Summary
Fine roots, and their functional traits, influence associated rhizosphere microorganisms via root exudation and root litter quality. However, little information is known about their relationship with rhizosphere microbial taxa and functional guilds.
We investigated the relationships of 11 fine root traits of 20 sub‐arctic tundra meadow plant species and soil microbial community composition, using phospholipid fatty acids (PLFAs) and high‐throughput sequencing. We primarily focused on the root economics spectrum, as it provides a useful framework to examine plant strategies by integrating the co‐ordination of belowground root traits along a resource acquisition–conservation trade‐off axis.
We found that the chemical axis of the fine root economics spectrum was positively related to fungal to bacterial ratios, but negatively to Gram‐positive to Gram‐negative bacterial ratios. However, this spectrum was unrelated to the relative abundance of functional guilds of soil fungi. Nevertheless, the relative abundance of arbuscular mycorrhizal fungi was positively correlated to root carbon content, but negatively to the numbers of root forks per root length.
Our results suggest that the fine root economics spectrum is important for predicting broader groups of soil microorganisms (i.e. fungi and bacteria), while individual root traits may be more important for predicting soil microbial taxa and functional guilds.
Keywords: fine root traits, fungi, plant–microorganism interactions, rhizosphere, tundra ecosystems
Introduction
Plant–microorganism interactions are important drivers of plant community composition and dynamics (Klironomos, 2002; Wardle et al., 2004). Plants influence soil microbial community composition by root exudation (Bais et al., 2006), symbiotic interactions, and by determining the quantity and quality of litter available for decomposers (Wardle et al., 2004; Veen et al., 2019). These interaction pathways are known to influence the presence and abundance of mutualistic (Bonfante & Genre, 2010) and antagonistic (Philippot et al., 2013) soil microorganisms. Depending on the nature of plant‐induced shifts in these groups of microbes, the overall soil microbial community can exert positive, neutral or negative feedbacks on their associated plant communities (Bever et al., 2012). These feedbacks can, in turn, have consequences for plant community composition, successional dynamics (Kardol et al., 2007) and plant species diversity (Mangan et al., 2010; Teste et al., 2017). Despite the possible direct influence of root traits on the abundance of microorganisms due to plant roots and soil microorganisms being tightly connected in space within the rhizosphere, there remains a huge knowledge gap about how plant roots and their chemical and morphological traits are associated with rhizosphere soil microbial taxa and functional guilds.
Plant fine root characteristics are increasingly being examined within trait‐based frameworks (Kattge et al., 2011; Iversen et al., 2017) in which trait values are used to indicate plant economic strategies, that is acquisitive vs conservative resource use (Freschet et al., 2010). For example, acquisitive trait values indicate higher plant resource investment into the rapid acquisition of nutrients and lower investment in defence, and include high specific root lengths and small fine root diameters (Cortois et al., 2016). By contrast, conservative trait values (e.g. lower specific root lengths and thicker root diameters) are associated with slower growth, well defended tissue and stronger dependence on mycorrhizal symbionts for nutrient uptake (Eissenstat et al., 2015; Cortois et al., 2016). Furthermore, higher concentrations of secondary defence compounds in plant tissues reduce the abundance of foliar and root pathogens (Tomova et al., 2005; Maupetit et al., 2018), and fine root litter polyphenol concentrations can modify saprotroph activity (Sun et al., 2018). In addition, the ratios of Gram‐positive to Gram‐negative bacteria and of fungi to bacteria are expected to respond to those fine root traits that are linked to root quality. This is because Gram‐negative bacteria utilise more labile plant‐derived carbon (Fanin et al., 2019a), while Gram‐positive bacteria utilise more recalcitrant soil‐derived carbon (Kramer & Gleixner, 2008), and increased litter recalcitrance promotes the dominance of fungi over bacteria (Strickland & Rousk, 2010). Hence, using a gradient of fine root trait values (i.e. across the fine root economics spectrum) will enable explicit testing of how fine root trait values affect the abundance of various rhizosphere soil microorganisms.
For the present study, we focused on the fine root trait relationships with soil microorganisms in the arctic tundra. These ecosystems are characterised by nutrient‐poor soils (Sundqvist et al., 2014), suggesting that their plants may be more dependent on tight root–microorganism associations for nutrient recycling than are those in other, more nutrient‐rich, ecosystems. In addition, the large fine root biomass and quantity of root litter often present in tundra ecosystems (Chapin et al., 2011; Iversen et al., 2015) is likely to support an extensive mycorrhizal network for nutrient uptake and a high abundance of saprotrophs (McCormack & Iversen, 2019). However, to our knowledge, no study has examined the variation in morphological and chemical root traits in tundra plants, although Björk et al. (2007) have studied the effect of warming on some morphological fine root traits within tundra plant communities. Instead, tundra root research to date has largely focused on total root biomass (Iversen et al., 2015) and phenology (Blume‐Werry et al., 2016). In addition, although tundra typically harbours a high abundance of cold‐tolerant fungi, including yeasts (Margesin et al., 2009; Treseder & Lennon, 2015), and mycorrhizal associations are common (Newsham et al., 2009), we know little about moulds, saprotrophic and pathogenic fungal taxa in these ecosystems.
Sub‐arctic tundra has two main vegetation types, that is meadow (dominated by graminoids and forbs) and heath (dominated by dwarf shrubs). Here, we focused on the sub‐arctic meadow vegetation because it has a large species pool, while heath does not (Sundqvist et al., 2011). We used a glasshouse experiment to test the effects of 11 morphological and chemical fine root traits, within and across 20 sub‐arctic tundra meadow plant species, on rhizosphere microbial community composition. We characterised the microbial community both at the level of broad microbial groups (phospholipid fatty acids, PLFAs) and at the level of fungal orders and functional guilds (through high‐throughput sequencing). Specifically, we tested the following hypotheses:
(1) Acquisitive root trait values will be associated with lower soil fungal to bacterial ratios and lower Gram‐positive to Gram‐negative bacterial ratios, relative to conservative root trait values.
This is because acquisitive root trait values are expected to be positively linked to bacterial decomposers (Wardle et al., 2004) but not to mycorrhizal fungi (Ma et al., 2018), and because Gram‐positive bacteria are expected to associate with recalcitrant litter from plants with conservative root traits (Fanin et al., 2019a).
(2) Conservative root traits will be associated with a higher relative abundance of mycorrhizal taxa in the fungal communities.
This is because roots of resource‐conservative plants are expected to have a higher spatial capacity for accommodating mycorrhizal hyphae and are associated with greater mycorrhizal nutrient foraging (Eissenstat et al., 2015; Chen et al., 2016).
(3) Acquisitive root trait values will be associated with a higher abundance of fungal saprotrophs, pathogens, yeasts and moulds.
This is because organs of plants with acquisitive traits are expected to be less defended, more active in exudation of simple organic carbon compounds and more nutrient‐rich, making them more susceptible to pathogens and more attractive to opportunists and saprotrophs (Tomova et al., 2005; Botha, 2006).
Materials and Methods
Study site
The site for sourcing soils and most of the seed material for our study is located in northern Sweden, c. 27 km north‐east of Abisko (68°21′N, 18°49′E), at the foothills of Mount Gearggečorrus at 700 m asl. The climate is sub‐arctic with a growing season of c. 3 months. The forests at lower altitudes in this mountainous landscape are dominated by mountain birch (Betula pubescens ssp. czerepanovii), with the treeline occurring at c. 500–600 m asl. The vegetation above the treeline is dominated by mosaics of heath (comprised primarily of ericaceous dwarf shrubs and Betula nana) and meadow (comprised primarily of graminoids and forbs). The mean annual precipitation from 2005–2017 at the Abisko Scientific Research Station was 340 mm and the mean temperature for the corresponding period was +13°C in July and −9.9°C in January (the Swedish Meteorological and Hydrological Institute).
Experimental design
We collected seeds from individuals of each of 17 meadow plant species, that is graminoids and forbs at the end of the growing season in the study area in August 2016, and sourced seeds from three additional meadow species (which also occur in the study area) from a local seed company (Pratensis, Lönashult, Sweden). Shrub species are not included in our study (see Supporting Information Fig. S1 for a list of species), as they are primarily found in heath within the sub‐arctic tundra (Sundqvist et al., 2011). All plant species except one (Luzula pilosa), which is nonmycorrhizal, are potential arbuscular mycorrhizal hosts (Soudzilovskaia et al., 2020). Soil was collected during late August 2016 from the rooting zone (= upper 10 cm) beneath individuals of all visible forb and graminoids species, during the expected period of peak root production (Blume‐Werry et al., 2016), bulked and stored in plastic bags at 4°C. The soil type is cryorthents (Darmody et al., 2000) and basic soil properties are as follows: pH 5.54, moisture 25.8 %, NH4 + 0.009 mg g−1 dry soil, and PO4 + < 0.000 mg g−1 dry soil (Veen et al., 2015). The bulk soil was transported to a glasshouse facility in Umeå in cool boxes, along with dried field‐collected seeds (40°C for 2 d) and stored at 22°C until the setup of the experiment in February 2017. Seeds were surface sterilised by soaking for 1 min in 1% sodium hypochlorite (De Long et al., 2015) and germinated on sterilised sand. To ensure that the seedlings were at a similar ontogenic stage at the time of planting, they were stored at 4°C with light (50%) as soon as they had reached a height of c. 3 cm, until the beginning of the experiment.
