Summary
Rice (Oryza sativa) is a short‐day (SD) plant originally having strong photoperiod sensitivity (PS), with SDs promoting and long days (LDs) suppressing flowering. Although the evolution of PS in rice has been extensively studied, there are few studies that combine the genetic effects and underlying mechanism of different PS gene combinations with variations in PS.
We created a set of isogenic lines among the core PS‐flowering genes Hd1, Ghd7 and DTH8 using CRISPR mutagenesis, to systematically dissect their genetic relationships under different day‐lengths. We investigated their monogenic, digenic, and trigenic effects on target gene regulation and PS variation.
We found that Hd1 and Ghd7 have the primary functions for promoting and repressing flowering, respectively, regardless of day‐length. However, under LD conditions, Hd1 promotes Ghd7 expression and is recruited by Ghd7 and/or DTH8 to form repressive complexes that collaboratively suppress the Ehd1‐Hd3a/RFT1 pathway to block heading, but under SD conditions Hd1 competes with the complexes to promote Hd3a/RFT1 expression, playing a tradeoff relationship with PS flowering. Natural allelic variations of Hd1, Ghd7 and DTH8 in rice populations have resulted in various PS performances.
Our findings reveal that rice PS flowering is controlled by crosstalk of two modules – Hd1–Hd3a/RFT1 in SD conditions and (Hd1/Ghd7/DTH8)–Ehd1–Hd3a/RFT1 in LD conditions – and the divergences of these genes provide the basis for rice adaptation to broad regions.
Keywords: flowering time, heading date, photoperiod sensitivity, rice, short‐day plant
Introduction
Photoperiod is an important environmental factor regulating plant growth and development. Many plant species sense day‐length to determine when to transition from vegetative growth to reproductive development. Plants are classified as long‐day (LD), short‐day (SD) and day‐neutral species according to how they sense photoperiod to induce flowering (Thomas & Vince‐Prue, 1997). Rice (Oryza sativa), a SD species, was domesticated from the wild rice species Oryza rufipogon in tropical and subtropical Asian regions, and has differentiated into two subspecies, indica and japonica (Huang et al., 2012). Many landraces in tropical and subtropical regions, such as O. rufipogon, have very strong photoperiod sensitivity (PS) for flowering (heading). This strong PS completely inhibits heading under LD conditions (i.e. in the early season) and induces heading only under SD conditions (i.e. in the late season in the tropical and subtropical regions). This gives the plants ample time for vegetative growth before flowering, a successful strategy to fully utilize light resource. However, the long‐term natural and artificial selection that occurred during rice domestication and breeding has resulted in the expanded rice cultivation from its primitive domesticated regions to wider regions (from 53ºN to 40ºS), and adaptation to different cropping systems (e.g., single‐ and double‐cropping systems, with a single rice planting per year in the mid‐season, and two rice plantings in the early and late seasons, respectively), which requires rice cultivars to have various degrees of PS that allows them to flower under different day‐lengths (Brambilla & Fornara, 2013; Brambilla et al., 2017).
In rice, a number of heading date genes have been cloned and functionally characterized, showing that rice possesses complicated flowering pathways that are both conservative and unique to those characterized in dicots (Song et al., 2015; Brambilla et al., 2017). An important regulator of photoperiod‐sensitive heading is Heading date 1 (Hd1), a homolog of Arabidopsis thaliana CONSTANS (CO) (Putterill et al., 1995; Yano et al., 2000). The tandem‐duplicated genes Heading date 3 (Hd3a) and RICE FLOWERING LOCUS T 1 (RFT1) are orthologs of the Arabidopsis florigen gene FLOWERING LOCUS (FT), and like FT, they function to induce flowering (Corbesier et al., 2007; Tamaki et al., 2007). In contrast to CO, which promotes flowering under LD conditions, Hd1 generally promotes heading under SD conditions but delays heading under LD by up‐ and downregulating Hd3a/RFT1 expression, respectively (Yano et al., 2000; Ishikawa et al., 2011; Nemoto et al., 2016).
Some aspects of the flowering pathways in rice differ from those discovered in Arabidopsis. For example, the rice heading date gene Early heading‐date 1 (Ehd1) encodes a B‐type response regulator promoting the expression of Hd3a and RFT1 (Doi et al., 2004; Zhao et al., 2015). In addition, several LD‐specific repressors of Ehd1 have been identified, such as Grain number, plant height and heading‐date7 (Ghd7; also named Heading date 4 (Hd4)), Days‐to‐heading on chromosome 8 (DTH8; also named Ghd8/Hd5) and Pseudo‐response regulator 37 (PRR37; also named Hd2/ Ghd7.1) (Xue et al., 2008; Wei et al., 2010; Yan et al., 2011; Koo et al., 2013; Yan et al., 2013; Gao et al., 2014). Hd1, Ghd7 and OsPRR37 encode CO, CO‐like and TOC1 (CCT)‐domain proteins, respectively (Yano et al., 2000; Xue et al., 2008; Koo et al., 2013), and DTH8 is a CCAAT‐box‐binding transcription factor, also named OsNF‐YB11 (Wei et al., 2010; Yan et al., 2011). These genes are central components in the LD‐suppression pathways that influence rice heading date and grain yield, and Ehd1 seems to function as a signal integrator for multiple regulatory pathways under LD conditions (Tsuji et al., 2011; Shrestha et al., 2014).
Research has been carried out to investigate the mutual regulation of Hd1, Ghd7, and DTH8 for flowering (Zhang et al., 2015; Li et al., 2015; Nemoto et al., 2016; Du et al., 2017; Z. Y. Zhang et al., 2017, 2019; B. Zhang et al., 2019). The Hd1 floral repressor activity basically requires Ghd7 in LD conditions in functional DTH8 background (Nemoto et al., 2016). The heading repression in LD by Hd1 also depends on DTH8 in functional Ghd7 background (Du et al., 2017). DTH8/Ghd8‐repressed heading depends on Ghd7 in functional Hd1 background (Wang et al., 2019). But this raises the question, are interactions of any two of the three genes, Hd1, Ghd7, and DTH8, especially in strong PS cultivars, the same when the third gene is nonfunctional? So, the comprehensive mechanism by which these genes interactively control the various extents of PS in the same genetic background, and how the dual activity of Hd1 is achieved, need to be further addressed. In this study, we identified Hd1, Ghd7 and DTH8 as major genetic factors controlling strong rice PS. We created a set of eight isogenic lines with all the combinations of the three genes, and investigated the heading date and expression of the heading‐related genes of these lines in different day‐length conditions. Our results revealed the basic functions of Hd1, Ghd7 and DTH8 and their collaborative roles. We demonstrate that the evolution of PS in rice populations is mainly attributed to natural allelic variations and genetic interactions of Hd1, Ghd7 and DTH8, which is critical for adaptation of rice to wider cultivation areas in different geographic regions.
Materials and Methods
Plant materials and growth conditions
Rice (Oryza sativa L.) materials used in this study included: the NHLD plants (with Hd1, Ghd7 and DTH8) and nhld plants (with hd1, Ghd7 and DTH8), which were selected from an F5 family from a cross between an indica landrace (accession no. I7) and the indica cv Qinghuazhan; Hd1‐transgenic lines with Hd1‐I7 from I7 and Hd1‐Nip from a japonica cv Nipponbare in the nhld background; Hd1‐, Ghd7‐ and DTH8‐knockout lines in the NHLD background and their crossing progenies; the wild rice and cultivated rice landraces. The rice plants examined under field conditions were grown at the Experimental Station of SCAU, Guangzhou (23.13°N, 113.27°E), China, in the normal rice‐growing early season with natural LD (NLD) conditions (13.5–14.0 h day‐lengths from mid‐May to mid‐July as photoperiod‐sensing stage), in the late season with natural SD (NSD) conditions (13.5–12.0 h day‐lengths from mid‐July to October), in artificial LD conditions (ALD, growing from mid‐March with supplemental lighting from July to mid‐November before dusk to reach a total daylight time of 14.0–14.5 h), and in artificial SD conditions (ASD, with 11.0–11.5 h day‐lengths by shading treatment in the late season). In all, 200 plants of the F5 family and 1049 F6 plants were used for primary and fine mapping.
Genomic fragments of 4.3 kb of Hd1 from I7 and Nipponbare, which harbored the coding region, 1.4 kb of upstream sequence, and 1 kb of downstream sequence, were amplified and cloned into the vector pCAMBIA‐1300. The constructs were transferred into nhld plants by Agrobacterium tumefaciens‐mediated transformation. For knockout of Hd1, Ghd7 and DTH8, the target sites were designed using clustered regularly interspaced short palindromic repeats (CRISPR)‐GE/targetDesign (http://skl.scau.edu.cn/) (Xie et al., 2017) and genome‐targeting constructs were prepared using the pYLCRISPR/Cas9Pubi‐H vector (Ma et al., 2015). The constructs were transferred into NHLD plants. The target sites were sequenced and analyzed using CRISPR‐GE/DSDecodeM (Liu et al., 2015; Xie et al., 2017).
Trait measurement and data analysis
Heading dates were scored as time (in d) from seed‐soaking to emergence of the first panicles. The net genetic effects (e) of monogenic Hd1, Ghd7, DTH8 in ALD, NSD and ASD were estimated by e ( H ) = D( Hgd‐7 )−D( hgd‐8 ), e ( G ) = D( hGd‐5 )−D( hgd‐8 ), and e ( D ) = D( hgD‐6 )−D( hgd‐8 ), respectively, where D indicates the average heading date. Their significances at α probability level were tested by the least significant difference (LSD) method (Yang et al., 2018) with the statistics (where Se 2 is the variance from experimental error, n is the average of sample size, and tα is the critical t‐value under α probability level and error freedom degree). To evaluate the high‐order digenic and trigenic genetic interactions between and among the genes, their e‐values were estimated as e ( HGd ) = D( HGd‐4 )−D( hgd‐8 )‐e ( H )‐e ( G ), e ( HgD ) = D( HgD‐4 )−D( hgd‐8 )‐e ( H )‐e ( D ), e ( hGD ) = D( hGD‐2 )−D( hgd‐8 )‐e ( G )‐e ( D ), and e ( HGD ) = D( HGD‐1 )−D( hgd‐8 )‐e ( H )‐e ( G )‐e ( D )‐e ( HGd )‐e ( HgD )‐e ( hGD ), which were tested by or (Yang et al., 2018). Correlation between flowering time and gene expression level was examined by Pearson’s correlation coefficient.
Subcellular localization and bimolecular fluorescence complementation (BiFC)
For subcellular localization assay, the Hd1coding sequence was cloned into the pLYd1GFP vector carrying the GFP tag. The constructs for Hd1‐GFP fusion and the nucleus marker TDR:RFP (transiently‐expressed TDR:RFP fusion protein) were transiently coexpressed in rice leaf protoplasts by polyethylene glycol treatment (Ji et al., 2013).
