Abstract
Bioactive peptides derived from milk proteins are an active research area. Exhibiting numerous positive physiological effects on digestive, cardiovascular, immune and nervous systems, these peptides thought to be one of the most promising ingredients for functional food. Generally, these peptides are inactive within the parent proteins and can be liberated during milk fermentation by the specific proteolytic systems of various Lactobacillus spp. Here we present the study of milk fermentation by Lactobacillus helveticus NK1, Lactobacillus rhamnosus F and Lactobacillus reuteri LR1 strains. It was demonstrated that the antioxidant activity of the milk fermented by these strains concomitantly increased with the strains’ proteolytic activity. For the angiotensin I-converting enzyme (ACE) inhibitory activity, the same tendency was not observed. Although the proteolytic activity of L. helveticus NK1 was two times higher than that of L. rhamnosus F, the milk fermented by these strains showed comparable ACE inhibition. The analysis of the peptide profiles of the fermented milk samples allowed us to hypothesize that some previously unreported peptides can be produced by L. rhamnosus F. In addition, it was demonstrated that these potential ACE-inhibiting peptides originated from the C-terminus of αS2-casein.
Keywords: Lactobacillus, milk fermentation, proteolytic activity, antioxidant activity, angiotensin-converting enzyme inhibitory activity (ACE-I), peptidomics, bioactive peptides
1. Introduction
Nutraceuticals are any compounds derived from food sources and possessing extra health benefits in addition to their basic nutritional value [1]. Typically, these compounds are present by non-specific biological agents used to promote general well-being, control symptoms and prevent malignant processes [2]. In this regards, nutraceuticals can be generally considered as an alternative to pharmaceuticals (specifically designed and usually chemically synthesized health-promoting and symptom-alleviating substances) [3].
Over the past decade, interest has risen in fermented dairy foods as a potent source of nutraceuticals [4]. The nutraceutical values of these foods had been mainly attributed to bioactive peptides encrypted within dairy proteins [5]. During fermentation these peptides are released by the specific sets of proteases—cell envelope proteinases, CEPs—possessed by lactic acid bacteria (LABs) [6]. Currently, the list of the proven health-promoting properties of dairy peptides includes, but is not limited to, antioxidant, antihypertensive, antimicrobial, antithrombotic and immunomodulatory properties [7].
Traditionally, the starter cultures for dairy fermentation are selected based only on the rheological, organoleptic and nutritional characteristics of the final product. However, the potential of LABs to produce bioactive peptides forced the reconsideration of these requirements. Nowadays, many scientists all over the world are involved in the search for new strains of LABs that can be either added to the traditional starters for their enrichment in bioactive peptides or used alone to produce bioactive premixes [8,9,10].
In our previous work, we demonstrated that Lactobacillus helveticus NK1, Lactobacillus rhamnosus F and Lactobacillus reuteri LR1 strains exhibit good safety and technological performance, as well as provide good probiotic properties to the final products [11,12]. In this article, the proteolytic activity of these LABs and their ability to release bioactive peptides with antioxidant and angiotensin I-converting enzyme inhibitory (ACE-I) properties were investigated.
2. Materials and Methods
2.1. Preparation of Fermented Milk Samples
L. helveticus NK1, L. rhamnosus F, and L. reuteri LR1 strains were obtained from the Microorganism Collection of the All-Russia Research Institute of the Dairy Industry (VNIMI, Moscow, Russia). The sequences of the 16S ribosomal RNA genes of these strains can be found at the GeneBank accessory numbers MT448799, MN994629 and MN994628 for L. helveticus NK1, L. rhamnosus F, and L. reuteri LR1, respectively.
For all fermentations, one liter of a reconstituted skim milk (RSM) was prepared by adding the appropriate amount of skim milk powder to water and heating at 85 °C for 30 min with stirring. The heat-treated milk was then cooled to approximately 40 °C. The sterile RSM was aseptically inoculated with 1% (v/v) of each strain (approximately 107 cells·mL−1) and incubated at 37 °C for 72 h. Samples were taken for analysis at 0, 6, 16, 24, 48, and 72 h of incubation and the change in pH was measured at each time point.
The cell population of each LAB was determined by counting the colony-forming units (CFU) on MRS agar after incubation at 37 °C for 48–72 h under anaerobic conditions using GasPak EZ anaerobe container systems (Becton, Dickinson and Company, Sparks, MD, USA).
All fermentations were performed at minimum in triplicate, and the obtained data are reported as mean ± SD.
2.2. Characterization of Fermented Milk Samples
To obtain water-soluble extracts (WSEs), the following procedure was performed. For the samples with a pH above 4.6, the pH was adjusted to 4.6 by adding 0.75% trichloroacetic acid (TCA), and the sample was centrifuged at 10,000× g for 20 min at 4 °C. The supernatant was filtered through a 0.45 µm syringe filter and stored at −80 °C until further analysis.
