Abstract
Background
Sirtuin 6 (SIRT6) is a NAD+-dependent deacetylase with key roles in cell metabolism. High SIRT6 expression is associated with adverse prognosis in breast cancer (BC) patients. However, the mechanisms through which SIRT6 exerts its pro-oncogenic effects in BC remain unclear. Here, we sought to define the role of SIRT6 in BC cell metabolism and in mouse polyoma middle T antigen (PyMT)-driven mammary tumors.
Methods
We evaluated the effect of a heterozygous deletion of Sirt6 on tumor latency and survival of mouse mammary tumor virus (MMTV)-PyMT mice. The effect of SIRT6 silencing on human BC cell growth was assessed in MDA-MB-231 xenografts. We also analyzed the effect of Sirt6 heterozygous deletion, of SIRT6 silencing, and of the overexpression of either wild-type (WT) or catalytically inactive (H133Y) SIRT6 on BC cell pyruvate dehydrogenase (PDH) expression and activity and oxidative phosphorylation (OXPHOS), including respiratory complex activity, ATP/AMP ratio, AMPK activation, and intracellular calcium concentration.
Results
The heterozygous Sirt6 deletion extended tumor latency and mouse survival in the MMTV-PyMT mouse BC model, while SIRT6 silencing slowed the growth of MDA-MB-231 BC cell xenografts. WT, but not catalytically inactive, SIRT6 enhanced PDH expression and activity, OXPHOS, and ATP/AMP ratio in MDA-MB-231 and MCF7 BC cells. Opposite effects were obtained by SIRT6 silencing, which also blunted the expression of genes encoding for respiratory chain proteins, such as UQCRFS1, COX5B, NDUFB8, and UQCRC2, and increased AMPK activation in BC cells. In addition, SIRT6 overexpression increased, while SIRT6 silencing reduced, intracellular calcium concentration in MDA-MB-231 cells. Consistent with these findings, the heterozygous Sirt6 deletion reduced the expression of OXPHOS-related genes, the activity of respiratory complexes, and the ATP/AMP ratio in tumors isolated from MMTV-PyMT mice.
Conclusions
Via its enzymatic activity, SIRT6 enhances PDH expression and activity, OXPHOS, ATP/AMP ratio, and intracellular calcium concentration, while reducing AMPK activation, in BC cells. Thus, overall, SIRT6 inhibition appears as a viable strategy for preventing or treating BC.
Supplementary Information
The online version contains supplementary material available at 10.1186/s40170-021-00240-1.
Keywords: SIRT6, Breast cancer, Cancer metabolism, Mammary tumorigenesis, Oxidative phosphorylation
Background
Breast cancer (BC) is the most frequently diagnosed malignancy, with 1.7 million new diagnoses/year (23% of cancer diagnoses) [1]. In 2018, there were more than six hundred thousand BC-related deaths globally, and it is estimated that this death toll will rise to one million deaths/year by 2040 without major advances in BC prevention or treatment [1]. Thus, there remains a crucial need to enhance our understanding of BC biology and to define new viable therapeutic targets for this condition.
Neoplastic transformation involves a metabolic reprogramming to support the biosynthesis of macromolecules that are needed for cell division and growth [2]. Such changes are similar to those occurring in highly proliferative normal cells during embryogenesis, wound healing, and immune response [3]. Cancer cells acquire mutations in oncogenes and in tumor suppressors that enhance glycolysis and anabolic metabolism in the absence of external signals (Warburg effect) [4]. In addition, in contrast to Warburg’s initial hypothesis, mitochondrial oxidative phosphorylation (OXPHOS) is also highly active and actually plays a key role in cancer cells. Specifically, while glycolysis represents a source of molecules necessary for biomass enhancement, OXPHOS appears to underlie anabolic metabolism, cell proliferation, cancer stemness, and metastasis [3].
Sirtuin 6 (SIRT6) is a member of the sirtuin family of NAD+-dependent deacetylases. In mammals, seven sirtuin isoforms exist (SIRT1-7), with different subcellular localization, catalytic activity, targets, and functions. SIRT6 is a multifunctional nuclear protein, involved in different physiological processes and pathological conditions, such as genome stability, longevity, glucose metabolism, neurodegenerative, heart and liver diseases, diabetes, inflammation, cancer, and bone disorders [5–8]. SIRT6 role in cancer was proposed to be tissue-dependent [9, 10]. In gastrointestinal tumors, SIRT6 was reported to have tumor-suppressive effects thanks to its ability to prevent HIF-1α and NF-κB activity [11]. However, in other types of malignancies, such as multiple myeloma, acute myeloid leukemia, and non-melanoma skin cancer, and in pancreatic cancer cells, SIRT6 was reported to have tumor-promoting effects thanks to its ability to enhance tumor DNA repair, secretion of pro-inflammatory and pro-angiogenic factors, Ca2+ signaling, cell migration, and cancer cell de-differentiation [12–15]. In BC patients, high tumor levels of SIRT6 were shown to enhance BC cell resistance to chemotherapeutics and to promote BC survival, migration, and invasion through the expression of cyclin D1, NF-κB, β-catenin, and matrix metalloproteinase 9 (MMP9) [16, 17].
Here, we focused on the effect of SIRT6 depletion on mammary tumorigenesis using the mammary tumor virus promoter (MMTV)-polyomavirus middle T antigen (PyMT) mouse BC model as well as MDA-MB-231 xenografts and assessed the role of SIRT6 in BC mitochondrial metabolism.
Methods
Mice
Six- to 8-week-old female BALB/c athymic nude mice (Hsd:Athymic Nude-Foxn1nu) were purchased from Envigo (Italy) and used to generate a xenograft mouse BC model (see the “Tumor models” section). Mice were housed in temperature- and light-controlled conditions (12-h light cycle) with food and water ad libitum. Mice were acclimatized for 2 weeks after their arrival. 129SvJ female mice heterozygous for Sirt6 (Sirt6+/−) were a kind gift from Dr. Raul Mostoslavsky (MGH Cancer Center, Boston, MA, USA) [18]; they were used to generate a transgenic model of spontaneous BC (see the “Tumor models” section). Male mice heterozygous for MMTV-PyMT (MMTV-PyMT+/−) in a 129/Ola genetic background were provided by Dr. Thorsten Berger (The Campbell Family Institute for Breast Cancer Research Ontario Cancer Institute) [19]. The MMTV-PyMT colony was maintained through heterozygous males because the heterozygous females develop tumors early and are therefore unable to breastfeed any puppies. Three- to 9-month-old Sirt6+/− female mice were bred with MMTV-PyMT+/− male mice to generate two different groups of interest: MMTV-PyMT+/−; Sirt6+/+ control mice and MMTV-PyMT+/−; Sirt6+/− experimental mice. The mammary glands were isolated from 12-week-old Sirt6+/+ and Sirt6+/− mice by standard procedures, flash-frozen, and stored at − 80 °C until utilization for RNA isolation, ATP, and AMP measurements and respiratory complex activity assays. All animal experiments were performed in accordance with the relevant guidelines and regulations (Italian 26/2014 and EU 2010/63/UE directives) and were approved by the Licensing and Ethical Committee (OPBA) of Ospedale Policlinico San Martino IRCCS and by the Italian Ministry of Health.
Cell lines
MCF7, MDA-MB-231, and Phoenix cell lines were purchased from ATCC and were cultured in RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum and penicillin-streptomycin (50 U/mL) (LifeTechnologies, Italy).
Viral production and transduction
The retroviral plasmids for silencing SIRT6 (short hairpin#2 - sh2) and for overexpressing wild-type (WT) or catalytically inactive (H133Y) SIRT6, and the respective vectors (PRS for the shRNA and pBABEpuro for the expression plasmids), were described previously [20, 21]. For retroviral transductions, 1 × 106 Phoenix cells were plated in 4 ml medium in 6-cm dishes and allowed to adhere for 24 h. Thereafter, cells were transfected with 4 μg of plasmid DNA using TransIT-293 (Mirus Bio, Madison, WI, USA) according to the manufacturer’s instructions. Viral supernatants were harvested after 36, 48, 60, and 72 h and used to infect MCF7 cells (5 × 105) and MDA-MB-231 (3 × 105) cells in 10-cm dishes in the presence of 5 μg/ml protamine sulfate. Successfully infected cells were selected using 1.5 μg/ml puromycin (MCF7) or 1 μg/ml puromycin (MDA-MB-231).
Western blot analysis
After transduction, 6 × 105 MCF7 and 4 × 105 MDA-MB-231 cells were plated in 10-cm dishes and allowed to adhere for 48 h. Thereafter, cells were lysed in ice-cold lysis buffer [50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% Nonidet P-40, Protease Inhibitor Cocktail, and Phosphatase Inhibitor Cocktail 2 from Sigma Aldrich], and the protein concentration was determined according to a standard Bradford assay. Proteins (35 μg) were separated by SDS-PAGE and transferred to a PVDF membrane (Immobilon-P, Millipore, Vimodrone, Italy). Proteins of interest were detected with the following antibodies: anti-SIRT6 (1:1000, #2590 Cell Signaling), anti-Phospho-AMPK (1:1000, PA5-17831 Thermo Scientific), anti-AMPK (1:1000, PA5-29679 Thermo Scientific), anti-pyruvate dehydrogenase (1:1000, #3205 Cell Signaling), anti-GAPDH (1:1000, #5174 Cell Signaling), anti-Vinculin H-300 (1:200, sc-5573 Santa Cruz Biotechnology), anti-SIRT6 (1:1000, LS-C49020 LifeSpan BioSciences; for detecting mouse Sirt6), and anti-αtubulin (1:1000, DM1A: sc-32293, Santa Cruz Biotechnology). Band intensities were quantified by the Quantity One SW software (Bio-Rad Laboratories, Inc.) using standard ECL.
Tumor masses were excised from xenograft mice and homogenized in ice-cold lysis buffer with an electric homogenizer. The samples were maintained in constant agitation for 2 h at 4 °C, on an orbital shaker in a cold room. Finally, the samples were centrifuged for 20 min at 12,000 rpm at 4 °C in a microcentrifuge, the supernatants were collected, and the protein content was evaluated by Bradford assay.
Quantitative real-time PCR
RNA extraction, cDNA synthesis, and quantitative real-time polymerase chain reaction (QPCR) were performed as described elsewhere [22]. Gene-specific primers were purchased from Sigma-Aldrich (Italy) or Thermo Fisher (Italy) and are listed in Supplementary Table 1. Comparisons in the gene expression were performed using the 2−ΔΔCt method. β-Actin was used as the housekeeping gene.
Mitochondrial mass determination
For mitochondrial mass determination, 2 × 105 MDA-MB-231 cells were stained with 200 nM Mitotracker deep red (M22426, Thermo Fisher, Italy) according to the manufacturer’s instructions and immediately analyzed by flow cytometry (FACS Aria, Becton Dickinson, Germany) by acquiring 10,000 events. The Mitotracker deep red mean fluorescence intensity (MFI) was also estimated. Changes in mitochondrial mass as a consequence of either SIRT6 overexpression or silencing were also evaluated by estimating mitochondrial DNA (mtDNA) to nuclear DNA (nDNA) ratio. To this end, 106 MDA-MB-231 cells were washed with PBS, centrifuged at 1.200 rpm for 5 min at 4 °C and resuspended in 100 μl lysis buffer (100 mM Tris-HCl pH 8, 200 mM NaCl, 5 mM EDTA, 0.2% SDS) with freshly added proteinase K (0.2 mg/ml). After 2 h incubation at 56 °C, samples were processed as described in "DNA extraction and mouse genotyping" (see below). One hundred nanograms of DNA was used to perform QPCR with SYBR green-based detection. Relative mtDNA:nDNA ratio was calculated using the 2−ΔΔCt method by targeting the nuclear-encoded gene, human B2M, and the mitochondrial-encoded gene, human tRNALeu. Gene-specific primers are listed in Supplementary Table 1.
