Abstract
Retinoblastoma (RB) is the most common ocular malignancy that occurs during childhood. Scavenger receptor class A member 5 (SCARA5) is considered to function as an anti-oncogene in several types of malignant tumor. The present study investigated the functional role and underlying mechanism of SCARA5 in human RB cells. Reverse transcription-quantitative PCR and western blotting were used to detect the relative expression levels of SCARA5 in four human RB cell lines. In addition, transfection was performed to either knockdown or induce overexpression of SCARA5 in human RB Y79 cells. The proliferation, migration and apoptosis of RB cells was then measured by Cell Counting Kit 8 assay, 5-ethynyl-2′-deoxyuridine assay, clone formation assay, Transwell assay, Hoechst staining and TUNEL staining, respectively. Western blotting was performed to detect changes in the expression levels of key proteins involved in the PI3K/AKT and apoptotic pathways. The present study revealed that SCARA5 was expressed at lower levels in four tumorigenic human RB cell lines compared with in a human retinal pigment non-tumorigenic cell line. Functional analysis demonstrated that overexpression of SCARA5 decreased the proliferation and migration, and promoted the apoptosis of human RB cells in vitro, whereas in vivo experiments revealed a decrease in RB progression following SCARA5 overexpression. In addition, overexpression of SCARA5 inhibited phosphorylated (p)-PI3K and p-AKT expression, and knockdown of SCARA5 increased p-PI3K and p-AKT expression; however, no changes in total PI3K and AKT expression were observed. Bcl-2 exhibited similar changes in expression to those displayed by p-PI3K and p-AKT, whereas Bax and cleaved caspase-3 exhibited trends in expression that were the opposite to those shown by p-PI3K and p-AKT. In conclusion, the present results demonstrated that SCARA5 could inhibit the proliferation and promote the apoptosis of RB cell lines by suppressing the PI3K/AKT signaling pathway, thus suggesting a novel strategy for treating RB.
Keywords: retinoblastoma, SCARA5, Y79, PI3K/AKT, apoptosis
Introduction
Retinoblastoma (RB) is a specific type of malignant tumor that is often characterized by features of occult seizure, rapid growth, and the ability for intracranial and distant metastasis (1,2). RB is the most common ocular malignancy diagnosed during childhood, and accounts for most intraocular tumors found in children (3,4). The mortality rate of patients with RB in developing countries is 50–70% (5,6). Children suffering from RB are also susceptible to other diseases, including RB cell invasion, malignant transformation of an intracranial neuroblastoma, and development of a heterochronogenous tumor (7). In recent years, various novel surgical techniques and individualized comprehensive therapies have demonstrated efficacy in the clinical treatment of RB. However, the survival rate of patients with RB remains very low, mainly because the disease is often diagnosed in its later stages (8). Moreover, the traditional therapies for RB, such as external beam radiotherapy and chemotherapy, often produce side effects, including infection and blindness (9). Thus, in-depth research into the molecular mechanisms underlying RB is of great clinical significance for controlling the growth of the tumor and improving the prognosis of patients.
Class A scavenger receptors (SRs) are essential cell surface receptors that can bind various types of ligands, such as nucleic acids and modified low-density lipoproteins (10). These receptors are known to have an important role in host defense and are characterized as containing a collagen-like domain (11). SR class A member 5 (SCARA5), a member of the SR family, is mainly expressed on epithelial cells associated with mucosal surfaces, suggesting that it participates in the innate immune response (11). Abnormally low expression levels of SCARA5 have been reported in various types of cancer, such as lung cancer, liver cancer, renal cancer and glioma (12–15). Inhibition of SCARA5 expression by Snail1 was previously shown to markedly attenuate the epithelial-to-mesenchymal transition-associated migration of human non-small cell lung carcinoma A549 cells (12). Another study reported that SCARA5 knockdown markedly enhanced human hepatocellular carcinoma tumorigenesis and metastasis by activating the FAK signaling pathway (13). Moreover, the proliferation of renal cell carcinoma cells with SCARA5 knockdown was promoted when Rho-associated coiled-coil forming protein kinase 2 (Rock2) was overexpressed (14). In addition, the systemic treatment of tumor-bearing mice with a SCARA5-cationic liposome complex not only reduced the growth of subcutaneous human glioma tumors, but also markedly suppressed the spontaneous formation of lung metastases (15). Consistent with these findings, SCARA5 knockdown was shown to promote cell cycle progression in hepatocellular carcinoma cells, whereas increased SCARA5 expression inhibited cell cycle progression (16). These findings suggested that SCARA5 may have an important role in cancer progression. However, to the best of our knowledge, its role in the progression and metastasis of RB remains to be fully elucidated.
The present study investigated the precise role of SCARA5 in RB tumor growth and metastasis, and clarified its potential mechanism of action in RB. The results of the present study may provide a novel therapeutic target for treating patients with RB.
Materials and methods
Cell lines and cell culture
The human retinal pigment epithelial cell line ARPE-19, and human RB cell lines HXO-Rb44, Y79, SO-RB50 and WERI-Rb-1, were purchased from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences. The cell lines (1×105 × cells) were cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin at 37°C in a 5% CO2 incubator with 95% humidity.
Vector construction and transfection
The pcDNA3.1 vector carrying full length of SCARA5 and the pGCSIL-GFP plasmid carrying a short hairpin (sh) RNA targeting SCARA5 were used for overexpression or knockdown of SCARA5, respectively (both from GenScript). Y79 cells were seeded in a six-well plate at a density of 3×105 cells/well prior to transfection. The pcDNA3.1-SCARA5 and sh-SCARA5 plasmids (2 µg) were transfected into Y79 cells when cell confluence reached 60–70% using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. Empty pcDNA3.1 vector and pGCSIL-GFP plasmid carrying a non-targeting control sh-RNA sequence (sh-control) were used as controls for SCARA5 overexpression and knockdown, respectively. Y79 cells were cultured with different treatment were cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin at 37°C in a 5% CO2 incubator with 95% humidity for 48 h. Transfection efficiency was detected at 48 h post-transfection using RT-qPCR and western blotting, and the transfected Y79 cells were collected. The short hairpin RNA (shRNA) used to knockdown SCARA5 expression was designed and synthesized by Shanghai GenePharma Co., Ltd. The SCARA5 and control shRNA sequences were as follows: SCARA5, 5′-GCUCCAUCUGUGAGGAUUCdTdT-3′; control, 5′-UUCCCGAACGUGUCACGUTT-3′.
