapid proliferation or maintenance of large abundances of ‘Ca. Aquarickettsia’ may predispose hosts to increased disease susceptibility via overconsumption of holobiont resources prior to the bleaching event, making it the trigger of the disease instead of the sole pathogen.
Mechanisms affecting ‘Ca. Aquarickettsia’ abundance in apparently healthy and bleached corals
Bleaching results in a breakdown of several aspects of symbiosis in the coral holobiont. During bleaching, less carbon production and transfer occurs between the algal symbiont and the host, resulting in temporary starvation of the holobiont unless coral heterotrophy is significantly increased. The production of reactive oxygen species by the symbiont and the subsequent diffusion of these free radicals into host tissue initiates a cascade of events ultimately leading to the loss of Symbiodiniaceae (Weis, 2008; Lesser, 2011). ‘Ca. Aquarickettsia’ and Symbiodiniaceae observed in fluorescent in situ hybridization imagery of A. cervicornis show colocalization of both microbes within the gastrodermis (Klinges et al., 2019), suggesting the possibility for the physical loss of ‘Ca. Aquarickettsia’ simultaneously with Symbiodiniaceae. Alternately, as ‘Ca. Aquarickettsia’ sp. are believed to be nutritionally dependent on the holobiont, coral bleaching may directly lead to the starvation and population decline of these parasites (Fig. 5).
Members of ‘Ca. Aquarickettsia’ sp. possess few genes involved in carbon or nitrogen metabolism or for the synthesis of amino acids, suggesting that like other members of Rickettsiales, they are dependent on their host for nutrients and metabolic byproducts (Klinges et al., 2019). While their acroporid hosts are unable to synthesize a full set of amino acids, Symbiodiniaceae synthesize all essential amino acids, including mycosporine‐like amino acids (Yellowlees et al., 2008; Rosic and Dove, 2011). We therefore hypothesize that ‘Ca. Aquarickettsia’ sp. may depend on the algal symbiont either by directly or by parasitizing amino acids from Acropora sp. after acquisition from Symbiodiniaceae (Fig. 5A). The loss of Symbiodiniaceae during bleaching would thus negatively affect ‘Ca. Aquarickettsia’ sp. and could contribute to a significant decline in these species after a thermal event (Fig. 5B). An alternate cause for significant loss of ‘Ca. Aquarickettsia’ sp. during heat stress could be the physical loss of ‘Ca. Aquarickettsia’ cells due to Symbiodiniaceae expulsion and degradation (Fig. 5B). Studies in Aiptasia pulchella and in corals have documented both the exocytosis of Symbiodiniaceae and the detachment of necrotic host cells containing Symbiodiniaceae during bleaching (Brown et al., 1995; Weis, 2008). As ‘Ca. Aquarickettsia’ were previously found in close proximity to Symbiodiniaceae in host tissue imagery (Klinges et al., 2019), they may be simultaneously eliminated in the same manner. ‘Ca. Aquarickettsia’ were previously detected in water samples despite their obligate parasitic lifestyle, supporting the hypothesis that they may be released into the water column after bleaching (Klinges et al., 2019).
Dysbiosis in disease‐susceptible genotypes after loss of ‘Ca. Aquarickettsia’ rohweri
While single‐taxon dominance is common in coral microbiomes, as the genus Endozoicomonas often exceeds 50% of the coral microbial community (Bayer et al., 2013; Pogoreutz et al., 2018; Maher et al., 2019), low diversity and single‐species dominance has been linked to disease in human systems (Kriss et al., 2018). Compounding this, long‐term dominance of a parasite likely has additional detrimental effects on host fitness as discussed above. Loss of this dominant microbial community member observed during heat stress may allow for the invasion of opportunists or primary pathogens (Fig. 5B). In fact, heat stress increased alpha diversity, consistent with previous results (McDevitt‐Irwin et al., 2017), but a significant change in individual community members with bleaching only occurred in disease‐susceptible genotypes (Figs. 2 and 3).
It was previously proposed that coral diseases may be the result of dysbiosis in the form of unchecked growth of normally commensal bacterial species triggered by changes in the environment such as increases in water temperature (Lesser et al., 2007; Muller and van Woesik 2012). Heat stress led to a sudden expansion of minor community members in disease‐susceptible genotypes, with members of the order Alteromonadales and other potentially opportunistic genera significantly increasing in these samples. While these genera were also found in disease‐resistant samples, their change in abundance with bleaching was insignificant (Fig. 2), suggesting these genotypes may harbour opportunistic species even when in a healthy state. Although not included in the present study, control samples during DNA extraction would be helpful in verifying the significance of minor or rare community members.
