Abstract
Background
Colorectal cancer (CRC) is one of the most common malignant cancers globally. Circular RNAs (circRNAs) have been implicated in the development of CRC. In this paper, we set to explore the precise action of circ_0067835 in CRC progression and radioresistance.
Methods
Quantitative real-time polymerase chain reaction (qRT-PCR) was used to evaluate the expression of circ_0067835, microRNA-296-5p (miR-296-5p) and insulin-like growth factor 1 receptor (IGF1R). Western blot was used to measure the level of IGF1R protein. Cell proliferation, cell cycle distribution and apoptosis were determined by Cell Counting Kit-8 (CCK-8), colony formation, flow cytometry and caspase-3 activity assays, respectively. The direct relationship between miR-296-5p and circ_0067835 or IGF1R was verified by dual-luciferase reporter assays. Additionally, in vivo assays were applied to confirm the role of circ_0067835 in vivo.
Results
Exosomal circ_0067835 was upregulated in the serum of CRC patients after radiotherapy. Exosome-mediated circ_0067835 knockdown repressed cell proliferation, cell cycle progression, and enhanced cell apoptosis and radiosensitivity in vitro. Circ_0067835 sponged miR-296-5p to regulate IGF1R expression in CRC cells. Moreover, the knockdown of circ_0067835 regulated CRC cell behaviors by up-regulating miR-296-5p and down-regulating IGF1R in vitro. Furthermore, circ_0067835 knockdown diminished tumor growth and promoted cell radiosensitivity in vivo.
Conclusion
Circ_0067835 knockdown suppressed CRC progression and enhanced CRC cell radiosensitivity partially by the miR-296-5p/IGF1R axis. The findings established a rationale that targeting circ_0067835 might be a promising point for improving CRC treatment.
Keywords: exosomes, circ_0067835, IGF1R, miR-296-5p, colorectal cancer
Introduction
Colorectal cancer (CRC) is one of the most common malignant cancers globally.1 Traditional therapeutic options against CRC include surgery, chemotherapy, radiotherapy and molecular targeted drugs treatment.2,3 Among these, radiotherapy has been widely used for CRC clinical treatment.4,5 However, the emergence of radiation resistance has become a major question for CRC treatment.6–8 Thus, understanding the molecular determinants underlying the regulation of CRC radioresistance is urgently needed.
Circular RNAs (circRNAs) are covalently closed RNA circles that are more stable than the corresponding linear mRNA.9 Interestingly, some circRNAs have been identified as post-transcriptional modulators of gene expression by sponging microRNAs (miRNAs).10,11 Recently, some circRNAs have been shown to be transferred by exosomes and thus involve in human carcinogenesis and radioresistance development.12,13 A previous report uncovered that circ_0067835 could inhibit cell proliferation and metastasis in endometrial carcinoma.14 Importantly, circ_0067835 contributed to CRC development by regulating miR-1236-3p/homeobox B7 (HOXB7) axis.15 However, the critical roles of circ_0067835 in CRC radioresistance largely remain to be elucidated.
Here, we identified that circ_0067835 could be transferred by exosomes in the serum of CRC patients. By combining functional analysis, we demonstrated that circ_0067835 knockdown repressed CRC progression and promoted radiosensitivity by modulating insulin-like growth factor 1 receptor (IGF1R) expression through sponging miR-1236-3p.
Materials and Methods
Clinical Tissue Specimens
Thirty-one healthy volunteers and 58 CRC patients were recruited to participate in this project which was approved by the Ethics Committee of Henan Provincial People’s Hospital. Among CRC patients, CRC tissues and adjacent normal tissues (ANT) were obtained from 39 patients who suffered CRC radical resection from Henan Provincial People’s Hospital. All participators signed the informed consent notice. Additionally, a total 19 blood samples were taken from another 19 CRC patients before or after radiation therapy.
Cell Culture
Fetal human colon cells (FHC), human colon cancer cell lines SW620 (derived from lymph node metastasis) and HCT116 were provided by American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco, Carlsbad, CA, USA) with 10% fetal bovine serum (PAN, Bavaria, Germany) and 1% penicillin-streptomycin (Gibco) at 37°C with 5% CO2.
Cell Transfection
shRNA-circ_0067835 (sh-circ_0067835), miR-296-5p mimics (miR-296-5p), IGF1R overexpression plasmid (IGF1R), miR-296-5p inhibitors (anti-miR-296-5p) and the corresponding negative controls (sh-NC, sh-NC-exo, miR-NC mimics, pcDNA, anti-NC) were commercially purchased from Lianmai Biological Engineering Co., Ltd (Shanghai, China). The transient transfections were performed with Lipofectamine 3000 transfection reagent (Invitrogen, Carlsbad, CA, USA).
