Summary
Viral infections are accompanied by a massive production of small interfering RNAs (siRNAs) of plant origin, such as virus‐activated (va)siRNAs, which drive the widespread silencing of host gene expression, and whose effects in plant pathogen interactions remain unknown.
By combining phenotyping and molecular analyses, we characterized vasiRNAs that are associated with typical mosaic symptoms of cauliflower mosaic virus infection in two crops, turnip (Brassica rapa) and oilseed rape (Brassica napus), and the reference plant Arabidopsis thaliana.
We identified 15 loci in the three infected plant species, whose transcripts originate vasiRNAs. These loci appear to be generally affected by virus infections in Brassicaceae and encode factors that are centrally involved in photosynthesis and stress response, such as Rubisco activase (RCA), senescence‐associated protein, heat shock protein HSP70, light harvesting complex, and membrane‐related protein CP5. During infection, the expression of these factors is significantly downregulated, suggesting that their silencing is a central component of the plant’s response to virus infections. Further findings indicate an important role for 22 nt long vasiRNAs in the plant’s endogenous RNA silencing response.
Our study considerably enhances knowledge about the new class of vasiRNAs that are triggered in virus‐infected plants and will help to advance strategies for the engineering of gene clusters involved in the development of crop diseases.
Keywords: 22‐nt sRNAs, CaMV, disease phenotype, oilseed rape, photosynthetic‐related genes, plant defense, turnip
Introduction
Small interfering RNAs (siRNAs) are main actors in the regulation of gene expression by RNA silencing. The biogenesis and function of siRNAs essentially involve the activity of a conserved group of enzymes. Dicer ribonucleases (DCLs) generate siRNAs from double‐stranded RNAs (dsRNAs). Argonaute endonucleases (AGOs) form the cores of RNA‐induced silencing complexes (RISCs), which, directed by the siRNA’s guide strands, mediate RNA silencing via endonucleolytic cleavage or translational inhibition of a target RNA. RNA‐dependent RNA polymerases (RDRs) generate dsRNA substrates, for example, from RISC cleavage products, which may be reused by the DCLs (Martínez de Alba et al., 2013).
In addition to central regulatory functions in tissue and organ development, differentiation, and the maintenance of genome integrity, siRNAs play a crucial role in the plant’s response to a wide range of environmental conditions, including abiotic and biotic stresses. In virus‐infected plants, viral siRNAs (vsiRNAs) 21–24 nt in length derive from RNAs of viral origin, such as transcripts, genomes, sub‐genomic RNAs, or satellites (Guo et al., 2019; Leonetti et al., 2020). Via these vsiRNAs, the plant silencing machinery restricts viral replication and virus spread. Furthermore, vsiRNAs may drive RNA silencing in trans by targeting host‐gene expression at the post‐transcriptional level (Pantaleo & Burgyán, 2008; Shimura et al., 2011; Zhu et al., 2011; Miozzi et al., 2013). As a counterdefense, viruses express viral suppressors of RNA silencing proteins (VSRs) that compromise antiviral silencing by, for example, binding vsiRNAs and/or by preventing their incorporation into RISCs, altering siRNA processing, or facilitating the decay of AGO proteins (Csorba et al., 2015). Depending on the state of infection, VSRs may also modulate the biogenesis and function of other endogenous small RNA (sRNA) classes, such as microRNAs (miRNAs; Mourrain et al., 2000; Shivaprasad et al., 2008; Kontra et al., 2016; Pertermann et al., 2018).
The unravelling of regulatory siRNA pathways is the subject of intensive research. Thus, in the Arabidopsis thaliana model plant, infections with members of two distinct positive‐strand RNA virus families (Cucumoviridae and Potyviridae) and antiviral RNA silencing were recently discovered to be accompanied by the generation of a new class of endogenous siRNAs, termed virus‐activated (va)siRNAs (Cao et al., 2014). Virus‐activated small interfering RNAs (vasiRNAs) are produced from host‐gene transcripts, and therefore differ from vsiRNAs, miRNA s, and other endogenous siRNAs. Incorporated into AGO1 and AGO2/RISC, 21 nt vasiRNAs were indicated to silence target messenger RNAs (mRNAs) of multiple host genes (hereafter denoted vasiRNA loci; Cao et al., 2014). The activity of vasiRNAs was further shown to be partly inhibited by VSRs. In fact, vasiRNAs were originally discovered in plants infected with cucumber mosaic virus (CMV), which lacks the 2b VSR (CMV‐Δ2b), and turnip mosaic virus (TuMV), whose VSR does not interfere with vasiRNA activity (Cao et al., 2014).
In our study, we compared the endogenous siRNA pattern of three members of the Brassicaceae family, namely of A. thaliana and the economically important crops Brassica rapa (turnip) and Brassica napus (oilseed rape (OSR)), that were infected by cauliflower mosaic virus (CaMV), a double‐stranded DNA (dsDNA) pararetrovirus of the Caulimoviridae family. By exploiting a novel machine‐learning approach capable of discriminating symptomatic from asymptomatic tissue, we identified a set of 15 orthologue coding sequences (CDSs) as prevalent vasiRNA loci in the infected tissue of all three species. Transcripts of these loci were suggested to generate 21 nt and 22 nt vasiRNAs, and gene expression was found to be downregulated in the infected crops. Interestingly, the set of loci identified, encoding central components of the photosynthetic machinery and factors sensitive to biotic and abiotic stimuli, closely corresponds to the one found earlier to be activated in CMV‐Δ2b and TuMV‐infected A. thaliana plants (Cao et al., 2014). Moreover, we obtained in vivo and in vitro evidence suggesting that 22 nt vasiRNAs represent major determinants of the endogenous RNA silencing response. Our report confirms the widespread silencing of host genes directed by vasiRNAs to be a general, broad‐spectrum defense mechanism of plant immunity.
Materials and Methods
Plant materials, virus inoculation, and phenotyping for tissue selection
Three plants of two species of the Brassica genus, B. rapa cv Purple To White Globe and B. napus cvr SariGol, were mechanically inoculated with CaMV strain NY8153 (accession no. M90541) as previously described (Schoelz et al., 1986). Phosphate buffer was used as a control in mock‐treated plants. Plants were maintained in a glasshouse at 22°C : 20°C, day : night, and photoperiod 16 h : 8 h, light : dark.
A digital image classifier was designed to identify plant leaves with mosaic symptoms that could be submitted to molecular analyses. The classifier was implemented according to the DeepLab v.3+ convolutional neural network (Chen et al., 2018), having ResNet‐18 as the backbone of (He et al., 2016). The classifier was trained by analyzing images of plant leaves of both turnip and OSR, either mock treated or CaMV infected, by running iterations over mini‐batches of eight images (three iterations per epoch), and by tuning and optimizing the weights of the network by means of the SGDM (stochastic gradient descent with momentum). A set of 30 images, with a resolution of 14 Mpx, captured by the Canon SX430 IS commercial camera (Canon Europe, Amsterdam, the Netherlands), defined a global population of over 420 000 000 px. In order to estimate the performance of the system, the following metrics were extracted: accuracy, intersection over union, and normalized confusion matrix. The accuracy (i.e. the ratio of the correct predictions, as they were labeled in the truth‐table, in each class to the total number of pixels belonging to that class) achieved with respect to the validation set was 0.95, 0.93 and 0.97 for the background, asymptomatic leaves, and symptomatic leaves, respectively.
RNA extraction, library generation, sequencing, microarray and bioinformatics analyses
In three biological repeats, total RNA was extracted 20 d post‐inoculation (dpi) from 10 collected leaves of mock‐treated and virus‐infected plants. The leaves of the infected plants were previously classified by the machine‐learning approach as either asymptomatic or symptomatic. RNA extraction was done using Trizol reagent (Invitrogen) according to the manufacturer’s instructions. A 5 µg sample of RNA was resolved in a 15% sodium dodecyl sulfate–polyacrylamide gel electrophoresis gel as previously described (Pirovano et al., 2014). RNA fraction replicates of length 20–30 nt were quantified and mixed in equimolar ratio. Libraries were obtained using high‐definition adapters according to a previous report (Sorefan et al., 2012). The quality of libraries was checked by BioAnalyzer High Sensitivity DNA Analysis (Agilent, Santa Clara, CA, USA) and then submitted to sequencing with a HiSeq 2500 Illumina platform (Illumina Inc., San Diego, CA, USA). Sequence quality checks, conversion into Fasta format, adapter removal, and size class selection were performed using the Fastx toolkit (http://hannonlab.cshl.edu/fastx_toolkit/index.html). sRNAs aligning to turnip and OSR CDSs (Wang et al., 2011; Chalhoub et al., 2014), ribosomal RNAs (Rfam database, www.sanger.ac.uk/Software/Rfam/), and transposon (Repbase, www.girinst.org) were identified by Bowtie alignment allowing zero mismatch (Langmead et al., 2009). miRNAs were filtered out from the sRNA data sets using miRProf tool (Stocks et al., 2012) with perfect matches to sequences from the miRNA repository (Kozomara & Griffiths‐Jones, 2014; Kozomara et al., 2019). vsiRNAs were identified by alignment to the CaMV genome (strain NY8153) as for the plant genome. sRNAs of mock‐treated or CaMV‐infected A. thaliana Col‐0 were collected from previous studies (Blevins et al., 2011; Seguin et al., 2014). Endogenous plant siRNAs that were specifically expressed in the presence or in absence of CaMV infection were obtained by comparison (compara tool, www.galaxy.org) of brassica genomes mapping data sets of mock‐treated vs virus‐infected plant tissues. Induced and repressed endogenous siRNAs were therefore ascribed to the corresponding annotated CDSs (ENSEMBL plant platform, https://plants.ensembl.org). Not yet annotated sRNA‐generating transcripts were tentatively annotated by BlastP or tBlastn search vs the A. thaliana transcriptome (Altschul et al., 1997) using the ENSEMBL platform. In the microarrays analyzed, the expression profiles of CaMV‐infected A. thaliana either wild type (Col‐0) or double mutants (dcl2‐dcl4, dcl2‐dcl3, and dcl4‐dcl3) were compared with not‐infected control plants. Raw data from Complete Arabidopsis Transcriptome Microarray (CATMA) assays were retrieved from Gene Expression Omnibus, GSE36457. CATMA probes specific for selected genes under analyses were searched and filtered out as means of the T (log2(FC), where FC is fold change) of expression values between case and control. Changes greater than two‐fold were considered significant.