Members of Alteromonadales have been found to increase in stressed or diseased coral microbiomes compared to healthy states, and at high levels may be pathogenic to corals (Sunagawa et al., 2009; Gignoux‐Wolfsohn and Vollmer, 2015). Exiguobacterium and Marivita may also be opportunistic pathogens: Exiguobacterium was found in White Syndrome lesions in Pacific Acropora species (Krediet et al., 2013) and Marivita was found in corals exposed to heat stress (Pootakham et al., 2019). A similar increase in opportunists in A. cervicornis exposed to heat stress was observed by Gignoux‐Wolfsohn et al. (2020), who observed lower abundance of Rickettsiales species in white band diseased tissue compared to apparently healthy tissue, and a correspondingly higher abundance of Alteromonadales species. The significant and sudden increase of previously minor microbial community members after bleaching may be an indicator of dysbiosis and may have contributed to increased disease susceptibility of these genotypes (Muller et al. 2018).
Diverse microbiomes may contribute to disease resistance
Samples of apparently healthy disease‐resistant genotypes possessed a diverse, even microbiome with no single member exceeding 11% of the community. While high microbial diversity in itself is not necessarily an indicator of host resilience, as disease‐susceptible genotypes increased in diversity after bleaching, higher diversity during non‐stressful conditions may provide a greater arsenal of antimicrobial defences provided by the microbial community itself, and a coral host that has encountered many different types of bacteria may be better equipped to combat future infections (Zilber‐Rosenberg and Rosenberg, 2008; West et al., 2019). A diverse, evenly distributed microbial community may also occupy the majority of available niches and exclude opportunistic parasites such as ‘Ca. Aquarickettsia’ sp. or other pathogens involved in disease. Furthermore, considering the demand a high level of parasitic Rickettsiales infection may pose upon the immune system of its host (Gillespie et al., 2012), it is likely that the resistant genotypes (with low abundance of ‘Ca. A. rohweri’) are better equipped to deal with the additional burden of exposure to a disease homogenate. The greater resistance of genotypes 7 and 3 to white band disease compared to other genotypes may then be due to a combination of these factors.
‘Ca. Aquarickettsia’ abundance as a metric of host potential for resilience
The role of coral‐associated Rickettsiales as an agent of coral disease has been debated as these species were identified in corals suffering from white band disease (type I) but are also highly prevalent community members in apparently healthy corals (Fig. 1, Casas et al., 2004; Miller et al., 2014; Gignoux‐Wolfsohn et al., 2020). While ‘Ca. Aquarickettsia’ infections are associated with reduced coral growth (Shaver et al., 2017), some antibiotics that effectively treated white band disease are ineffective against intracellular pathogens (Kline and Vollmer, 2011; Sweet et al., 2014). As species of Vibrio were also identified as putative white band disease pathogens, it has been suggested that disease signs may be a product of a bacterial consortium instead of a single pathogen (Kline and Vollmer, 2011; Gignoux‐Wolfsohn and Vollmer, 2015; Klinges et al., 2019). Genomic evidence suggests ‘Ca. Aquarickettsia’ sp. possess parasitic capabilities and in vivo data suggest that these species detrimentally affect their hosts when present at high levels (Shaver et al., 2017). Chronic ‘Ca. Aquarickettsia’ infections in otherwise healthy corals may increase susceptibility to opportunistic infections. In bleached corals, ‘Ca. Aquarickettsia’ loss may open up available niche space for the proliferation of pathogens. Regardless of bleaching status, the diverse microbiome of disease‐resistant genotypes could provide protection against opportunistic pathogens in the form of antibacterial defences in addition to reducing available niche space for invaders. As disease‐resistant A. cervicornis genotypes are rare, with only two genotypes thus far shown to maintain resistance during thermal stress, the application of robust quantitative metrics such as qPCR to quickly characterize disease resilience potential of other genotypes is essential.