RNA Isolation and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted from tissues, cells and serums by TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA). Reverse transcription was performed by complementary DNA (cDNA) reverse transcription kit (Applied Biosystems, Foster City, CA, USA). The expression of interest genes was determined by SYBR Green Master Mix kit (Takara, Dalian, China) with ABI7500 Real time PCR system (Applied Biosystems). Relative circ_0067835, IGF1R and miR-296-5p levels were calculated by the method of 2−ΔΔCT. The primers (5ʹ-3ʹ) in this study were presented as below: circ_0067835 F, CACAACCTCATGAGCATCCCA, circ_0067835 R, TAGTCCAGCCCACACAGCTT; IGF1R F, GACCTCTTCCCGAACCTC, IGF1R R, TGTAGTTATTGGACACCGCAT; miR-296-5p F, GTATCCAGTGCAGGGTCCGA, miR-296-5p R, CGACGAGGGCCCCCCCT; U6 F, GCTTGCTTCGGCAGCACATATAC, U6 R, TGCATGTCATCCTTGCTCAGGG; GAPDH F, GATATTGTTGCCATCAATGAC, GAPDH R, TTGATTTTGGAGGGATCTCG. U6 or GAPDH was served as the reference gene.
Exosome Isolation and Identification
The exosomes were separated from CRC patients’ serum or the cell culture supernatant by ultracentrifugation method. To eliminate the effect of debris on the isolation of exosomes, the samples including serums and cell culture supernatant were orderly centrifuged at 300 ×g for 10 min, 2000 ×g for 10 min and 10,000 ×g for 30 min by using cryogenic high-speed centrifuge (Thermo Fisher scientific). Then, the collected supernatants were centrifuged at 120,000 ×g for 90 min to isolate the precipitate that contained the exosomes. The isolated precipitate was purified by particle-free PBS (pH 7.4, Sigma-Aldrich, St. Louis, MO, USA). After that, the appearance of exosomes was visualized by transmission electron microscopy (TEM, JEM-2000EX TEM, JEOL Ltd., Tokyo, Japan) at aproportional scale of 0.2 μm. Furthermore, the size and concentration of exosomes were detected by using a nanoparticle tracking analysis (NTA, Malvern, Malvern, UK).
Western Blot
Total protein was isolated from tissues, cells and exosomes by Radio-Immunoprecipitation Assay (RIPA) buffer (Cell Signaling Technology, Boston, MA, USA). The protein samples were resolved on sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE, Beyotime, Shanghai, China) and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica, MA, USA). After that, the membranes were fixed by non-fat milk and washed by 1×Tris-buffered saline tween-20 (TBST, Sigma-Aldrich). Then, the membranes were incubated with the primary antibodies including anti-CD63 (1:500, Sigma-Aldrich), anti-TSG101 (1:500, Sigma-Aldrich), anti-IGF1R (1:500, Sigma-Aldrich), anti-β-actin loading control (1:5000, Sigma-Aldrich). The protein bands were further incubated with horseradish peroxidase-conjugated goat anti-rabbit (1:100000, Sigma-Aldrich). Enhanced chemiluminescence (ECL) detection kit (Santa Cruz Biotechnology, Santa Cruz, CA, USA) was employed to detect the protein complexes.
Cell Counting Kit-8 (CCK-8) Assay
Cell Counting Kit-8 (CCK-8, Dojindo, Tokyo, Japan) assay was applied to test the viability of CRC cells. Briefly, the transfected cells were cultured in 96-well plates 12 h before incubation with 10 μL of CCK-8 solution (Dojindo) in each well for 2 h at 37°C. Then, the absorbance at 450 nm was detected by a MultiMode Microplate Reader (Thermo Fisher Scientific).
Colony Formation Assay
CRC cells were cultured in six-well plates supplemented with the DMEM (Gibco) for 24 h at a density of 500 cells per well. Then, cells were treated with 0 Gy, 1 Gy, 2 Gy, 4 Gy, or 8 Gy dose IR (Faxitron MultiRad 225, Tucson, AZ, USA) and then grew in 60 mm dishes for 14 days. Cells were washed with PBS (Sigma-Aldrich), fixed with methanol (Sigma-Aldrich), and stained with 0.1% crystal violet (Sigma-Aldrich) for 5 min. The colonies were recorded by using a digital camera (Nikon, Tokyo, Japan).
Flow Cytometry Assay
The apoptosis and cell cycle distribution were assessed by flow cytometry assay. For the assessment of apoptosis, the Annexin V-Fluorescein isothiocyanate (FITC)/Propidium lodide (PI) Apoptosis Kit (BD Biosciences, San Jose, CA, USA) was used. Briefly, the transfected cells treated with a 4 Gy dose of IR were collected and washed with PBS (Sigma-Aldrich) for three times. Then, cells were incubated with a 200 µL binding buffer containing 10 µL Annexin V-FITC and 5 µL PI (Sigma-Aldrich). The FACS Canto™ II flow cytometer (BD Biosciences) was used to analyze the data of apoptosis. For the determination of cell cycle distribution, the transfected cells treated with 4 Gy dose of IR were harvested and washed by PBS (Sigma-Aldrich) for three times. Then, cells were incubated with PI at 37°C for 30 min. The distribution of G1, S, and G2/M phases in cells were inspected by BD FACS Canto™ II flow cytometer (BD Biosciences).
Caspase-3 Activity Assay
The caspase-3 activity was assessed by caspase-3 colorimetric assay kit (Sigma-Aldrich). The transfected cells were lysed by RIPA buffer (Beyotime). The lysates were centrifuged at 20,000 ×g, 4°C for 15 min to isolate the supernatant which was further reacted with caspase-3 substrate (Ac-DEVD-pNA, Sigma-Aldrich) buffer in 96-well plates for 4 h. A microplate reader (Bio-Rad Laboratories, Hercules, CA, USA) was applied to determine the absorbance at 405 nm.