The protein content of WSEs was determined using the Pierce BSA Protein Assay Kit (ThermoFisher, Rockford, IL, USA).
The proteolytic activity was quantified by the measurement of the amount of released amino groups in WSEs using the 2,4,6-trinitrobenzenesulfonic acid solution (TNBS, Sigma-Aldrich, St. Louis, MO, USA) method. The optical density at 340 nm was measured using a Synergy 2 microplate photometer–fluorimeter (BioTek, Winooski, VT, USA). A calibration curve was prepared using L-leucine (L-Leu) as a standard (0.1–2.0 mM). The results were expressed as L-Leu molar equivalents (mM (Leu)).
The in vitro antioxidant activity in WSEs was determined by the oxygen radical absorbance capacity fluorescence method (ORAC) using a Synergy 2 microplate photometer–fluorometer as described in Ref. [13]. The peroxyl radical was generated directly in the reaction medium during the thermal decomposition of the azo compound 2,2′-azobis (2-methylpropionamidine) dihydrochloride (AAPH, Sigma-Aldrich, St. Louis, MO, USA), initiated by incubation at 37 °C for 10 min. The antioxidant activity was expressed as the amount of Trolox (Sigma-Aldrich, St. Louis, MO, USA) molar equivalents (µM TE).
ACE-I activity in WSEs was determined by their ability to inhibit ACE (Sigma-Aldrich, St. Louis, MO, USA). o-Aminobenzoyl-Phe-Arg-Lys(dinitrophenyl)-Pro (Sigma-Aldrich, St. Louis, MO, USA) was used as a substrate with internal fluorescence quenching, as described in Ref. [13]. The 96-well, black, nonbinding polypropylene microplates (Greiner Bio One, Frickenhausen, Germany) were used. The measurements were carried out on a Synergy 2 microplate photometer–fluorometer. The half maximal inhibitory concentration (IC50) was expressed as mg of protein per mL.
2.3. Peptide Profile Analysis
The peptide profile of the WSEs was analyzed by liquid chromatography–tandem mass spectrometry (LC-MS/MS) in a system consisting of an Agilent 1100 chromatograph (Agilent Technologies, Santa Clara, CA, USA) and an LTQ-FT Ultra mass spectrometer (Thermo Scientific, Waltham, MA, USA) as described in Ref. [13]. The data are available in Supplementary Materials, Table S1.
The set analysis of peptidomics data was performed using “UpSetR” and “eulerr” R packages. The heat maps of peptidomics data were constructed using Peptigram—a web-based application for peptidomics data visualization [14]. The search for peptides with previously reported antioxidant and ACE-I activities was performed in the Milk Bioactive Peptide Database [15] and BIOPEP-UWM Database [16].
3. Results and Discussion
3.1. Growth Performance, Acidification Capability and Proteolytic Activity
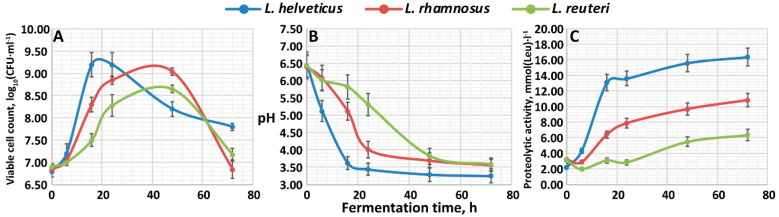
To assess the growth performance during milk fermentation of L. helveticus NK1, L. rhamnosus F and L. reuteri LR1, milk was inoculated with corresponding LAB cultures and fermented for 72 h. The dynamics of changes in the viable cell count are shown in Figure 1A. During the fermentation process, L. helveticus NK1 demonstrated the highest growth rate as well as the highest maximally attainable viable cell count, (1.58 ± 0.15) × 109 CFU·mL−1 (at 24 h), among all studied LABs. The exponential growth phase for this strain lasted until 16 h, and the stationary phase until 24 h. For the L. rhamnosus F, having an intermediate growth rate, the exponential growth phase lasted approximately until 24 h, and the stationary phase until 48 h; the maximally attainable viable cell count comprised (1.10 ± 0.11) × 109 CFU·mL−1 (at 48 h). The lowest growth rate was demonstrated by L. reuteri LR1. This strain grew exponentially until 48 h, when it attained the maximal viable cell count of (4.50 ± 0.19) × 108 CFU·mL−1; there was no pronounced stationary growth phase observed for this LAB (the values at 24 and 48 h were significantly different according to the Student’s t-test, p < 0.05).
Figure 1.
Fermentation characteristics of the L. helveticus NK1, L. rhamnosus F and L. reuteri LR1: (A)—the dynamics of change in the viable cell count; (B)—the dynamics of change in the pH value; (C)—the dynamics of change in the proteolytic activity.