Conclusion: ‘Ca. Aquarickettsia’ abundance as a metric of host potential for disease susceptibility
Acropora cervicornis has declined precipitously over the last 50 years and is now the primary species used for large‐scale coral restoration efforts throughout the Florida reef tract. However, disturbances such as chronic disease events limit long‐term population recovery (Miller et al., 2014; Ware et al., 2020). As bleaching events become increasingly frequent worldwide, the extent of coral disease may also increase as bleaching reduces disease resistance (Muller et al., 2018). The results of the present study, however, suggest there may be a microbial signature to identify disease‐resistant genotypes (i.e., low abundances of ‘Ca. Aquarickettsia’ species). The presence of these parasites leads to nutritional deficiencies in both the coral host and the algal symbiont, contributing to a compromised health state of the holobiont. Long‐term parasitism by these species may have chronic effects, and based on the results of this study ‘Ca. Aquarickettsia’ are associated with increased disease risk after bleaching. This potential ‘biomarker’ could be integrated into the decision‐making framework associated with coral restoration initiatives and is a potential intervention tool to increase the resilience of restored populations.
Experimental procedures
In August 2015, two replicate fragments from 16 genotypes of Acropora cervicornis were collected from the Mote Marine Laboratory in situ coral nursery. In September, another set of two fragments from 15 of the same genotypes were collected (one genotype, G20, was not available due to high mortality rates). By September, the nursery corals had been experiencing anomalously high water temperatures reaching approximately ~2°C above historical averages. For further details, see Supplementary Methods SI and Muller et al. (2018).
All samples were flash frozen in liquid nitrogen and stored in at −80°C until processing. A total of two polyps were scraped from each replicate using a sterile razor blade, and total DNA was extracted from polyp tissue using the MoBio Powersoil kit. The microbiome of each sample was assessed using 16S rRNA Illumina sequencing on the MiSeq platform. Amplification of the 16S rRNA gene was conducted using the 515F‐806R primer set, which targets the V4 region of the 16S rRNA, with barcodes on the forward primer (Apprill et al., 2015). Polymerase chain reaction (PCR) and library preparation protocols are detailed in the Supporting Information Methods SI. A PCR negative control was included in library preparation but did not produce a viable library. Paired‐end sequencing was performed at MR DNA (www.mrdnalab.com, Shallowater, TX, USA) using a single flow cell on a MiSeq following the manufacturer's guidelines.
Demultiplexing and barcode removal was performed using sabre (v1.0) (Copyright© Nikhil Joshi, UC Davis), during which reads with no barcode match were discarded from the initial read pool (Supporting Information Table S5). Reads were subsequently processed using DADA2 (v1.16) (Callahan et al., 2016) in R (v1.1.383) (R Development Core Team, 2017). Supporting Information Table S5 presents reads lost during quality control, performed using parameters detailed in the Supporting Information. After quality control, amplicon sequence variants (ASVs) were inferred from unique reads and paired‐end reads were subsequently merged. Two‐parent chimeras (bimeras) were removed and taxonomy was assigned at 100% sequence identity using the Silva reference database (v132) (Quast et al., 2012). The resulting ASV table contained 2979 unique ASVs across 62 samples and was imported into phyloseq (v1.30.0) (McMurdie and Holmes, 2013). ASVs that were annotated as mitochondrial or chloroplast sequences as well as rare ASVs (total counts in the bottom quartile) were removed resulting in a total of 2133 unique ASVs (Supporting Information Table S5).
Alpha and beta diversity metrics were analysed with and without the dominant taxon, genus ‘Ca. Aquarickettsia’, at rarefaction levels of 8663 and 823 reads respectively. Genotype 20 was not sampled during bleaching and was therefore not included in statistical analyses. Differences observed in Simpson's diversity index (Heip et al., 2001) between sample types were tested with the Kruskal‐Wallis chi‐squared test and the Pairwise Wilcoxon Rank Sum Test with FDR correction for multiple testing. Using Bray–Curtis distances, permutational multivariate analysis of variance (PERMANOVA; Anderson, 2001) and permutational analysis of multivariate dispersions (PERMDISP; Anderson, 2006) were conducted to test for differences in the beta diversity with the package vegan (v2.5.5) (Oksanen et al., 2019). Due to the potential bias of an unbalanced design with these tests (Anderson & Walsh, 2013), we performed a down‐sampling procedure with 1000 permutations to verify our results (Supporting Information SI). Statistical analyses determined differences among groups including resistance to white band disease (resistant/susceptible), bleaching status (apparently healthy/bleached) and the interaction of the two groups. Metadata is included in the Supporting Information Table S6. An analysis of composition of microbiomes (ANCOM) (Mandal et al., 2015) was performed to identify key genera discriminating the microbiomes of susceptible and resistant Acropora genotypes before and during bleaching using an unrarefied ASV table summarized to the genus level and taxa with at least 10 counts in 20% of samples. A 16S rRNA phylogeny of ASVs within ‘Ca. Aquarickettsia’ was produced by aligning ASV sequences with six other described members of the genus including the type species ‘Ca. A. rohweri’ using cmalign (part of the INFERNAL 1.1.1 package, Nawrocki and Eddy, 2013). PhyML v3.1 (Guindon et al., 2010) was used for phylogenetic analysis, with parameters selected using jModelTest 2 v0.1.10 (Supporting Information Table S7) (Darriba et al., 2012).