Dual-Luciferase Reporter Assay
Analysis of the targeted miRNAs of circ_0067835 and miR-296-5p targets was performed by the Circinteractome and Starbase softwares, respectively. The segments of circ_0067835 and IGF1R 3ʹUTR harboring the miR-296-5p-binding sequences or mutant target regions were individually cloned into the pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega, Madison, WI, USA). Each reporter construct was transfected into cells together with miR-NC mimics or miR-296-5p mimics using Lipofectamine 2000 (Invitrogen). The Dual-Luciferase Reporter detection System (Promega) was used to detect the luciferases activity.
In vivo Experiments
Animal studies were approved by the Animal Ethics Committee of Henan Provincial People’s Hospital. Animal studies were performed in compliance with the ARRIVE guidelines and the Basel Declaration. BABL/c male nude mice (n=24, 4 week old) were provided by the Slaccas Shanghai Laboratory Animal Co., Ltd. (Shanghai, China). All mice were divided into four groups (n = 6 per group) and subcutaneously inoculated 5 × 106 SW620 cells which were transfected with sh-NC or sh-circ_0067835. Additionally, the mice were further treated with or without 4 Gy IR every 5 days. Tumor volume was recorded every 5 days after 5 days implantation. Twenty-five days later, the tumors were excised from the mice.
Statistical Analysis
GraphPad Prism version 7.0 (GraphPad Software, Inc., La Jolla, CA, USA) was used to analyze the data. Data were expressed as mean ± standard deviation (SD) from at least three replications. Pearson Correlation Coefficient was applied to evaluate the correlation between miR-296-5p and circ_0067835 or IGF1R expression in the serum of CRC patients. The differences were analyzed by Student’s t-test or one-way Analysis of Variance (ANOVA). Receiver operating characteristic (ROC) curve was plotted to analyze the area under the ROC curve (AUC). P<0.05 was considered as the significant difference.
Results
Serum Exosomal Circ_0067835 Was Upregulated in CRC Patients After Radiotherapy
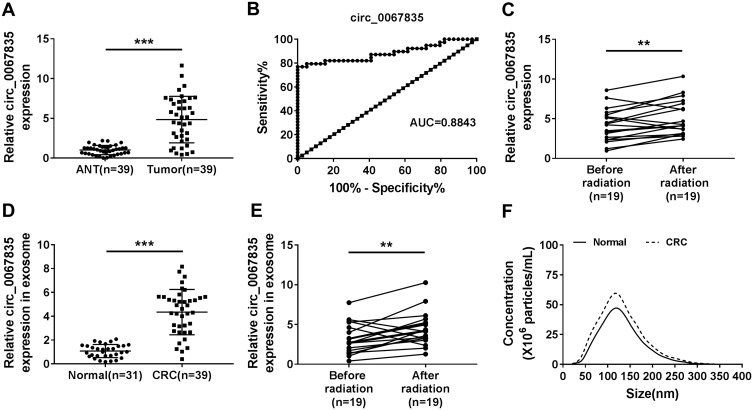
In order to explore the involvement of circ_0067835 in CRC radioresistance, we firstly used qRT-PCR to gauge its expression in CRC tissues. The upregulation of circ_0067835 expression was found in CRC tissues compared with adjacent normal tissues (ANT) (Figure 1A). The curve of ROC showed that the AUC value of circ_0067835 was 0.8843, suggesting the sensitivity and specificity of circ_0067835 in CRC (Figure 1B). Moreover, serum circ_0067835 was upregulated in CRC patients after radiotherapy compared with the controls untreated with radiotherapy (Figure 1C). To confirm whether circ_0067835 could be transferred by exosomes, we then isolated the exosomes from the serum of CRC patients. TEM showed the image of exosomes and Western blot validated the expression of exosome-related proteins (CD63 and TSG101) (Supplementary Figure 1A and 1B). Interestingly, circ_0067835 expression in exosomes was markedly increased in the serum of CRC patients (Figure 1D). Radiotherapy led to a striking increase in circ_0067835 expression in exosomes in the serum of CRC patients (Figure 1E). Additionally, NTA analysis revealed that the diameters of the most particles were within the range of 70–160 nm, and CRC patients showed a higher concentration of serum exosome compared with normal patients (Figure 1F).
Figure 1.
Circ_0067835 was upregulated in CRC. (A) The expression of circ_0067835 was detected by qRT-PCR in CRC tissues (n=39) and adjacent normal tissues (ANT) (n=39). (B) ROC curve and AUC values were analyzed for circ_0067835 in CRC patients. (C) The expression of circ_0067835 was detected by qRT-PCR in the serum of CRC patients with (n=19) or without (n=19) radiation treatment. (D) The expression of circ_0067835 in exosome was detected by qRT-PCR in the serum of CRC patients (n=39) and normal patients (n=31). (E) The expression of circ_0067835 in exosome was detected by qRT-PCR in the serum of CRC patients with (n=19) or without (n=19) radiation treatment. (F) The size of cellular exosome diameters was detected by NTA. **P<0.01, ***P<0.001.