For BiFC assay, the Ghd7 and DTH8 coding sequences were cloned into the pVN and pVC vectors fusing with the N‐terminal or C‐terminal sequence of YFP to generate Ghd7‐YFPn and DTH8‐YFPc, respectively (Chen et al., 2006). The plasmids were transfected into rice protoplasts followed by an incubation in darkness at 30°C for 15 h, and then their fluorescence was imaged with an Olympus BX61 epifluorescence microscope.
Transcriptional activation activity assay
This assay was performed using the Matchmaker GAL4 Two‐Hybrid System 3 (Clontech, Mountain View, CA, USA). Plasmids containing the GAL4 DNA‐binding domain fused with Hd1‐truncated fragments were individually cotransformed with pGADT7 into yeast strain AH109. Subsequently, the cells were grown on −2 Synthetic Dropout medium lacking ‐Leu‐Trp, and Ade and −4 Synthetic Dropout medium lacking ‐Leu‐Trp‐His‐Ade.
RNA extraction and analysis of gene expression
Fifty‐eight‐day‐old plants grown in ASD, NSD and ALD conditions were used for expression analysis of flowering‐time genes. Leaves were harvested every 4 h within 1 d with three biological replicates. Total RNAs were extracted using Trizol (Invitrogen). Quantitative reverse transcription polymerase chain reaction (qRT‐PCR) assays were carried out as described previously (Zhao et al., 2015), using Actin1 as the internal control.
In vitro protein pulldown
The coding sequences of Hd1, Ghd7 and DTH8 were cloned into the pET‐32‐a, pGET‐4T‐2 and pMAL‐p5X vectors to express the His‐Hd1/maltose‐binding protein (MBP)‐Hd1, glutathione S‐transferase (GST)‐Ghd7 and MBP‐DTH8 proteins, respectively, in Escherichia coli Rosetta. The cells were cultured to OD600nm = 0.8–1, then induced with 0.5 mM isopropyl‐β‐d‐thiogalactopyranoside at 23°C for 6 h. Total proteins were subsequently in a phosphate‐buffered solution (2 mM KH2PO4, 8 mM Na2HPO4, and 200 mM NaCl, pH 8.0) by sonication. Total protein concentration was determined by the Bio‐Rad protein assay (Bio‐Rad).
For in vitro pulldown assay, GST‐Ghd7 and MBP‐DTH8 as baits were immobilized to glutathione agarose beads at 4°C for 2 h. The resins were washed five times with the binding buffer at 300 g 4°C for 2 min. Then MBP‐Hd1, His‐Hd1 and MBP‐DTH8 as prey proteins were added and incubated with bait proteins at 4°C for 2 h. After washing and denaturing, these proteins were pulled down and loaded onto a 10% sodium dodecyl sulfate–polyacrylamide gel, electrophoresed and detected by Western blot with anti‐MBP (1 : 5000, cat. no. HT701; TransGen Biotech, Beijing, China or cat. no. M1321; Sigma), anti‐GST (1 : 5000, cat. no. HT601; TransGen Biotech, Beijing, China), and anti‐His (1 : 5000, cat. no. HT501; TransGen Biotech).
Yeast two‐hybrid (Y2H)
The Hd1 coding sequence and its N‐terminal (encoding B‐box domain, 1–116 amino acids (aa)) and C‐terminal (encoding CCT domain, 277–407 aa) parts were cloned into pGBKT7 as prey. The full‐length coding sequence of DTH8 was cloned into the pGADT7 vector and used as a bait. These constructs were cotransformed into yeast Y187 cells. Interactions between the bait and prey were observed on −3 Synthetic Dropout medium lacking ‐Leu‐Trp‐His, −4 Synthetic Dropout medium lacking Leu‐Trp‐His‐Ade. Plates were incubated for 4 d at 30°C.
Yeast one‐hybrid (Y1H)
The 2.8 kb promoter sequence of Ghd7 was divided into 10 fragments and cloned into the pAbAi vector; these were transferred into the yeast Y1H Gold strain. The Hd1 sequence cloned in pGADT7 was used to transform Y1H Gold that contained the pAbAi constructs with the Ghd7 promoter fragments. Positive interactions were confirmed by Y1H Gold yeast grown on Synthetic Dropout ‐Ura medium at 30℃.
The primers used are listed in Supporting Information Table S3.
Results
Hd1 is the factor suppressing heading under LD conditions
To study the molecular genetic basis of PS for heading in rice, we generated a rice recombinant inbred line population derived from a cross between an indica landrace (accession no. I7 with very strong PS) and another indica variety Qinghuazhan (with weak PS). From this population we identified an F5 family that segregated in a 3 : 1 ratio (χ2 = 0.24, P > 0.5) for dominant late‐heading (153 plants) and recessive early‐heading (47 plants) phenotypes, grown in Guangzhou, China under NLD conditions (in early season) (Fig. S1). When this segregating population was grown in sustained ALD conditions, the dominant plants (and the parent I7) showed a nonheading phenotype even after growing for > 250 d, while the recessive plants headed at 115 d. We named this dominant phenotype NonHeading in LD (NHLD), and the recessive phenotype nhld. When grown in the late season under NSD conditions (Fig. S1), however, the NHLD and nhld plants headed at 75.6 and 82.9 d, respectively (Fig. S2a,b). These observations indicated that the segregation of PS heading in this population is controlled by a single Mendelian genetic factor.
Genetic mapping using the segregating families (F5 and F6) grown in ALD conditions narrowed down the locus controlling the NHLD and nhld phenotypes to a region on chromosome 6, where the heading date gene Hd1 is located (Fig. S2c). Sequencing analysis indicated that NHLD plants (and I7) carry a functional allele, Hd1‐I7, but nhld plants (and Qinghuazhan) have a mutant allele, hd1, with a 4 bp deletion in the second exon, which results in disruption of the conserved CCT domain of the protein (Fig. S2c). We prepared two binary constructs carrying the genomic fragments of Hd1 alleles from I7 (Hd1‐I7) and a japonica variety (Nipponbare) (Hd1‐Nip), which has a 36 bp deletion in the first exon (see later) and introduced them into the nhld plants. Under ALD conditions, the Hd1‐I7‐containing transgenic (T2) lines did not flower even when grown for > 250 d (Fig. S2d,e). In NSD conditions, these T2 lines headed at 103.3 d as compared with 83.5 d in the nhld plants. We also observed that the Hd1‐Nip‐containing transgenic T2 line grown in ALD conditions headed at 141.9 d, 31 d later than the nhld plants (Fig. S2d,e). These results indicate that the Hd1‐I7 allele acts as a strong flowering repressor under LD conditions, and that Hd1‐Nip is a weaker allele.
Hd1 is a nuclear protein with transcriptional activation activity
Hd1 contains two B‐box motifs and a CCT motif, which are highly conserved in plants and predicted to be involved in protein–protein interactions (Takahashi et al., 2009; Gangappa & Botto, 2014). However, the protein characterization of Hd1 is still largely unknown. We investigated the subcellular localization of Hd1 through fusing the full‐length coding sequence of Hd1‐I7 to the 5′‐end of the enhanced green fluorescent protein gene (eGFP) driven by the CaMV 35S promoter. The transiently expressed Hd1:eGFP fusion protein in rice protoplasts colocalized with the nuclear marker TDR:RFP (Ji et al., 2013), indicating that Hd1 is a nuclear protein (Fig. 1a). This result is consistent with a previous report that transient expression of Hd1 was accumulated in the nucleus of tobacco epidermal cells (Goretti et al., 2017).
Fig. 1.
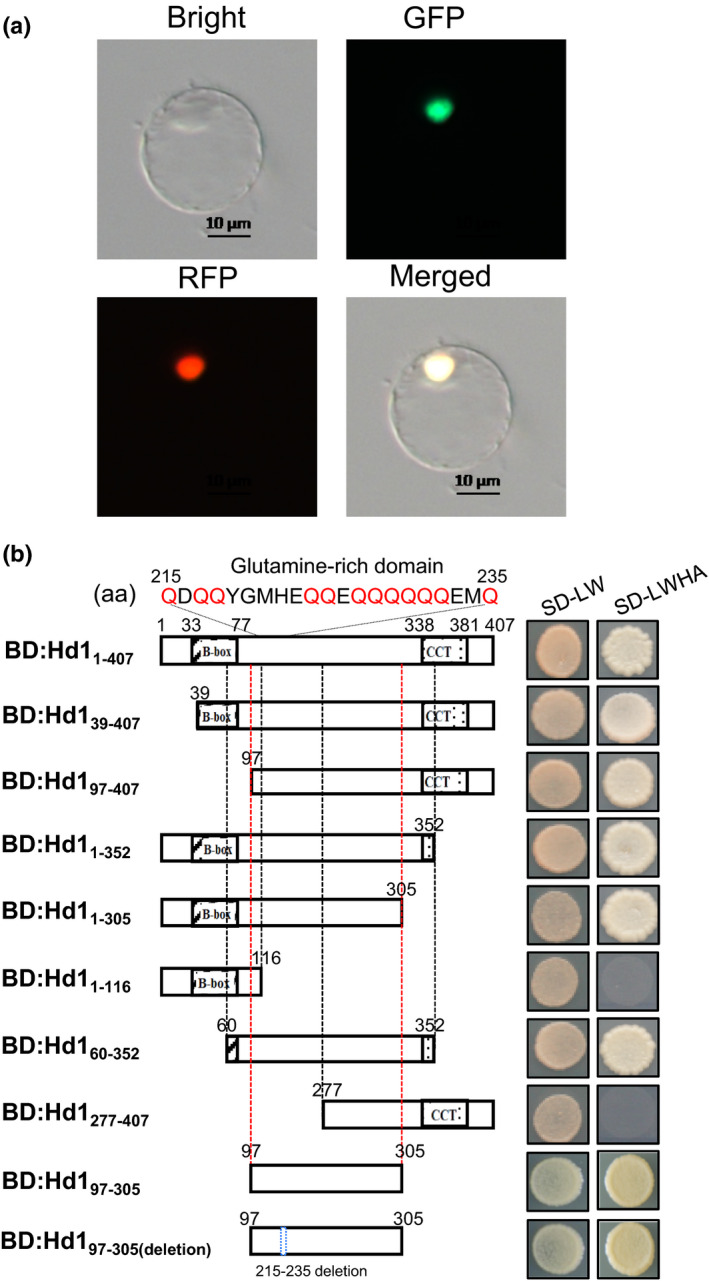
Characterization of Hd1 protein. (a) Subcellular localization of Hd1‐eGFP fusion protein in rice protoplasts. The TDR‐RFP fusion protein served as the nuclear marker. Bars, 10 μm. (b) Transcriptional activation assays of Hd1 and its truncated derivatives in the yeast GAL4 system. BD is the GAL4‐DNA‐binding domain. The red Q indicates glutamine (Gln). SD‐LW, SD‐Leu‐Trp. SD‐LWHA, SD‐Leu‐Trp‐His + Ade.
