Abstract
Background
In cancer, kappa B-interacting protein (IKBIP) has rarely been reported. This study aimed at investigating its expression pattern and biological function in brain glioma at the transcriptional level.
Methods
We selected 301 glioma patients with microarray data from CGGA database and 697 glioma patients with RNAseq data from TCGA database. Transcriptional data and clinical data of 998 samples were analyzed. Statistical analysis and figure generating were performed with R language.
Results
We found that IKBIP expression showed positive correlation with WHO grade of glioma. IKBIP was increased in isocitrate dehydrogenase (IDH) wild type and mesenchymal molecular subtype of glioma. Gene ontology analysis demonstrated that IKBIP was profoundly associated with extracellular matrix organization, cell–substrate adhesion and response to wounding in both pan-glioma and glioblastoma. Subsequent gene set enrichment analysis revealed that IKBIP was particularly correlated with epithelial-to-mesenchymal transition (EMT). To further elucidate the relationship between IKBIP and EMT, we performed gene set variation analysis to screen the EMT-related signaling pathways and found that IKBIP expression was significantly associated with PI3K/AKT, hypoxia and TGF-β pathway. Moreover, IKBIP expression was found to be synergistic with key biomarkers of EMT, especially with N-cadherin, vimentin, snail, slug and TWIST1. Finally, higher IKBIP indicated significantly shorter survival for glioma patients.
Conclusions
IKBIP was associated with more aggressive phenotypes of gliomas. Furthermore, IKBIP was significantly involved in EMT and could serve as an independent prognosticator in glioma.
Keywords: glioma, IKBIP, epithelial-to-mesenchymal transition, prognosis
1. Introduction
Gliomas account for the most common and aggressive primary brain cancers among adult patients [1]. Despite great advances in diagnosis and treatment, the prognosis for glioma patients remains unfavorable. Especially for those with glioblastoma (GBM), the most devastating type, the median survival time is only about 15 months [2,3]. Epithelial-to-mesenchymal transition (EMT) has been widely reported as a key mechanism in promoting migration, invasion and tumor progression in glioma [4]. Identification of novel EMT-related markers is of great necessity.
I kappa B kinase interacting protein (IKBIP), also known as IKIP, is on human chromosome 12. Researchers have paid little attention to this gene. Currently, we know that this gene promotes the function of apoptosis. IKBIP was found to be one of the target genes of p53 and plays a crucial role in proapoptotic function [5]. Recently, IKBIP was identified as a vital modulator of inflammation [6]. Heretofore, the biological function of IKBIP in malignancies has been rarely reported. Only one study [7], through weighted gene co-expression network analysis (WGCNA), preliminarily revealed IKBIP as a potential hub gene in gliomagenesis. However, the role of IKBIP in glioma still remains largely unclear. In the present study, we took advantage of 998 glioma patients with transcriptome data to investigate the clinical significance, molecular characterization and biological function of IKBIP in glioma.
2. Materials and methods
2.1. Sample and data collection
Transcriptome and clinical data of glioma patients were available in Chinese Glioma Genome Atlas (CGGA) website (http://www.cgga.org.cn/) and TCGA website (http://cancergenome.nih.gov/). In total, 998 glioma patients, including 301 CGGA microarray data (GeneSpring GX 11.0 normalization) and 697 TCGA RNAseq data (RSEM normalization, level 3), were enrolled. The baseline characteristics of patients in both cohorts were described in Table S1.
First, we took advantage of CGGA microarray data to explore the IKBIP expression status in pan-glioma and GBM. Then TCGA RNAseq data were analyzed in parallel to further validate what we have revealed in CGGA data set and found that the consistency of results between two cohorts was fairly satisfying.
2.2. Statistical analysis
For TCGA cohort, RSEM RNAseq data were log2 transformed. For CGGA cohort, microarray data (already normalized and centered by data provider) were directly analyzed. Statistical analysis was performed with R language. Multiple R packages, including ggplot2, pROC [8], pheatmap, corrgram, circlize [9], gene set variation analysis (GSVA), as well as survival, were used to generate figures. The biological processes of IKBIP-related genes were annotated using Metascape [10] (https://metascape.org). Hallmark gene sets were downloaded from gene set enrichment analysis (GSEA) website (http://software.broadinstitute.org/) for GSEA [11] and GSVA [12]. All statistical tests were two sided, and a p value of <0.05 was considered statistically significant.