Author Contributions
E.M. designed and performed the bleaching experiment, R.L.M. and J.G.K. performed analyses, and all authors contributed to data interpretation and writing of the manuscrip
Supporting information
Appendix S1: Supplementary Methods.
Figure S1 Supplementary Figures
Table S1 Results from ANCOM analysis for each of four pairwise contrasts. An unrarefied ASV table was first summarized to the genus level, then was filtered to contain only taxa with at least 10 counts in 20% of the samples. A significance level at W = 0.6 was used in which the null hypothesis for a given taxon was rejected in 60% of the tests and p‐values were corrected with Benjamini‐Hochberg FDR. MD3‐55 and HIMB11 are the genus identifications in Silva for Ca. Aquarickettsia and Roseobacter, respectively.
Table S2 Kruskal Wallis tests for differences in Simpson's diversity with and without Ca. Aquarickettsia rohwerii sequences with pairwise comparisons
Table S3 Results of PERMANOVA tests for differences between groups with and without Ca. Aquarickettsia rohwerii sequences and with pairwise comparisons. Perm column presents the percentage of 1000 tests that produced a p < 0.05 after permutational down‐sampling.
Table S4 Results of PERMDISP tests for differences in group dispersion with and without Ca. Aquarickettsia rohwerii Sequences. Perm column presents the percentage of 1000 tests that produced a p < 0.05 after permutational down‐sampling.
Table S5 Reads and corresponding ASVs lost through quality control processing in sabre, DADA2 and Phyloseq.
Table S6 Sample metadata with sequencing depth after removal of rare ASVs and ASVs annotating to mitochondria or chloroplasts.
Acknowledgements
This work was funded by an NSF CAREER award to E.M. (#1452538‐OCE), an NSF Biological Oceanography grant to R.V.T. and E.M. (#1923836) as well as NSF Graduate Fellowships to both J.G.K. (#1840998‐DGE) and R.L.M. (#1314109‐DGE). The authors would also like to thank Caitlin Lustic (The Nature Conservancy) for authorizing the use of nursery grown staghorn corals under permit #FKNMS‐2011‐150‐A3 and Erich Bartels (Mote Marine Laboratory) for propagating and providing the corals for research.
Data accessibility
Raw sequence data were deposited into the NCBI Sequence Read Archive (SRA) under accession number PRJNA639601. All scripts involved in the preparation and analysis of this dataset are available at https://github.com/maherrl/Aquarickettsia_bleaching_dynamics.
References
- Acropora Biological Review Team . Atlantic Acropora Status Review Document. Report to National Marine Fisheries Service, Southeast Regional Office, (2005).
- Ahmed, H.I. , Herrera, M. , Liew, Y.J. , and Aranda, M. (2019) Long‐term temperature stress in the coral model Aiptasia supports the “Anna Karenina principle” for bacterial microbiomes. Front Microbiol 10: (Article 975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, M.J. , and Walsh, D.C.I. (2013). PERMANOVA, ANOSIM, and the Mantel test in the face of heterogeneous dispersions: What null hypothesis are you testing?. Ecological Monographs 83: 557–574. 10.1890/12-2010.1 [DOI] [Google Scholar]
- Anderson, M.J. (2001) A new method for non‐parametric multivariate analysis of variance. Austral Ecol 26: 32–46. [Google Scholar]
- Anderson, M.J. (2006) Distance‐based tests for homogeneity of multivariate dispersions. Biometrics 62: 245–253. [DOI] [PubMed] [Google Scholar]
- Apprill, A. , McNally, S. , Parsons, R. , and Weber, L. (2015) Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat Microb Ecol 75: 129–137. [Google Scholar]
- Apprill, A. (2017) Marine animal microbiomes: toward understanding host–microbiome interactions in a changing ocean. Front Mar Sci 4: (Article 222). [Google Scholar]
- Aronson, R.B. , and Precht, W.F. (2001) White‐band disease and the changing face of Caribbean coral reefs In The Ecology and Etiology of Newly Emerging Marine Diseases. Dordrecht: Springer, pp. 25–38. 10.1007/978-94-017-3284-0_2. [DOI] [Google Scholar]
- Aronson, R.B. , and Precht, W.F. (2006) Conservation, precaution, and Caribbean reefs. Coral Reefs 25: 441–450. [Google Scholar]
