Summary
The specification of the meristem/organ boundary is critical for plant development. Here, we investigate two previously uncharacterized NAC transcription factors: the first, OsCUC1, which is negatively regulated by osa‐miR164c, dimerizes with the second, OsCUC3, and functions partially redundantly in meristem/organ boundary specification in rice (Oryza sativa).
We produced knockout lines for rice OsCUC1 (the homolog of Arabidopsis CUC1 and CUC2) and OsCUC3 (the homolog of Arabidopsis CUC3), as well as an overexpression line for osa‐miR164c, to study the molecular mechanism of boundary specification in rice.
A single mutation in either OsCUC1 or OsCUC3 leads to defects in the establishment of the meristem/organ boundary, resulting in reduced stamen numbers and the fusion of leaves and filaments, and the defects are greatly enhanced in the double mutant. Transgenic plants overexpressing osa‐miR164c showed a phenotype similar to that of the OsCUC1 knockout line. In addition, knockout of OsCUC1 leads to multiple defects, including dwarf plant architecture, male sterility and twisted‐rolling leaves. Further study indicated that OsCUC1 physically interacts with leaf‐rolling related protein CURLED LEAF AND DWARF 1 (CLD1) and stabilizes it in the nucleus to control leaf morphology.
This work demonstrated that the interplay of osa‐miR164c, OsCUC1 and OsCUC3 controls boundary specification in rice.
Keywords: boundary specification, CLD1, degradation, miR164, partially redundant, CUP‐SHAPED COTYLEDON genes, twisted‐rolling leaves
Introduction
Most of the aerial parts of higher plants are derived from the shoot apical meristem (SAM). In the SAM, the creation of a boundary that separates a meristem/organ primordium from its surroundings is critical for organ formation. In Arabidopsis, the CUP‐SHAPED COTYLEDON (CUC) genes CUC1 and CUC2, which encode a paralogous pair of NAC transcription factors and are negatively regulated by the microRNA miR164, have been shown to be involved in embryonic SAM formation and boundary specification (Aida & Tasaka, 2006). Since CUC1 and CUC2 are functionally redundant, neither the cuc1 nor cuc2 single mutant displays a severe seedling phenotype, while the cuc1 cuc2 double mutant shows severe cotyledon fusion and the absence of a SAM (Aida et al., 1997, 1999; Takada et al., 2001). Another NAC gene family member, CUC3, which is a homolog of CUC1 and CUC2, also participates in the establishment of the cotyledon boundary and the shoot meristem redundantly with CUC1 and CUC2 (Vroemen et al., 2003; Hibara et al., 2006).
Systematic sequence analysis revealed 151 putative NAC or NAC‐like genes in rice (Oryza sativa L.) (Nuruzzaman et al., 2010). Some of these genes have been demonstrated to be involved in different rice developmental processes. For example, NAC29/30 regulates cellulose synthesis (Huang et al., 2015); O. sativa NAC‐like activated by apetala3/pistillata (OsNAP) positively regulates leaf senescence and serves as a link between abscisic acid (ABA) and leaf senescence (Chen et al., 2014; Liang et al., 2014); OsNAC2 not only affects plant height, shoot branching, and thus, yield (Mao et al., 2007; Chen et al., 2015; Jiang et al., 2018), but also promotes leaf senescence via ABA biosynthesis (Mao et al., 2017); ONAC020, ONAC023 and ONAC026 heteromerize and play important roles in seed development (Mathew et al., 2016). In addition, many rice NAC genes have been shown to be involved in responses to various abiotic and biotic stresses, including ONAC048 (OsNAC6), ONAC017 (OsNAC111), ONAC122, ONAC131, ONAC054 (RICE DWARF VIRUS MULTIPLICATION 1) and ONAC068 (OsNAC4) in defense against pathogen infection (Nakashima et al., 2007; Kaneda et al., 2009; Yoshii et al., 2009; Sun et al., 2012; Yokotani et al., 2014), and ONAC022, ONAC002 (STRESS‐RESPONSIVE NAC 1/OsNAC9), ONAC048 (SNAC2/OsNAC6), ONAC009 (OsNAC5), ONAC122 (OsNAC10), ONAC045, ONAC058 (OsNAP), ONAC004, ONAC060, ONAC011 and ONAC104 in abiotic stress tolerance (Hu et al., 2006, 2008; Nakashima et al., 2007; Zheng et al., 2009; Jeong et al., 2010, 2013; Song et al., 2011; Redillas et al., 2012; Chen et al., 2014; Fang et al., 2014; Liang et al., 2014; Hong et al., 2016).
Although many NAC genes have been characterized in rice, whether they are involved in meristem or organ boundary specification remains unknown. To clarify this question, we knocked out the homologues of CUC genes in rice using CRISPR/Cas9. Our results indicated that OsCUC1, which is negatively regulated by osa‐miR164c, together with OsCUC3, is involved in rice meristem/organ boundary specification in a partially redundant manner. In addition, OsCUC1 physically interacts with a previously known leaf rolling related protein CURLED LEAF AND DWARF 1 (CLD1) and stabilizes it in the nucleus to control leaf morphology.
Materials and Methods
Plant materials and growth conditions
Rice (Oryza sativa L.) plants were grown in a glasshouse at 30°C (for daytime)/25°C (for nighttime) under a 13.5 h : 8.5 h, light : dark photoperiod (>3000 lux) with 60% humidity. The genetic background of all transgenic plants used was Nipponbare. omtn4 ZH11 and omtn6 ZH11 were obtained from Biogle GeneTech (Hangzhou Biogle Co. Ltd, Hangzhou, Zhejiang, China)
Phylogenetic tree and sequence alignment
Rice OsCUC1, OsCUC3 and 70 homologues from 13 species were chosen for phylogenetic analysis. Protein sequences were obtained from NCBI (https://www.ncbi.nlm.nih.gov/) according to their accession number. The sequence alignment was performed with clustalx and the phylogenetic tree was constructed using mega7 with the neighbor‐joining method and bootstrap analysis (1000 replicates). The accession numbers are listed in Supporting Information Table S1.
Vector construction
To knock out OsCUC1 and OsCUC3, target sites were designed online (http://cbi.hzau.edu.cn/crispr/), and the optimal target sites with low off‐target scores and high sgRNA scores were selected. We prepared the CRISPR/Cas9 binary constructs as described previously (Ma et al., 2015). To overexpress osa‐miR164c, a polymerase chain reaction (PCR) fragment amplified from Nipponbare genomic DNA using primers OE‐miR164cF and OE‐miR164cR was cloned into the PstI/SpeI sites of binary vector pOX between the maize Ubi1 promoter and Nos terminators.
Generation of pOsCUC1::GUS was achieved by amplifying a 2 kb OsCUC1 promoter from Nipponbare genomic DNA using primers OsCUC1pgus‐F and OsCUC1pgus‐R, which was then inserted into pCAMBIA1300‐GN at the HindIII/XbaI site. To generate pOsCUC1::OsCUC1‐GUS, a PCR fragment harboring the 2 kb upstream promoter and gDNA (without a stop codon) of OsCUC1 was amplified from Nipponbare genomic DNA using primers pro‐cds‐gus‐F and pro‐cds‐gus‐R, and was then inserted into pCAMBIA1300‐GN at the SalI/XbaI site. To generate pOsCUC1::mOsCUC1‐GUS, two overlapped fragments harboring the 2 kb upstream promoter and gDNA (without a stop codon) of OsCUC1 were amplified using two primer sets: pro‐cds‐gus‐F/mOsCUC1‐R (carrying the 7 bp mutations) and mOsCUC1‐F (carrying the 7 bp mutations)/pro‐cds‐gus‐R, respectively. After purification, the two fragments were mixed in equal molar ratios and used as the PCR templates to amplify a fragment containing the 2 kb upstream promoter and gDNA (without a stop codon) of OsCUC1, into which the osa‐miR164c‐resistant mutation was introduced, using pro‐cds‐gus‐F and pro‐cds‐gus‐R. After sequencing, the fragment was inserted into pCAMBIA1300‐GN at the SalI/XbaI site.
Primer sequences for the constructions are listed in Table S2.
Pollen viability test
Mature pollen grains from the unopened flowers were collected and immediately put on a microscope slide. A drop of IKI (iodine potassium iodide) solution was deposited onto the pollen, and the slide was covered with a coverslip. Pollen viability counts were made 5 min after the IKI solution was added to the pollen. Pollen grains exhibiting dark staining (dark red or black color) were counted as viable.
Histochemical analysis and β‐glucuronidase (GUS) assay
Samples of the transgenic plants were incubated with GUS staining solution (50 mM sodium phosphate at pH 7.2, 10 mM ethylenediaminetetraacetic acid (EDTA), 0.1% Triton X‐100, 2 mM of X‐Gluc, 2 mM potassium ferricyanide, 2 mM potassium ferrocyanide) overnight at 37°C. After staining, the tissues were rinsed several times with ethanol, then mounted on slides and photographed.
For histochemical analysis, the samples were fixed with formaldehyde alcohol acetic acid (FAA) fixation solution at 4°C overnight, followed by dehydration and embedding in paraffin (Paraplast Plus, Sigma). They were then cut into 7 μm sections with a microtome, and stained with safranine and fast green FCF. Sections were observed under bright field with a microscope (CX31; Olympus, Tokyo, Japan).
Yeast‐two‐hybrid assay
The Matchmaker yeast‐two‐hybrid system (Clontech, Kusatsu, Japan) was used to study the interaction of OsCUC1, OsCUC3, OMTN4 and OMTN6 with CLD1. The deduced amino acid sequences of OsCUC1, OsCUC3, OMTN4 and OMTN6 were separately cloned into pGADT7 (AD) vectors, while CLD1 was inserted into pGBKT7 (BD). Yeast strain Y2HGold was transformed with bait plasmid and strain Y187 was transformed with prey plasmid. Co‐transformants were plated on synthetic defined (SD)/–Leu/–Trp/–His/–Ade medium plates and SD/–Leu/–Trp/–His/–Ade/X‐α‐Gal medium plates for examination of growth. Primer sequences for the constructions are listed in Table S2.
Transactivation assay
The deduced amino acid sequences of OsCUC1 and OsCUC3 were separately cloned into pGBKT7 (BD), resulting in fusions with the GAL4 binding domain. The fusion plasmids pBD‐OsCUC1 and pBD‐OsCUC3 were transformed into yeast strain AH109 and plated on SD/–Trp/–His/–Ade and SD/–Trp/–His/–Ade/X‐α‐Gal medium plates for examination of growth. Primer sequences for the constructions are listed in Table S2.