CO is a transcription factor, which has a glutamine‐rich activation domain located between the B‐box and CCT motifs (Tiwari et al., 2003, 2010). Hd1 also contains a glutamine‐rich region from aa 215–235 (Fig. 1b). We used a Y2H system to test whether this region has activation activity by fusing Hd1 and a set of deletion fragments to the GAL4‐binding domain. The assay showed that Hd1 has strong transcriptional activation activity in yeast, and a region from 97 to 305 aa, but not including the B‐boxes or the CCT domain, is essential for the activation activity (Fig. 1b). The 97–305 aa region is still active after deleting the glutamine‐rich region (215–235 aa). These results suggest that Hd1 may activate the expression of downstream target gene(s) through its 97–305 aa region.
Genetic interactions among and between Hd1, Ghd7 and DTH8 determine differential PS
To test whether other heading‐date regulatory genes contribute to the differential PS between NHLD and nhld plants, we sequenced some heading date genes. NHLD and nhld plants harbor functional alleles of Ghd7, DTH8, Hd18, Ehd1, Hd3a and RFT1 (identical to the alleles in Nipponbare; Kojima et al., 2002; Doi et al., 2004; Zhao et al., 2015; Zhang et al., 2015; Shibaya et al., 2016); functional PRR37 (identical to the alleles in MH63; Gao et al., 2014); functional DTH3 (identical to the alleles in Dianjingyou 1, Bian et al., 2011); and weakly functional Ehd4 (identical to the alleles in 93‐11; Gao et al., 2013). These results confirmed that except for the difference in the Hd1 and hd1 alleles, NHLD and nhld plants are homozygous for most (if not all) of the other major heading‐date genes, consistent with the single‐factor segregation for PS heading in this population.
Ghd7 and DTH8 are important suppressors of LD flowering. Hd1 is reported to interact with Ghd7 and DTH8 at both the genetic level and the molecular level, and the complexes downregulate the expression of Ehd1 and/or Hd3a/RFT1 (Xue et al., 2008; Wei et al., 2010; Nemoto et al., 2016; Du et al., 2017). To systematically analyze the genetic relationship between and among these three genes in controlling differential PS in the same genetic background, we then used the CRISPR/CRISPR‐associated protein 9 (Cas9) genome editing system (Ma et al., 2015) to knock out Hd1, Ghd7 and DTH8 in NHLD plants. We obtained a number of knockout plants for these genes, and identified several homozygous loss‐of‐function mutant lines for each gene (Figs S3a–c, S4a,b). As expected, under ALD conditions the Hd1‐knockout plants showed an early‐heading phenotype (at 109.4 d), similar to that (at 113.2 d) of nhld plants. Under NSD conditions the heading dates of Hd1‐knockout and NHLD plants were similar (at 78.6 and 77.2 d, respectively). Similarly, under ALD conditions, the DTH8‐knockout plants headed at 132.0 d, and in NSD conditions they headed at 69.7 d. We also observed an early‐heading phenotype (90.2 and 63.0 d in ALD and NSD conditions, respectively) for the Ghd7‐knockout plants; this is notably earlier than those of NHLD, nhld, Hd1‐knockout plants and DTH8‐knockout plants in the corresponding day‐length conditions (Fig. S4a,b). These results demonstrated that Hd1, Ghd7 and DTH8 are all required for the very strong PS in the NHLD (or I7) background, as loss of function of any one of these genes negates this PS.
Furthermore, we made several crosses with the NHLD line (with a genotype of Hd1Hd1/Ghd7Ghd7/DTH8DTH8, for short HGD), a Hd1‐knockout line hd1‐6 (hGD), a Ghd7‐knockout line ghd7‐2 (HgD), and a DTH8‐knockout line dth8‐3 (HGd) to generate various combinations (eight in total) of double and triple mutant lines in the same genetic background: the wild‐type NHLD line (HGD‐1), single mutant lines (hGD‐2, HgD‐3 and HGd‐4), double mutant lines (hGd‐5, hgD‐6 and Hgd‐7) and a triple mutant line (hgd‐8). Then we examined their heading date (and expression; see later) under ASD, NSD and ALD conditions during the same late season in Guangzhou.
We found that under ALD conditions, three lines carrying the functional Ghd7 allele with a combination of functional or nonfunctional Hd1 and/or DTH8 (HGD‐1, hGD‐2 and HGd‐4) showed obviously delayed heading as compared with the other lines (Fig. 2a,b). When all eight lines were grown under ASD conditions, four lines carrying functional Hd1 (Hdg‐7, HgD‐3, HGD‐1 and HGd‐4) headed earlier than the hd1‐carrying lines (hGD‐2, hGd‐5, hgD‐6 and hgd‐8) (Fig. 2a,b). These observations imply that Ghd7 is the major heading‐repressor gene under LD conditions but it requires Hd1 and/or DTH8, while Hd1 is the heading‐promoter gene under SD conditions.
Fig. 2.
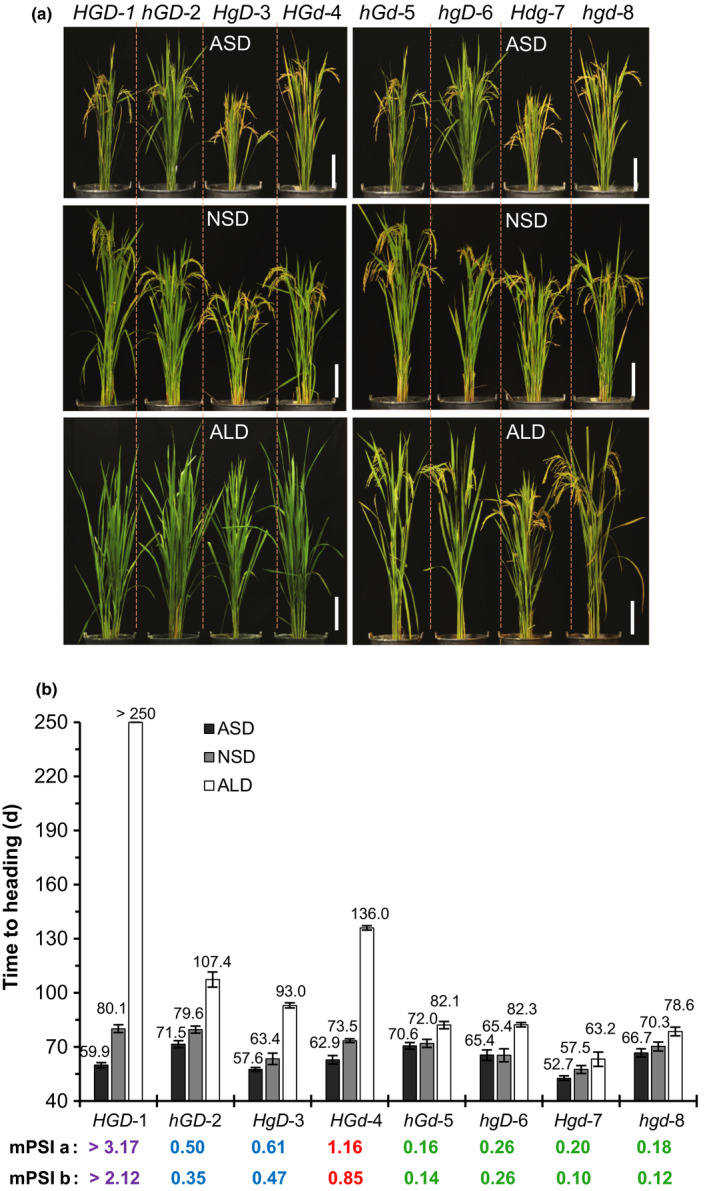
Eight isogenic lines with combinations of the Hd1, Ghd7 and DTH8 alleles generated by gene knockout and crossing. (a) Phenotypes of the lines (97‐d‐old plants) under artificial short‐day (ASD), natural short‐day (NSD) and artificial long‐day (ALD) conditions in Guangzhou. Bars, 25 cm. In NHLD background (HGD‐1), which has strong photoperiod sensitivity (PS), Hd1, Ghd7 and DTH8 were knocked out, respectively, via CRISPR/Cas9 to generate mutant lines (hGD‐2, HgD‐3, HGd‐4). Then a set of eight isogenic lines with all combinations of Hd1, Ghd7 and DTH8 alleles were developed by crossing among hGD‐2, HgD‐3 and HGd‐4 to generate hGd‐5, hgD‐6, Hgd‐7 and hgd‐8”. (b) Heading dates of the lines in the different day‐length conditions, and their modified PS index (mPSI) values based on ASD (a) or NSD conditions (b). Data are means ± SD (n = 50). The degrees of PS are classified into very strong (purple), strong (red), moderate (blue), and weak (green) based on the mPSI values.
The PS index (PSI), previously defined as PSI = [DTHLD – DTHSD]/DTHLD (DTHLD and DTHSD represent daus to heading under LD and SD conditions, respectively), is an indicator for quantitatively measuring PS in rice (Immark et al., 1997). In this study, we observed the heading date of these lines grown in the same period under ASD, NSD and ALD conditions. Since NHLD plants (HGD‐1) were nonheading in ALD conditions, the previous PSI definition is not suitable for this genotype. Therefore, we used modified PSI formulas (mPSIa = (DTHALDc –DTHASD)/DTHASD, or mPSIb = (DTHALD – DTHNSD)/DTHNSD) in this study.
HGD‐1 plants had the highest mPSI (> 3.17 based on ASD conditions or > 2.12 based on NSD conditions), HGd‐4 plants had a relatively higher mPSI (1.16 or 0.85), and two lines (hGD‐2 and HgD‐3) exhibited moderate mPSI values (0.50 and 0.61, respectively) (Fig. 2b). The lines carrying one or none of these functional genes (hGd‐5, hgD‐6, Hgd‐7 and hgd‐8) had low mPSI values (0.16, 0.26, 0.20 and 0.18, respectively). The finding of the hgd‐8 triple mutant having a low mPSI value suggests that other photoperiod‐related heading date gene(s), if any, can only weakly affect PS independent of Hd1, Ghd7 and DTH8.
We calculated the net genetic effect and carried out significance analysis to quantify the monogenic, digenic and trigenic effects among Hd1, Ghd7 and DTH8 on heading date under different day‐lengths. We first analyzed the genetic effect of single genes in regulating heading. We found that in the ghd7/dth8 background (Hgd‐7 vs hgd‐8), Hd1 promoted heading by 14 and 15 d in ASD and ALD conditions, respectively, with significant net genetic effect (e) on heading (Fig. 2b; Table 1), while in the hd1/dth8 background (hGd‐5 vs hgd‐8), Ghd7 suppressed heading by 4 d in both ASD and ALD conditions (Fig. 2b, Table 1). In the hd1/ghd7 background (hgD‐6 vs hgd‐8), DTH8 also slightly suppressed heading (by 4 d) under ALD conditions (Fig. 2b; Table 1).