- Bayer, T. , Neave, M.J. , Alsheikh‐Hussain, A. , Aranda, M. , Yum, L.K. , Mincer, T. , et al (2013) The microbiome of the Red Sea coral Stylophora pistillata is dominated by tissue‐associated Endozoicomonas bacteria. Appl Environ Microbiol 79: 4759–4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt, M.E. , and McManus, J.W. (2009) Disease incidence is related to bleaching extent in reef‐building corals. Ecology 90: 2859–2867. [DOI] [PubMed] [Google Scholar]
- Brown, B. , Le Tissier, M. , and Bythell, J. (1995) Mechanisms of bleaching deduced from histological studies of reef corals sampled during a natural bleaching event. Mar Biol 122: 655–663. [Google Scholar]
- Bruno, J.F. (2015) The coral disease triangle. Nat Clim Change 5: 302–303. [Google Scholar]
- Callahan, B.J. , McMurdie, P.J. , Rosen, M.J. , Han, A.W. , Johnson, A.J.A. , and Holmes, S.P. (2016) DADA2: high‐resolution sample inference from Illumina amplicon data. Nat Methods 13: 581–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas, V. , Kline, D.I. , Wegley, L. , Yu, Y. , Breitbart, M. , and Rohwer, F. (2004) Widespread association of a Rickettsiales‐like bacterium with reef‐building corals. Environ Microbiol 6: 1137–1148. [DOI] [PubMed] [Google Scholar]
- Chu, N.D. , and Vollmer, S.V. (2016) Caribbean corals house shared and host‐specific microbial symbionts over time and space. Environ Microbiol Rep 8: 493–500. [DOI] [PubMed] [Google Scholar]
- Croquer, A. , Cavada‐Blanco, F. , Zubillaga, A.L. , Agudo‐Adriani, E.A. , and Sweet, M. (2016) Is Acropora palmata recovering? A case study in los Roques national park Venezuela. PeerJ 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba, D. , Taboada, G.L. , Doallo, R. , and Posada, D. (2012) jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9: 772–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lauro, S. (2015) Time‐series evaluation of suspect rickettsiales‐like bacteria presence in Acropora cervicornis off of Broward County from years 2001–2012. MSc Thesis. Fort Lauderdale, FL: Nova Southeastern University.
- Drury, C. , Manzello, D. , and Lirman, D. (2017) Genotype and local environment dynamically influence growth, disturbance response and survivorship in the threatened coral, Acropora cervicornis . PLoS One 12: e0174000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmunds, P.J. (2014). Large numbers of Acropora palmata grow in shallow water in St. John, US Virgin Islands. Bulletin of Marine Science, 90: 999–1000. 10.5343/bms.2014.1027 [DOI] [Google Scholar]
- Gardner, T.A. , Côté, I.M. , Gill, J.A. , Grant, A. , and Watkinson, A.R. (2003) Long‐term region‐wide declines in Caribbean corals. Science 301: 958–960. [DOI] [PubMed] [Google Scholar]
- Gignoux‐Wolfsohn, S.A. , Aronson, F.M. , and Vollmer, S.V. (2017). Complex interactions between potentially pathogenic, opportunistic, and resident bacteria emerge during infection on a reef‐building coral. FEMS Microbiol Ecol, 93: 10.1093/femsec/fix080 [DOI] [PubMed] [Google Scholar]
- Gignoux‐Wolfsohn, S. , Precht, W. , Peters, E. , Gintert, B. , and Kaufman, L. (2020) Ecology, histopathology, and microbial ecology of a white‐band disease outbreak in the threatened staghorn coral Acropora cervicornis . Dis Aquat Organ 137: 217–237. [DOI] [PubMed] [Google Scholar]
- Gignoux‐Wolfsohn, S.A. , and Vollmer, S.V. (2015) Identification of candidate coral pathogens on White band disease‐infected Staghorn coral. PLOS ONE 10: e0134416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, J.A. , Quinn, R.A. , Debelius, J. , Xu, Z.Z. , Morton, J. , Garg, N. , et al (2016) Microbiome‐wide association studies link dynamic microbial consortia to disease. Nature 535: 94–103. [DOI] [PubMed] [Google Scholar]
- Gillespie, J.J. , Joardar, V. , Williams, K.P. , Driscoll, T. , Hostetler, J.B. , Nordberg, E. , et al (2012) A Rickettsia genome overrun by mobile genetic elements provides insight into the acquisition of genes characteristic of an obligate intracellular lifestyle. J Bacteriol 194: 376–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasl, B. , Smith, C.E. , Bourne, D.G. , and Webster, N.S. (2019) Disentangling the effect of host‐genotype and environment on the microbiome of the coral Acropora tenuis . PeerJ 7: e6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon, S. , Dufayard, J.F. , Lefort, V. , Anisimova, M. , Hordijk, W. , and Gascuel, O. (2010) New algorithms and methods to estimate maximum‐likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59: 307–321. [DOI] [PubMed] [Google Scholar]
