Abstract
Across the water sector, Escherichia coli is the preferred microbial water quality indicator and current guidance upholds that it indicates recent faecal contamination. This has been challenged, however, by research demonstrating growth of E. coli in the environment. In this study, we used whole genome sequencing to investigate the links between E. coli and recent faecal contamination in drinking water. We sequenced 103 E. coli isolates sampled from 9 water supplies in rural Kitui County, Kenya, including points of collection (n = 14) and use (n = 30). Biomarkers for definitive source tracking remain elusive, so we analysed the phylogenetic grouping, multi-locus sequence types (MLSTs), allelic diversity, and virulence and antimicrobial resistance (AMR) genes of the isolates for insight into their likely source. Phylogroup B1, which is generally better adapted to water environments, is dominant in our samples (n = 69) and allelic diversity differences (z = 2.12, p = 0.03) suggest that naturalised populations may be particularly relevant at collection points with lower E. coli concentrations (<50 / 100mL). The strains that are more likely to have originated from human and/or recent faecal contamination (n = 50), were found at poorly protected collection points (4 sites) or at points of use (12 sites). We discuss the difficulty of interpreting health risk from E. coli grab samples, especially at household level, and our findings support the use of E. coli risk categories and encourage monitoring that accounts for sanitary conditions and temporal variability.
Introduction
The gram-negative coliform bacterial species, Escherichia coli, was discovered in 1885 during an investigation of the microbial life of the human gastrointestinal tract [1]. On the basis of their association with the gastrointestinal tract, E. coli have been recommended as indicators of microbial contamination risk in water since 1892 [2]. For decades, they were studied primarily in lab and clinical settings that further emphasised their association with the gastrointestinal tract and, therefore, the faeces of humans and other vertebrates. But their ecological niche is much broader than initially realised, and the species is now recognised as ‘hardy generalist’ rather than gastrointestinal tract specialist [3]. This has consequences for its use as a water quality indicator.
E. coli occupy two habitats: the primary [gastrointestinal tract) and secondary [water, sediment, soil, flora). Initially, they were thought to have a net negative growth rate in the secondary habitat, implying short-term extra-host persistence [4]. But since the early 2000s, the assumption of negative growth rate in secondary habitats has increasingly been challenged by recognition of naturalised E. coli populations. E. coli have a core genome of about 2,000 genes, but roughly half the genes in any E. coli bacterium are contained in the ‘accessory genome’–which exhibits large genotypic and phenotypic diversity across strains, allowing for diverse adaptive paths [5, 6]. This adaptability enables E. coli to contend with many stresses in environmental habitats [7, 8], which could include nutrient deprivation; sub-optimal temperature, salinity, moisture, and substrate texture ranges; exposure to solar radiation; competition with other microbes; and predation by protozoans [9, 10]. E. coli have been found to grow in solutions of pH ranging from 4.5 to 9 [11]. They are able to acquire energy in versatile ways and grow in both anaerobic and aerobic conditions with temperatures ranging from 7.5 to 49°C [10].
Studies have demonstrated E. coli persisting on algae and in soils, sediment, and sand in tropical, subtropical and temperate environments [10], as well as in handpumps removed from boreholes [12]. Furthermore, long-term growth of E. coli has been demonstrated in a diverse array of source environments including lakes, rivers, sediments, beaches, soils, and aquatic plants and animals [13]. Several investigations have described genetically distinct populations of naturalised E. coli [14–20], including environmentally associated cryptic clade phylogroups [13]. And a recent meta-analysis representing the phylogenetic diversity of more than 5000 (mostly) non-clinical isolates from Australia associated genetic backgrounds with specific habitats, including freshwater [21].
Such a meta-analysis is not yet possible for Kenya, nor the African continent more broadly, because genetic studies of non-clinical E. coli are relatively sparse. The majority of E. coli isolates sequenced in Kenya are from clinical, livestock, or food hygiene sampling [22], so there is limited information on the genetic background of E. coli sampled from the environment, including water systems. In general, few studies have undertaken whole genome sequencing of E. coli in drinking water supplies, although a characterisation of 28 isolates from chlorinated water supplies in Australia suggested that 9 isolates were naturalised E. coli that were unlikely to represent human health risk [23].