Table 1.
Net genetic effect (e) on heading date of single genes and their combinatorial effects of Hd1, Ghd7 and DTH8 in different day‐length conditions.
| Lines | Monogenic e | Digenic e | Trigenic e | ||||||
|---|---|---|---|---|---|---|---|---|---|
| ASD | NSD | ALD | ASD | NSD | ALD | ASD | NSD | ALD | |
| H gd‐7 | −14.1** | −12.8** | −15.4** | ||||||
| h G d‐5 | +3.8** | +1.7** | +3.5** | ||||||
| hg D ‐6 | −1.3* | −4.9** | +3.7** | ||||||
| HG d‐4 | +6.3** | +14.2** | +69.3** | ||||||
| H g D ‐3 | +6.2** | +10.8** | +26.1** | ||||||
| h GD ‐2 | +2.2* | +12.5** | +21.6** | ||||||
| HGD ‐1 | −10.0** | −11.8** | > +62.7** | ||||||
ASD, artificial short‐day conditions; ALD, artificial long‐day conditions; NSD, natural short‐day conditions.
‘−’ indicates the promotion effect on heading from functional gene. ‘+’ indicates the inhibition effect on heading from functional gene. * , **, significance at 5% and 1% levels, respectively.
The single‐gene effects, for example, the effect of Hd1, were calculated by subtracting the basic effect of the hgd genotype by e(H) = D(Hgd‐7) – D(hgd‐8), to clarify the original function of Hd1.
The double‐gene effects, the effect of the Hd1/Ghd7 combination, were calculated by subtracting the effects of the hgd genotype and the single‐gene effects, e ( HGd ) = D( HGd‐4 ) – D( hgd‐8 )‐e ( H )‐e ( G ).
The effect of the Hd1/Ghd7/DTH8 combination, the basic effect of the hgd genotype, the single‐gene effects, and the double‐gene effects are all subtracted, e ( HGD ) = D( HGD‐1 ) – D( hgd‐8 )‐e ( H )‐e ( G )‐e ( D )‐e ( HGd )‐e ( HgD )‐e ( hGD ).
Next, we evaluated the effects of two‐gene combinations and compared them to the effects of single genes. Under ALD conditions, the Hd1/Ghd7 combination (HGd‐4) showed relatively stronger heading suppression effect (delayed heading by 54 d as compared with the hGd‐5 line). Under ASD conditions, the heading date of the HGd‐4 line was 8 d earlier than that of the hGd‐5 line, but 10 d later than that of the Hgd‐7 line (Fig. 2b). Under ALD conditions the suppression genetic effect of the Hd1‐DTH8 interaction (in HgD‐3) was moderate (delayed by 30 d) as compared with Hgd‐7, and was weak (delayed heading by 10 d) when compared with the hgD‐6 line. The Hd1/Ghd7 and Hd1/DTH8 combinations were detected with significant digenic genetic effects (as compared with the responding single gene effects) in ALD, NSD and ASD conditions; for example, Hd1/Ghd7 had an e‐value of 69.3 under ALD conditions (Table 1). The Ghd7–DTH8 interaction in hGD‐2 also had a moderate suppression effect (delayed heading by 25 d) under ALD conditions as compared with the hGd‐5 and hgD‐6 lines (with an e‐value of 21.6), respectively (Fig. 2b; Table 1), confirming the presence of high‐order genetic interaction among the three genes, in addition to the accumulative effects of single gene and double‐gene combinations.
We noticed that the presence of the functional Ghd7 or DTH8 alleles changed the primary function of Hd1 from promotion to suppression of heading under ALD conditions, while in the ASD condition, the promotion effect of Hd1 alone (Hgd‐7 vs. hgd‐8) with 14 d was competed and partially impaired by Ghd7 (inHGd‐4) or DTH8 (in HgD‐3), leading to only promoted heading for 8 d, asHGd‐4, HgD‐3 compared to the hGd‐5, hgD‐6 respectively (Fig. 2b; Table 1). Here, we confirmed again that the Hd1‐Ghd7‐DTH8 trigenic interaction in HGD‐1 produced the very strong inhibition effect (with an e‐value > 62.7) under ALD conditions (Fig. 2b; Table 1).
Taken together, our results suggest that Hd1 promotes flowering and Ghd7 represses flowering regardless of day‐length. The Ghd7/DTH8 combination can enhance the suppression under LD conditions. The Hd1/DTH8, Hd1/Ghd7 and Hd1/Ghd7/DTH8 combinations have incrementally enhanced LD‐suppression roles that convert the Hd1 effect from promotion to great delays of heading, but they compete the promotion effect of Hd1 to different extents in SD conditions, which determine the degrees of PS in rice.
Hd1, Ghd7 and DTH8 regulate Ehd1 and Hd3a/RFT1 expression in a complex manner
Ehd1 is the essential integrator gene that regulates the expression of the florigen genes Hd3a and RFT1 under different day‐lengths (Zhao et al., 2015). We analyzed their expression in the eight lines discussed earlier, to understand the molecular effects of Hd1, Ghd7 and DTH8 and their combinations. We found that the overall expression levels of Ehd1, Hd3a and RFT1 in these lines were roughly consistent with their heading dates in the ALD and ASD conditions (Fig. S5a,b), showing significant Pearson’s correlation coefficients between the heading date and gene expression level under ALD conditions at most of the time‐points (Table S1). We also compared the expression of Ehd1, Hd3a and RFT1 under ALD and ASD conditions in the eight lines. The results showed that the lines with high mPSI values had larger differences in the expression of these genes between the ALD and ASD conditions. For the hgd‐7 and hgd‐8 lines with weak PSI values, the RNA levels of Ehd1, Hd3a and RFT1 were very similar (Fig. S6a–h).
We then analyzed the regulation of Ehd1, Hd3a and RFT1 expression in the hGd‐5, hgD‐6 and Hgd‐7 lines. Under both ASD and ALD conditions, the expression levels of Hd3a and RFT1 in the Hgd‐7 line, mainly at Zeitgeber time 20 (ZT20) and ZT24, were higher than in the hgd‐8 line (Fig. 3a). In the hd1/ghd7 background (hgD‐6 vs hgd‐8), DTH8 slightly suppressed heading in ALD by repressing the expression of Ehd1, Hd3a and RFT1 in the morning (Fig. 3b). In the hd1/dth8 background (hGd‐5 vs hgd‐8), Ghd7 weakly suppressed heading in ALD by repressing the expression of the Ehd1‐Hd3a/RFT1 pathway from dawn to morning. While under ASD conditions, Ghd7 also weakly suppressed heading, but the transcript abundances of Ehd1/RFT1/Hd3a were similar to those of hgd‐8 (Fig. 3c).
Fig. 3.
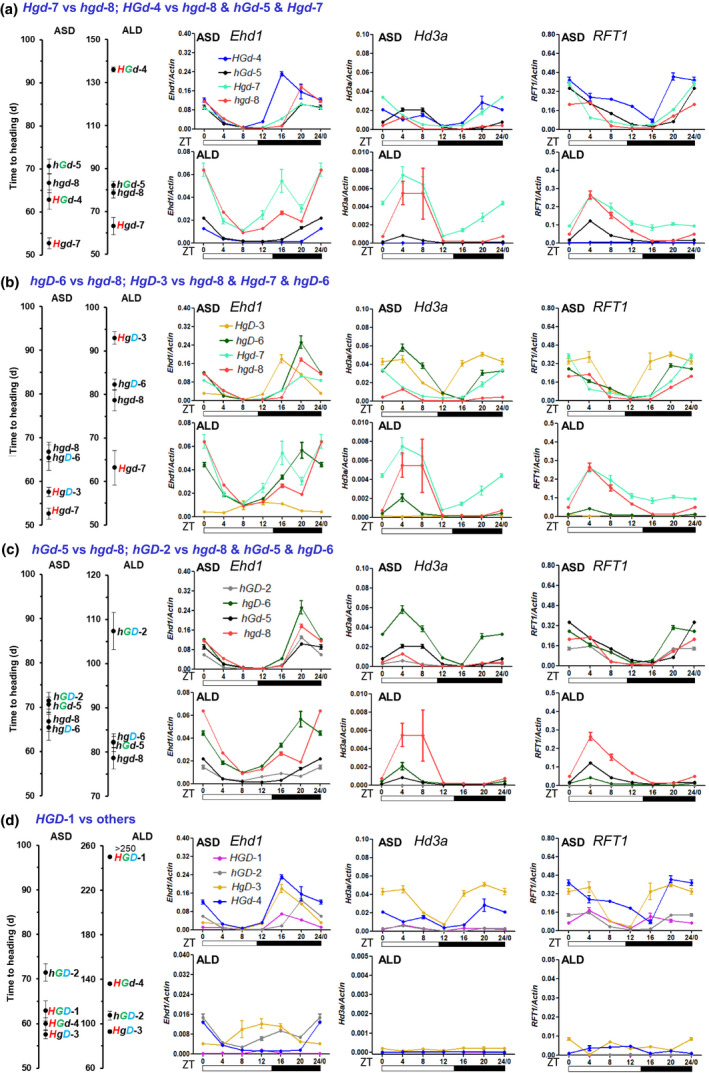
Heading dates and expression of Ehd1, Hd3a and RFT1 in the eight isogenic lines under artificial short‐day (ASD) and artificial long‐day (ALD) conditions. Fifty‐eight‐day‐old plants were analyzed. (a) Comparisons of the effects with and without Hd1 (Hgd‐7 vs hgd‐8), and with and without the Hd1‐Ghd7 genetic interaction (HGd‐4 vs hgd‐8 & hGd‐5 & Hgd‐7). (b) Comparisons of the effects with and without DTH8 (hgD‐6 vs hgd‐8), and with and without the Hd1‐DTH8 genetic interaction (HgD‐3 vs hgd‐8 &Hgd‐7 & hgD‐6). (c) Comparisons of the effects with and without Ghd7 (hGd‐5 vs hgd‐8), and with and without the Ghd7‐DTH8 genetic interaction (hGD‐2 vs hgd‐8 & hGd‐5 & hgD‐6). (d) Comparisons of the effects with and without the Hd1‐Ghd7‐DTH8 genetic interaction (HGD‐1 vs others). ZT, Zeitgeber time. Error bars represent SD of three biological replicates.