The field‐collected soil was sieved (10‐mm mesh size) to remove stones and large roots, homogenised and the volume split equally into six blocks. Soil from each block was then mixed with autoclaved sand (soil : sand, 3 : 1) to facilitate better drainage. To enable acclimatisation of the soil microbial community to glasshouse conditions, we left the bulk soil at room temperature for 2 d (20°C) until the time of planting. We filled 1.4 L pots (9 × 9 × 20 cm) with 0.4 L of warm‐water‐washed gravel, followed by the homogenised bulk soil–sand mixture. This resulted in a soil depth of c. 10 cm, which is similar to the soil depth at the site where we collected the soil. We planted two seedlings of the same species in each pot (top right and bottom left), resulting in 20 species × 6 blocks = 120 pots and 12 plants per species. We randomised the location of the pots within the block every 2 wk to account for microclimatic variation in the glasshouse. Plants were grown in the glasshouse (18°C : 13°C, day : night temperature; 80% humidity, and 18 h : 6 h, light : dark regime) for 12 wk, which is similar to the length of one growing season in the Abisko region. We watered the plants every 2 d. During the first 2 wk of the experiment, we replaced any dead seedlings with live seedlings.
Harvest
We harvested the experimental units block‐wise after 84 d. As soil moisture could influence the volume of soil adhering to the roots, we maintained a consistent watering and harvesting regime between blocks. Harvested blocks were stored at 4°C in the dark for a maximum of 3 d and the harvesting order of individual pots within blocks was random. After carefully removing the plants from the pots by tapping the pot, plants were gently shaken (Saj et al., 2009) over trays layered with aluminium foil to obtain the rhizosphere soil surrounding the plant roots (i.e. soil stuck to plant roots) (Smalla et al., 2001; Carrillo et al., 2017). Soil that fell away from roots while removing plants from pots was not considered rhizosphere soil, and thus was not used in the study. We replaced the aluminium sheets in the trays between samples to avoid cross‐contamination. We used this method to collect rhizosphere soil, as opposed to brushing (Clemensson‐Lindell & Persson, 1992; Clemmensen et al., 2008), as we wanted to minimise fine root damage (Fitz et al., 2006). We bulked and transferred the rhizosphere soil from both plants within each pot into clean plastic bags and homogenised the combined sample by shaking the closed bags. Subsamples of c. 500 mg (dry weight equivalent) of all rhizosphere soil samples were immediately frozen at −20°C and after 1 wk freeze dried and ground using a roller grinder for subsequent PLFA and DNA analyses.
Root trait measurements
Before measuring root traits, we washed roots according to protocols outlined in Pérez‐Harguindeguy et al., (2013). Briefly, we carefully washed the plant roots over a sieve (4 mm) and a collection tray. Between successive rinses, we poured the flow‐through from the tray over a 2 mm sieve to recover any roots that were accidentally broken during rinsing. Any remaining soil particles on the roots were washed off with light water spraying. A representative fine root (≤ 1 mm diameter) subsample was collected from the clean root system of the biggest plant of each pair from each pot (or the smaller plant whenever the bigger plant was diseased or yellowing). We used a graduated ruler to determine root diameters ≤ 1 mm. Subsamples were scanned in transparent trays (15 × 20 cm) with cold tap water using WinRhizo 2016 with a flatbed scanner (Epson Perfection V800/V850 1.9 V3.93 3.9.3.2) with the following settings: dust removal on; resolution 800 d post inoculation; root and background detection on; 128 grey detection level and 100 pixels; detection of very pale roots on; overhead lights on. We standardised the quantity of fine roots in each root subsample using the transparent tray. We selected roots until the tray was filled, while ensuring the roots did not overlap within the tray. For small plants, we scanned all fine roots ≤ 1 mm diameter. One root subsample from each pot was scanned. After each scan, we confirmed that all fine roots had the desired diameter threshold by examining the colour‐coded diameter images generated by the winrhizo software (Regent Instruments Inc., Quebec, Canada).
We selected a suite of morphological and chemical root traits from the FRED database (Iversen et al., 2017) based on their potential relevance for root‐associated microbial communities (see Table 1). We measured the following morphological root traits for each root subsample: number of root tips, total root length, root branching, average diameter and root surface area. After scanning, we recorded the fresh weight of each scanned sample and the dry weight following drying at 60°C for 2 d. The remaining roots were dried using the same protocol as used for the scanned root samples and the biomass was recorded. We recorded the total biomass of each scanned root subsample, and calculated specific root length (cm mg−1), specific root tip abundance (tips mg−1), specific root area (cm2 mg−1) and root dry matter content (dry mass per unit fresh mass; mg mg−1). Root dry matter content has been shown to be a good proxy for root tissue density (Birouste et al., 2014), and is a commonly used trait in belowground studies (Henneron et al., 2019).
Table 1.
Fine root traits measured in this study with their units, as well as their ecological relevance.
| Root trait | Unit | Ecological relevance | |
|---|---|---|---|
| Chemical | Carbon (C) content | % | Carbon source for microbes |
| Nitrogen (N) content | % | Nutrients for microorganisms | |
| C : N ratio | – | Tissue quality | |
| Phenol : N ratio | mg g−1 | Investment in defence | |
| Total phenol | mg g−1 | Investment in defence | |
| Morphological | Average diameter | mm | Space for mycorrhizal hyphae |
| Dry matter content | mg mg−1 | Tissue density and quality | |
| Forks per root length | forks cm−1 | Soil exploration | |
| Specific root tip abundance | tips mg−1 | Soil exploration | |
| Specific root area | cm2 mg−1 | Nutrient absorption | |
| Specific root length | cm mg−1 | Soil exploration |
To obtain chemical root trait data we manually ground a subsample (c. 150 mg) of the remaining fine root material from each of the same individuals from which we obtained the scanned root sample. When necessary, we ground the roots with a Retch MM400 ball mill. We measured total root phenol concentrations from 50 ± 5 mg dry ground roots using the method of Stern et al. (1996). Total root carbon (C) and nitrogen (N) concentrations were analysed by dry combustion using an elemental analyser (Flash EA 2000; Thermo Fisher Scientific, Bremen, Germany).
Phospholipid fatty acid analysis
PLFAs were extracted from each freeze‐dried ground rhizosphere soil sample according to Frostegård et al. (1991). The absolute abundance of PLFAs was expressed in nmol g−1 organic matter (OM). We used i14:0, 14:0, i‐15:0, a‐15:0, 15:0, i‐16:0, 16:1ω9, 16:1ω7c, 16:1ω7t, i‐17:0, a‐17:0, 17:1ω8, cy‐17:0, 17:0, 18:1ω7, and cy‐19:0 as indicators for bacteria (Frostegård & Bååth, 1996) and 18:2ω6,9 for fungi (Kaiser et al., 2010). PLFAs i‐15:0, a‐15:0, i‐16:0, i‐17:0, and a‐17:0 represented Gram‐positive bacteria, whereas cy‐17:0, 18:1ω7, and cy‐19:0 represented Gram‐negative bacteria (Wardle et al., 2013). Branched chained fatty acids 10me16:0, 10me17:0 and 10me18:0 were used as indicators of actinobacteria (Maaroufi et al., 2019). The PLFA data were used to calculate the ratios of fungi to bacteria and Gram‐positive to Gram‐negative bacteria. PLFA ratios are commonly used in ecological studies to explain the response of microbial groups to plant tissue quality (Fanin et al., 2019b).
Fungal community
Fungal community composition for each freeze‐dried ground rhizosphere soil sample was assessed using high‐throughput sequencing of amplified ITS2 markers (Clemmensen et al., 2016). We extracted DNA from 300 ± 10 mg of freeze‐dried rhizosphere soil using the Nucleospin® Soil Kit (Macherey‐Nagel, Düren, Germany) following the manufacturer’s instructions. We performed 50 µl polymerase chain reactions (PCRs) with 25 µl diluted DNA extracts (1 ng µl−1) using a 5 µl mixture of 8‐bp extended sample‐specific tagged forward primer gITS7 (Ihrmark et al., 2012) and reverse primer ITS4 (White et al., 1990) to amplify the ITS2 region and 20 µl PCR master mixture. PCR master mixtures were prepared by mixing 8.25 µl distilled water, 0.25 µl DNA polymerase (DreamTaq Green; Thermo Fisher Scientific, Waltham, MA, USA), 5 µl reaction buffer, 5 µl dNTPs (2 mM each of dATP, dCTP, dGTP and dTTP) and 1.5 µl MgCl2.
The PCRs were run under the following conditions: 5 min at 95°C; 26–30 cycles of 30 s at 95°C, 57 s at 57°C, 30 s at 72°C; and 7 min at 72°C. We optimised the number of PCR cycles for each sample (based on preliminary PCRs), to interrupt PCRs in the exponential phase, when amplification is quantitatively more accurate. Negative controls (both for DNA extraction and PCRs) were included and PCR products were checked by gel electrophoresis. We then purified amplicons using the Agencourt AMPure kit (Beckman Coulter, Beverly, MA, USA) and measured the concentrations fluorometrically (Qubit Fluorometer, Invitrogen, Carlsbad, CA, USA). Equimolar mixtures of amplicons from each sample were pooled into three composite samples for sequencing. Sequencing of amplicons were performed using the Pacific Biosciences Sequel Technology Platform (one single‐molecule real‐time (SMRT) sequencing cell per composite sample) at SciLifeLab, Uppsala, Sweden. Raw sequences were uploaded to the Sequence Read Archive of the NCBI database under accession no. PRJNA664687.