3. Results
3.1. IKBIP expression was correlated with aggressive phenotypes of glioma
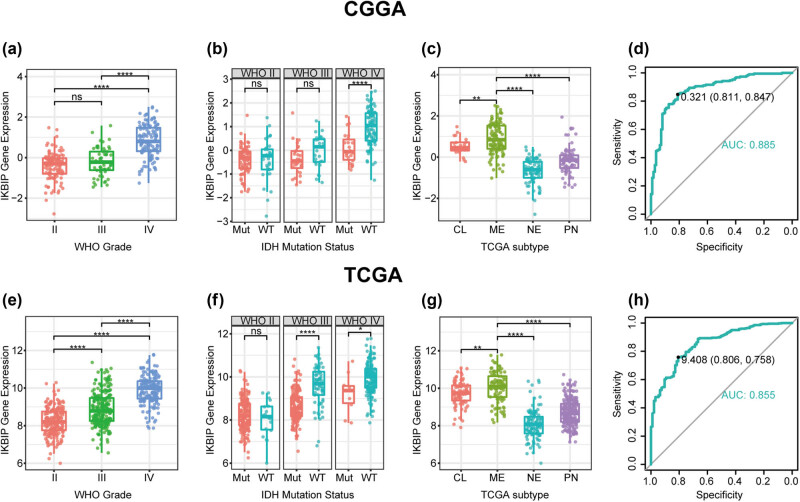
IKBIP expression levels were compared across different WHO grades. The results of both CGGA and TCGA cohorts consistently showed a significant positive correlation between IKBIP expression and WHO grade, except for comparison between WHO II and WHO III in CGGA, which also showed an apparent trend (Figure 1a and e). In addition, when patients were subclassified with respect to isocitrate dehydrogenase (IDH) mutation status, IDH wild type was found to be more associated with an increased pattern of IKBIP expression in both data sets, even though no statistical significance was reached in some groups (Figure 1b and f). These results suggested that higher IKBIP was paralleled with higher malignancy in glioma. Moreover, the correlation between IKBIP and TCGA molecular subtype was further examined. As shown in Figure 1c and g, IKBIP expression in mesenchymal subtype was significantly upregulated compared to the other subtypes, suggesting that IKBIP expression could contribute as a specific marker for mesenchymal subtype. The ROC curves were subsequently performed to evaluate the performance of IKBIP in distinguishing mesenchymal subtype. Areas under curves were 88.5% in CGGA and 85.5% in TCGA (Figure 1d and h).
Figure 1.
IKBIP expression in CGGA and TCGA data set according to the WHO grade (a and e), IDH mutation status (b and f), TCGA molecular subtype (c and g) and ROC curves (d and h) for distinguishing mesenchymal subtype. Mut, mutation; WT, wild type; CL, classical; ME, mesenchymal; NE, neural; PN, proneural. * indicates p value < 0.05, **indicates p value < 0.01, *** indicates p value < 0.001, **** indicates p value < 0.0001.
3.2. IKBIP-related biological process
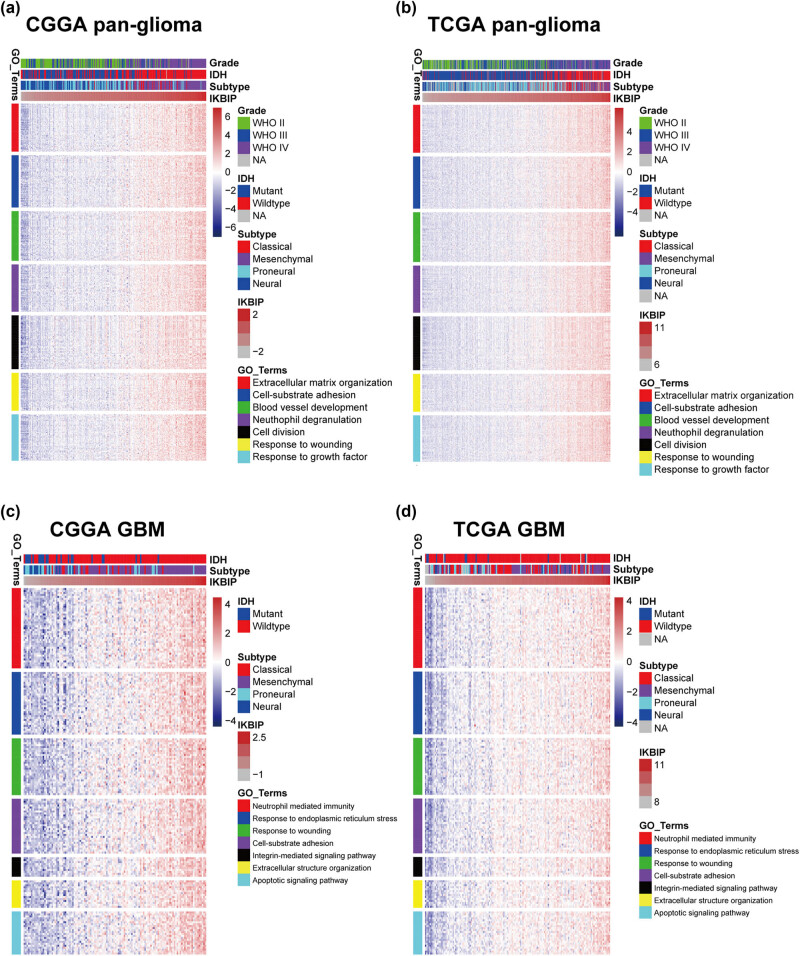
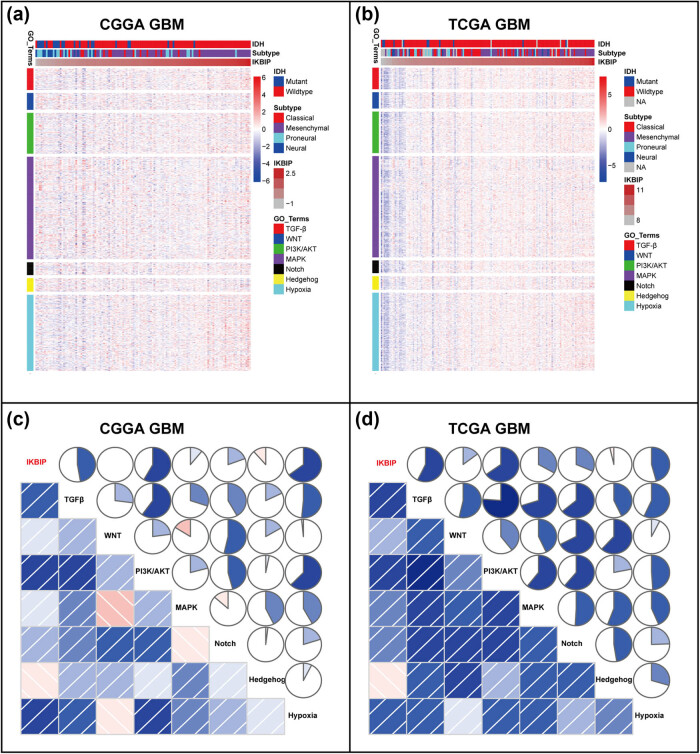