- Heip, C.H.R. , Herman, P.M.J. , and Soetaert, K. (2001) Indices of diversity and evenness. Ocánis 24: 61–87. [Google Scholar]
- Holt, C.C. , Bass, D. , Stentiford, G.D. , and van der Giezen, M. (2020) Understanding the role of the shrimp gut microbiome in health and disease. J Invertebr Pathol: 107387 10.1016/j.jip.2020.107387. [DOI] [PubMed] [Google Scholar]
- Honda, K. , and Littman, D.R. (2012) The microbiome in infectious disease and inflammation. Annu Rev Immunol 30: 759–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline, D.I. , and Vollmer, S.V. (2011) White band disease (type I) of endangered Caribbean Acroporid corals is caused by pathogenic bacteria. Sci Rep 1(7), [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinges, J.G. , Rosales, S.M. , McMinds, R. , Shaver, E.C. , Shantz, A.A. , Peters, E.C. , et al (2019) Phylogenetic, genomic, and biogeographic characterization of a novel and ubiquitous marine invertebrate‐associated Rickettsiales parasite, Candidatus Aquarickettsia rohweri, gen. Nov., sp. nov. ISME J 13: 2938–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krediet, C.J. , Ritchie, K.B. , Alagely, A. , and Teplitski, M. (2013) Members of native coral microbiota inhibit glycosidases and thwart colonization of coral mucus by an opportunistic pathogen. ISME J 7: 980–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriss, M. , Hazleton, K.Z. , Nusbacher, N.M. , Martin, C.G. , and Lozupone, C.A. (2018) Low diversity gut microbiota dysbiosis: drivers, functional implications and recovery. Curr Opin Microbiol 44: 34–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesser, M.P. (2011) Coral bleaching: causes and mechanisms In Coral Reefs: An Ecosystem in Transition, Dubinsky Z., and Stambler N. (eds). Netherlands: Springer, pp. 405–419. 10.1007/978-94-007-0114-4_23. [DOI] [Google Scholar]
- Lesser, M.P. , Bythell, J.C. , Gates, R.D. , Johnstone, R.W. , and Hoegh‐Guldberg, O. (2007) Are infectious diseases really killing corals? Alternative interpretations of the experimental and ecological data. J Exp Mar Biol Ecol 346: 36–44. [Google Scholar]
- Lewis, J.D. , Chen, E.Z. , Baldassano, R.N. , Otley, A.R. , Griffiths, A.M. , Lee, D. , et al (2015) Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in Pediatric Crohn's disease. Cell Host Microbe 18: 489–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libro, S. , and Vollmer, S.V. (2016) Genetic signature of resistance to White band disease in the Caribbean Staghorn coral Acropora cervicornis . PLoS One 11: e0146636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher, R.L. , Rice, M.M. , McMinds, R. , Burkepile, D.E. , and Vega Thurber, R. (2019) Multiple stressors interact primarily through antagonism to drive changes in the coral microbiome. Sci Rep 9: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal, S. , van Treuren, W. , White, R.A. , Eggesbø, M. , Knight, R. , and Peddada, S.D. (2015) Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb Ecol Health Dis 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao‐Jones, J. , Ritchie, K.B. , Jones, L.E. , and Ellner, S.P. (2010) How microbial community composition regulates coral disease development. PLoS Biol 8: e1000345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDevitt‐Irwin, J.M. , Baum, J.K. , Garren, M. , and Thurber, V. (2017) L, R. responses of coral‐associated bacterial communities to local and global stressors. Front Mar Sci 4: (Article 262). [Google Scholar]
- McMurdie, P.J. , and Holmes, S. (2013) Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8: e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, M.W. , Lohr, K.E. , Cameron, C.M. , Williams, D.E. , and Peters, E.C. (2014) Disease dynamics and potential mitigation among restored and wild staghorn coral, Acropora cervicornis . PeerJ 2: e541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagna, M. , Sassera, D. , Epis, S. , Bazzocchi, C. , Vannini, C. , Lo, N. , et al (2013) “Candidatus Midichloriaceae” fam. Nov. (Rickettsiales), an ecologically widespread clade of intracellular Alphaproteobacteria. Appl Environ Microbiol 79: 3241–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, E.M. , Rogers, C.S. , Spitzack, A.S. , and van Woesik, R. (2008) Bleaching increases likelihood of disease on Acropora palmata (Lamarck) in Hawksnest Bay, St John, US Virgin Islands. Coral Reefs 27: 191–195. [Google Scholar]