Reverse transcription quantitative-PCT (RT-qPCR)
Total RNA was extracted from cells or tumor tissues using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. Subsequently, 2 µg total RNA was reverse transcribed into cDNA using a PrimeScript RT Reagent kit (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. RT-qPCR was performed using an ABI PRISM 7900 Sequence Detector (Applied Biosystems; Thermo Fisher Scientific, Inc.) and a SYBR Green PCR kit (Applied Biosystems Biosystems; Thermo Fisher Scientific, Inc.). GAPDH served as an internal control. Thermocycling parameters for the amplification were as follows: 95°C, 10 min; followed by 95°C, 15 sec and 60°C, 1 min for 40 cycles. Relative expression levels of targeted genes were calculated by 2−ΔΔCt method as previously described (17). The primer sequences used were as follows: SCARA5 forward, 5′-AAAGCTATGTACCTACACACCGT-3′ and reverse, 5′-CCGCCGTTTGTGACATGGA-3′; GAPDH forward, 5′-TGTTCGTCATGGGTGTGAAC-3′ and reverse, 5′-ATGGCATGGACTGTGGTCAT-3′.
Western blotting
Total proteins were extracted from cells or tumor tissues using RIPA lysis buffer containing 1% protease inhibitor cocktail (cat. no. R0278-50ML, Sigma-Aldrich; Merck KGaA). The total protein concentration in each extract was quantified using a bicinchoninic acid protein assay kit. Subsequently, 20 µg protein from each sample was mixed with protein loading buffer and heated at 100°C for 5 min. The protein samples were then separated by SDS-PAGE on 10–12% gels and the separated protein bands were transferred onto PVDF membranes (EMD Millipore). Membranes were subsequently blocked with 5% fat-free buffered milk for 2 h at room temperature and were incubated with primary antibodies against PI3K (1:1,000; rat monoclonal; cat. no. ABS1856; Sigma-Aldrich; Merck KGaA), phosphorylated (p)-PI3K (1:1,000; cat. no. 04–1138; rat monoclonal; Sigma-Aldrich; Merck KGaA), AKT (1:1,000; rabbit polyclonal; cat. no. 4685; Cell Signaling Technology, Inc.), p-AKT (1:1,000; rabbit polyclonal; cat. no. 4060; Cell Signaling Technology, Inc.), SCARA5 (1:1,000; rabbit polyclonal; cat. no. ab106439; Abcam), Bcl-2 (1:2,000 rabbit polyclonal; cat. no. ab182858; Abcam), Bax (1:3,000; rabbit polyclonal; cat. no. ab182734; Abcam), cleaved caspase-3 (1:1,000; rabbit polyclonal; cat. no. ab49822; Abcam) and GAPDH (1:2,000; rabbit polyclonal; cat. no. ab9485; Abcam) overnight at 4°C. Subsequently, the membranes were incubated with an anti-rabbit IgG secondary antibody (1:20,000; cat. no. ab6721; Abcam) or anti-rat IgG (1:20,000; cat. no. ab125900; Abcam) for 2–3 h at room temperature; after which, the immunostained protein bands were detected using an ECL reagent (Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. Densitometric analysis was performed by ImageJ V1.8.0 software (National Institutes of Health).
Immunofluorescence assay
Immunofluorescence assays were performed as previously described (18). Briefly, 1×103 cells were plated onto slides that had been treated with polylysine and were incubated at 37°C for 4 h. Subsequently, the cells were fixed with 10% formaldehyde and then blocked with 5% bovine serum albumin (cat. no. 810784; Sigma-Aldrich; Merck KGaA) in PBS for 2 h at room temperature. Subsequently, the cells were stained with rabbit anti-human polyclonal antibodies against SCARA5 (1:200; cat. no. ab106439; Abcam) at 4°C overnight, and were incubated with a Cy3-conjugated (red) anti-rabbit IgG antibody (1:1,000; cat. no. ab6939; Abcam) at 4°C for 2 h. After repeated rinsing in PBS, the cells were observed under an inverted fluorescence microscope.
Cell proliferation assay
The cells were incubated for 5 h with 5-ethynyl-2′-deoxyuridine (EdU; Guangzhou Ribobio Co., Ltd.) according to the manufacturer's protocol. Subsequently, cells were washed three times with PBS and were then treated with 300 µl 1X Apollo reaction cocktail for 30 min; after which, they were stained with 100 µl EdU (5 µg/ml) for 30 min at room temperature, and their DNA contents were detected with a fluorescence microscope. Cell proliferation was also measured using a Cell Counting Kit 8 (CCK8) kit (Sigma-Aldrich; Merck KGaA) according to the manufacturer's instructions. Briefly, the cells were seeded onto 96-well plates (Corning, Inc.) and cultured in medium at a concentration of 1×105 cells/ml. Subsequently, the cells were transfected with vectors for 24, 48 and 72 h. CCK8 (10 µl) was then added to the cells and the cells were cultured at 37°C for 4 h; after which, cell proliferation was assessed with a microplate reader (Thermo Fisher Scientific, Inc.) at a wavelength of 450 nm.
Transwell assay
Cell migration was evaluated using the Transwell assay as previously described (19). Briefly, 5×104 cells suspended in serum-free medium were placed into the upper chamber of a Transwell plate (pore size, 8 µm; EMD Millipore), which was not precoated with Matrigel, and medium containing 10% FBS was added to the lower chamber. After 24 h, the cells which had migrated into the lower chamber were fixed with 100% methanol and stained with 0.1% crystal violet for 24 h at 37°C; after which, they were observed and counted with an IX71 inverted light microscope (Olympus Corporation). Each experiment was performed three times.
Clone formation assay
Cells were seeded into the wells of 12-well plates and cultured in complete medium for 2 weeks. The cells were then fixed with 4% paraformaldehyde and stained with 0.1% crystal violet for 15 min at 37°C. The number of colonies per well was then counted.
Apoptosis assay
Apoptosis assays were performed using TUNEL and Hoechst staining. TUNEL analysis was performed using a TUNEL detection kit (cat. no. KGA702; Nanjing KeyGen Biotech Co., Ltd.) according to the manufacturer's instructions. Briefly, 1×106 cells were suspended, permeabilized with 0.1% Triton X-100 (Beyotime Institute of Biotechnology) for 30 min and incubated in 50 mM TUNEL reaction mixture for 2 h at room temperature. Slides were counterstained with DAPI for 10 min at room temperature for nuclear staining. Images were captured using a fluorescence microscope (Olympus Corporation). Apoptosis in tumor tissues was also detected using a TUNEL assay according to the manufacturer's instructions. Slides were stained with 10 µl hematoxylin (cat. no. ab220365; Abcam) for 3 min at room temperature for nuclear staining. Images were captured using a XSP-36 light microscope (Boshida Optical Co., Ltd.). Each experiment was performed three times. For Hoechst staining, 6×104 cells transfected cells were inoculated into the wells of a 6-well plate. After removing the culture medium, the culture plate was rinsed 3–5 times with PBS. Subsequently, the cells in each well were fixed with 4% paraformaldehyde at 4°C for 20 min, and then rinsed with PBS. Approximately 1 ml Hoechst staining buffer was then added to each well and the plate was incubated for 20 min at room temperature. After being repeatedly rinsed with PBS, the plate was observed under an inverted fluorescence microscope, and the results were recorded for subsequent statistical analysis.