The utility of E. coli as a faecal contamination indicator in water systems is challenged by the existence of naturalised E. coli populations [24]. It can be difficult to interpret E. coli sampling results, particularly when water system safety controls have not been validated and when sampling is infrequent [25]. Thus, the severity and immediacy of the potential threat indicated by an E. coli positive sample is unclear. Despite this uncertainty, E. coli remains the preferred indicator of microbial water safety, including for rural community managed supplies [26]. The World Health Organisation drinking-water quality guidelines do not account for naturalised E. coli populations in source waters or distribution systems; they continue to assure that “the presence of E. coli should be considered as evidence of recent faecal contamination” [26 p57].
In tracking progress towards the drinking-water Sustainable Development Goal (SDG 6.1), E. coli is used to assess whether water is free from faecal contamination, a prerequisite of being considered ‘safely managed’. As a result, E. coli testing is included in large-scale household survey programmes in more than thirty countries [27]. These survey programmes are testing water both at point of collection (PoC) and household-level point of use (PoU). That concentrations of E. coli often increase between PoC and PoU is well-established in the literature, evinced by studies from both urban and rural areas in Africa, South-East Asia, South and Central America, and the Caribbean [28–30]. However, the health implications of this increase are debated, as are the implications for where water quality monitoring and treatment efforts should focus [30]. Increases in E. coli concentration between PoC and PoU may result from contamination introduced during transport and household storage, which is influenced by sanitation, hygiene, and water handling practices; however, it may also result from growth of E. coli that was in the water supply, or from sloughing of biofilms in storage containers–neither of which would normally represent an increased health hazard [31, 32].
Thus, use of E. coli in rural water quality monitoring involves two key uncertainties. First, to what extent is E. coli at PoC linked to recent faecal contamination? Second, what are the health hazard implications of changes in E. coli concentration between PoC and PoU? These long-standing [33] and strongly context dependant uncertainties are not easily resolved. In this study, we sought insight for managing them in the context of water supply in rural Kitui County, Kenya. We isolated E. coli from drinking water supplies and household storage and used whole genome sequencing to characterise each isolate to investigate the diversity of phylogroups, multi-locus sequence types, and virulence and antibiotic resistance genes within and between water supplies and household water storage. The results contribute to understanding of non-clinical E. coli populations in Kenya and provide insight into the occurrence of E. coli in rural water systems, allowing comparison between PoC and PoU and informing on the utility of E. coli as a faecal contamination indictor in these settings.
Materials and methods
Ethics statement
Field research was conducted with permission from the Kenyan National Commission for Science, Technology and Innovation, License No: NACOSTI/P/18/22793/24854. Ethical approval was obtained from the University of Oxford’s Central University Research Ethics Committee. All participation was informed and uncompensated. Engagement with participants was contingent upon consent from participants after they were informed of the study process and objective verbally. Personal identifiers were stored only for the duration of the study and in a secured platform.
Water sampling
Nine water supply systems were selected from among those that were sampled by a rural water maintenance service provider in northern Kitui County, Kenya as part of a monitoring programme that had commenced 7 months prior to this study. The systems were chosen to include multiple supply types with varying degrees of protection against contamination (piped schemes sourcing water from boreholes or reservoirs and point sources including boreholes and hand-dug wells with or without handpumps). To be selected, the system had to be a main or alternative source of drinking-water with above zero median E. coli concentration from the monitoring programme results. Most of the selected systems are community managed and are either point sources or small (<5 km) piped distribution systems, all of which include one or more storage tanks. Only one of them (S5) is managed by a formal water service provider (utility). The water in this system is clarified and chlorinated in a water treatment facility and then piped through a 66 km distribution network. The utility produces about 3000 m3/day on average, but due to the size of the distribution system and high demand for water, the supply at the PoC is intermittent (approximately 2 days / week).