Analysis of mitochondrial morphology by Mitotracker deep red
The analysis of mitochondrial morphological features was performed as described elsewhere [23]. In brief, 4 × 104 MDA-MB-231 cells were plated on glass coverslips (Thermo Scientific Nunc Lab-Tek II chamber slide system, Italy) and allowed to adhere overnight. Thereafter, cells were stained with Mitotracker deep red (Thermo Fisher, Italy) according to the manufacturer’s instructions and analyzed by confocal microscopy. Images were collected using a three-channel TCS SP2 laser-scanning confocal microscope (Leica Microsystems, Wetzlar, Germany). Mitochondrial solidity was defined as the fraction of pixels contained with a convex polygon (fitted around a mitochondrion) that is also mitochondrial pixels. Low solidity (close to 0.0) tends to describe highly tortuous mitochondria that are not uniform in shape, while high solidity values (close to 1.0) tend to describe mitochondria that are more uniform in shape and do not contain a high level of branching. Mitochondrial perimeter was defined as the number of exterior mitochondrial pixels multiplied by the length of the pixels, in microns. Circularity was calculated by the following formula: circularity = 4π × (area/perimeter2). Mitochondria exhibiting a perfect circular shape have a circularity value close to 1.0, whereas more elongated mitochondria have a circularity value that is close to 0.0.
Tumor models
To generate the MDA-MB-231 BC xenograft model, 2 × 106 MDA-MB-231 cells engineered with the PRS vector (VECTOR) or with a SIRT6-targeting shRNA (SIRT6-sh) were subcutaneously injected into both flanks of female BALB/c athymic nude mice. Tumor growth was monitored over time measuring tumor sizes with a manual caliper, and the tumor volume was registered twice a week. Mice were sacrificed when the tumor reached a volume of about 1.5 cm3 [the maximal tumor volume permitted by our Institutional Animal Care and Use Committee (IACUC)]. A genetically modified model that spontaneously develops breast tumors was also used (see above). MMTV-PyMT+/−; Sirt6+/+ and MMTV-PyMT+/−; Sirt6+/− mice were monitored palpating the mammary glands: when the tumors appeared as palpable masses, the age of the mice was recorded to study tumor latency and mouse survival. Tumor volume was calculated using the following formula: tumor volume = (w2 × W) × π/6, where w and W are “minor side” and “major side” (in mm), respectively. Blood glucose was measured in blood samples that were obtained from the tail of 3-month-old Sirt6+/+, Sirt6+/−, MMTV-PyMT+/−; Sirt6+/+, and MMTV-PyMT+/−; Sirt6+/− mice using Glucomen Areo 2k (Menarini, Italy). Organs [heart, lung, kidney, spleen, and mesenteric visceral adipose tissue (VAT)] from 4-week-old MMTV-PyMT+/−; Sirt6+/+ and MMTV-PyMT+/−; Sirt6+/− mice were collected by standard procedures, flash-frozen, and stored at − 80 °C until utilization for RNA isolation.
DNA extraction and mouse genotyping
DNA extraction from mouse tails was performed according to the following protocol: approximately 4 mm mouse tail was digested with 500 μl tail lysis buffer [5 mM EDTA, 0.2% sodium dodecyl sulfate (SDS), 200 mM NaCl, in 100 mM Tris-HCl, pH 8.0] containing 200 μg/ml proteinase K, in a 1.5-ml tube at 56 °C overnight with agitation (1,000 rpm on a heated shaker). The day after, samples were centrifuged at 14,000 rpm for 10 min at room temperature for debris removal. The supernatant was transferred to a clean tube with 500 μl isopropanol and inverted until DNA precipitation was observed. Subsequently, samples were centrifuged at 14,000 rpm for 5 min at RT, the supernatant was discarded, and the DNA pellet was washed by adding 300 μl of 70% ethanol and centrifuged at 14,000 rpm for 5 min at RT. After centrifugation, the ethanol solution was removed, and the DNA pellet was dried at RT or in a desiccator. Finally, the DNA pellet was resuspended in 35–100 μl DNase-free water. Genotyping was performed using Multiplex PCR Master Mix 2x (BR0200801, Biotechrabbit, Germany) to determine the presence of WT or KO allele of Sirt6 and of PyMT genes. For Sirt6 gene, three oligonucleotides were used, two specific for WT or KO allele and one in common for the two different alleles (WT: TTTCGTATAAGTCCAAGCCC, KO: GCAATAGCATCACAAATTTCAC, COMMON: GGAAGGACCTGGACAAG). WT allele primer pair (WT and COMMON) amplifies a fragment of 422 bp spanning across exon 8, while KO allele primer pair (KO and COMMON) amplifies a fragment of 300 bp spanning across exon 8. For PyMT gene, two oligonucleotides were used (PyMT 1: GGAAGCAAGTACTTCACAAGG and PyMT 2: GGAAAGTCACTAGGAGCAGGG) to amplify a fragment of 600 bp. PCR products were analyzed by gel electrophoresis with 2% precast agarose gels (54813, 2% seakem gold agarose, Lonza, Italy) at 100 V. The amplification products were visualized with a ChemiDoc XRS (BioRad, Italy) instrument.
Mammary tissue morphologic analysis
To prepare whole mounts, the fourth inguinal mammary gland was excised from 30-day-old female MMTV-PyMT+/−; Sirt6+/+ and MMTV-PyMT+/−; Sirt6+/− mice and spread out on a pre-cleaned microscope slide. The gland was fixed in Carnoy’s solution (75% ethanol–25% glacial acetic acid) overnight at RT. The day after, the slides were hydrated in 70% ethanol and water and stained overnight with carmine alum (07070, STEMCELL Technologies, Italy). After staining, the slides were rinsed in water, dehydrated in increasing concentrations of ethanol, and cleared in histolemon (454915, Carlo Erba, Italy). Photographic images of the whole mounts were acquired by Nikon SMZ1270 microscope using the X-Entry software. Terminal end buds (TEB) were counted in the whole mammary gland.
Histopathological and immunohistochemical analysis of mammary tumors
After being sacrificed by cervical dislocation, MMTV-PyMT+/−; Sirt6+/+ and MMTV-PyMT+/−; Sirt6+/− mice were refrigerated and immediately sent to the pathological anatomy laboratory. Each animal was fixed by immersion in 10% buffered formalin for 12–18 h, after inoculation of the fixative intraperitoneally and intrathoracically. Subsequently, each animal was eviscerated en-block and the organs seriated with consecutive sections in the craniocaudal sense. Bilateral paramedian incisions along the breast lines were also performed to sample mammary tumors. All samples were routinely processed and paraffin-embedded to obtain 3-μm-thick histological sections stained with hematoxylin-eosin. Microvessel density was determined by CD34 staining (anti-CD34, clone QBEnd/10, Ventana) as described in [24].
Cell homogenate preparation for ATP and AMP measurements and respiratory complex activity assays
To remove the growth medium, cells were centrifuged at 1000g for 2 min. The pellet was suspended in phosphate-buffered saline (PBS) buffer and sonicated on ice, 2 times for 10 s, with a 30-s break. The total protein concentration was estimated by the Bradford method [25].
Evaluation of intracellular ATP and AMP content
Quantification of ATP and AMP was based on the enzyme coupling method [26]. Twenty micrograms of total proteins was used. Briefly, ATP was assayed spectrophotometrically at 340 nm, following NADP+ reduction, at 25 °C. The reaction mixture contained the following: 1 mM NADP+, 10 mM MgCl2, 5 mM glucose, and 100 mM Tris-HCl, pH 7.4, in 1 ml final volume. Samples were analyzed before and after the addition of purified hexokinase and glucose-6-phosphate dehydrogenase (4 μg; HK+G6PD, HKG6PDH-RO, Sigma-Aldrich, Italy). AMP was assayed spectrophotometrically at 340 nm, following NADH oxidation. The reaction mixture contained the following: 75 mM KCl, 5 mM MgCl2, 0.2 mM ATP, 0.5 mM phosphoenolpyruvate, 0.2 mM NADH, 10 IU adenylate kinase (AK, M3003, Sigma-Aldrich, Italy), 25 IU pyruvate kinase plus 15 IU lactate dehydrogenase (PK+LDH, Sigma-Aldrich, Italy), and 100 mM Tris-HCl pH 8.0.
Oxymetric analysis
Oxygen consumption was measured with an amperometric oxygen electrode (Unisense) in a closed chamber, magnetically stirred, at 37 °C. For each assay, 2 × 105 cells were used. After cell permeabilization with 0.03 mg/ml digitonin for 10 min, samples were suspended in 137 mM NaCl, 5 mM KH2PO4, 5 mM KCl, 0.5 mM EDTA, 3 mM MgCl2, and 25 mM Tris-HCl, pH 7.4. To activate the pathway composed of complexes I, III, and IV, 5 mM pyruvate and 2.5 mM malate were added. To activate the pathway composed of complexes II, III, and IV, 20 mM succinate was used [27].
Evaluation of F1Fo-ATP synthase activity
F1Fo-ATP synthase (ATP synthase) activity was detected by measuring the ATP production by the highly sensitive luciferin/luciferase method. The assays were conducted at 37 °C, for 2 min, and data were collected every 30 s. Cells (1 × 105) were added to the incubation medium (0.1 ml final volume), which contained 50 mM KCl, 1 mM EGTA, 2 mM EDTA, 5 mM KH2PO4, 2 mM MgCl2, 0.6 mM ouabain, 1 mM P1,P5-Di(adenosine-5′) pentaphosphate, 0.040 mg/ml ampicillin, 0.2 mM adenosine diphosphate (ADP), 10 mM Tris-HCl pH 7.4, and the metabolic substrates (5 mM pyruvate + 2.5 mM malate or 20 mM succinate). Cells were equilibrated for 10 min at 37 °C, and then ATP synthesis was induced by the addition of 0.2 mM ADP. ATP synthesis was measured using the luciferin/luciferase ATP bioluminescence assay kit CLSII (11699695001, Roche) and a Luminometer (GloMax® 20/20 Luminometer, Promega). ATP standard solutions in the concentration range 10−10–10−7 M were used for calibration [27].
Respiratory complex activity assay
The activity of the redox complexes I, III, and IV was measured, in a double beam spectrophotometer (UNICAM UV2, Analytical S.n.c.), at 25 °C. For each assay, 50 μg total proteins were employed, and the reaction was followed for 5 min, collecting data every 1 min [28]. The enzymatic activity was expressed as mIU/mg total protein (nanomoles/min/mg protein). Complex I (NADH-ubiquinone oxidoreductase) was assayed following the reduction of ferrocyanide, in the presence of NADH, at 420 nm; the reaction mixture contained 30 mM NADH, 40 mM potassium ferrocyanide, and 40 μM antimycin A in 10 mM phosphate buffer pH 7.2. Complex III (cytochrome c reductase) activity was measured at 550 nm following the reduction of oxidized cytochrome c. The reaction mixture contained 10 mM phosphate buffer pH 7.2, 0.03% oxidized cytochrome c, and 0.5 mM KCN. Complex IV (cytochrome c oxidase) was assayed following the oxidation of ascorbate-reduced cytochrome c at 550 nm, in a solution containing 10 mM phosphate buffer pH 7.2, 0.03% reduced cytochrome c, and 40 μM antimycin A. In both assays, the cytochrome c extinction coefficient was considered 19.1 × 10−3 M−1 cm−1, at 550 nm.