Scanning electron microscopy (SEM)
The samples were fixed in FAA (50% ethanol/acetic acid/formaldehyde, 9 : 0.5 : 0.5), and then dehydrated in an ethanol series and dried by supercritical fluid drying with CO2. The dried samples were mounted on copper supports and sputter‐coated with gold, and then observed under SEM (S‐3400N; Hitachi, Tokyo, Japan).
Bi‐molecular fluorescence complementation (BiFC) assay
To study the interactions of OsCUC1, OsCUC3, OMTN4, OMTN6, CLD1, CUC1 CUC2, CUC3, mCUC1 and mCUC2, the coding sequences of these genes were correspondingly cloned into p35S‐Vn and p35S‐Vc. The plasmids were co‐expressed in rice/Arabidopsis protoplasts. The protein–protein interaction was evaluated using a confocal laser scanning microscope (TCS‐SP8MP; Leica, Wetzlar, Germany). Primer sequences for the constructions are listed in Table S2.
Subcellular localization assay
The coding sequences of OsCUC1, OsCUC3 and CLD1 (without stop codons) from Nipponbare were fused with green fluorescent protein (GFP) to generate the fusion protein. The fusion protein driven by the cauliflower mosaic virus (CaMV) 35S promoter was transcribed in rice protoplasts. The subcellular localization was evaluated using a confocal laser‐scanning microscope (TCS‐SP8MP; Leica). OsRac3‐mCherry and Ghd7‐mCherry were used as the membrane localization marker (Chen et al., 2010) and nuclear localization marker (Xue et al., 2008), respectively. Primer sequences for the constructions are listed in Table S2.
RNA extraction and quantitative real time (RT) polymerase chain reaction assay
To evaluate gene expression, total RNA was extracted using the RNeasy Plant Mini Kit (cat. no. 74904; Qiagen, Germany) following the manufacturer's instructions. First‐strand cDNA was synthesized from 1 μg of total RNA using the PrimeScript RT Reagent Kit (cat. no. RR047A; Takara, Kusatsu, Japan) according to the manufacturer’s instructions. ChamQ SYBR qPCR Master Mix (cat. no. Q311‐01; Vazyme, Nanjing, Jiangsu, China) and a LightCycler 480II (Roche) were used for qRT‐PCR, according to the manufacturers’ instructions. Rice Ubiquitin (Os03g0234200) was used as the internal reference, and the level of gene expression was normalized to the Ubiquitin level. To evaluate the expression level of osa‐miR164, RNA extraction and stem‐loop qRT‐PCR were performed as described previously (Wu et al., 2007). Rice miRNA U6 was used as the internal reference and the osa‐miR164 level was normalized to U6. Primer sequences for the qRT‐PCR are listed in Table S2.
In situ hybridization
To prepare the probe, a 500‐bp fragment of OsCUC1‐specific cDNA and a 500‐bp fragment of OsCUC3‐specific cDNA were amplified. The probes were labeled using a DIG RNA Labeling Kit (Roche). The in situ hybridization experiments were carried out as described previously (Brewer et al., 2006). Primer sequences for the in situ hybridization are listed in Table S2.
Co‐immunoprecipitation (Co‐IP) assay
To confirm the protein–protein interactions of OsCUC1, OsCUC3 and CLD1, OsCUC1‐GFP, OsCUC1‐MYC, OsCUC3‐GFP and CLD1‐Flag were artificially synthesized (GenScript Biotech Corp., Nanjing, Jiangsu, China) and transcribed under the control of the cauliflower mosaic virus (CaMV) 35S promoter. Twelve hours after co‐transformation of the plasmids, the rice protoplasts were harvested by centrifugation at 400 g for 5 min. The collected protoplasts were homogenized in CelLytic immunoprecipitation (IP) buffer (B7345; Sigma‐Aldrich), and incubated for 30 min on ice and then centrifuged at 15 000 g for 10 min at 4°C to remove aggregates. Next, 40 μl of Protein A/G Agarose Beads (nProtein A Sepharose 4 Fast Flow; GE Healthcare, Uppsala, Sweden) were added into the protein extract, which was diluted by IP buffer. The mixture was incubated for 3 h at 4°C with gentle shaking (40–50 rpm). After centrifugation at 14 000 g for 5 min at 4°C, the supernatant was separated from the beads, and the antibodies were added to the supernatant. After overnight incubation at 4°C, 40 μl of protein A‐Sepharose was added and incubated for a further 2–3 h at 4°C. The beads were collected by centrifugation at 100 g for 3 min at 4 °C, and then washed five times with ice‐cold washing buffer (25 mM Tris‐HCl, pH 7.5, 150 mM NaCl, 0.5% Triton X‐100, 1 mM EDTA, and 1% protease inhibitor). The proteins were eluted from the beads by boiling in sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS‐PAGE) sample buffer for 5 min and analysed by Western blotting. The original blot images are shown in Fig. S1. Primer sequences for the constructions are listed in Table S2.
In vitro pull‐down assay
To confirm the protein–protein interaction of OsCUC1 and OsCUC3, the coding sequences of OsCUC1 and OsCUC3 were amplified and inserted into pGEX‐6P‐1 (in frame fusion with a glutathione‐S‐transferase (GST) tag) and pRSFDuet‐MBP (in frame fusion with a maltose binding protein (MBP) tag), respectively, and then GST‐OsCUC1 and MBP‐OsCUC3 were amplified and cloned into pTNT (Promega). The recombinant GST‐OsCUC1 and MBP‐OsCUC3 were synthesized using the TNT SP6 High‐Yield Wheat Germ Protein Expression System (Promega) following the manufacturer’s instructions. The recombinant GST‐OsCUC1 fusion protein was immobilized on GST‐Binding Resin (MagneGST Glutathione Particles, Promega) and incubated with MBP‐OsCUC3 for 4 h at 4°C. After incubation, the beads were washed five times in washing buffer (4.2 mM Na2HPO4, 2 mM KH2PO4, 140 mM NaCl and 10 mM KCl), and subsequently eluted using elution buffer (50 mM Tris‐HCl, 50 mM glutathione). The supernatant was subjected to immunoblotting analysis with anti‐MBP (TransGen, Beijing, China) and anti‐GST (TransGen) antibodies.
To confirm the protein–protein interaction of OsCUC1 and CLD1, the coding sequence of CLD1 was amplified and inserted into pRSFDuet‐His (in frame fusion with a His tag), and then His‐CLD1 was amplified and cloned into pTNT (Promega). The recombinant His‐CLD1 was synthesized using the TNT SP6 High‐Yield Wheat Germ Protein Expression System (Promega) following the manufacturer’s instructions. The recombinant GST‐OsCUC1 fusion protein was immobilized on GST‐Binding Resin (MagneGST™ Glutathione Particles; Promega) and incubated with His‐CLD1 for 4 h at 4°C. After incubation, the beads were washed five times in washing buffer (4.2 mM Na2HPO4, 2 mM KH2PO4, 140 mM NaCl and 10 mM KCl), and subsequently eluted using elution buffer (50 mM Tris‐HCl, 50 mM glutathione). The supernatant was subjected to immunoblotting analysis with anti‐His (TransGen) and anti‐GST (TransGen) antibodies.
The original blot images are shown in Fig. S1. Primer sequences for the constructions are listed in Table S2.
Protein extraction
For an in planta CLD1 accumulation assay, total proteins were extracted from 0.2 g of 12‐d‐old fresh leaves of non‐KO1OsCUC1 and oscuc1‐KO1. The leaves were collected and ground in liquid nitrogen, then homogenized in protein extraction buffer (50 mM Tris‐HCl, pH7.5, 150 mM NaCl, 4 M urea and 1 mM phenylmethylsulfonyl fluoride). Extracts were incubated at 4°C for 1 h, then centrifuged for 30 min at maximum speed. Supernatants were collected for further analysis. To obtain the nuclear‐enriched fraction, samples were collected in the same quantity (0.2 g of 12‐d‐old fresh leaves), and the nuclear proteins were extracted as described previously (Kaufmann et al., 2010).
For the CLD1‐GFP accumulation assay, to obtain total proteins, rice protoplasts were collected and treated with lysis buffer (cat. no. B7345; Sigma‐Aldrich). After incubation at 4°C for 1 h, the extracts were centrifuged for 30 min at maximum speed. Supernatants were collected for further analysis. The nuclear proteins of the rice protoplasts were extracted using a Minute Cytoplasmic and Nuclear Kit (Invent Biotechnologies, Eden Prairie, MN, USA) according to the manufacturer's instructions.
Protein degradation assay
The recombinant His‐CLD1 was synthesized using the TNT SP6 High‐Yield Wheat Germ Protein Expression System (Promega) according to the manufacturer’s instructions. The equivalent amount of His‐CLD1 was added to 120 μl crude total proteins (20 µg µl–1) extracted from indicated plants (non‐KO1OsCUC1 or oscuc1‐KO1) using reaction buffer (50 mM Tris‐HCl, pH 7.8, 100 mM NaCl, 0.1% (v/v) Tween 20, 10% (v/v) glycerol and 20 mM β‐mercaptoethanol). The mixture was incubated at 25°C and sampled at indicated time points (0, 3, 6 and 9 h). The abundance of remaining His‐CLD1 in these samples was detected by immunoblotting using an anti‐His (TransGen) antibody. The original blot images are shown in Fig. S1.
Statistical analysis
Statistical analysis was performed by Student's t‐test. P‐values of < 0.05 were considered to indicate statistical significance. P‐values of < 0.01 were considered to indicate statistically high significance. Statistical calculations were performed using Microsoft Excel 2010.
Results
Knocking out OsCUC1 and OsCUC3 in rice leads to defects in meristem/organ boundary specification
To identify the OsCUC genes, we searched for homologs of CUC1, CUC2 and CUC3 in rice. Sequence alignment indicated that LOC_Os06g23650 (Os06g0344900) is the closest homolog of both CUC1 and CUC2, hereafter referred to as OsCUC1; LOC_Os08g40030 (Os08g0511200) shares the greatest similarity with CUC3, and we therefore named it OsCUC3. Both genes were previously uncharacterized. OsCUC1 encodes a 373 amino acid (aa) protein with a typical NAC domain; while OsCUC3 encodes a 340‐aa NAC protein (Fig. S2a). Both OsCUC1 and OsCUC3 belong to the NAM subgroup of the NAC gene family (Fang et al., 2008; Nuruzzaman et al., 2010). A phylogenetic tree analysis showed that OsCUC1 is the homologue of Arabidopsis CUC1, and it is also very similar to Arabidopsis CUC2, petunia NAM, tomato GOBLET, strawberry FveCUC2a, FveCUC2b, and FveCUC2c. OsCUC3 is the homologue of CUC3 in Arabidopsis and FvH4_5g12090 in strawberry (Fig. S2c).