To explore the biological process of IKBIP in glioma, Pearson correlation test was performed between IKBIP and other genes. With the criteria of Pearson coefficient |r| > 0.6, we identified 711 IKBIP positively correlated genes and 462 IKBIP negatively correlated genes in CGGA, and 938 IKBIP positively correlated genes and 17 IKBIP negatively correlated genes in TCGA. To ensure accuracy, IKBIP significantly correlated genes that were overlapped between both data sets were selected for Gene Ontology (GO) analysis. A Venn diagram (Figure S1a) was constructed, illustrating an overlap of 376 IKBIP positively correlated genes (Table S2), which were subsequently annotated with GO analysis. We found that IKBIP positively correlated genes were mainly involved in EMT-related biological processes, including extracellular structure organization, cell–substrate adhesion, blood vessel development, response to wounding, and response to growth factor. Other biological processes included neutrophil degranulation and cell division, pointing toward an association between IKBIP and regulation of immune response and cell cycle, respectively (Figure 2a and b).
Figure 2.
Gene ontology analysis for IKBIP in pan-glioma (a and b) and GBM (c and d). NA, not available.
In view of GBM as a distinct subgroup of glioma, we then conducted an independent GO enrichment analysis in this group. In GBM of both data sets, Venn diagram (Figure S1b) exhibited an overlap of 191 IKBIP positively correlated genes (Table S3). Subsequent GO analysis revealed that these genes were also significantly associated with EMT-related biological processes, including response to endoplasmic reticulum stress [13], response to wounding, cell–substrate adhesion, integrin-mediated signaling pathway [14] and extracellular structure organization. Besides, IKBIP seemed to be more associated with neutrophil-mediated immunity, suggesting that IKBIP upregulation was accompanied by immunosuppression of GBM, which indicated a more malignant characteristic in glioma. While it should be noted that IKBIP showed positive correlation with apoptotic-signaling pathway, enlightening us that IKBIP might also act as a proapoptotic factor in GBM [5] (Figure 2c and d).