For each system, we sampled one or more PoCs. For each PoC, we sampled multiple PoUs. We asked water system managers and users to help us find homes with stored water. The goal was to sample at least three PoUs per PoC, but it was sometimes difficult to find households with stored water from the PoC of interest due to multiple source use and mixing water from multiple sources in home storage. As indicated in Table 1, a total of 44 sites were sampled including 14 PoCs and 30 PoUs (more details on site protection and storage length and location are available for each site in S1 Table). We refer to a system and its associated PoCs and PoUs as a set, and each sampling site is labelled according to its corresponding set, PoC and PoU (S#C#U#). The sampling sites are located within 1,400 km2, with the distance between any two PoCs from different systems ranging from 50 km to 15 m apart.
Table 1. List of sampling sets with site IDs and source, system, collection, transport and storage types.
| Set | Source and system | Collection type | Site ID | Transport type | Storage type |
|---|---|---|---|---|---|
| 1 | Reservoir piped | Standpipe | S1C1 | ||
| S1C1U1 | Motorbike | Jerrican | |||
| S1C1U2 | Donkey | Drum | |||
| S1C1U3 | Donkey | Drum | |||
| S1C1U4 | Donkey | Jerrican | |||
| S1C1U5 | Donkey | Jerrican | |||
| S1C1U6 | Donkey | Jerrican | |||
| Standpipe | S1C2 | ||||
| S1C2U1 | Donkey | Drum | |||
| S1C2U2 | Donkey | Jerrican | |||
| S1C2U3 | Donkey | Jerrican | |||
| 2 | Borehole piped | Standpipe | S2C1 | ||
| S2C1U1 | Wheelbarrow | Drum | |||
| S2C1U2 | Self-carry | Jerrican | |||
| Standpipe | S2C2 | ||||
| S2C2U1 | Motorbike | Jerrican | |||
| S2C2U2 | Motorbike | Drum | |||
| S2C2U3 | Wheelbarrow | Druma | |||
| School standpipe | S2C3b | ||||
| 3 | Borehole piped2 | Standpipe | S3C1 | ||
| S3C1U1 | Self-carry | Jerrican | |||
| S3C1U2 | Self-carry | Drum | |||
| S3C1U3 | Donkey | Drum | |||
| S3C1U4 | Self-carry | Jerrican | |||
| School concrete tank | S3C2b | ||||
| 4 | Borehole piped | Plastic tank | S4C1 | ||
| S4C1U1 | Self-carry | Jerrican | |||
| S4C1U2 | Wheelbarrow | Jerrican | |||
| Concrete tank | S4C2c | ||||
| 5 | Reservoir pipedd | Plastic tank | S5C1 | ||
| S5C1U1 | Donkey | Jerrican | |||
| S5C1U2 | Donkey | Drum | |||
| S5C1U3 | Donkey | Jerrican | |||
| 6 | Borehole point | Handpump | S6C1 | ||
| S6C1U1 | Donkey | Jerrican | |||
| S6C1U2 | Donkey | Drum | |||
| S6C1U3 | Donkey | Jerrican | |||
| 7 | Borehole point | Handpump | S7C1 | ||
| S7C1U1 | Donkey | Jerrican | |||
| S7C1U2 | Self-carry | Jerrican | |||
| 8 | Dug well point | Bucket draw | S8C1 | ||
| S8C1U1 | Donkey | Jerrican | |||
| S8C1U2 | Donkey | Jerrican | |||
| 9 | Borehole piped | School concrete tank | S9C1b |
aThis storage drum had a dispensing tap.
bThe PoCs in schools (n = 3) were not associated with PoU storage because students drank directly from the standpipes or tank taps.
cThis PoC was sampled twice, two weeks apart, but no PoU water was available either time.
dChlorine is used for treatment in this system, but dosing is inconsistent and chlorine residual testing is not done.