The dynamics of changes in the pH values are shown in Figure 1B. As expected, the acidification capabilities of all studied LABs negatively correlated with their growth patterns, and the faster the strain grew, the greater acidification of its medium was observed. While L. helveticus NK1 showed a pH decrease of approximately three units in a 24 h timespan (reaching the final pH of 3.25 ± 0.20 at 72 h), L. rhamnosus F and L. reuteri LR1 decreased pH by the same amount only at the end of the fermentation. It should be noted that all performed fermentations eventually led to the coagulation of milk proteins, because of casein precipitation at a pH lower than 4.6 [17].
The degree of hydrolysis of milk proteins, which characterizes the proteolytic activity of LABs, was measured as the concentration of free amino groups expressed in L-leucine equivalents. The dynamics of changes of the proteolytic activity for the studied LABs are shown in Figure 1C. Among all studied LABs, the highest proteolytic activity was demonstrated by L. helveticus NK1. For this LAB, an active growth of the proteolytic activity was observed in the first 16 h of fermentation, when the proteolytic activity reached 13.07 ± 1.05 mM (Leu). Then, during the period of 16–24 h, the proteolytic activity remained constant, after which a slight increase up to 16.36 ± 1.15 mM (Leu) was observed. For the L. rhamnosus F and L. reuteri LR1, the dynamics of the proteolytic activity were different. For both of these LABs, a slight decrease in the proteolytic activity was observed in the first 6 h of fermentation—from 3.17 ± 0.22 mM (Leu) to 2.89 ± 0.19 mM (Leu) for L. rhamnosus F and from 3.08 ± 0.37 mM (Leu) to 1.98 ± 0.22 mM (Leu) for L. reuteri LR1. This can indicate that the process of the utilization of peptides, which were already present in milk, prevailed over the process of the hydrolysis of milk proteins by LAB’s proteases. The most active increase (up to 7.87 ± 0.63 and 2.86 ± 0.34 mM (Leu) for L. rhamnosus F and L. reuteri LR1, respectively) in proteolytic activity was observed from 6 to 24 h of fermentation, after which the proteolysis went slower. At the end of the fermentation, the proteolytic activities comprised 10.81 ± 0.86 mM (Leu) and 6.35 ± 0.76 mM (Leu) for the L. rhamnosus F and L. reuteri LR1, respectively.
While the strain L. helveticus NK1 demonstrated comparable proteolytic activity to those previously reported for this LAB species [18,19], the strain L. rhamnosus F studied in this work showed a higher proteolytic activity than those described in the literature—the L. rhamnosus PRA331 strain after 72 h of fermentation on milk had a proteolytic activity of 6.4 mM (Leu) (pH 4.0) [20]. Unfortunately, no comparable data on the proteolytic activity of L. reuteri have been previously published. The low proteolytic activity of L. reuteri LR1 can be explained by its lifestyle. Apart from the L. helveticus and L. rhamnosus, which are nomadic species, persisting for a limited time in the gut, the L. reuteri is strongly host-associated [21]. As such, this species does not require a potent proteolytic system on its own. Indeed, while two and one CEPs were identified in the genomes of L. helveticus and L. rhamnosus [6,22], respectively, there were no CEPs reported in the genomes of L. reuteri [23].
3.2. The Development of Antioxidant and Antihypertensive Properties
The development of antioxidant activity during the fermentation of milk by the studied LABs is shown in Figure 2A. The antioxidant activity of milk fermented by L. helveticus NK1 actively increased up to 657 ± 53 µM TE in the first 16 h of fermentation, after which it remained at the same level until 24 h. Further, a secondary increase up to 1045 ± 73 µM TE in antioxidant activity was observed. For milk fermented by L. rhamnosus F and L. reuteri LR1, the growth of antioxidant activity was not observed in the first 6 h of fermentation. From 6 to 24 h, the milk fermented by L. rhamnosus F demonstrated an intense increase in antioxidant activity up to 751 ± 60 µM TE, after which it continued to linearly grow until the end of fermentation, reaching 855 ± 60 µM TE. As for the milk fermented by L. reuteri LR1, the antioxidant activity increased up to 415 ± 46 µM TE at 24 h, after which it remained the same until the end of fermentation.
Figure 2.
The development of antioxidant and angiotensin I-converting enzyme inhibitory (ACE-I) activities in the milk fermented by L. helveticus NK1, L. rhamnosus F and L reuteri LR1: (A)—the development of antioxidant activity; (B)—the development of ACE-I activity.