We then deduced the interactive effects of the two functional‐allele combinations. For the Hd1/Ghd7 combination (in HGd‐4), under ASD conditions, it showed higher levels of Ehd1 and Hd3a/RFT1 expression than the hgd‐8 line and earlier heading, while under ALD conditions the expression of the Ehd1‐Hd3a/RFT1 pathway was strongly suppressed mainly at ZT20 and ZT24 compared with hgd‐8 and hGd‐5, leading to very late heading (Fig. 3a). The genetic effect of the Hd1/DTH8 combination (HgD‐3 vs hgd‐8) was similar to that of the Hd1/Ghd7 combination (HGd‐4) in the ALD and ASD conditions (Fig. 3b). Under ALD conditions the Ghd7/DTH8 combination (hGD‐2 vs. hgd‐8 and hGd‐5) showed stronger suppression effects to the Ehd1‐Hd3a/RFT1 pathway in the morning, but under ASD conditions it showed only weak suppression to this pathway (Fig. 3c). We again confirmed that the combination of Hd1/Ghd7/DTH8 (HGD‐1) completely suppressed the Ehd1‐Hd3a/RFT1 pathway at all time‐points under ALD conditions, but moderately repressed this pathway under ASD conditions (Fig. 3d). These data support the idea that the primary function of Ghd7 is to suppress the expression of the Ehd1‐Hd3a/RFT1 pathway, whereas the primary function of Hd1 is to promote Hd3a/RFT1 expression, regardless of day‐length. In LD conditions, the Ehd1‐Hd3a/RFT1 pathway is progressively suppressed by these combinations – Hd1/DTH8, Ghd7/DTH8, Hd1/Ghd7 and Hd1/Ghd7/DTH8 – while in SD conditions, the expression patterns of Ehd1 and Hd3a/RFT1 are regulated in a more complex manner.
We also analyzed the expression of Hd1, Ghd7 and DTH8 in these lines and explored their possible regulatory relationships based on their genetic effects. We observed that under ASD conditions, the expression levels of Ghd7 in the HGD‐1, HGd‐2, hGD‐4 and hGd‐5 lines were very low, but the Ghd7 mRNA levels were increased in the daytime under ALD conditions. This is consistent with the notion that Ghd7 exerts a repressive effect mainly in LD conditions. Under ALD conditions, the Ghd7 mRNA levels at ZT4 in the lines with Hd1 (HGD‐1 and HGd‐2) were higher than those in the hGD‐4 and hGd‐5 lines (Fig. S7a). DTH8 has peak expression during the darkness in ASD, and higher expression during the dusk and darkness in ALD conditions (Fig. S7b). In ASD conditions, the Hd1 mRNA levels in the HGD‐1, HgD‐3, HGd‐4 and Hgd‐7 lines were similar, but under ALD conditions the Hd1 mRNA levels in the HGD‐1 and HGd‐4 lines were higher than those in the HgD‐3 and Hgd‐7 lines (Fig. S7c). Considering that the Hd1/Ghd7 and Hd1/Ghd7/DTH8 genotypes have stronger repression of flowering in LD conditions, we speculated that Hd1 might play a role in promoting Ghd7 expression, thus enhancing the function of Ghd7. Indeed, using Y1H assays we detected direct binding of the Hd1 protein to the promoter region of Ghd7 (Fig. S8a,b), supporting the presence of a protein–DNA interaction between these two genes.
Hd1, Ghd7 and DTH8 form functional complexes
Previous reports showed that pairwise interactions exist between Hd1, Ghd7 and DTH8. (Nemoto et al., 2016; Du et al., 2017; Goretti et al., 2017; Cai et al., 2019). In this study we also detected their interactions using different methods. GST‐tagged Ghd7 could bind MBP‐tagged Hd1 (Fig. 4a), MBP‐tagged DTH8 could pull down HIS‐tagged Hd1 (Fig. 4b), and GST‐tagged Ghd7 could pull down MBP‐tagged DTH8 (Fig. 4c). Y2H assays also confirmed that DTH8 interacted with both the B‐box and the CCT domain of Hd1, and BiFC assay verified that Ghd7‐YFPn and DTH8‐YFPc interacted in the nuclei of rice protoplasts (Fig. S9a,b). Finally, we further used a three‐protein pulldown assay to verify that MBP‐Hd1 and MBP‐DTH8 were simultaneously pulled down by GST‐Ghd7 (Fig. 4d). Moreover, in this experiment regardless of the binding prioritization of Hd1 and DTH8 by sequentially or simultaneously adding the proteins to the reactions, they could equally bind to Ghd7 without biased competition (Fig. 4d), indicating that the three proteins interacted with each other though different binding domains to form a complex (the HGD complex).
Fig. 4.
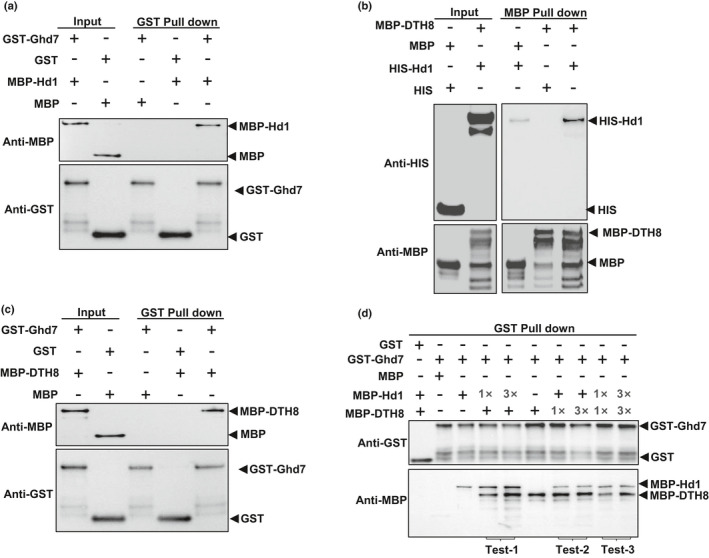
In vitro pulldown assays for interactions between and among Hd1, Ghd7 and DTH8. (a) Hd1 interacted with Ghd7. GST‐Ghd7 is used as bait. (b) Hd1 interacted with DTH8. MBP‐DTH8 is used as bait. (c) DTH8 interacted with Ghd7. GST‐Ghd7 is used as bait. (d) Interactions among Hd1, Ghd7 and DTH8. GST‐Ghd7 was used as a bait. In Test‐1, the prey MBP‐Hd1 protein was first added to the bait GST‐Ghd7. From 1× to 3×, the intensity of the anti‐MBP hybridization band of MBP‐Hd1 gradually increased, indicating that more MBP‐Hd1 was pulled down by GST‐Ghd7. After washing, the second prey, MBP‐DTH8, was added, but the amount was constant. The intensity of the hybridization band of MBP‐DTH8 was also increased, which implies that DTH8 was more likely to bind the increased Hd1, proving the formation of a three‐protein complex of Ghd7/Hd1/DTH8. In Test‐2, the prey MBP‐DTH8 was added first to the bait GST‐Ghd7. From 1× to 3×, the anti‐MBP hybridization band of MBP‐DTH8 remained unchanged, indicating that 1× of DTH8 was enough to bind to all the bait GST‐Ghd7. After washing, the second prey MBP‐Hd1 was added with a constant amount. Its hybridization band was basically unchanged, implying that Hd1 bound to Ghd7 and DTH8 with a different domain, forming a three‐protein complex. In Test‐3, 1× of prey MBP‐DTH8 and MBP‐Hd1, or 3× of prey MBP‐DTH8 and MBP‐Hd1 were added simultaneously to the reaction. It was observed that the hybridization bands of MBP‐DTH8 and MBP‐Hd1 had similar intensities. Test‐1, Test‐2 and Test‐3 together explained that Ghd7/DTH8/Hd1 formed a three‐protein complex at a ratio of 1 : 1 : 1, instead of independent pairwise interactions. It is not the case that the amount of GST‐Ghd7 was excessive, and thus a part of it bound to MBP‐Hd1 and another part of it interacted with MBP‐DTH8.
Natural variations of Hd1, Ghd7 and DTH8 confer PS evolution
To explore the relationship between the natural variation of these genes and PS evolution during rice domestication/breeding, we sequenced the coding region of Hd1, Ghd7 and DTH8 from a germplasm collection, including 65 landraces (native cultivars), 61 modern varieties, and an O. rufipogon accession, and classified these genes into functional and nonfunctional haplotypes based on the prediction of deleterious effect of amino acid changes through the SIFT website. We also observed the heading dates of these materials in the same late season under ASD (or NSD) and ALD conditions, or under early season NLD and late season in Guangzhou (Table S2). Then we analyzed the relationship between the different combinations of the gene types and the degree of PS.
Thirteen Hd1 haplotypes (types) were detected in these cultivated rice lines (and O. rufipogon); seven types are functional and six types are nonfunctional. For Ghd7, 17 haplotypes were detected among these materials, of which two (types 15 and 16) are nonfunctional. Nine haplotypes of DTH8 were identified and five of them are functional (Fig. S10a–c; Table S2). These functional haplotypes might include normal‐ or reduced‐function variations, such as the weakly functional type 14 present in Nipponbare verified by genetic transformation (Fig. S2d,e).
The perennial wild rice O. rufipogon contains functional alleles of Hd1 (Type 13), Ghd7 (type 17) and DTH8 (type 5) (Fig. S10d); this genotype confers very strong PS (e.g. heading only in October in typical SD conditions). Among the 65 landraces, which are mainly distributed in subtropical regions, 41 lines had the HGD‐type combinations, and most of them (36 lines) showed strong or very strong PS (no‐heading in ALD conditions). The other 21 lines had nonHGD‐type combinations with weak or even no PS (Fig. 5a,c; Table S2). Strikingly, most of the modern indica varieties cultivated in South China were the hGd‐type with low mPSI values (Fig. 5a,c; Table S2), suggesting that weak PS had been selected in breeding of the modern indica rice varieties. In the modern japonica rice varieties, those (11 varieties) mainly cultivated in Middle China contained the HGD‐type combination and exhibited relatively strong PS, while those with other genotypes producing weak PS are cultivated in North and Northeast China (Fig. 5b; Table S2).
Fig. 5.