Xenograft tumor experiment
A total of 15 male Balb/c nude mice (weight, 20–25 g; age, 3–4 weeks) were purchased from the Animal Center of Central South University (Changsha, China). All of the nude mice were bred in a specific-pathogen-free animal laboratory with 60–65% humidity at 22–25°C under a 12-h light/dark cycle, and the experiment were performed after 1 week of free access to food and water. Nude mice were randomly divided into 3 groups with 5 in each group, including Blank, Over-SCARA5, sh-SCARA5 groups. A xenograft tumor model was created by subcutaneously injecting 3×106 cells (untransfected, SCARA5-overexpressing or SCARA5-silenced Y79 cells) into the flanks of nude mice. The size of each tumor was recorded every 7 days, and images of each tumor were captured after 4 weeks. At the end of the experiment, the nude mice were anesthetized by an intraperitoneal injection of 3% sodium pentobarbital (50 mg/kg) and in accordance with Hawkins P's animal experiment guidance, mice were euthanized with 100% CO2, with a flow rate of 20% chamber volume per minute (20). During the present study, the Guide for the Care and Use of Laboratory Animals (NRC 2011) of AAALAC was adhered to, in order to minimize animal pain and discomfort (21). All study protocols involving animals were approved by the Ethics Committee of Central South University and were conducted at the Experimental Animal Center of Central South University.
Immunohistochemistry
Tumor samples were fixed with 10% formalin for 24 h at room temperature, embedded with paraffin and then cut into 4-µm serial sections, which were used for immunohistochemistry. Subsequently, deparaffinized and rehydrated sections were incubated with 3% H2O2 in methanol for 10 min at room temperature to block endogenous peroxidase activity. The sections were then subjected to antigen retrieval for 10 min in a pressure cooker containing sodium citrate, and were incubated in 5% normal goat serum (cat. no. DXT-50197Z; Invitrogen; Thermo Fisher Scientific, Inc.) for 20 min at 37°C, permeabilized in PBS-Triton solution and incubated with a primary antibody against SCARA5 (1:200; cat. no. ab106439; Abcam) at 4°C overnight. The sections were then incubated with a horseradish peroxidase-conjugated mouse anti-rabbit secondary antibody (1:1,000; cat. no. ab6728; Abcam) at 37°C for 1 h.
For Ki67 staining, immunohistochemical analyses were performed using anti-Ki67 (1:400; cat. no. SP6; Zytomed) which incubated for 1 h at room temperature. The HRP-labeled goat anti-rabbit IgG secondary antibody (1:1,000; cat. no. ab6721; Abcam) was added to the sample, which was incubated for 10 min at 37°C. The sections were then counterstained with hematoxylin for 5 min at room temperature and finally dehydrated and covered with a coverslip. The sections were observed under a XSP-36 light microscope (Boshida Optical Co., Ltd.). Each experiment was performed three times.
Statistical analysis
All statistical analyses were performed using GraphPad Prism 6 software (GraphPad Software, Inc.) and results are shown as the mean ± SEM. Unpaired Student's t-tests and one-way ANOVA followed by the Student-Newman-Keuls post hoc test were performed to compare the two groups and multiple groups, respectively. P<0.05 was considered to indicate a statistically significant difference.
Results
SCARA5 is downregulated in RB cell lines
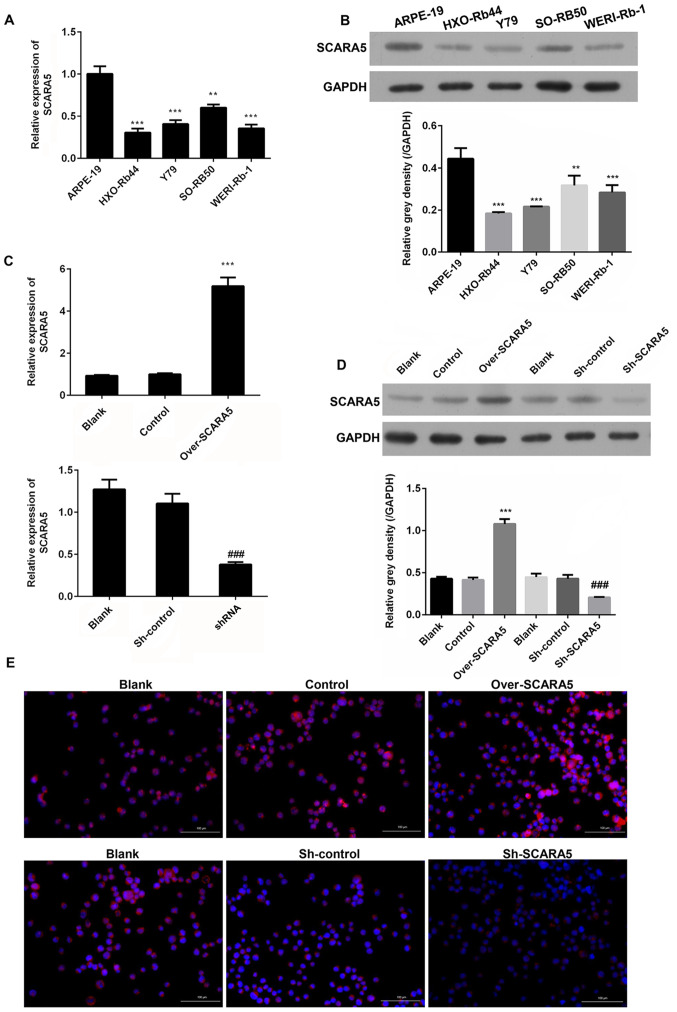
In order to determine the role played by SCARA5 in the development and progression of RB, RT-qPCR was performed to measure the mRNA expression levels of SCARA5 in RB cells. The results revealed that SCARA5 mRNA was expressed at lower levels in the RB cell lines (HXO-Rb44, Y79, SO-RB50 and WERI-Rb-1) compared with in the healthy human retinal pigment epithelial cell line (ARPE-19) (Fig. 1A). Consistent with the difference in mRNA expression levels, western blot analysis revealed that the protein expression levels of SCARA5 were also downregulated in the RB cell lines compared with those in the healthy retinal pigment epithelial cell line (Fig. 1B). These results suggested that SCARA5 may participate in RB development as a tumor suppressor. As SCARA5 expression was markedly downregulated in the Y79 cell line compared with that in the ARPE-19 cell line, and due to the convenience to construct transfected Y79 cells, this cell line was selected for subsequent experiments.