The development of ACE-I activity is shown in Figure 2B. For milk fermented by all studied LABs, the most active increase in ACE-I activity (i.e., decrease in IC50) was observed in the first 24 h of fermentation. At this time, for milk fermented by L. helveticus NK1 and L. rhamnosus F, the IC50 reached the values of 0.60 ± 0.07 and 0.95 ± 0.1 mg·mL−1, respectively. For milk fermented by L. reuteri LR1, the IC50 at 24 h of fermentation was 2.63 ± 0.29 mg·mL−1. For all studied LABs, after 24 h the IC50 decreased slowly and reached 0.18 ± 0.02, 0.21 ± 0.03 and 1.07 ± 0.13 mg·mL−1 for L. helveticus NK1, L. rhamnosus F and L. reuteri LR1 at 72 h of fermentation, respectively.
The observed differences between the patterns of development of the antioxidant and ACE-I activities for studied LABs can be explained by the general requirements for peptides to possess such activities. In the case of antioxidant activity, peptides should be short (up to 20 amino acids) and contain a high proportion of hydrophobic amino acids [24,25,26]. Since many peptides can satisfy these minimal requirements, any LAB can potentially produce several of them as a part of the total peptide pool. Therefore, the bigger the total pool of peptides (i.e., degree of proteolysis) in the fermented product, the greater the amount of antioxidant peptides it will contain, and the higher the antioxidant activity it will possess. In our case, the proteolytic activity decreased in the series L. helveticus NK1 > L. rhamnosus F > L. reuteri LR1, and the same was observed for antioxidant activity. It should be noted that the same correlation of proteolytic and antioxidant activities was previously reported in Refs. [27,28].
In contrast to antioxidant peptides, ACE-I peptides are very sequence-specific, since they have to perform competitive inhibition at the catalytic site of ACE [26]. Therefore, their production is strongly dependent on the ability of the LAB’s proteolytic system to cut at specific sequences. Consequently, only a small fraction, if any, of peptides will possess this activity independently of the degree of hydrolysis. In our study, we observed that at 24 h; the ACE-I activities of L. helveticus NK1 and L. rhamnosus F became roughly the same, despite the almost two times greater degree of proteolysis for L. helveticus NK1. Most probably, L. rhamnosus F produces more active ACE-I peptides unique to this LAB.
3.3. Peptide Profile
To get more insight into the process of the development of antioxidant and ACE-I activities, for each LAB, biological replicates from the 16 h of fermentation were pooled together and the peptide profile was determined using the LC-MS/MS technique. As expected, all detected peptides were mainly released from the milk’s caseins—β-, αS1-, αS2- and κ-casein; only these peptides were subjected to further analysis. The performed analysis allowed for identifying 104, 149, 98 and 128 peptides in the original milk and the milk fermented by L. helveticus NK1, L. rhamnosus F and L. reuteri LR1, respectively. In total, 331 unique peptides were identified in all studied samples.
As can be seen from Figure 3, all the studied LABs significantly altered the peptide composition of the original milk. From the 104 peptides originally present in the milk, 31 peptides were totally absent in all fermentations. The greatest alteration was observed in the milk fermented by L. helveticus NK1. Among all peptides detected in the fermentation with this LAB, 113 were unique to it; 9 and 5 peptides were uniquely shared with L. rhamnosus F and L. reuteri LR1, respectively. For L. rhamnosus F, 44 peptides were unique; 9 and 6 peptides were uniquely shared with L helveticus NK1 and L. reuteri LR1, respectively. Compared to other LABs, the fermentation with L. reuteri LR1 contained the highest amount of the peptides originally present in the milk; however, the number of unique peptides—47—was comparable with those detected in L. rhamnosus F. Interestingly, L. helveticus NK1 mainly released unique peptides from β-casein (82 peptides), while L. rhamnosus F and L. reuteri LR1 mainly released unique peptides from αS1-casein (18 and 26 peptides for L. rhamnosus F and L. reuteri LR1, respectively).
Figure 3.
Set analysis of peptidomics data: UpSet plot, and area-proportional Euler diagrams. Unfermented milk was used as a control.
To illustrate the proteolysis patterns of the casein fractions, all detected peptides were mapped on the β-, αS1-, αS2- and κ-casein sequences (Figure 4). Generally, all studied LABs were able to cleave the β-casein along almost the entire length of its sequence; the αS1-casein was predominantly cleaved at its N- and C-termini, and the κ-casein at its C-terminus. In the case of αS2-casein, the different LABs demonstrated the most different patterns of cleavage, which is especially apparent at the protein’s termini: L. helveticus NK1 did not produce peptides from both termini of the protein; L. rhamnosus F was able to cleave both N- and C-termini; and L. reuteri LR1 performed cleavages only at the C-terminus.
Figure 4.
Heat map of peptidomics data. The color intensity is proportional to the normalized per protein ion intensities of peptides, with dark green indicating high peptide intensity and light green indicating low peptide intensity.