Exosome-Mediated Circ_0067835 Knockdown Inhibited Cell Proliferation, Cell Cycle Progression, and Enhanced Apoptosis and Radiosensitivity in vitro
The data of qRT-PCR showed a significant upregulation of circ_0067835 in SW620 and HCT116 CRC cells compared with FHC normal cells (Figure 2A). To elucidate the functional role of circ_0067835, we performed loss-of-function analysis using sh-circ_0067835 in SW620 and HCT116 cells (Figure 2B). We then used the exosomes (sh-circ_0067835-exo) from sh-circ_0067835-transducing cells to treat the corresponding parental cells (SW620 and HCT116). Interestingly, sh-circ_0067835-exo treatment induced a significant suppression in cell viability compared with sh-NC-exo in CRC cells (Figure 2C and D). Moreover, sh-circ_0067835-exo treatment led to a decrease in cell survival fraction under IR treatment, demonstrating that sh-circ_0067835-exo promoted cell radiosensitivity (Figure 2E and F). Furthermore, sh-circ_0067835-exo treatment repressed cell cycle progression (Figure 2G), enhanced caspase-3 activity (Figure 2H), and promoted cells apoptosis (Figure 2I) in the two cell lines with or without IR treatment.
Figure 2.
Exosome-mediated circ_0067835 knockdown inhibited cell proliferation, cell cycle progression, and enhanced apoptosis and radiosensitivity in vitro. (A) Circ_0067835 expression in SW620 and HCT116 cells and the normal colon epithelial cells (FHC) was detected by qRT-PCR. (B) qRT-PCR analyzed circ_0067835 expression in SW620 and HCT116 cells transfected with sh-circ_0067835 or sh-NC. (C and D) The viability of SW620 and HCT116 cells treated with sh-circ_0067835-exo or negative control was detected by CCK-8 assay. (E and F) The survival fraction of SW620 and HCT116 cells treated with 0 Gy, 1 Gy, 2 Gy, 4 Gy, 8 Gy IR and sh-circ_0067835-exo or negative control was detected by colony formation assay. (G) Cell cycle distribution was measured by flow cytometry assay in SW620 and HCT116 cells treated with or without 4 Gy IR and sh-circ_0067835-exo or negative control. (H) The caspase-3 activity was tested by caspase-3 activity assay in SW620 and HCT116 cells treated with or without 4 Gy IR and sh-circ_0067835-exo or negative control. (I) Flow cytometry assay was used to detect the apoptosis rate in SW620 and HCT116 cells treated with or without 4 Gy IR and sh-circ_0067835-exo or negative control. *P<0.05, **P<0.01, ***P<0.001.
Circ_0067835 Targeted miR-296-5p and miR-296-5p Overexpression Suppressed Cell Proliferation, Cell Cycle Progression, and Promoted Apoptosis and Radiosensitivity
Given the function of circ_0067835 in CRC, we further investigated its regulatory mechanisms using Circinteractome online database. The predicted data showed a putative pairing sequence for miR-296-5p in circ_0067835 (Figure 3A). To validate this, we performed dual-luciferase reporter assays. The transfection efficiency of miR-296-5p mimics was confirmed by qRT-PCR (Figure 3B). The transfection of miR-296-5p mimics significantly reduced the luciferase activity of circ_0067835 luciferase reporter (circ_0067835 WT) but did not affect the luciferase activity of the site-directed mutation in the target region (Figure 3B). Additionally, miR-296-5p was downregulated in CRC tissues compared with ANT (Figure 3C). Radiation therapy led to a significant decrease in miR-296-5p expression in the serum of CRC patients (Figure 3D). Interestingly, a negative relationship between the expression of circ_0067835 and miR-296-5p was found in the serum of CRC patients (Figure 3E). Importantly, circ_0067835 downregulation resulted in a marked increase in miR-296-5p expression in CRC cells (Figure 3F). These data together indicated that circ_0067835 directly targeted miR-296-5p.
Figure 3.
Circ_0067835 targeted miR-296-5p and miR-296-5p overexpression suppressed cell proliferation, cell cycle progression, and promoted apoptosis and radiosensitivity. (A) The miRNA containing the binding sites of circ_0067835 was predicted by online database Circinteractome. (B) Dual-luciferase reporter assays analyzed the relationship between miR-296-5p and circ_0067835. (C) Relative expression level of miR-296-5p in CRC tissues and adjacent normal tissues (ANT) was detected by qRT-PCR. (D) MiR-296-5p expression in the serum of CRC patients with (n=19) or without (n=19) radiation treatment was detected by qRT-PCR. (E) Pearson Correlation Coefficient analyzed the relationship between the expression of miR-296-5p and circ_0067835. (F) MiR-296-5p expression was detected by qRT-PCR in SW620 and HCT116 cells transfected with sh-circ_0067835 or sh-NC. (G) CCK-8 assay was used to detect the viability of SW620 and HCT116 cells transfected with miR-296-5p mimics or miR-NC mimics. (H) Colony formation assay was applied to determine the survival fraction of SW620 and HCT116 cells treated with 0 Gy, 1 Gy, 2 Gy, 4 Gy, 8 Gy IR and transfected with miR-296-5p or miR-NC. (I and K) Flow cytometry assay was used to assess cell cycle distribution in SW620 and HCT116 cells treated with 4 Gy IR and transfected with miR-296-5p or miR-NC. (J) The caspase-3 activity was detected by caspase-3 activity assay in SW620 and HCT116 cells treated with 4 Gy IR and transfected with miR-296-5p or miR-NC. *P<0.05, **P<0.01, ***P<0.001.