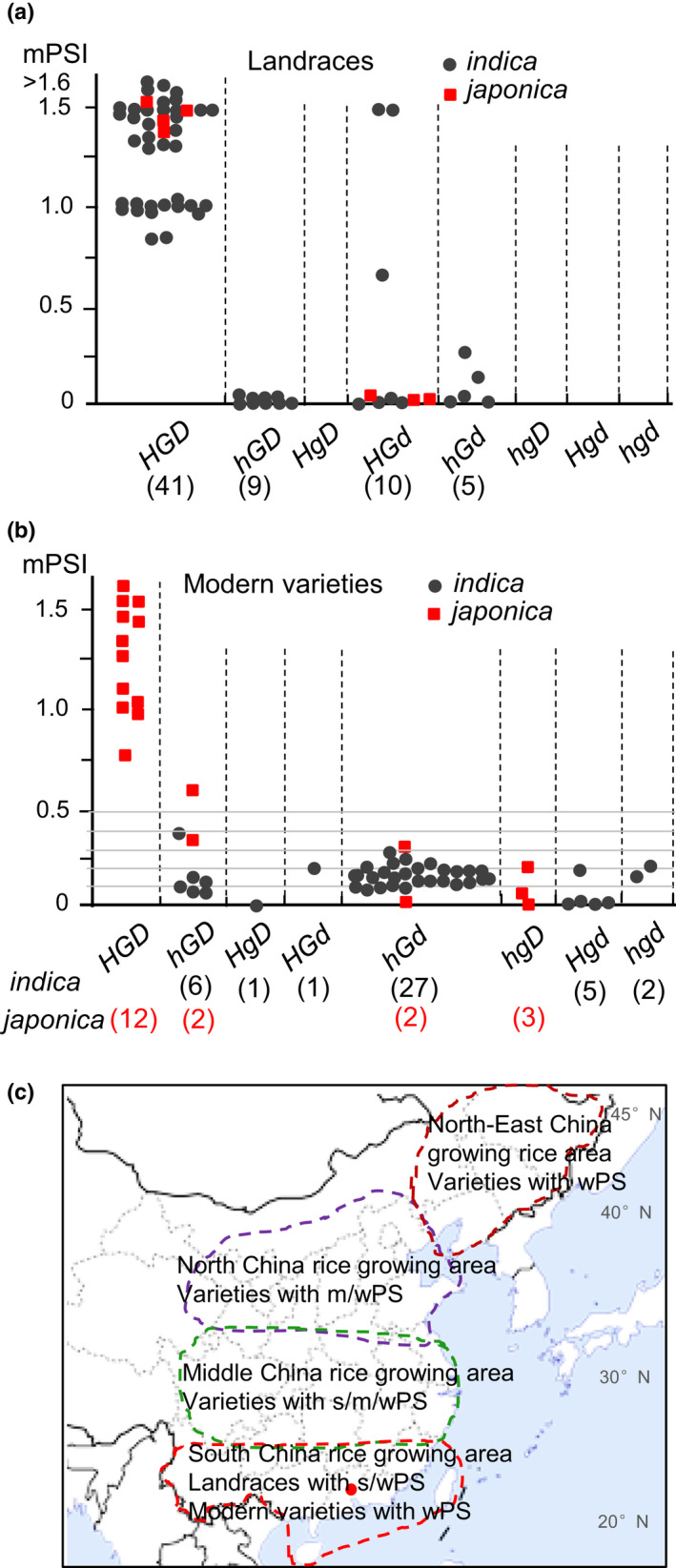
Hd1, Ghd7 and DTH8 allele combinations confer variations in rice photoperiod sensitivity (PS) and contribute to geographical adaptation. (a, b) Modified PS index (mPSI) of landraces (a) and modern indica varieties (b), which are mainly distributed in South China, and modern japonica varieties (b) in Middle and North/Northeast China. The functional types of the genes (H, G, D) include various haplotypes that may have normal‐ or reduced‐function variations. The nonfunctional alleles (h, g, d) were derived from mutations causing frame‐shift. The mPSI values in (a) were measured with heading dates under artificial long‐day (ALD) and natural short‐day (NSD) conditions, and those in (b) were based on heading dates under ALD and artificial short‐day (ASD) conditions for japonica varieties and NLD and NSD conditions for indica varieties. (c) Adaptation of rice varieties to different day‐lengths and cropping systems in the main rice‐growing areas in China. In the South China (Guangzhou is indicated with a red dot) rice‐growing area where mainly indica varieties are cultivated, those landraces (carrying the HGD‐type) with strong photoperiod sensitivity (sPS) are planted in the late season (with SD conditions), and some landraces and most modern varieties (mainly containing the hGd‐type) having weak PS (wPS) are planted in the early season (with LD conditions) and the late season. In Middle China rice‐growing areas where both indica and japonica varieties are cultivated, indica varieties having wPS are planted in the early season (with LD conditions) and the late season (with SD conditions), or in the mid‐season (with SD conditions, heading in late August to early September). And japonica varieties having sPS or moderate PS (mPS) (mainly containing the HGD‐type or hGD‐type) are planted the in the mid‐season (with SD conditions, heading in early September). In North and Northeast China rice‐growing areas, only japonica varieties having wPS are planted in the mid‐season (with LD conditions, heading in early to mid‐August).
Discussion
In this study, we systematically analyzed the genetic effects and the regulatory mechanism of the Hd1, Ghd7 and DTH8 alleles and their combinations for controlling PS variation in rice by using a set of eight isogenic lines with single, dual or triple knockout of these genes, and a collection of landraces and modern cultivars. Based on the results, we proposed a molecular model of this regulatory system (Fig. 6a,b). Hd1 has a primary function for promoting the expression of Hd3a/RFT1 and heading regardless of the day‐length, while Ghd7 (hGd) or DTH8 (hgD) alone has only a weak effect on suppressing heading in LD conditions. However, in the lines with different combinations of Hd1, Ghd7 and DTH8, Hd1 is recruited by Ghd7 and/or DTH8 to form repressive complexes (Hd1/DTH8, Ghd7/DTH8, Hd1/Ghd7 or Hd1/Ghd7/DTH8), which possess gradually enhanced effects to suppress the Ehd1‐Hd3a/RFT1 pathway, thus repressing heading in increasing degrees. In SD conditions, the repressive functions of these complexes decrease, and Hd1 plays its original promotion role to compete the repressive complexes. These characters exhibit a collaborative effect in LD conditions and an antagonistic effect in SD conditions between Hd1 and Ghd7/DTH8. The genetic interactions of these alleles mediated by the opposing actions of Hd1 enlarge the differences of heading date between LD and SD conditions, thus conferring various degrees of PS in rice populations (Fig. 6).
Fig. 6.
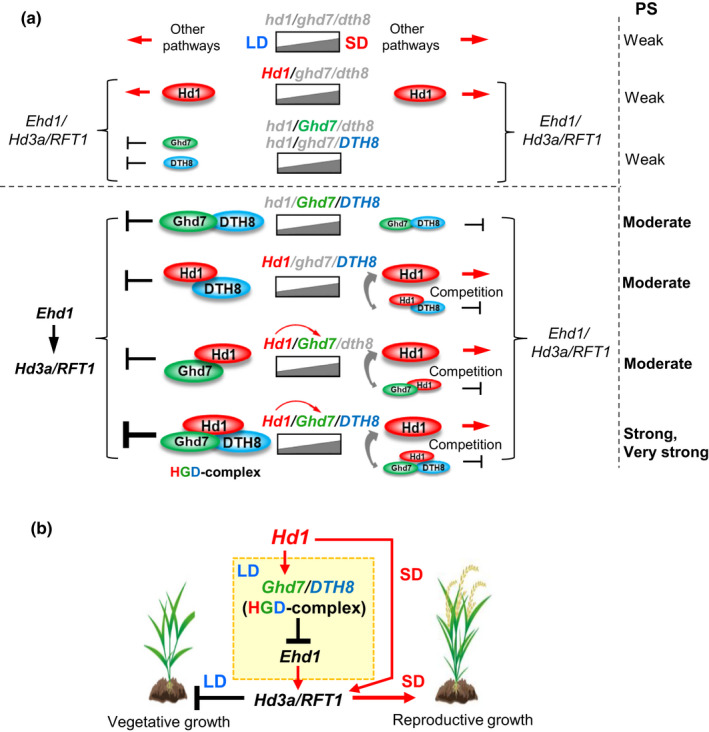
A working model of the interactions among Hd1, Ghd7 and DTH8 for regulating heading date and photoperiod sensitivity (PS) in rice. (a) In the hd1/ghd7/dth8 genotype, the basal expressions of Ehd1 and Hd3a/RFT1 are regulated by other pathways. ‘Ehd1/Hd3a/RFT1’ means that expression of these genes is regulated independently by the upstream regulator(s), or Ehd1 is first regulated and then Hd3a/RFT1 are regulated by Ehd1, or both ways. In the absence of functional Ghd7 and DTH8 alleles, Hd1 shows a primary role of promoting Hd3a/RFT1 expressions and heading under both short‐day (SD) and long‐day (LD) conditions. Ghd7 (hGd) or DTH8 (hgD) alone have weak effect on suppressing heading, mainly in LD conditions. In various combinations among the Hd1, Ghd7 and DTH8 alleles, under LD conditions Hd1 promotes Ghd7 expression and is also recruited by Ghd7 and/or DTH8 to form repressive complexes that have differently enhanced suppression effects on the Ehd1‐Hd3a/RFT1 pathway for repressing heading. In SD conditions, owing to the weakened suppression effect of Ghd7, the repressive functions of these complexes are decreased, and Hd1 competes with the complexes to promoting heading with reduced extents. The interactions of the different gene combinations (including multiple allelic variations of the genes) produce various extents of suppression and promotion effects on heading in response to different day‐lengths, and thus control different heading dates and confer various degrees of PS in rice populations. The different sizes of the marks for the proteins and promotion and repression indicate their relatively different levels of effect. Notably, the HGD working model we proposed for the PS control is based on the presence of other possibly essential factor(s) that are involved in photoperiodic flowering regulation in rice. (b) Simplified rice flowering regulatory pathways mediated commonly by Hd1 in LD and SD conditions. Other known factors related to these two pathways are omitted.
Our results clarified several questions. First, why doesn’t Hd1 always promote flowering under SD conditions and suppress it under LD conditions. The primary functions of Hd1, in the ghd7/dth8 background, is for promoting flowering, regardless of day‐length. In the HGD lines, when the day‐length is shorter than a certain threshold, the competition between the Hd1 promotion and HGD‐type repression reaches a balance: Hd1 may neither promote nor repress heading (Fig. 2b). Second, why do Ghd7 or DTH8 have large differences of LD‐suppression effects in different background? We revealed that the LD‐suppression effect of Ghd7 (hGd) or DTH8 (hgD) alone is weak in plants lacking functional Hd1, but this effect is largely enhanced upon genetic interactions with Hd1. Third, we demonstrate that Hd1‐H3da/RFT1 and (Ghd7/DTH8)‐Ehd1‐Hd3a/RFT1 are not simply two independent pathways for controlling heading date as previously proposed (Song et al., 2015). In fact, Hd1 has bidirectional roles in mediating the crosstalk between the SD‐ and LD‐related molecular pathways Hd1‐Hd3a/RFT1 and (Hd1/Ghd7/DTH8)‐Ehd1‐H3da/RFT1, respectively (Fig. 6b).
Importantly, our results (Table 1) clearly indicate that the net genetic effect of the HGD‐1 line under LD conditions (the genetic effect of the three genes after subtracting those of all the single genes and two‐gene combinations) was much larger than zero (> 62.7 d), meaning that the genetic effect of the three genes is not simply an accumulative and replacing effect of the single genes and two‐gene interactions, but there must be a high‐order genetic/molecular interaction among the three (and possibly more other(s)) genes, which produces a much stronger LD‐suppression effect than the total single‐ and two‐gene effects.