Evaluation of pyruvate dehydrogenase expression and activity
The expression and activity of pyruvate dehydrogenase (PDH) were performed using the “Pyruvate dehydrogenase (PDH) Combo (Activity + Profiling) Microplate Assay Kit” provided by Abcam (ab110671, Abcam), following the manufacturer’s instructions. For each sample, 4 × 106 MDA-MB-231 cells were used.
Quantification and statistical analysis
Statistical analyses were performed with the GraphPad Prism software version 5 (GraphPad Software, San Diego, USA). Data are shown as mean ± SD. All parameters were tested by two-tailed Student’s t test (in all in vitro experiments), by two-way ANOVA followed by Bonferroni post hoc test (in vivo xenografts experiments), or log rank test (in vivo survival and latency experiments). P values less than 0.05 were considered significant.
Results
SIRT6 downregulation slows BC progression in MMTV-PyMT mouse model
To define the role of SIRT6 in mammary carcinogenesis, we utilized transgenic mice expressing the PyMT under the MMTV promoter, which is an established animal model of human BC [29, 30]. Specifically, we crossed MMTV-PyMT+/− male mice with Sirt6+/− female animals and compared tumor latency and mouse survival between MMTV-PyMT+/−; Sirt6+/+ and MMTV-PyMT+/−; Sirt6+/− mice. This approach was chosen since complete Sirt6 knockout results in postnatal lethality (Sirt6−/− mice are typically only viable until weaning) [18]. Thus, since the typical tumor latency in MMTV-PyMT 129 mice is between 10 and 12 weeks [19], the use of Sirt6−/− mice would not have allowed us to assess the effect of Sirt6 depletion on mammary tumor development. Conversely, a heterozygous Sirt6 deletion (Sirt6+/−) was shown to be compatible with mouse survival [18] and yet to produce biologically relevant effects, including enhancing adhesion molecule expression and exacerbating atherosclerosis [31].
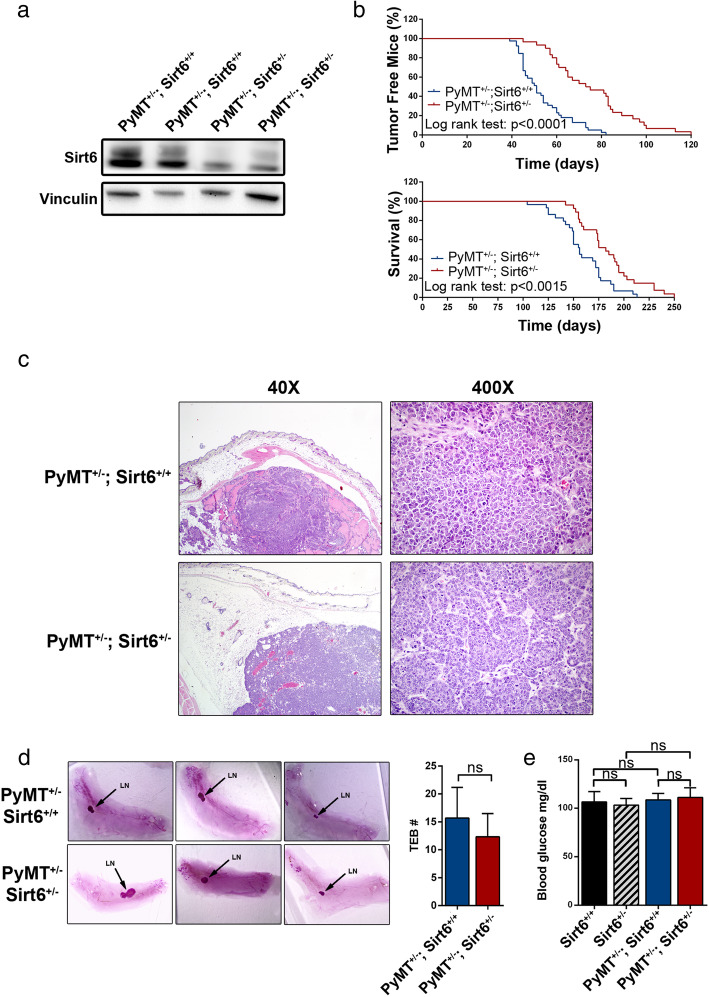
In line with a previous study [31], Sirt6+/− mice, which we typically maintained for several months, failed to show any adverse or debilitating phenotype. They remained fully viable and active up to more than 21 months of age (which is the longest observation period we recorded) and were phenotypically indistinguishable from the Sirt6+/+ animals. MMTV-PyMT+/−; Sirt6+/+ and MMTV-PyMT+/−; Sirt6+/− animals were born at the expected Mendelian rate [49.7% MMTV-PyMT+/−; Sirt6+/+ mice (n = 149) and 50.7% MMTV-PyMT+/−; Sirt6+/− (n = 152)]. The heterozygous Sirt6 deletion resulted in a decreased Sirt6 protein expression in mammary tumor masses isolated from MMTV-PyMT+/−; Sirt6+/− animals as compared to the control mice (Fig. 1a and Additional file 1: Fig. S1A). Mice carrying the heterozygous Sirt6 deletion exhibited a marked increase in tumor latency and a consistent increase in their overall survival (Fig. 1b). No difference in terms of histology between mammary tumors from MMTV-PyMT+/−; Sirt6+/− vs. MMTV-PyMT+/−; Sirt6+/+ mice was detected. In both cases, our pathology assessments demonstrated multifocal, highly fibrotic, adenocarcinomas involving the entire mammary fat pad (Fig. 1c). No difference between tumors from MMTV-PyMT+/−; Sirt6+/− vs. MMTV-PyMT+/−; Sirt6+/+ mice was observed in terms of the number of mitoses [41.2 ± 16.6 and 33.5 ± 18.4 in MMTV-PyMT+/−; Sirt6+/+ (n = 6) and in MMTV-PyMT+/−; Sirt6+/− (n = 4) mice, respectively; P = 0.53] and of microvessels [as detected by CD34 staining; 10.3 ± 8.5 and 12.8 ± 9.5 in MMTV-PyMT+/−; Sirt6+/+ (n = 6) and in MMTV-PyMT+/−; Sirt6+/− (n = 4) mice, respectively; P = 0.7] per field. A non-significant trend towards an increase in the number of apoptotic tumor cells per field was observed as a result of Sirt6 heterozygous deletion [5.7 ± 1 and 7.3 ± 1 in MMTV-PyMT+/−; Sirt6+/+ (n = 6) and in MMTV-PyMT+/−; Sirt6+/− (n = 4) mice, respectively; P = 0.13].
Fig. 1.
Sirt6 heterozygous deletion increases tumor latency and enhances survival in the MMTV-PyMT mammary tumor model. a Tumor masses were excised from MMTV-PyMT+/−; Sirt6+/+ and MMTV-PyMT+/−; Sirt6+/− mice; protein lysates were generated, and Sirt6 and vinculin levels were detected by Western blot. b Tumor latency (upper panel) and overall survival (lower panel) of MMTV-PyMT+/−; Sirt6+/+ (n = 39) and of MMTV-PyMT+/−; Sirt6+/− (n = 30) mice were monitored over time. c Histopathological analysis of the mammary glands from MMTV-PyMT+/−; Sirt6+/+ and MMTV-PyMT+/−; Sirt6+/− mice (hematoxylin-eosin; × 40 and × 400) shows poorly differentiated (G3) murine ductal carcinoma of the breast consisting in dysmetrical elements with solid growth, brisk mitotic activity, and scattered areas of necrosis in both types of tumor. d Mammary glands from 4-week-old MMTV-PyMT+/−; Sirt6+/+ (n = 3) and MMTV-PyMT+/−; Sirt6+/− (n = 3) mice were stained by carmine alum (left) and used to quantify the TEB (right). LN, lymph node. Data are presented as mean ± SD; ns, not statistically significant. e Blood glucose levels in Sirt6+/+ (n = 6), Sirt6+/− (n = 3), MMTV-PyMT+/−; Sirt6+/+ (n = 3), and MMTV-PyMT+/−; Sirt6+/− (n = 6) mice. Data are presented as mean ± SD. ns, not statistically significant
Since previous studies indicated that the heterozygous deletion of another sirtuin family member, Sirt1, increases mammary tumor latency by interfering with the normal development of the mammary gland [32], we monitored the mammary gland development in MMTV-PyMT+/−; Sirt6+/+ and MMTV-PyMT+/−; Sirt6+/− mice. In particular, we quantified the number of TEB, the key structures that regulate elongation and branching of the mammary gland into the fat pad and thereby drive ductal morphogenesis. Comparisons of 4-week-old MMTV-PyMT+/−; Sirt6+/+ and MMTV-PyMT+/−; Sirt6+/− mice showed no significant difference in terms of number and size of TEB (Fig. 1d). Thus, the observed effect of Sirt6 deletion on mammary tumorigenesis did not reflect impaired ductal morphogenesis in MMTV-PyMT+/−; Sirt6+/− mice.
A homozygous deletion of Sirt6 was previously reported to lower blood glucose levels due to increased expression of glucose transporters, such as GLUT4, and of increased glycolytic enzymes in many tissues [18]. Thus, because reduced circulating glucose levels could in principle account for the antitumor effect of SIRT6 depletion, we assessed whether the heterozygous Sirt6 deletion would also lower blood sugar levels in mice. However, we could not detect any reduction in blood glucose levels in MMTV-PyMT+/−; Sirt6+/− as compared to MMTV-PyMT+/−; Sirt6+/+ animals or in Sirt6+/− compared to Sirt6+/+ mice (Fig. 1e). Consistent with this finding, we failed to detect increased Glut4 and hexokinase 2 (Hk2) expression in several tissues, such as heart, lung, spleen, kidney, and mesenteric VAT of MMTV-PyMT+/−; Sirt6+/− mice as compared to MMTV-PyMT+/−; Sirt6+/+ animals (Additional file 1: Fig. S1B). Therefore, reduced circulating glucose levels did not account for the anticancer effect observed in response to the heterozygous deletion of Sirt6 in this BC model.
Overall, our results clearly indicate that reducing Sirt6 expression slows mammary cancer development in the mouse MMTV-PyMT BC model. These results are also consistent with those studies attributing an adverse prognostic significance to high SIRT6 levels in BC [16, 17].