RNA transcripts quantification by real‐time quantitative PCR
For quantification of gene expression, cDNA was synthesized from 1 μg of total RNA using the QuantiTect Reverse Transcription Kit (Qiagen) with random hexamers following the manufacturer’s instructions. Quantitative PCR (qPCR) was carried out by means of a StepOnePlus real‐time system (Applied Biosystems Inc., Foster City, CA, USA). All PCR reactions were performed three times in independent experiments. Gene expression was calculated using the Delta–Delta cycle threshold method (Livak & Schmittgen, 2001; Bustin et al., 2009). Relative quantitative results were calculated by normalization to UBC10 (GenBank accession no. XM_013859290.2; Han et al., 2017).
Oligonucleotides denoted as LHCB1.3_770b_rev and LHCB1.3_6b_for were used to generate complementary DNAs (cDNAs) from sense or antisense LHCB1.3 transcripts, respectively, in mock‐inoculated and CaMV‐infected plants. Strand‐specific LHCB1.3 cDNA synthesis was coupled with the constitutive UBC10 cDNA synthesis by using oligonucleotide UBC10_rev for both turnip and OSR in the same reaction at the same final concentration of 0.5 µM. cDNAs were synthesized from 1 μg of DNase‐treated (Ambion) total RNA and using SuperScript II (Invitrogen), following manual instructions. qPCR for sense and antisense transcript quantification normalized to constitutive UBC10 expression was carried out using the StepOnePlus real‐time PCR system in three different regions of the entire gene with the strategy detailed in Supporting Information Fig. S7(b). The oligonucleotides used for qPCR are listed in Table S1.
Small interfering RNAs and in vitro slicer assay
The 22 nt siRNAs used in the cleavage assay corresponded to the 2 nt 3′‐overhang duplexes that were found to be most represented in the data sets, and 21 nt were the shorter versions of the selected 22 nt. Single‐stranded synthetic RNAs were purchased from Biomers (Ulm, Germany). To produce RNA duplexes, complementary RNA molecules corresponding to the guide and passenger strands as listed in Fig. 4(a) (see later) were heated for 1 min at 90°C and annealed for 60 min at 37°C in annealing buffer (30 mM HEPES–potassium hydroxide, pH 7.4, 100 mM potassium acetate, 2 mM magnesium acetate).
Fig. 4.
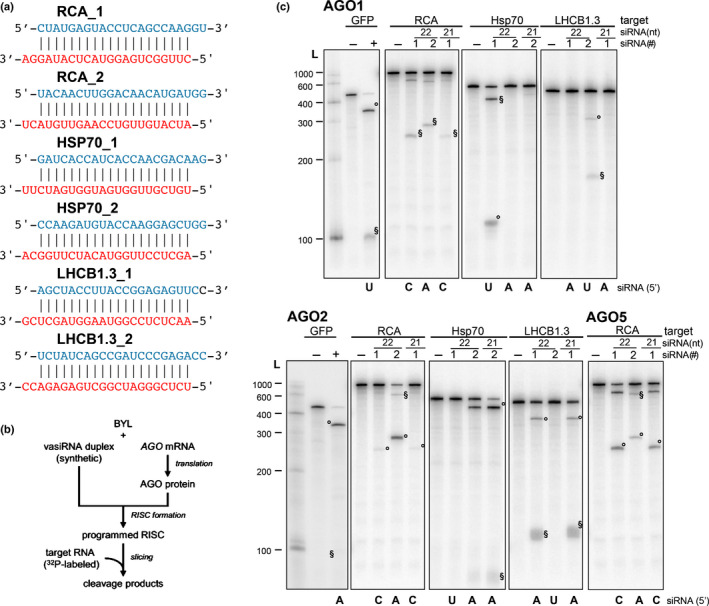
Slicer activity of 22 nt virus‐activated small interfering RNAs (vasiRNAs). (a) List of 22 nt vasiRNA duplexes used in the slicer activity assays. The guide and passenger strands are depicted in red and blue, respectively. The 21 nt long versions are shortened by one nucleotide at each strand (i.e. 3′ at the guide strand and 5′ at the passenger strand). (b) Schematic representation of the ‘slicer assay’. Argonaute (AGO)1, AGO2, or AGO5 was generated by in vitro translation in the Bright Yellow lysate (BYL) in the presence of synthetic vasiRNA duplexes generating programmed RNA‐induced silencing complexes (RISCs). Subsequently, phosphorus‐32 (32P)‐labeled, in vitro transcribed messenger RNA (mRNA) fragments containing the respective vasiRNA target sites were added and analyzed for RISC‐mediated cleavage by denaturing polyacrylamide gel electrophoresis and autoradiography. (c) Arabidopsis thaliana AGO1, AGO2, or AGO5 mRNA was translated in BYL in the presence of the individual indicated vasiRNAs to form AGO/RISC. The 5′ and 3′ cleavage products are indicated as ‘°’ and ‘§’, respectively. L, radiolabeled 100 nt RNA ladder; vasiRNAs duplexes listed in (a) are indicated with their identifying number, #1 or #2. In the negative controls (−), no small interfering RNA (siRNA) was included in the cleavage assay. Cleavage of a fragment of the mRNA of green fluorescent protein (GFP) and a GFP‐specific siRNA was used as a positive control (+).
To generate the target transcripts by T7 in vitro transcription, amplicons of the target RNAs were generated by reverse transcription PCR using specific oligonucleotides (listed in Table S1). The PCR products were resolved by 1.2% agarose, 1× Tris–borate–EDTA gel electrophoresis (Fig. S4, see later), eluted (Kit Promega), and sequenced using the reverse oligonucleotide (Fig. S4, see later). Culturing of Nicotiana tabacum Bright Yellow (BY)‐2 cells, the preparation of BY lysates, in vitro translation of AGO1 and AGO2, in vitro transcriptions of the RNA targets, and cleavage assays were performed as described previously (Schuck et al., 2013; Gago‐Zachert et al., 2019).
Statistics and small interfering RNA plots
Changes in read counts were evaluated with fold change (FC) and with a χ2 test, adjusted for multiple testing with false discovery rate. An absolute FC > 1.5 and an adjusted P‐value of < 0.05 were considered significant.
In order to graphically represent the possible arrangement in phases of the sRNA detected in the sequencing, we developed a script in R that produces plots for each gene, according to the following steps. First of all, RNA sequencing (RNA‐Seq) reads were aligned to the reference genome, to know their position on the chromosomes and on specific genes, and to know from which strand they derive. The reads were then distinguished by length n and by strand. Since each phase involves all the reads spaced by n nucleotides, we obtained n possible phases that included the reads with a starting point in , where i varies from zero to the maximum number of reads that can be originated by a phase in the gene, and j varies in . For each j, we obtained a horizontal line in the plot where we aligned the reads on the positive strand starting from and reads on the antisense strand starting from .
Results
Cauliflower mosaic virus infection significantly affects diversity of endogenous small RNAs in A. thaliana, turnip, and oilseed rape
Virus‐infected plants usually show an uneven distribution of mosaic symptoms and yellowing of the leaves. The symptom pattern may vary among different plants and even on the same plant or the same symptomatic leaf, a phenomenon thought to be associated with heterogeneous virus‐induced reprogramming of gene expression in infected tissues (Medzihradszky et al., 2019). Infections with CaMV typically generate mosaic symptoms in turnip and OSR. Symptoms in OSR are often milder, and plants typically show recovery as a consequence of silencing (Shepherd, 1979; Saunders et al., 1990). Based on the incidence of these symptoms, occurring approximately 20 dpi, turnip and OSR were classified as being highly sensitive and semi‐sensitive to CaMV infection, respectively. Since symptom heterogeneity considerably complicates the characterization of the causal molecular determinants, we classified the symptomatic leaves of CaMV‐infected beets or rape for subsequent RNA analyses. In order to achieve the greatest accuracy possible, a novel approach based on deep learning has been appositely designed, oriented to classify tissues as mock‐treated and CaMV‐infected, through automatic semantic segmentation of their photographs (see Materials and Methods section). Fig. 1(a) shows indicative examples of the semantic segmentations performed; the architecture of the artificial neural network is provided in Fig. S1. Plant leaves were sampled from mock‐treated or CaMV‐infected plants using this phenotype‐based classification approach and then subjected to RNA HiSeq (see the Materials and Methods section; Fig. 1b; Table S2).
Fig. 1.
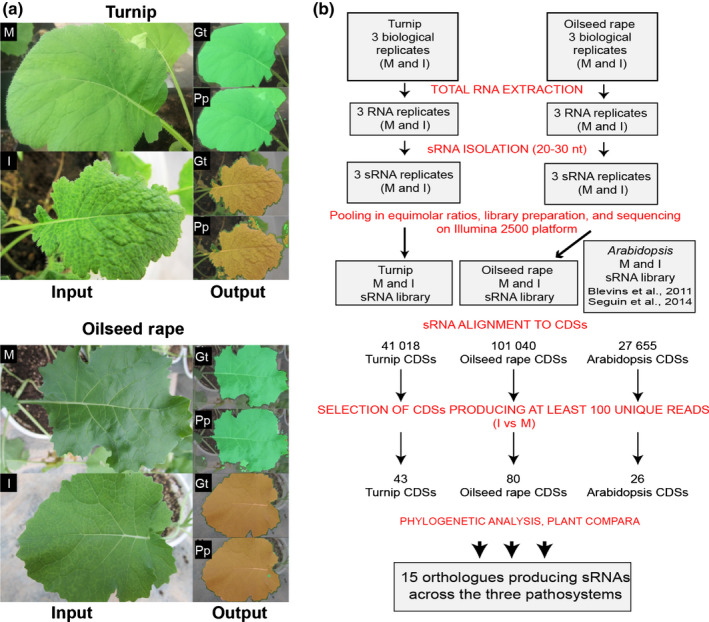
Tissue selection and flowchart describing the identification and analysis of small RNAs (sRNAs) that are originated during cauliflower mosaic virus (CaMV) infections of different Brassicaceae. (a) Images of leaves from either mock‐treated (M) or CaMV‐infected (I) turnip and oilseed rape were submitted to a novel machine‐learning process using the architecture of a DeepLab v.3+ convolutional neural‐network‐based classifier (see also Supporting Information Fig. S1) in order to classify the leaves as symptomatic or asymptomatic. Examples of asymptomatic and symptomatic leaves, related ground truth (Gt) accuracy and performed predictions (Pp) (green, asymptomatic; brown, symptomatic; black, background). (b) Flowchart of the analyses ranging from RNA extraction of selected leaves to Ensembl (Plant Compara), leading finally to the characterization of endogenous sRNAs and the identification of the loci of origin. CDSs, protein‐coding sequences.
