Summary
Advances in omics technologies have generated exponentially larger volumes of biological data; however, their analyses and interpretation are limited to computationally proficient scientists. We created OmicsVolcano, an interactive open-source software tool to enable visualization and exploration of high-throughput biological data, while highlighting features of interest using a volcano plot interface. In contrast to existing tools, our software and user-interface design allow it to be used without requiring any programming skills to generate high-quality and presentation-ready images.
Subject areas: Bioinformatics, Genomics, RNA-seq, Proteomics
Graphical Abstract
Highlights
-
•
A free and open-source tool for interactive exploration of omics data
-
•
Immediate visualization of gene and protein changes on a volcano plot
-
•
Visualization and generation of figures with gene ontologies linked to cellular processes
-
•
Data exploration and visualization of user-defined cellular and molecular processes
Advances in omics technologies have generated exponentially larger volumes of biological data; however, their analyses and interpretation are limited to computationally proficient scientists. We created OmicsVolcano, an interactive open-source software tool to enable visualization and exploration of high-throughput biological data, while highlighting features of interest using a volcano plot interface. In contrast to existing tools, our software and user-interface design allow it to be used without requiring any programming skills to generate high-quality and presentation-ready images.
Before you begin
Significant advances have been made in biological sciences through the generation of data from genomic, transcriptomic, proteomic and metabolomic analyses - jointly referred to as omics technologies (Sandhu et al., 2018). The amount of data varies depending on the type of omics platform and recently there have been concerted efforts toward integrating omics datasets to provide global insights into cellular function (Yan et al. 2017; Cambiaghi et al. 2016). The major limitation of omics technologies has become the interpretation and visualization of the analyzed datasets in a coherent and user-friendly manner that would be suitable for users from diverse fields without any computational skills. Volcano plots effectively visualize significantly increased and reduced changes in entire omics datasets. There are two major limitations in the use of volcano plots: the requirement for computational skills in R to summarize and visualize the data in volcano plots; and the lack of flexibility to interactively highlight or discover specific sets of changes or processes. Furthermore, each addition or change of a highlighted gene or protein requires manual generation of a new plot, making the process of producing volcano plots time consuming for computational biologists.
Existing tools to date, such as EnhancedVolcano, VolcanoR, and msVolcano (Blighe et al. 2019; Naumov et al., 2017; Singh et al. 2016) are capable of plotting omics data as volcano plots and have specific functionalities. These allow genes of choice to be labeled, and some of the programs - such as msVolcano - enable significance testing. Additional software tools, such as DEIVA, are capable of carrying out enrichment analyses. This functionality is also part of software packages such as DAVID (Huang et al. 2008; Huang et al. 2009; Harshbarger et al. 2017). We sought to build upon these approaches and provide interactive volcano plots where sets of genes or proteins that have been classified as part of specific cellular processes could be selectively highlighted. Our goal was to enable biologists without any computational expertise to view, visualize, and explore specific cellular or molecular processes.
OmicsVolcano has been designed for biologists without any programing experience and provides an easy-to-use web-based tool. One of OmicsVolcano’s strengths is its capability to allow interactive exploration of omics data that allows users to focus on the biological aspects of the data. The interactive graphics that are part of OmicsVolcano enhance the impact of the findings and put them into a physiological context. Those include the ability of the user to highlight primary changes, to visualize a group of genes and proteins that are related to a specific cellular process or multiple cellular processes, and to examine their cellular localizations. In each case information about selected genes or proteins is presented in a table below the graph. OmicsVolcano generates high-quality and publication-ready images in scalable vector graphic (SVG) format.
Overview
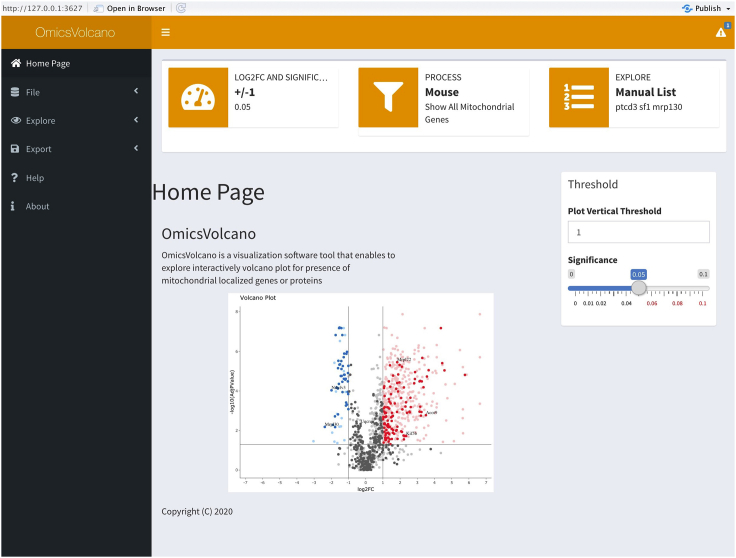
The OmicsVolcano software consists of a set of scripts, functions, and modules which search for the presence of duplicated gene names in the input data; add numerical extensions to duplicated gene names; visualize the data as interactive volcano plots; and filter data to significant values based on information provided in the input file. The input file is provided by the user, and consists of five columns that represent identification numbers (IDs), gene symbols, gene descriptions, log fold changes, and adjusted p values. The software package also contains reference files for mitochondrial processes and cellular compartments for the human and mouse genomes. The software uses the input data and processes or cellular compartment information to create volcano plots in a user-friendly way (Figure 1). The software tool allows an interactive interpretation of the data through a web interface, which is easy and intuitive to use.
Figure 1.