To understand the biological function of OsCUC1 and OsCUC3, we employed CRISPR/Cas9 to produce two individual knockout lines for each of these two genes (Figs 1a, 2a). For OsCUC1, a 1 bp deletion for oscuc1‐KO1 and a 1 bp insertion for oscuc1‐KO2 (at different sites) lead to a frame‐shift with premature transcription termination (Fig. S3a), while both oscuc3‐KO1 and oscuc3‐KO2 harbor a 1 bp insertion at different sites of the coding region resulting in a premature stop codon (Fig. S3b).
Fig. 1.
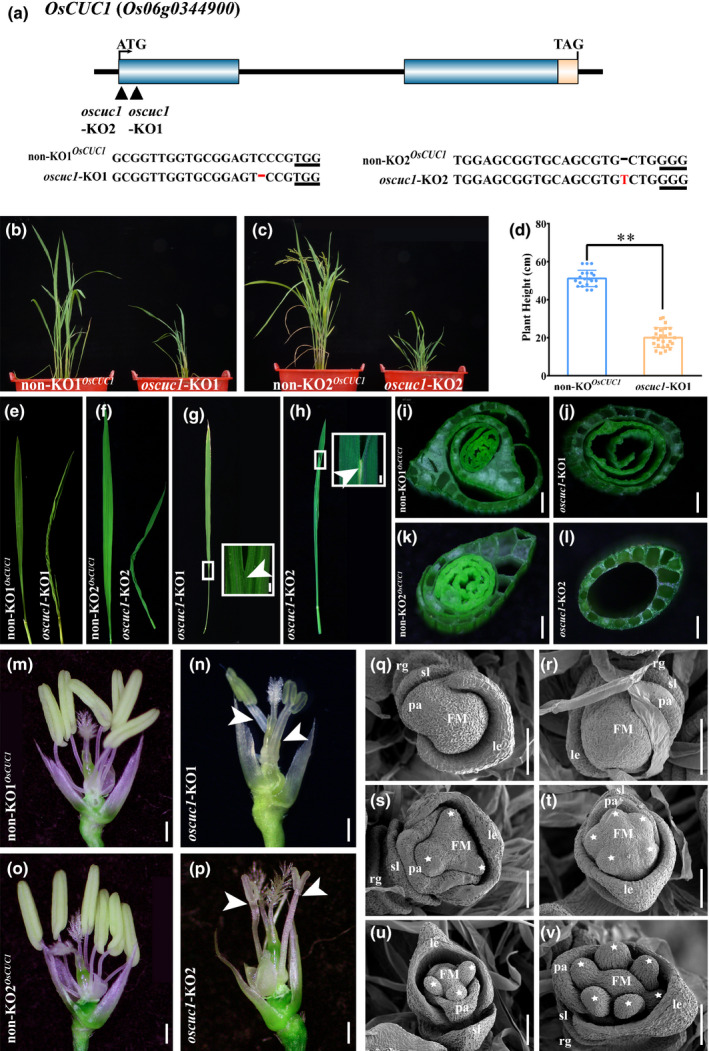
Appearance of rice oscuc1 knockout mutants. (a) Schematic diagram of OsCUC1 gene structure and the CRISPR/Cas9 target sites. The black triangles indicate the two individual target sites for knockout of OsCUC1. The PAM site is underlined. The mutation site is shown in red. (b, c) The whole plant phenotypes of oscuc1‐KO1 and oscuc1‐KO2. (d) The height of mature non‐KO1OsCUC1 and oscuc1‐KO1 plants. Values are shown as means ± SD (20 non‐KO1OsCUC1 plants and 25 oscuc1‐KO1 were used for the statistical analysis). **, P < 0.01 (Student's t‐test). (e, f) The twisted‐rolling leaves in oscuc1‐KO1 and oscuc1‐KO2. (g, h) Fusion of leaf blade in oscuc1‐KO1 and oscuc1‐KO2. The fusion regions outlined by small white rectangles are shown in detail on the right‐hand side of their respective panels. The arrows indicate the fusion of the leaf blades. (i–l) Cross‐section of the leaf sheath of oscuc1‐KO1, oscuc1‐KO2 and their non‐KO control. (m–p) Knocking out OsCUC1 leads to fusion in filaments and reduction in stamens. The arrows indicate the fusion of the filaments. (q–v) Scanning electron micrographs of early‐arising non‐KO1OsCUC1 and oscuc1‐KO1 floret. FM, flora meristem; le, lemma; pa, palea; sl, sterile lemma; rg, rudimentary glumes. The white asterisks represent the primordia of the stamens. Bars: (g–p) 500 μm; (q–v) 100 μm.
Fig. 2.

Appearance of rice oscuc3 knockout mutants. (a) Schematic diagram of OsCUC3 gene structure and the CRISPR/Cas9 target sites. The black triangles indicate the two individual target sites for knockout of OsCUC3. The PAM site is underlined. The mutation site is shown in red. (b, c) The whole plant phenotype of oscuc3‐KO1 and oscuc3‐KO2. (d–g) Cross‐section of the leaf sheath of oscuc3‐KO1, oscuc3‐KO2 and their non‐KO control. (h, i) Fusion of leaf blade in oscuc3‐KO1 and oscuc3‐KO2. The fusion region outlined by the small white rectangle is shown in detail in the bottom right corner. The arrows indicate the fusion of the leaf blades. (j–m) Knocking out OsCUC3 leads to fusion in filaments. The arrow indicates the fusion of two filaments of oscuc3‐KO2. Bars, 500 μm.
In comparison with the non‐KO control line, which harbored the intact OsCUC1, both homozygous oscuc1‐KO lines showed multiple defects in rice development, including the fusion of leaves and filaments, decreased stamen numbers, reduced plant height and twisted‐rolling leaves (Fig. 1b–p; Table S3). During the vegetative growth stage, fusion could be observed in the leaf blade (Figs 1g,h, S4a,b) as well as in the whole leaf sheath (Figs 1i–l, S4c,d) of the oscuc1‐KO plants. The tube‐like leaf sheath physically prevented the outgrowth of the new leaf or the panicle. During the reproductive growth stage, we also observed defects in floral development in oscuc1‐KO, in which the florets displayed fusions of two or more filaments (Fig. 1m–p). This fusion in the mutants indicated the function of OsCUC1 in organ boundary specification. At the same time, most of the florets showed reduced stamen numbers (Fig. 1m–p; Table S3). An SEM assay showed that not all of the stamen primordia correctly formed in the early floral developmental stage in the oscuc1‐KO plants, indicating that OsCUC1 also functions in meristem boundary specification (Fig. 1q–v). We did not obtain any seeds from any of the individual T0 homozygous oscuc1‐KO plants, while the non‐KO plants displayed normal fertility. This result may be attributable to the development of aberrant stamens, which may lead to pollen defects in the mutant plants. The pollen viability test indicated that almost no pollen was produced in the KO plants (Fig. S5). Furthermore, pollen from wild‐type plants (Nipponbare) was used to pollinate the gynoecia of oscuc1‐KO1. In contrast with the self‐pollinated florets, the cross‐pollinated florets maintained normal development in later stages and yielded fertile seeds (Fig. S6a–e). We further investigated whether these seeds could develop normally. No defects were detected in any developmental stage in these F1 progeny (oscuc1‐KO1/+). In the F2 population, the homozygous oscuc1‐KO1 plants showed the same phenotype as the T0 transgenic plants, while the non‐KO plants and heterozygous mutants developed normally (Fig. S6f,g).
Both of the oscuc3‐KO lines exhibited defects in meristem/organ boundary specification in the vegetative growth stage as well as in the reproductive growth stage, including the fusion of leaves and filaments, and defects in stamen identification (Fig. 2b–m; Table S3), which were highly similar to those of the oscuc1‐KO lines. However, knocking out OsCUC3 did not lead to dwarf plant stature or twisted‐rolling leaves. In addition, in contrast to the infertility of the oscuc1‐KO plants, fertile seeds could be obtained from both of the oscuc3‐KO lines. Similar to the oscuc1‐KO1/+ plants, the heterozygous OsCUC3 mutant plants showed no defects during any developmental stages (Fig. S6h,i).
The expression profiles of OsCUC1 and OsCUC3
NAC family proteins have been shown to function as transcription factors (Xie et al., 1999; Vroemen et al., 2003). Since OsCUC1 and OsCUC3 encode NAC domain proteins, we used GFP fluorescence to determine their subcellular localization by transiently expressing OsCUC1‐GFP or OsCUC3‐GFP in‐frame fusion proteins in rice protoplasts. The results revealed that both OsCUC1 and OsCUC3 are localized to the nucleus (Fig. 3a). In addition, to determine whether these genes have transactivation activity, OsCUC1 and OsCUC3 were fused to the GAL4 DNA binding domain and transformed into the yeast strain AH109. The yeast strains containing pBD‐OsCUC1 or pBD‐OsCUC3 grew in Trp‐, His‐ and Ade‐deficient SD medium while the negative control did not grow in triple‐deficient SD medium (Fig. 3b), which indicated that both genes have transactivation activity. Taken together, these data indicated that OsCUC1 and OsCUC3 function as transcription factors.
Fig. 3.