CaMV replication leads to the synthesis of genomic RNAs, and structured parts of this are known to be processed by the plant DCLs into vsiRNAs. Thus, in Arabidopsis, a massive amount of vsiRNAs, which are mainly 24 nt long, was reported to derive from the highly structured leader sequence of the CaMV RNA (Blevins et al., 2011). In accordance with this, we also found 21 nt, 22 nt and 24 nt vsiRNAs that derived from the entire CaMV genomic RNA in turnip and OSR (Fig. S2). Unique 21 and 22 nt vsiRNAs were at similar levels, whereas total (redundant) 22 nt vsiRNAs were more abundant than 21 nt vsiRNAs (see histograms in Fig. S2). No or only a few siRNAs aligning to the CaMV genomic RNA were detected in mock‐treated plant tissues as selected by the machine‐learning method, supporting the reliability of the approach used.
We performed an alignment of sRNAs from A. thaliana (Blevins et al., 2011), turnip, and OSR (present work) to the annotated loci available for the three plant species at Ensemble Plants (https://plants.ensembl.org/index.html); that is, orthologue mRNA CDSs, miRNA‐coding regions, ribosomal RNA (rRNA)‐coding regions, and other loci. The results showed that, in all three plants, the CaMV infection caused a significant variation in the diversity of 21 nt, 22 nt and 24 nt sRNAs that could be mapped to these loci (Fig. 2a). In the three pathosystems, no univocal trend was observed for sRNAs that mapped to rRNA or other RNAs encoding genomic loci (blue and gray bars in Fig. 2a). sRNAs, which mapped to miRNA‐encoding loci, in particular 21 nt sRNA predicted to correspond to the actual miRNAs, showed an apparent decline in diversity in the cases of the infected turnip and OSR (green bars in Fig. 2a). In contrast to all infected tissues tested, we observed an obvious increase of the classes of 21 nt, 22 nt and 24 nt sRNAs that mapped to protein CDSs (red bars in Fig. 2a; CaMV infected vs mock treated).
Fig. 2.
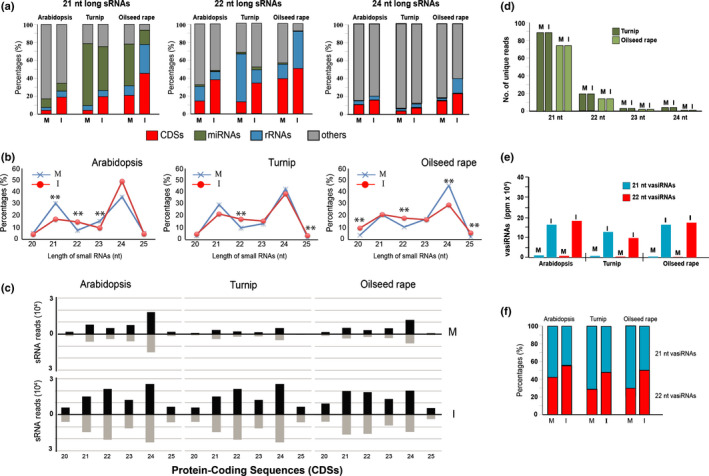
Origin of small RNAs (sRNAs) in Arabidopsis thaliana, turnip, and oilseed rape infected by cauliflower mosaic virus (CaMV) and 21 nt and 22 nt virus‐activated small interfering RNA (vasiRNA) in infected plants. (a) Incidence of 21, 22, and 24 nt total sRNAs according to their different origin. (b) Profile of unique sRNAs according to their lengths, with statistical evaluation of changes (**, adjusted P < 0.01, χ 2 test). (c) Length distribution and abundance (reads per million × 104) of the sRNAs unique reads derived from protein‐coding sequences (CDSs) in mock‐treated (M) or CaMV‐infected (I) plants. (d) Size distribution profile of sRNAs from microRNA (miRNA) precursor transcripts. (e) The 21 nt and 22 nt unique vasiRNAs from 15 orthologous transcripts. The values are given in parts per million (ppm) referring to all unique 20–25 nt sRNAs. (f) Ratio of 21 nt and 22 nt vasiRNAs (21 nt + 22 nt = 100). M, mock‐treated; I, CaMV infected; CDSs, protein‐coding DNA sequences; rRNAs, ribosomal RNAs; length of sRNAs is in nucleotides (nt).
As previously explained, infections with different RNA viruses had been observed to trigger the production of vasiRNAs in A. thaliana (Cao et al., 2014). Accordingly, the following analysis was aimed at understanding whether this was also the case for turnip and OSR infected with the entirely unrelated pararetrovirus CaMV. Thus, we performed a comparative analysis of the sRNA populations considering solely distinct sequences (hereafter designated as ‘unique’), in contrast to the total number of sequences termed ‘redundant’, which included all reads (Fig. 2a), Interestingly, in all three pathosystems these data confirmed a statistically significant increase in the overall number of sRNAs with a size of 22 nt (Fig. 2b, red lines; Table S3).
Subsequently, by further dissecting the origin of the sRNAs that were induced in the infected plants, we found all sRNA size classes that mapped to CDSs, showing a significant increase in number (Fig. 2c; CaMV infected vs mock treated). This was in contrast to the situation found with rRNA and transposon loci (Fig. S3). However, in agreement with the earlier findings, the accumulation of 22 nt sRNAs was particularly evident; that is, the number of 22 nt sRNAs increased by five, eight, and six‐fold and the number of 21 nt sRNAs increased by two, five, and three‐fold in the infected A. thaliana, turnip and OSR, respectively (Fig. 2c). Importantly, the CDS‐derived 21 nt and 22 nt sRNAs were equally represented with sense and antisense strands.
Taken together, these findings indicated that the effects on CaMV‐infected turnip and OSR crops together with A. thaliana plants is very similar to what was previously observed in RNA virus infections of A. thaliana (Cao et al., 2014). In each case, the production of sRNAs from CDS‐derived transcripts was considerably increased, suggesting that these RNAs were indeed vasiRNAs. However, contrary to the situation described with the RNA virus‐infected plants, in CaMV‐infected samples, the production of unique 22 nt long siRNAs was significantly and actively induced more than that of 21 nt siRNAs.
Virus infections activate common target loci in different Brassicaceae
In the next step of our investigation, we wanted to identify the transcripts that generated the sRNAs detected in the three CaMV‐infected plant species. Considering the different genome sizes of A. thaliana, turnip, and OSR, this required a comparative qualitative analysis capable of handling 27 655, 41 018 and 101 040 transcripts, respectively (Fig. 1b). Note that B. napus is an allopolyploid species (AACC, 2n = 4X = 38). It is believed to have been formed by hybridization of two diploid species, B. rapa (AA, 2n = 2X = 20) and Brassica oleracea (CC, 2n =2X = 18) (Lu et al., 2019).
First, by comparing mock‐treated and CaMV‐infected plants of the three species, we selected the transcripts that produced > 95 unique sRNAs of 21 nt and 22 nt in length, respectively. Subsequently, by using the phylogenetic trees of the Ensemble Plants Compara platform (https://plants.ensembl.org/index.html), we pinpointed the orthologues of A. thaliana, turnip, and OSR within two speciation nodes, thus distinguishing the three plant species under analysis (see flowchart applied in Fig. 1b). The results obtained are summarized in Table 1; the complete list of sRNAs associated with each transcript of each species is available in the Gene Expression Omnibus (GEO) DataSet (accession no. GSE119398). Thus, we defined 15 CDS loci in all three Brassicaceae species whose transcripts were considered to generate unique amounts of sRNAs in diseased leaves, but only a few or none in asymptomatic leaves (Table 1).
Table 1.