Under LD conditions, the Hd1/Ghd7/DTH8 and Hd1/Ghd7 combinations completely and largely inhibited the Ehd1‐H3da/RFT1 pathway, respectively. We speculate that this is partly a result of the promoted Ghd7 expression by Hd1 (Fig. S7a). On the other hand, besides the expression regulation of the genes, the protein complexes with Hd1, Ghd7 and/or DTH8 may possess largely enhanced or new biochemical functions compared with the single proteins. Hd1 and Ghd7 are CCT‐domain‐containing proteins (Yano et al., 2000; Xue et al., 2008), while DTH8 is a subunit of the NF‐Y complex (Wei et al., 2010). The NF‐Y complex is a sequence‐specific heterotrimeric transcription factor (Mantovani, 1999). Many CCT‐domain proteins, which are involved in flowering regulation in plants, interact with NF‐Y subunits to regulate transcription of target genes (Song et al., 2015; Brambilla & Fornara, 2017). The Ghd7/Hd1 complex can directly bind the Ehd1 promoter (Nemoto et al., 2016). Therefore, in LD conditions, Hd1, Ghd7 and DTH8 may form active NF‐Y complexes that undergo dynamic changes in composition and/or protein modifications in response to different day‐lengths. In SD conditions, the expression of Ghd7 is decreased and Hd1 may form a complex with a blue light‐responsive flavin mononucleotide‐binding protein OsHAL3, which binds to the promoter region of Hd3a (and possibly also to that of RFT1) for promoting its expression (Su et al., 2016).
The Arabidopsis CO protein accumulates specifically in the daytime under LD conditions, which induces the expression of FT to promote flowering (Song et al., 2012). CO accumulation is regulated by the GI/FKF1–CDF1 module, and the different effects of day‐length on flowering (LD promotes flowering and SD has no effect) are determined by the coincident and staggered expression phases of GI and FKF1 in LD and SD conditions, respectively (Sawa et al., 2007). We found that in rice, the CO homolog Hd1 promotes the expression of the FT orthologs Hd3a/RFT1 in both SD and LD conditions. This indicates that the basic CO–FT and Hd1–Hd3aRFT1 regulatory pathways are functionally conserved in LD and SD plants, but rice has specifically evolved the (Hd1/Ghd7/DTH8)–Ehd1–H3da/RFT1 pathway, which recruited the conserved Hd1 to a novel repressive function.
Moreover, the differential effects of day‐length on rice heading are determined by the conflict between the SD‐promotion Hd1‐Hd3aRFT1 pathway and the LD‐repression (Hd1/Ghd7/DTH8)–Ehd1–H3da/RFT1 pathway, indicating the genetic diversity of flowering‐control mechanisms between monocot and dicot species. Homologous genes of Hd1, Ghd7, DTH8 and Ehd1 exist in maize (Zea mays) and sorghum (Sorghum bicolor) (Brambilla et al., 2017), suggesting that the PS‐flowering mechanism in rice may also be conserved in other tropical‐origin monocot SD plants.
Hd1, Ghd7 and DTH8 show abundant natural variations (Fig. S10; Table S2), which greatly increased the genetic complexity in heading‐date control and PS in rice. We show that most rice landraces, like O. rufipogon, contain functional Hd1, Ghd7 and DTH8 alleles and have strong or very strong PS. In the past, such landraces could only be cultivated in NSD conditions in tropical and subtropical regions. Our results demonstrate that the evolution of rice PS from very strong in the wild rice and landraces to various reduced degrees in the modern varieties during rice domestication and breeding mainly involved natural allelic variations (including functional and nonfunctional haplotypes) of Hd1, Ghd7 and DTH8 and their genetic interactions (Figs 5, S10). For example, many modern indica varieties were bred to possess the hGd‐type and hGD‐type of the genes, having low sensitivity to day‐length (Fig. 5b), which are suitable for broad cultivation in both NLD and NSD conditions in South China and Middle China areas with different day‐length conditions. The modern japonica varieties with strong or moderate PS (mainly having the HGD‐type) are suitable for planting in the mid‐season (single‐cropping system) in Middle China areas (heading in late August to early September under SD conditions) (Fig. 5b,c). However, those modern japonica varieties with weak PS were bred for planting in the mid‐season (heading in early to mid‐August under LD conditions) in North and North‐east China areas (Fig. 5b,c). Because functional Ghd7 and DTH8 alleles are also contribute to high grain yield (Xue et al., 2008; Wei et al., 2010; Yan et al., 2011), existing varieties with inappropriately strong PS (having the HGD‐type) can be improved for weaker PS by CRISPR/Cas9 editing of Hd1. Therefore, our findings provide valuable information for breeding of high‐yield rice varieties with certain genotypes of these genes and appropriate PS degrees as described earlier.
Author contributions
JG and Y‐GL conceived this work, analyzed data and wrote the manuscript; WZ and DR performed most of the experiments; MH, KS, JF, JZ, DX, WX, SL, HZ, RQ, WT, RY, HC, XX and LC performed some of the experiments and data analysis. WZ and DR contributed equally to this work.
Supporting information
Fig. S1 Relationship between day‐length and latitude in different months in several locations (cities) of China.
Fig. S2 Identification of Hd1 as the factor conferring the nonheading phenotype under sustained long‐day conditions in a segregating population.
Fig. S3 CRISPR/Cas9‐mediated targeted mutagenesis of Hd1, Ghd7 and DTH8 in NHLD plants.
Fig. S4 Phenotypes of the CRISPR/Cas9‐based knockout lines of Hd1, Ghd7 and DTH8.
Fig. S5 Expression levels of Ehd1, Hd3a and RFT1 of the eight isogenic lines (58‐d‐old plants) under ALD and ASD conditions.
Fig. S6 Comparison of the expression of Ehd1, Hd3a and RFT1 in the eight isogenic lines (58 day‐old plants) between ASD and ALD.
Fig. S7 Expression of Ghd7, DTH8 and Hd1 in the seven isogenic lines (58‐d‐old plants) under ASD and ALD conditions.
Fig. S8 Test of the interaction of Hd1 with the Ghd7‐promoter region.
Fig. S9 Verification of the interactions among Hd1, DTH8 and Ghd7.
Fig. S10 Haplotypes (type) of Hd1, Ghd7 and DTH8 in rice (and O. rufipogon), and a summary of haplotypes and PS in several representative cultivated and wild rice lines.
Table S1 Pearson’s correlation coefficient between heading time and gene expression level.
Table S2 Information of 65 landraces, 61 modern indica and japonica varieties, and a wild rice accession analyzed in the study.
Table S3 Primers used in this study.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
We thank Prof. Xiangdong Liu, Prof. Jun Fang, Prof. Qingyao Shu, Prof Shiyong Song, Prof Dekai Wang and Junmin Wang for providing rice materials, and Prof. Guifu Liu and Dr Zifeng Yang for helping with data analysis. This work was supported by grants from the National Key Research and Development Program of China (2016YFD0100903), the Major Program of Guangdong Basic and Applied Research (2019B030302006), and the National Natural Science Foundation of China (31871596, 31921004). The authors declare there are no competing interests.
Contributor Information
Yao‐Guang Liu, Email: ygliu@scau.edu.cn.
Jingxin Guo, Email: jingxinguo@scau.edu.cn.
References
- Bian XF, Liu X, Zhao ZG, Jiang L, Gao H, Zhang YH, Zheng M, Chen LM, Liu SJ, Zhai HQ et al 2011. Heading date gene, dth3 controlled late flowering in O. Glaberrima Steud. by down‐regulating Ehd1 . Plant Cell Reports 30: 2243–2254. [DOI] [PubMed] [Google Scholar]
- Brambilla V, Fornara F. 2013. Molecular control of flowering in response to day length in rice. Journal of Integrative Plant Biology 55: 410–418. [DOI] [PubMed] [Google Scholar]
- Brambilla V, Fornara F. 2017. Y flowering? Regulation and activity of CONSTANS and CCT‐domain proteins in Arabidopsis and crop species. Biochimica et Biophysica Acta (BBA) ‐ Gene Regulatory Mechanisms 1860: 655–660. [DOI] [PubMed] [Google Scholar]
- Brambilla V, Gomez‐Ariza J, Cerise M, Fornara F. 2017. The importance of being on time: regulatory networks controlling photoperiodic flowering in cereals. Frontiers in Plant Science 8: 665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai MH, Chen SH, Wu MM, Zheng TH, Zhou L, Li CN, Zhang H, Wang JC, Xu XY, Chai JT et al 2019. Early heading 7 interacts with DTH8, and regulates flowering time in rice. Plant Cell Reports 38: 521–532. [DOI] [PubMed] [Google Scholar]
- Chen SB, Tao LZ, Zeng LR, Vega‐Sanchez ME, Umemura KJ, Wang GL. 2006. A highly efficient transient protoplast system for analyzing defence gene expression and protein‐protein interactions in rice. Molecular Plant Pathology 7: 417–427. [DOI] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C et al 2007. FT protein movement contributes to long‐distance signaling in floral induction of Arabidopsis . Science 316: 1030–1033. [DOI] [PubMed] [Google Scholar]
- Doi K, Izawa T, Fuse T, Yamanouchi U, Kubo T, Shimatani Z, Yano M, Yoshimura A. 2004. Ehd1, a B‐type response regulator in rice, confers short‐day promotion of flowering and controls FT‐like gene expression independently of Hd1 . Genes & Development 18: 926‐936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du AP, Tian W, Wei MH, Yan W, He H, Zhou D, Huang X, Li SG, Ouyang XH. 2017. The DTH8‐Hd1 module mediates day‐length‐dependent regulation of rice flowering. Molecular Plant 10: 948–961. [DOI] [PubMed] [Google Scholar]
- Gangappa SN, Botto JF. 2014. The BBX family of plant transcription factors. Trends in Plant Science 19: 460–470. [DOI] [PubMed] [Google Scholar]