As a result of the search for the determined peptides in publicly available databases and published literature, 22 complete matches were found (Table 1). While 14 peptides with proven ACE-I activity were determined in the milk fermented by L. helveticus NK1, for L. rhamnosus F and L. reuteri LR1 this number was only 4 and 10 peptides, respectively. Although, the smaller number of ACE-I peptides in L. reuteri LR1 well explained its smaller ACE-I activity, for L. rhamnosus F, having an ACE-I comparable to L. helveticus NK1, the data again suggested the presence of unique, previously unreported peptides.
Table 1.
Bioactive peptides identified in milk samples fermented by Lactobacillus spp. strains after 16 h of milk fermentation *.
| Peptide | Protein Fragment | MS Extracted Ion Intensity | Activity | Ref. | |||
|---|---|---|---|---|---|---|---|
| Control | Lr LR1 | Lrh F | Lhel NK1 | ||||
| DKIHPF | β-CN f (47–52) | nd | nd | 1.34 × 106 | 3.33 × 106 | ACE-I/Antioxidant | [29] |
| VVPPFLQPE | β-CN f (83–91) | nd | nd | nd | 1.44 × 106 | ACE-I | [30] |
| YPFPGPIPN | β-CN f (60–68) | nd | 2.77 × 105 | nd | nd | ACE-I | [31] |
| NIPPLTQTPV | β-CN f (73–82) | nd | nd | nd | 6.56 × 106 | ACE-I | [29] |
| TQTPVVVPPFLQPE | β-CN f (78–91) | nd | nd | nd | 1.25 × 106 | Antioxidant | [32] |
| DVENLHLPLPLLQSWM | β-CN f (129–144) | nd | nd | nd | 8.21 × 105 | ACE-I | [33] |
| LHLPLPLLQSW | β-CN f (133–143) | nd | nd | nd | 1.29 × 105 | ACE-I | [29,30] |
| SLSQSKVLPVPQK | β-CN f (164–176) | nd | nd | nd | 9.27 × 106 | Antioxidant | [32] |
| KVLPVPQ | β-CN f (169–175) | nd | nd | nd | 2.17 × 105 | ACE-I | [34] |
| LLYQEPVLGPVRGPFPIIV | β-CN f (191–209) | 8.89 × 106 | 8.52 × 105 | nd | 1.28 × 108 | ACE-I/Antioxidant | [32] |
| YQEPVLGPVRGPFP | β-CN f (193–206) | nd | 3.42 × 106 | 1.73 × 105 | nd | ACE-I | [32] |
| QEPVLGPVRGPFPIIV | β-CN f (194–209) | nd | 1.8 × 106 | 3.04 × 107 | 1.13 × 108 | ACE-I/Antioxidant | [32] |
| EPVLGPVRGPFP | β-CN f (195–206) | nd | 4.17 × 105 | nd | nd | ACE-I | [35] |
| GPVRGPFPIIV | β-CN f (199–209) | 1.17 × 106 | 7.49 × 106 | 2.66 × 104 | 2.42 × 107 | ACE-I | [30] |
| RPKHPIKHQ | αS1-CN f (1–9) | nd | 4.09 × 104 | nd | 6.06 × 106 | ACE-I | [31] |
| EVLNENLLRF | αS1-CN f (14–23) | nd | 1.46 × 105 | nd | nd | ACE-I | [32] |
| FVAPFPEVFGKE | αS1-CN f (24–35) | nd | nd | nd | 3.72 × 105 | ACE-I/Antioxidant | [30] |
| VAPFPEVFGKE | αS1-CN f (25–35) | nd | nd | nd | 4.45 × 105 | ACE-I | [32] |
| LYQGPIVLNPWDQVK | αS2-CN f (99–113) | nd | nd | nd | 3.9 × 105 | ACE-I | [32] |
| NAVPITPT | αS2-CN f (115–122) | 6.11 × 105 | 5.81 × 105 | nd | nd | ACE-I | [32] |
| KYIPIQYVL | κ-CN f (30–38) | nd | nd | nd | 4.47 × 105 | Antioxidant | [33] |
| VQVTSTAV | κ-CN f (162–169) | 3.21 × 105 | 3.15 × 105 | nd | nd | ACE-I | [30] |
* Unfermented milk was used as a control. Abbreviations: β-CN—β-casein; αS1-CN—αS1-casein; αS2-CN—αS2-casein; κ-CN—κ-casein. Lr LR1—L. reuteri LR1; Lrh F—L. rhamnosus F; Lhel NK1—L. helveticus NK1.
To identify potential ACE-I peptides from L. rhamnosus F, its unique peptides were searched for specific ACE-I patterns. Previously, it was shown that peptides that possess in their sequence an aromatic residue at the C-terminal position are better ACE inhibitors [36]. Moreover, the location of aliphatic, basic and aromatic residues in the penultimate position, and proline, aromatic and aliphatic residues in the last position, was proposed as the strong ACE-I pattern [37]. In our case, five peptides satisfying these requirements were identified (Table 2). Interestingly, all these peptides originated from the C-terminus of αS2-casein.