Figure 1.
Comparison of SCARA5 expression levels in four human retinoblastoma cell lines (HXO-Rb44, Y79, SO-RB50, WERI-Rb-1) and a human retinal pigment epithelial cell line (ARPE-19). (A) RT-qPCR analysis of SCARA5 expression. (B) Western blot analysis of SCARA5 expression. **P<0.01, ***P<0.001 vs. ARPE-19. (C) RT-qPCR analysis of SCARA5 expression in Y79 cells with SCARA5 overexpression or knockdown. (D) Western blot analysis of SCARA5 expression in Y79 cells with SCARA5 overexpression or knockdown. Data are presented as the mean ± SEM (n=3 with three repeats). ***P<0.001 vs. Control; ###P<0.001 vs. sh-control. (E) Immunofluorescence analysis of SCARA5 expression in Y79 cells with SCARA5 overexpression or knockdown. Blue signal indicated the nuclei stained by 4,6-diamino-2-phenyl indole (DAPI); Pink signal was produced by the red immunofluorescence (Cy3) signal merging with blue signal stained by DAPI. Scale bar, 100 µm. SCARA5, scavenger receptor class A member 5; RT-qPCR, reverse transcription-quantitative PCR; NC, negative control; shRNA/sh, short hairpin RNA.
Overexpression and knockdown of SCARA5 in Y79 cells
Y79 cells were used for transfection experiments designed to investigate the role played by SCARA5 in RB progression. Y79 cells were transfected with recombinant vectors that induced overexpression or knockdown of SCARA5, and the cellular expression levels of SCARA5 mRNA and protein were detected by RT-qPCR and western blotting, respectively. As shown in Fig. 1C and D, when compared with the blank control group, transfection with the SCARA5 vector significantly upregulated the mRNA and protein expression levels of SCARA5, whereas transfection with the SCARA5 knockdown vector produced the opposite result. Consistent with these findings, immunofluorescence analyses of SCARA5 protein expression revealed that SCARA5 protein expression levels were upregulated or downregulated after the cells had been transfected with the recombinant SCARA5 vector or SCARA5 knockdown vector, respectively (Fig. 1E). These results confirmed that SCARA5 was successfully enhanced or suppressed by transfection with the respective recombinant vectors.
Overexpression of SCARA5 inhibits the proliferation and migration of Y79 cells
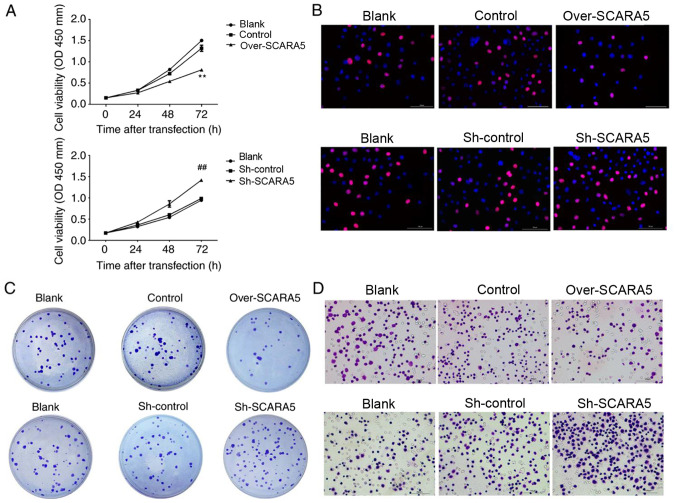
To assess the biological effect of SCARA5 on RB cell proliferation, CCK8 assays were performed to explore the effect of SCARA5 overexpression or knockdown on RB cell proliferation. As shown in Fig. 2A, when compared with the control group, overexpression of SCARA5 significantly inhibited the proliferation of Y79 cells, whereas knockdown of SCARA5 significantly promoted the proliferation of Y79 cells at 72 h. Furthermore, Edu assay results confirmed that overexpression of SCARA5 inhibited the growth rate of Y79 cells, when compared with non-transfected Y79 cells (Fig. 2B). Consistent with these findings, the colony formation assays revealed that compared with in the control group, the number of cell colonies formed in the SCARA5 knockdown group was increased. Conversely, SCARA5 overexpression decreased the number of cell colonies that formed (Fig. 2C). The migratory abilities of Y79 cells were evaluated by Transwell assay. The results revealed that overexpression of SCARA5 reduced the migratory capabilities of Y79 cells when compared with control cells, whereas those migratory abilities were increased in the knockdown cells when compared with the control cells (Fig. 2D). These results demonstrated that SCARA5 suppressed the proliferation and migration of RB cells.
Figure 2.
Effect of SCARA5 overexpression on Y79 cell proliferation and migration. (A) Cell Counting Kit 8 assay curve for the proliferation of Y79 cells with SCARA5 overexpression or knockdown. Data are presented as the mean ± SEM (n=3 with three repeats). **P<0.01 vs. Control; ##P<0.01 vs. Sh-control. (B) Proliferative ability of Y79 cells with SCARA5 overexpression or knockdown, as determined by 5-ethynyl-2′-deoxyuridine assay. Blue signal indicates the nuclei stained with DAPI. Red indicates the EdU signal. (C) Colony formation assay results for Y79 cells with SCARA5 overexpression or knockdown. (D) Transwell assay results for Y79 cells with SCARA5 overexpression or knockdown. Scale bar, 100 µm. SCARA5, scavenger receptor class A member 5; sh, short hairpin RNA.