SIRT6 silencing reduces tumor growth in MDA-MB-231 xenografts
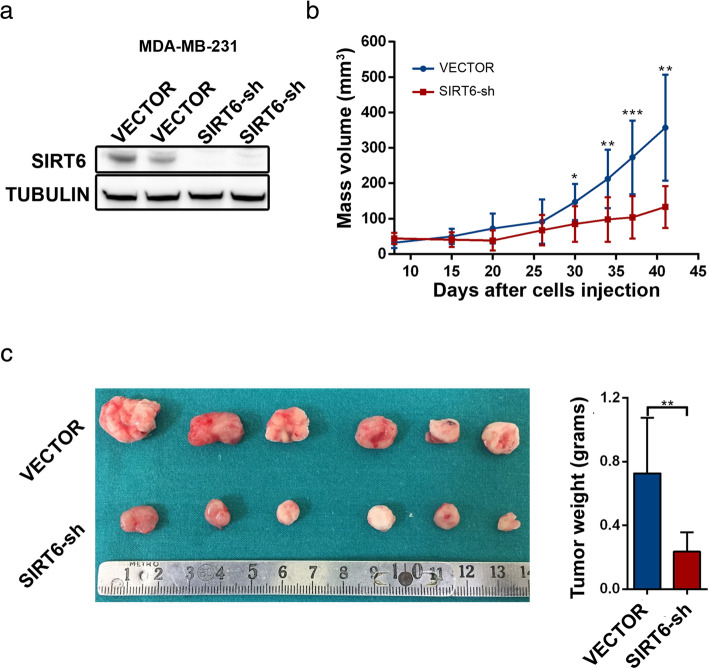
Since in MMTV-PyMT+/−; Sirt6+/− mice, Sirt6 deletion affects all bodily tissues, the observed delay in tumor development and the corresponding enhancement of mouse survival could in principle reflect non-cell-autonomous anticancer effects (i.e., effects unrelated to Sirt6 deletion in tumor cells). To address this possibility, we silenced SIRT6 by RNA interference in the BC cell line MDA-MB-231 (Fig. 2a), injected these cells (or cells harboring a control shRNA) subcutaneously into both flanks of nude mice, and monitored tumor growth. As shown in Fig. 2b, tumor masses developed from SIRT6-silenced MDA-MB-231 cells exhibited a markedly reduced growth, which was also confirmed by isolating the tumor masses at the end of the experiment and by measuring their size and weight (Fig. 2c). These results are consistent with reduced SIRT6 levels slowing BC growth in a cell-autonomous fashion, although in principle, the possibility that Sirt6 depletion also causes non-cell-autonomous anti-tumor effects in the MMTV-PyMT mammary tumor model cannot be excluded.
Fig. 2.
SIRT6 silencing reduces tumor growth in MDA-MB-231 xenografts. a, b 2 × 106 MDA-MB-231 BC cells transduced with either a SIRT6-shRNA or a control vector were injected subcutaneously in both flanks of BALB/c athymic nude mice (Hsd:Athymic Nude-Foxn1nu) (VECTOR, n = 8 and SIRT6-sh, n = 10). In a, mice were sacrificed when the tumors became palpable; tumors were used for protein lysate generation, and SIRT6 and α-tubulin were detected by Western blot. In b, tumor volume was monitored over time from the day of tumor cell inoculation. Data are presented as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001 (statistical values refer to the comparison between tumors with silenced SIRT6 and control tumors at the same time point). c Fifty days after tumor cell inoculation, mice were sacrificed, and masses were collected, imaged, and weighted (VECTOR: n = 6; SIRT6-sh: n = 6). Data are presented as mean ± SD. **P < 0.01
SIRT6 enhances OXPHOS in mouse mammary tumors and in human BC cells
Studies showed that tumor growth requires active mitochondrial aerobic metabolism to support cancer cell proliferation, cancer stem cell survival, and metastasis [3, 33]. Consistent with this notion, agents targeting mitochondrial respiratory complexes, such as metformin (which inhibits complex I activity) show promising anticancer properties [26, 34].
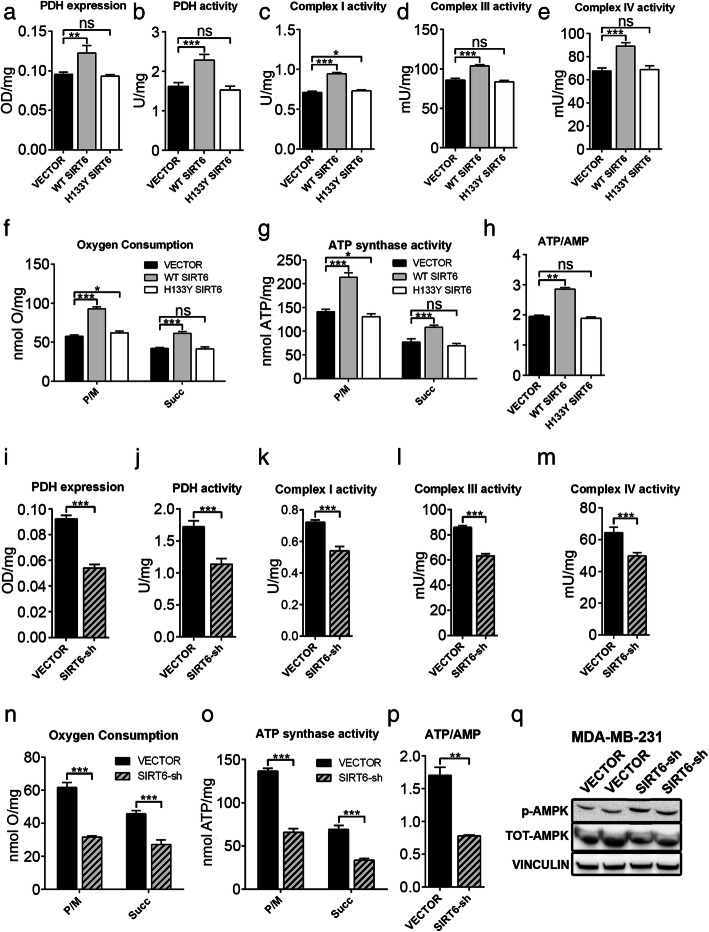
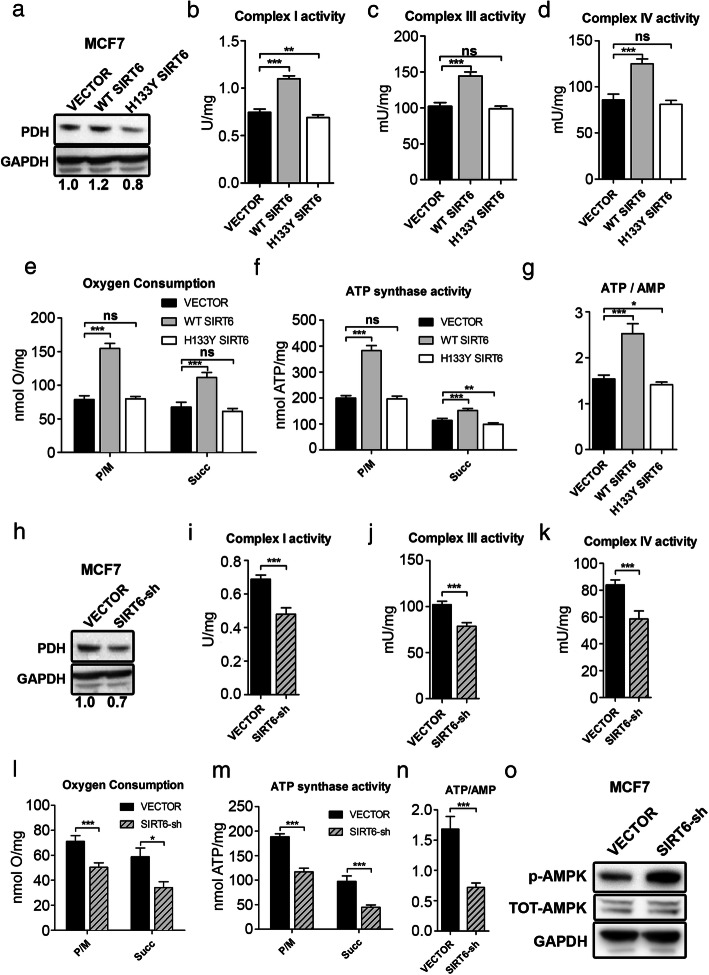
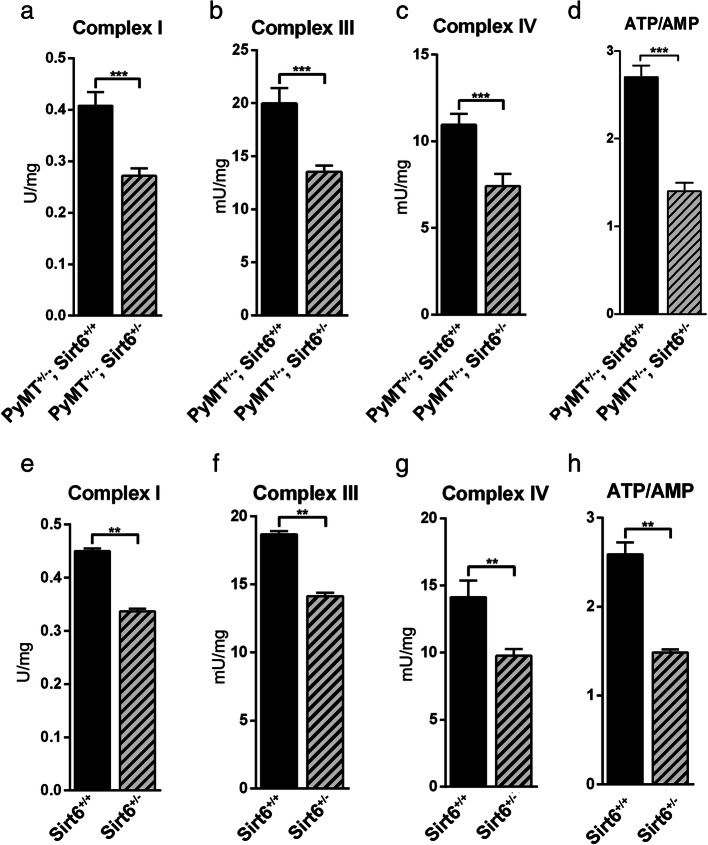
Previous studies suggested that OXPHOS may be reduced as a consequence of decreased SIRT6 levels [18, 35]. Thus, we hypothesized that blunting SIRT6 expression could affect OXPHOS and cellular energy status in BC cells. To address this hypothesis, we used MDA-MB-231 and MCF7 cells, after transducing them with either WT or catalytically inactive (H133Y) SIRT6 (Additional file 2: Fig. S2A, B). In these engineered cells, we measured the expression and activity of PDH, the activity of respiratory complexes I, III, and IV; O2 consumption; ATP synthase activity; and ATP/AMP ratio. Overexpression of WT, but not of catalytically inactive, SIRT6 enhanced the expression and activity of PDH, the activity of respiratory complexes, O2 consumption, and ATP synthesis, determining a consistent increase of the ATP/AMP ratio (Figs. 3a–h and 4a–g) in both cell lines. Opposite effects were obtained by SIRT6 silencing in MDA-MB-231 and in MCF7 cells (Figs. 3i–p and 4 h–n and Additional file 2: Fig. S2C, D). Consistent with these data obtained in human BC cell lines, mammary tumors from MMTV-PyMT+/−; Sirt6+/− mice exhibited decreased complex I, III, and IV activity as well as reduced ATP/AMP ratio as compared to tumors from MMTV-PyMT+/−; Sirt6+/+ mice (Fig. 5a–d). We also isolated the mammary glands from regular Sirt6+/+ or Sirt6+/− 129/Ola mice which had not been crossed with MMTV-PyMT animals and subjected them to the same biochemical analyses. Decreased respiratory complex activity and ATP/AMP ratio were also detected in healthy, non-tumor-bearing mammary glands from Sirt6+/− mice as compared to the glands from Sirt6+/+ animals (Fig. 5e–h). Therefore, these data indicate that the reduction in OXPHOS through SIRT6 depletion is not restricted to transformed mammary tissue. Rather, it appears to be also present in healthy mammary glands, likely contributing to the delay in the development of PyMT-induced tumors.