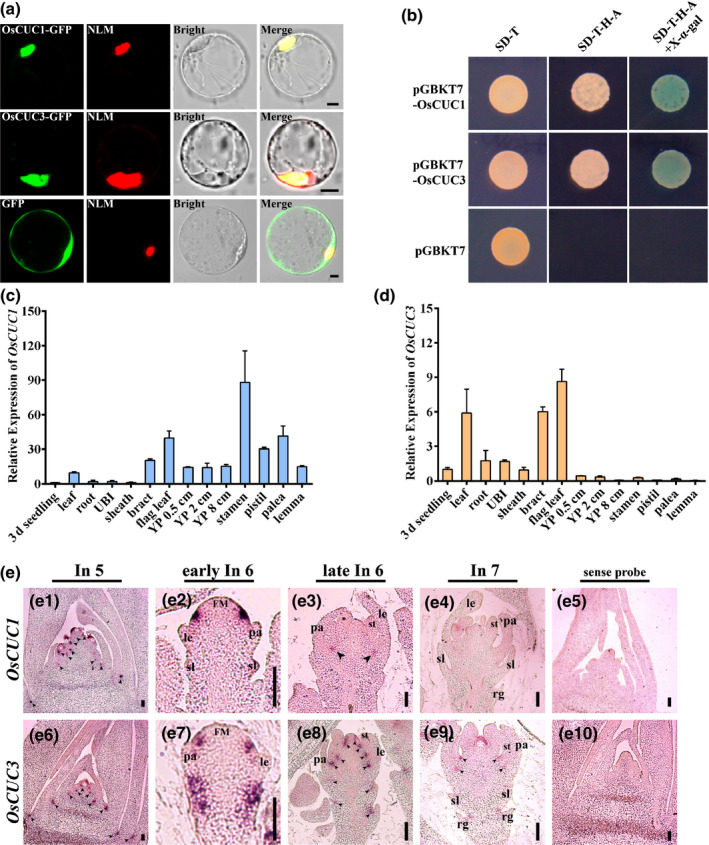
The expression patterns of OsCUC1 and OsCUC3 in rice. (a) Subcellular localization of OsCUC1 and OsCUC3. NLM, nuclear localization marker (Ghd7‐mCherry). Bars, 5 μm. (b) Transcriptional activity analysis of OsCUC1 and OsCUC3. The empty vector pGBKT7 was used as a negative control. (c, d) Relative expression of OsCUC1 and OsCUC3 in seedling (3 d after germination), leaf, root, UBI (unelongated basal internode), leaf sheath, bract, flag leaf, YP 0.5 cm (young panicle 0.5 cm), YP 2 cm, YP 8 cm, stamen, pistil, palea and lemma. Values are shown as means ± SD (n = 3). (e) In situ hybridization of OsCUC1 and OsCUC3. (e1–e4) Accumulation of OsCUC1 in wild‐type plants. (e6–e9) Accumulation of OsCUC3 in wild‐type plants. (e5, e10) Negative controls with sense probes. The black arrows indicate the accumulation regions of the mRNA. FM, flora meristem; le, lemma; pa, palea; rg, rudimentary glumes; sl, sterile lemma; st, stamens. Bars, 50 μm.
We used qRT‐PCR to study the expression patterns of OsCUC1 and OsCUC3 throughout plant development. OsCUC1 was highly expressed in the leaf, bract, flag leaf, and reproductive organs, especially in the stamens (Fig. 3c). In comparison to that of OsCUC1, the expression level of OsCUC3 was lower in all tested organs. However, OsCUC3 transcripts accumulated more abundantly in the vegetative organs than in the reproductive organs (Fig. 3d). To obtain the details of the expression patterns of these two genes, we carried out an in‐situ hybridization assay (Fig. 3e). In the early floral developmental stage (In 5), OsCUC1 and OsCUC3 exhibited highly overlapping expression patterns, with both mRNAs accumulated mainly in the boundaries (between the leaves and between the meristems) as well as within the meristems (Fig. 3e1,e6). In early In 6 stage, OsCUC1 mRNA was strongly and specifically accumulated at the region where the stamen primordia will arise (Fig. 3e2). Afterward, the expression of OsCUC1 became diffused and uniform in the flower organs (Fig. 3e3,e4). For OsCUC3, in early In 6 stage, its mRNA was detected in the same region where OsCUC1 mRNA accumulated (Fig. 3e7); in addition, OsCUC3 was also detected in the primordia of sterile lemmas and rudimentary glumes (Fig. 3e7). In the later stages (late In 6 and In 7), in contrast to OsCUC1, OsCUC3 was mainly detected in the boundaries between the flower organs (Fig. 3e8,e9).
OsCUC1 and OsCUC3 dimerize and function in a partially redundant manner in boundary specification and SAM activity maintenance
To understand the relationship between OsCUC1 and OsCUC3, we first studied the protein–protein interaction of these two proteins. A bimolecular fluorescence complementation (BiFC) assay showed that OsCUC1 and OsCUC3 could form homodimers or heterodimers (Fig. 4a,b). An in vivo co‐immunoprecipitation (Co‐IP) assay and an in vitro pull‐down assay further confirmed the dimerization between OsCUC1 and OsCUC3 (Fig. 4c,d). In Arabidopsis, due to the miR164 function in negatively regulating CUC1 and CUC2, the interaction between the CUC proteins cannot be detected in protoplasts by BiFC. However, we detected that the miR164‐resistant versions of CUC1 and CUC2 (mCUC1 and mCUC2) formed homodimers, while mCUC1‐mCUC2, mCUC1‐CUC3 and mCUC2‐CUC3 formed heterodimers (Fig. S7). These results were largely consistent with previous findings (Rubio‐Somoza et al., 2014; Gonçalves et al., 2015), indicating the conserved protein–protein interaction behavior of CUC proteins in both rice and Arabidopsis.
Fig. 4.
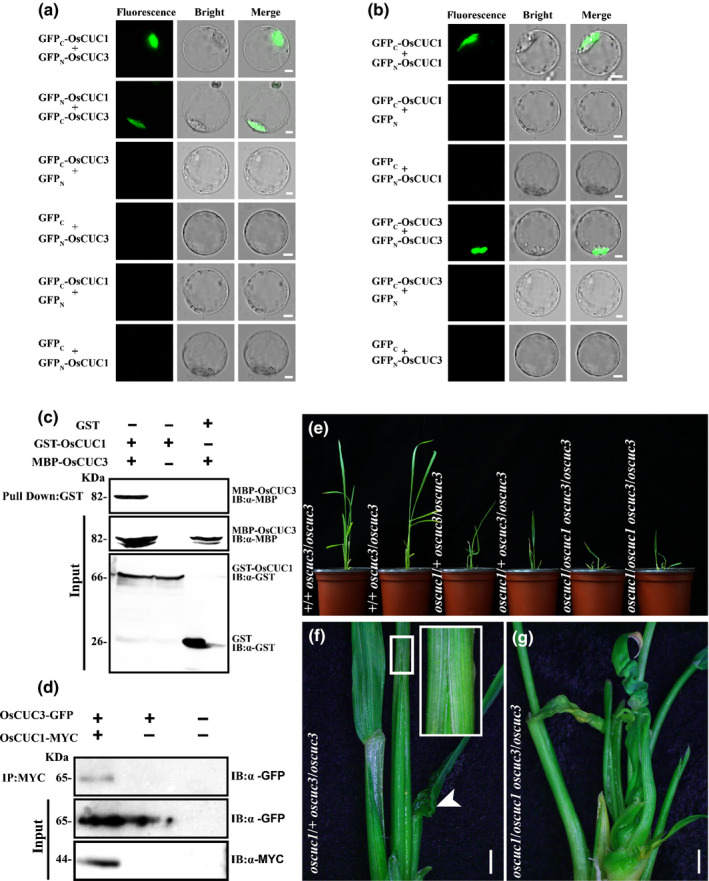
OsCUC1 and OsCUC3 dimerize and function redundantly in rice. (a, b) BiFC assays showing the heterodimerization and homodimerization of OsCUC1 and OsCUC3. Co‐expression of GFPC‐OsCUC3 plus GFPN, GFPC plus GFPN‐OsCUC3, GFPC‐OsCUC1 plus GFPN, and GFPC plus GFPN‐OsCUC1 were used as negative controls. Bars, 5 μm. (c) Interaction between OsCUC1 and OsCUC3, analysed by in vitro pull‐down assay. Recombinant GST‐OsCUC1 and MBP‐OsCUC3 proteins were used for the pull‐down assay. IB, immunoblot. (d) Interaction between OsCUC1 and OsCUC3, analysed by in vivo CoIP assay. After the co‐transformation of OsCUC1‐MYC and OsCUC3‐GFP in rice protoplasts, total proteins of protoplasts were immunoprecipitated using an anti‐MYC antibody and were detected with anti‐GFP and anti‐MYC antibodies. IB, Immunoblot; IP, immunoprecipitation. (e) The phenotypes of +/+ oscuc3/oscuc3, oscuc1/+ oscuc3/oscuc3 and oscuc1/oscuc1 oscuc3/oscuc3. (f) The fusion of leaf sheath and twisted‐rolling leaf in oscuc1/+ oscuc3/oscuc3. The fusion region of the leaf sheath outlined by a small white rectangle in the main image is shown in detail in the top right corner; the arrow indicates the twisted‐rolling leaf. Bar, 500 μm. (g) The enhanced defects in the oscuc1/oscuc1 oscuc3/oscuc3 double mutant. Bar, 500 μm.
Furthermore, we developed double mutant plants for these two genes. In contrast with the single mutant plants, which showed no defects during the early seedling stage (Fig. S6f, h), the oscuc1/+ oscuc3/oscuc3 plants exhibited leaf sheath fusion and twisted‐rolling leaves; these defects were greatly enhanced in the oscuc1/oscuc1 oscuc3/oscuc3 plants (Fig. 4e–g). Although loss of function of OsCUC1 and OsCUC3 did not lead to defects in SAM initiation (Fig. S8a–d), the maintenance of SAM activity was markedly affected – the development of the double mutant plants was arrested in the early seedling stage, and they eventually died soon after that (Fig. S8e–l). Together, these results indicate that OsCUC1 and OsCUC3 dimerize and function in a partially redundant manner in boundary specification and SAM activity maintenance.
osa‐miR164c is involved in organ boundary specification and leaf development by negatively regulating OsCUC1
In Arabidopsis, it has been demonstrated that miR164 negatively regulates CUC1 and CUC2 (Baker et al., 2005; Nikovics et al., 2006; Sieber et al., 2007; Raman et al., 2008). To test whether OsCUC1 is regulated by microRNA, we searched for the corresponding microRNA using OsCUC1 as a putative target in the Plant Non‐coding RNA Database, (PNRD http://structuralbiology.cau.edu.cn/PNRD/index.php) (Yi et al., 2014). The results indicated that OsCUC1 may be regulated by all members of the rice microRNA osa‐miR164 family (osa‐miR164a to osa‐miR164f). These findings implied that osa‐miR164 may also be involved in organ boundary specification and leaf development by negatively regulating OsCUC1. The bioinformatics analysis by PNRD indicated that OsCUC1 is considered to be the target of osa‐miR164c with a high expectation value; moreover, only four genes, including OsCUC1, are predicted to be the potential targets of osa‐miR164c, while the other osa‐miR164 targets more genes. Thus, osa‐miR164c was selected for further study. The results from RT‐PCR followed by Sanger sequencing indicated that the real existence of the osa‐miR164c is supported by transcript evidence (Fig. S9a). Moreover, the qRT‐PCR results indicated that osa‐miR164c was accumulated highly in leaf tissues, but almost no expression could be detected in the mature flower organs, in which OsCUC1 is relatively highly expressed (Fig. S9b). To verify the functions of osa‐miR164c in regulating boundary specification and leaf morphology, we overexpressed osa‐miR164c in the genetic background of Nipponbare. In comparison to the control plants carrying the empty vector, OsCUC1 was significantly downregulated in both the vegetative and reproductive organs of OE‐osa‐miR164c plants (Fig. 5a–c). Similar to oscuc1‐KO, the OE‐osa‐miR164c plants also showed defects in organ boundary separation and leaf development (Fig. 5d–h). However, unlike in oscuc1‐KO, the stamens developed normally in OE‐osa‐miR164c, indicating that a low expression level of OsCUC1 is sufficient for maintaining normal organ separation in the reproductive growth stage but not in the vegetative growth stage. Notably, two transgenic lines with higher expression of osa‐miR164c (OE‐osa‐miR164c‐4 and OE‐osa‐miR164c‐15) showed more severe defects than the others, and no seeds could be obtained from these two lines.