The OmicsVolcano home page
The software requires four steps to generate the interactive omics-data visualization plots. The “file” option allows the upload of the input data as an ASCII file which contains IDs, gene names, gene descriptions, log2 fold changes, and adjusted p values. The “explore” option provides several main core functionalities of the software: plot, custom gene or protein selection, mitochondrial processes, multiple process visualization, and cellular compartment visualization. Customization of the statistical significance and threshold of the y-axis, are performed by adjustments of the slider widgets. Additional options, e.g., “upload a gene file”, “insert a list of genes”, “select organism”, and “show mitochondrial process” widgets allow the customization of the data exploration processes. The “export” option enables the export of data as tables or graphics in various pre-defined formats. A manual is available through the “help” option, and the software package version including additional information about the software can be found in the “about” option.
Software download and prerequisites
A pre-installed R software environment is required to use OmicsVolcano. The easiest option to run OmicsVolcano is to use RStudio. Both, R and RStudio can be downloaded from https://www.r-project.org/ and https://rstudio.com/products/rstudio/download/, respectively. We encountered some rare cases of R installation environments, where it is required to adjust the file extension settings either to capitalized or lower key letters. In these cases, the extension of source files should be either capitalized or set to lower key letters (e.g., as “config.R” instead of “config.r”).
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| RNA sequencing data | GEO | GSE105406, GSE111228 |
| Proteomics data | PRIDE | PXD015521 |
| Software and algorithms | ||
| R version 3.6.1 and 4.02 | Team, R.C. 2019 R: A language and environment for statistical computing | https://www.R-project.org/ |
| shiny version 1.4.0 | Chang et al., 2019. shiny: Web Application Framework for R | https://CRAN.R-project.org/package=shiny. |
| shinydashboard version 0.7.1 | Chang and Ribeiro, 2018. shinydashboard: Create Dashboards with “Shiny” | https://CRAN.R-project.org/package=shinydashboard |
| shinydashboardPlus version 0.7.1 | Granjon, 2020. shinydashboardPlus: Add More “AdminLTE2” Components to “shinydashboard” | https://CRAN.R-project.org/package=shinydashboardPlus |
| shinyWidgets version 0.5.0 | (Perrier et al., 2020). shinyWidgets: Custom Inputs Widgets for Shiny | https://CRAN.R-project.org/package=shinyWidgets |
| shinythemes version 1.1.2 | Chang, 2018. shinythemes: Themes for Shiny | https://CRAN.R-project.org/package=shinythemes. |
| shinyjs version 2.0.0 | Attali, 2020. shinyjs: Easily Improve the User Experience of Your Shiny Apps in Seconds | https://CRAN.R-project.org/package=shinyjs |
| dplyr version 0.8.3 | Wickham et al., 2019. dplyr: A Grammar of Data Manipulation | https://CRAN.R-project.org/package=dplyr |
| plotly version 4.9.1 | Sievert, 2018. plotly for R | https://plotly-r.com |
| ggplot2 version 3.2.1 | Wickham, 2016. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag New York | https://ggplot2.tidyverse.org |
| crosstalk version 1.0.0 | Cheng, 2016. crosstalk: Inter-Widget Interactivity for HTML Widgets | https://CRAN.R-project.org/package=crosstalk |
| DT version 0.12 | Xie et al., 2020. DT: A Wrapper of the JavaScript Library “DataTables” | https://CRAN.R-project.org/package=DT |
| svglite version 1.2.3 | Wickham et al., 2020. svglite: An “SVG” Graphics Device | https://CRAN.R-project.org/package=svglite |
| stringr version 1.4.0 | Wickham, 2019. stringr: Simple, Consistent Wrappers for Common String Operations | https://CRAN.R-project.org/package=stringr |
| config version 0.3 | Allaire, 2018. config: Manage Environment Specific Configuration Values | https://CRAN.R-project.org/package=config |
| colourpicker version 1.1.0 | https://cran.r-project.org/web/packages/colourpicker/index.html | https://CRAN.R-project.org/package=colourpicker |
| gridExtra version 2.3 | https://cran.r-project.org/web/packages/gridExtra/index.html | https://cran.r-project.org/web/packages/gridExtra/index.html |
| OmicsVolcano | https://github.com/IrinaVKuznetsova/OmicsVolcano | this manuscript |
Materials and equipment
OmicsVolcano is available as open-source software and is hosted at GitHub: https://github.com/IrinaVKuznetsova/OmicsVolcano. The software implementation is written in R (3.6.1) using the following packages: shiny (1.4.0) enabling the creation of an intuitive and interactive web application; ggplot2 (3.2.1) and plotly (4.9.1) enabling the creation of the interactive volcano plot graphics; dplyr (0.8.3) enabling the dataset aggregation and analysis; DT (0.11) enabling the display of the R data objects as tables in HTML format; and crosstalk (1.0.0) providing interactions between R objects (Wickham et al. 2019; Chang et al. 2019; Sievert 2018; Xie et al. 2020; Wickham 2016; Team 2019; Cheng 2016), and colourpicker enabling a color palette (https://cran.r-project.org/web/packages/colourpicker/index.html). A full list of utilized packages can be found in the Key resources table. The application is hosted as an open-source package in the GitHub repository.
The test input data consisted of RNA or protein fold changes and adjusted p values from RNA sequencing and proteomic analyses that were processed as described previously (Rudler et al. 2019; Siira et al. 2018; Perks et al., 2018). The input file columns were designated as: “ID,” “GeneSymbol,” “Description,” “Log2FC,” and “AdjPValue” (an example format of the input file is attached to the software browser widget and also shown in the OmicsVolcano GitHub web-page). Specific information about mitochondrial processes was based on our previous studies (Kühl et al. 2017), MitoXplorer (Yim et al. 2019) and combined with MitoCarta 2.0 (Calvo et al. 2015; Pagliarini et al. 2008). Cellular compartment localization information was retrieved from the Human Protein Atlas database available from http://www.proteinatlas.org (Thul et al., 2017).
The software is based on R and uses R packages shown in the Key resources table.
CRITICAL: The software has been tested for both R version 3.6.1 and version 4.0.0. It also has been tested on Windows and Mac OS platforms (Table 1).