3.3. IKBIP was associated with EMT
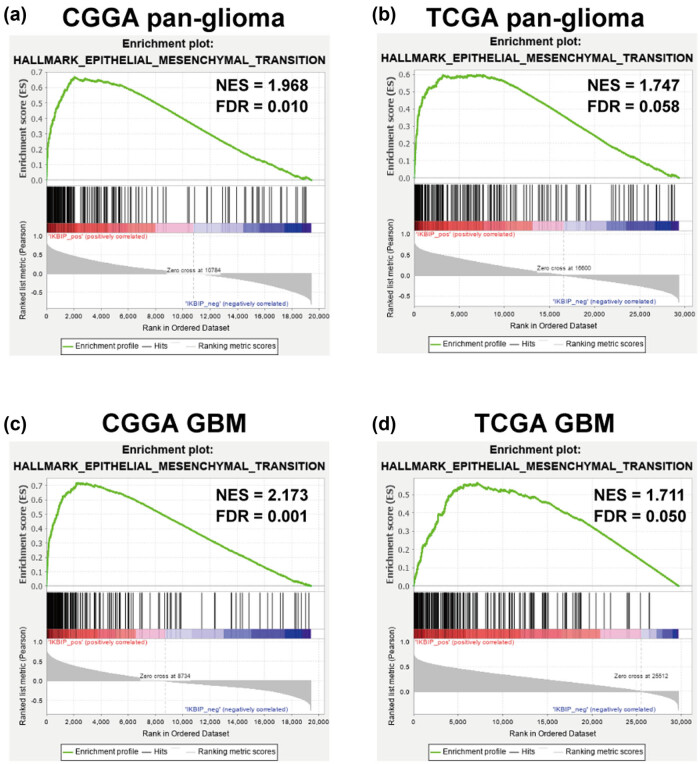
GSEAs were performed in both CGGA and TCGA data sets, and it turned out that IKBIP was significantly correlated with EMT in CGGA (normalized enrichment score (NES) = 1.968, false discovery rate (FDR) = 0.010; Figure 3a), which was further validated in TCGA (NES = 1.747, FDR = 0.058; Figure 3b). Furthermore, in GBM, IKBIP showed an even higher association with EMT in both cohorts (Figure 3c and d). These results indicated that IKBIP could be profoundly associated with EMT phenotype in glioma.
Figure 3.
GSEA for enrichment of EMT according to IKBIP expression in pan-glioma (a and b) and GBM (c and d). NES, normalized enrichment score; FDR, false discovery rate.
3.4. IKBIP interacted with EMT-related signaling pathways in glioma
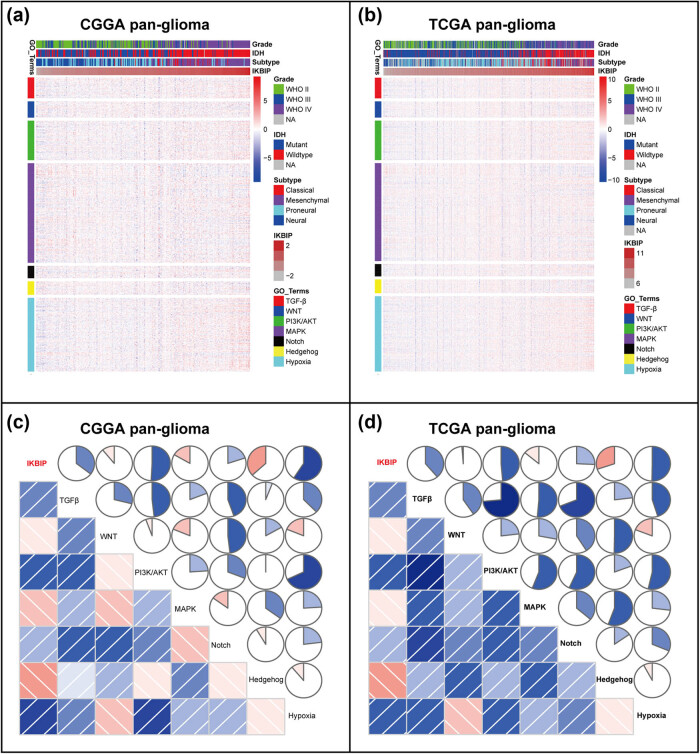
To further investigate the relationship between IKBIP and EMT, we downloaded seven gene sets from GSEA website (Table S4), which were subsequently transformed into metagenes, representing different EMT-related signaling pathways, summarized by Gonzalez et al. [15]. As shown in Figure 4a and b, three clusters, including TGF-β-, PI3K/AKT-, and hypoxia-signaling pathways, were significantly associated with IKBIP expression. To quantify what we observed in clusters, GSVA was performed to generate seven metagenes based on the corresponding genes of seven EMT-related signaling pathways. According to the Pearson r value between IKBIP and seven metagenes, Corrgrams were generated to evaluate their interrelations (Figure 4c and d). IKBIP showed a robust correlation with TGF-β, PI3K/AKT and hypoxia signaling pathway, while only showed a weak correlation with WNT, MAPK, NOTCH and HEDGEHOG pathway, in consistent with what we observed in Figure 4a and b. Moreover, a similar pattern of EMT-related signaling pathways was observed in GBM of both CGGA and TCGA data set (Figure 5).
Figure 4.
Cluster (a and b) and GSVA (c and d) of IKBIP-related EMT-signaling pathways in pan-glioma. NA, not available.
Figure 5.
Cluster (a and b) and GSVA (c and d) of IKBIP-related EMT-signaling pathways in GBM. NA, not available.