Fig. 5.
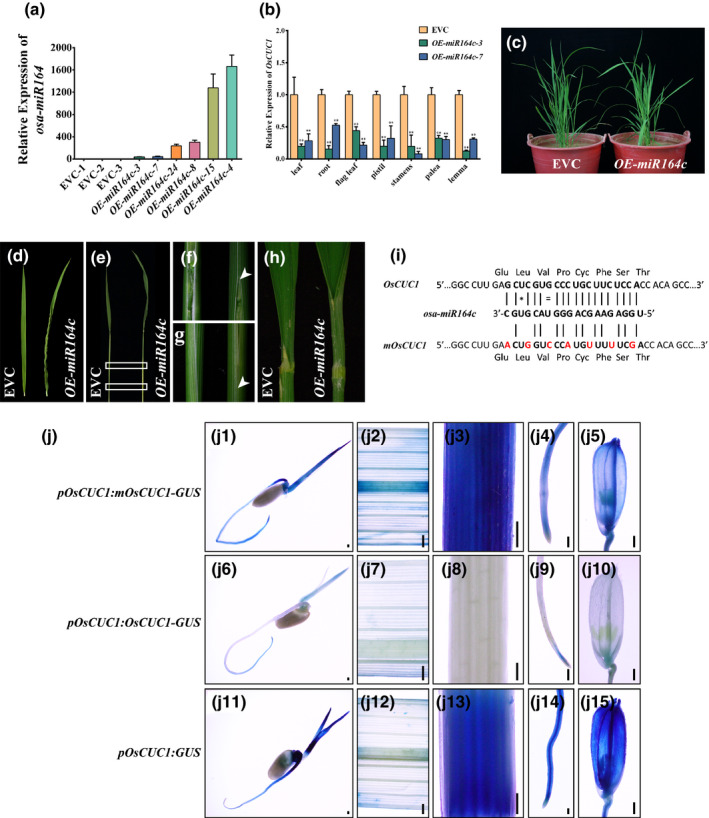
osa‐miR164c is involved in boundary specification and leaf development by negatively regulating OsCUC1 in rice. (a) Relative expression of osa‐miR164c in different individual OE‐osa‐miR164c lines and empty‐vector control lines (EVC). Values are shown as means ± SD (n = 3). (b) Relative expression of OsCUC1 in EVC and two OE‐osa‐miR164c lines in different tissues. Values are shown as means ± SD (n = 3). **, P < 0.01 (Student's t‐test). (c) The phenotype of OE‐osa‐miR164c and EVC lines. (d) The twisted‐rolling leaf in OE‐osa‐miR164c. (e) Overexpressing osa‐miR164c leads to fusion in leaf sheaths. (f, g) Detailed images of the fusion regions depicted in (e); the arrows indicate the fusion regions on the leaf sheath. (h) Overexpressing osa‐miR164c leads to fusion in the leaf blade. (i) Schematic diagram depicting the construction of mOsCUC1. The red letters indicate mutant nucleotides introduced into the osa‐miR164 target site in the mOsCUC1‐GUS transgene, which interrupts the osa‐miR164 target site without changing the amino acid residues. Watson–Crick base pairing between the mRNA and osa‐miR164c is indicated by black lines. Mismatches and G:U wobbles are indicated by a star or equals sign, respectively. (j) Gus staining for pOsCUC1::mOsCUC1‐GUS, pOsCUC1::OsCUC1‐GUS and pOsCUC1::GUS in seedling (j1, j6, j11), leaf blade (j2, j7, j12), leaf sheath (j3, j8, j13), root (j4, j9, j14), and floret (j5, j10, j15). Bars, 200 μm.
OsCUC1 contains an osa‐miR164‐target sequence in the second exon. To further test the effects of osa‐miR164 on OsCUC1, we generated transgenic lines carrying an OsCUC1 in‐frame fusion with the β‐glucuronidase (GUS) gene driven by its endogenous promoter with or without synonymous mutations in the osa‐miR164‐targeted site (pOsCUC1::OsCUC1‐GUS and pOsCUC1::mOsCUC1‐GUS) (Fig. 5i). The pOsCUC1::mOsCUC1‐GUS lines showed a relatively high level of GUS staining in most of the organs (Fig. 5j1–j5), while only minor staining was observed in the pOsCUC1::OsCUC1‐GUS lines (Fig. 5j6–j10). These results strongly suggest that osa‐miR164c functions by dampening the transcription level of OsCUC1. In agreement with this idea, the transcriptional reporter of OsCUC1 (pOsCUC1::GUS) showed a strong expression pattern, as expected (Fig. 5j11–j15).
osa‐miR164 targeting OMTN4 and OMTN6 may not be involved in meristem/organ boundary specification or leaf development
In addition to OsCUC1, five other NAC (OMTN) genes, OMTN1 (ONAC027), OMTN2 (ONAC004/OsNAC2), OMTN3 (ONAC060), OMTN4 (ONAC011) and OMTN6 (ONAC104), are considered to be putative targets of osa‐miR164 (Fang et al., 2014). We have shown that osa‐miR164c plays a critical role in boundary specification and leaf development by downregulating OsCUC1. Our qRT‐PCR results indicated that only OsCUC1 – and not the other five putative osa‐miR164 targets – was significantly downregulated in all the organs tested in OE‐osa‐miR164c plants (Figs 5b, S10). In a previous study, OMTN4‐RNAi (RNA interference) and OMTN6‐RNAi transgenic plants were reported to show severe abnormal phenotypes, such as twisted‐rolling leaves and fused organs, which were similar to those of the oscuc1‐KO plants (Fang et al., 2014). To verify whether OMTN4 and OMTN6 function in controlling organ specification and leaf development, we knocked out these two genes using CRISPR/Cas9 in the Nipponbare genetic background. We did not observe any fused or twisted‐rolling leaves for the homozygous knockout (KO) mutants of these two genes during the vegetative growth stage, and in the reproductive stage, defects in the florets were barely detected. Moreover, no significant defects could be observed from two other mutant lines with the Zhonghua11 background (omtn4 ZH11 and omtn6 ZH11) either (Fig. S11; Table S4). Therefore, the defects in the two RNAi lines may be caused by RNAi off‐target effects. Thus, we postulated that the targets of osa‐miR164, OMTN4 and OMTN6 may not respond to meristem/organ boundary specification or leaf development as OsCUC1 does.
OsCUC1 physically interacts with CLD1 and maintains CLD1 stability in the nucleus to control leaf morphology
Knocking out OsCUC1 in rice gives rise to defects in leaf development, which resemble the phenotype of the curled leaf and dwarf 1 (cld1) mutants (Li et al., 2017). However, the expression level of CLD1 showed no significant change in the KO plants (Fig. S12). Thus, we posited that OsCUC1 might interact with CLD1 and function in maintaining the stability of the CLD1 protein. In previous research, it has been demonstrated that CLD1, also known as Semi‐Rolled Leaf 1 (SRL1), encodes a glycophosphatidylinositol (GPI)‐anchored membrane protein, which localizes predominantly at the plasma membrane and modulates leaf development (Xiang et al., 2012; Li et al., 2017). To verify whether CLD1 can interact with OsCUC1, we first tested whether CLD1 is located in the nucleus, as OsCUC1 is. We transiently co‐expressed CLD1‐GFP in‐frame fusion proteins with a nuclear localization marker or membrane localization marker in rice protoplasts. Green fluorescence protein fluorescence indicated that CLD1 was located in both the membrane and the nucleus (Fig. 6a). Moreover, CLD1‐GFP was detected in the nuclei‐enriched fraction by immunoblotting (Fig. 6b). This result further confirmed the nuclear localization of CLD1. We then studied the protein–protein interactions between CLD1 and the NAC proteins; BiFC, yeast‐two‐hybrid (Y2H), in vivo Co‐IP and in vitro pull‐down assays revealed the interactions between CLD1 and OsCUC1 (Fig. 6c–f). By contrast, we did not detect any interactions between CLD1 and OsCUC3 or two other NAC proteins (the putative osa‐miR164 targets OMTN4 and OMTN6) by either BiFC or Y2H assays (Fig. S13). In the in vitro assay conditions, recombinant His‐CLD1 was stable in the crude protein extract from non‐KO1OsCUC1. By contrast, His‐CLD1 proteins were rapidly degraded after the addition of crude protein extract from oscuc1‐KO1, while the degradation of His‐CLD1 was greatly slowed down when recombinant GST‐OsCUC1 was first mixed with His‐CLD1 (Fig. 7a). Moreover, we carried out an assay for CLD1 stability in planta. Immunoblotting assays revealed that CLD1 was not detected in the nucleus‐enriched fraction from oscuc1‐KO plants, indicating that the nuclear accumulation of CLD1 is dependent on OsCUC1 functioning (Fig. 7b). These results further confirmed the role of OsCUC1 in the stabilization of CLD1. Overall, the protein–protein interaction between OsCUC1 and CLD1 may stabilize CLD1 in the nucleus; loss of function of OsCUC1 may lead to acceleration of CLD1 degradation in the nucleus, resulting in a phenotype with twisted‐rolling leaves, which is similar to that of the cld1 mutant.
Fig. 6.