Overexpression of SCARA5 promotes the apoptosis of Y79 cells
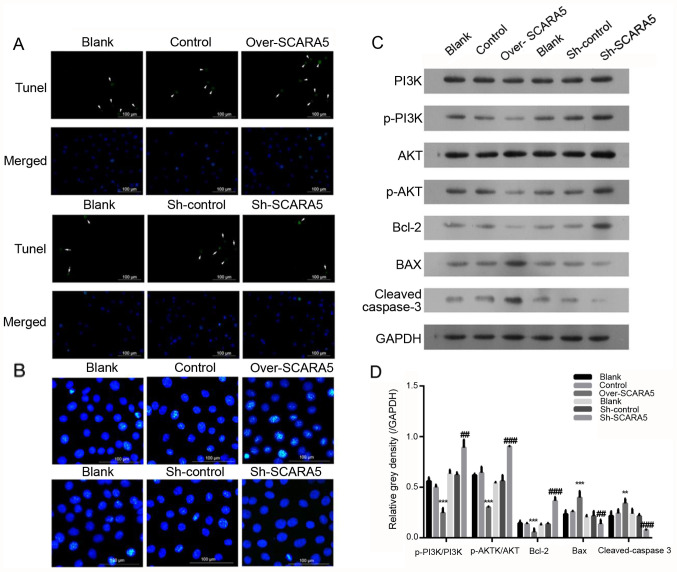
To assess the effect of SCARA5 on apoptosis of RB cells, Hoechst 33342 and TUNEL staining were used to explore the effect of SCARA5 overexpression or knockdown on Y79 cell apoptosis. The present results demonstrated that the percentage of apoptotic cells was increased in the SCARA5 overexpression group compared with that in the control group (Fig. 3A). Conversely, SCARA5 knockdown decreased the proportion of apoptotic cells, as evidenced by decreased fluorescence intensity in the knockdown group compared with in the control group. In addition, compared with in the control group, overexpression of SCARA5 promoted apoptosis of Y79 cells whereas knockdown of SCARA5 suppressed cell apoptosis (Fig. 3B).
Figure 3.
Effect of SCARA5 overexpression on Y79 cell apoptosis. (A) TUNEL analysis of apoptosis in Y79 cells. The arrows indicate the apoptotic cells with green signal; (B) Hoechst 33342 staining of apoptotic Y79 cells with SCARA5 overexpression or knockdown. The blue signals indicate the apoptotic cells. Scale bar, 100 µm. (C and D) Western blot analysis of PI3K, p-PI3K, AKT, p-AKT, Bax, Bcl-2 and cleaved caspase-3 expression in Y79 cells with SCARA5 overexpression or knockdown. Data are presented as the mean ± SEM (n=3 with three repeats). **P<0.01, ***P<0.001 vs. Control; ##P<0.01, ###P<0.001 vs. Sh-control. SCARA5, scavenger receptor class A member 5; sh, short hairpin RNA; p-, phosphorylated.
Overexpression of SCARA5 affects the expression of proteins related to the PI3K/AKT pathway and apoptotic signaling pathway in Y79 cells
The PI3K/AKT signaling pathway serves essential roles in the apoptosis, proliferation and migration of tumor cells (22). The expression levels of key proteins involved in the PI3K/AKT pathway were detected in RB cells. The results revealed that the expression levels of p-PI3K/PI3K and p-AKT/AKT were decreased in the SCARA5 overexpression group compared with their expression levels in the control group (Fig. 3C and D). Meanwhile, when compared with the control group, the expression levels of p-PI3K/PI3K and p-AKT/AKT were markedly increased in the SCARA5 knockdown group (Fig. 3C and D). Furthermore, the expression levels of key proteins involved in the regulation of cell apoptosis were detected; the protein expression levels of Bcl-2 were decreased, whereas the protein expression levels of Bax and cleaved caspase-3 were increased in the SCARA5 overexpression group compared with those in the control group (Fig. 3C and D). Consistent with these findings, knockdown of SCARA5 increased the expression levels of Bcl-2, and decreased the expression levels of Bax and cleaved-caspase 3 (Fig. 3C and D), which was consistent with the results of the cell apoptosis analysis.
Effect of SCARA5 on RB carcinogenesis in vivo
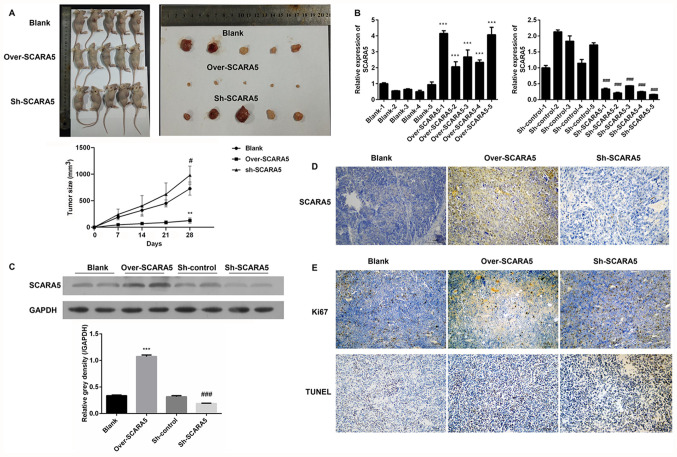
A xenograft tumor model was used to examine the role played by SCARA5 in the development of RB in vivo. Y79 cells transfected with the SCARA5 overexpression vector or SCARA5 knockdown vector were subcutaneously injected into nude mice. A subsequent examination revealed that the subcutaneous tumors in the SCARA5 overexpression group were smaller in size compared with those in the control group and SCARA5 knockdown group, demonstrating that overexpression of SCARA5 inhibited tumor growth (Fig. 4A). Subsequently, the expression levels of SCARA5 were examined by RT-qPCR, western blotting and immunohistochemistry. As shown in Fig. 4B, mice injected with SCARA5-overexpressing cells exhibited an increased expression of SCARA5, whereas mice injected with the SCARA5 knockdown cells exhibited decreased levels of SCARA5 expression. The changes in SCARA5 protein expression levels were consistent with the changes in mRNA expression, suggesting that the xenograft tumor model with SCARA5 overexpression or suppression had been successfully established (Fig. 4C and D). The results of immunohistochemistry revealed that mice injected with SCARA5-overexpressing cells had decreased numbers of Ki67-positive cells and increased numbers of TUNEL-positive cells when compared with the control and SCARA5 knockdown groups (Fig. 4E). These results were consistent with the present in vitro results, suggesting that overexpression of SCARA5 could inhibit proliferation and promote apoptosis of Y79 Rb cells.
Figure 4.
Effect of SCARA5 overexpression on carcinogenesis. (A) Subcutaneous tumor volumes in mice injected with SCARA5 overexpression or knockdown cells. **P<0.01, Over-SCARA5 vs. Blank; #P<0.05, sh-SCARA5 vs. Blank. (B) Reverse transcription-quantitative PCR analysis of SCARA5 expression in mice injected with SCARA5 overexpression or knockdown cells. ***P<0.001 vs. Blank, ###P<0.001 vs. Sh-control. (C) Western blot analysis of SCARA5 expression in mice injected with SCARA5 overexpression or knockdown cells. Data are presented as the mean ± SEM (n=3 with three repeats). ***P<0.001 vs. Blank, ###P<0.001 vs. Sh-control. (D) Immunohistochemical analysis of SCARA5 expression in mice injected with SCARA5 overexpression or knockdown cells. (E) Immunohistochemical analysis of Ki67-positive cells and TUNEL-positive cells in mice injected with SCARA5 overexpression or knockdown cells. Scale bar, 100 µm. SCARA5, scavenger receptor class A member 5; sh, short hairpin RNA; NC, negative control.