Table 1.
-
•Recommended hardware: minimal 4 Gb memory. Memory requirements may increase with input data size.
-
•Processors: 1 required, 2 recommended.
-
•Example data are provided with the software package. User input files for omics datasets should be formatted as a tab or as a semicolon separated file in ASCII/text format. The file should contain five columns with the column names as shown in Table 2: ID, GeneSymbol, Description, Log2FC, AdjPValue.
-
•Column names are case-sensitive and require a header row for the input file. Thus, when preparing input files for OmicsVolcano, it is essential to provide a header row. The following rows contain various values, which will be processed by the OmicsVolcano software.
| Operating System | Version |
|---|---|
| Windows | Windows 10 |
| Mac OS | Mojave version 10.14.6 |
Table 2.
Input file example
| ID | Gene Symbol | Description | Log2FC | AdjPValue |
|---|---|---|---|---|
| Q4U4S6 | Xirp2 | Xin actin-binding repeat-containing protein 2 OS=Mus musculus OX=10090 GN=Xirp2 PE=1 SV=1 | 6.64 | 1.33E-08 |
| Q497D7 | Rpl30fo | Rpl30 protein OS=Mus musculus OX=10090 GN=Rpl30 PE=2 SV=1 | 2.14 | 0.8 |
| Q9CPP6 | Ndufa5 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5 OS=Mus musculus OX=10090 GN=Ndufa5 PE=1 SV=3 | -1.52 | 6.24E-08 |
| P09055 | Itgb1 | Integrin beta-1 OS=Mus musculus OX=10090 GN=Itgb1 PE=1 SV=1 | 0.08 | 6.29E-08 |
| … | … | … | ... | … |
Step-by-step method details
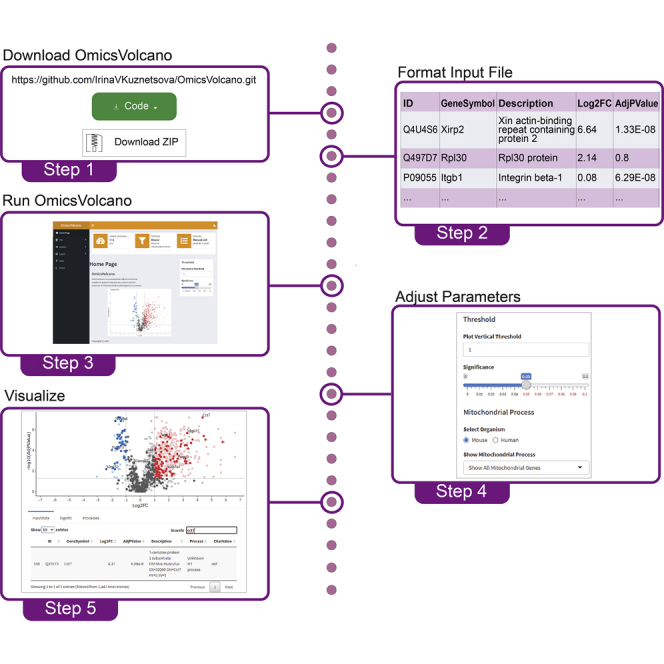
Step 1: Installing and initializing the OmicsVolcano software tool
Timing: <3 min
-
1.
Download the OmicsVolcano software from the GitHub repository as zip folder.
-
2.
Unzip the folder into a directory of your choice, such as onto the Desktop.
-
3.
Open RStudio.
-
4.
Open the R script OmicsVolcano_App.r through RStudio, by selecting “File,” then select “Open File” and select OmicsVolcano_App.r that will be loaded into the RStudio interface.
-
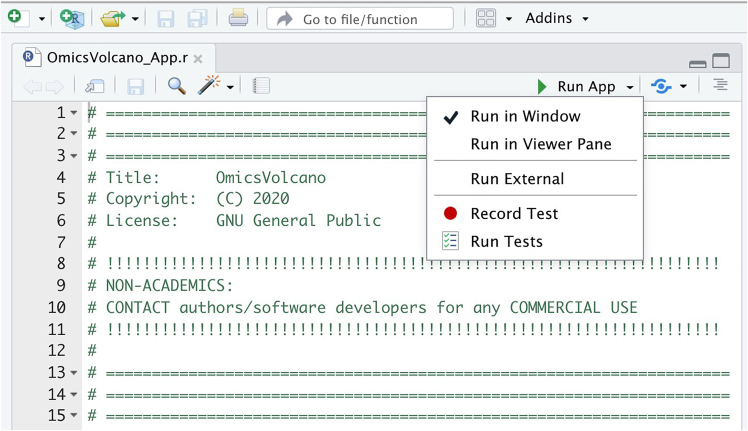
5.
Once the OmicsVolcano_App.r is loaded on the interface press the “Run App” button to load the OmicsVolcano software. Ensure that the “Run in Window” or "Run External" selection is checked when you press “Run App” (Figure 2).
CRITICAL: Check that RStudio is working correctly. Troubleshooting 1
CRITICAL: Mac OS users are required to install the XQuartz app from https://www.xquartz.org.
Note: Once the software is downloaded and functional, steps 1–5 do not need to be repeated for future and continued use of the OmicsVolcano software.
Figure 2.
Initializing the OmicsVolcano software in R studio
Screen capture indicating how to select the run app in R studio to open the software.
Step 2: Visualizing omics data
Timing: 5 min
There are three menu options under the Home Page heading that enable the visualization and exploration of the data.
-
6.Press the File – Open… tab to upload a user provided input file in a .txt format, not applicable (NA) or empty fields are allowed.Note: A demo file can be used for a test run. The demo file designated demofile.txt can be downloaded by selecting File – Open… - Download demo file to local PC. This demo file will be automatically downloaded in the user-selected folder and then used to visualize the data.