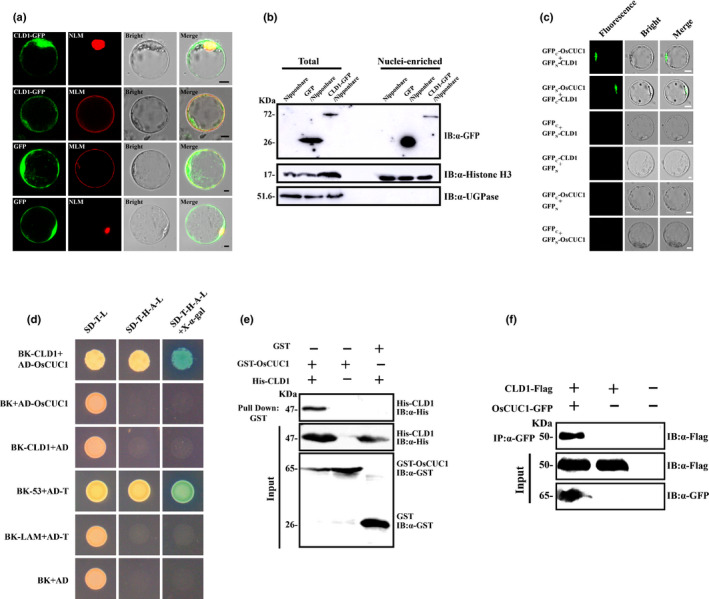
OsCUC1 interacts with CLD1 in rice. (a) CLD1 is localized to the cell membrane as well as the nucleus. Upper row: co‐localization of CLD1‐GFP and an mCherry‐tag nuclear localization marker. Upper middle row: co‐localization of CLD1‐GFP and an mCherry‐tag membrane localization marker. Lower middle row: co‐localization of green fluorescent protein (GFP) and an mCherry‐tag membrane localization marker as a control. Lower row: co‐localization of GFP and an mCherry‐tag nuclear localization marker as a control. MLM, membrane localization marker (OsRac3‐mCherry); NLM, nuclear localization marker (Ghd7‐mCherry). Bars, 5 μm. (b) Accumulation of CLD1‐GFP in the nucleus. Rice (Nipponbare) protoplast transiently expressed GFP or CLD‐GFP. CLD1‐GFP proteins were detected using anti‐GFP antibodies in total proteins of the rice protoplast or in the nuclei‐enriched fraction only. Histone H3 served as a nuclear marker and UGPase (UDP‐glucose pyrophosphorylase) as a cytoplasmic marker. (c) BiFC assay. Co‐expression of GFPC plus GFPN‐CLD1, GFPC‐CLD1 plus GFPN, GFPC‐OsCUC1 plus GFPN, and GFPC plus GFPN‐OsCUC1 were used as negative controls. Bars, 5 μm. (d) Y2H assay. The combination of BK‐53 plus AD‐T was used as a positive control, while BK‐CLD1 plus AD, BK‐LAM plus AD‐T, and BK plus AD were used as negative controls. (e) In vivo co‐immunoprecipitation (CoIP) assay. After the co‐transformation of OsCUC1‐GFP and CLD1‐Flag in rice protoplasts, total proteins of protoplasts were immunoprecipitated using an anti‐GFP antibody and were detected with anti‐GFP and anti‐Flag antibodies. IB, immunoblot; IP, immunoprecipitation. (f) In vitro pull‐down assay. Recombinant GST‐OsCUC1 and His‐CLD1 proteins were used for the pull‐down assay. IB, immunoblot.
Fig. 7.
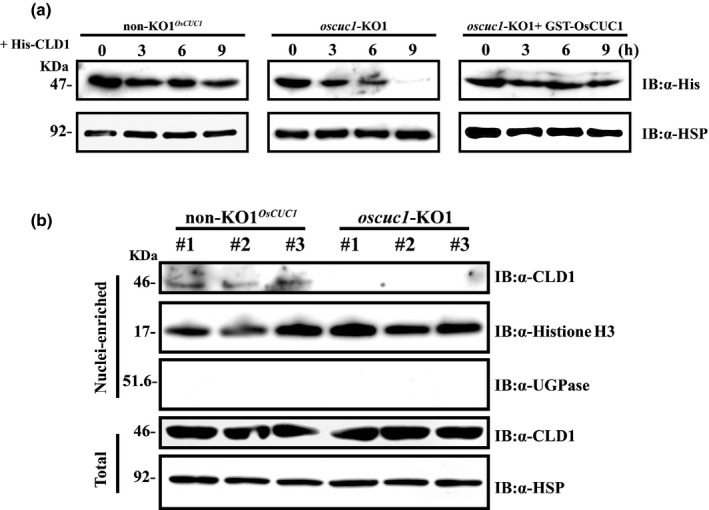
OsCUC1 stabilizes CLD1 in the nucleus in rice. (a) In vitro protein degradation assay. Recombinant His‐CLD1 protein was incubated with total crude extract from non‐KO1OsCUC1 (left) and oscuc1‐KO1 (middle, without GST‐OsCUC1; right, with GST‐OsCUC1). HSP (Hsp90) was used as a loading control. (b) The accumulation of CLD1 in the nucleus depends on OsCUC1 functioning; accumulation of CLD1 in non‐KO1OsCUC1 and oscuc1‐KO1 plants is shown. CLD1 was detected with an anti‐CLD1 antibody. Histone H3 served as a nuclear marker, and UGPase (UDP‐glucose pyrophosphorylase) served as a cytoplasmic marker. HSP (Hsp90) was used as a loading control. Three independent plants of each line were used for the assay.
Discussion
In plants, members of CUP‐SHAPED COTYLEDON (CUC)⁄NO APICAL MERISTEM (NAM), a group of plant‐specific NAC transcription factors, play critical roles in boundary specification, including CUC1, 2 and 3 in Arabidopsis, CUPULIFORMIS (CUP) in Antirrhinum majus, GOBLET (GOB) in tomato (Solanum lycopersicum), NO APICAL MERISTEM (NAM) in Petunia and MtNAM in Medicago truncatula (Souer et al., 1996; Aida et al., 1997; Takada et al., 2001; Vroemen et al., 2003; Weir et al., 2004; Berger et al., 2009; Cheng et al., 2012). On the other hand, miR164 is involved in multiple developmental processes by negatively regulating its targets, which are always NAC transcription factors. In Arabidopsis, miR164 regulates boundary specification, leaf margin serration, lateral root development, pathogen‐induced cell death, age‐dependent cell death and the salt stress response by targeting different NAC genes (Guo et al., 2005; He et al., 2005; Nikovics et al., 2006; Sieber et al., 2007; Raman et al., 2008; Kim et al., 2009; Lee et al., 2017); in kiwifruit (Actinidia spp.), an interplay between ade‐miR164 and AdNAC6/7 regulates fruit ripening (Wang et al., 2019); in strawberry (Fragaria vesca), the miR164‐CUC2 module regulates leaf and flower development (Zheng et al., 2019); in M. truncatula, miR164‐MtNAC1 pathway is involved in root and symbiotic nodule development (D'haeseleer et al., 2011); and in wheat (Triticum aestivum), TaNAC21/22, the targets of tae‐miR164, play an important role in regulating the resistance of host plants to stripe rust (Feng et al., 2014). In rice, overexpression of osa‐miR164b led to dwarf plant architecture and small panicles (Jiang et al., 2018). Transgenic plants overexpressing the osa‐miR164‐targeted genes, including OMTN2 (ONAC004/OsNAC2), OMTN3 (ONAC060), OMTN4 (ONAC011) and OMTN6 (ONAC104) increased the sensitivity to drought stress at the reproductive growth stage, indicating that these genes appear to be associated with the response to abiotic stresses (Fang et al., 2014).
The role of OsCUC1 and OsCUC3 in SAM formation and maintenance
In dicots, it has been demonstrated that NAM in Petunia, MtNAM in Medicago, CUP in Antirrhinum and CUCs in Arabidopsis are involved in SAM formation. Mutations in NAM, MtNAM and CUP lead to fusion in cotyledons, and no apical meristem can be formed. In Medicago, the development of the strong mutant of MtNAM is arrested in the fused cotyledon stage (Cheng et al., 2012). By contrast, in Petunia, the nam mutant occasionally produces escape shoots, which develop normally in the vegetative stage, but show defects in flower development (Souer et al., 1996). In Antirrhinum, the cup mutant always produces escape shoots; however, these escape shoots display defects during all developmental stages (Weir et al., 2004). In Arabidopsis, CUC1 and CUC2 function redundantly in SAM formation, and seedlings of the cuc1 cuc2 double mutant, which completely lacks a SAM, exhibit a cup‐shaped cotyledon (Aida et al., 1997). Further study indicated that cuc2 cuc3 double mutant seedlings, but not cuc1 cuc3 seedlings, have no functional SAM. Thus, only cuc2 in combination with cuc1 and/or cuc3 leads to the absence of the SAM (Vroemen et al., 2003).
In our study, we found that the oscuc1 oscuc3 double mutant produces aberrant organs at the seedling stage, and the development of the double mutant seedling is arrested. These results indicated that, in addition to OsCUC1 and OsCUC3, other factor(s) may be involved in the regulation of SAM initiation, and loss of function of these two genes alone is not sufficient to prevent SAM formation. However, OsCUC1 and OsCUC3 are critical for maintaining normal SAM activity. Plants without functional OsCUC1 and OsCUC3 products stop developing and ultimately die at the seedling stage.
Functional divergence between OsCUC1 and OsCUC3
Although OsCUC1 and OsCUC3 function together in boundary specification and SAM maintenance, their functions remain different in other developmental processes. Loss of function of OsCUC1 leads to a dwarf plant structure, pollen defection and twisted‐rolling leaves, while oscuc3 mutants show no defects in these related developmental processes. Our study indicates that only OsCUC1 – but not OsCUC3 or the other two NAC proteins (OMTN4 and OMTN6) – interacts with the leaf‐rolling related protein CLD1. These results are consistent with the fact that oscuc1 mutants but not oscuc3 or omtn4/6 mutants show defects in leaf development. Thus, the different interaction behaviors of OsCUC1 and OsCUC3 with the other gene may be partly responsible for the functional divergence of these two proteins.
Despite divergence, the mechanisms of meristem/organ boundary specification between Arabidopsis and rice are conserved
In Arabidopsis, due to the redundant function of CUC1 and CUC2, neither the cuc1 nor the cuc2 single mutation results in a severe phenotype, but the cuc1 cuc2 double mutation causes a complete lack of embryonic shoot meristem formation. The regenerated cuc1 cuc2 mutant exhibits abnormal flowers, in which all the sepals and most of the stamens are severely fused (Aida et al., 1997, 1999; Takada et al., 2001). In rice, the oscuc1 single mutation causes severe defects in meristem/organ boundary specification, resulting in a reduction in stamen numbers and fusion of leaves/filaments. Consistent with this, systematic sequence analysis revealed that OsCUC1 is the only homolog of both CUC1 and CUC2 (Kondou et al., 2009), suggesting that the function of OsCUC1 may be executed redundantly by CUC1 and CUC2 in Arabidopsis. Despite the divergence between OsCUC1 and CUC1/2, the mechanisms of meristem/organ boundary specification are conserved between the dicot Arabidopsis and the monocot O. sativa (Fig. 8): one or more microRNA (osa‐miR164/miR164) target(s) (OsCUC1/CUC1&CUC2) function redundantly with another NAC gene family member (OsCUC3/CUC3) in meristem/organ boundary specification. In addition, OsCUC1 is involved in leaf development by affecting the stability of CLD1 in the nucleus.