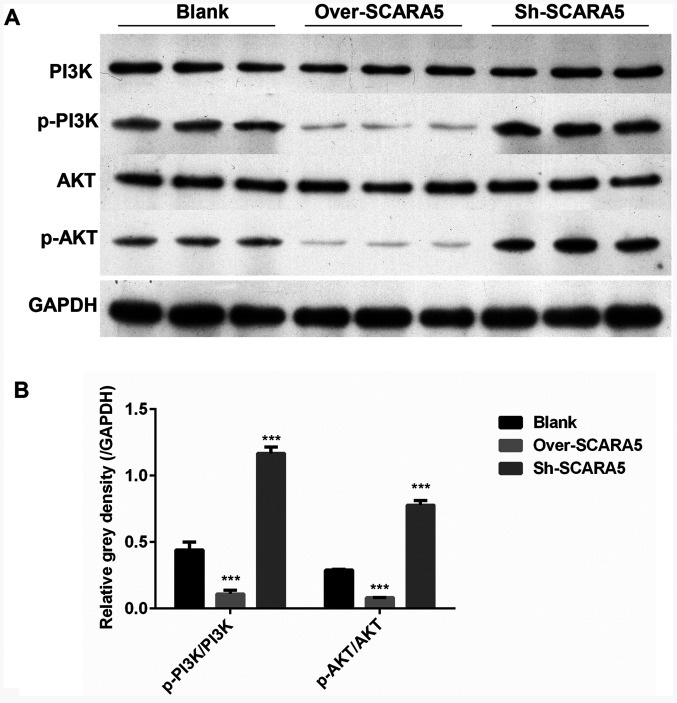
Overexpression of SCARA5 inhibits p-PI3K and p-AKT expression in vivo
In order to determine how expression of the PI3K/AKT signaling pathway was affected by SCARA5 in vivo, the in vivo expression levels of several key proteins involved in the PI3K/AKT pathway were detected. As shown in Fig. 5A and B, the protein expression levels of p-PI3K/PI3K and p-AKT/AKT were decreased in the SCARA5 overexpression group compared with their levels in the control group. Conversely, the protein expression levels of p-PI3K/PI3K and p-AKT/AKT were markedly increased in the SCARA5 knockdown group. These results were consistent with the present in vitro results.
Figure 5.
Western blot analysis of PI3K, p-PI3K, AKT and p-AKT expression in mice injected with SCARA5 overexpression or knockdown cells. (A) Protein expression of PI3K, p-PI3K, AKT and p-AKT detected by western blot. GAPDH was used as a control. (B) Gray density (relative to GAPDH) of p-PI3K/PI3K and p-AKT/AKT; Data are presented as the mean ± SEM (n=3 with three repeats). ***P<0.001 vs. Blank. SCARA5, scavenger receptor class A member 5; sh, short hairpin RNA; p-, phosphorylated.
Discussion
RB is one of the most malignant tumors derived from embryonic retinal cells. RB is almost exclusively diagnosed in children and a number of children who survive the disease lose their vision (23). Approximately 70–80% of children with RB are diagnosed at an age of 3 years or younger, and RB is very uncommon in children >6 years. Notably, the prevalence of RB is much higher in developing countries compared with in developed countries (24). In addition, it has been reported that the presence of human papilloma virus and poor socioeconomic conditions are suspected risk factors for RB. Current treatment methods include conservative treatment or removal of the eyeball (25); however, the prognosis of patients with RB remains unsatisfactory. Therefore, novel targets for treating RB need to be identified.
SRs are an important pattern recognition receptor family; this family includes a large subfamily of members classified by their multidomain protein structure (26). SRs were initially thought to contribute to various lipid metabolism disorders by recognizing acetylated low-density lipoproteins (27). Numerous studies have focused on SR class A members, due to the critical roles they play in innate immunity (27–29). SCARA5 is a class A SR. A previous study suggested that SCARA5 may participate in iron delivery (30). Moreover, SCARA5 was revealed to be highly expressed in white adipose tissue and was suggested to serve an important role in the differentiation of pluripotent stem cells into adipocytes (31). Numerous studies have revealed that SCARA5 expression is markedly decreased in numerous types of cancer cells; moreover, increased SCARA5 expression has been reported to inhibit the growth of several types of cancer cells (12–14,16). Consistent with these reports, the present study demonstrated that overexpression of SCARA5 could inhibit the proliferation and promote the apoptosis of RB cells. The invasive and migratory capabilities of tumor cells are essential indicators of tumor cell aggressiveness and metastatic ability (32). The majority of patients with RB already show signs of distant metastasis and tissue invasion at the time of initial diagnosis, and tumor metastasis is a major cause of death among pediatric patients with RB (33). Therefore, a reduction in the mortality rate of patients with RB might be achieved by reducing tumor cell migration and/or invasion (34). The present study demonstrated that overexpression of SCARA5 inhibited the migratory abilities of Y79 cells, indicating the crucial role of SCARA5 in RB metastasis.
It is well known that the PI3K/AKT signaling pathway regulates a series of cellular processes, including proliferation, invasion and metastasis (35). As the initiator of the PI3K/AKT signaling pathway, P13K serves important roles in cancer onset and development by regulating the proliferation and survival of tumor cells (36). Aberrant P13K activity has been reported to be associated with several processes that contribute to tumor development, such as the induction of malignant transformation, the migration and adhesion of tumor cells, and degradation of the extracellular matrix (37). It has been demonstrated that PI3K can regulate the activity of multifunctional proteins associated with cell proliferation and apoptosis, which are critical activities associated with cancer progression (38). In addition, AKT can activate or inhibit numerous downstream signaling molecules, including Bcl-2 and Bax, by directly targeting the PI3K protein (39). It has been suggested that suppression of PI3K/AKT signaling may aid in inhibiting the proliferation of RB cells (40). Thus, the PI3K/AKT pathway has been considered an attractive target for treating RB. Previously, the PI3K inhibitor NVP-BEZ235 was shown to effectively induce apoptosis of RB cells (41); furthermore, gingerol exhibited potent anticancer effects in RB355 human RB cells by modulating PI3K/AKT signaling (42). The results of the present study revealed that the protein expression levels of p-PI3K and p-AKT were decreased in RB cells that overexpressed SCARA5, but were increased in cells with SCARA5 knockdown. Moreover, these changes were accompanied by altered expression levels of the key apoptosis-associated proteins Bcl-2 and Bax. Thus, the present results indicated that the PI3K/AKT signaling pathway may be involved in regulating the effects of SCARA5 on the induction of apoptosis in RB cells. However, the direct effects of the PI3K/AKT signaling pathway on the proliferation and migration of human RB cells were not assessed. Therefore, PI3K/AKT or inhibition of their phosphorylation in cell or animal models should be studied to confirm the role of the PI3K/AKT signaling pathway in the cell proliferation and migration of human RB cells. In addition, further research is required to investigate whether other crucial proteins associated with cell proliferation and migration are affected by SCARA5 in RB cells.