We processed sequences using the bioinformatics pipeline Scata (http://scata.mykopat.slu.se/) and carried out quality filtering and clustering with parameters as in Fanin et al. (2017). After discarding sequences with missing primer sequences, and missing or mismatching identification tags, 1687 418 out of a total of 2523 609 sequences (67%) passed the initial quality control. In total, 1648 502 high quality sequences were assembled into 7474 Species Hypotheses (SHs; Kõljalg et al ., (2013)), after removal of singletons. For this study, we focused on the relative abundances of the 100 most abundant fungal SHs. We assigned fungal SHs to functional guilds, whenever possible, by comparison with the UNITE Database and the annotated International Nucleotide Sequence Database (INSD) using the massBlaster option (https://unite.ut.ee/) (Kõljalg et al., 2013; Nilsson et al., 2019). We considered the 10 best‐matching references for annotation. In cases in which no reliable taxon name was available, we ran manual Blastn searches against INSDC and used 98.0%, 90.0%, 85.0%, 80.0% and 75.0% similarity as criteria for assigning SHs to species, genus, family, order and class, respectively. Blastn search results of ˂e−50 were considered reliable for assigning sequences to the fungal kingdom, whereas those of > e−20 were considered ‘unknown’, as proposed by Tedersoo et al. (2014).
We categorised fungi identified to species or genera with a well known life‐form (i.e. according to published literature) into guilds: saprotrophs and pathogens, ectomycorrhizal fungi, arbuscular mycorrhizal fungi, other root‐associated (e.g. dark septate endophytes), yeasts and moulds, and other (e.g. mycoparasites). Fungi in supported clades (based on phylogenetic trees produced by Blastn searches of INSDC) that either belonged to higher taxonomic ranks with known life‐form or matched with environmental database sequences derived from a specific substrate (e.g. surface‐sterilised roots of ectomycorrhizal hosts) were categorised into putative functional guilds (Clemmensen et al., 2013). Pathogens and saprotrophs were treated as one guild, as many pathogens and saprotrophs are capable of switching strategies from one to the other (Olson et al., 2012; Zanne et al., 2020). Yeasts and moulds were also merged into one composite functional guild, because they both have a limited capacity to utilise more complex C substrates (Treseder & Lennon, 2015). Clusters with insufficient information for classification into a putative functional guild were assigned as ‘unknown’.
Data analyses
All statistical analyses were carried out using R 3.4.0 (R Core Team 2018, Vienna, Austria), with the following packages: nlme v.3.1‐149 (Pinheiro et al., 2020), lme4 v.1.1‐23 (Bates et al., 2015), car v.3.0‐8 (Fox & Weisberg, 2019), mass v.7.3‐51.6 (Venables & Ripley, 2002), tidyverse v.1.3.0 (Wickham et al., 2019), vegan v.2.5‐6 (Oksanen et al., 2019), ape v.5.4 (Paradis & Schliep, 2019), xlsx v.0.6.3 (Dragulescu & Cole, 2019), and phytools v.0.7‐47 (Revell, 2012). Analyses were conducted using total trait variation across all experimental units, and thus included all variation (i.e. both intraspecific and interspecific).
Fine root traits and fungal rhizosphere functional guilds
The assessment of fine root trait variation and the establishment of the root economics spectrum across the 20 plant species was performed using principal component analysis (PCA). In order to fulfil the normality assumption for data analysis, data were log‐transformed, and samples were removed from the analysis if they were either dead (n = 5 across the whole experiment) or had insufficient biomass for root chemical analyses (n = 4). Furthermore, we excluded one outlier (value was 132 times larger than the mean value, possibly due to a measurement error). The axes scores of the first two PCA principal components were later used as predictors in linear mixed models to represent a conservative–acquisitive root economics spectrum.
Hypothesis 1
We used general linear mixed effects models to test for the effect of the fine root economics spectrum on the ratios of fungi to bacteria and Gram‐positive to Gram‐negative bacteria PLFAs, using the axis scores from the first two PCA axes as fixed factors, and block as a random factor. We selected our final models based on the lowest Akaike Information Criterion (AIC) and therefore excluded the model that included species as a random factor (AIC for model with block and species as random factors = −420.44; AIC for model with block as a random factor = −422.43). Furthermore, to determine the relationships between fine root traits and the microbial groups assessed by PLFA, we first conducted a canonical correspondence analysis (CCA) with all log‐transformed fine root traits as explanatory variables. As this was significant (F = 2.16; P = 0.03), we then performed post hoc Spearman’s rank correlations to assess relationships between individual fine root traits and the microbial PLFA groups.
Hypotheses 2 and 3
Similar to hypothesis 1, we first conducted a CCA to determine the relationship between fine root traits and the fungal functional guilds, which was significant (F = 1.67; P = 0.04). We then performed Spearman’s rank correlations to further assess the relationship between fine root traits and the various fungal guilds. In addition, we tested the effects of individual fine root traits on the relative abundances of fungal orders by performing a partial CCA (pCCA) with the 100 most abundant SHs and the fine root traits, with block included as a random variable (Ramette, 2007). For this analysis, the SHs were grouped at the order‐level (24 orders in total and the functional guild ‘unknown’). We tested for correlation significance using permutation tests.
Before the pCCA analysis, the relative abundances of the fungal orders were square‐root‐transformed, which is equivalent to a Hellinger transformation (Ramette, 2007), as it reduces the weighting of rare OTUs in ordinations (Legendre & Gallagher, 2001). We first conducted automated model selection (i.e. ‘forward’, ‘backward’ and ‘ordistep’ as available in the vegan package) and variables that were significant in any of the models were selected for inclusion into our final model. This resulted in the inclusion of root forks (ramifications) per root length, and root C and N content as explanatory variables and block as a random variable. In addition, closely related plant species possess similar trait values due to their common ancestry (Adams, 2014) and this could potentially influence root trait associations with arbuscular mycorrhizal fungi. Therefore, we further tested for phylogenetic signal of the root traits found to be associated with arbuscular mycorrhizal fungi by calculating Blomberg’s K values (Adams, 2014) and determining its significance with 1000 permutations using the function ‘phylosig’ in the package phytools.
Finally, we used generalised linear mixed effects models to test for the effects of the root economics spectrum on fungal functional guilds, with the axis scores from the first two PCA axes as explanatory variables and block as a random factor. We assumed a binomial distribution for our models, because our response variables were relative abundances.
Results
Fine root traits
The PCA resulted in the alignment of plant root trait values to form a conservative–acquisitive root economics trait spectrum, with the first and second PCA axes explaining 43% and 23% of the total variation in the data set, respectively (Fig. 1). Morphological root traits explained most of the variation along the first PCA axis, while the chemical root traits (except for the phenol to N ratio) and root dry matter content explained most of the variation along the second PCA axis. As such, along the first PCA axis, average diameter was negatively correlated with specific root length and specific root area. Furthermore, there was considerable variation among the 20 species for some traits (e.g. specific root length and specific root tip abundance), but not for others (e.g. average diameter and root C content; see Table S1).
Fig. 1.
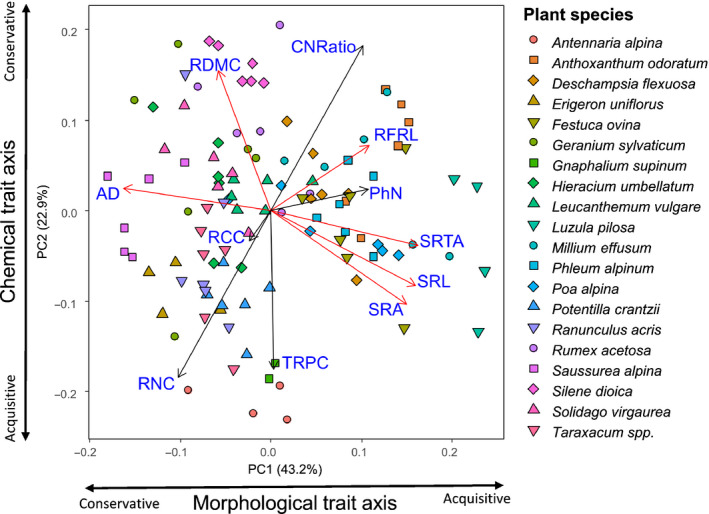
Principal component analysis of fine root traits across all species. Black arrows represent chemical root traits and red arrows represent morphological root traits. Fine root traits corresponding to the bi‐plot arrows are shown in blue font: average diameter (AD); root carbon content (RCC); root nitrogen content (RNC); root phenol content (TRPC); specific root area (SRA); specific root length (SRL); specific root tip abundance (SRTA); root phenol to nitrogen ratio (PhN); root forks per root length (RFRL); root carbon to nitrogen ratio (CNRatio) and root dry matter content (RDMC).
PLFA markers and fine root traits
We found a significant association between the second PCA root traits axis (but not the first) and for the ratios of fungi to bacteria (F = 4.497; P = 0.036; coefficient = 0.004) and Gram‐positive to Gram‐negative bacteria (F = 3.961; P = 0.049; coefficient = −0.007). We further found significant, but rather weak, negative correlations between root dry matter content and the concentrations of all PLFA biomarkers except Gram‐negative bacteria (r = −0.21 with bacteria, r = −0.21 with fungi, r = −0.22 with Gram‐positive bacteria and r = −0.24 with actinobacteria; Fig. 2a). Furthermore, fungal to bacterial ratios were positively correlated with root forks per root length (r = 0.19) and root C‐to‐N ratio (r = 0.20) and negatively with root N content (r = −0.23). In addition, the ratio of Gram‐positive to Gram‐negative bacteria was positively and negatively correlated with root N content (r = 0.20) and root C‐to‐N ratio (r = −0.20), respectively (Fig. 2a). However, the remaining seven fine root traits were not strongly correlated with any of the PLFA biomarkers.