Fig. 8.
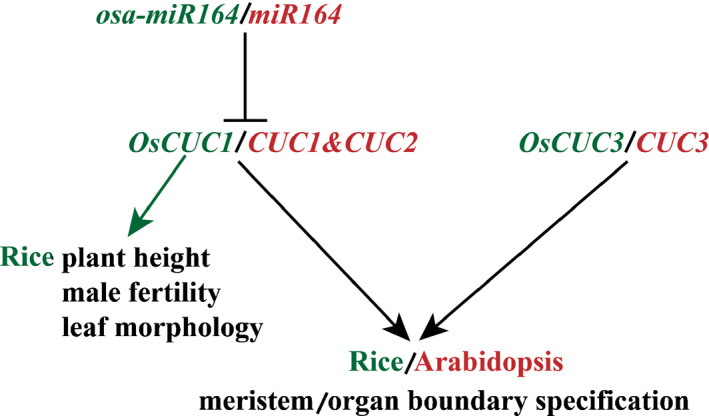
Interplay of a microRNA and CUP‐SHAPED COTYLEDON genes controls meristem/organ boundary specification in rice and Arabidopsis. The microRNA (osa‐miR164/miR164) negatively regulates the CUP‐SHAPED COTYLEDON genes (OsCUC1/CUC1&CUC2), which function redundantly with another NAC gene family member (OsCUC3/CUC3) in meristem/organ boundary specification in both rice and Arabidopsis. In addition, OsCUC1 controls plant height, male fertility and leaf morphology in rice. Red represents Arabidopsis, and green represents rice.
Author contributions
JJ planned and designed the research. JW, JB, BZ, ML and XL performed experiments. JJ wrote the manuscript. All authors commented on the manuscript. JW and JB contributed equally to this work.
Supporting information
Fig. S1 Original blot images from this study.
Fig. S2 Sequence analysis for OsCUC1 and OsCUC3.
Fig. S3 Loss‐of‐function of OsCUC1 and OsCUC3 generated by CRISPR/Cas9 in rice.
Fig. S4 Boundary specification defects in the vegetative growing stage of the rice oscuc1‐KO1 mutant.
Fig. S5 Pollen defects in the rice oscuc1‐KO1 mutant.
Fig. S6 Phenotypes of heterozygous mutants of OsCUC1 and OsCUC3 in rice.
Fig. S7 Dimerization of the Arabidopsis CUC proteins.
Fig. S8 The development of rice oscuc1 oscuc3 homozygous double mutants is arrested at the seedling stage.
Fig. S9 Transcript evidence and expression pattern for rice osa‐miR64c.
Fig. S10 The expression patterns for the other five osa‐miR164 targets in rice.
Fig. S11 Knocking out OMTN4 or OMTN6 does not lead to defects either in boundary specification or leaf development in rice.
Fig. S12 The CLD1 expression level does not significantly change in the rice oscuc1 mutant.
Fig. S13 CLD1 does not interact with OsCUC3, OMTN4 or OMTN6 in rice.
Table S1 The accession numbers of the proteins listed in the phylogenetic tree.
Table S2 The primer sequences used in this study.
Table S3 Percentages of aberrant florets of oscuc1‐KO and oscuc3‐KO plants.
Table S4 Percentages of aberrant florets of omtn4‐KO and omtn6‐KO plants.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
We thank Prof. Daiyin Chao, Prof. Wei Huang and Prof. Liangfa Ge for critical reading of the manuscript. This research was supported by the National Natural Science Foundation of China (Grant No. 31560381), the Project of High Level Innovation Team and Outstanding Scholar in Guangxi Colleges and Universities (Third batch, 2016) and the Project of Scientific Research in Guangxi Colleges and Universities (KY2015ZD004).
References
- Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M. 1997. Genes involved in organ separation in Arabidopsis: an analysis of the cup‐shaped cotyledon mutant. Plant Cell 9: 841–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida M, Ishida T, Tasaka M. 1999. Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: interaction among the CUP‐SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development 126: 1563–1570. [DOI] [PubMed] [Google Scholar]
- Aida M, Tasaka M. 2006. Genetic control of shoot organ boundaries. Current Opinion in Plant Biology 9: 72–77. [DOI] [PubMed] [Google Scholar]
- Baker CC, Sieber P, Wellmer F, Meyerowitz EM. 2005. The early extra petals1 mutant uncovers a role for microRNA miR164c in regulating petal number in Arabidopsis. Current Biology 15: 303–315. [DOI] [PubMed] [Google Scholar]
- Berger Y, Harpaz‐Saad S, Brand A, Melnik H, Sirding N, Alvarez JP, Zinder M, Samach A, Eshed Y, Ori N. 2009. The NAC‐domain transcription factor GOBLET specifies leaflet boundaries in compound tomato leaves. Development 136: 823–832. [DOI] [PubMed] [Google Scholar]
- Brewer PB, Heisler MG, Hejátko J, Friml J, Benková E. 2006. In situ hybridization for mRNA detection in Arabidopsis tissue sections. Nature Protocols 1: 1462–1467. [DOI] [PubMed] [Google Scholar]
- Chen L, Shiotani K, Togashi T, Miki D, Aoyama M, Wong HL, Kawasaki T, Shimamoto K. 2010. Analysis of the Rac/Rop Small GTPase family in rice: expression, subcellular localization and role in disease resistance. Plant & Cell Physiology 51: 585–595. [DOI] [PubMed] [Google Scholar]
- Chen X, Lu S, Wang Y, Zhang X, Lv B, Luo L, Xi D, Shen J, Ma H, Ming F. 2015. OsNAC2 encoding a NAC transcription factor that affects plant height through mediating the gibberellic acid pathway in rice. The Plant Journal 82: 302–314. [DOI] [PubMed] [Google Scholar]
- Chen X, Wang Y, Lv B, Li J, Luo L, Lu S, Zhang X, Ma H, Ming F. 2014. The NAC family transcription factor OsNAP confers abiotic stress response through the ABA pathway. Plant & Cell Physiology 55: 604–619. [DOI] [PubMed] [Google Scholar]
- Cheng X, Peng J, Ma J, Tang Y, Chen R, Mysore KS, Wen J. 2012. NO APICAL MERISTEM (MtNAM) regulates floral organ identity and lateral organ separation in Medicago truncatula . New Phytologist 195: 71–84. [DOI] [PubMed] [Google Scholar]
- D’haeseleer K, Den Herder G, Laffont C, Plet J, Mortier V, Lelandais‐Brière C, De Bodt S, De Keyser A, Crespi M, Holsters M et al 2011. Transcriptional and post‐transcriptional regulation of a NAC1 transcription factor in Medicago truncatula roots. New Phytologist 191: 647–661. [DOI] [PubMed] [Google Scholar]
- Fang Y, Xie K, Xiong L. 2014. Conserved miR164‐targeted NAC genes negatively regulate drought resistance in rice. Journal of Experimental Botany 65: 2119–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, You J, Xie K, Xie W, Xiong L. 2008. Systematic sequence analysis and identification of tissue‐specific or stress‐responsive genes of NAC transcription factor family in rice. Molecular Genetics and Genomics 280: 547–563. [DOI] [PubMed] [Google Scholar]
- Feng H, Duan X, Zhang Q, Li X, Wang B, Huang L, Wang X, Kang Z. 2014. The target gene of tae‐miR164, a novel NAC transcription factor from the NAM subfamily, negatively regulates resistance of wheat to stripe rust. Molecular Plant Pathology 15: 284–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves B, Hasson A, Belcram K, Cortizo M, Morin H, Nikovics K, Vialette‐Guiraud A, Takeda S, Aida M, Laufs P et al 2015. A conserved role for CUP‐SHAPED COTYLEDON genes during ovule development. The Plant Journal 83: 732–742. [DOI] [PubMed] [Google Scholar]
- Guo H‐S, Xie Q, Fei J‐F, Chua N‐H. 2005. MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell 17: 1376–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X‐J, Mu R‐L, Cao W‐H, Zhang Z‐G, Zhang J‐S, Chen S‐Y. 2005. AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. The Plant Journal 44: 903–916. [DOI] [PubMed] [Google Scholar]
- Hibara K‐I, Karim MR, Takada S, Taoka K‐I, Furutani M, Aida M, Tasaka M. 2006. Arabidopsis CUP‐SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation. Plant Cell 18: 2946–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y, Zhang H, Huang L, Li D, Song F. 2016. Overexpression of a stress‐responsive NAC transcription factor gene ONAC022 improves drought and salt tolerance in rice. Frontiers in Plant Science 7: 419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, Xiong L. 2006. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proceedings of the National Academy of Sciences, USA 103: 12987–12992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, You J, Fang Y, Zhu X, Qi Z, Xiong L. 2008. Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Molecular Biology 67: 169–181. [DOI] [PubMed] [Google Scholar]
- Huang D, Wang S, Zhang B, Shang‐Guan K, Shi Y, Zhang D, Liu X, Wu K, Xu Z, Fu X et al 2015. A gibberellin‐mediated DELLA‐NAC signaling cascade regulates cellulose synthesis in rice. Plant Cell 27: 1681–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong JS, Kim YS, Baek KH, Jung H, Ha S‐H, Do Choi Y, Kim M, Reuzeau C, Kim J‐K. 2010. Root‐specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiology 153: 185–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong JS, Kim YS, Redillas MCFR, Jang G, Jung H, Bang SW, Choi YD, Ha S‐H, Reuzeau C, Kim J‐K. 2013. OsNAC5 overexpression enlarges root diameter in rice plants leading to enhanced drought tolerance and increased grain yield in the field. Plant Biotechnology Journal 11: 101–114. [DOI] [PubMed] [Google Scholar]
- Jiang D, Chen W, Dong J, Li J, Yang F, Wu Z, Zhou H, Wang W, Zhuang C. 2018. Overexpression of miR164b‐resistant OsNAC2 improves plant architecture and grain yield in rice. Journal of Experimental Botany 69: 1533–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneda T, Taga Y, Takai R, Iwano M, Matsui H, Takayama S, Isogai A, Che F‐S. 2009. The transcription factor OsNAC4 is a key positive regulator of plant hypersensitive cell death. EMBO Journal 28: 926–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K, Wellmer F, Muiño JM, Ferrier T, Wuest SE, Kumar V, Serrano‐Mislata A, Madueño F, Krajewski P, Meyerowitz EM et al 2010. Orchestration of floral initiation by APETALA1. Science 328: 85–89. [DOI] [PubMed] [Google Scholar]