Seven months of monitoring data were available for the selected PoC sites, which had been sampled either weekly or monthly (with some gaps due to breakdowns and dry periods). Monitoring included in situ measurement of pH and conductivity using a HACH multimeter (HQ 40D), which was calibrated weekly, and E. coli sampling using an IDEXX Quanti-Tray2000 system with weekly field blank and duplicate samples. No E. coli was detected in the blank samples and duplicates had an average relative percent difference of 25.6% with 88% of duplicate pairs indicating the same risk category, as defined by the World Health Organisation [26]. Surveys and interviews were conducted alongside the monitoring activities to understand water use practices and track managers’ responses to the test results.
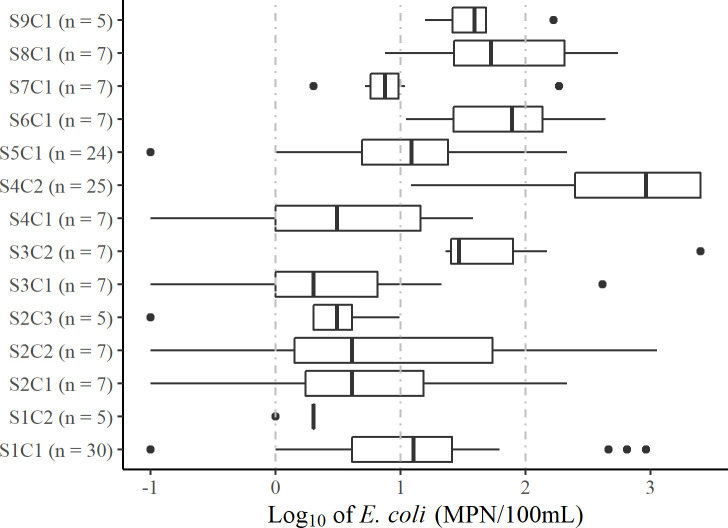
The median pH for each site ranged from 6.6 to 8.6 and median conductivity ranged from 90 to 1600 μS/cm except for Sets 4 and 7, which had median conductivity of 5000 and 10,500 μS/cm, respectively. Due to the high conductivity, which is linked to salinity, the managers reported that water from these sites is only used for drinking when better alternatives are unavailable due to dry periods, breakdowns, affordability, and intermittent supply from the utility. Box-and-whisker plots for each PoC are available for pH (S1 Fig) and conductivity (S2 Fig). The median E. coli concentrations ranged from 1 to 920 MPN/100mL, with all PoCs having variable results over the monitoring period (Fig 1).
Fig 1. Box-and-whisker plot of log10-transformed monitoring programme E. coli results for selected PoCs.
The boxes show median values and span lower to upper quartiles, the whiskers show the lowest and highest datums within 1.5 times the interquartile range. Data are censored by a lower bound of 0 MPN/100 mL and an upper bound of > 2419.6 MPN/100mL. Results at the lower bound were converted to 0.1 (log = -1) and results at the upper bound were converted to 2500 (log = 3.4). Vertical dashed grey lines show the WHO recommended risk category cut-offs [26] with E. coli equal to 1, 10, and 100 MPN/100mL (corresponding to log values of 0, 1, and 2). The risk categories are low (<1), intermediate (1–9), high (10–99), and very high (>99).
No previous sampling was available for the PoU sites. During the sampling visits for this study, short surveys were conducted with the household water managers (as distinct from household heads who are not always aware of the details of water management in the home). Respondents reported that PoU water was transported by donkey, motorbike, wheelbarrow, or self-carry to be stored in either plastic drums (approximately 200L) or jerricans (20L) located either inside the main house, a separate shelter, or outside in a private or communal area; at the time of sampling, the water was reported to have been stored between 0 and 4 days, with 1 or 2 days being most common (S1 Table). Respondents said that jerricans were occasionally cleaned with sand; they did not approximate a washing schedule but said the decision to clean depends on the colour inside the jerrican. Although the first household in set 8 reported occasional boiling and the third in set 6 reported occasional chlorination, none had treated the water that we sampled. PoU water was sampled in line with standard practice by asking the respondent to provide us with a cup of water that they would normally drink from.