Fig. 2.
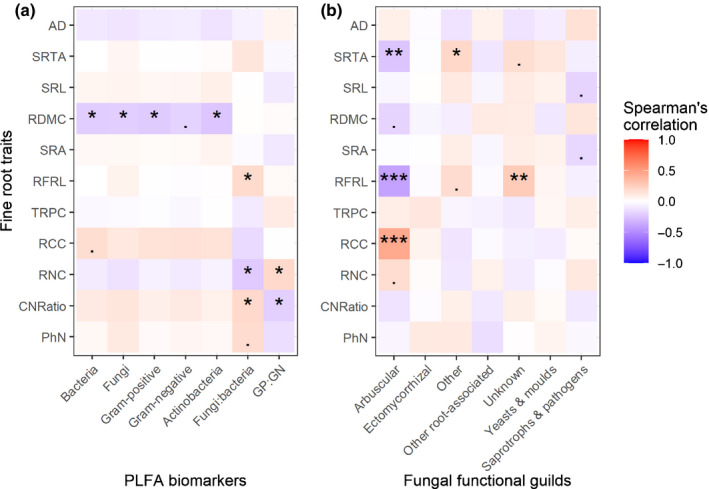
Spearman’s rank correlation matrices between fine root traits and absolute abundances of PLFA biomarkers, as well as for relative abundances of fungal guilds across all experimental units. (a) Fine root traits vs PLFA biomarkers. (b) Fine root traits vs fungal functional guilds. The 11 root traits tested are root phenol to nitrogen ratio (PhN), root carbon to nitrogen ratio (CNRatio), root nitrogen content (RNC), root carbon content (RCC), root phenol content (TRPC), root forks per root length (RFRL), specific root area (SRA), root dry matter content (RDMC), specific root length (SRL), specific root tip abundance (SRTA) and average diameter (AD). GP : GN means the ratio of Gram‐positive to Gram‐negative bacteria. Asterisks indicate statistical significance (***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05; ‘.’ indicates 0.05 < P ≤ 0.1).
Fine root traits and fungal community composition
Across all samples, saprotrophs–pathogens were the most abundant guild (mean relative abundance = 37.2% ± 5.3 SD), followed by yeasts and moulds (mean = 24.1% ± 6.6 SD), root‐associated fungi (mean = 19.0% ± 6.5 SD), fungi with unknown functions (mean = 12.9% ± 2.8 SD), ectomycorrhizal fungi (mean = 3.0% ± 1.0 SD), fungi with other functions (mean = 2.8% ± 1.8 SD) and arbuscular mycorrhizal fungi (mean = 1.2% ± 1.1 SD) (see Fig. S2).
We found no evidence for a relationship between the fine root economics spectrum (represented by PCA axis scores) and the relative abundances of saprotrophs–pathogens, yeasts and moulds, and arbuscular mycorrhizal fungal functional guilds (see Table S2; Fig. 2b). However, the relative abundance of arbuscular mycorrhizal fungi was significantly and positively correlated with root C content (r = 0.44; Fig. 2b), and negatively correlated with root forks per root length (r = −0.39). In addition, specific root tip abundance was negatively correlated with the relative abundance of arbuscular mycorrhizal fungi (r = −0.25; Fig. 2b). Furthermore, root forks per root length and the relative abundance of the fungal guild ‘unknown’ were positively correlated (r = 0.27; Fig. 2b). Correlations between all individual fine root traits and the relative abundances of yeasts and moulds, ectomycorrhizal fungi and other root‐associated fungi were not significant (Fig. 2b).
Overall, the fine root traits in the pCCA explained almost 10% of the variation in the relative abundances of fungal orders (Fig. 3). After accounting for block effects (accounting for 7.5% of the variation), two root traits, that is root forks per root length and root C content, significantly explained the variation in the relative abundance of fungal orders (F = 5.79; P = 0.001 and F = 4.45; P = 0.001, respectively), and the effects of a third trait (root N content) was not significant at α = 0.05 (F = 1.62; P = 0.082). The arbuscular mycorrhizal orders Archaeosporales and Glomerales were positively associated with root C content and negatively associated with root forks per root length. We found no phylogenetic signal for root C content (Blomberg’s K = 0.16; P = 0.87) and root forks per root length (Blomberg’s K = 0.30; P = 0.15). Conversely, the order Diaporthales, consisting of known pathogens, saprobes and endophytes, was associated with higher values of root forks per root length. Mucorales (moulds) was associated with higher values of root N content. Meanwhile, Sporidiobolales (yeasts) was associated with lower values of root N content, but its relative abundance did not vary with any of the morphological root traits. The orders Pezizales and Xylariales (which in our dataset consisted of saprotrophs–pathogens) were associated with higher and lower values of root C content and root N content, respectively.
Fig. 3.
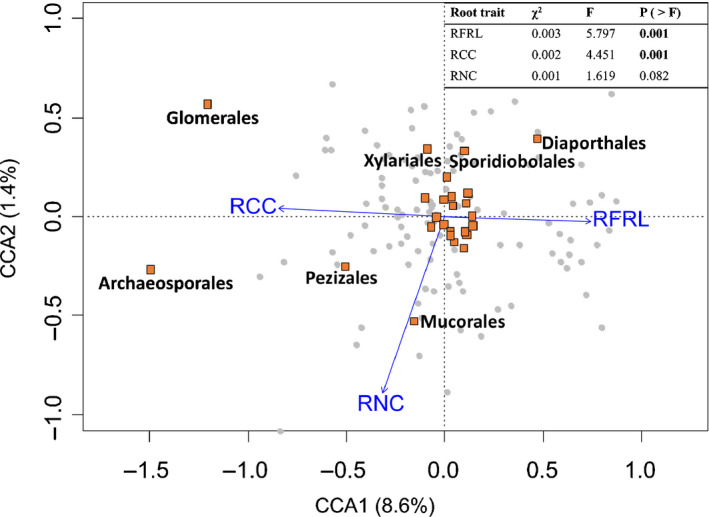
Partial canonical correspondence analysis of fungal orders constrained by fine root traits across all experimental units. The blue arrows are fine root traits: root carbon content (RCC), root forks per root length (RFRL) and root nitrogen content (RNC). Grey dots represent the experimental units. Open squares are the fungal orders. Black text labels correspond to fungal orders with co‐ordinates larger than ±0.25 on either axis. Plot is displayed with symmetric scaling. Fungal orders Archaeosporales and Glomerales are arbuscular mycorrhizal symbionts (Morton & Redecker, 2001; Helgason et al., 2003). Diaporthales consists of known pathogens, saprobes and endophytes (Rossman et al., 2007). Pezizales and Xylariales consist of species with various functions, but in our study the taxa for these orders are all saprotrophs–pathogens. Mucorales and Sporidiobolales are moulds and yeasts, respectively. The table in the upper right corner shows the statistical results of the model, with bold numbers indicating statistical significance at α = 0.05.
Discussion
We showed that fine root traits of sub‐arctic meadow plant species are associated with the relative abundance of various soil microbial groups and fungal guilds, but that the strength of these associations varied and were not necessarily particularly strong and/or did not apply to all measured root traits. We also found that the chemical trait axis of the fine root economics spectrum (but not the morphological trait axis) is a useful predictor of the ratios of broad microbial groups as measured by PLFA analysis. Conversely, the relative abundances of major fungal functional guilds were not associated with the root economics spectrum. However, more detailed analyses of the taxonomic composition of fungi showed that the relative abundance of several fungal orders were related to some chemical and morphological fine root traits. Below, we discuss these findings (see overview in Fig. 4) in relation to our hypotheses.
Fig. 4.
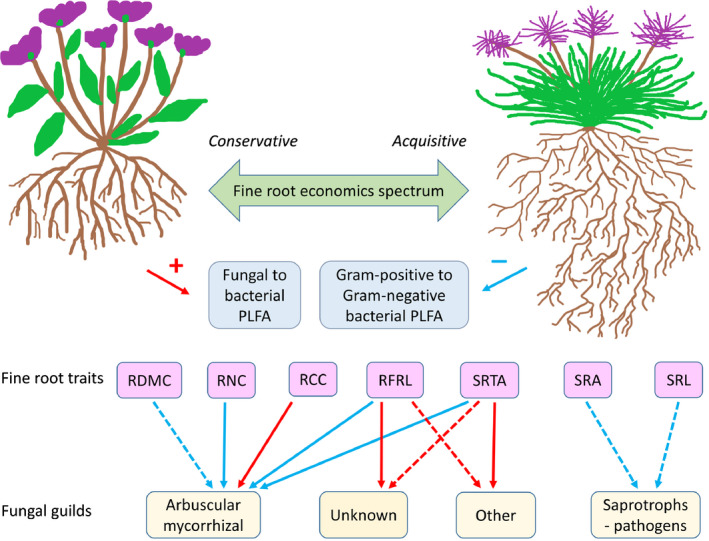
Conceptual figure synthesising the new insights from this study into observed associations between fine roots and soil microorganisms. The fine root economics spectrum consists primarily of chemical root traits. Only fine root traits with significant or nearly significant associations with any fungal functional guild at α = 0.05 are included: root nitrogen content (RNC), root carbon content (RCC), root forks per root length (RFRL), specific root area (SRA), root dry matter content (RDMC), specific root length (SRL), and specific root tip abundance (SRTA). Red arrows indicate positive relationships, while blue arrows indicate negative relationships. Solid arrows indicate a significant association and broken arrows a nearly significant association at α = 0.05.