Fig. 3.
SIRT6 enhances OXPHOS and energy status in MDA-MB-231 cells. a–h Expression (a) and activity (b) of pyruvate dehydrogenase (PDH), activity of mitochondrial complexes (c–e), oxygen consumption (f), activity of Fo-F1 ATP synthase (g), and energy status, expressed as ATP/AMP ratio (h), were measured in MDA-MB-231 cells transduced with human WT or catalytically inactive (H133Y) SIRT6, or with a control vector (VECTOR). Data are presented as mean ± SD of three different experiments. *P < 0.05, **P < 0.01, ***P < 0.001; ns, not statistically significant. i–p Expression (i) and activity (j) of PDH, activity of mitochondrial complexes (k–m), oxygen consumption (n), activity of Fo-F1 ATP synthase (o), and energy status, expressed as ATP/AMP ratio (p) were measured in MDA-MB-231 cells transduced with a short hairpin RNA targeting SIRT6 or with a control vector (VECTOR). Data are presented as mean ± SD of three different experiments. **P < 0.01, ***P < 0.001. q 2 × 106 MDA-MB-231 BC cells transduced with either a SIRT6-shRNA or a control vector (VECTOR) were injected subcutaneously in both flanks of BALB/c athymic nude mice. Mice were sacrificed, and tumors were excised 50 days after cell inoculation. Proteins were extracted, and phosphorylated AMPK (Thr183, Thr172), total AMPK, and vinculin were detected by Western blot. One representative experiment out of two is presented
Fig. 4.
SIRT6 enhances OXPHOS and energy status in MCF7 cells. a Western blot analysis of PDH expression in MCF7 cells transduced with human WT or catalytically inactive (H133Y) SIRT6 or with a control vector (VECTOR). PDH expression was quantified following normalization to GAPDH. b-g Activity of mitochondrial complexes (b–d), oxygen consumption (e), activity of Fo-F1 ATP synthase (f), and energy status, expressed as ATP/AMP ratio (g), were measured in MCF7 cells transduced with human WT or catalytically inactive (H133Y) SIRT6 or with a control vector (VECTOR). Data are presented as mean ± SD of three different experiments. *P < 0.05, **P < 0.01, ***P < 0.001; ns, not statistically significant. h Western blot analysis of PDH expression in MCF7 cells transduced with an shRNA targeting SIRT6 (SIRT6-sh) or with a control vector (VECTOR). PDH expression was quantified following normalization to GAPDH. i–n Activity of mitochondrial complexes (i–k), oxygen consumption (l), activity of Fo-F1 ATP synthase (m), and energy status, expressed as ATP/AMP ratio (n), were measured in MCF7 cells transduced with an shRNA targeting SIRT6 (SIRT6-sh) or with a control vector (VECTOR). Data are presented as mean ± SD of three different experiments. *P < 0.05, ***P < 0.001. o Protein lysates were generated from MCF7 that were engineered with a control vector (VECTOR) or with an shRNA targeting SIRT6 (SIRT6-sh)\. Phosphorylated (Thr183, Thr172) and total AMPK as well as GAPDH were detected by Western blot (one representative experiment out of three is presented)
Fig. 5.
Sirt6 heterozygous deletion decreases OXPHOS and ATP/AMP ratio in MMTV-PyMT mammary tumors and in healthy mammary glands. a–d Mitochondrial complex activity (a–c) and energy status, expressed as ATP/AMP ratio (d), were measured in tumor masses that were isolated from 12-week-old MMTV-PyMT+/−; Sirt6+/+ (n = 5) and MMTV-PyMT+/−; Sirt6+/− (n = 4) mice. Data are presented as mean ± SD. ***P < 0.001. e–h Mitochondrial complex activity (e–g) and energy status, expressed as ATP/AMP ratio (h), were measured in the mammary glands that were isolated from 12-week-old Sirt6+/+ (n = 4) and Sirt6+/− (n = 4) mice. Data are presented as mean ± SD. **P < 0.01
Homozygous Sirt6 deletion was previously reported to reduce the expression of the OXPHOS-related genes ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 (Uqcrfs1), cytochrome c oxidase subunit 5B (Cox5b), NADH:ubiquinone oxidoreductase subunit B8 (Ndufb8), ubiquinol-cytochrome c reductase core protein 2 (Uqcrc2), and succinate dehydrogenase complex iron sulfur subunit B (Sdh) in mouse skeletal muscle tissue [36]. Thus, we evaluated whether similar changes would also occur in BC cells in response to reduced SIRT6, possibly contributing to the observed effects of SIRT6 modulation on OXPHOS. UQCRFS1, COX5B, and NDUFB8 expression was blunted by both SIRT6 silencing in MDA-MB-231 cells and by the Sirt6 heterozygous deletion in mouse mammary tumors (Additional file 3: Fig. S3A, B). UQCRC2 expression was reduced by SIRT6 silencing in MDA-MB-231, but not in Sirt6+/− mouse tumor masses. Finally, SDH was not affected by either SIRT6 silencing in MDA-MB-231 cells or by Sirt6 deletion in mouse mammary neoplasms. Therefore, these data are consistent with SIRT6 also promoting the expression of genes encoding for respiratory chain proteins, such as UQCRFS1, COX5B, NDUFB8, and UQCRC2, in BC cells.
We also evaluated whether the changes in OXPHOS we observed in response to different SIRT6 levels in BC cells would reflect differences in mitochondrial mass or in mitochondrial morphology. To this end, we first stained MDA-MB-231 with the mitochondrial dye Mitotracker deep red and analyzed them by flow cytometry (Additional file 3: Fig. S3C). However, according to this approach, SIRT6 overexpression and SIRT6 silencing failed to increase and to decrease, respectively, the mitochondrial mass. Similar results were obtained by monitoring the ratio between mitochondrial and nuclear DNA in MDA-MB-231 cells: neither SIRT6 overexpression nor its silencing affected such ratio (Additional file 3: Fig. S3D). In subsequent experiments, MDA-MB-231 cells overexpressing SIRT6 or silenced for this protein were stained with Mitotracker deep red and analyzed by confocal microscopy. Here, again, we failed to detect significant changes in any of the morphological parameters that we analyzed, including mitochondrial area, perimeter, circularity, and solidity in response to different SIRT6 levels (Additional file 3: Fig. S3E and data not shown). Therefore, overall, these data indicate that the effects of SIRT6 overexpression and of its silencing on OXPHOS in BC cells are not justified by corresponding changes in mitochondrial mass or morphological features.
Finally, in the attempt to define the mechanism underlying the decreased PDH activity in tumors from MMTV-PyMT+/−; Sirt6+/− mice, we investigated whether this would reflect an increased expression of piruvate dehydrogenase kinase 1 (PDK1) or piruvate dehydrogenase kinase 4 (PDK4) (which both catalyze inhibitory phosphorylation of PDH), as previously reported as a consequence of reduced Sirt6 in intestinal cancer and in the heart, respectively [11, 37]. However, neither of these two genes was increased in its expression by the heterozygous Sirt6 deletion (Additional file 4: Fig. S4A). Similarly, no increase in PDK1 and PDK4 expression was detected in MDA-MB-231 cells as a result of SIRT6 silencing (data not shown). On the other hand, we found that, consistent with our previous data in pancreatic cancer cells [14], SIRT6 overexpression increased, while SIRT6 silencing decreased, intracellular Ca2+ concentration in MDA-MB-231 cells (Additional file 4: Fig. S4B). Thus, since intracellular Ca2+ enhances PDH activity [38, 39], SIRT6’s ability to regulate these cation levels could account (together with the increased PDH expression) for the enhanced PDH function we observed in SIRT6-overexpressing BC cells.
Overall, these findings indicate that SIRT6 depletion reduces OXPHOS and energy status in BC cells of a murine and human source and point to a probable mechanism for the reduced tumor growth occurring in response to SIRT6 depletion.
SIRT6 regulates AMPK activity in BC cells
The ATP/AMP ratio is the main determinant of AMP-activated protein kinase (AMPK) activation [40]. In turn, activated AMPK is responsible for orchestrating a tumor-suppressive response that includes a mammalian target of rapamycin (mTOR) inhibition and autophagy initiation [41]. We found that MCF7 and MDA-MB-231 cells with silenced SIRT6 exhibited increased levels of AMPK phosphorylation (Fig. 4o and Additional file 5: Fig. S5A). Accordingly, higher AMPK activation was also observed in xenografts of MDA-MB-231 with silenced SIRT6 as compared to xenografts of control cells (Fig. 3q and Additional file 5: Fig. S5B). Therefore, these findings are in line with SIRT6 depletion causing energetic stress in BC cells and consequently increasing AMPK activity.
Discussion
Here, we show that a heterozygous deletion of the Sirt6 gene increases tumor latency and improves overall survival in the MMTV-PyMT mouse model. Such an effect is recapitulated by SIRT6 silencing in MDA-MB-231 xenografts, indicating a cell-autonomous anticancer effect of SIRT6 depletion in BC cells. We show that SIRT6 boosts OXPHOS and enhances energy status in BC cells and that this translates into dampened AMPK activity (Additional file 6: Fig. S6). Our in vivo data, showing delayed mammary tumor development and increased survival in MMTV-PyMT+/−; Sirt6+/− mice as compared to MMTV-PyMT+/−; Sirt6+/+ animals, are in line with the previously reported adverse prognostic significance of high SIRT6 expression in BC [16, 17]. These two studies attributed the “pro-oncogenic” role of SIRT6 in BC not only to the ability of SIRT6 to promote DNA repair in BC cells and, consequently, to mediate resistance to chemotherapeutics, but also to the increased expression of MMP9, β-catenin, CCND1, and NF-κB and to enhanced BC cell migration and invasiveness. Our data point out a new “metabolic” function of this NAD+-dependent deacetylase in BC cells, which consists of SIRT6’s ability to enhance PDH expression and activity, OXPHOS, and ATP availability in BC cells. Quite remarkably, SIRT6 overexpression also increased oxygen consumption and ATP synthase activity in MDA-MB-231 cells, which is normally considered to be a glycolysis-dependent cell line, as opposed to other BC cells, which primarily rely on OXPHOS for their metabolism, including MCF7 cells [42].
Studies show that, despite early models proposing that cancer cells would primarily rely on aerobic glycolysis for their survival, mitochondrial function is actually decisive in many neoplasms. Loss of the tumor suppressor RB1, but also several proto-oncogenes, such as mitochondrial STAT3, FER, and its variant, FerT, and CHCHD2, induces OXPHOS [3]. Accordingly, OXPHOS inhibitors, such as metformin, tigecycline, or salinomycin, hold promise for preventing or treating different forms of cancer, including BC. Our study indicates SIRT6 as a new, potentially druggable target [43–46] to be exploited for interfering with OXPHOS in BC, thereby achieving cancer-preventive but also, possibly, therapeutic effects.