-
(a)If the file data is not formatted according to the instructions described in the Materials and equipment then an error message will be displayed: “File Loading Error in Data Input File! Please check the number of columns or select the correct field separator character. Alternatively, review the Help Page for the file input format requirements.”
-
(b)If the data has duplicates, for example in proteomic data sets when there are multiple isoforms of proteins, the default option of the software is to add a numerical extension for each duplicate in order of their appearance in the file provided. This is done so that duplicate names of proteins are not lost when they are visualized. However, if there are no duplicates in the data then the “Check for Duplicates” option can be unselected in the File - Open… - Check for Duplicates.
-
(a)
-
7.
Press Open…, select input file, and the data will appear in a table with the headings used in the input file. The headings can be toggled to order the data alphabetically, in an ascending or descending order of log2 fold changes, or based on the adjusted p values. Troubleshooting 2
-
8.
Select File Separator. Troubleshooting 3
-
9.
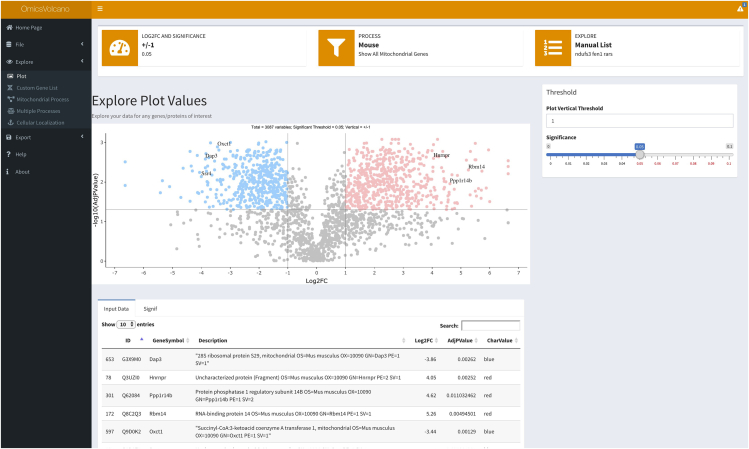
To visualize the data select Explore tab and then select Plot. This generates a volcano plot where the significantly increased and reduced data points are colored in red and blue, respectively. Non-significant data points are shown in gray (Figure 3). Any data point can be labeled by clicking on the plotted data points (see 12). Troubleshooting 4
-
10.
The significance and vertical thresholds can be adjusted from the right-side panel named Threshold.
Optional: The software offers the option to choose statistical significance, which is recommended to be either 0.05 or 0.01. Another option is to adjust the log2 fold change threshold, which is set to +/-1 as the default value.
-
11.
Select a data point on the volcano plot by pressing on it within the plot to provide a label for the identity of the gene or protein. Multiple data points can be labeled by holding Shift in Windows or Command in Mac platforms. The information for the selected data point can be seen in the table below the volcano plot. Troubleshooting 5
-
12.
The table below the volcano plot has two tabs, the Input Data tab shows all the data and the Signific tab lists all the significantly changing data. The Search box within the table enables the user to query specific gene or protein names.
CRITICAL: The data input file has to follow the format shown in Table 2 above. Troubleshooting 6 and 7
Note: The Help tab provides a brief guide to the use of the software. The About tab provides information about the authors, license, used packages and their versions, and how to cite the software.
Figure 3.
A volcano plot example with specific interactively selected gene labels
Transcriptomic data are visualized where significantly increased transcripts are shown in light red and significantly decreased transcripts in light blue. Non-significant transcripts are shown in gray. Three increased and three decreased transcripts are indicated in the plot as examples, showing the software’s capability to label individual transcripts of interest.
Step 3: Exploring omics data
Timing: 5–30 min
The Explore tab has four additional features that enable interactive searches for specific genes or processes linked to specific genes.
-
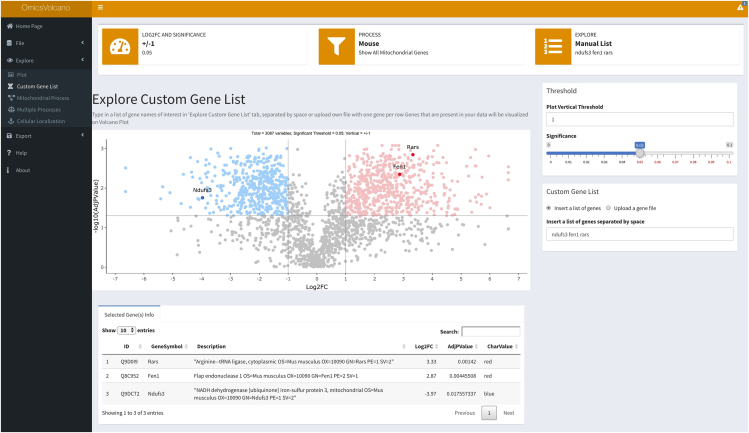
13.Select Custom Gene List feature:
-
a.Inserting a list of genes enables the user to manually input a user-specified list of genes that will appear on the volcano plot with gene or protein labels and their information in the table below the plot (Figure 4).
-
b.The Upload a gene file option enables the user to provide their own gene ontology lists from an enrichment analysis to highlight specific genes in a biological process, molecular function, or cellular compartment or a user-specified list of genes within a file. The file is rather simple, and each line of the file represents a gene name used for a process (see list below). The file does not require a header row.
-
a.
| Example: | Ndufs2 |
| Gatc | |
| Cox7a1 | |
| lmnb1 | |
| Ndufa8 |
-
14.