3.5. IKBIP interacted with EMT-related key biomarkers in glioma
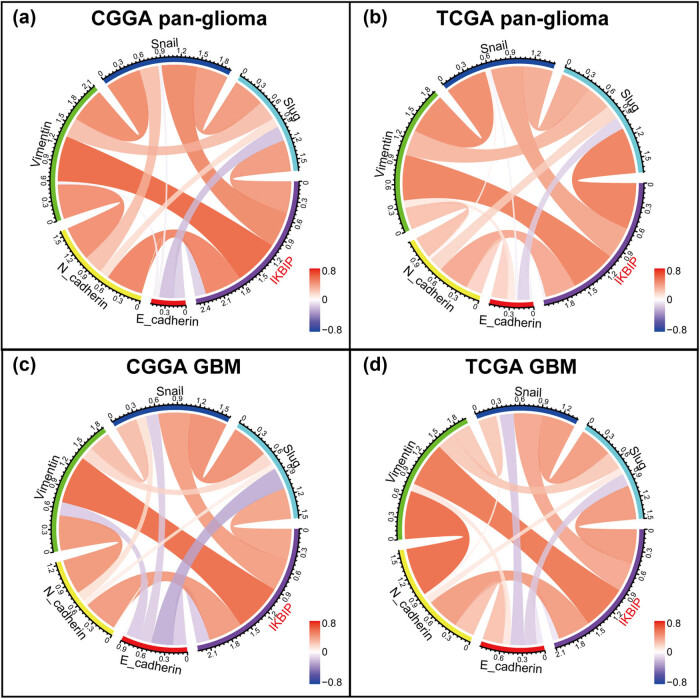
To further validate the role of IKBIP in EMT-related signaling pathways, we examined the correlation between IKBIP and EMT-related key biomarkers, including E-cadherin, N-cadherin, vimentin, snail and slug. Circos plots revealed that IKBIP expression was significantly associated with N-cadherin, vimentin, snail and slug (Figure 6a and b). To further demonstrate the interaction of these markers in GBM, Pearson correlation tests were additionally performed. As shown in Figure 6c and d, the correlation between IKBIP and these markers in GBM was also very robust in both data sets, indicating synergistic effects of these members during glioma EMT. The correlation between IKBIP and E-cadherin was very weak, and this might be deemed as a noise.
Figure 6.
Correlation of IKBIP and EMT key biomarkers.
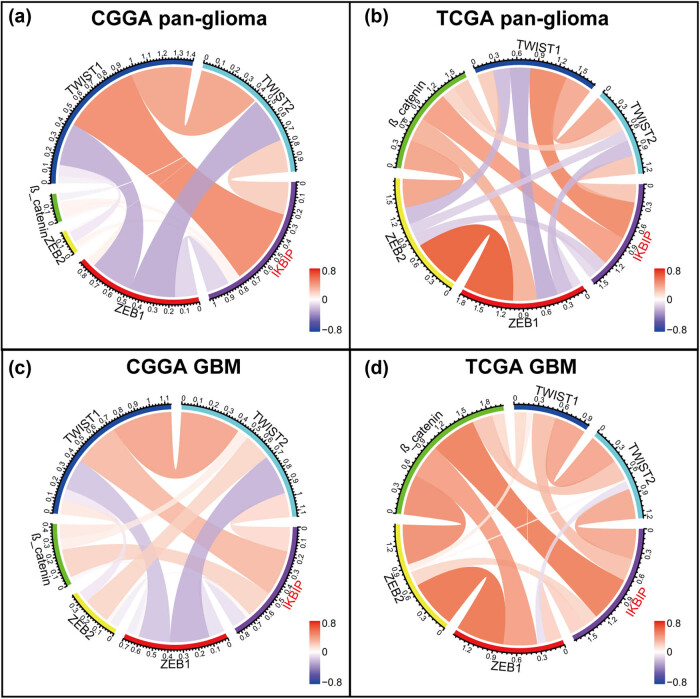
In EMT, many other molecules have been identified as EMT-related key biomarkers [16]. We additionally enrolled EMT-related markers including ZEB1/2, β-catenin and TWIST1/2 and put them into analysis together with IKBIP. Subsequent Circos plots in both CGGA and TCGA congruently revealed that IKBIP expression was especially correlated with TWIST1 in both pan-glioma and GBM (Figure 7).
Figure 7.
Correlation of IKBIP and other EMT key biomarkers.
3.6. Higher IKBIP predicts shorter survival for glioma
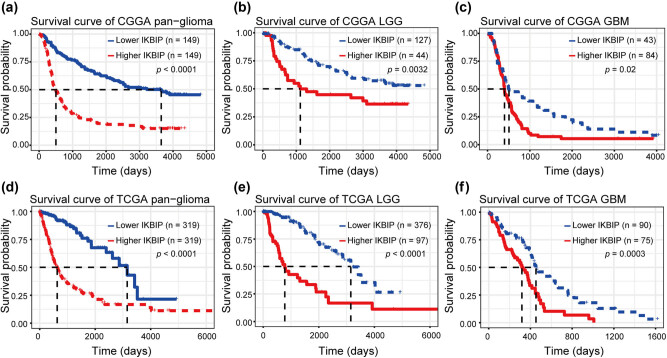
Kaplan–Meier (KM) survival analyses were performed to examine the prognostic value of IKBIP in glioma. According to IKBIP expression, pan-glioma samples were divided into two groups in each data set. As shown in Figure 8a and d, a higher level of IKBIP expression predicted a significantly shorter survival. Moreover, a similar pattern of the KM survival curve was observed among patients with lower grade glioma (LGG) (Figure 8b and e) and GBM (Figure 8c and f).