To further understand the role of miR-296-5p, we overexpressed its level in both cell lines using miR-296-5p mimics. By contrast, miR-296-5p overexpression induced a remarkable decrease in cell viability (Figure 3G). Moreover, miR-296-5p overexpression significantly reduced the survival fraction of CRC cells under IR treatment (Figure 3H), indicating the enhancement of miR-296-5p overexpression on cell radiosensitivity. Furthermore, miR-296-5p overexpression dramatically repressed cell cycle progression (Figure 3I), and promoted caspase-3 activity (Figure 3J), as well as enhanced cell apoptosis (Figure 3K).
MiR-296-5p Targeted IGF1R and Circ_0067835 Modulated IGF1R Expression Through miR-296-5p
Given the direct relationship between circ_0067835 and miR-296-5p, we further studied the downstream effectors. Using Starbase online tool, IGF1R mRNA harbored a putative complementary for miR-296-5p in its 3ʹUTR (Figure 4A). Dual-luciferase reporter assays showed that the overexpression of miR-296-5p significantly reduced the luciferase activity of IGF1R 3ʹUTR luciferase reporter (WT-IGF1R 3ʹUTR) but not affect the luciferase activity of the site-directed mutation (MUT-IGF1R 3ʹUTR) in the target region (Figure 4B). Our data also showed that IGF1R mRNA and protein levels were highly expressed in CRC tissues and cells compared with their counterparts (Figure 4C–F). Importantly, radiation therapy led to a significant upregulation of IGF1R mRNA expression in the serum of CRC patients (Figure 4G). A strong inverse relationship between the expression of IGF1R mRNA and miR-296-5p was found in the serum of CRC patients (Figure 4H). To assess whether miR-296-5p regulated IGF1R expression, we manipulated miR-296-5p expression using miR-296-5p mimics or miR-296-5p inhibitor (anti-miR-296-5p). The transfection efficiency of anti-miR-296-5p was detected by qRT-PCR (Figure 4I). As wound be expected, IGF1R mRNA and protein levels were reduced by miR-296-5p overexpression and elevated by miR-296-5p depletion in both cell lines (Figure 4J and K). More importantly, the downregulation of circ_0067835 led to a remarkable decrease in the expression of IGF1R mRNA and protein, and this impact was markedly abolished by anti-miR-296-5p (Figure 4L and M).
Figure 4.
MiR-296-5p targeted IGF1R and circ_0067835 modulated IGF1R expression through miR-296-5p. (A) StarBase online tool was applied to predict the genes possessing the binding sequence of miR-296-5p. (B) Dual-luciferase reporter assays analyzed the relationship between IGF1R and miR-296-5p in SW620 and HCT116 cells. (C and D) Relative expression of IGF1R at both mRNA and protein levels in CRC tissues and adjacent normal tissues (ANT) were respectively detected by qRT-PCR and Western blot. (E and F) Relative mRNA and protein levels of IGF1R in SW620 and HCT116 cells and normal cells (FHC) were respectively quantified by qRT-PCR and Western blot. (G) IGF1R mRNA expression in the serum of CRC patients with (n=19) or without (n=19) radiation treatment was detected by qRT-PCR. (H) The relationship between the expression of miR-296-5p and IGF1R was analyzed by Pearson Correlation Coefficient in CRC tissues. (I) MiR-296-5p expression was determined by qRT-PCR in SW620 and HCT116 cells transfected with anti-miR-296-5p or anti-NC. (J and K) The expression of IGF1R at both mRNA and protein levels was respectively determined by qRT-PCR and Western blot in SW620 and HCT116 cells transfected with miR-296-5p mimics, anti-miR-296-5p or negative controls. (L and M) The IGF1R mRNA and protein expression were respectively detected by qRT-PCR and Western blot in SW620 and HCT116 cells transfected with sh-circ_0067835 or sh-NC, sh-circ_0067835+anti-miR-296-5p or sh-circ_0067835+anti-NC. **P<0.01, ***P<0.001.
Knockdown of Circ_0067835 Regulated Cell Proliferation, Cell Cycle Progression, Apoptosis and Radiosensitivity in vitro by Up-Regulating miR-296-5p
To explore whether miR-296-5p was a downstream effector of circ_0067835 function in CRC progression, we reduced miR-296-5p expression with anti-miR-296-5p in sh-circ_0067835-transducing cells (Figure 5A). Further analysis showed that the inhibition of miR-296-5p strikingly counteracted sh-circ_0067835-induced suppression on cell proliferation (Figure 5B), enhancement on cell radiosensitivity (Figure 5C), and repression on cell cycle progression (Figure 5D), as well as promotion on cell apoptosis (Figure 5E and F) in the two CRC cell lines.
Figure 5.