- Kim JH, Woo HR, Kim J, Lim PO, Lee IC, Choi SH, Hwang D, Nam HG. 2009. Trifurcate feed‐forward regulation of age‐dependent cell death involving miR164 in Arabidopsis. Science 323: 1053–1057. [DOI] [PubMed] [Google Scholar]
- Kondou Y, Higuchi M, Takahashi S, Sakurai T, Ichikawa T, Kuroda H, Yoshizumi T, Tsumoto Y, Horii Y, Kawashima M et al 2009. Systematic approaches to using the FOX hunting system to identify useful rice genes. The Plant Journal 57: 883–894. [DOI] [PubMed] [Google Scholar]
- Lee M‐H, Jeon HS, Kim HG, Park OK. 2017. An Arabidopsis NAC transcription factor NAC4 promotes pathogen‐induced cell death under negative regulation by microRNA164. New Phytologist 214: 343–360. [DOI] [PubMed] [Google Scholar]
- Li W‐Q, Zhang M‐J, Gan P‐F, Qiao L, Yang S‐Q, Miao H, Wang G‐F, Zhang M‐M, Liu W‐T, Li H‐F et al 2017. CLD1/SRL1 modulates leaf rolling by affecting cell wall formation, epidermis integrity and water homeostasis in rice. The Plant Journal 92: 904–923. [DOI] [PubMed] [Google Scholar]
- Liang C, Wang Y, Zhu Y, Tang J, Hu B, Liu L, Ou S, Wu H, Sun X, Chu J et al 2014. OsNAP connects abscisic acid and leaf senescence by fine‐tuning abscisic acid biosynthesis and directly targeting senescence‐associated genes in rice. Proceedings of the National Academy of Sciences, USA 111: 10013–10018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y et al 2015. A Robust CRISPR/Cas9 system for convenient, high‐efficiency multiplex genome editing in monocot and dicot plants. Molecular Plant 8: 1274–1284. [DOI] [PubMed] [Google Scholar]
- Mao C, Ding W, Wu Y, Yu J, He X, Shou H, Wu P. 2007. Overexpression of a NAC‐domain protein promotes shoot branching in rice. New Phytologist 176: 288–298. [DOI] [PubMed] [Google Scholar]
- Mao C, Lu S, Lv B, Zhang B, Shen J, He J, Luo L, Xi D, Chen X, Ming F. 2017. A rice NAC transcription factor promotes leaf senescence via ABA biosynthesis. Plant Physiology 174: 1747–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew IE, Das S, Mahto A, Agarwal P. 2016. Three rice NAC transcription factors heteromerize and are associated with seed size. Frontiers in Plant Science 7: 2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Tran L‐SP, Van Nguyen D, Fujita M, Maruyama K, Todaka D, Ito Y, Hayashi N, Shinozaki K, Yamaguchi‐Shinozaki K. 2007. Functional analysis of a NAC‐type transcription factor OsNAC6 involved in abiotic and biotic stress‐responsive gene expression in rice. The Plant Journal 51: 617–630. [DOI] [PubMed] [Google Scholar]
- Nikovics K, Blein T, Peaucelle A, Ishida T, Morin H, Aida M, Laufs P. 2006. The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell 18: 2929–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuruzzaman M, Manimekalai R, Sharoni AM, Satoh K, Kondoh H, Ooka H, Kikuchi S. 2010. Genome‐wide analysis of NAC transcription factor family in rice. Gene 465: 30–44. [DOI] [PubMed] [Google Scholar]
- Raman S, Greb T, Peaucelle A, Blein T, Laufs P, Theres K. 2008. Interplay of miR164, CUP‐SHAPED COTYLEDON genes and LATERAL SUPPRESSOR controls axillary meristem formation in Arabidopsis thaliana . The Plant Journal 55: 65–76. [DOI] [PubMed] [Google Scholar]
- Redillas MCFR, Jeong JS, Kim YS, Jung H, Bang SW, Choi YD, Ha S‐H, Reuzeau C, Kim J‐K. 2012. The overexpression of OsNAC9 alters the root architecture of rice plants enhancing drought resistance and grain yield under field conditions. Plant Biotechnology Journal 10: 792–805. [DOI] [PubMed] [Google Scholar]
- Rubio‐Somoza I, Zhou C‐M, Confraria A, Martinho C, von Born P, Baena‐Gonzalez E, Wang J‐W, Weigel D. 2014. Temporal control of leaf complexity by miRNA‐regulated licensing of protein complexes. Current Biology 24: 2714–2719. [DOI] [PubMed] [Google Scholar]
- Sieber P, Wellmer F, Gheyselinck J, Riechmann JL, Meyerowitz EM. 2007. Redundancy and specialization among plant microRNAs: role of the MIR164 family in developmental robustness. Development 134: 1051–1060. [DOI] [PubMed] [Google Scholar]
- Song S‐Y, Chen Y, Chen J, Dai X‐Y, Zhang W‐H. 2011. Physiological mechanisms underlying OsNAC5‐dependent tolerance of rice plants to abiotic stress. Planta 234: 331–345. [DOI] [PubMed] [Google Scholar]
- Souer E, van Houwelingen A, Kloos D, Mol J, Koes R. 1996. The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 85: 159–170. [DOI] [PubMed] [Google Scholar]
- Sun L, Zhang H, Li D, Huang L, Hong Y, Ding XS, Nelson RS, Zhou X, Song F. 2012. Functions of rice NAC transcriptional factors, ONAC122 and ONAC131, in defense responses against Magnaporthe grisea . Plant Molecular Biology 81: 41–56. [DOI] [PubMed] [Google Scholar]
- Takada S, Hibara K, Ishida T, Tasaka M. 2001. The CUP‐SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development 128: 1127–1135. [DOI] [PubMed] [Google Scholar]
- Vroemen CW, Mordhorst AP, Albrecht C, Kwaaitaal MACJ, de Vries SC. 2003. The CUP‐SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell 15: 1563–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WQ, Wang J, Wu YY, Li DW, Allan AC, Yin XR. 2019. Genome‐wide analysis of coding and non‐coding RNA reveals a conserved miR164‐ NAC regulatory pathway for fruit ripening. New Phytologist 225: 1618–1634. [DOI] [PubMed] [Google Scholar]
- Weir I, Lu J, Cook H, Causier B, Schwarz‐Sommer Z, Davies B. 2004. CUPULIFORMIS establishes lateral organ boundaries in Antirrhinum . Development 131: 915–922. [DOI] [PubMed] [Google Scholar]
- Wu R‐M, Wood M, Thrush A, Walton EF, Varkonyi‐Gasic E. 2007. Real‐Time PCR Quantification of plant miRNAs using Universal ProbeLibrary technology. Biochemica 2: 1–4. [Google Scholar]
- Xiang J‐J, Zhang G‐H, Qian Q, Xue H‐W. 2012. Semi‐rolled leaf1 encodes a putative glycosylphosphatidylinositol‐anchored protein and modulates rice leaf rolling by regulating the formation of bulliform cells. Plant Physiology 159: 1488–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q, Sanz‐Burgos AP, Guo H, García JA, Gutiérrez C. 1999. GRAB proteins, novel members of the NAC domain family, isolated by their interaction with a geminivirus protein. Plant Molecular Biology 39: 647–656. [DOI] [PubMed] [Google Scholar]
- Xue W, Xing Y, Weng X, Zhao Y, Tang W, Wang L, Zhou H, Yu S, Xu C, Li X et al 2008. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nature Genetics 40: 761–767. [DOI] [PubMed] [Google Scholar]
- Yi X, Zhang Z, Ling Y, Xu W, Su Z. 2014. PNRD: a plant non‐coding RNA database. Nucleic Acids Research 43: D982–D989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokotani N, Tsuchida‐Mayama T, Ichikawa H, Mitsuda N, Ohme‐Takagi M, Kaku H, Minami E, Nishizawa Y. 2014. OsNAC111, a blast disease‐responsive transcription factor in rice, positively regulates the expression of defense‐related genes. Molecular Plant–Microbe Interactions 27: 1027–1034. [DOI] [PubMed] [Google Scholar]
- Yoshii M, Shimizu T, Yamazaki M, Higashi T, Miyao A, Hirochika H, Omura T. 2009. Disruption of a novel gene for a NAC‐domain protein in rice confers resistance to Rice dwarf virus. The Plant Journal 57: 615–625. [DOI] [PubMed] [Google Scholar]
- Zheng G, Wei W, Li Y, Kan L, Wang F, Zhang X, Li F, Liu Z, Kang C. 2019. Conserved and novel roles of miR164 ‐CUC2 regulatory module in specifying leaf and floral organ morphology in strawberry. New Phytologist 224: 480–492. [DOI] [PubMed] [Google Scholar]
- Zheng X, Chen B, Lu G, Han B. 2009. Overexpression of a NAC transcription factor enhances rice drought and salt tolerance. Biochemical and Biophysical Research Communications 379: 985–989. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Original blot images from this study.
Fig. S2 Sequence analysis for OsCUC1 and OsCUC3.
Fig. S3 Loss‐of‐function of OsCUC1 and OsCUC3 generated by CRISPR/Cas9 in rice.
Fig. S4 Boundary specification defects in the vegetative growing stage of the rice oscuc1‐KO1 mutant.
Fig. S5 Pollen defects in the rice oscuc1‐KO1 mutant.
Fig. S6 Phenotypes of heterozygous mutants of OsCUC1 and OsCUC3 in rice.
Fig. S7 Dimerization of the Arabidopsis CUC proteins.
Fig. S8 The development of rice oscuc1 oscuc3 homozygous double mutants is arrested at the seedling stage.
Fig. S9 Transcript evidence and expression pattern for rice osa‐miR64c.
Fig. S10 The expression patterns for the other five osa‐miR164 targets in rice.
Fig. S11 Knocking out OMTN4 or OMTN6 does not lead to defects either in boundary specification or leaf development in rice.
Fig. S12 The CLD1 expression level does not significantly change in the rice oscuc1 mutant.
Fig. S13 CLD1 does not interact with OsCUC3, OMTN4 or OMTN6 in rice.
Table S1 The accession numbers of the proteins listed in the phylogenetic tree.
Table S2 The primer sequences used in this study.
Table S3 Percentages of aberrant florets of oscuc1‐KO and oscuc3‐KO plants.
Table S4 Percentages of aberrant florets of omtn4‐KO and omtn6‐KO plants.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.