Figure 8.
Survival analysis for IKBIP in pan-glioma (a and d), LGG (b and e) and GBM (c and f).
4. Discussion
In the present study, we investigated the transcriptional expression profiles of IKBIP in 998 glioma patients and revealed that IKBIP expression showed significant positive correlation with the WHO grade of glioma. Furthermore, higher IKBIP expression was usually accompanied by a more aggressive and malignant phenotype in glioma, including GBM, IDH wild type and mesenchymal subtype. Moreover, higher IKBIP expression indicated a significantly shorter survival for patients with glioma, across different WHO grades. These findings suggested that IKBIP played a vital role in the malignant progression of gliomas, in line with the results of a previous WGCNA study [7]. Understanding the molecular mechanism of IKBIP in glioma may provide a novel therapeutic target to overcome this fatal disease.
To elucidate the biological function of IKBIP in glioma, GO analysis was performed, and it turned out that IKBIP was highly associated with a series of EMT-related biological processes, including extracellular matrix organization, cell–substrate adhesion and response to wounding in both pan-glioma and GBM. Subsequent GSEA analysis revealed remarkable evidence because IKBIP was particularly correlated with EMT, which had been extensively confirmed to play a key role not only in glioma migration/invasion but also in glioma recurrence and therapeutic resistance [17,18,19]. These results enlightened us that IKBIP might promote tumorigenesis and progression of glioma mainly by means of EMT induction, which has yet been previously reported. Besides, GO analysis also revealed that IKBIP played a crucial role in tumor-induced immune and inflammatory response in glioma, especially in GBM, in line with the results presented by Wu et al. [6]. They demonstrated that IKBIP played an inhibitory role in immune and inflammatory response through negative regulation of NF-κB pathway. Based on these, we concluded that apart from being a key molecule for EMT induction, IKBIP might contribute as an immune suppressor in glioma as well, which further validated its oncogenic role in glioma. Meanwhile, it was noteworthy that IKBIP showed robust correlation with apoptotic-signaling pathway in GBM, suggesting a potential proapoptotic function [5]. As a result, we speculated that IKBIP might have a dualistic nature in gliomagenesis, and the robust protumoral effect through EMT induction and immune inhibition overwhelmed the antitumoral effect through proapoptotic function.
To further validate the pro-EMT effect of IKBIP in glioma, we selected a series of EMT-related signaling pathways and biomarkers, which were then analyzed to determine their interaction with IKBIP and found that IKBIP showed robust correlation with PI3K/AKT-, hypoxia- and TGF-β-signaling pathway, suggesting that IKBIP might promote EMT process through these pathways. Moreover, most of EMT biomarkers, including N-cadherin, snail, slug, vimentin and TWIST1 were significantly associated with IKBIP, indicating that IKBIP interacted synergistically with these key molecules of EMT. These results further validated the involvement of IKBIP in glioma EMT. Thus, our findings might bring a novel EMT target for potential glioma treatment.
In conclusion, IKBIP expression was associated with more aggressive phenotypes of glioma and predicted much worse survival for patients. Moreover, IKBIP was significantly associated with EMT process and interacted synergistically with EMT-related signaling pathways and key biomarkers. However, a limitation of the current study was that no experimental validation was performed. Further in vitro and in vivo studies are needed to validate its role in glioma.
Acknowledgments
We appreciate the generosity of CGGA and TCGA projects for sharing data.
Footnotes
Funding: This work was supported by Medical Scientific Research Foundation of Shenzhen Health Commission (szfz2018022), Shenzhen Science and Technology Innovation Foundation (JCYJ20190806150005453) and Futian Public Welfare Scientific Research Project (FTWS2020099).
Author contributions: Ying Yang performed the analysis and wrote the manuscript. Jin Wang and Shihai Xu designed the study and reviewed the manuscript. Wen Lv, Fei Shi and Aijun Shan provided technical support and analyzed the data. Ying Yang and Shihai Xu provided financial support. All of the authors read and approved the final manuscript.
Conflict of interest: Authors state no conflict of interest.
Contributor Information
Ying Yang, Email: 15914141979@139.com.
Jin Wang, Email: szph3022@szhospital.com.
Shihai Xu, Email: heykojnu@163.com.