Sites within a set were sampled on the same day between July 18 and August 2, 2019, which is dry season in Kitui. This season was chosen to control for rain events, which would differentially impact the quality of PoC water versus PoU water that was stored for multiple days. Two samples were collected at each site in sterile 100mL Whirl-pak bags with sodium thiosulphate to neutralise residual chlorine. At the PoCs, taps and handpump spouts were disinfected by flame and at least 20L of water was pumped or flushed prior to sample collection.
Sample filtration, culturing, and preservation
Samples were transported in a cooler box with icepacks to a field-lab where they underwent membrane filtration. Based on previous E. coli monitoring results, multiple dilutions were used for each site to maximise the chance of growing well-isolated colonies. Following filtration, the samples were incubated with m-ColiBlue24 broth (EPA Approved Hach Co.: 10029 method), which indicates E. coli colonies by blue colouration resulting from hydrolysis of 5-bromo-4-chloro-3-indolyl-β-D-glucuronide (BCIG). In all cases, the time between sample collection and filtration was less than 6 hours. Between filtration and incubation, samples had resuscitation time of 1 to 4 hours. Samples were incubated at 44.5°C for 18–24 hours. The incubation temperature was chosen to discourage growth of non-thermotolerant coliforms and improve isolation of E. coli colonies. We considered that incubation at 44.5°C could disadvantage naturalised E. coli, but studies have shown that although environmental strains grow better than enteric strains at low temperatures, their maximal growth rate and optimal temperature for growth are not distinct from enteric strains [34, 35].
Up to 6 colonies per site were selected for streaking on agar plates (ReadyPlate CHROM Chromocult Coliform Agar ISO 9308–1:2014), which inhibit growth of non-coliforms and distinguish E. coli based on β-glucuronidase activity. The plates were incubated at 37.5°C for 18–24 hours. To increase the likelihood of selecting single strain E. coli colonies, selected colonies were well isolated and consistent in colour and morphology. The goal was to select 6 colonies per site, although this was not possible in a few sites that had low E. coli concentrations. With 6 colonies selected, a strain that composes 25% of the population of E. coli in the sample is 80% likely to be selected at least once. Increasing the number of selected colonies gives diminishing returns in terms of the likelihood of sampling at least one isolate of a strain with a given prevalence (see S3 Fig). Two key factors constrained out total sample size: 1) we were limited to sampling one set per day because of the long distances between sites and the need to walk to many of the homesteads for collecting PoU samples, and 2) our field lab was not equipped for freezing samples nor doing clean DNA extractions, thus limiting the window of time available to us before samples had to be transported to a better-equipped lab to proceed with DNA extraction.
Following incubation, E. coli growth on the agar was carefully scraped with a sterile inoculation loop, with care taken to minimise inclusion of agar and off-colour growth (pink growth observed in 12% of samples and colourless growth in 3% of samples). The E. coli growth was mixed into 1 mL of DNA Shield (Zymo Research R1100) for preservation in a sterile microcentrifuge tube and stored in a fridge before transport to the Kenya Medical Research Institute (KEMRI) lab in Kilifi.
DNA extraction and whole genome sequencing
DNA were extracted within 1 to 3 weeks of sampling using a Zymo Research Quick-DNA Mini-prep kit. We adjusted the recommended protocol for monolayer cells to suit E. coli that is already lysed by preservation in DNA Shield. Thus, we combined 175 μL of sample lysate with 525 μL of genomic lysis buffer and then proceeded with the monolayer cell protocol as recommended by Zymo Research. Samples were normalised to 5 ng following DNA quantification using a Qubit dsDNA HS Assay Kit and Qubit 3.0 Fluorometer (Life Technologies). We proceeded to library preparation with Illumina Nextera XT DNA Sample Preparation kit as per manufacturer instructions with half reaction alteration.
Following tagmentation and indexing, we did a size selection bead clean-up using 0.6x AMPure XP beads (Beckman Coulter) to select for >500 bp fragments. The libraries were then quantified using Qubit and size distribution was determined on a 2100 Bioanalyser using the High Sensitivity DNA Kit (Agilent Technologies). We proceeded to manual normalization bringing all samples to 2 nm and thereafter pooling the libraries. The pooled libraries were then denatured, spiked with 8% Phix, and run on an Illumina MiSeq platform using the 600 cycles v3 reagent kit with an output of 2 x 200 bp. We did two runs, with 59 libraries pooled in the first run and 68 in the second. One library that was poor quality in the first run, was sequenced again in the second.