Sequencing read numbers of unique small RNAs 21 nt and 22 nt long mapping to the orthologous coding sequences of Arabidopsis thaliana, turnip, and oilseed rape during cauliflower mosaic virus infection.
| Gene name | Description | A. thaliana | Turnip | Oilseed rape | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Locus | Mock | CaMV | Locus | Mock | CaMV | Locus | Mock | CaMV | ||
| SAP | Senescence‐associated protein | AT5G20700.1* | 19 | 272 | Bra020123.1 | 16 | 601 | BnaA02g05040D‐1 | 7 | 415 |
| ABCG36 | ABC transporter G family member 36 | AT1G59870.1* | 3 | 113 | Bra003527.1 | 4 | 180 | BnaA07g19610D‐1 | 1 | 230 |
| ACT2 | Actin‐2 | AT3G18780.1* | 6 | 66 | Bra037560.1 | 6 | 273 | BnaA01g26500D‐1 | 3 | 1214 |
| CP5 | P‐loop membrane related protein | AT1G64720 § | 11 | 107 | Bra017435 | 0 | 69 | BnaC09g20830D‐1 | 1 | 97 |
| RPT2 | Root phototropism protein 2 | AT2G30520.1 § | 4 | 97 | Bra018309.1 | 2 | 105 | BnaA05g11930D‐1 | 0 | 77 |
| CAT3 | Catalase | AT1G20620.4* | 15 | 366 | Bra016459.1 | 0 | 107 | BnaA08g21730D‐1 | 0 | 159 |
| GUN5 | Magnesium chelatase | AT5G13630.2 § | 6 | 153 | Bra006208.1 | 9 | 101 | BnaA03g04440D‐1 | 1 | 160 |
| FBA1 | Fructose‐bisphosphate aldolase 1 | AT2G21330.2 § | 5 | 68 | Bra030303.1 | 9 | 718 | BnaA04g12130D‐1 | 2 | 721 |
| HSP70‐1 | Heat shock protein 70‐1 | AT5G02500.1* | 12 | 245 | Bra035909.1 | 7 | 112 | BnaA09g05850D‐1 | 1 | 109 |
| LHCA1 | Chl a/b binding protein, chloroplastic | AT3G54890.2 § | 12 | 61 | Bra003198.1 | 9 | 134 | BnaA07g16530D‐1 | 1 | 191 |
| LHCB1.3 | Chl a/b binding protein 1, chloroplastic | AT1G29930.1* | 56 | 818 | Bra010807.1 | 104 | 1157 | BnaC03g59520D‐1 | 8 | 1368 |
| LHCB1.4 | Chl a/b‐binding protein, chloroplastic | AT2G34430.1* | 15 | 294 | Bra005425.1 | 27 | 567 | BnaA05g09410D‐1 | 1 | 614 |
| LOS1 | Translation elongation factor 2‐like protein | AT1G56070.3* | 15 | 107 | Bra020542.1 | 5 | 243 | BnaA02g31580D‐1 | 1 | 140 |
| NFXL1 | NF‐X‐like | AT1G10170.1* | 0 | 228 | Bra018458.1 | 0 | 71 | BnaC08g14030D‐1 | 0 | 153 |
| RCA | Ribulose bisphosphate carboxylase/oxygenase activase, chloroplastic | AT2G39730.3 § | 30 | 85 | Bra017055.1 | 24 | 198 | BnaC04g46560D‐1 | 2 | 725 |
Interestingly, of the 15 CDS transcripts identified, 13 were previously defined in A. thaliana to be specific targets of RDR1‐dependent vasiRNAs (Cao et al., 2014). Transcripts of nine genes – AT5G20700, ABCG36, ACT2, CAT3, HSP70‐1, LHCB1.3, LHCB1.4, LOS1 and NFXL1 – were previously identified in A. thaliana plants infected with either CMV‐Δ2b or TuMV, and transcripts of six genes – CP5, RPT2, GUN5, FBA1, LHCA1 and RCA – were identified in A. thaliana plants infected with CMV‐Δ2b (Cao et al., 2014).
Accordingly, these data revealed that the sRNA‐associated response of all three Brassicaceae plants investigated to infections of members of taxonomically distant viruses (i.e. positive‐strand RNA viruses or dsDNA viruses) is remarkably conserved (see Discussion section). Moreover, our results support the notion that the infection‐induced sRNAs detected are vasiRNAs, which derive from transcripts encoded by a common set of ‘vasiRNA loci’.
Size profile of virus‐activated small interfering RNAs in turnip and oilseed rape
Our earlier data revealed that CDS‐derived 21 nt and 22 nt sRNAs were generally enriched in infected A. thaliana, turnip, and OSR. In addition, we detected a significant increase in the number of 22 nt sRNAs as a unique feature of all pathosystems investigated. To further evaluate these observations and exclude any biases/anomalies in the sequencing or bioinformatics analyses, we followed two experimental lines. First, we reanalyzed the previously obtained unique RNA‐Seq reads to determine the 21 nt and 22 nt profiles of miRNAs, because miRNAs represent the best‐characterized class of endogenous sRNAs. miRNA primary transcripts are processed by DCL1 (Rajagopalan et al., 2006), which generates predominantly 21 nt but also 22 nt long species (Shivaprasad et al., 2012; Zhao et al., 2012; Shen et al., 2014). However, we found that the number of unique sequence reads of miRNAs remained essentially constant during CaMV infection. In other words, regardless of whether the plants were mock treated or CaMV infected, the tissues analyzed exhibited 89 unique miRNA‐based sRNA species of 21 nt and 19 of 22 nt in turnip and 74 unique sRNA species of 21 nt and 14 of 22 nt in OSR (Fig. 2d, mock treated vs CaMV infected).
In the second approach, we analyzed the profiles of the unique reads of 21 nt and 22 nt sRNAs, which derived from the orthologous transcripts shown in Table 1 and were predicted to correspond to vasiRNAs. Thus, we confirmed a significant increase of unique 21 nt and 22 nt species, which was exclusively measurable in the tissues of CaMV‐infected plants (Fig. 2e); that is, in A. thaliana, turnip, and OSR, unique 21 nt sRNAs increased 14, 13 and 26‐fold, respectively (blue bars in Fig. 2e, CaMV infected vs mock treated), whereas unique 22 nt sRNAs increased 22, 29 and 68‐fold, respectively (red bars in Fig. 2e, CaMV infected vs mock treated). Comparing the ratio of 21 nt and 22 nt sRNAs in mock‐treated and in CaMV‐infected plants, the fraction of the 22 nt species was found to increase from 42% to 53% in infected A. thaliana, from 28% to 47% in infected turnip, and from 29% to 51% in infected OSR, and the proportion of 21 nt RNAs decreased correspondingly (red and blue bars in Fig. 2f, CaMV infected vs mock treated). Further analyses revealed that the number of unique 22 nt sRNAs that derived from the CDS transcripts (Table 1) exceeded in most cases that of the 21‐nt species (i.e. with CP5, ACT2, RPT2, CAT3, GUNS5, LHCA1, RCA, SAP, ABCG36 and LOS1) (Table S4).
Accordingly, CaMV infections in the Brassicaceae crops were further confirmed to promote the production of endogenous sRNAs, in particular of 22 nt sRNAs. As these sRNAs were definitely not processed from primary miRNA transcripts, this finding further confirmed the opinion that these sRNAs represent vasiRNAs.
Transcripts encoded by the identified virus‐activated small interfering RNA loci were significantly downregulated in cauliflower mosaic virus‐infected tissues in turnip and oilseed rape
Previous studies in A. thaliana have shown that, for example, in the case of a CMV‐Δ2b infection leading to the production of vasiRNAs, the expression of the corresponding vasiRNA loci (i.e. CP5, HSP70‐1, RCA, RBCS‐1A, LHCB1.3 and AT5G20700) is downregulated (Cao et al., 2014). To understand if this was also the case in the CaMV‐infected plants, we carried out transcript‐specific expression analyses of the cluster of CDS genes identified (Table 1). Thus, in both CaMV‐infected crops, we confirmed that RCA, SAP, HSP70‐1, LHCB1.3, and CP5 mRNAs were significantly downregulated in the infected turnip and OSR relative to mock to by < 10%, 30%, c. 50%, 40% and < 10%, respectively (Fig. 3a–e).
Fig. 3.
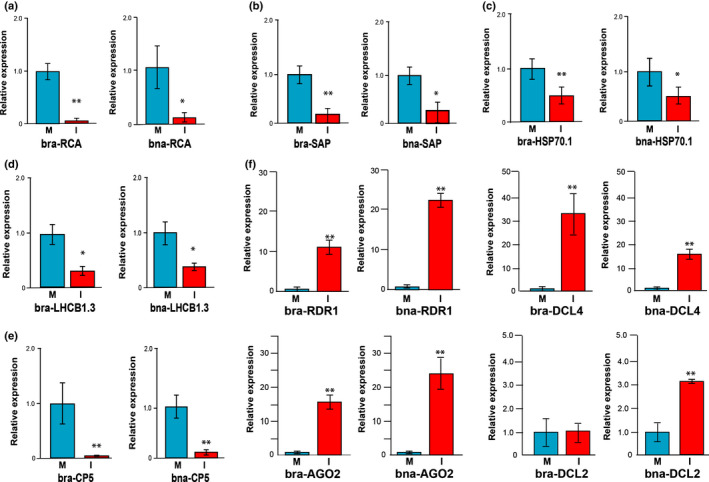
Expression analysis of target transcripts in turnip and oilseed rape. Relative expression levels of (a) Rubisco activase (RCA) transcripts, (b) senescence‐associated protein (SAP) transcript, (c) heat shock protein‐70 (HSP70‐1) transcripts, (d) light harvesting complex protein 1.3 (LHCB1.3), and (e) CP5 transcripts in selected tissues of mock‐treated (M) or cauliflower mosaic virus (CaMV)‐infected turnip and oilseed rape plants. (f) Relative expression levels of transcripts of major RNA silencing factors, RNA‐dependent RNA polymerase 1 (RDR1), Dicer‐like 4 (DCL4), Argonaute 2 (AGO2), and Dicer‐like 2 (DCL2) in selected tissues of mock‐treated or CaMV‐infected turnip and oilseed rape plants. Data are shown as the mean value ± SD from three independent assays using Student’s t‐test (*, P < 0.05; **, P < 0.01). bra, Brassica rapa; bna, Brassica napus.
The earlier report of Cao et al. (2014) revealed an evident downregulation of selected mRNAs (i.e. LHCB1.3) in the case of virus‐infected Arabidopsis wild‐type and dcl2 single mutants but not in the case of dcl4 single mutant and dcl2‐dcl4 double mutants, thus concluding that DCL4‐dependent 21 nt vasiRNAs are responsible for mRNA silencing. To evaluate the situation during CaMV infections, we analyzed microarray data (see Materials and Methods section) focusing on available CATMA probes that addressed the genes that are summarized in Table 1. In striking agreement with our qPCR results (Fig. 3b–e), we found a downregulation of CP5, HSP70‐1, and SAP (which is statistically significant in the last two genes) in CaMV‐infected vs mock‐treated Arabidopsis Col‐0, and the opposite situation was found when infected dcl2‐dcl4 double mutants were compared with Col‐0 (Table S5). When analyzing mutants where the biogenesis of either 21 nt or of 22 nt was abolished (i.e. mutants including only dcl4 or dcl2, respectively) we did not find significant changes in the relative expression levels of the respective genes (Table S5). These data suggest that both DCL4 and DCL2‐dependent 21 nt and 22 nt vasiRNAs are triggered and functional in the case of a CaMV infection.