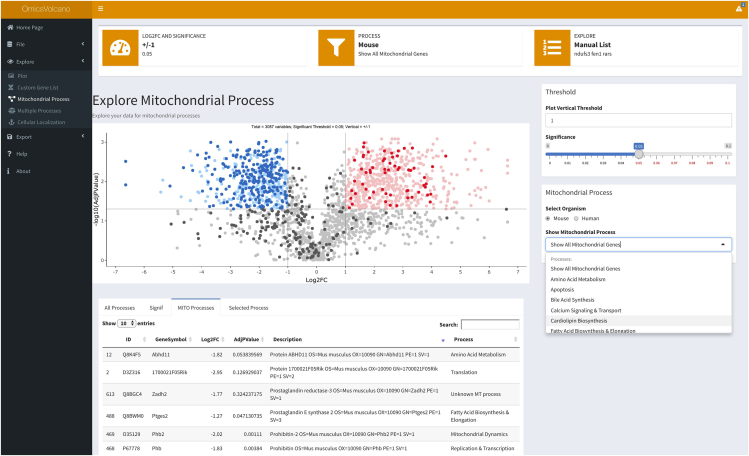
The Mitochondrial Processes feature highlights all mitochondria specific genes or proteins in either bright red for the upregulated genes or proteins, or bright blue for the downregulated gene or proteins. Non-mitochondrial genes or proteins are shown as faded red or blue for those that are significantly increased or decreased (Figure 5). Limitations
-
15.
An additional feature of theMitochondrial Processes dropdown menu enables the exploration of 40 different processes linked to mitochondrial function present in the user-provided data. Selection of a specific process highlights a set of genes or proteins related to that mitochondrial process and shows them in bright red for the upregulated genes or proteins, or bright blue for the downregulated gene or proteins. This feature enables a fast identification of specific processes and genes or proteins that are significantly altered in the entire dataset (Figure 5).
Note: The Custom Gene List features enables users to expand the scope of cellular and molecular processes that can be added to the software in addition to Mitochondrial Processes. The Mitochondrial Processes feature is an example of curated gene ontologies linked to MitoCarta 2.0 genes. Troubleshooting 8
-
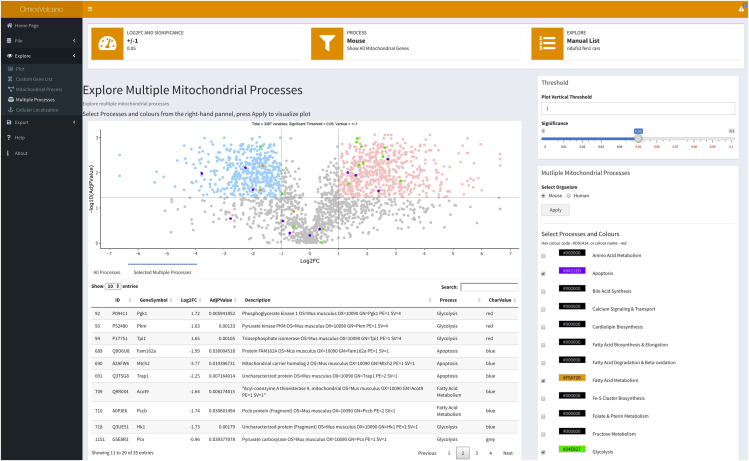
16.
The Multiple Mitochondrial Processes function enables the selection and user-defined color selection of multiple processes to highlight all the changes in the dataset that are of interest to the user (Figure 6). The user should check the process of interest and select the color for the process and repeat this until all processes required by the user are selected then press “Apply” to show them on the plot as shown in Figure 6. The right-hand panel shows the figure legend revealing the process linked to each selected color and the table below lists all the genes and their descriptions along with the colors representing them on the plot. Troubleshooting 9
-
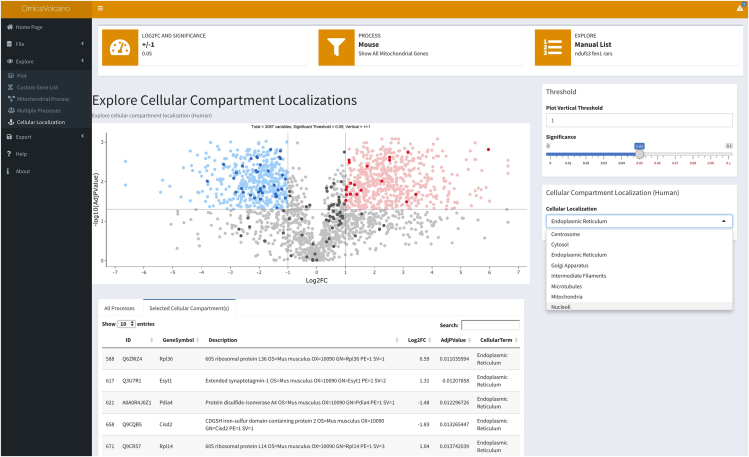
17.
TheCellular Localization dropdown menu enables the exploration of different cellular compartments related to the changes in the user-provided data. This feature enables a fast identification of genes or proteins that are significantly altered in the entire dataset and related to a specific cellular location (Figure 7). In the example, we identified that the loss of a specific gene with unknown function caused the downregulation of genes involved in endoplasmic reticulum function, providing a valuable and fast insight into the role of our gene of interest (Figure 7). This can be applied to any dataset to quickly identify changes in different cellular compartments. Troubleshooting 9
Figure 4.
Volcano custom plot example including user-defined searches
Transcriptomic data are visualized with significantly increased transcripts shown in light red and significantly decreased transcripts shown in light blue. Non-significant transcripts are shown in gray. User-specified transcripts or proteins can be searched in the right-hand box under “Custom Gene list” or uploaded in a user-defined file. The file-based import is useful when a large number of transcripts or proteins are to be searched and visualized in the plot. Dark red color indicates if they are significantly increased, and dark blue if they are significantly reduced within the dataset.
Figure 5.
Volcano plot showing mitochondrial processes
Transcriptomic data are visualized and mitochondrial transcripts are highlighted in dark red, dark blue, and dark gray, if they are significantly increased, decreased, or unchanged, respectively. Specific processes can be selected from the dropdown menu and these will be highlighted in either dark red or dark blue depending on their change within the dataset.
Figure 6.