- Gao H, Jin MN, Zheng XM, Chen J, Yuan DY, Xin YY, Wang MQ, Huang DY, Zhang Z, Zhou KN et al 2014. Days to heading 7, a major quantitative locus determining photoperiod sensitivity and regional adaptation in rice. Proceedings of the National Academy of Sciences, USA 111: 16337–16342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Zheng XM, Fei GL, Chen J, Jin MN, Ren YL, Wu WX, Zhou KN, Sheng PK, Zhou F et al 2013. Ehd4 encodes a novel and Oryza‐Genus‐Specific regulator of photoperiodic flowering in rice. PLoS Genetics 9: e1003281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goretti D, Martignago D, Landini M, Brambilla V, Gómez‐Ariza J, Gnesutta N, Galbiati F, Collani S, Takagi H, Terauchi R et al 2017. Transcriptional and post‐transcriptional mechanisms limit heading date 1 (Hd1) function to adapt rice to high latitudes. PLoS Genetics 13: e1006530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XH, Kurata N, Wei XH, Wang ZX, Wang AH, Zhao Q, Zhao Y, Liu KY, Lu HY, Li WJ et al 2012. A map of rice genome variation reveals the origin of cultivated rice. Nature 490: 497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immark S, Mitchell JM, Jongdee B, Boonwite C, Somrith B, Polvatana A, Fukai S. 1997. Determination of phenology development in rainfed lowland rice in Thailand and Lao PDR. Breeding Strategies for Rainfed Lowland Rice in Drought‐Prone Environments 77: 89–97. [Google Scholar]
- Ishikawa R, Aoki M, Kurotani K, Yokoi S, Shinomura T, Takano M, Shimamoto K. 2011. Phytochrome B regulates Heading date 1 (Hd1)‐mediated expression of rice florigen Hd3a and critical day length in rice. Molecular Genetics & Genomics 285: 461–470. [DOI] [PubMed] [Google Scholar]
- Ji CH, Li HY, Chen LB, Xie M, Wang FP, Chen YL, Liu YG. 2013. A novel rice bHLH transcription factor, DTD, acts coordinately with TDR in controlling tapetum function and pollen development. Molecular Plant 6: 1715–1718. [DOI] [PubMed] [Google Scholar]
- Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, Araki T, Yano M. 2002. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short‐day conditions. Plant and Cell Physiology 43: 1096–1105. [DOI] [PubMed] [Google Scholar]
- Koo BH, Yoo SC, Park JW, Kwon CT, Lee BD, An G, Zhang ZY, Li JJ, Li ZC, Paek NC. 2013. Natural variation in OsPRR37 regulates heading date and contributes to rice cultivation at a wide range of latitudes. Molecular Plant 6: 1877–1888. [DOI] [PubMed] [Google Scholar]
- Li XF, Liu HZ, Wang MQ, Liu HL, Tian XJ, Zhou WJ, Lü TX, Wang ZY, Chu CC, Fang J et al 2015. Combinations of Hd2 and Hd4 genes determine rice adaptability to Heilongjiang Province, northern limit of China. Journal of Integrative Plant Biology 57: 698–707. [DOI] [PubMed] [Google Scholar]
- Liu WZ, Xie XR, Ma XL, Li J, Chen JH, Liu YG. 2015. DSDecode: a web‐based tool for decoding of sequencing chromatograms for genotyping of targeted mutations. Molecular Plant 8: 1431–1433. [DOI] [PubMed] [Google Scholar]
- Ma XL, Zhang QY, Zhu QL, Liu W, Chen Y, Qiu R, Wang B, Yang ZF, Li HY, Lin YR et al 2015. A robust CRISPR/Cas9 system for convenient, high‐efficiency multiplex genome editing in monocot and dicot plants. Molecular Plant 8: 1274–1284. [DOI] [PubMed] [Google Scholar]
- Mantovani R. 1999. The molecular biology of the CCAAT‐binding factor NF‐Y. Gene 239: 15–27. [DOI] [PubMed] [Google Scholar]
- Nemoto Y, Nonoue Y, Yano M, Izawa T. 2016. Hd1, a CONSTANS ortholog in rice, functions as an Ehd1 repressor through interaction with monocot‐specific CCT‐domain protein Ghd7. The Plant Journal 86: 221–233. [DOI] [PubMed] [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, Coupland G. 1995. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80: 847–857. [DOI] [PubMed] [Google Scholar]
- Sawa M, Nusinow DA, Kay SA, Imaizumi T. 2007. FKF1 and GIGANTEA complex formation is required for day‐length measurement in Arabidopsis . Science 318: 261–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibaya T, Hori K, Ogiso‐Tanaka E, Yamanouchi U, Shu K, Kitazawa N, Shomura A, Ando T, Ebana K, Wu JZ et al 2016. Hd18, encoding histone acetylase related to Arabidopsis FLOWERING LOCUS D, is involved in the control of flowering time in rice. Plant & Cell Physiology 57: 1828–1838. [DOI] [PubMed] [Google Scholar]
- Shrestha R, Gómez‐Ariza J, Brambilla V, Fornara F. 2014. Molecular control of seasonal flowering in rice, Arabidopsis and temperate cereals. Annals of Botany 114: 1445–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Shim JS, Kinmonth‐Schultz HA, Imaizumi T. 2015. Photoperiodic flowering: time measurement mechanisms in leaves. Annual Review of Plant Biology 66: 441–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Smith RW, To BJ, Millar AJ, Imaizumi T. 2012. FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336: 1045–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su L, Shan JX, Gao JP, Lin HX. 2016. OsHAL3, a blue light‐responsive protein, interacts with the floral regulator Hd1 to activate flowering in rice. Molecular Plant 9: 233–244. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Teshima KM, Yokoi S, Innan H, Shimamoto K. 2009. Variations in Hd1 proteins, Hd3a promoters, and Ehd1 expression levels contribute to diversity of flowering time in cultivated rice. Proceedings of the National Academy of Sciences, USA 106: 4555–4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K. 2007. Hd3a protein is a mobile flowering signal in rice. Science 316: 1033–1036. [DOI] [PubMed] [Google Scholar]
- Thomas B, Vince‐Prue D. 1997. Daylength perception in short‐day plants Photoperiodism in plants. London: Academic Press, 85–117. [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle T. 2003. The roles of auxin response factor domains in auxin‐responsive transcription. The Plant Cell 15: 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Shen Y, Chang HC, Hou Y, Harris A, Ma SF, McPartland M, Hymus GJ, Adam L, Marion C et al 2010. The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis‐element. New Phytologist 187: 57–66. [DOI] [PubMed] [Google Scholar]
- Tsuji H, Taoka K, Shimamoto K. 2011. Regulation of flowering in rice: two florigen genes, a complex gene network, and natural variation. Current Opinion in Plant Biology 14: 45–52. [DOI] [PubMed] [Google Scholar]
- Wang P, Gong R, Yang Y, Yu SB. 2019. Ghd8 controls rice photoperiod sensitivity by forming a complex that interacts with Ghd7. BMC Plant Biology 19: 462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei XJ, Xu JF, Guo HN, Jiang L, Chen SH, Yu CY, Zhou ZL, Hu PS, Zhai HQ, Wan JM. 2010. DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiology 153: 1747–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie XR, Ma XL, Zhu QL, Zeng DC, Li GS, Liu YG. 2017. CRISPR‐GE: a convenient software toolkit for CRISPR‐based genome editing. Molecular Plant 10: 1246–1249. [DOI] [PubMed] [Google Scholar]
- Xue WY, Xing YZ, Weng XY, Zhao Y, Tang WJ, Wang L, Zhou HJ, Yu SB, Xu CG, Li XH et al 2008. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nature Genetics 40: 761–767. [DOI] [PubMed] [Google Scholar]
- Yan WH, Liu HY, Zhou XC, Li QP, Zhang J, Lu L, Liu T, Liu HJ, Zhang CJ, Zhang ZY et al 2013. Natural variation in Ghd7.1 plays an important role in grain yield and adaptation in rice. Cell Research 23: 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan WH, Wang P, Chen HX, Zhou HJ, Li QP, Wang CR, Ding ZH, Zhang YS, Yu SB, Xing YZ et al 2011. A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Molecular Plant 4: 319–330. [DOI] [PubMed] [Google Scholar]
- Yang ZF, Jin LL, Zhu HT, Wang SK, Zhang GQ, Liu GF. 2018. Analysis of Epistasis among QTLs on Heading Date based on Single Segment Substitution Lines in Rice. Scientific Reports 8: 3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y et al 2000. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS . The Plant Cell 12: 2473–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Liu HY, Qi FX, Zhang ZY, Li QP, Han ZM, Xing YZ. 2019. Genetic interactions among Ghd7, Ghd8, OsPRR37 and Hd1 contribute to large variation in heading date in Rice. Rice 12: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zhou XC, Yan WH, Zhang ZY, Lu L, Han ZM, Zhao H, Liu HY, Song P, Hu Y et al 2015. Combinations of the Ghd7, Ghd8 and Hd1 genes largely define the ecogeographical adaptation and yield potential of cultivated rice. New Phytologist 208: 1056–1066. [DOI] [PubMed] [Google Scholar]
- Zhang ZY, Hu W, Shen GJ, Liu HY, Hu Y, Zhou XC, Liu TM, Xing YZ. 2017. Alternative functions of Hd1 in repressing or promoting heading are determined by Ghd7 status under long‐day conditions. Scientific Reports 7: 5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZY, Zhang B, Qi FX, Wu H, Li ZX, Xing YZ. 2019. Hd1 function conversion in regulating heading is dependent on gene combinations of Ghd7, Ghd8, and Ghd7.1 under long‐day conditions in rice. Molecular Breeding 39: 92. [Google Scholar]
- Zhao J, Chen HY, Ren D, Tang HW, Qiu R, Feng JL, Long YM, Niu BX, Chen DP, Zhong TY et al 2015. Genetic interactions between diverged alleles of Early heading date 1 (Ehd1) and Heading date 3a (Hd3a)/ RICE FLOWERING LOCUS T1 (RFT1) control differential heading and contribute to regional adaptation in rice (Oryza sativa). New Phytologist 208: 936–948. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Relationship between day‐length and latitude in different months in several locations (cities) of China.
Fig. S2 Identification of Hd1 as the factor conferring the nonheading phenotype under sustained long‐day conditions in a segregating population.
Fig. S3 CRISPR/Cas9‐mediated targeted mutagenesis of Hd1, Ghd7 and DTH8 in NHLD plants.
Fig. S4 Phenotypes of the CRISPR/Cas9‐based knockout lines of Hd1, Ghd7 and DTH8.
Fig. S5 Expression levels of Ehd1, Hd3a and RFT1 of the eight isogenic lines (58‐d‐old plants) under ALD and ASD conditions.
Fig. S6 Comparison of the expression of Ehd1, Hd3a and RFT1 in the eight isogenic lines (58 day‐old plants) between ASD and ALD.
Fig. S7 Expression of Ghd7, DTH8 and Hd1 in the seven isogenic lines (58‐d‐old plants) under ASD and ALD conditions.
Fig. S8 Test of the interaction of Hd1 with the Ghd7‐promoter region.
Fig. S9 Verification of the interactions among Hd1, DTH8 and Ghd7.
Fig. S10 Haplotypes (type) of Hd1, Ghd7 and DTH8 in rice (and O. rufipogon), and a summary of haplotypes and PS in several representative cultivated and wild rice lines.
Table S1 Pearson’s correlation coefficient between heading time and gene expression level.
Table S2 Information of 65 landraces, 61 modern indica and japonica varieties, and a wild rice accession analyzed in the study.
Table S3 Primers used in this study.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.