Previous studies attributed SIRT6’s ability to enhance OXPHOS to SIRT6-mediated reduction of PDK1 and PDK4 expression, which in turn would increase PDH activity [11, 37]. As opposed to these studies, in mammary tumors from MMTV-PyMT+/−; Sirt6+/− mice and in MDA-MB-231 cells with silenced SIRT6, we did not detect increased PDK1 or PDK4 expression. However, we did find SIRT6 to regulate PDH expression. Specifically, SIRT6 overexpression increased, while its silencing decreased, PDH levels. In addition, consistent with a previous study by our group in pancreatic cancer cells [14], we found SIRT6 to increase the intracellular concentration of Ca2+, which is a known enhancer of PDH function [38, 39], in MDA-MB-231 cells. Thus, we propose that SIRT6 may promote PDH activity in BC cells via at least two mechanisms: (i) by increasing PDH levels and (ii) by enhancing the intracellular Ca2+ concentration. Conceivably, by blunting PDH expression and activity, SIRT6 depletion [and possibly SIRT6 inhibition via small molecules [45]] could lead to lower levels of acetyl-CoA and TCA cycle intermediates in BC cells. Thus, it is possible that cancer cells with low SIRT6 will show increased uptake of other mitochondrial fuels, such as glutamine and fatty acids. Future studies should address this possibility as a way to identify metabolic liabilities of cancer cells with low SIRT6 expression or in which SIRT6 is pharmacologically inhibited.
A previous study also reported that the expression of the OXPHOS-related genes Uqcrfs1, Cox5b, Ndufb8, Uqcrc2, and Sdh was markedly reduced in Sirt6−/− skeletal muscle tissue [36]. In line with this study, we found UQCRFS1, COX5B, and NDUFB8 expression to be reduced by both SIRT6 silencing (in MDA-MB-231 cells) and Sirt6 heterozygous deletion (in tumors from MMTV-PyMT+/−; Sirt6+/− mice). UQCRC2 expression was also lower in MDA-MB-231 cells with silenced SIRT6 as compared with the control cells. SDH was neither reduced by SIRT6 silencing in MDA-MB-231 cells nor in tumors from MMTV-PyMT+/−; Sirt6+/− mice. Thus, these data indicate that SIRT6-mediated regulation of OXPHOS in BC cells probably also relies on SIRT6 ability to regulate the amounts of respiratory chain proteins in addition to its effect on PDH expression and activity. Finally, we previously reported that SIRT6-overexpressing MCF7 cells have higher levels of NAD(H) and of NADPH, which are both important coenzymes participating in various energy metabolism pathways [44]. These findings are also in line with the results of this study and suggest that the enhanced energy status of SIRT6-overexpressing BC cells could also reflect higher NAD(P)(H) availability.
The heterozygous deletion of Sirt6 also reduced complex I, III, and IV activity and ATP/AMP ratio in healthy (non-PyMT-transformed) mammary glands, indicating that the effect of reduced SIRT6 on mammary gland cell metabolism is independent of whether the tissue is transformed or not. Based on these data, one could argue that reduced ATP availability in the mammary gland may hinder tumor development starting from its very early stages, thus justifying the marked extension in tumor latency observed in MMTV-PyMT+/−; Sirt6+/− mice.
In our hands, SIRT6 depletion resulted in increased AMPK activation as detected by its phosphorylation on threonines 172 and 183. These findings, which we attribute to the low ATP/AMP ratio that we observed in response to reduced SIRT6, are consistent with those of Ming and colleagues, who also found such an increase in AMPK phosphorylation upon SIRT6 silencing in skin cancer cells [47]. On the other hand, Elhanati and colleagues found that, in the liver, SIRT6 overexpression, rather than its downregulation, decreases ATP/AMP ratio and activates AMPK [48]. This raises the interesting possibility that SIRT6 effects on AMPK activity may vary between different tissues and reflect different regulatory mechanisms.
Curiously, a previous study evaluated the effect of SIRT6 overexpression (using Sirt6BAC mice) in the MMTV-PyMT mammary tumor model and found that SIRT6 overexpression also has the ability to hamper tumor progression in mice [49]. According to this study, the antitumor activity of SIRT6 overexpression would be restricted to BC with constitutively active PI3K and result from SIRT6 ability to reduce PI3K signaling as well as BC stem cell-like characteristics. In addition, the anticancer activity of overexpressed SIRT6 was shown to be independent of SIRT6 deacetylase activity. Our data complement and extend these findings and indicate that both SIRT6 depletion and SIRT6 overexpression probably result in suboptimal conditions for BC development. SIRT6 depletion has anticancer effects in BC very likely through its ability to reduce the expression and activity of PDH and OXPHOS and to cause energy stress in cancerous and pre-cancerous lesions. These effects are dependent on SIRT6 deacetylase activity, but independent of PI3KCA mutational status, since SIRT6 depletion had the same effect on OXPHOS-related parameters in MDA-MB-231 cells, which have WT PI3K, and in MCF7, which have a PIK3CA activating mutation [50]. On the other hand, in breast tumors with mutated PI3KCA, SIRT6 overexpression inhibits PI3K signaling in a deacetylase activity-independent fashion and thereby affects the cancer stem cell compartment.
Conclusion
Although other mechanisms, such as impaired DNA repair ability and reduced β-catenin, cyclin D1 or NF-κB activity could also have contributed to the antitumor effects of low SIRT6 in our BC models, as suggested by earlier reports [16, 17], our findings complement these previous studies and depict a novel, key role for SIRT6 in BC metabolism. These new insights strengthen the rationale for targeting this deacetylase as a way to prevent or treat BC.
Supplementary Information
Additional file 1: Fig. S1. Effect of Sirt6 heterozygous deletion on Sirt6, Glut4 and Hk2 expression in mouse mammary tumors and tissues. A, B, Mammary tumors and healthy tissues [heart, lung, spleen, kidney and visceral adipose tissue (VAT)] were isolated from twelve-week-old and from four-week-old MMTV-PyMT+/-; Sirt6+/+ (n = 3) and MMTV-PyMT+/-; Sirt6+/- (n = 3) mice, respectively, and used for protein lysate generation and for RNA extraction. In A, Sirt6 and vinculin levels were assessed by Western blot in mammary tumors. In B, Hk2 and Glut4 expression in bodily tissues was detected by QPCR. Data are presented as mean ± SD. ns: not statistically significant.
Additional file 2: Fig. S2. SIRT6 silencing and overexpression in MDA-MB-231 and MCF7 cell lines. A, B, Western blot analysis of MDA-MB-231 (A) and MCF7 (B) cell lines overexpressing wild type (WT) or catalytically inactive (H133Y) SIRT6 with respect to control cells (VECTOR). C, D, Western blot analysis of MDA-MB-231 (C) and MCF7 (D) cell lines silenced for SIRT6 (SIRT6-sh) with respect to control cells (VECTOR). A-D, One representative experiment out of three is presented.
Additional file 3: Fig. S3. Effect of SIRT6 deletion and overexpression on the expression of genes encoding for respiratory chain proteins and on mitochondrial mass and features. A, B, RNA was extracted from MDA-MB-231 cells engineered with a control vector (VECTOR) or a SIRT6-targeting shRNA (SIRT6-sh) (A) and from MMTV-PyMT+/-; Sirt6+/+ (n = 6) and MMTV-PyMT+/-; Sirt6+/- (n = 5) mice (B) and UQCRFS1, COX5B, NDUFB8, UQCRC2 and SDH expression was determined by QPCR. In A, data are presented as mean ± SD of three different experiments. *p<0.05, **p<0.01, ns: not statistically significant. C, D, MDA-MB-231 cells were engineered to overexpress WT SIRT6 (or a control vector) or to express a SIRT6-shRNA (or a control vector). Thereafter, cells were stained with Mitotracker deep red and analyzed by flow cytometry to detect their mitochondrial mass (C) or used for DNA extraction (D). In D, the amounts of DNA coding for tRNALeu and B2M were used to quantify mitochondrial and nuclear DNA, respectively, and their ratio was calculated. In C, one representative experiment out of three is presented. In D, data are presented as mean ± SD of three different experiments. ns: not statistically significant. (E) MDA-MB-231 cells that were engineered to overexpress WT SIRT6 (or a control vector) or a SIRT6-shRNA (or a control vector) were stained with Mitotracker deep red and analyzed by confocal microscopy, estimating mitochondrial area and circularity. Panel on the left shows one representative experiment out of three. In the histograms, data are presented as mean ± SD of at least three biological replicates. ns: not statistically significant.
Additional file 4: Fig. S4. SIRT6 levels do not affect Pdk1/4 expression but regulate intracellular calcium concentration in breast cancer cells. A, RNA was extracted from mammary tumors from twelve-week-old MMTV-PyMT+/-; Sirt6+/+ (n = 6) and MMTV-PyMT+/-; Sirt6+/- (n = 5) mice and Pdk1 and Pdk4 expression was determined by QPCR. B, MDA-MB-231 were engineered to overexpress WT SIRT6 (or a control vector) or to express a SIRT6-targeting shRNA (or a control vector). Thereafter, cells were used to measure intracellular calcium concentration. Data are presented as mean ± SD of three separate experiments. *p<0.05, ns: not statistically significant.
Additional file 5: Fig. S5. SIRT6 silencing enhances AMPK phosphorylation in MDA-MB-231 xenografts. A, MDA-MB-231 cells were engineered to express a SIRT6-shRNA (or a control vector). Thereafter, cells were used for protein lysate generation and phosphorylated AMPK (Thr183, Thr172), total AMPK and GAPDH were detected by Western blot. One representative experiment out of three is presented. B, MDA-MB-231 BC cells transduced with either a SIRT6-shRNA or with a control vector were injected subcutaneously into both flanks of BALB/c athymic nude mice. Animals were sacrificed 50 days after cell inoculation; tumors were used for protein lysate generation and phosphorylated and total AMPK, SIRT6 and vinculin were detected by Western blot. In the right panel, the intensity of the phospho-AMPK bands was normalized to that of the total AMPK bands and the phospho-AMPK/total-AMPK ratio in tumors with silenced SIRT6 was compared to that detected in control tumors. Data are presented as mean ± SD. *p<0.05.
Additional file 6: Fig. S6. Putative model for the metabolic, pro-oncogenic role of SIRT6 in breast tumorigenesis. SIRT6 increases expression and activity of PDH, as well as the levels of mitochondrial respiratory chain proteins. This results in increased OXPHOS and in a higher ATP/AMP ratio in BC cells. Ultimately, as a result of SIRT6 activity, AMPK activation is prevented and more ATP is available for cancer cells to grow.
Additional file 7: Supplementary Table 1. QPCR primer list.
Acknowledgements
We thank Dr. Raul Mostoslavsky (MGH Cancer Center, Boston, MA, USA) for providing us with the Sirt6+/− mice (129SvJ background), Dr. Thorsten Berger (Campell Family Institute for Breast Cancer Research, Ontario Cancer Institute, University Health Network, Toronto, Ontario, Canada) for providing us with the MMTV-PyMT mice in the 129 background, Dr. Katrin Chua (Department of Medicine - Med/Endocrinology, University of Stanford, Stanford, CA, USA) for providing us with the retroviral plasmids for silencing and for overexpressing SIRT6, and Dr. Gabriele Zoppoli and Dr. Gabriella Cirmena (Department of Internal Medicine and Medical Specialties, University of Genoa, Genoa, Italy) for their help with the gene expression studies of mammary tumor masses.