Bioinformatics
There are no known biomarkers that can definitively distinguish whether an E. coli isolate comes from a naturalised population or other source [13], so we used multiple characteristics including phylogroup, sequence type, allelic diversity, and presence of virulence and antibiotic resistance genes as suggestive evidence of likely isolate origin. The sequencing reads were processed using the Nullarbor pipeline [36]. Reads were filtered and trimmed using Trimmomatic [37] and only reads that were >100 bp with PHRED quality score >20 were retained. Kraken2 was used for species identification [38] and SPAdes v3.13.1 was used for de novo genome assembly [39].
The assembled genomes were assigned as E. coli sensu stricto (phylogroups A, B1, B2, C, D, F, E, or G), Escherichia cryptic clades I-V, E. fergusonii, or E. albertii using the ClermonTyping in silico approach based on standard PCR assays and Mash genome distance estimation [40, 41]. The threshold for minimal nucleotides for a contig to be included in the analysis was set at 100. Twelve of the assembled genomes were overlarge (ranging from 5.8 to 11.9 Mbp) so, suspecting chimeric genomes, we conducted Benchmarking Universal Single-Copy Ortholog (BUSCO) assessment [42, 43] to check the assembled genomes for completeness and duplication. For the non-chimeric genomes, we used Roary [44] for pan-genome analysis. With Roary, genes that are present once in every isolate are combined in a multiple FASTA alignment, enabling phylogenetic tree construction from the core genes. We used FastTree version 2.1.10 SSE3 [45], with generalised time reversible model for nucleotide alignment, to construct an approximately-maximum-likelihood phylogenetic tree. We used GrapeTree [46] to visualise the tree including metadata. For comparison with our sample strains, we included 14 strains from the ClemonTyping Mash database [40] in our analysis, chosen to reflect the diversity of Escherichia spp.
Multi-locus sequence typing was done through Nullarbor using the PubMLST database, and virulence and antimicrobial resistance (AMR) gene identification was done using the Abricate package [47] by screening contigs against the VFDB [48] and NCBI AMRFinderPlus [49] databases, respectively. For multi-locus sequence typing, we chose the Achtman MLST scheme [50], which uses genes adk, fumC, gyrB, icd, mdh, purA, and recA, because it is most congruent with an established phylogeny for E. coli that is based on whole genome sequencing [51]. In addition to the Nullarbor typing, the raw sequencing reads were also run through the MLST screening tool hosted by the Centre for Genomic Epidemiology (CGE) [52] for confirmation of the allele identifications.
Statistics
To investigate possible relationships between allelic diversity and sample source, we segregated our sampling sites into groups based on location (PoC or PoU) and E. coli concentrations (lower or higher) on the day that the isolates were sampled, as shown in Table 2. The cut-off between ‘lower’ or ‘higher’ was set at 50 CFU/100mL to balance the number of isolates in each group as evenly as possible. Focussing on the 7 Achtman MLST scheme genes, we used an approach developed for estimating average population heterozygosity from a small number of individuals [53] by calculating the genetic diversity (H) of each group based on the diversity of alleles (hj) as:
| (1) |
where pi is the frequency of allele i at locus j and n is the number of isolates. Genetic diversity (H) was then calculated as the average diversity of alleles using:
| (2) |
where m is the total number of loci. We assessed the significance of differences in the diversity of the groups using permutation tests derived from the Strasser-Weber framework for conditional inference procedures [54] and performed using the ‘coin’ package in R version 3.6.1 [55]. We used the general independence test function with asymptotic null distribution as computed by the randomised quasi-Monte Carlo method [56].