Table 2.
The αS2-casein (αS2-CN) peptides identified in milk samples fermented by L. rhamnosus F strains after 16 h of milk fermentation.
| Peptide * | Protein Fragment | MS Extracted Ion Intensity |
|---|---|---|
| QHQKAMKPW | αS2-CN f (200–208) | 1.88 × 104 |
| PWIQPKTKVIPYVRYL | αS2-CN f (207–222) | 1.66 × 105 |
| IQPKTKVIPYVRYL | αS2-CN f (209–222) | 1.14 × 107 |
| IQPKTKVIPY | αS2-CN f (209–218) | 1.44 × 105 |
| KVIPYVRYL | αS2-CN f (214–222) | 7.53 × 104 |
* Typical for ACE-I peptides amino acid residues are shown in bold.
4. Conclusions
In the current work, different abilities of LABs to develop antioxidant and ACE-inhibitory properties during milk fermentation were observed. For all studied LABs, the development of antioxidant activity was correlated with the degree of milk proteolysis. The strain demonstrating the highest proteolytic as well as antioxidant activities was L. helveticus NK1, followed by L. rhamnosus F and L. reuteri LR1. Although the proteolytic activity of L. helveticus NK1 was two times higher than that for L. rhamnosus F, the milk fermented by these strains showed comparable ACE inhibition after 24 h of fermentation. The analysis of the peptide profile of the obtained products of fermentation allowed us to hypothesize that some previously unreported peptides should be produced by L. rhamnosus F. The detailed analysis of the L. rhamnosus F peptide profile allowed us to suggest that, most probably, its ACE-inhibitory peptides originated from the C-terminus of the αS2-casein. These peptides are promising candidates for the further testing of their antihypertensive properties in vivo using an appropriate pure line of rats (e.g., spontaneously hypertensive rats, SHR).
Supplementary Materials
The following are available online at https://www.mdpi.com/2304-8158/10/1/17/s1, Table S1: Results of the peptidomics analysis.
Author Contributions
T.V.F. and K.V.M.; investigation, A.V.B., O.S.S., O.A.G., K.V.M., I.V.R. and T.V.F.; data curation, T.V.F. and I.V.R.; writing—original draft preparation, T.V.F., A.V.B.; writing—review and editing, K.V.M., O.A.G., I.V.R. and T.V.F.; visualization, K.V.M. and A.V.B.; supervision, T.V.F. and I.V.R.; project administration, T.V.F.; funding acquisition, T.V.F. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Russian Science Foundation, grant number 16-16-00094.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article or supplementary material.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Nasri H., Baradaran A., Shirzad H., Rafieian-Kopaei M. New Concepts in Nutraceuticals as Alternative for Pharmaceuticals. Int. J. Prev. Med. 2014;5:1487–1499. [PMC free article] [PubMed] [Google Scholar]
- 2.Mine Y., Li-Chan E.C., Jiang B. Bioactive Proteins and Peptides as Functional Foods and Nutraceuticals. Wiley; Hoboken, NJ, USA: 2010. Biologically Active Food Proteins and Peptides in Health: An Overview; pp. 3–11. [Google Scholar]
- 3.Chanda S., Tiwari R.K., Kumar A., Singh K. Nutraceuticals Inspiring the Current Therapy for Lifestyle Diseases. Adv. Pharmacol. Sci. 2019;2019:1–5. doi: 10.1155/2019/6908716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beltrán-Barrientos L., Hernández-Mendoza A., Torres-Llanez M., González-Córdova A., Vallejo-Cordoba B. Invited review: Fermented milk as antihypertensive functional food. J. Dairy Sci. 2016;99:4099–4110. doi: 10.3168/jds.2015-10054. [DOI] [PubMed] [Google Scholar]
- 5.Séverin S., Wenshui X. Milk Biologically Active Components as Nutraceuticals: Review. Crit. Rev. Food Sci. Nutr. 2005;45:645–656. doi: 10.1080/10408690490911756. [DOI] [PubMed] [Google Scholar]
- 6.Brown L., Pingitore E.V., Mozzi F., Saavedra L., Villegas J.M., Hebert E. Lactic Acid Bacteria as Cell Factories for the Generation of Bioactive Peptides. Protein Pept. Lett. 2017;24:146–155. doi: 10.2174/0929866524666161123111333. [DOI] [PubMed] [Google Scholar]
- 7.Sánchez A., Vázquez A. Bioactive peptides: A review. Food Qual. Saf. 2017;1:29–46. doi: 10.1093/fqs/fyx006. [DOI] [Google Scholar]