References
- [1].Meng X, Zhao Y, Han B, Zha C, Zhang Y, Li Z, et al. Dual functionalized brain-targeting nanoinhibitors restrain temozolomide-resistant glioma via attenuating EGFR and MET signaling pathways. Nat Commun. 2020;11(1):594. [DOI] [PMC free article] [PubMed]; Meng X, Zhao Y, Han B, Zha C, Zhang Y, Li Z. et al. Dual functionalized brain-targeting nanoinhibitors restrain temozolomide-resistant glioma via attenuating EGFR and MET signaling pathways. Nat Commun. 2020;11(1):594. doi: 10.1038/s41467-019-14036-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Yang W, Wu PF, Ma JX, Liao MJ, Wang XH, Xu LS, et al. Sortilin promotes glioblastoma invasion and mesenchymal transition through GSK-3β/β-catenin/twist pathway. Cell Death Dis. 2019;10(3):208. [DOI] [PMC free article] [PubMed]; Yang W, Wu PF, Ma JX, Liao MJ, Wang XH, Xu LS. et al. Sortilin promotes glioblastoma invasion and mesenchymal transition through GSK-3β/β-catenin/twist pathway. Cell Death Dis. 2019;10(3):208. doi: 10.1038/s41419-019-1449-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Wei J, Ouyang X, Tang Y, Li H, Wang B, Ye Y, et al. ER-stressed MSC displayed more effective immunomodulation in RA CD4(+)CXCR5(+)ICOS(+) follicular helper-like T cells through higher PGE2 binding with EP2/EP4. Mod Rheumatol. 2019;30(3):509–16. [DOI] [PubMed]; Wei J, Ouyang X, Tang Y, Li H, Wang B, Ye Y. et al. ER-stressed MSC displayed more effective immunomodulation in RA CD4(+)CXCR5(+)ICOS(+) follicular helper-like T cells through higher PGE2 binding with EP2/EP4. Mod Rheumatol. 2019;30(3):509–16. doi: 10.1080/14397595.2019.1651446. [DOI] [PubMed] [Google Scholar]
- [4].Ma YS, Wu ZJ, Bai RZ, Dong H, Xie BX, Wu XH, et al. DRR1 promotes glioblastoma cell invasion and epithelial-mesenchymal transition via regulating AKT activation. Cancer Lett. 2018;423:86–94. [DOI] [PubMed]; Ma YS, Wu ZJ, Bai RZ, Dong H, Xie BX, Wu XH. et al. DRR1 promotes glioblastoma cell invasion and epithelial-mesenchymal transition via regulating AKT activation. Cancer Lett. 2018;423:86–94. doi: 10.1016/j.canlet.2018.03.015. [DOI] [PubMed] [Google Scholar]
- [5].Hofer-Warbinek R, Schmid JA, Mayer H, Winsauer G, Orel L, Mueller B, et al. A highly conserved proapoptotic gene, IKIP, located next to the APAF1 gene locus, is regulated by p53. Cell Death Differ. 2004;11(12):1317–25. [DOI] [PubMed]; Hofer-Warbinek R, Schmid JA, Mayer H, Winsauer G, Orel L, Mueller B. et al. A highly conserved proapoptotic gene, IKIP, located next to the APAF1 gene locus, is regulated by p53. Cell Death Differ. 2004;11(12):1317–25. doi: 10.1038/sj.cdd.4401502. [DOI] [PubMed] [Google Scholar]
- [6].Wu H, Liu H, Zhao X, Zheng Y, Liu B, Zhang L, et al. IKIP negatively regulates NF-kappaB activation and inflammation through inhibition of IKKalpha/beta phosphorylation. J Immunol. 2020;204(2):418–27. [DOI] [PubMed]; Wu H, Liu H, Zhao X, Zheng Y, Liu B, Zhang L. et al. IKIP negatively regulates NF-kappaB activation and inflammation through inhibition of IKKalpha/beta phosphorylation. J Immunol. 2020;204(2):418–27. doi: 10.4049/jimmunol.1900626. [DOI] [PubMed] [Google Scholar]
- [7].Chen TY, Liu Y, Chen L, Luo J, Zhang C, Shen XF. Identification of the potential biomarkers in patients with glioma: a weighted gene co-expression network analysis. Carcinogenesis. 2019;41(6):743–50. [DOI] [PMC free article] [PubMed]; Chen TY, Liu Y, Chen L, Luo J, Zhang C, Shen XF. Identification of the potential biomarkers in patients with glioma: a weighted gene co-expression network analysis. Carcinogenesis. 2019;41(6):743–50. doi: 10.1093/carcin/bgz194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinforma. 2011;12:77. [DOI] [PMC free article] [PubMed]; Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC. et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinforma. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Gu Z, Gu L, Eils R, Schlesner M, Brors B. Circlize implements and enhances circular visualization in R. Bioinformatics. 2014;30(19):2811–2. [DOI] [PubMed]; Gu Z, Gu L, Eils R, Schlesner M, Brors B. Circlize implements and enhances circular visualization in R. Bioinformatics. 2014;30(19):2811–2. doi: 10.1093/bioinformatics/btu393. [DOI] [PubMed] [Google Scholar]