As already outlined in the Introduction section, vasiRNAs are genetically distinct from other plant endogenous siRNAs that have been characterized to date. Instead, vasiRNAs resemble vsiRNAs by requiring DCL4 and RDR1 for their biogenesis, and AGO1 or AGO2 to execute silencing. Viral infections usually activate the expression of key factors of RNA silencing pathways at the transcriptional level, thereby enhancing antiviral RNA silencing. For example, the expression of AGO1, AGO2, or RDR1 is induced by infections with diverse viruses (Zhang et al., 2006; Havelda et al., 2008; Várallyay et al., 2010; Harvey et al., 2011). Considering this scenario, we also evaluated the accumulation of the transcripts encoding DCL4, DCL2, AGO2, and RDR1. Similar to previous observations, the levels of RDR1, DCL4, and AGO2 mRNAs were significantly upregulated in both CaMV‐infected turnip and OSR, whereas the mRNA level of DCL2 increased only in the CaMV‐infected OSR (Fig. 3f).
In sum, these data suggest that the siRNAs that are generated from the transcripts of the orthologues identified can be considered as bona fide CaMV‐induced endogenous vasiRNAs in turnip and OSR. These vasiRNAs are apparently capable of post‐transcriptionally silencing the expression of these genes in plants.
Slicer activity of 22 nt virus‐activated small interfering RNAs
The earlier report of Cao et al. (2014), which describes the discovery of vasiRNAs in RNA virus‐infected A. thaliana plants, revealed that the CDS‐generated vasiRNAs consisted predominantly of 21 nt RNAs and only a small fraction of 22 nt RNAs. Corresponding ratios of 21 nt and 22 nt RNAs were incorporated into AGO1 and AGO2. Furthermore, the 21 nt vasiRNAs binding to AGO1 exhibited a strong bias for a 5′‐terminal U‐residue, and those binding preferentially to AGO2 had a strong bias for a 5′‐terminal A‐residue. The binding preferences of AGOs for vasiRNAs are consistent with other classes of sRNAs (Mi et al., 2008; Schuck et al., 2013). Therefore, the observed widespread silencing of transcripts of the vasiRNA loci was attributed to the activity of 21 nt RNAs rather than to that of 22 nt RNAs (Cao et al., 2014).
As previously outlined, in all three Brassicaceae species that were infected with the pararetrovirus CaMV, 22 nt vasiRNAs were the predominant species generated from CDS‐derived transcripts (Fig. 2). Moreover, with these transcripts we observed an evident target downregulation in the infected tissues (Fig. 3a–e). Accordingly, we considered it important to evaluate the AGO‐mediated silencing potential of these 22 nt RNAs. For this purpose, we identified from the sRNA pools of turnip and OSR the two most abundant 22 nt and 3′‐overhang vasiRNA duplexes that derived from the RCA, HSP70‐1, and LHCB1.3 mRNAs (Fig. 4a).
To test these vasiRNA duplexes, we applied synthetic versions of these two 22 nt vasiRNAs (termed ‘_1’ and ‘_2’ here) to a well‐established in vitro RNA silencing assay (Fig. 4b), which was performed with fragments of the respective CDS‐derived transcripts (see also Figs S4, S5) (Iki et al., 2010; Schuck et al., 2013). This assay enables the functional evaluation of different types of sRNAs in terms of their capacity to (1) recruit AGO1, AGO2, or other AGO/RISCs and (2) to support AGO‐catalyzed sequence‐specific cleavage (‘slicing’) of their canonical target RNA transcripts. Thus, with AGO1/RISC, we found readily detectable cleavages of the vasiRNA loci transcripts with vasiRNA RCA_1 or _2, HSP70_1 and LHCB1.3_2 (Fig. 4c). With AGO2/RISC, we observed cleavages with vasiRNA RCA_1 or _2, HSP70_2 and LHCB1.3_1, respectively (Fig. 4c). Accordingly, from each of the AGO1 or AGO2‐loaded vasiRNAs, either one or both candidates tested supported AGO1 or AGO2‐catalyzed slicing of the canonical target RNA, albeit with different efficiencies. On the one hand, and similar to the situation with 21 nt siRNAs, cleavage was determined by the type of 5′‐terminal nucleotide (Mi et al., 2008; Schuck et al., 2013); for example, the most efficient cleavages were observed with AGO1/RISC and HSP70_1 and LHCB1.3_2 (guide strands initiating with a 5′U) and with AGO2/RISC and RCA_2 and HSP70_2 (guide strands initiating with a 5′A). An experiment performed with AGO5, which is also shown in Fig. 4(c), showed a congruent result. AGO5 had a preference in binding siRNAs with a guide‐strand 5′‐terminal C‐residue (Mi et al., 2008; Schuck et al., 2013); accordingly, the RCA_1 22 nt vasiRNA was found to be highly active with this AGO/RISC. Interestingly, and in agreement with previous observations (Schuck et al., 2013), we found that 22 nt siRNA‐directed slicing did not exclusively follow these rules. Thus, RCA‐1, having a 5′‐C (Fig. 4a), was also found to be active with AGO1 and AGO2/RISC, and RCA_2 having a 5′A also showed cleavage with AGO1/RISC (Fig. 4c). To compare the cleavage efficacy of 22 nt and 21 nt vasiRNAs targeting the same mRNA, we also performed the assay with selected shorter versions of 21 nt having the same nucleotide at 5′‐ends (Fig. 4c). Although, in most cases, we did not observe remarkable differences in the slicing of the target, the 21 nt vasiRNA appeared less efficient in cleaving the RCA target than the corresponding 22 nt vasiRNA did (Fig. 4c). Conversely, with the HSP70‐1 target, the 21 nt version had a slightly higher cleavage efficiency than the 22 nt vasiRNA under the conditions applied (Fig. 4c).
Distribution of 21 nt and 22 nt virus‐activated small interfering RNAs on the originating transcripts
In a final approach, we aimed at gaining further insights into the characteristics of the vasiRNAs that were associated with CaMV infections. For this purpose, we reanalyzed the 21 nt and 22 nt vasiRNA data sets with regard to the following three aspects: (1) distribution of the vasiRNAs along the originating CDSs; (2) vasiRNA strands as a meaning of ratio of sense and antisense orientation referred to the CDSs; and (3) a potential phasing during processing. For these purposes, a specific R script was designed to align all 21 nt and 22 nt reads to the CDSs in Table 1 and graphically represent the distribution along CDSs by length and by strand. The script was optimized to obtain and represent all possible phases that include the reads from putative starting points. Fig. 5 displays the analyses carried out on the emblematic case of LHCB1.3 CDS, which is the most represented in term of vasiRNAs; the data obtained with other selected vasiRNAs loci are supplied in Fig. S6.
Fig. 5.
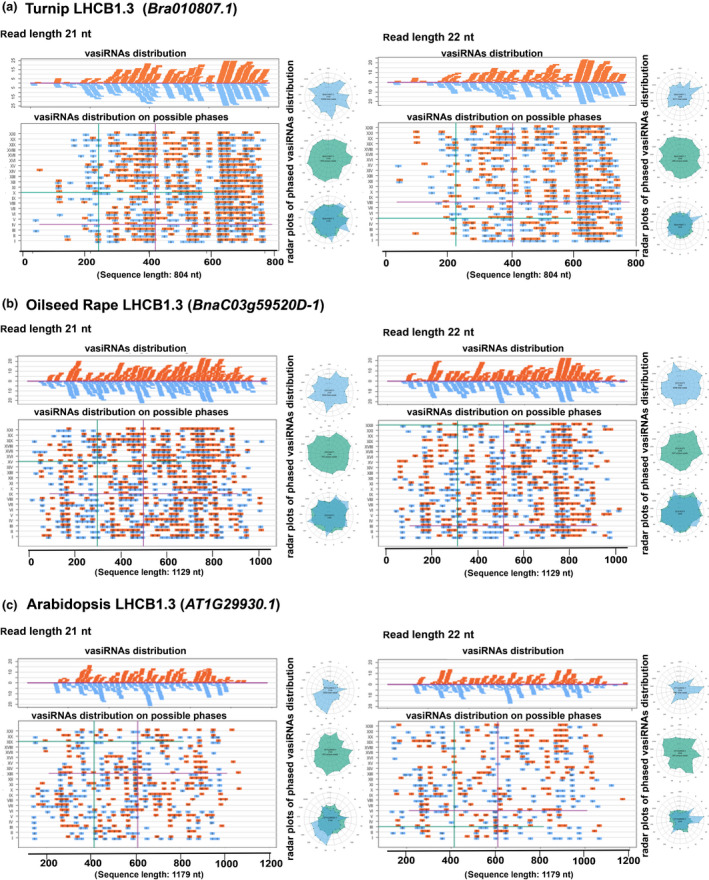
Distribution and phasing of 21 nt and 22 nt virus‐activated small interfering RNAs (vasiRNAs) on the light harvesting complex protein 1.3 (LHCB1.3) transcripts of turnip, oilseed rape, and Arabidopsis thaliana. (a) The upper panels show the distribution of unique 21 and 22 nt vasiRNAs along the whole LHCB1.3 transcript of turnip (Bra010807.1; 804 nt in length). In the lower panels, vasiRNAs were distributed along each putative phasing pattern; that is, I to XXI (21 nt long vasiRNAs) or I to XXII (22 nt long vasiRNAs). Radar plots represent phased unique vasiRNA (light blue, top), redundant (pale green, middle), and a superimposition of the two (bottom). These analyses were performed with 21 nt (left) and 22 nt (right) vasiRNAs, respectively. (b) The analogous analyses as in (a) but performed with the whole LHCB1.3 transcript of oilseed rape (BnaC03g59520D‐1, 1129 nt in length). (c) The analogous analyses as in (a) and (b) but performed with the whole LHCB1.3 transcript of A. thaliana (AT1G29930.1, 1179 nt in length). vasiRNAs of sense orientation are represented in orange; vasiRNAs of 5′ to 3′ of negative antisense orientation are represented in light blue. Numbers inside vasiRNAs indicate redundancy.