- Muller, E.M. , Bartels, E. , and Baums, I.B. (2018) Bleaching causes loss of disease resistance within the threatened coral species Acropora cervicornis . Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, E.M. , and Woesik, R.v. (2012) Caribbean coral diseases: primary transmission or secondary infection? Glob Chang Biol 18: 3529–3535. [Google Scholar]
- Nawrocki, E.P. , and Eddy, S.R. (2013) Infernal 1.1: 100‐fold faster RNA homology searches. Bioinformatics 29: 2933–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neave, M.J. , Apprill, A. , Ferrier‐Pagès, C. , and Voolstra, C.R. (2016) Diversity and function of prevalent symbiotic marine bacteria in the genus Endozoicomonas . Appl Microbiol Biotechnol 100: 8315–8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, T.M. , Apprill, A. , Mann, J. , Rogers, T.L. , and Brown, M.V. (2015) The marine mammal microbiome: current knowledge and future directions. Microbiol Aust 36: 8–13. [Google Scholar]
- Oksanen, J. , Blanchet, F.G. , Friendly, M. , Kindt, R. , Legendre, P. , McGlinn, D. , et al. (2019) vegan: community Ecology Package.
- Pogoreutz, C. , Rädecker, N. , Cárdenas, A. , Gärdes, A. , Wild, C. , and Voolstra, C.R. (2018) Dominance of Endozoicomonas bacteria throughout coral bleaching and mortality suggests structural inflexibility of the Pocillopora verrucosa microbiome. Ecol Evol 8: 2240–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pootakham, W. , Mhuantong, W. , Yoocha, T. , Putchim, L. , Jomchai, N. , Sonthirod, C. , et al (2019) Heat‐induced shift in coral microbiome reveals several members of the Rhodobacteraceae family as indicator species for thermal stress in Porites lutea . Microbiol Open 8: e935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast, C. , Pruesse, E. , Yilmaz, P. , Gerken, J. , Schweer, T. , Yarza, P. , et al (2012) The SILVA ribosomal RNA gene database project: improved data processing and web‐based tools. Nucleic Acids Res 41: D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie, K.B. (2011) Bacterial symbionts of corals and symbiodinium In Beneficial Microorganisms in Multicellular Life Forms, Rosenberg E., and Gophna U. (eds). Berlin, Heidelberg: Springer, pp. 139–150. 10.1007/978-3-642-21680-0_9. [DOI] [Google Scholar]
- R Core Team (2017). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/ [Google Scholar]
- Rogers, C.S. , and Muller, E.M. (2012) Bleaching, disease and recovery in the threatened scleractinian coral Acropora palmata in St. John, US Virgin Islands: 2003–2010. Coral Reefs 31: 807–819. [Google Scholar]
- Rosales, S.M. , Miller, M.W. , Williams, D.E. , Traylor‐Knowles, N. , Young, B. , and Serrano, X.M. (2019) Microbiome differences in disease‐resistant vs. susceptible Acropora corals subjected to disease challenge assays. Sci Rep 9: 18279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosic, N.N. , and Dove, S. (2011) Mycosporine‐like amino acids from coral Dinoflagellates. Appl Environ Microbiol 77: 8478–8486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter, S.J. , Cox, M.J. , Turek, E.M. , Calus, S.T. , Cookson, W.O. , Moffatt, M.F. , et al (2014) Reagent and laboratory contamination can critically impact sequence‐based microbiome analyses. BMC Biol 12: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaver, E.C. , Shantz, A.A. , McMinds, R. , Burkepile, D.E. , Vega Thurber, R.L. , and Silliman, B.R. (2017) Effects of predation and nutrient enrichment on the success and microbiome of a foundational coral. Ecology 98: 830–839. [DOI] [PubMed] [Google Scholar]
- Sunagawa, S. , DeSantis, T. , Piceno, Y.M. , Brodie, E.L. , DeSalvo, M. , Voolstra, C.R. , et al (2009) Bacterial diversity and White plague disease‐associated community changes in the Caribbean coral Montastraea faveolata . ISME J 3: 512–521. [DOI] [PubMed] [Google Scholar]