PLFA biomarkers and fine root traits
We found partial support for our first hypothesis that acquisitive fine root trait values of sub‐arctic tundra meadow plant species are associated with lower ratios of fungi to bacteria and of Gram‐positive to Gram‐negative bacteria. The second PCA axis, which comprised primarily chemical traits (i.e. total polyphenol content and root C‐to‐N ratios) rather than morphological traits, was most important for explaining these ratios. Lower C : N ratios (and therefore more acquisitive trait values) may favour bacteria over fungi, because fungi have lower nutrient requirements (Fanin et al., 2013) and possess a broader suite of enzymes for decomposition (Strickland & Rousk, 2010; Soares & Rousk, 2019). Conversely, the negative association that we found between the ratio of Gram‐positive to Gram‐negative bacteria and fine roots with higher C : N ratios may be due to a higher need by Gram‐positive bacteria for nutrients to enable decomposition of more complex C forms (Orwin et al., 2018; Fanin et al., 2019a). Prior studies have shown differential responses of Gram‐positive bacteria and fungi to N addition (Maaroufi et al., 2015). Meanwhile, the morphological root trait axis was not associated with either ratios of soil fungi to bacteria or Gram‐positive to Gram‐negative bacteria, except for the positive correlation between root forks per root length and fungal to bacterial ratios. Nevertheless, root dry matter content could be potentially important for influencing the soil microbial biomass, because it was negatively correlated with the total abundance of all PLFA biomarkers except Gram‐negative bacteria. Together, these relationships suggest that both some chemical components of the fine root economics spectrum, as well as root dry matter content, can predict the biomass of broad soil microbial groups and their ratios within the sub‐arctic tundra meadow. Hence, within the cold and nutrient‐poor tundra, these broad microbial groups may respond to tissue stoichiometry and nutrients rather than morphological root traits.
Fine root traits and fungal functional guilds
Contrary to our second hypothesis, neither the chemical nor the morphological trait axes of the fine root economics spectrum explained the relative abundances of any of the fungal guilds. This was unexpected because the allocation of plant resources to fine roots is expected to be linked to nitrogen acquisition (Reich, 2014), which is coupled to fungal guilds involved in nutrient cycling. In particular, the morphological trait axis, which is similar to the collaborative gradient presented in Bergmann et al. (2020), was expected to be important for predicting the relative abundance of arbuscular mycorrhizal fungi. However, some individual fine root traits did explain a significant proportion of the variation in fungal taxa and functional guilds, which indicates that the fine root economics spectrum may not necessarily be the ‘holy grail’ when examining root–microorganism interactions. For example, the relative abundances of arbuscular mycorrhizal fungal orders had strong positive associations with root C content. This could be due to the production of C‐rich cellular structures by conservative plants that depend more on mycorrhiza for nutrient uptake under the nutrient‐limited conditions (Li et al., 2015) that characterise the sub‐arctic tundra. However, caution should be taken with this interpretation, because some mycorrhizal hyphae, and the lipids that they store in their vesicles (Luginbuehl et al., 2017), may be included in the measurements of total root C content, potentially elevating estimates of root C content. Nevertheless, as the biomass of plant roots is several magnitudes greater than that of the arbuscular fungi in the roots (Hart & Reader, 2002), it is unlikely that this could explain the relationship between root C content and the relative abundance of arbuscular mycorrhizal fungi.
Furthermore, we found a significant negative correlation between the relative abundance of arbuscular mycorrhizal fungi and root forks per root length. This is likely to have been because roots colonised with arbuscular mycorrhizal fungi can have lower branching intensity (Comas et al., 2014) due to their increased dependence on mycorrhizal foraging (Eissenstat et al., 2015; McCormack et al., 2015). However we did not find average fine root diameter to be associated with the relative abundance of arbuscular mycorrhizal fungi, despite previous studies showing a positive relationship between average root diameter and mycorrhizal root colonisation (Ma et al., 2018; McCormack & Iversen, 2019). This is likely to have been due to the focus of our study on colonisation of arbuscular mycorrhizal fungi in the rhizosphere soil (i.e. the soil zone surrounding roots; Bais et al., 2006), as opposed to in the fine roots themselves. Furthermore, although arbuscular mycorrhizal associations within arctic ecosystems are ubiquitous, there is a high variation in fine root colonisation across species (Gardes & Dahlberg, 1996; Newsham et al., 2009). As such, the plant‐mediated factors contributing to arbuscular mycorrhizal colonisation rates in arctic ecosystems require further investigation.
Fine root traits, saprotrophs‐pathogens and yeasts and moulds
By contrast with our expectations, the saprotroph–pathogen guild was not significantly associated with any fine root trait, despite it being the most abundant one in our study. Other studies have found that high specific root length is associated with a shorter root life span (Weemstra et al., 2020), which could in turn stimulate oligotrophic saprotrophs through a greater production of high quality root litter. However, faster root turnover could also serve as a mechanism to achieve a lower abundance of pathogens (Dietze & Matthes, 2014). In addition, the nutrient resorption rates for sub‐arctic plant species are high (Freschet et al., 2010), which should reduce the quality of root litter inputs and in turn negatively affect the relative abundance of copiotrophic decomposers. In addition, we found evidence that the biomass of bacteria, which are largely saprotrophic, is favoured by acquisitive root trait values. This suggests that saprotrophic bacteria may have stronger links than fungi to fine root traits (particularly those related to root N content), which could be of particular importance in sub‐arctic meadows (rather than heath communities) where there may be a high abundance of bacteria relative to fungi (Sundqvist et al., 2011).
We further showed that the relative abundance of yeasts and moulds in arctic soil is high (24%), compared with 0.4–14.3% found in temperate forests (Mašínová et al., 2017), probably due to their tolerance for cold and stressful environments (Treseder & Lennon, 2015). Yeasts and moulds primarily derive their C from simple C compounds exuded by plant roots (Botha, 2006; Pawłowska et al., 2019). While little studied, there is evidence that yeasts may have positive effects on nutrient recycling, plant growth and soil structure (Botha, 2011). Although we found no significant associations between the fine root economics spectrum and the relative abundances of yeasts and moulds, there was a strong positive relationship between root nitrogen content and the mould order Mucorales, and a negative association between root nitrogen content and the yeast order Sporidiobolales. The positive association between root nitrogen content (RNC) and Mucorales suggests a high N requirement for maintaining their rapid growth rates (Mehrotra, 1967). Conversely, the negative association between RNC and the yeast order Sporidiobolales suggests that they may be less reliant on freshly available plant‐derived N.
Conclusions and future outlook
Overall, our study showed that the chemical trait axis of the fine root economics spectrum is a useful predictor of the relative abundance of major microbial groups, such as the ratios of soil fungi to bacteria and Gram‐positive to Gram‐negative bacteria. Conversely, individual fine root traits may be more important for understanding relationships with fungal community composition, that is, soil fungal guilds and at the taxonomic order level, although this might be influenced by root or fungal carbon storage. This suggests that it may be more useful to focus on individual fine root traits when research is aimed at understanding the effects on specific fungal guilds. Together, these findings (summarised in Fig. 4) provide new insights into the relationships between fine root traits and rhizosphere microbial communities. However, our study was focused on sub‐arctic tundra meadow and not heath vegetation, which typically has higher fungal to bacterial ratios (Sundqvist et al., 2011). Hence, the effects of fine root traits on fungal guilds may potentially be stronger in sub‐arctic heath than in sub‐arctic meadows. Based on these new insights, below we present some lessons learnt and suggest considerations for future research related to plant–microorganism interactions within the sub‐arctic tundra.
First, the fine root economics spectrum represents a simplistic one‐dimensional spectrum (i.e. acquisitive vs conservative), whereas there may be several dimensions of resource use (Kramer‐Walter et al., 2016; Kong et al., 2019) that could each be linked to different subsets of the fungal community. For example, Bergmann et al. (2020) identified two main axes of root trait variation related to plant strategies for nutrient acquisition with implications for plant–microorganism interactions (‘do it yourself vs outsourcing’ and ‘slow vs fast turnover’). In our study, the chemical trait axis predicted the ratios of broad microbial groups, while the morphological trait axis did not predict the abundance of microorganisms. Therefore, in order to understand fine root–microorganism interactions, the use of individual fine root traits may be more informative. However, although there are large numbers of potential fine root traits (Iversen et al., 2017), the selection of fine root traits in studies related to plant–microorganism associations should be based on a priori knowledge of their ecological relevance. For example, in our study, root traits were selected a priori based on ecological relevance (Table 1) and we found associations between six out of 11 selected root traits and various soil microbial guilds. In addition, at larger scales, other factors not addressed in our study (e.g. soil OM quality) may also be important drivers of soil microbial groups in the tundra (Eskelinen et al., 2009).