In conclusion, the present study revealed that SCARA5 expression was markedly downregulated in RB cells when compared with its expression in healthy retinal pigment epithelial cells. By performing cell transfection, it was demonstrated that overexpression of SCARA5 could markedly inhibit the proliferation and migration of Y79 cells, and promote apoptosis of Y79 cells. Conversely, knockdown of SCARA5 promoted proliferation and migration, but inhibited apoptosis of Y79 cells. Moreover, in vivo studies conducted using a xenograft tumor model further confirmed the in vitro findings and suggested that SCARA5 may function as a tumor suppressor gene in preventing the occurrence and development of RB.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JT conceived and designed the study, and provided administrative support. JW, SW and LC performed the experiments. JW and SW analyzed and interpretated the data. JT and JW wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All study protocols involving animals were approved by the Ethics Committee of Central South University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Gao Y, Lu X. Decreased expression of MEG3 contributes to retinoblastoma progression and affects retinoblastoma cell growth by regulating the activity of Wnt/β-catenin pathway. Tumour Biol. 2016;37:1461–1469. doi: 10.1007/s13277-015-4564-y. [DOI] [PubMed] [Google Scholar]
- 2.Chantada G, Schaiquevich P. Management of retinoblastoma in children: Current status. Paediatr Drugs. 2015;17:185–198. doi: 10.1007/s40272-015-0121-9. [DOI] [PubMed] [Google Scholar]
- 3.Zhou D, Liu P, Sun DW, Chen ZJ, Hu J, Peng SM, Liu YL. USP22 down-regulation facilitates human retinoblastoma cell aging and apoptosis via inhibiting TERT/P53 pathway. Eur Rev Med Pharmacol Sci. 2017;21:2785–2792. [PubMed] [Google Scholar]
- 4.Yang Q, Tripathy A, Yu W, Eberhart CG, Asnaghi L. Hypoxia inhibits growth, proliferation, and increases response to chemotherapy in retinoblastoma cells. Exp Eye Res. 2017;162:48–61. doi: 10.1016/j.exer.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 5.Shields CL, Shields JA. Retinoblastoma management: Advances in enucleation, intravenous chemoreduction, and intra-arterial chemotherapy. Curr Opin Ophthalmol. 2010;21:203–212. doi: 10.1097/ICU.0b013e328338676a. [DOI] [PubMed] [Google Scholar]
- 6.Shields CL, Shields JA. Basic understanding of current classification and management of retinoblastoma. Curr Opin Ophthalmol. 2006;17:228–234. doi: 10.1097/01.icu.0000193079.55240.18. [DOI] [PubMed] [Google Scholar]
- 7.Jabbour P, Chalouhi N, Tjoumakaris S, Gonzalez LF, Dumont AS, Chitale R, Rosenwasser R, Bianciotto CG, Shields C. Pearls and pitfalls of intraarterial chemotherapy for retinoblastoma. J Neurosurg Pediatr. 2012;10:175–181. doi: 10.3171/2012.5.PEDS1277. [DOI] [PubMed] [Google Scholar]
- 8.Zhang H, Zhong J, Bian Z, Fang X, Peng Y, Hu Y. Long non-coding RNA CCAT1 promotes human retinoblastoma SO-RB50 and Y79 cells through negative regulation of miR-218-5p. Biomed Pharmacother. 2017;87:683–691. doi: 10.1016/j.biopha.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 9.Lin P, O'Brien JM. Frontiers in the management of retinoblastoma. Am J Ophthalmol. 2009;148:192–198. doi: 10.1016/j.ajo.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 10.Peiser L, Gordon S. The function of scavenger receptors expressed by macrophages and their role in the regulation of inflammation. Microbes Infect. 2001;3:149–159. doi: 10.1016/S1286-4579(00)01362-9. [DOI] [PubMed] [Google Scholar]
- 11.Jiang Y, Oliver P, Davies KE, Platt N. Identification and characterization of murine SCARA5, a novel class A scavenger receptor that is expressed by populations of epithelial cells. J Biol Chem. 2006;281:11834–11845. doi: 10.1074/jbc.M507599200. [DOI] [PubMed] [Google Scholar]
- 12.Liu J, Hu G, Chen D, Gong AY, Soori GS, Dobleman TJ, Chen XM. Suppression of SCARA5 by Snail1 is essential for EMT-associated cell migration of A549 cells. Oncogenesis. 2013;2:e73. doi: 10.1038/oncsis.2013.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang J, Zheng DL, Qin FS, Cheng N, Chen H, Wan BB, Wang YP, Xiao HS, Han ZG. Genetic and epigenetic silencing of SCARA5 may contribute to human hepatocellular carcinoma by activating FAK signaling. J Clin Invest. 2010;120:223–241. doi: 10.1172/JCI38012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu Z, Hong Z, Ma M, Liu X, Chen L, Zheng C, Xi X, Shao J. Rock2 promotes RCC proliferation by decreasing SCARA5 expression through β-catenin/TCF4 signaling. Biochem Biophys Res Commun. 2016;480:586–593. doi: 10.1016/j.bbrc.2016.10.097. [DOI] [PubMed] [Google Scholar]
- 15.Yan N, Zhang S, Yang Y, Cheng L, Li C, Dai L, Dai L, Zhang X, Fan P, Tian H, et al. Therapeutic upregulation of Class A scavenger receptor member 5 inhibits tumor growth and metastasis. Cancer Sci. 2012;103:1631–1639. doi: 10.1111/j.1349-7006.2012.02350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu H, Hu J, Wei R, Zhou L, Pan H, Zhu H, Huang M, Luo J, Xu W. SPAG5 promotes hepatocellular carcinoma progression by downregulating SCARA5 through modifying β-catenin degradation. J Exp Clin Cancer Res. 2018;37:229. doi: 10.1186/s13046-018-0891-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Onishi T, Matsuda S, Nakamura Y, Kuramoto J, Tsuruma A, Sakamoto S, Suzuki S, Fuchimoto D, Onishi A, Chikaki S, et al. Magnetically promoted rapid immunofluorescence staining for frozen tissue sections. J Histochem Cytochem. 2019;67:575–587. doi: 10.1369/0022155419841023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thayanithy V, O'Hare P, Wong P, Zhao X, Steer CJ, Subramanian S, Lou E. A transwell assay that excludes exosomes for assessment of tunneling nanotube-mediated intercellular communication. Cell Commun Signal. 2017;15:46. doi: 10.1186/s12964-017-0201-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hawkins P, Playle L, Golledge H, Leach M, Banzett R, Coenen A, Cooper J, Danneman P, Flecknell R, Kirkden R, et al. Newcastle consensus meeting on carbon dioxide euthanasia of laboratory animals. 2006:1–17. [Google Scholar]