Concerning aspect (1), the analyses revealed no apparent differences in the distribution of the 21 nt and 22 nt vasiRNAs along the CDSs in A. thaliana, turnip, or OSR, indicating a rather similar processing pattern (Fig. 5a–c, left and right upper panels). One possible interpretation is that nascent mRNAs from LHCB1.3 are readily processed by either DCL4 or DCL2, thus producing 21 nt and 22 nt vasiRNAs in CaMV‐infected tissues. This idea is consistent with the observed upregulation of DCL4 transcripts in both turnip and OSR and DCL2 transcripts in OSR leaf tissue (Fig. 3f). Concerning the distribution along the originating CDSs (aspect 2), both polarities were equally represented, and vasiRNAs were found to be distributed almost mirror inverted along the CDS as one would expect this to be the case with full‐length dsRNAs processed by DCLs (Fig. 5a–c, orange and light blue bars in the upper panels). In line with these findings, we monitored the presence of antisense LHCB1.3 RNA by strand‐specific PCR (Fig. S7). Though antisense LHCB1.3 transcripts were also detectable in mock‐treated plant tissues (Fig. S7a), we could reveal by strand‐specific qPCR (Fig. S7b) that its level was considerably increased in infected tissues (blue bars in Fig. S7c,d). In the case of turnip, the increase of antisense transcript was particularly evident when analyzing the 3′‐region of LHCB1.3 mRNA (Fig. S7c, block 3). In the context of OSR (Fig. S7d, blocks 1–3), this was clearly evident for all three regions of the transcript investigated. This supports the earlier interpretation of the data from (1) as well as the notion that vasiRNAs arise primarily via DCL processing of dsRNA generated from overlapping transcription units. The generation of dsRNA may be promoted, for example, by the activity of RDR1, whose expression is also upregulated in CaMV‐infected tissues (Fig. 3f) (see also Discussion section).
Concerning aspect (3), it is important to note that, in the study of Cao et al. (2014), the 21 nt vasiRNAs that were detected in CMV‐Δ2b and TuMV‐infected A. thaliana derived from almost the entire sequence of the vasiRNA loci transcripts. In other words, the authors obtained no hints of a phasing pattern. Phasing implies a processive activity of DCLs on dsRNAs, as reported earlier either for vegetative or reproductive phasiRNAs (Xie et al., 2005; Xia et al., 2019). With DCL4 and DCL2, this would lead to the accumulation of vasiRNAs processed at regular intervals (i.e. 21 nt or 22 nt, respectively) along the originating CDS transcripts. To explore this option, we evaluated the placement of all vasiRNAs in all conceivable 21 nt or 22 nt phases on the turnip, OSR, and A. thaliana LHCB1.3 CDSs (see roman numbers I to XXII in the middle panels and in the radar plots in Fig. 5a–c). The analyses did not reveal for either 21 nt or 22 nt vasiRNAs and in all conceivable frames a statistically significant phasing in all three pathosystems. In other words, the LHCB1.3 CDS was found to be uniformly covered by the selected 21 or 22 nt vasiRNAs, suggesting that the processing of CDS dsRNAs occurs at different sites.
Discussion
When investigating populations of sRNAs in A. thaliana plants after the induction of antiviral silencing by members of two RNA virus supergroups, Cao et al. (2014) identified vasiRNAs as a new, distinct class of secondary, endogenous, and ‘virus‐activated’ siRNAs that do not act antivirally but silence the expression of host genes. The sequences of these RNAs map to exon regions of the host genome (denoted vasiRNA loci here), and their biogenesis was shown to derive from host‐cell transcripts requiring RDR1 and DCL4 (Cao et al., 2014).
In the present study, actually representing the second report on vasiRNAs, we characterized secondary siRNA loci in three Brassicaceae species, two of which – turnip and OSR – representing important crops, were infected by CaMV. CaMV is one of the most widely studied plant viruses of the DNA virus supergroups, which multiplies in the nucleus similar to a mini‐chromosome (Saunders et al., 1990). Applying a suitably designed and trained machine‐learning algorithm, we were able to accurately select tissues for molecular analyses in search of specific pathogenesis‐associated patterns, despite the heterogeneity of symptoms caused by virus infections. Each pixel of several 30 Mpx images, in fact, was manually segmented and classified by an expert virologist in three classes: background pixels (‘bg’), symptomatic pixels (‘CaMV’), and asymptomatic pixels (‘mock’), thus creating the ground truth to be used for training and validating the classifier (see the Materials and Methods section; Figs 1, S1). Combining sRNA‐Seq results from CaMV‐inoculated turnip and OSR plants with previously released data in A. thaliana (Blevins et al., 2011), we identified a discrete and conserved list of orthologous CDSs that lead to the massive genesis of vasiRNAs (Fig. 2). This promotes confidence that our data describe a fundamental response to virus infection and not a serendipitous effect that is specific to a single host plant species. Interestingly, the 15 vasiRNA loci that were defined during our analyses (Table 1) match perfectly with the origins of vasiRNAs that were earlier defined by Cao et al. (2014) in RNA virus‐infected A. thaliana plants. Accordingly, as a major finding of our study, all three Brassicaceae species investigated display a highly conserved vasiRNA induction response to infections of taxonomically unrelated viruses. Furthermore, we were able to demonstrate that the production of bona fide CaMV‐induced vasiRNA actually led to a switch‐off of host‐gene expression in the virus‐infected plants; that is, in most target genes we observed a significant post‐transcriptional downregulation of up to 90% compared with mock‐treated plants (Fig. 3).
Arabidopsis 22 nt endogenous siRNAs have been recently described as specifically triggered to play regulatory functions of specific genes in response to environmental stresses (Wu et al., 2020); in line with this, we also provide evidence that CaMV infection promotes similar plant responses. While analyzing the 15 vasiRNA loci, we interestingly found a higher incidence of 22 nt than 21 nt vasiRNAs. These observations differ from the earlier total vasiRNA analyses by Cao et al. (2014), which revealed an apparent excess of 21 nt vasiRNAs in RNA virus‐infected A. thaliana. However, we demonstrate herein that abundant 22 nt siRNA duplexes are capable of efficient sequence‐specific RNA silencing of transcripts via different AGO/RISCs, like AGO1 and AGO2/RISCs, as well as AGO5/RISC (Fig. 4). Our data confirm that the sorting of the 22 nt siRNAs into the respective AGO/RISCs is determined by the 5′‐terminal residue of the guide RNA (Mi et al., 2008). On the other hand, this process was also suggested to depend on other, undefined criteria. As a result, the association of 22 nt vasiRNAs with AGO‐containing RISCs appears to be more adaptable, as is the case with 21 nt siRNAs (Fig. 4; Schuck et al., 2013), implicating that one vasiRNA may activate several types of AGO/RISC. For example, the vasiRNA RCA_1 functions in AGO5/RISC as well as in AGO1 and AGO2/RISC (Fig. 4). The observation that all vasiRNAs identified, when binding to one or more AGO/RISCs, support the cleavage of their cognate mRNA targets suggests that post‐transcriptional regulation of gene expression by these 22 nt vasiRNAs is an overall efficient process. The striking silencing efficacy of host‐gene expression that was observed in the plant experiments shown in Fig. 3 clearly supports the notion that 22 nt vasiRNAs should be considered as highly active components of CaMV‐activated host‐gene silencing. It is noteworthy that, in A. thaliana, CaMV infection was shown to alter the profile of endogenous siRNAs by inhibiting the production of 21 nt DCL4 and RDR6‐dependent siRNAs and by increasing the general level of 22 nt siRNAs (Shivaprasad et al., 2008). Because DCL2 is known to rescue antiviral silencing (if DCL4 is inactivated or suppressed) by producing 22 nt siRNAs (Bouché et al., 2006), this could explain the observed increase in accumulation of 22 nt vasiRNAs (Fig. 2). Considering that all experiments were performed with unmodified wild‐type virus, it can be concluded that, similar to the TuMV Hc‐Pro VSR, the CaMV P6 does not impair the biogenesis of vasiRNAs (Cao et al., 2014). It will be an important future task to understand the molecular features and the biological relevance of these diverging characteristics of viral VSRs. Our studies have revealed that vasiRNAs derive from most, if not all, regions of the originating transcripts (Fig. 5). Theoretically, all vasiRNAs, in turn, should be capable of targeting these areas of the cognate CDS; this could be another explanation for the highly significant downregulation reported in Fig. 3. Moreover, we obtained initial evidence suggesting that vasiRNAs are processed by either DCL2 or DCL4 from fully dsRNAs. This idea is supported by the detection of long antisense RNAs that are complementary to the vasiRNA loci (Fig. S7) and the similar distribution of sRNAs across the CDS derived from both sense and antisense strands (Figs 5, S6).
The study by Cao et al. (2014) suggests a DCL4‐RDR1‐AGO2 pathway mediating vasiRNA generation and silencing. Our data support this idea, in that the expression of all three enzymes was found to be considerably induced during a CaMV infection (Fig. 3f). However, our study further suggests a significant role of DCL2 in vasiRNA generation, since DCL2 is the only DICER protein in plants known to produce 22 nt siRNAs from dsRNA precursors. This has been demonstrated, for example, for trans‐acting siRNA loci in DCL4‐defective A. thaliana (Gasciolli et al., 2005). Finally, in view of the scenario where AGO1 and AGO5 can operate with vasiRNAs, other AGO/RISCs may be considered to be part of this silencing response as well.