Second, the resolution at which microbial communities are examined may influence the observed fine root–microbe relationships. For example, bulking of fungal taxa into functional guilds may obscure existing and ecologically relevant patterns, because some fungal taxa within a particular guild may have stronger associations with fine root traits than might others, and because different taxa could have opposing relationships with the same traits. In addition, our understanding of fine root–microorganism interactions may be obscured by a lack of knowledge about the function of several soil microbial taxa in general, but particularly in arctic ecosystems. For example, in our study there was a high relative abundance of dark septate endophytes, yeasts and other fungi with unknown functions that were associated with fine roots. Their functions and benefits to plants are currently still not well understood. Thus, a more detailed investigation of the ecological functions of these understudied organisms will be important for advancing the field of plant–microorganism–soil interactions within arctic and other ‘cold’ ecosystems.
Finally, understanding root trait–microbe relationships is an essential foundation for predicting responses of C cycling to global change. For example, the positive effects on arbuscular mycorrhizal fungi colonisation in fine roots that is expected with elevated temperature (Bennett & Classen, 2020) could in turn affect C sequestration (Drigo et al., 2010). In addition, responses of root traits to global change (Parts et al., 2019) could influence the relative abundance of various fungal guilds that contribute in varying degrees to soil C cycling. Taken together, it is important to understand root–microorganism interactions as a means towards understanding implications for future ecosystem C balance in arctic ecosystems.
Author contributions
CMS, PK and DAW designed the study. CMS and PK collected soils and seeds. CMS conducted the glasshouse study and laboratory work. BL supervised the molecular laboratory work. CMS conducted the fungal annotations, with supervision from BL and NF. CMS performed the statistical computations. NF verified the analytical methods. CMS took the lead in writing the manuscript. CMS, BL, DAW, MKS, MJG, NF and PK discussed the results and contributed to each draft of the manuscript.
Supporting information
Fig. S1 Phylogenetic tree with the 20 tundra plant species included in the experiment and their functional group assignment.
Fig. S2 Boxplot showing relative read abundances of fungal functional guild across all experimental units.
Table S1 Mean fine root trait values for all plant species with standard deviations.
Table S2 Summary statistics of generalised linear mixed effects models with relative abundances of fungal functional guilds as explained by the root economics spectrum.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
We thank Rémy Beugnon and Tania Maxwell for assistance in the glasshouse and the laboratory. We also thank the Department of Forest Mycology and Plant Pathology, SLU, for providing workspace for DNA extractions. The authors would like to acknowledge the support of the National Genomics Infrastructure (NGI)/Uppsala Genome Centre at UPPMAX for providing assistance in massive parallel sequencing and its computational infrastructure. Work performed at NGI/Uppsala Genome Centre was funded by RFI/VR and Science for Life Laboratory, Sweden. This project was funded by a grant (grant no. 2015‐04214) from the Swedish Research Council (Vetenskapsrådet) to PK.
References
- Adams DC. 2014. A generalized K statistic for estimating phylogenetic signal from shape and other high‐dimensional multivariate data. Systematic Biology 63: 685–697. [DOI] [PubMed] [Google Scholar]
- Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM. 2006. The role of root exudates in rhizosphere interactions with plants and other organisms. Annual Review of Plant Biology 57: 233–266. [DOI] [PubMed] [Google Scholar]
- Bates D, Maechler M, Bolker B, Walker S. 2015. Fitting linear mixed‐effects models using lme4. Journal of Statistical Software 67: 1–48. [Google Scholar]
- Bennett AE, Classen AT. 2020. Climate change influences mycorrhizal fungal–plant interactions, but conclusions are limited by geographical study bias. Ecology 101: 1–11. [DOI] [PubMed] [Google Scholar]
- Bergmann J, Weigelt A, Van Der Plas F, Laughli DC, Kuype TW, Guerrero‐Ramirez N, Valverde‐Barrantes OJ, Bruelheide H, Fresche GT, Iversen CM et al 2020. The fungal collaboration gradient dominates the root economics space in plants. Science Advances 6: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bever JD, Platt TG, Morton ER. 2012. Microbial population and community dynamics on plant roots and their feedbacks on plant communities. Annual Review of Microbiology 66: 265–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birouste M, Zamora‐Ledezma E, Bossard C, Pérez‐Ramos IM, Roumet C. 2014. Measurement of fine root tissue density: a comparison of three methods reveals the potential of root dry matter content. Plant and Soil 374: 299–313. [Google Scholar]
- Björk RG, Majdi H, Klemedtsson L, Lewis‐Jonsson L, Molau U. 2007. Long‐term warming effects on root morphology, root mass distribution, and microbial activity in two dry tundra plant communities in northern Sweden. New Phytologist 176: 862–873. [DOI] [PubMed] [Google Scholar]
- Blume‐Werry G, Wilson SD, Kreyling J, Milbau A. 2016. The hidden season: growing season is 50% longer below than above ground along an arctic elevation gradient. New Phytologist 209: 978–986. [DOI] [PubMed] [Google Scholar]
- Bonfante P, Genre A. 2010. Mechanisms underlying beneficial plant‐fungus interactions in mycorrhizal symbiosis. Nature Communications 1: 1–11. [DOI] [PubMed] [Google Scholar]
- Botha A. 2006. Yeasts in soil In: Rosa CA, Gábor P, eds. The yeast handbook‐ biology and ecophysiology of yeasts. Heidelberg/Berlin, Germany: Springer, 222–240. [Google Scholar]
- Botha A. 2011. The importance and ecology of yeasts in soil. Soil Biology and Biochemistry 43: 1–8. [Google Scholar]
- Carrillo Y, Bell C, Koyama A, Canarini A, Boot CM, Wallenstein M, Pendall E. 2017. Plant traits, stoichiometry and microbes as drivers of decomposition in the rhizosphere in a temperate grassland. Journal of Ecology 105: 1750–1765. [Google Scholar]
- Chapin FS, Matson PA, Vitousek PM. 2011. Principles of terrestrial ecosystem ecology. New York, NY, USA: Springer. [Google Scholar]
- Chen W, Koide RT, Adams TS, DeForest JL, Cheng L, Eissenstat DM. 2016. Root morphology and mycorrhizal symbioses together shape nutrient foraging strategies of temperate trees. Proceedings of the National Academy of Sciences, USA 113: 8741–8746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemensson‐Lindell A, Persson H. 1992. Effects of freezing on rhizosphere and root nutrient content using two soil sampling methods. Plant and Soil 139: 39–45. [Google Scholar]
- Clemmensen KE, Bahr A, Ovaskainen O, Dahlberg A, Ekblad A, Wallander H, Stenlid J, Finlay RD, Wardle DA, Lindahl BD. 2013. Roots and associated fungi drive long‐term carbon sequestration in boreal forest. Science 340: 1615–1618. [DOI] [PubMed] [Google Scholar]
- Clemmensen KE, Ihrmark K, Brandström‐Durling M, Lindahl BD. 2016. Sample preparation for fungal community analysis by high‐throughput sequencing of barcode amplicons In: Martin FM, Uroz S, eds. Microbial environmental genomics (MEG). New York, NY, USA: Humana Press, 61–88. [Google Scholar]
- Clemmensen KE, Sorensen LP, Michelsen A, Jonasson S, Ström L. 2008. Site‐dependent N uptake from N‐form mixtures by arctic plants, soil microbes and ectomycorrhizal fungi. Oecologia 155: 771–783. [DOI] [PubMed] [Google Scholar]
- Comas LH, Callahan HS, Midford PE. 2014. Patterns in root traits of woody species hosting arbuscular and ectomycorrhizas: Implications for the evolution of belowground strategies. Ecology and Evolution 4: 2979–2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortois R, Schröder‐Georgi T, Weigelt A, van der Putten WH, De Deyn GB. 2016. Plant–soil feedbacks: role of plant functional group and plant traits. Journal of Ecology 104: 1608–1617. [Google Scholar]
- Darmody RG, Thorn CE, Dixon JC, Schlyter P. 2000. Soils and landscapes of Kärkevagge, Swedish Lapland. Soil Science Society of America Journal 64: 1455–1466. [Google Scholar]
- De Long JR, Kardol P, Sundqvist MK, Veen GF (Ciska), Wardle DA. 2015. Plant growth response to direct and indirect temperature effects varies by vegetation type and elevation in a subarctic tundra. Oikos 124: 772–783. [Google Scholar]
- Dietze MC, Matthes JH. 2014. A general ecophysiological framework for modelling the impact of pests and pathogens on forest ecosystems. Ecology Letters 17: 1418–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragulescu A, Cole A. 2019. xlsx: read, write, format excel 2007 and excel 97/2000/XP 2003 files. R package version 0.6.3. [WWW document] URL https://CRAN.R‐Project.org/package=xlsx [accessed 1 February 2020].