Abbreviations
- SIRT6
Sirtuin 6
- BC
Breast cancer
- PyMT
Polyoma middle tumor-antigen
- MMTV-PyMT
Mouse mammary tumor virus-polyoma middle tumor-antigen
- ATP
Adenosine triphosphate
- AMP
Adenosine monophosphate
- AMPK
AMP-activated protein kinase
- OXPHOS
Oxidative phosphorilation
- CCND1
Cyclin D1
- MMP9
Matrix metalloproteinase 9
- WT
Wild-type
- DNA
Deoxyribonucleic acid
- SDS-PAGE
Sodium dodecyl sulpahte-polyacrilamide gel electrophoresis
- PVDF
Polyvinylidene fluoride
- GAPDH
Glyceraldehyde 3-phosphate dehydrogenase
- EDTA
Ethylenediaminetetraacetic acid
- RT
Room temperature
- QPCR
Quantitative real-time polymerase chain reaction
- cDNA
Complementary DNA
- VAT
Visceral adipose tissue
- FACS
Fluorescence-activated cell sorting
- MFI
Mean fluorescence intensity
- mtDNA
Mitochondrial DNA
- nDNA
Nuclear DNA
- KO
Knock out
- PBS
Phosphate-buffered saline
- NADP+
Nicotinamide adenine dinucleotide phosphate
- HK
Hexokinase
- G6PD
Glucose-6-phosphate dehydrogenase
- NADH
Nicotinamide adenine dinucleotide reduced
- AK
Adenylate kinase
- PK
Pyruvate kinase
- LDH
Lactate dehydrogenase
- ADP
Adenosine diphosphate
- PDH
Pyruvate dehydrogenase
- STAT3
Signal transducer and activator of transcription 3
- FER
Feline encephalitis virus-related kinase
- FerT
FER variant
- CHCHD2
Coiled-coil-helix-coiled-coil-helix domain containing 2
- PDK
Pyruvate dehydrogenase kinase
- Uqcrfs1
Ubiquinol-cytochrome C reductase
- COX5B
Cytochrome C oxidase subunit 5B
- Ndufb8
NADH:ubiquinone oxidoreductase subunit B8
- Uqcrc2
Ubiquinol-cytochrome C reductase core protein 2
- PI3K
Phosphatidylinositol-4,5-bisphosphate 3-kinase
- HIF-1α
Hypoxia-inducible factor 1 subunit alpha
- NF-κB
Nuclear factor kappa B
- ECL
Enhanced chemiluminescence
- sh
Short hairpin
Authors’ contributions
Conceptualization, funding acquisition, supervision: A.N. Methodology: P.B. and I.C. Investigation: P.B., I.C., P.D., V.V., A.B., T.B., D.R., M.P., and S.R. Writing—original draft: P.B., F.P., and A.N. Writing—review and editing: A.N., S.B., S.R., F.P., I.C., A.K., M.G., A.G., L.T., M.S., A.P., F.M. and M.C. Visualization: P.B., F.P. The author(s) read and approved the final manuscript.
Funding
This work was supported in part by the Associazione Italiana per la Ricerca sul Cancro (AIRC; IG#17736 and #22098 to A.N.), the Italian Ministry of Health (GR-2011-02347192 to A.N.), the Seventh Framework Program projects PANACREAS (#256986) and ATHERO-B-CELL (#602114), the 5×1000 2014 Funds to the IRCCS Ospedale Policlinico San Martino (to A.N.), the BC161452P1 grants of the Breast Cancer Research Program (US Department of Defense, to A.N.), and the Associazione Italiana contro le Leucemie-linfomi e Mieloma (AIL), Sezione Liguria (to A.N.). In addition, this project has received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement No 813284 (to A.N. and S.B.).
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Ethics approval and consent to participate
Animal studies were conducted in accordance with the laws and institutional guidelines for animal care and were approved by the Institutional Animal Care and Use Committee of the Scientific Institute for Research and Healthcare (IRCCS) University Hospital San Martino – National Institute for Cancer Research (IST, Genoa, Italy; protocol #453).
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Pamela Becherini, Irene Caffa and Francesco Piacente contributed equally to this work.
Contributor Information
Silvia Ravera, Email: silvia.ravera@unige.it.
Alessio Nencioni, Email: alessio.nencioni@unige.it.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Metallo CM, Vander Heiden MG. Understanding metabolic regulation and its influence on cell physiology. Mol Cell. 2013;49(3):388–398. doi: 10.1016/j.molcel.2013.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zacksenhaus E, Shrestha M, Liu JC, Vorobieva I, Chung PED, Ju Y, et al. Mitochondrial OXPHOS induced by RB1 deficiency in breast cancer: implications for anabolic metabolism, stemness, and metastasis. Trends Cancer. 2017;3(11):768–779. doi: 10.1016/j.trecan.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 4.Warburg O, Wind F, Negelein E. The metabolism of tumors in the body. J Gen Physiol. 1927;8(6):519–530. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khan RI, Nirzhor SSR, Akter R. A review of the recent advances made with SIRT6 and its implications on aging related processes, major human diseases, and possible therapeutic targets. Biomolecules. 2018;8(3):44. doi: 10.3390/biom8030044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Gool F, Galli M, Gueydan C, Kruys V, Prevot PP, Bedalov A, et al. Intracellular NAD levels regulate tumor necrosis factor protein synthesis in a sirtuin-dependent manner. Nat Med. 2009;15(2):206–210. doi: 10.1038/nm.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawahara TL, Michishita E, Adler AS, Damian M, Berber E, Lin M, et al. SIRT6 links histone H3 lysine 9 deacetylation to NF-κB-dependent gene expression and organismal life span. Cell. 2009;136(1):62–74. doi: 10.1016/j.cell.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sugatani T, Agapova O, Malluche HH, Hruska KA. SIRT6 deficiency culminates in low-turnover osteopenia. Bone. 2015;81:168–177. doi: 10.1016/j.bone.2015.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Etchegaray JP, Zhong L, Mostoslavsky R. The histone deacetylase SIRT6: at the crossroads between epigenetics, metabolism and disease. Curr Top Med Chem. 2013;13(23):2991–3000. doi: 10.2174/15680266113136660213. [DOI] [PubMed] [Google Scholar]
- 10.Nahalkova J. The protein-interaction network with functional roles in tumorigenesis, neurodegeneration, and aging. Mol Cell Biochem. 2016;423(1-2):187–196. doi: 10.1007/s11010-016-2836-5. [DOI] [PubMed] [Google Scholar]
- 11.Sebastian C, Zwaans BM, Silberman DM, Gymrek M, Goren A, Zhong L, et al. The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell. 2012;151(6):1185–1199. doi: 10.1016/j.cell.2012.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cea M, Cagnetta A, Adamia S, Acharya C, Tai YT, Fulciniti M, et al. Evidence for a role of the histone deacetylase SIRT6 in DNA damage response of multiple myeloma cells. Blood. 2016;127(9):1138–1150. doi: 10.1182/blood-2015-06-649970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cagnetta A, Soncini D, Orecchioni S, Talarico G, Minetto P, Guolo F, et al. Depletion of SIRT6 enzymatic activity increases acute myeloid leukemia cells’ vulnerability to DNA-damaging agents. Haematologica. 2018;103(1):80–90. doi: 10.3324/haematol.2017.176248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bauer I, Grozio A, Lasiglie D, Basile G, Sturla L, Magnone M, et al. The NAD+-dependent histone deacetylase SIRT6 promotes cytokine production and migration in pancreatic cancer cells by regulating Ca2+ responses. J Biol Chem. 2012;287(49):40924–40937. doi: 10.1074/jbc.M112.405837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lefort K, Brooks Y, Ostano P, Cario-Andre M, Calpini V, Guinea-Viniegra J, et al. A miR-34a-SIRT6 axis in the squamous cell differentiation network. EMBO J. 2013;32(16):2248–2263. doi: 10.1038/emboj.2013.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bae JS, Park SH, Jamiyandorj U, Kim KM, Noh SJ, Kim JR, et al. CK2α/CSNK2A1 phosphorylates SIRT6 and is involved in the progression of breast carcinoma and predicts shorter survival of diagnosed patients. Am J Pathol. 2016;186(12):3297–3315. doi: 10.1016/j.ajpath.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 17.Khongkow M, Olmos Y, Gong C, Gomes AR, Monteiro LJ, Yague E, et al. SIRT6 modulates paclitaxel and epirubicin resistance and survival in breast cancer. Carcinogenesis. 2013;34(7):1476–1486. doi: 10.1093/carcin/bgt098. [DOI] [PubMed] [Google Scholar]
- 18.Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124(2):315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 19.Berger T, Cheung CC, Elia AJ, Mak TW. Disruption of the Lcn2 gene in mice suppresses primary mammary tumor formation but does not decrease lung metastasis. Proc Natl Acad Sci U S A. 2010;107(7):2995–3000. doi: 10.1073/pnas.1000101107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCord RA, Michishita E, Hong T, Berber E, Boxer LD, Kusumoto R, et al. SIRT6 stabilizes DNA-dependent protein kinase at chromatin for DNA double-strand break repair. Aging. 2009;1(1):109–121. doi: 10.18632/aging.100011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tasselli L, Xi Y, Zheng W, Tennen RI, Odrowaz Z, Simeoni F, et al. SIRT6 deacetylates H3K18ac at pericentric chromatin to prevent mitotic errors and cellular senescence. Nat Struct Mol Biol. 2016;23(5):434–440. doi: 10.1038/nsmb.3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caffa I, D’Agostino V, Damonte P, Soncini D, Cea M, Monacelli F, et al. Fasting potentiates the anticancer activity of tyrosine kinase inhibitors by strengthening MAPK signaling inhibition. Oncotarget. 2015;6(14):11820–11832. doi: 10.18632/oncotarget.3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salani B, Ravera S, Fabbi P, Garibaldi S, Passalacqua M, Brunelli C, et al. Glibenclamide mimics metabolic effects of metformin in H9c2 cells. Cell Physiol Biochem. 2017;43(3):879–890. doi: 10.1159/000481638. [DOI] [PubMed] [Google Scholar]
- 24.Weidner N. Current pathologic methods for measuring intratumoral microvessel density within breast carcinoma and other solid tumors. Breast Cancer Res Treat. 1995;36(2):169–180. doi: 10.1007/BF00666038. [DOI] [PubMed] [Google Scholar]
- 25.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 26.Marini C, Ravera S, Buschiazzo A, Bianchi G, Orengo AM, Bruno S, et al. Discovery of a novel glucose metabolism in cancer: the role of endoplasmic reticulum beyond glycolysis and pentose phosphate shunt. Sci Rep. 2016;6:25092. doi: 10.1038/srep25092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ravera S, Aluigi MG, Calzia D, Ramoino P, Morelli A, Panfoli I. Evidence for ectopic aerobic ATP production on C6 glioma cell plasma membrane. Cell Mol Neurobiol. 2011;31(2):313–321. doi: 10.1007/s10571-010-9624-7. [DOI] [PubMed] [Google Scholar]
- 28.Bartolucci M, Ravera S, Garbarino G, Ramoino P, Ferrando S, Calzia D, et al. Functional expression of electron transport chain and FoF1-ATP synthase in optic nerve myelin sheath. Neurochem Res. 2015;40(11):2230–2241. doi: 10.1007/s11064-015-1712-0. [DOI] [PubMed] [Google Scholar]