- Sweet, M.J. , Croquer, A. , and Bythell, J.C. (2014) Experimental antibiotic treatment identifies potential pathogens of white band disease in the endangered Caribbean coral Acropora cervicornis . Proc R Soc B 281: 20140094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurber, R.V. , Willner‐Hall, D. , Rodriguez‐Mueller, B. , Desnues, C. , Edwards, R.A. , Angly, F. , et al (2009) Metagenomic analysis of stressed coral holobionts. Environ Microbiol 11: 2148–2163. [DOI] [PubMed] [Google Scholar]
- Vollmer, S.V. , and Kline, D.I. (2008) Natural disease resistance in threatened Staghorn corals. PLoS ONE 3: e3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware, M. , Garfield, E.N. , Nedimyer, K. , Levy, J. , Kaufman, L. , Precht, W. , et al (2020) Survivorship and growth in staghorn coral (Acropora cervicornis) outplanting projects in the Florida keys National Marine Sanctuary. PLoS ONE 15: e0231817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis, V. (2008) Cellular mechanisms of cnidarian bleaching: stress causes the collapse of symbiosis. J Exp Biol 211: 3059–3066. [DOI] [PubMed] [Google Scholar]
- West, A.G. , Waite, D.W. , Deines, P. , Bourne, D.G. , Digby, A. , McKenzie, V.J. , and Taylor, M.W. (2019) The microbiome in threatened species conservation. Biol Conserv 229: 85–98. [Google Scholar]
- Yellowlees, D. , Rees, T.A.V. , and Leggat, W. (2008) Metabolic interactions between algal symbionts and invertebrate hosts. Plant Cell Environ 31: 679–694. [DOI] [PubMed] [Google Scholar]
- Zaneveld, J.R. , Burkepile, D.E. , Shantz, A.A. , Pritchard, C.E. , McMinds, R. , Payet, J.P. , et al (2016) Overfishing and nutrient pollution interact with temperature to disrupt coral reefs down to microbial scales. Nat Commun 7: 11833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaneveld, J.R. , McMinds, R. , and Thurber, R.V. (2017) Stress and stability: applying the Anna Karenina principle to animal microbiomes. Nat Microbiol 2: 17121. [DOI] [PubMed] [Google Scholar]
- Ziegler, M. , Grupstra, C.G.B. , Barreto,, M.M. , Eaton, M. , BaOmar, J. , Zubier, K. , et al. (2019). Coral bacterial community structure responds to environmental change in a host‐specific manner. Nat Commun 10: 10.1038/s41467-019-10969-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilber‐Rosenberg, I. , and Rosenberg, E. (2008) Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution. FEMS Microbiol Rev 32: 723–735. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1: Supplementary Methods.
Figure S1 Supplementary Figures
Table S1 Results from ANCOM analysis for each of four pairwise contrasts. An unrarefied ASV table was first summarized to the genus level, then was filtered to contain only taxa with at least 10 counts in 20% of the samples. A significance level at W = 0.6 was used in which the null hypothesis for a given taxon was rejected in 60% of the tests and p‐values were corrected with Benjamini‐Hochberg FDR. MD3‐55 and HIMB11 are the genus identifications in Silva for Ca. Aquarickettsia and Roseobacter, respectively.
Table S2 Kruskal Wallis tests for differences in Simpson's diversity with and without Ca. Aquarickettsia rohwerii sequences with pairwise comparisons
Table S3 Results of PERMANOVA tests for differences between groups with and without Ca. Aquarickettsia rohwerii sequences and with pairwise comparisons. Perm column presents the percentage of 1000 tests that produced a p < 0.05 after permutational down‐sampling.
Table S4 Results of PERMDISP tests for differences in group dispersion with and without Ca. Aquarickettsia rohwerii Sequences. Perm column presents the percentage of 1000 tests that produced a p < 0.05 after permutational down‐sampling.
Table S5 Reads and corresponding ASVs lost through quality control processing in sabre, DADA2 and Phyloseq.
Table S6 Sample metadata with sequencing depth after removal of rare ASVs and ASVs annotating to mitochondria or chloroplasts.
Data Availability Statement
Raw sequence data were deposited into the NCBI Sequence Read Archive (SRA) under accession number PRJNA639601. All scripts involved in the preparation and analysis of this dataset are available at https://github.com/maherrl/Aquarickettsia_bleaching_dynamics.