- Drigo B, Pijl AS, Duyts H, Kielak AM, Gamper HA, Houtekamer MJ, Boschker HTS, Bodelier PLE, Whiteley AS, Van Veen JA et al 2010. Shifting carbon flow from roots into associated microbial communities in response to elevated atmospheric CO2. Proceedings of the National Academy of Sciences, USA 107: 10938–10942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenstat DM, Kucharski JM, Zadworny M, Adams TS, Koide RT. 2015. Linking root traits to nutrient foraging in arbuscular mycorrhizal trees in a temperate forest. New Phytologist 208: 114–124. [DOI] [PubMed] [Google Scholar]
- Eskelinen A, Stark S, Männistö M. 2009. Links between plant community composition, soil organic matter quality and microbial communities in contrasting tundra habitats. Oecologia 161: 113–123. [DOI] [PubMed] [Google Scholar]
- Fanin N, Fromin N, Buatois B, Hättenschwiler S. 2013. An experimental test of the hypothesis of non‐homeostatic consumer stoichiometry in a plant litter‐microbe system. Ecology Letters 16: 764–772. [DOI] [PubMed] [Google Scholar]
- Fanin N, Gundale MJ, Farrell M, Ciobanu M, Baldock JA, Nilsson M, Kardol P, Wardle DA. 2017. Consistent effects of biodiversity loss on multifunctionality across contrasting ecosystems. Nature Ecology & Evolution 2: 1–36. [DOI] [PubMed] [Google Scholar]
- Fanin N, Kardol P, Farrell M, Nilsson M, Gundale MJ, Wardle DA. 2019a. The ratio of Gram‐positive to Gram‐negative bacterial PLFA markers as an indicator of carbon availability in organic soils. Soil Biology and Biochemistry 128: 111–114. [Google Scholar]
- Fitz WJ, Puschenreiter M, Wenzel WW. 2006. Growth systems In: Luster J, Finlay R, eds. Handbook of methods used in rhizosphere research. Birmensdorf, Switzerland: Swiss Federal Research Institute WSL, 9–15. [Google Scholar]
- Fox J, Weisberg S. 2019. An R companion to applied regression. Thousand Oaks, CA, USA: Sage. [Google Scholar]
- Freschet GT, Cornelissen JHC, van Logtestijn RSP, Aerts R. 2010. Evidence of the ‘ plant economics spectrum ’ in a subarctic flora. Journal of Ecology 98: 362–373. [Google Scholar]
- Frostegård A, Bååth E. 1996. The use of phospholipid fatty acid analysis to estimate bacterial and fungal biomass in soil. Biology and Fertility of Soils 22: 59–65. [Google Scholar]
- Frostegård A, Tunlid A, Bååth E. 1991. Microbial biomass measured as total lipid phosphate in soils of different organic matter content. Journal of Microbial Methods 14: 151–163. [Google Scholar]
- Gardes M, Dahlberg A. 1996. Mycorrhizal diversity in arctic and alpine tundra: an open question. New Phytologist 133: 147–157. [Google Scholar]
- Hart MM, Reader RJ. 2002. Taxonomic basis for variation in the colonization strategy of arbuscular mycorrhizal fungi. New Phytologist 153: 335–344. [Google Scholar]
- Helgason T, Watson IJ, Young JPW. 2003. Phylogeny of the Glomerales and Diversisporales (Fungi: Glomeromycota) from actin and elongation factor 1‐alpha sequences. FEMS Microbiology Letters 229: 127–132. [DOI] [PubMed] [Google Scholar]
- Henneron L, Fontaine S, Cros C, Picon C, Vida C. 2019. Plant economic strategies of grassland species control soil carbon dynamics through rhizodeposition. Journal of Ecology 108: 528–545. [Google Scholar]
- Ihrmark K, Bödeker ITM, Cruz‐Martinez K, Friberg H, Kubartova A, Schenck J, Strid Y, Stenlid J, Brandström‐Durling M, Clemmensen KE et al 2012. New primers to amplify the fungal ITS2 region ‐ evaluation by 454‐sequencing of artificial and natural communities. FEMS Microbiology Ecology 82: 666–677. [DOI] [PubMed] [Google Scholar]
- Iversen CM, McCormack ML, Powell AS, Blackwood CB, Freschet GT, Kattge J, Roumet C, Stover DB, Soudzilovskaia NA, Valverde‐Barrantes OJ et al 2017. A global Fine‐Root Ecology Database to address below‐ground challenges in plant ecology. New Phytologist 215: 15–26. [DOI] [PubMed] [Google Scholar]
- Iversen CM, Sloan VL, Sullivan PF, Euskirchen ES, Mcguire AD, Norby RJ, Walker AP, Warren JM, Wullschleger SD. 2015. The unseen iceberg: plant roots in arctic tundra. New Phytologist 205: 34–58. [DOI] [PubMed] [Google Scholar]
- Kaiser C, Frank A, Wild B, Koranda M, Richter A. 2010. Negligible contribution from roots to soil‐borne phospholipid fatty acid fungal biomarkers 18:2ω6,9 and 18:1ω9. Soil Biology and Biochemistry 42: 1650–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardol P, Cornips NJ, van Kempen MML, Bakx‐Schotman JMT, van der Putten WH. 2007. Microbe‐mediated plant‐soil feedback causes historical contingency effects in plant community assembly. Ecological monographs 77: 147–162. [Google Scholar]
- Kattge J, Díaz S, Lavorel S, Prentice IC, Leadley P, Bönisch G, Garnier E, Westoby M, Reich PB, Wright IJ et al 2011. TRY ‐ a global database of plant traits. Global Change Biology 17: 2905–2935. [Google Scholar]
- Klironomos JN. 2002. Feedback with soil biota contributes to plants rarity and invasiveness in communities. Nature 417: 67–69. [DOI] [PubMed] [Google Scholar]
- Kõljalg U, Nilsson RH, Taylor AFS, Bates ST, Thomas D, Bengtsson‐palme J, Callaghan TM, Douglas B, Griffith GW, Ucking RL et al 2013. Towards a unified paradigm for sequence‐based identification of fungi. Molecular Ecology 22: 5271–5277. [DOI] [PubMed] [Google Scholar]
- Kong D, Wang J, Wu H, Valverde‐Barrantes OJ, Wang R, Zeng H, Kardol P, Zhang H, Feng Y. 2019. Nonlinearity of root trait relationships and the root economics spectrum. Nature Communications 10: 2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer C, Gleixner G. 2008. Soil organic matter in soil depth profiles: Distinct carbon preferences of microbial groups during carbon transformation. Soil Biology and Biochemistry 40: 425–433. [Google Scholar]
- Kramer‐Walter KR, Bellingham PJ, Millar TR, Smissen RD, Richardson SJ, Laughlin DC. 2016. Root traits are multidimensional: specific root length is independent from root tissue density and the plant economic spectrum. Journal of Ecology 104: 1299–1310. [Google Scholar]
- Legendre P, Gallagher ED. 2001. Ecologically meaningful transformations for ordination of species data. Oecologia 129: 271–280. [DOI] [PubMed] [Google Scholar]
- Li W, Jin C, Guan D, Wang Q, Wang A, Yuan F, Wu J. 2015. The effects of simulated nitrogen deposition on plant root traits: A meta‐analysis. Soil Biology and Biochemistry 82: 112–118. [Google Scholar]
- Luginbuehl LH, Menard GN, Kurup S, Van Erp H, Radhakrishnan GV, Breakspear A, Oldroyd GED, Eastmond PJ. 2017. Fatty acids in arbuscular mycorrhizal fungi are synthesized by the host plant. Science 356: 1175–1178. [DOI] [PubMed] [Google Scholar]
- Ma Z, Guo D, Xu X, Lu M, Bardgett RD, Eissenstat DM, McCormack ML, Hedin LO. 2018. Evolutionary history resolves global organization of root functional traits. Nature 555: 94–97. [DOI] [PubMed] [Google Scholar]
- Maaroufi NI, Forsmark B, Gundale MJ, Nordin A, Palmqvist K, Hasselquist NJ, Rosenstock NP, Wallander H. 2019. Anthropogenic nitrogen enrichment enhances soil carbon accumulation by impacting saprotrophs rather than ectomycorrhizal fungal activity. Global Change Biology 25: 2900–2914. [DOI] [PubMed] [Google Scholar]
- Maaroufi NI, Nordin A, Hasselquist NJ, Bach LH, Palmqvist K, Gundale MJ. 2015. Anthropogenic nitrogen deposition enhances carbon sequestration in boreal soils. Global Change Biology 21: 3169–3180. [DOI] [PubMed] [Google Scholar]
- Mangan SA, Schnitzer SA, Herre EA, MacK KML, Valencia MC, Sanchez EI, Bever JD. 2010. Negative plant‐soil feedback predicts tree‐species relative abundance in a tropical forest. Nature 466: 752–755. [DOI] [PubMed] [Google Scholar]
- Margesin R, Jud M, Tscherko D, Schinner F. 2009. Microbial communities and activities in alpine and subalpine soils. FEMS Microbiology Ecology 67: 208–218. [DOI] [PubMed] [Google Scholar]
- Mašínová T, Bahnmann BD, Větrovský T, Tomšovský M, Merunková K, Baldrian P. 2017. Drivers of yeast community composition in the litter and soil of a temperate forest. FEMS Microbiology Ecology 93: 1–10. [DOI] [PubMed] [Google Scholar]
- Maupetit A, Larbat R, Pernaci M, Andrieux A, Guinet C, Boutigny AL, Fabre B, Frey P, Halkett F. 2018. Defense compounds rather than nutrient availability shape aggressiveness trait variation along a leaf maturity gradient in a biotrophic plant pathogen. Frontiers in Plant Science 9: e1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack ML, Dickie IA, Eissenstat DM, Fahey TJ, Fernandez CW, Guo D, Helmisaari HS, Hobbie EA, Iversen CM, Jackson RB et al 2015. Redefining fine roots improves understanding of below‐ground contributions to terrestrial biosphere processes. New Phytologist 207: 505–518. [DOI] [PubMed] [Google Scholar]
- McCormack ML, Iversen CM. 2019. Physical and functional constraints on viable belowground acquisition strategies. Frontiers in Plant Science 10: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrotra BR. 1967. Nitrogen nutrition of two species of mucor. Flora oder Allgemeine Botanische Zeitung, Abt. A, Physiologie und Biochemie 158: 560–568. [Google Scholar]