Viruses are obligated parasites, and their replication requires considerable energy sources. It is widely accepted that viral infections provoke readjustments of the main physiological energy production processes (Souza et al., 2019; Cui et al., 2020). In this sense, the congruent data of Cao et al. (2014) together with ours show a virus‐activated, but ultimately host‐mediated, modulation of the expression of stress and photosynthesis‐related genes (PRGs) A vasiRNA‐mediated downregulation of PRGs may thus be part of a prompt and powerful response restricting viral attacks by reducing light harvesting and, with this, energy available for viral functions. The broad‐spectrum immunity of plants to viruses is often linked to the induced regulation of PRGs. This regulation is directly mediated by miRNAs, which, as a result of viral infection, effectively cleave PRG mRNAs and also trigger secondary siRNA production. The latter can induce several cascade silencing mechanisms (Chiumenti et al., 2018; Cui et al., 2020). In‐depth studies can verify whether there are any miRNAs or endogenous siRNAs that are not yet known that can be involved in a novel strategy used to modulate the balance between plant growth and defense against pathogens. During studies addressing the functional dissection of the photosystem II genes, members of the lhcb family have been extensively investigated in Arabidopsis (Caffarri et al., 2004). More recently, the Lhcb1.3 gene has been specifically deactivated by artificial (a)miRNAs or Cas9. When grown under controlled conditions, amiLhcb1 or Cas9‐Lhcb1 mutants were shown to grow more slowly than the wild‐type plant and the leaves were slightly smaller and paler (yellow) (Pietrzykowska et al., 2014; Ordon et al., 2020). The significant reduction in lhcb1 transcripts that we observed in CaMV‐infected tissues and the coexistence of a diseased phenotype (mosaic) is consistent with PRG involvement in symptom development.
During various stages of their life cycle, viruses hijack the host's transport, transcription, and translation engines (e.g. for genome replication and systemic invasion). Our data are in line with earlier findings that have suggested virus‐induced remodeling of the host's transcriptome as another mode to support the viral infection process (Pesti et al., 2019). Within this framework, an important future perspective is expected in the modulation of photosynthesis. This may be achieved, for example, by fine‐tuning the expression of core components of the light harvesting complex, such as the LHC proteins. LHC proteins, representing the most abundant proteins on Earth (Pietrzykowska et al., 2014), play pivotal roles in defining the structure of plant chloroplasts and in optimizing and balancing their functions. Because the expression of at least three of these proteins was found to be downregulated by vasiRNAs in virus‐infected plants (Table 1), our study may point the way towards engineered crop plants, such as turnip and OSR, endowed with increased resilience to viruses and other biotic stresses resulting from a sustainable balance with photosynthesis efficiency.
Author contributions
VP and AG conceived the experiments of RNA‐seq analysis. AG carried out plant experiments and virus inoculation. VP, AG, and AC performed the bioinformatics analyses. TG performed cleavage assays. PL performed gene expression analyses and applied the machine learning approach. VP, PL, and SEB wrote the manuscript. All authors reviewed the manuscript.
Supporting information
Fig. S1 Schematic representation of the machine learning structure for image analyses and tissues selection.
Fig. S2 Size distribution profile of unique and redundant CaMV‐derived siRNAs (vsiRNAs), in turnip and oilseed rape.
Fig. S3 Origin of sRNAs in Arabidopsis thaliana, turnip and oilseed rape infected by CaMV.
Fig. S4 PCR amplicons for T7 in vitro transcripts used in cleavage assays.
Fig. S5 Alignments of the Sanger‐sequenced amplicons (RCA, HSP70‐1, LHCB1.3 ) obtained from oilseed rape with the orthologues in turnip.
Fig. S6 21‐ and 22‐nt vasiRNAs distribution and phasing in Arabidopsis thaliana, turnip and oilseed rape.
Fig. S7 Detection of antisense LHCB1.3 transcripts.
Table S1 Primers used in this study.
Table S2 Statistic of small RNAs from Arabidopsis thaliana and Brassica sp. related to Fig.1(b).
Table S3 χ2 test and fold change evaluation for unique read counts, grouped by length.
Table S4 Output of unique 21‐nt, 22‐nt vasiRNA analysis in mock‐treated and CaMV‐infected tissues, referred to each gene in Table 1.
Table S5 Microarray comparison of expression profiles of CP5, HSP70‐1 and SAP.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
We thank Professor James Schoelz from the University of Missouri, Columbia for providing us with CaMV strain NY8153, and Dr Mikhael Pooggin for providing us with siRNA data of the A. thaliana–CaMV pathosystem. We thank Eng. Nicola Altini, PhD candidate in computer engineering at the Polytechnic University of Bari (Italy), for his helpful contribution in implementing and running the machine‐learning architecture. In addition, we thank Antonia Antonacci for technical assistance and Michela Chiumenti for plotting vsiRNAs. The study is in the frame of Project ‘Photosintesis 2.0’ and ‘NUTR‐AGE’ (FOE‐2019, DSBAD004.271) to Dipartimento di scienze Bio‐AgroAlimentari (DiSBA) of the National Research Council, Italy. VP was a recipient of a STM CNR 2018 fellowship. AG was recipient of a fellowship by the Ministry of Sciences Research and Technology of Iran for a period of study at CNR, Italy. AG currently holds a scholarship in the program ‘Determinants of Plant Performance (DPP)’ of the International Graduate Schools in Agricultural and Polymer Science (AGRIPOLY) financed via the European Social Fund by the state of Saxony Anhalt.
Data availability
The sRNA data sets have been deposited in GEO under entry code GSE119398.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped Blast and PSI‐Blast: a new generation of protein database search programs. Nucleic Acids Research 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins T, Rajeswaran R, Aregger M, Borah BK, Schepetilnikov M, Baerlocher L, Farinelli L, Meins F Jr, Hohn T, Pooggin MM. 2011. Massive production of small RNAs from a non‐coding region of cauliflower mosaic virus in plant defense and viral counter‐defense. Nucleic Acids Research 39: 5003–5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouché N, Lauressergues D, Gasciolli V, Vaucheret H. 2006. An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO Journal 25: 3347–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL et al 2009. The MIQE guidelines: minimum information for publication of quantitative real‐time PCR experiments. Clinical Chemistry 55: 611–622. [DOI] [PubMed] [Google Scholar]
- Caffarri S, Croce R, Cattivelli L, Bassi R. 2004. A look within LHCII: differential analysis of the Lhcb1‐3 complexes building the major trimeric antenna complex of higher‐plant photosynthesis. Biochemistry 43: 9467–9476. [DOI] [PubMed] [Google Scholar]
- Cao M, Du P, Wang X, Yu YQ, Qiu YH, Li W, Gal‐On A, Zhou C, Li Y, Ding SW. 2014. Virus infection triggers widespread silencing of host genes by a distinct class of endogenous siRNAs in Arabidopsis . Proceedings of the National Academy of Sciences, USA 111: 14613–14618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalhoub B, Denoeud F, Liu S, Parkin IA, Tang H, Wang X, Chiquet J, Belcram H, Tong C, Samans B et al 2014. Plant genetics. Early allopolyploid evolution in the post‐Neolithic Brassica napus oilseed genome. Science 345: 950–953. [DOI] [PubMed] [Google Scholar]
- Chen LC, Zhu Y, Papandreou G, Schroff F, Adam H. 2018. Encoder–decoder with atrous separable convolution for semantic image segmentation In: Ferrari V, Hebert M, Sminchisescu C, Weiss Y, eds. Computer Vision – ECCV 2018. Lecture Notes in Computer Science, vol. 11211 Cham, Switzerland: Springer, 833–851. [Google Scholar]
- Chiumenti M, Catacchio CR, Miozzi L, Pirovano W, Ventura M, Pantaleo V. 2018. A short indel‐lacking‐resistance gene triggers silencing of the photosynthetic machinery components through TYLCSV‐associated endogenous siRNAs in tomato. Front Plant Sci 9: e1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csorba T, Kontra L, Burgyán J. 2015. Viral silencing suppressors: tools forged to fine‐tune host–pathogen coexistence. Virology 479–480: 85–103. [DOI] [PubMed] [Google Scholar]
- Cui C, Wang JJ, Zhao JH, Fang YY, He XF, Guo HS, Duan CG. 2020. A Brassica miRNA regulates plant growth and immunity through distinct modes of action. Molecular Plant 13: 231–245. [DOI] [PubMed] [Google Scholar]
- Gago‐Zachert S, Schuck J, Weinholdt C, Knoblich M, Pantaleo V, Grosse I, Gursinsky T, Behrens SE. 2019. Highly efficacious antiviral protection of plants by small interfering RNAs identified in vitro . Nucleic Acids Research 47: 9343–9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasciolli V, Mallory AC, Bartel DP, Vaucheret H. 2005. Partially redundant functions of Arabidopsis DICER‐like enzymes and a role for DCL4 in producing trans‐acting siRNAs. Current Biology 15: 1494–1500. [DOI] [PubMed] [Google Scholar]
- Guo Z, Li Y, Ding SW. 2019. Small RNA‐based antimicrobial immunity. Nature Reviews Immunology 19: 31–44. [DOI] [PubMed] [Google Scholar]
- Han T, Dong H, Cui J, Li M, Lin S, Cao J, Huang L. 2017. Genomic, molecular evolution, and expression analysis of genes encoding putative classical AGPs, lysine‐rich AGPs, and AG peptides in Brassica rapa . Frontiers in Plant Science 8: e397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey JJ, Lewsey MG, Patel K, Westwood J, Heimstadt S, Carr JP, Baulcombe DC. 2011. An antiviral defense role of AGO2 in plants. PLoS ONE 6: e14639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havelda Z, Várallyay E, Válóczi A, Burgyán J. 2008. Plant virus infection induced persistent host gene down regulation in systemically infected leaves. The Plant Journal 55: 278–288. [DOI] [PubMed] [Google Scholar]
- He K, Zhang X, Ren S, Sun J. 2016. Deep residual learning for image recognition. Proceedings: 29th IEEE Conference on Computer Vision and Pattern Recognition – CVPR 2016. Los Alamitos, CA, USA: IEEE Computer Society. [Google Scholar]
- Iki T, Yoshikawa M, Nishikiori M, Jaudal MC, Matsumoto‐Yokoyama E, Mitsuhara I, Meshi T, Ishikawa M. 2010. In vitro assembly of plant RNA‐induced silencing complexes facilitated by molecular chaperone HSP90. Molecular Cell 39: 282–291. [DOI] [PubMed] [Google Scholar]