Knockdown of circ_0067835-regulated cell proliferation, cell cycle progression, apoptosis and radiosensitivity in vitro by up-regulating miR-296-5p. (A) MiR-296-5p expression was determined by qRT-PCR in SW620 and HCT116 cells transfected with sh-circ_0067835 or sh-NC, sh-circ_0067835+anti-miR-296-5p or sh-circ_0067835+anti-NC. (B) CCK-8 assay was employed to evaluate the viability of SW620 and HCT116 cells transfected with sh-circ_0067835 or sh-NC, sh-circ_0067835+anti-miR-296-5p or sh-circ_0067835+anti-NC. (C) Colony formation assay was used to assess the survival fraction of SW620 and HCT116 cells treated with 0 Gy, 1 Gy, 2 Gy, 4 Gy, 8 Gy IR and transfected with sh-circ_0067835 or sh-NC, sh-circ_0067835+anti-miR-296-5p or sh-circ_0067835+anti-NC. (D) Flow cytometry assay was used to detect the cell cycle distribution in SW620 and HCT116 cells transfected with sh-circ_0067835 or sh-NC, sh-circ_0067835+anti-miR-296-5p or sh-circ_0067835+anti-NC. (E) Caspase-3 activity assay was used to detect the caspase-3 activity in SW620 and HCT116 cells transfected with sh-circ_0067835 or sh-NC, sh-circ_0067835+anti-miR-296-5p or sh-circ_0067835+anti-NC. (F) Flow cytometry assay was used to detect cell apoptosis rate in SW620 and HCT116 cells transfected with sh-circ_0067835 or sh-NC, sh-circ_0067835+anti-miR-296-5p or sh-circ_0067835+anti-NC. **P<0.01, ***P<0.001.
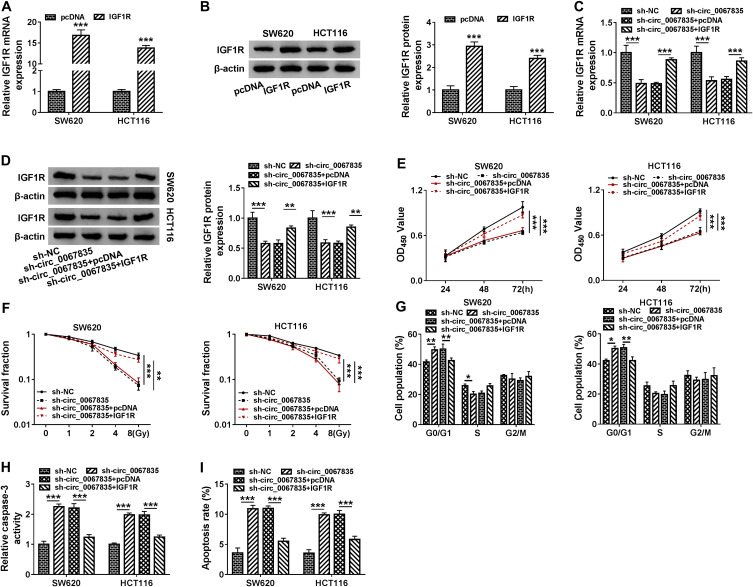
IGF1R Was an Important Mediator of Circ_0067835 Function in CRC Cells
Based on the above, we further determined whether circ_0067835-regulated CRC cell progression by IGF1R. IGF1R mRNA and protein expression were significantly upregulated by the transfection of IGF1R overexpression plasmid in both cell lines (Figure 6A and B). Moreover, sh-circ_0067835-mediated suppression on IGF1R mRNA and protein expression was abolished by IGF1R overexpression plasmid in CRC cells (Figure 6C and D). Importantly, IGF1R overexpression reversed the impact of circ_0067835 depletion on cell proliferation (Figure 5E), radiosensitivity (Figure 5F), cell cycle progression (Figure 5G) and cell apoptosis (Figure 5H and I) in both SW620 and HCT116 cells.
Figure 6.
IGF1R was an important mediator of circ_0067835 function in CRC cells. (A and B) The expression of IGF1R mRNA and protein was tested by qRT-PCR and Western blot in SW620 and HCT116 cells transfected with IGF1R overexpression plasmid (IGF1R) or pcDNA. (C and D) The expression of IGF1R mRNA and protein was respectively detected by qRT-PCR and Western blot in SW620 and HCT116 cells transfected with sh-circ_0067835, sh-circ_0067835+IGF1R overexpression plasmid or negative controls. (E) CCK-8 assay was used to evaluate the viability of SW620 and HCT116 cells transfected with sh-circ_0067835, sh-circ_0067835+IGF1R or negative controls. (F) Colony formation assay was employed to assess the proliferation of SW620 and HCT116 cells treated with 0 Gy, 1 Gy, 2 Gy, 4 Gy, 8 Gy IR and transfected with sh-circ_0067835, sh-circ_0067835+IGF1R or negative controls. (G) Flow cytometry assay was used to detect the cell cycle distribution in SW620 and HCT116 cells transfected with sh-circ_0067835, sh-circ_0067835+IGF1R or negative controls. (H) Caspase-3 activity assay was used to detect the caspase-3 activity in SW620 and HCT116 cells transfected with sh-circ_0067835, sh-circ_0067835+IGF1R or negative controls. (I) Flow cytometry assay was used to detect cell apoptosis rate in SW620 and HCT116 cells transfected with sh-circ_0067835, sh-circ_0067835+IGF1R or negative controls. *P<0.05, **P<0.01, ***P<0.001.