- 21.National Research Council (US) Committee for the Update of the Guide for the Care and Use of Laboratory Animals, corp-author. 8th ed. National Academies Press (US); Washington (DC): 2011. Guide for the Care and Use of Laboratory Animals. [Google Scholar]
- 22.Martini M, De Santis MC, Braccini L, Gulluni F, Hirsch E. PI3K/AKT signaling pathway and cancer: An updated review. Ann Med. 2014;46:372–383. doi: 10.3109/07853890.2014.912836. [DOI] [PubMed] [Google Scholar]
- 23.Ancona-Lezama D, Dalvin LA, Shields CL. Modern treatment of retinoblastoma: A 2020 review. Indian J Ophthalmol. 2020;68:2356–2365. doi: 10.4103/ijo.IJO_721_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rossukon K, Duangnate R. Retinoblastoma: Etiology, modeling, and treatment. Cancers (Basel) 2020;12:2304. doi: 10.3390/cancers12082304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sweid A, El Naamani K, Sajja KC, Hammoud B, Knapp MD, Moylan DD, Joffe D, Morse CE, Habbal D, Weinberg JH, et al. Incidence and predictors of ophthalmic artery occlusion in intra-arterial chemotherapy for retinoblastoma. J Neurointerv Surg. doi: 10.1136/neurintsurg-2020-016759. doi: http://dx.doi.org/10.1136/neurintsurg-2020-016759. [DOI] [PubMed] [Google Scholar]
- 26.Chao H, Walsh CE. RNA repair for haemophilia A. Expert Rev Mol Med. 2006;8:1–8. doi: 10.1017/S1462399406010337. [DOI] [PubMed] [Google Scholar]
- 27.Pluddemann A, Neyen C, Gordon S. Macrophage scavenger receptors and host-derived ligands. Methods. 2007;43:207–217. doi: 10.1016/j.ymeth.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 28.Peiser L, Mukhopadhyay S, Gordon S. Scavenger receptors in innate immunity. Curr Opin Immunol. 2002;14:123–128. doi: 10.1016/S0952-7915(01)00307-7. [DOI] [PubMed] [Google Scholar]
- 29.Areschoug T, Gordon S. Scavenger receptors: Role in innate immunity and microbial pathogenesis. Cell Microbiol. 2009;11:1160–1169. doi: 10.1111/j.1462-5822.2009.01326.x. [DOI] [PubMed] [Google Scholar]
- 30.Li JY, Paragas N, Ned RM, Qiu A, Viltard M, Leete T, Drexler IR, Chen X, Sanna-Cherchi S, Mohammed F, et al. Scara5 is a ferritin receptor mediating non-transferrin iron delivery. Dev Cell. 2009;16:35–46. doi: 10.1016/j.devcel.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee H, Lee YJ, Choi H, Seok JW, Yoon BK, Kim D, Han JY, Lee Y, Kim HJ, Kim JW. SCARA5 plays a critical role in the commitment of mesenchymal stem cells to adipogenesis. Sci Rep. 2017;7:14833. doi: 10.1038/s41598-017-12512-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De la Fuente IM, Lopez JI. Cell motility and cancer. Cancers (Basel) 2020;12:2177. doi: 10.3390/cancers12082177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kashyap S, Singh L, Kumar N, Singh MK, Pushker N, Bakhshi S, Sen S, Lomi N, Meel R, Chawla B. Combined association of massive choroidal and optic nerve invasion as a prognostic relevance in primary retinoblastoma: A 10-year study. Asia Pac J Clin Oncol. 2020 doi: 10.1111/ajco.13348. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 34.Bock AJ, Nymoen DA, Brenne K, Kaern J, Davidson B. SCARA3 mRNA is overexpressed in ovarian carcinoma compared with breast carcinoma effusions. Hum Pathol. 2012;43:669–674. doi: 10.1016/j.humpath.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 35.Cao Y, Xia F, Wang P, Gao M. MicroRNA935p promotes the progression of human retinoblastoma by regulating the PTEN/PI3K/AKT signaling pathway. Mol Med Rep. 2018;18:5807–5814. doi: 10.3892/mmr.2018.9573. [DOI] [PubMed] [Google Scholar]
- 36.Fransecky L, Mochmann LH, Baldus CD. Outlook on PI3K/AKT/mTOR inhibition in acute leukemia. Mol Cell Ther. 2015;3:2. doi: 10.1186/s40591-015-0040-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Faes S, Dormond O. PI3K and AKT: Unfaithful partners in cancer. Int J Mol Sci. 2015;16:21138–21152. doi: 10.3390/ijms160921138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Follo MY, Manzoli L, Poli A, McCubrey JA, Cocco L. PLC and PI3K/Akt/mTOR signalling in disease and cancer. Adv Biol Regul. 2015;57:10–16. doi: 10.1016/j.jbior.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 39.Ferenc P, Solar P, Kleban J, Mikes J, Fedorocko P. Down-regulation of Bcl-2 and Akt induced by combination of photoactivated hypericin and genistein in human breast cancer cells. J Photochem Photobiol B. 2010;98:25–34. doi: 10.1016/j.jphotobiol.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 40.Wu S, Ai N, Liu Q, Zhang J. MicroRNA448 inhibits the progression of retinoblastoma by directly targeting ROCK1 and regulating PI3K/AKT signalling pathway. Oncol Rep. 2018;39:2402–2412. doi: 10.3892/or.2018.6302. [DOI] [PubMed] [Google Scholar]
- 41.Xie C, Freeman MJ, Lu H, Wang X, Forster CL, Sarver AL, Hallstrom TC. Retinoblastoma cells activate the AKT pathway and are vulnerable to the PI3K/mTOR inhibitor NVP-BEZ235. Oncotarget. 2017;8:38084–38098. doi: 10.18632/oncotarget.16970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meng B, Ii H, Qu W, Yuan H. Anticancer effects of gingerol in retinoblastoma cancer cells (RB355 Cell Line) are mediated via apoptosis induction, cell cycle arrest and upregulation of PI3K/Akt signaling pathway. Med Sci Monit. 2018;24:1980–1987. doi: 10.12659/MSM.914733. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.