Volcano plot showing specific selection and color coding of multiple mitochondrial processes
This dropdown menu allows the user to select specific processes and custom colors for each process and visualizes them on the plot. This feature enables additional multiple processes to be visualized at the same time.
Figure 7.
Volcano plot showing cellular localizations
This feature enables the user to explore changes in specific cellular locations by selecting specific cellular compartments from the dropdown menu. The selected cellular compartments are visualized depending on the changes (increased in dark red and decreased in dark blue) in the input file related to the selected cellular compartment (in this case the endoplasmic reticulum).
Step 4: Exporting visualized data
Timing: <1 min
The final features of our software are the download options of the completed volcano plot and associated tables. All interactive plots can be downloaded in a vector format as SVG files directly from the plot by pressing the camera-like icon. The image is saved to the default PC location. Static plots generated by Custom Gene List can be exported in numerous different formats. Tables associated with the generated volcano plot can be downloaded in txt and csv formats depending on the requirements of the end-user.
-
18.Select the Export function and choose either Plot or Table.
-
a.Select Plot and in the first dropdown menu Custom Gene List can be exported as png, jpeg, or tiff for an image and SVG or pdf for a vector file.
-
b.Select Table and in the first dropdown menu and then select the type of table to export. In the second dropdown menu choose to save the tabulated data in csv or txt format.
-
a.
Note: Hovering the cursor over the volcano plot reveals a camera icon in the top right corner of the volcano plot. Selecting the camera icon enables a shortcut to download the volcano plot in SVG format to the PC default location. Selecting the camera icon enables downloading of volcano plots from each feature on OmicsVolcano. Users using the Ubuntu operating system can use this feature to download their plots.
Expected outcomes
We created OmicsVolcano to provide a simple to use tool for biologists to explore and highlight changes in genes, transcripts and proteins in an interactive manner and visualize them for presentations and publications (Figure 1). To date, visualization of omics datasets has been restricted to computational experts, while our software by-passes any requirements for computational skills - empowering scientists from any discipline and skill level to explore their data.
The software requires minimal data input consisting of gene or protein IDs, fold change, and adjusted p values to visualize the data in an interactive volcano plot. The input file can be provided by the user in a simple text format. Once the input file is uploaded in the software, two simple point-and-click features enable the user to choose the plot threshold and to define the changes based on their significance, typically users would choose 0.01 or 0.05, however, the toggle provides users the freedom to select the significance of their choice. These functions enable immediate visualization of the changes on a volcano plot, where the significantly downregulated genes or proteins are highlighted in blue and the significantly upregulated genes, transcripts or proteins are highlighted in red, compared to the remaining genes, transcripts or proteins that are not significantly changed that are shown in gray.
The software provides a table below the volcano plot with additional information on the gene or protein ID, gene symbol, log fold change, adjusted p value and description of the gene/protein function for the entire dataset. The volcano plot is interactive such that each point shown on the plot can be selected, highlighted with its related gene or protein name and the information related to that specific point can be viewed in the table below. Conversely, any gene or protein can be searched in the table and selecting it from the table will highlight it in a specific color on the volcano plot and provide its name on the plot. These features are unique compared to static volcano plots graphed in R where the users cannot identify which gene is related to specific point on the plot unless they have computational expertise to use R to select specific genes or proteins to highlight, making it laborious and time consuming to re-plot the volcano graph whenever a new set of genes, transcripts or proteins need to be examined.
The software functionalities named “Plot” and “Custom Gene List” are broadly applicable to explore datasets of any genome, transcriptome or proteome. The “Plot” - enables exploration of the entire volcano plot with the ability to select and label any point of interest. The “Custom Gene List” function enables users to type in or upload a file with their choice of gene, transcript or protein names in a text format, enabling users from diverse fields to explore processes specific to their research interests.
The “Mitochondrial Process” function enables users to explore changes in mouse or human mitochondria. For this function we have provided a built-in option to analyze 40 different processes related to mitochondrial function as an example, to demonstrate practically how effective our software is at interactively highlighting changes in genes, transcripts or proteins in a specific process. The selection of a specific mitochondrial function highlights the changes in genes, transcripts or proteins linked to the specified process on the plot. The table below the volcano plot provides information on the names and descriptions of the highlighted genes or proteins. To add an additional layer of information within the “processes” selection we highlight the up- and downregulated genes, transcripts or proteins in bright red and blue colors from the specified process, while the remaining significantly up- or downregulated genes, transcripts or proteins remain shown in faded red and blue. This feature highlights the genes, transcripts or proteins in the specified process that are significantly changed compared to the overall identified changes. The “Mitochondrial Process” function exemplifies the flexibility of our software and users can upload their own user-defined cellular and molecular processes associated with specified genes, RNAs or proteins to enable exploration of other cellular processes in OmicsVolcano. The “Multiple Mitochondrial Processes” are an additional feature providing users with the ability to choose multiple processes and color code them specifically. The “Cellular Localization” feature enables the exploration of entire datasets to reveal changes in gene expression or protein levels related to specific compartments within the cell. This enables users to quickly draw conclusions about their findings related to specific parts of the cell. Finally, the generated volcano plots can be downloaded in vector format, that can be further edited if required, whereas the tables can be downloaded in txt or csv format that are publication ready.
In summary, the main features of OmicsVolcano are: (1) a free and open-source software under GNU General Public license available from GitHub as an interactive web application to be run from RStudio; (2) interactive exploration of omics-generated datasets; (3) a data export functionality enabling further examination using other software tools or production of graphics for reports and scientific publications; (4) exploration of processes linked to mitochondrial function or cellular compartment; (5) an intuitive and easy-to-use interface for biologists of any field and (6) broad applicability to any cellular process or organism by providing customizable list of genes, transcripts, proteins, or metabolites associated with specific processes. The software interface enables scientists without any computational skills to independently analyze their data, highlight specific cellular processes, genes, transcripts or proteins of interest and visualize them within minutes. The software is designed to be extendable, so that additional functionalities can easily be added in future releases.