Circ_0067835 Knockdown Weakened Tumor Growth and Enhanced Cell Radiosensitivity in vivo
To further confirm the role of circ_0067835 in CRC progression in vivo, we implanted the sh-circ_0067835-transduced or sh-NC-infected SW620 cells into the nude mice. The transduction of sh-circ_0067835 led to a significant decrease in circ_0067835 expression in the xenograft tumors (Figure 7A). As expected, tumor volume was strikingly suppressed by circ_0067835 knockdown or radiation treatment, and a more significant repression of tumor growth was found when simultaneous sh-circ_0067835 transduction and radiation treatment (Figure 7B and C).
Figure 7.
Circ_0067835 knockdown weakened tumor growth and enhanced cell radiosensitivity in vivo. (A) The expression of circ_0067835 was examined by qRT-PCR in sh-circ_0067835- or sh-NC-transduced SW620 tumors. (B) Tumor volumes were monitored every 5 days after injection of SW620 cells transfected with sh-circ_0067835 or sh-NC and treatment with or without 4 Gy IR. (C) The effects between circ_0067835 silencing and radiation treatment on tumor weight was measured. ***P<0.001.
Discussion
CircRNAs function as crucial regulators in the biological behaviors of cancer cells, including the proliferation, apoptosis and metastasis.16 The transmittal of circRNAs is partly realized by exosomes which are secreted by almost all cells.17 Exosome-transmitted circRNAs have been implicated in human carcinogenesis and radioresistance development.15,18 Our results reported here showed that circ_0067835 upregulation was associated with CRC development and radioresistance. Circ_0067835 could be transmitted by exosomes. Moreover, the knockdown of circ_0067835-repressed CRC cell progression and sensitized CRC cells to radiotherapy in vitro and in vivo. In addition, our results provided a novel molecular explanation for the critical roles of circ_0067835 in CRC development and radioresistance.
MiR-296-5p has been identified as a tumor suppressor in various cancers, such as hepatocellular carcinoma and thyroid cancer.19,20 Moreover, several studies reported that miR-296-5p suppressed CRC development.21,22 Here, we validated that circ_0067835 directly targeted miR-296-45p, and the knockdown of circ_0067835-repressed CRC development and enhanced cell radiosensitivity in vitro by up-regulating miR-296-5p. Similarly, Wang et al demonstrated that circ_0000512 silencing exerted an inhibitory effect in CRC development through sponging miR-296-5p.22
In this paper, IFG1R might be a strong candidate as a downstream effector of miR-296-5p function because of its oncogenic property in various cancers, including cervical cancer and small intestinal carcinoma.23,24 Our data first showed that miR-296-5p directly targeted IFG1R, and IGF1R was an important mediator of circ_0067835 function in CRC cells. However, the direct evidence between the miR-296-5p/IFG1R axis and circ_0067835-mediated CRC development and radiosensitivity in vivo was absent, which was expected to be investigated in further work. Previous work identified that IFG1R enhanced the proliferation of endometrial cancer cells by the PI3K/Akt and MAPK pathways,25 which were associated with CRC progression.26,27 A future challenge will be to identify whether these pathways could function as the downstream effectors of the circ_0067835/miR-296-5p/IFG1R axis in CRC development and radiosensitivity.
To summarize, our findings demonstrated that circ_0067835 knockdown suppressed CRC progression and enhanced CRC cell radiosensitivity partially by modulating GF1R expression through sponging miR-1236-3p. These findings established a rationale that targeting circ_0067835 might be a promising point for improving CRC treatment.
Funding Statement
There is no funding to report.
Disclosure
The authors declare that they have no financial or non-financial conflicts of interest.
References
- 1.Cummings OW. Pathology of the adenoma-carcinoma sequence: from aberrant crypt focus to invasive carcinoma. Semin Gastrointest Dis. 2000;11(4):229–237. [PubMed] [Google Scholar]
- 2.Dekker E, Tanis PJ, Vleugels JLA, et al. Colorectal cancer. Lancet. 2019;394(10207):1467–1480. doi: 10.1016/S0140-6736(19)32319-0 [DOI] [PubMed] [Google Scholar]
- 3.Ohhara Y, Fukuda N, Takeuchi S, et al. Role of targeted therapy in metastatic colorectal cancer. World J Gastrointest Oncol. 2016;8(9):642–655. doi: 10.4251/wjgo.v8.i9.642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clifford R, Govindarajah N, Parsons JL, et al. Systematic review of treatment intensification using novel agents for chemoradiotherapy in rectal cancer. Br J Surg. 2018;105(12):1553–1572. doi: 10.1002/bjs.10993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cousins A, Thompson SK, Wedding AB, et al. Clinical relevance of novel imaging technologies for sentinel lymph node identification and staging. Biotechnol Adv. 2014;32(2):269–279. doi: 10.1016/j.biotechadv.2013.10.011 [DOI] [PubMed] [Google Scholar]
- 6.Munakata K, Uemura M, Tanaka S, et al. Cancer stem-like properties in colorectal cancer cells with low proteasome activity. Clin Cancer Res. 2016;22(21):5277–5286. doi: 10.1158/1078-0432.CCR-15-1945 [DOI] [PubMed] [Google Scholar]