- 8.Raveschot C., Cudennec B., Coutte F., Flahaut C., Fremont M., Drider D., Dhulster P. Production of Bioactive Peptides by Lactobacillus Species: From Gene to Application. Front. Microbiol. 2018;9:2354. doi: 10.3389/fmicb.2018.02354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tagliazucchi D., Martini S., Solieri L. Bioprospecting for Bioactive Peptide Production by Lactic Acid Bacteria Isolated from Fermented Dairy Food. Fermentation. 2019;5:96. doi: 10.3390/fermentation5040096. [DOI] [Google Scholar]
- 10.Liong M.-T., editor. Beneficial Microorganisms in Food and Nutraceuticals. Vol. 27. Springer International Publishing; Cham, Switzerland: 2015. [DOI] [Google Scholar]
- 11.Fedorova T.V., Vasina D.V., Begunova A.V., Rozhkova I.V., Raskoshnaya T.A., Gabrielyan N.I. Antagonistic Activity of Lactic Acid Bacteria Lactobacillus spp. against Clinical Isolates of Klebsiella pneumoniae. Appl. Biochem. Microbiol. 2018;54:277–287. doi: 10.1134/S0003683818030043. [DOI] [Google Scholar]
- 12.Begunova A.V., Rozhkova I.V., Zvereva E.A., Glazunova O.A., Fedorova T.V. Lactic and Propionic Acid Bacteria: The Formation of a Community for the Production of Functional Products with Bifidogenic and Hypotensitive Properties. Appl. Biochem. Microbiol. 2019;55:660–669. doi: 10.1134/S0003683819060048. [DOI] [Google Scholar]
- 13.Torkova A.A., Ryazantseva K.A., Agarkova E.Y., Kruchinin A.G., Tsentalovich M.Y., Fedorova T.V. Rational design of enzyme compositions for the production of functional hydrolysates of cow milk whey proteins. Appl. Biochem. Microbiol. 2017;53:669–679. doi: 10.1134/S0003683817060138. [DOI] [Google Scholar]
- 14.Manguy J., Jehl P., Dillon E.T., Davey N.E., Shields D.C., Holton T.A. Peptigram: A web-based application for peptidomics data visualization. J. Proteome Res. 2017;16:712–719. doi: 10.1021/acs.jproteome.6b00751. [DOI] [PubMed] [Google Scholar]
- 15.Nielsen S.D., Beverly R.L., Qu Y., Dallas D.C. Milk bioactive peptide database: A comprehensive data-base of milk protein-derived bioactive peptides and novel visualization. Food Chem. 2017;232:673–682. doi: 10.1016/j.foodchem.2017.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Minkiewicz P., Iwaniak A., Darewicz M. BIOPEP-UWM database of bioactive peptides: Current opportunities. Int. J. Mol. Sci. 2019;20:5978. doi: 10.3390/ijms20235978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raak N., Rohm H., Jaros D. Enzymatic Cross-Linking of Casein Facilitates Gel Structure Weakening Induced by Overacidification. Food Biophys. 2017;16:261–268. doi: 10.1007/s11483-017-9483-6. [DOI] [Google Scholar]
- 18.Giacometti Cavalheiro F., Parra Baptista D., Domingues Galli B., Negrão F., Nogueira Eberlin M., Lúcia Gigante M. High protein yogurt with addition of Lactobacillus helveticus: Peptide profile and angiotensin-converting enzyme ACE-inhibitory activity. Food Chem. 2020;333:127482. doi: 10.1016/j.foodchem.2020.127482. [DOI] [PubMed] [Google Scholar]
- 19.Pihlanto A., Virtanen T., Korhonen H.J.T. Angiotensin I converting enzyme (ACE) inhibitory activity and antihypertensive effect of fermented milk. Int. Dairy J. 2010;20:3–10. doi: 10.1016/j.idairyj.2009.07.003. [DOI] [Google Scholar]
- 20.Solieri L., De Vero L., Tagliazucchi D. Peptidomic study of casein proteolysis in bovine milk by Lactobacillus casei PRA205 and Lactobacillus rhamnosus PRA331. Int. Dairy J. 2018;85:237–246. doi: 10.1016/j.idairyj.2018.06.010. [DOI] [Google Scholar]
- 21.Duar R.M., Lin X.B., Zheng J., Martino M.E., Grenier T., Pérez-Muñoz M.E., Leulier F., Gänzle M., Walter J. Lifestyles in transition: Evolution and natural history of the genus Lactobacillus. FEMS Microbiol. Rev. 2017;41:S27–S48. doi: 10.1093/femsre/fux030. [DOI] [PubMed] [Google Scholar]
- 22.Sadat-Mekmene L., Genay M., Atlan D., Lortal S., Gagnaire V. Original features of cell-envelope pro-teinases of Lactobacillus helveticus. A review. Int. J. Food Microbiol. 2011;146:1–13. doi: 10.1016/j.ijfoodmicro.2011.01.039. [DOI] [PubMed] [Google Scholar]