- Morton JB, Redecker D. 2001. Two new families of Glomales, Archaeosporaceae and Paraglomaceae, with two new genera Archaeospora and Paraglomus, based on concordant molecular and morphological characters. Mycologia 93: 181–195. [Google Scholar]
- Newsham KK, Upson R, Read DJ. 2009. Mycorrhizas and dark septate root endophytes in polar regions. Fungal Ecology 2: 10–20. [DOI] [PubMed] [Google Scholar]
- Nilsson RH, Larsson KH, Taylor AFS, Bengtsson‐Palme J, Jeppesen TS, Schigel D, Kennedy P, Picard K, Glöckner FO, Tedersoo L et al 2019. The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Research 47: D259–D264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos P et al 2019. vegan: community ecology package. R package v.2.5‐6. [WWW document] URL https://CRAN.R‐Project.org/package=vegan [accessed 10 March 2020]. [Google Scholar]
- Olson Å, Aerts A, Asiegbu F, Belbahri L, Bouzid O, Broberg A, Durling M, Gonthier P, Grimwood J, Fossdal CG et al 2012. Insight into trade‐off between wood decay and parasitism from the genome of a fungal forest pathogen. New Phytologist 194: 1001–1013. [DOI] [PubMed] [Google Scholar]
- Orwin KH, Dickie IA, Holdaway R, Wood JR. 2018. A comparison of the ability of PLFA and 16S rRNA gene metabarcoding to resolve soil community change and predict ecosystem functions. Soil Biology and Biochemistry 117: 27–35. [Google Scholar]
- Paradis E, Schliep K. 2019. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35: 526–528. [DOI] [PubMed] [Google Scholar]
- Parts K, Tedersoo L, Schindlbacher A, Sigurdsson BD, Leblans NIW, Oddsdóttir ES, Borken W, Ostonen I. 2019. Acclimation of fine root systems to soil warming: comparison of an experimental setup and a natural soil temperature gradient. Ecosystems 22: 457–472. [Google Scholar]
- Pawłowska J, Okrasińska A, Kisło K, Aleksandrzak‐Piekarczyk T, Szatraj K, Dolatabadi S, Muszewska A. 2019. Carbon assimilation profiles of mucoralean fungi show their metabolic versatility. Scientific Reports 9: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez‐Harguindeguy N, Díaz S, Garnier E, Lavorel S, Poorter H, Jaureguiberry P, Bret‐Harte MS, Cornwell WK, Craine JM, Gurvich DE et al 2013. New handbook for standardised measurement of plant functional traits worldwide. Australian Journal of Botany 64: 715–716. [Google Scholar]
- Philippot L, Raaijmakers JM, Lemanceau P, Van Der Putten WH. 2013. Going back to the roots: The microbial ecology of the rhizosphere. Nature Reviews Microbiology 11: 789–799. [DOI] [PubMed] [Google Scholar]
- Pinheiro J, Bates D, DebRoy S, Sarkar D, RC Team . 2020. nlme: linear and nonlinear mixed effects models. R package v.3.1‐145. [WWW document] URL https://CRAN.R‐Project.org/package=nlme [accessed 13 April 2020]. [Google Scholar]
- Ramette A. 2007. Multivariate analyses in microbial ecology. FEMS Microbiology Ecology 62: 142–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich PB. 2014. The world‐wide ‘fast‐slow’ plant economics spectrum: a traits manifesto. Journal of Ecology 102: 275–301. [Google Scholar]
- Revell LJ. 2012. phytools: An R package for phylogenetic comparative biology (and other things). Methods in Ecology and Evolution 3: 217–223. [Google Scholar]
- Rossman AY, Farr DF, Castlebury LA. 2007. A review of the phylogeny and biology of the Diaporthales. Mycoscience 48: 135–144. [Google Scholar]
- Saj S, Mikola J, Ekelund F. 2009. Species‐specific effects of live roots and shoot litter on soil decomposer abundances do not forecast plant litter‐nitrogen uptake. Oecologia 161: 331–341. [DOI] [PubMed] [Google Scholar]
- Smalla K, Wieland G, Buchner A, Zock A, Parzy J, Kaiser S, Roskot N. 2001. Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: plant‐dependent enrichment and seasonal shifts revealed. Applied and Environmental Microbiology 67: 4742–4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares M, Rousk J. 2019. Microbial growth and carbon use efficiency in soil: links to fungal‐bacterial dominance, SOC‐quality and stoichiometry. Soil Biology and Biochemistry 131: 195–205. [Google Scholar]
- Soudzilovskaia NA, Vaessen S, Barcelo M, He J, Rahimlou S, Abarenkov K, Brundrett MC, Gomes SIF, Merckx V, Tedersoo L. 2020. FungalRoot: global online database of plant mycorrhizal associations. New Phytologist 227: 955–966. [DOI] [PubMed] [Google Scholar]
- Stern JL, Hagerman AE, Steinberg PD, Winter FC, Estes JA. 1996. A new assay for quantifying brown algal phlorotannins and comparisons to previous methods. Journal of Chemical Ecology 22: 1273–1293. [DOI] [PubMed] [Google Scholar]
- Strickland MS, Rousk J. 2010. Considering fungal: bacterial dominance in soils ‐ Methods, controls, and ecosystem implications. Soil Biology and Biochemistry 42: 1385–1395. [Google Scholar]
- Sun T, Hobbie SE, Berg B, Zhang H, Wang Q, Wang Z. 2018. Contrasting dynamics and trait controls in first‐order root compared with leaf litter decomposition. Proceedings of the National Academy of Sciences, USA 115: 10392–10397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundqvist MK, Giesler R, Graae BJ, Wallander H, Fogelberg E, Wardle DA. 2011. Interactive effects of vegetation type and elevation on aboveground and belowground properties in a subarctic tundra. Oikos 120: 128–142. [Google Scholar]
- Sundqvist MK, Wardle DA, Vincent A, Giesler R. 2014. Contrasting nitrogen and phosphorus dynamics across an elevational gradient for subarctic tundra heath and meadow vegetation. Plant and Soil 383: 387–399. [Google Scholar]
- Tedersoo L, Bahram M, Põlme S, Kõljalg U, Yorou NS, Wijesundera R, Ruiz LV, Vasco‐palacios AM, Thu PQ, Suija A et al 2014. Global diversity and geography of soil fungi. Science 346: 1078–1089. [DOI] [PubMed] [Google Scholar]
- Teste FP, Kardol P, Turner BL, Wardle DA, Zemunik G, Renton M, Laliberté E. 2017. Plant‐soil feedback and the maintenance of diversity in Mediterranean‐climate shrublands. Science 355: 173–176. [DOI] [PubMed] [Google Scholar]
- Tomova L, Braun S, Flückiger W. 2005. The effect of nitrogen fertilization on fungistatic phenolic compounds in roots of beech (Fagus sylvatica) and Norway spruce (Picea abies). Forest Pathology 35: 262–276. [Google Scholar]
- Treseder KK, Lennon T. 2015. Fungal traits that drive ecosystem dynamics on land. Microbiology and Molecular Biology Reviews 79: 243–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veen GF(Ciska), Sundqvist MK, Metcalfe D, Wilson SD. 2015. Above‐ground and below‐ground plant responses to fertilization in two subarctic ecosystems. Arctic, Antarctic, and Alpine Research 47: 693–702. [Google Scholar]
- Veen GFC, Fry EL, Hooven FC, Kardol P, Morriën E. 2019. The role of plant litter in driving plant‐soil feedbacks. Frontiers in Environmental Science 7: 1–10. [Google Scholar]
- Venables WN, Ripley BD. 2002. Modern applied statistics with S. New York, NY, USA: Springer US. [Google Scholar]
- Wardle DA, Bardgett RD, Klironomos JN, Setala H, van der Putten WH, Wall DH. 2004. Ecological linkages between aboveground and belowground biota. Science 304: 1629–1633. [DOI] [PubMed] [Google Scholar]
- Wardle DA, Gundale MJ, Jäderlund A, Nilsson MC. 2013. Decoupled long‐term effects of nutrient enrichment on aboveground and belowground properties in subalpine tundra. Ecology 94: 904–919. [Google Scholar]
- Weemstra M, Kiorapostolou N, van Ruijven J, Mommer L, de Vries J, Sterck F. 2020. The role of fine‐root mass, specific root length and life span in tree performance: a whole‐tree exploration. Functional Ecology 34: 575–585. [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics In: Innis MA, Gelfand DH, Sninsky JH, White TJ, eds. PCR protocols: a guide to methods and applications. New York, NY, USA: Academic Press, 315–322. [Google Scholar]
- Wickham H, Averick M, Bryan J, Chang W. 2019. Welcome to the tidyverse. Journal of Open Source Software 4: e1686. [Google Scholar]
- Zanne AE, Abarenkov K, Afkhami ME, Aguilar‐trigueros CA, Bates S, Bhatnagar JM, Busby PE, Christian N, Cornwell WK, Crowther TW et al 2020. Fungal functional ecology: bringing a trait‐based approach to plant‐associated fungi. Biological Reviews 95: 409–433. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Phylogenetic tree with the 20 tundra plant species included in the experiment and their functional group assignment.
Fig. S2 Boxplot showing relative read abundances of fungal functional guild across all experimental units.
Table S1 Mean fine root trait values for all plant species with standard deviations.
Table S2 Summary statistics of generalised linear mixed effects models with relative abundances of fungal functional guilds as explained by the root economics spectrum.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.