- Kontra L, Csorba T, Tavazza M, Lucioli A, Tavazza R, Moxon S, Tisza V, Medzihradszky A, Turina M, Burgyan J. 2016. Distinct effects of p19 RNA silencing suppressor on small RNA mediated pathways in plants. PLoS Pathogens 12: e1005935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Birgaoanu M, Griffiths‐Jones S. 2019. miRBase: from microRNA sequences to function. Nucleic Acids Research 47: D155–D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths‐Jones S. 2014. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Research 42: D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory‐efficient alignment of short DNA sequences to the human genome. Genome Biology 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonetti P, Miesen P, van Rij RP, Pantaleo V. 2020. Viral and subviral derived small RNAs as pathogenic determinants in plants and insects. Advances in Virus Research 107: 1–36. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real‐time quantitative RT‐PCR and the 2−ΔΔCT method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Lu K, Wei L, Li X, Wang Y, Wu J, Liu M, Zhang C, Chen Z, Xiao Z, Jian H et al 2019. Whole‐genome resequencing reveals Brassica napus origin and genetic loci involved in its improvement. Nature Communications 10: e1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez de Alba AE, Elvira‐Matelot E, Vaucheret H. 2013. Gene silencing in plants: a diversity of pathways. Biochimica et Biophysica Acta 1829: 1300–1308. [DOI] [PubMed] [Google Scholar]
- Medzihradszky A, Gyula P, Sós‐Hegedűs A, Szittya G, Burgyán J. 2019. Transcriptome reprogramming in the shoot apical meristem of CymRSV‐infected Nicotiana benthamiana plants associates with viral exclusion and the lack of recovery. Molecular Plant Pathology 20: 1748–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C et al 2008. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133: 116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miozzi L, Gambino G, Burgyan J, Pantaleo V. 2013. Genome‐wide identification of viral and host transcripts targeted by viral siRNAs in Vitis vinifera . Molecular Plant Pathology 14: 30–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourrain P, Béclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, Jouette D, Lacombe AM, Nikic S, Picault N et al 2000. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101: 533–542. [DOI] [PubMed] [Google Scholar]
- Ordon J, Bressan M, Kretschmer C, Dall'Osto L, Marillonnet S, Bassi R, Stuttmann J. 2020. Optimized Cas9 expression systems for highly efficient Arabidopsis genome editing facilitate isolation of complex alleles in a single generation. Functional & Integrative Genomics 20: 151–162. [DOI] [PubMed] [Google Scholar]
- Pantaleo V, Burgyán J. 2008. Cymbidium ringspot virus harnesses RNA silencing to control the accumulation of virus parasite satellite RNA. Journal of Virology 82: 11851–11858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertermann R, Tamilarasan S, Gursinsky T, Gambino G, Schuck J, Weinholdt C, Lilie H, Grosse I, Golbik RP, Pantaleo V et al 2018. A viral suppressor modulates the plant immune response early in infection by regulating miRNA activity. MBio 9: e00419‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesti R, Kontra L, Paul K, Vass I, Csorba T, Havelda Z, Várallyay E. 2019. Differential gene expression and physiological changes during acute or persistent plant virus interactions may contribute to viral symptom differences. PLoS ONE 14: e0216618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrzykowska M, Suorsa M, Semchonok DA, Tikkanen M, Boekema EJ, Aro EM, Jansson S. 2014. The light‐harvesting chlorophyll a/b binding proteins Lhcb1 and Lhcb2 play complementary roles during state transitions in Arabidopsis . The Plant Cell 26: 3646–3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirovano W, Miozzi L, Boetzer M, Pantaleo V. 2014. Bioinformatics approaches for viral metagenomics in plants using short RNAs: model case of study and application to a Cicer arietinum population. Frontiers in Microbiology 5: e790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. 2006. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana . Genes & Development 20: 3407–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders K, Lucy AP, Covey SN. 1990. Susceptibility of Brassica species to cauliflower mosaic virus infection is related to a specific stage in the virus multiplication cycle. Journal of General Virology 71: 1641–1647. [DOI] [PubMed] [Google Scholar]
- Schoelz J, Shepherd RJ, Daubert S. 1986. Region VI of cauliflower mosaic virus encodes a host range determinant. Molecular and Cellular Biology 6: 2632–2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck J, Gursinsky T, Pantaleo V, Burgyán J, Behrens SE. 2013. AGO/RISC‐mediated antiviral RNA silencing in a plant in vitro system. Nucleic Acids Research 41: 5090–5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seguin J, Rajeswaran R, Malpica‐López N, Martin RR, Kasschau K, Dolja VV, Otten P, Farinelli L, Pooggin MM. 2014. De novo reconstruction of consensus master genomes of plant RNA and DNA viruses from siRNAs. PLoS ONE 9: e88513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen D, Suhrkamp I, Wang Y, Liu S, Menkhaus J, Verreet JA, Fan L, Cai D. 2014. Identification and characterization of microRNAs in oilseed rape (Brassica napus) responsive to infection with the pathogenic fungus Verticillium longisporum using Brassica AA (Brassica rapa) and CC (Brassica oleracea) as reference genomes. New Phytologist 204: 577–594. [DOI] [PubMed] [Google Scholar]
- Shepherd R. 1979. DNA plant viruses. Annual Review of Plant Physiology 30: 405–426. [Google Scholar]
- Shimura H, Pantaleo V, Ishihara T, Myojo N, Inaba J, Sueda K, Burgyán J, Masuta C. 2011. A viral satellite RNA induces yellow symptoms on tobacco by targeting a gene involved in chlorophyll biosynthesis using the RNA silencing machinery. PLoS Pathogens 7: e1002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaprasad PV, Chen HM, Patel K, Bond DM, Santos BA, Baulcombe DC. 2012. A microRNA superfamily regulates nucleotide binding site‐leucine‐rich repeats and other mRNAs. The Plant Cell 24: 859–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaprasad PV, Rajeswaran R, Blevins T, Schoelz J, Meins F Jr, Hohn T, Pooggin MM. 2008. The CaMV transactivator/viroplasmin interferes with RDR6‐dependent trans‐acting and secondary siRNA pathways in Arabidopsis. Nucleic Acids Research 36: 5896–5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorefan K, Pais H, Hall AE, Kozomara A, Griffiths‐Jones S, Moulton V, Dalmay T. 2012. Reducing ligation bias of small RNAs in libraries for next generation sequencing. Silence 3: e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza PFN, Garcia‐Ruiz H, Carvalho FEL. 2019. What proteomics can reveal about plant–virus interactions? Photosynthesis‐related proteins on the spotlight. Theoretical and Experimental Plant Physiology 31: 227–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocks MB, Moxon S, Mapleson D, Woolfenden HC, Mohorianu I, Folkes L, Schwach F, Dalmay T, Moulton V. 2012. The UEA sRNA workbench: a suite of tools for analysing and visualizing next generation sequencing microRNA and small RNA datasets. Bioinformatics 28: 2059–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Várallyay E, Válóczi A, Agyi A, Burgyán J, Havelda Z. 2010. Plant virus‐mediated induction of miR168 is associated with repression of ARGONAUTE1 accumulation. EMBO Journal 29: 3507–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, Bai Y, Mun JH, Bancroft I, Cheng F et al 2011. The genome of the mesopolyploid crop species Brassica rapa . Nature Genetics 43: 1035–1039. [DOI] [PubMed] [Google Scholar]
- Wu H, Li B, Iwakawa HO, Pan Y, Tang X, Ling‐Hu Q, Liu Y, Sheng S, Feng L, Zhang H et al 2020. Plant 22‐nt siRNAs mediate translational repression and stress adaptation. Nature 581: 89–93. [DOI] [PubMed] [Google Scholar]
- Xia R, Chen C, Pokhrel S, Ma W, Huang K, Patel P, Wang F, Xu J, Liu Z, Li J et al 2019. 24‐nt reproductive phasiRNAs are broadly present in angiosperms. Nature Communications 10: e627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Allen E, Wilken A, Carrington JC. 2005. DICER‐LIKE 4 functions in trans‐acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana . Proceedings of the National Academy of Sciences, USA 102: 12984–12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yuan YR, Pei Y, Lin SS, Tuschl T, Patel DJ, Chua NH. 2006. Cucumber mosaic virus‐encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes & Development 20: 3255–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao YT, Wang M, Fu SX, Yang WC, Qi CK, Wang XJ. 2012. Small RNA profiling in two Brassica napus cultivars identifies microRNAs with oil production and development‐correlated expression and new small RNA classes. Plant Physiology 158: 813–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Duan CG, Hou WN, Du QS, Lv DQ, Fang RX, Guo HS. 2011. Satellite RNA‐derived small interfering RNA satsiR‐12 targeting the 3′ untranslated region of Cucumber mosaic virus triggers viral RNAs for degradation. Journal of Virology 85: 13384–13397. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Schematic representation of the machine learning structure for image analyses and tissues selection.
Fig. S2 Size distribution profile of unique and redundant CaMV‐derived siRNAs (vsiRNAs), in turnip and oilseed rape.
Fig. S3 Origin of sRNAs in Arabidopsis thaliana, turnip and oilseed rape infected by CaMV.
Fig. S4 PCR amplicons for T7 in vitro transcripts used in cleavage assays.
Fig. S5 Alignments of the Sanger‐sequenced amplicons (RCA, HSP70‐1, LHCB1.3 ) obtained from oilseed rape with the orthologues in turnip.
Fig. S6 21‐ and 22‐nt vasiRNAs distribution and phasing in Arabidopsis thaliana, turnip and oilseed rape.
Fig. S7 Detection of antisense LHCB1.3 transcripts.
Table S1 Primers used in this study.
Table S2 Statistic of small RNAs from Arabidopsis thaliana and Brassica sp. related to Fig.1(b).
Table S3 χ2 test and fold change evaluation for unique read counts, grouped by length.
Table S4 Output of unique 21‐nt, 22‐nt vasiRNA analysis in mock‐treated and CaMV‐infected tissues, referred to each gene in Table 1.
Table S5 Microarray comparison of expression profiles of CP5, HSP70‐1 and SAP.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Data Availability Statement
The sRNA data sets have been deposited in GEO under entry code GSE119398.