Limitations
The software is limited to presenting omics data in volcano plots, however, these plots have become the visualization method of choice in the omics field due to their intuitive representation of complex data. Our software provides an interactive interface that enables users to select and highlight as many or as few data points of interest without having to generate new plots every time a new data point is selected – opening up the use of volcano plots to all interested users.
The software can be adapted to highlight specific gene ontologies by providing user-provided input files, which is an added advantage. The software enables visualization of one dataset compared to another but not multidimensional analyses. This is a common feature of volcano plots and most comparisons in biological datasets, however, alternative methods should be used for visualizing data that require multidimensional comparisons. The software has built-in human and mouse reference files that will be updated by the software developers annually.
Troubleshooting
Problem 1
Software versions and operating system specific requirements.
Potential solution
Make sure that R and RStudio versions are 3.6.1 or greater.
Mac OS users need to install the XQuartz app from https://www.xquartz.org.
Problem 2
Wrong source file formatting, typos, and incorrect column names.
Potential solution
In the case of incorrect source file formatting, the software will display an error message to notify that there is a typographical error or incorrect column name. One typical example message for incorrectly formatted source files is: "Please check the name of the 2nd column. It is GeneSymbol, not Genesymbol."
Problem 3
Incorrect field separator characters or incorrect number of columns in source files.
Potential solution
In File, check that the correct field separator is chosen in the File Separator tab.
OmicsVolcano will display the error message: "Please check number of columns or select correct field separator character. Alternatively, review the Help Page for the file input format."
Problem 4
A gene or protein is present in the input source file but not visualized by the software.
Potential solution
This gene or protein has no numeric value assigned and is represented as NA or empty field in the “LogFC” or “AdjPValue” columns. Therefore, it cannot be shown on the plot.
Problem 5
No blue and/or red values in the plot – only gray values are visualized.
Potential solution
The data may not have significantly changing genes or proteins. Also, verify and carefully choose significance and vertical thresholds.
Problem 6
Gray values are located below the significance threshold and are not visualized.
Potential solution
The input file should contain all values. Do not prefilter the input file for significantly changing genes or proteins only.
Problem 7
Frozen browser page.
Potential solution
Re-fresh (reload) the browser page.
Problem 8
Undo selected genes.
I have changed my mind and would like to undo selected gene(s). Note, this is only relevant for the features “Plot","Mitochondrial Process” and “Cellular Localization.”
Potential solution
Double click on the plot image and select desired gene(s) again or re-fresh (reload) browser page and select desired gene(s) again.
Alternatively, visualize genes of interest with the Custom Gene List feature located at the Plot tab. Create a file that contains the gene names that should be visualized, upload this file in the Custom Gene List located on the right-hand side.
Problem 9
Visualize multiple genes and identify their location.
I would like to visualize multiple genes, but I do not know where they are located on the plot. Note, this is relevant for the features “Plot” and “Mitochondrial Process” only.
Potential solution
Use the Search tab in the table below the plot. Write the required gene name and press the row with the gene information. The table row will be highlighted in blue, and the selected gene label will appear on the plot. Continue this way, as many times as required.
Alternatively, use the feature “Custom Gene List.” Create a file that contains gene names that have to be visualized, upload this file in the “Custom Gene List” located on the right-hand side.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Aleksandra Filipovska (aleksandra.filipovska@uwa.edu.au).
Materials availability
The software generated in this study is freely available from: https://github.com/IrinaVKuznetsova/OmicsVolcano
Data and code availability
The example datasets and code generated during this study are available at.: https://github.com/IrinaVKuznetsova/OmicsVolcano. All data generated or analyzed during this study are included in this published article.
Acknowledgments
This project was supported by fellowships and project grants from the National Health and Medical Research Council (APP1058442, APP1045677, APP1041582, APP1023460, APP1005030, APP1043978 to A.F. and O.R.), the Australian Research Council (DP180101656 to A.F. and O.R.), and the Cancer Council of Western Australia (to A.F. and O.R.). I.K. is supported by a UWA Postgraduate Scholarship. The funders had no role in the design of the study, collection, analysis, and interpretation of data or in the preparation of the manuscript. We thank members of the Filipovska and Rackham laboratories for testing the software and providing valuable advice.
Author contributions
I.K. contributed to the algorithm development, script implementation, software design, and manuscript writing. A.L. contributed to the supervision of the software design, software implementation, and manuscript writing. O.R. supervised the software design and manuscript writing. A.F. acted as a principal supervisor of the project and contributed to each stage of the project and manuscript writing. All authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Irina Kuznetsova, Email: irina.kuznetsova@perkins.org.au.
Aleksandra Filipovska, Email: aleksandra.filipovska@uwa.edu.au.