- 29.Lin EY, Jones JG, Li P, Zhu L, Whitney KD, Muller WJ, et al. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am J Pathol. 2003;163(5):2113–2126. doi: 10.1016/S0002-9440(10)63568-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol. 1992;12(3):954–961. doi: 10.1128/MCB.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu S, Yin M, Koroleva M, Mastrangelo MA, Zhang W, Bai P, et al. SIRT6 protects against endothelial dysfunction and atherosclerosis in mice. Aging. 2016;8(5):1064–1082. doi: 10.18632/aging.100975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clark-Knowles KV, Dewar-Darch D, Jardine KE, McBurney MW. SIRT1 catalytic activity has little effect on tumor formation and metastases in a mouse model of breast cancer. PLoS One. 2013;8(11):e82106. doi: 10.1371/journal.pone.0082106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones RA, Robinson TJ, Liu JC, Shrestha M, Voisin V, Ju Y, et al. RB1 deficiency in triple-negative breast cancer induces mitochondrial protein translation. J Clin Invest. 2016;126(10):3739–3757. doi: 10.1172/JCI81568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bruno S, Ledda B, Tenca C, Ravera S, Orengo AM, Mazzarello AN, et al. Metformin inhibits cell cycle progression of B-cell chronic lymphocytic leukemia cells. Oncotarget. 2015;6(26):22624–22640. doi: 10.18632/oncotarget.4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhong L, D'Urso A, Toiber D, Sebastian C, Henry RE, Vadysirisack DD, et al. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell. 2010;140(2):280–293. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cui X, Yao L, Yang X, Gao Y, Fang F, Zhang J, et al. SIRT6 regulates metabolic homeostasis in skeletal muscle through activation of AMPK. Am J Physiol Endocrinol Metab. 2017;313(4):E493–E505. doi: 10.1152/ajpendo.00122.2017. [DOI] [PubMed] [Google Scholar]
- 37.Khan D, Sarikhani M, Dasgupta S, Maniyadath B, Pandit AS, Mishra S, et al. SIRT6 deacetylase transcriptionally regulates glucose metabolism in heart. J Cell Physiol. 2018;233(7):5478–5489. doi: 10.1002/jcp.26434. [DOI] [PubMed] [Google Scholar]
- 38.Chiang PK, Sacktor B. Control of pyruvate dehydrogenase activity in intact cardiac mitochondria. Regulation of the inactivation and activation of the dehydrogenase. J Biol Chem. 1975;250(9):3399–3408. doi: 10.1016/S0021-9258(19)41529-9. [DOI] [PubMed] [Google Scholar]
- 39.Pettit FH, Roche TE, Reed LJ. Function of calcium ions in pyruvate dehydrogenase phosphatase activity. Biochem Biophys Res Commun. 1972;49(2):563–571. doi: 10.1016/0006-291X(72)90448-2. [DOI] [PubMed] [Google Scholar]
- 40.Steinberg GR, Kemp BE. AMPK in health and disease. Physiol Rev. 2009;89(3):1025–1078. doi: 10.1152/physrev.00011.2008. [DOI] [PubMed] [Google Scholar]
- 41.Wang Z, Wang N, Liu P, Xie X. AMPK and cancer. Experientia Suppl. 2016;107:203–226. doi: 10.1007/978-3-319-43589-3_9. [DOI] [PubMed] [Google Scholar]
- 42.Theodossiou TA, Ali M, Grigalavicius M, Grallert B, Dillard P, Schink KO, et al. Simultaneous defeat of MCF7 and MDA-MB-231 resistances by a hypericin PDT-tamoxifen hybrid therapy. NPJ Breast Cancer. 2019;5:13. doi: 10.1038/s41523-019-0108-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Damonte P, Sociali G, Parenti MD, Soncini D, Bauer I, Boero S, et al. SIRT6 inhibitors with salicylate-like structure show immunosuppressive and chemosensitizing effects. Bioorg Med Chem. 2017;25(20):5849–5858. doi: 10.1016/j.bmc.2017.09.023. [DOI] [PubMed] [Google Scholar]
- 44.Sociali G, Grozio A, Caffa I, Schuster S, Becherini P, Damonte P, et al. SIRT6 deacetylase activity regulates NAMPT activity and NAD(P)(H) pools in cancer cells. FASEB J. 2019;33(3):3704–3717. doi: 10.1096/fj.201800321R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sociali G, Galeno L, Parenti MD, Grozio A, Bauer I, Passalacqua M, et al. Quinazolinedione SIRT6 inhibitors sensitize cancer cells to chemotherapeutics. Eur J Med Chem. 2015;102:530–539. doi: 10.1016/j.ejmech.2015.08.024. [DOI] [PubMed] [Google Scholar]
- 46.Sinha S, Patel S, Athar M, Vora J, Chhabria MT, Jha PC, et al. Structure-based identification of novel sirtuin inhibitors against triple negative breast cancer: an in silico and in vitro study. Int J Biol Macromol. 2019;140:454–468. doi: 10.1016/j.ijbiomac.2019.08.061. [DOI] [PubMed] [Google Scholar]
- 47.Ming M, Han W, Zhao B, Sundaresan NR, Deng CX, Gupta MP, et al. SIRT6 promotes COX-2 expression and acts as an oncogene in skin cancer. Cancer Res. 2014;74(20):5925–5933. doi: 10.1158/0008-5472.CAN-14-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elhanati S, Kanfi Y, Varvak A, Roichman A, Carmel-Gross I, Barth S, et al. Multiple regulatory layers of SREBP1/2 by SIRT6. Cell Rep. 2013;4(5):905–912. doi: 10.1016/j.celrep.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 49.Ioris RM, Galie M, Ramadori G, Anderson JG, Charollais A, Konstantinidou G, et al. SIRT6 suppresses cancer stem-like capacity in tumors with PI3K activation independently of its deacetylase activity. Cell Rep. 2017;18(8):1858–1868. doi: 10.1016/j.celrep.2017.01.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bachman KE, Argani P, Samuels Y, Silliman N, Ptak J, Szabo S, et al. The PIK3CA gene is mutated with high frequency in human breast cancers. Cancer Biol Ther. 2004;3(8):772–775. doi: 10.4161/cbt.3.8.994. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Fig. S1. Effect of Sirt6 heterozygous deletion on Sirt6, Glut4 and Hk2 expression in mouse mammary tumors and tissues. A, B, Mammary tumors and healthy tissues [heart, lung, spleen, kidney and visceral adipose tissue (VAT)] were isolated from twelve-week-old and from four-week-old MMTV-PyMT+/-; Sirt6+/+ (n = 3) and MMTV-PyMT+/-; Sirt6+/- (n = 3) mice, respectively, and used for protein lysate generation and for RNA extraction. In A, Sirt6 and vinculin levels were assessed by Western blot in mammary tumors. In B, Hk2 and Glut4 expression in bodily tissues was detected by QPCR. Data are presented as mean ± SD. ns: not statistically significant.
Additional file 2: Fig. S2. SIRT6 silencing and overexpression in MDA-MB-231 and MCF7 cell lines. A, B, Western blot analysis of MDA-MB-231 (A) and MCF7 (B) cell lines overexpressing wild type (WT) or catalytically inactive (H133Y) SIRT6 with respect to control cells (VECTOR). C, D, Western blot analysis of MDA-MB-231 (C) and MCF7 (D) cell lines silenced for SIRT6 (SIRT6-sh) with respect to control cells (VECTOR). A-D, One representative experiment out of three is presented.
Additional file 3: Fig. S3. Effect of SIRT6 deletion and overexpression on the expression of genes encoding for respiratory chain proteins and on mitochondrial mass and features. A, B, RNA was extracted from MDA-MB-231 cells engineered with a control vector (VECTOR) or a SIRT6-targeting shRNA (SIRT6-sh) (A) and from MMTV-PyMT+/-; Sirt6+/+ (n = 6) and MMTV-PyMT+/-; Sirt6+/- (n = 5) mice (B) and UQCRFS1, COX5B, NDUFB8, UQCRC2 and SDH expression was determined by QPCR. In A, data are presented as mean ± SD of three different experiments. *p<0.05, **p<0.01, ns: not statistically significant. C, D, MDA-MB-231 cells were engineered to overexpress WT SIRT6 (or a control vector) or to express a SIRT6-shRNA (or a control vector). Thereafter, cells were stained with Mitotracker deep red and analyzed by flow cytometry to detect their mitochondrial mass (C) or used for DNA extraction (D). In D, the amounts of DNA coding for tRNALeu and B2M were used to quantify mitochondrial and nuclear DNA, respectively, and their ratio was calculated. In C, one representative experiment out of three is presented. In D, data are presented as mean ± SD of three different experiments. ns: not statistically significant. (E) MDA-MB-231 cells that were engineered to overexpress WT SIRT6 (or a control vector) or a SIRT6-shRNA (or a control vector) were stained with Mitotracker deep red and analyzed by confocal microscopy, estimating mitochondrial area and circularity. Panel on the left shows one representative experiment out of three. In the histograms, data are presented as mean ± SD of at least three biological replicates. ns: not statistically significant.
Additional file 4: Fig. S4. SIRT6 levels do not affect Pdk1/4 expression but regulate intracellular calcium concentration in breast cancer cells. A, RNA was extracted from mammary tumors from twelve-week-old MMTV-PyMT+/-; Sirt6+/+ (n = 6) and MMTV-PyMT+/-; Sirt6+/- (n = 5) mice and Pdk1 and Pdk4 expression was determined by QPCR. B, MDA-MB-231 were engineered to overexpress WT SIRT6 (or a control vector) or to express a SIRT6-targeting shRNA (or a control vector). Thereafter, cells were used to measure intracellular calcium concentration. Data are presented as mean ± SD of three separate experiments. *p<0.05, ns: not statistically significant.
Additional file 5: Fig. S5. SIRT6 silencing enhances AMPK phosphorylation in MDA-MB-231 xenografts. A, MDA-MB-231 cells were engineered to express a SIRT6-shRNA (or a control vector). Thereafter, cells were used for protein lysate generation and phosphorylated AMPK (Thr183, Thr172), total AMPK and GAPDH were detected by Western blot. One representative experiment out of three is presented. B, MDA-MB-231 BC cells transduced with either a SIRT6-shRNA or with a control vector were injected subcutaneously into both flanks of BALB/c athymic nude mice. Animals were sacrificed 50 days after cell inoculation; tumors were used for protein lysate generation and phosphorylated and total AMPK, SIRT6 and vinculin were detected by Western blot. In the right panel, the intensity of the phospho-AMPK bands was normalized to that of the total AMPK bands and the phospho-AMPK/total-AMPK ratio in tumors with silenced SIRT6 was compared to that detected in control tumors. Data are presented as mean ± SD. *p<0.05.
Additional file 6: Fig. S6. Putative model for the metabolic, pro-oncogenic role of SIRT6 in breast tumorigenesis. SIRT6 increases expression and activity of PDH, as well as the levels of mitochondrial respiratory chain proteins. This results in increased OXPHOS and in a higher ATP/AMP ratio in BC cells. Ultimately, as a result of SIRT6 activity, AMPK activation is prevented and more ATP is available for cancer cells to grow.
Additional file 7: Supplementary Table 1. QPCR primer list.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.