- 23.Liu M., Bayjanov J.R., Renckens B., Nauta A., Siezen R.J. The proteolytic system of lactic acid bacteria revisited: A genomic comparison. BMC Genom. 2010;11:36. doi: 10.1186/1471-2164-11-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sarmadi B.H., Ismail A. Antioxidative peptides from food proteins: A review. Peptides. 2010;31:1949–1956. doi: 10.1016/j.peptides.2010.06.020. [DOI] [PubMed] [Google Scholar]
- 25.Pihlanto A. Antioxidative peptides derived from milk proteins. Int. Dairy J. 2006;16:1306–1314. doi: 10.1016/j.idairyj.2006.06.005. [DOI] [Google Scholar]
- 26.Udenigwe C.C., E Aluko R. Food Protein-Derived Bioactive Peptides: Production, Processing, and Potential Health Benefits. J. Food Sci. 2011;77:R11–R24. doi: 10.1111/j.1750-3841.2011.02455.x. [DOI] [PubMed] [Google Scholar]
- 27.Ramesh V., Kumar R., Singh R.R.B., Kaushik J.K., Mann B. Comparative evaluation of selected strains of lactobacilli for the development of antioxidant activity in milk. Dairy Sci. Technol. 2011;92:179–188. doi: 10.1007/s13594-011-0048-z. [DOI] [Google Scholar]
- 28.Virtanen T., Pihlanto A., Akkanen S., Korhonen H.J. Development of antioxidant activity in milk whey during fermentation with lactic acid bacteria. J. Appl. Microbiol. 2007;102:106–115. doi: 10.1111/j.1365-2672.2006.03072.x. [DOI] [PubMed] [Google Scholar]
- 29.Gobbetti M., Ferranti P., Smacchi E., Goffredi F., Addeo F. Production of angiotensin-I-converting-enzyme-inhibitory peptides in fermented milks started by Lactobacillus delbrueckii subsp. bulgaricus SS1 and Lactococcus lactis subsp. cremoris FT4. Appl. Environ. Microbiol. 2000;66:3898–3904. doi: 10.1128/AEM.66.9.3898-3904.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Contreras M.D.M., Carrón R., Montero M.J., Ramos M., Recio I. Novel casein-derived peptides with antihypertensive activity. Int. Dairy J. 2009;19:566–573. doi: 10.1016/j.idairyj.2009.05.004. [DOI] [Google Scholar]
- 31.Saito T., Nakamura T., Kitazawa H., Kawai Y., Itoh T. Isolation and structural analysis of antihypertensive peptides that exist naturally in gouda cheese. J. Dairy Sci. 2000;83:1434–1440. doi: 10.3168/jds.S0022-0302(00)75013-2. [DOI] [PubMed] [Google Scholar]
- 32.Ali E., Nielsen S.D., Aal S.A.-E., El-Leboudy A., Saleh E., Lapointe G. Use of Mass Spectrometry to Profile Peptides in Whey Protein Isolate Medium Fermented by Lactobacillus helveticus LH-2 and Lactobacillus acidophilus La-5. Front. Nutr. 2019;6:152. doi: 10.3389/fnut.2019.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abdel-Hamid M., Romeih E., Gamba R.R., Nagai E., Suzuki T., Koyanagi T., Enomoto T. The biological activity of fermented milk produced by Lactobacillus casei ATCC 393 during cold storage. Int. Dairy J. 2019;91:1–8. doi: 10.1016/j.idairyj.2018.12.007. [DOI] [Google Scholar]
- 34.Maeno M., Yamamoto N., Takano T. Identification of an antihypertensive peptide from casein hydrolysate produced by a proteinase from Lactobacillus helveticus CP790. J. Dairy Sci. 1996;79:1316–1321. doi: 10.3168/jds.S0022-0302(96)76487-1. [DOI] [PubMed] [Google Scholar]
- 35.Hayes M., Stanton C., Slattery H., O’Sullivan O., Hill C., Fitzgerald G.F., Ross R.P. Casein Fermentate of Lactobacillus animalis DPC6134 Contains a Range of Novel Propeptide Angiotensin-Converting Enzyme Inhibitors. Appl. Environ. Microbiol. 2007;73:4658–4667. doi: 10.1128/AEM.00096-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ong L., Shah N. Release and identification of angiotensin-converting enzyme-inhibitory peptides as influenced by ripening temperatures and probiotic adjuncts in Cheddar cheeses. LWT. 2008;41:1555–1566. doi: 10.1016/j.lwt.2007.11.026. [DOI] [Google Scholar]
- 37.Pihlanto-Leppälä A., Koskinen P., Piilola K., Tupasela T., Korhonen H. Angiotensin I-converting enzyme inhibitory properties of whey protein digests: Concentration and characterization of active peptides. J. Dairy Res. 2000;67:53–64. doi: 10.1017/S0022029999003982. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is contained within the article or supplementary material.