References
- Allaire J.J. https://CRAN.R-project.org/package=config; 2018. config: Manage Environment Specific Configuration Values. [Google Scholar]
- Attali Dean. https://CRAN.R-project.org/package=shinyjs; 2020. shinyjs: Easily Improve the User Experience of Your Shiny Apps in Seconds. [Google Scholar]
- Blighe K., Rana S., Lewis M. EnhancedVolcano: publication-ready volcano plots with enhanced colouring and labeling. 2019. https://github.com/kevinblighe/EnhancedVolcano
- Calvo S.E., Clauser K.R., Mootha V.K. MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 2015;44(D1):D1251–D1257. doi: 10.1093/nar/gkv1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambiaghi A., Ferrario M., Masseroli M. Analysis of metabolomic data: tools, current strategies and future challenges for omics data integration. Brief. Bioinform. 2016;18:498–510. doi: 10.1093/bib/bbw031. [DOI] [PubMed] [Google Scholar]
- Chang W., Cheng C., Allaire J.J., Xie Y., McPherson J., Otto M., Thornton J., Farkas A., Jehl S., Petre S. shiny: web application framework for R. 2019. https://cran.r-project.org/web/packages/shiny/index.html
- Chang W. https://CRAN.R-project.org/package=shinythemes; 2018. shinythemes: Themes for Shiny. [Google Scholar]
- Chang W., Ribeiro B.B. https://CRAN.R-project.org/package=shinydashboard; 2018. shinydashboard: Create Dashboards with ’Shiny’. [Google Scholar]
- Sandhu C., Qureshi A., Emili A. Panomics for precision medicine. Trends Mol. Med. 2018;24:85–101. doi: 10.1016/j.molmed.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J. crosstalk: inter-widget interactivity for HTML widgets. 2016. https://CRAN.R-project.org/package=crosstalk
- Granjon D. https://CRAN.R-project.org/package=shinydashboardPlus; 2020. shinydashboardPlus: Add More ’AdminLTE2’ Components to ’shinydashboard. [Google Scholar]
- Harshbarger J., Kratz A., Carninci P. DEIVA: a web application for interactive visual analysis of differential gene expression profiles. BMC Genomics. 2017;18 doi: 10.1186/s12864-016-3396-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D.W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2008;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Kühl I., Miranda M., Atanassov I., Kuznetsova I., Hinze Y., Mourier A., Filipovska A., Larsson N.G. Transcriptomic and proteomic landscape of mitochondrial dysfunction reveals secondary coenzyme Q deficiency in mammals. eLife. 2017;6:e30952. doi: 10.7554/eLife.30952. A. Chacinska. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumov V., Balashov I., Lagutin V., Borovikov P., Alexeev A. VolcanoR - web service to produce volcano plots and do basic enrichment analysis. bioRxiv. 2017 https://www.biorxiv.org/content/early/2017/07/18/165100 [Google Scholar]
- Pagliarini D.J., Calvo S.E., Chang B., Sheth S.A., Vafai S.B., Ong S.E., Walford G.A., Sugiana C., Boneh A., Chen W.K. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perks K.L., Rossetti G., Kuznetsova I., Hughes L.A., Ermer J.A., Ferreira N., Busch J.D., Rudler D.L., Spahr H., Schöndorf T. PTCD1 is required for 16S rRNA maturation complex stability and mitochondrial ribosome assembly. Cell Rep. 2018;23 doi: 10.1016/j.celrep.2018.03.033. [DOI] [PubMed] [Google Scholar]
- Perrier V., Meyer F., Granjon D., Fellows I., Davis W., Matthews S. https://cran.r-project.org/web/packages/shinyWidgets/index.html
- Rudler D.L., Hughes L.A., Perks K.L., Richman T.R., Kuznetsova I., Ermer J.A., Abudulai L.N., Shearwood A.J., Viola H.M., Hool L.C. Fidelity of translation initiation is required for coordinated respiratory complex assembly. Sci. Adv. 2019;5:eaay2118. doi: 10.1126/sciadv.aay2118. https://advances.sciencemag.org/content/5/12/eaay2118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievert C. plotly for R. 2018. https://plotly-r.com
- Siira S.J., Rossetti G., Richman T.R., Perks K., Ermer J.A., Kuznetsova I., Hughes L., Shearwood A.J., Viola H.M., Hool L.C. Concerted regulation of mitochondrial and nuclear non-coding RNAs by a dual-targeted RNase Z. EMBO Rep. 2018;19:e46198. doi: 10.15252/embr.201846198. https://www.embopress.org/doi/abs/10.15252/embr.201846198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S., Hein M.Y., Stewart A.F. msVolcano: A flexible web application for visualizing quantitative proteomics data. Proteomics. 2016;16:2491–2494. doi: 10.1002/pmic.201600167. https://onlinelibrary.wiley.com/doi/abs/10.1002/pmic.201600167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team R.C. R Foundation for Statistical Computing; 2019. R: A Language and Environment for Statistical Computing.https://www.R-project.org/ [Google Scholar]
- Wickham H., François R., Henry L., Müller K. dplyr: A Grammar of Data Manipulation. 2019. https://cran.r-project.org/web/packages/dplyr/index.html
- Thul P.J., Åkesson L., Wiking M., Mahdessian D., Geladaki A., Ait Blal H., Alm T., Asplund A., Björk L., Breckels L.M. A subcellular map of the human proteome. Science. 2017;356 doi: 10.1126/science.aal3321. In press. [DOI] [PubMed] [Google Scholar]
- Wickham H. Springer-Verlag; 2016. ggplot2: Elegant Graphics for Data Analysis.https://ggplot2.tidyverse.org [Google Scholar]
- Wickham H. https://CRAN.R-project.org/package=stringr; 2019. stringr: Simple, Consistent Wrappers for Common String Operations. [Google Scholar]
- Wickham H., Henry L., Lin Pedersen T., Luciani T.J., Decorde M., Lise V. https://CRAN.R-project.org/package=svglite; 2020. svglite: An ‘SVG’ Graphics Device. [Google Scholar]
- Xie Y., Cheng J., Tan X. DT: A Wrapper of the JavaScript Library ’DataTables. 2020. https://CRAN.R-project.org/package=DT
- Yan J., Risacher S.L., Shen L., Saykin A.J. Network approaches to systems biology analysis of complex disease: integrative methods for multi-omics data. Brief. Bioinform. 2017;19:1370–1381. doi: 10.1093/bib/bbx066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim A., Koti P., Bonnard A., Marchiano F., Dürrbaum M., Garcia-Perez C., Villaveces J., Gamal S., Cardone G., Perocchi F. mitoXplorer, a visual data mining platform to systematically analyze and visualize mitochondrial expression dynamics and mutations. Nucleic Acids Res. 2019;48:605–632. doi: 10.1093/nar/gkz1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The example datasets and code generated during this study are available at.: https://github.com/IrinaVKuznetsova/OmicsVolcano. All data generated or analyzed during this study are included in this published article.