- 7.Kelley K, Ruhl R, Rana S, et al. Understanding and resetting radiation sensitivity in rectal cancer. Ann Surg. 2017;266(4):610–616. doi: 10.1097/SLA.0000000000002409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dearling J, Qureshi U, Begent R, et al. Combining radioimmunotherapy with antihypoxia therapy 2-deoxy-D-glucose results in reduction of therapeutic efficacy. Clin Cancer Res. 2007;13(6):1903–1910. doi: 10.1158/1078-0432.CCR-06-2094 [DOI] [PubMed] [Google Scholar]
- 9.Yin Y, Long J, He Q, et al. Emerging roles of circRNA in formation and progression of cancer. J Cancer. 2019;10(21):5015–5021. doi: 10.7150/jca.30828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhong Y, Du Y, Yang X, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17(1):79. doi: 10.1186/s12943-018-0827-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang ZJ, Zhang YH, Qin XJ, et al. Circular RNA circDENND4C facilitates proliferation, migration and glycolysis of colorectal cancer cells through miR-760/GLUT1 axis. Eur Rev Med Pharmacol Sci. 2020;24(5):2387–2400. doi: 10.26355/eurrev_202003_20506 [DOI] [PubMed] [Google Scholar]
- 12.Du S, Zhang P, Ren W, et al. Circ-ZNF609 accelerates the radioresistance of prostate cancer cells by promoting the glycolytic metabolism through miR-501-3p/HK2 axis. Cancer Manag Res. 2020;12:7487–7499. doi: 10.2147/CMAR.S257441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin Y, Su Z, Sheng H, et al. Circ_0086720 knockdown strengthens the radiosensitivity of non-small cell lung cancer via mediating the miR-375/SPIN1 axis. Neoplasma. 2020. doi: 10.4149/neo_2020_200331N333 [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, Chang Y, Cai Y. Circ_0067835 sponges miR-324-5p to induce HMGA1 expression in endometrial carcinoma cells. J Cell Mol Med. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng W, Gong H, Wang Y, et al. circIFT80 functions as a ceRNA of miR-1236-3p to promote colorectal cancer progression. Mol Ther Nucleic Acids. 2019;18:375–387. doi: 10.1016/j.omtn.2019.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Yu CY, Kuo HC. The emerging roles and functions of circular RNAs and their generation. J Biomed Sci. 2019;26(1):29. doi: 10.1186/s12929-019-0523-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tintut Y, Demer L. Exosomes: nanosized cellular messages. Circ Res. 2015;116(8):1281–1283. doi: 10.1161/CIRCRESAHA.115.306324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu D, Kang H, Gao M, et al. Exosome-transmitted circ_MMP2 promotes hepatocellular carcinoma metastasis by upregulating MMP2. Mol Oncol. 2020;14(6):1365–1380. doi: 10.1002/1878-0261.12637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi DM, Li LX, Bian XY, et al. miR-296-5p suppresses EMT of hepatocellular carcinoma via attenuating NRG1/ERBB2/ERBB3 signaling. J Exp Clin Cancer Res. 2018;37(1):294. doi: 10.1186/s13046-018-0957-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Y, Gao H, Li Y. Inhibition of LncRNA FOXD3-AS1 suppresses the aggressive biological behaviors of thyroid cancer via elevating miR-296-5p and inactivating TGF-β1/Smads signaling pathway. Mol Cell Endocrinol. 2020;500:110634. doi: 10.1016/j.mce.2019.110634 [DOI] [PubMed] [Google Scholar]
- 21.Ma C, Ma N, Qin L, et al. DICER1-AS1 promotes the malignant behaviors of colorectal cancer cells by regulating miR-296-5p/STAT3 axis. Cancer Manag Res. 2020;12:10035–10046. doi: 10.2147/CMAR.S252786 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Wang L, Wu H, Chu F, et al. Knockdown of circ_0000512 inhibits cell proliferation and promotes apoptosis in colorectal cancer by regulating miR-296-5p/RUNX1 axis. Onco Targets Ther. 2020;13:7357–7368. doi: 10.2147/OTT.S250495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma Z, Cai Y, Zhang L, et al. LINC00319 promotes cervical cancer progression via targeting miR-147a/IGF1R pathway. Cancer Biother Radiopharm. 2020. doi: 10.1089/cbr.2020.3722 [DOI] [PubMed] [Google Scholar]
- 24.Shin HC, Bae YK, Gu MJ, et al. Expression of insulin-like growth factor 1 and insulin-like growth factor 1 receptor is associated with the favorable clinicopathologic parameters in small intestinal carcinomas. Pathobiology. 2013;80(5):265–270. doi: 10.1159/000350309 [DOI] [PubMed] [Google Scholar]
- 25.Mendivil A, Zhou C, Cantrell LA, et al. AMG 479, a novel IGF-1-R antibody, inhibits endometrial cancer cell proliferation through disruption of the PI3K/Akt and MAPK pathways. Reprod Sci. 2011;18(9):832–841. doi: 10.1177/1933719111398501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bahrami A, Khazaei M, Hasanzadeh M, et al. Therapeutic potential of targeting PI3K/AKT pathway in treatment of colorectal cancer: rational and progress. J Cell Biochem. 2018;119(3):2460–2469. doi: 10.1002/jcb.25950 [DOI] [PubMed] [Google Scholar]
- 27.Song G, Xu S, Zhang H, et al. TIMP1 is a prognostic marker for the progression and metastasis of colon cancer through FAK-PI3K/AKT and MAPK pathway. J Exp Clin Cancer Res. 2016;35(1):148. doi: 10.1186/s13046-016-0427-7 [DOI] [PMC free article] [PubMed] [Google Scholar]