- [10].Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. [DOI] [PMC free article] [PubMed]; Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–50. [DOI] [PMC free article] [PubMed]; Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinforma. 2013;14:7. [DOI] [PMC free article] [PubMed]; Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinforma. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Shin HS, Ryu ES, Oh ES, Kang DH. Endoplasmic reticulum stress as a novel target to ameliorate epithelial-to-mesenchymal transition and apoptosis of human peritoneal mesothelial cells. Lab Invest. 2015;95(10):1157–73. [DOI] [PubMed]; Shin HS, Ryu ES, Oh ES, Kang DH. Endoplasmic reticulum stress as a novel target to ameliorate epithelial-to-mesenchymal transition and apoptosis of human peritoneal mesothelial cells. Lab Invest. 2015;95(10):1157–73. doi: 10.1038/labinvest.2015.91. [DOI] [PubMed] [Google Scholar]
- [14].Park H, Kim D, Kim D, Park J, Koh Y, Yoon SS. Truncation of MYH8 tail in AML: a novel prognostic marker with increase cell migration and epithelial-mesenchymal transition utilizing RAF/MAPK pathway. Carcinogenesis. 2019;41(6):817–27. [DOI] [PubMed]; Park H, Kim D, Kim D, Park J, Koh Y, Yoon SS. Truncation of MYH8 tail in AML: a novel prognostic marker with increase cell migration and epithelial-mesenchymal transition utilizing RAF/MAPK pathway. Carcinogenesis. 2019;41(6):817–27. doi: 10.1093/carcin/bgz146. [DOI] [PubMed] [Google Scholar]
- [15].Gonzalez DM, Medici D. Signaling mechanisms of the epithelial-mesenchymal transition. Sci Signal. 2014;7(344):re8. [DOI] [PMC free article] [PubMed]; Gonzalez DM, Medici D. Signaling mechanisms of the epithelial-mesenchymal transition. Sci Signal. 2014;7(344):re8. doi: 10.1126/scisignal.2005189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Xu J, Zhang Z, Qian M, Wang S, Qiu W, Chen Z, et al. Cullin-7 (CUL7) is overexpressed in glioma cells and promotes tumorigenesis via NF-κB activation. J Exp Clin Cancer Res. 2020;39(1):59. [DOI] [PMC free article] [PubMed]; Xu J, Zhang Z, Qian M, Wang S, Qiu W, Chen Z. et al. Cullin-7 (CUL7) is overexpressed in glioma cells and promotes tumorigenesis via NF-κB activation. J Exp Clin Cancer Res. 2020;39(1):59. doi: 10.1186/s13046-020-01553-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Li C, Zheng H, Hou W, Bao H, Xiong J, Che W, et al. Long non-coding RNA linc00645 promotes TGF-β-induced epithelial-mesenchymal transition by regulating miR-205-3p-ZEB1 axis in glioma. Cell Death Dis. 2019;10(10):717. [DOI] [PMC free article] [PubMed] [Retracted]; Li C, Zheng H, Hou W, Bao H, Xiong J, Che W. et al. Long non-coding RNA linc00645 promotes TGF-β-induced epithelial-mesenchymal transition by regulating miR-205-3p-ZEB1 axis in glioma. Cell Death Dis. 2019;10(10):717. doi: 10.1038/s41419-019-1948-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [18].Li H, Li J, Chen L, Qi S, Yu S, Weng Z, et al. HERC3-mediated SMAD7 ubiquitination degradation promotes autophagy-induced EMT and chemoresistance in glioblastoma. Clin Cancer Res. 2019;25(12):3602–16. [DOI] [PubMed]; Li H, Li J, Chen L, Qi S, Yu S, Weng Z. et al. HERC3-mediated SMAD7 ubiquitination degradation promotes autophagy-induced EMT and chemoresistance in glioblastoma. Clin Cancer Res. 2019;25(12):3602–16. doi: 10.1158/1078-0432.CCR-18-3791. [DOI] [PubMed] [Google Scholar]
- [19].Li H, Li J, Zhang G, Da Q, Chen L, Yu S, et al. HMGB1-induced p62 overexpression promotes snail-mediated epithelial-mesenchymal transition in glioblastoma cells via the degradation of GSK-3β. Theranostics. 2019;9(7):1909–22. [DOI] [PMC free article] [PubMed]; Li H, Li J, Zhang G, Da Q, Chen L, Yu S. et al. HMGB1-induced p62 overexpression promotes snail-mediated epithelial-mesenchymal transition in glioblastoma cells via the degradation of GSK-3β. Theranostics. 2019;9(7):1909–22. doi: 10.7150/thno.30578. [DOI] [PMC free article] [PubMed] [Google Scholar]