Table 2. Site groupings based on source type and concentration of E. coli on the day that the isolates were sampled.
| Groups | Sites Included in Each Group | No. of Sets | No. of Samples | No. of MLSTs | No. of Isolates | Median E. coli* | Min E. coli* | Max E. coli* |
|---|---|---|---|---|---|---|---|---|
| All | all sites | 9 | 25 | 46 | 108+ | 45 | 1 | 2420 |
| PoU | PoU sites | 7 | 15 | 30 | 59 | 16 | 1 | 2000 |
| PoC | PoC sites | 7 | 10 | 24 | 49 | 64 | 2 | 2420 |
| Higher | sites with [>50] | 9 | 11 | 26 | 57 | 460 | 50 | 2420 |
| Lower | sites with [<50] | 5 | 14 | 22 | 51 | 13 | 1 | 45 |
| PoU-H | S1C2U1, S2C1U1, S5C1U3, S7C1U1, S8C1U1, S8C1U2 | 5 | 6 | 15 | 31 | 125 | 50 | 2000 |
| PoU-L | S1C1U2, S2C1U2, S2C2U1, S2C2U2, S2C2U3, S3C1U3, S5C1U2, S6C1U1, S6C1U3 | 5 | 9 | 16 | 28 | 10 | 1 | 17 |
| PoC-H | S3C2, S4C2-A/B, S8C1, S9C1 | 4 | 5 | 14 | 26 | 1120 | 84 | 2420 |
| PoC-L | S1C1, S2C1, S2C2, S2C3, S6C1 | 3 | 5 | 10 | 23 | 42 | 2 | 45 |
*Units are MPN/100mL.
+This includes 5 chimeric isolates that contained only one match for each MLST gene with perfect identity matches for each allele as explained in the following sequencing overview section.
Results and discussion
Sequencing overview
The sampling effort targeted 6 isolates for each of the 44 sites, but fewer were sequenced for 16 sites and none were sequenced for 20 sites. This was because of low or no concentration of E. coli in the sample water (S1C1, S1C1U1/U3-6, S1C2, S1C2U2-3, S2C2U1, S2C3, S3C1, S3C1U1-2/4, S4C1, S4C1U1-2, S5C1, S5C1U1, S6C1, S7C1) or high concentration of thermotolerant coliforms (TTCs) preventing clean selection of 6 colonies (S1C2U1, S2C2U3, S3C1U3, S4C2-A, S5C1U2, S5C1U3, S6C1U2, S7C1U2). Other issues preventing sequencing of isolates included TTC contamination of the agar plate (1 isolate each from S3C2 and S7C1U1); limited growth on the agar plate (1 isolate from S2C1U2); inadequate DNA extraction (1 isolate from S4C2-B); and sequencing library preparation failure (1 isolate from S2C1U1).
A total of 125 libraries were successfully sequenced (see S2 Table for the full list), including 4 duplicates. The duplicates were consistent in phylogroup and sequence type. They are not otherwise included in the results. Six of the libraries were contaminated with non-Escherichia DNA, five from S1C1U2 contained Cronobacter sakazakii and one from S5C1U2 contained Klebsiella pneumoniae. These libraries were removed from further analysis. The remaining 115 libraries had 628,219 reads per library on average (SD = 145 199), with mean read length of 177 bp (SD = 7), mean PHRED quality score of 31 (SD = 0.7), and mean depth of 23 (SD = 6). BUSCO assessment of the genomes assembled from these 115 libraries identified 12 chimeric genomes. Excluding the chimeric genomes, the reads from each library assembled into an average of 246 contigs (SD = 141), with mean genome size of 4.8 Mbp (SD = 0.2).
Four of the chimeric genomes have multiple perfect allele matches for at least one of the Achtman MLST genes, confirming them as chimeras of multiple MLSTs. A further three of them have imperfect identity matches for multiple alleles (ranging from 86.5% to 99.8% identity match) and do not correspond to known sequence types. These 7 chimeras with multiple MLSTs or imperfect allele matches were excluded from further analysis. The remaining 5 chimeric genomes contain only one match for each MLST gene with perfect identity matches for each allele. These single MLST chimeric genomes are included in the ClermonTyping and MLST results (total of 108 isolates) but were excluded from the pangenome and virulence and AMR analyses (total of 103 isolates).
Pan-genome and phylogeny
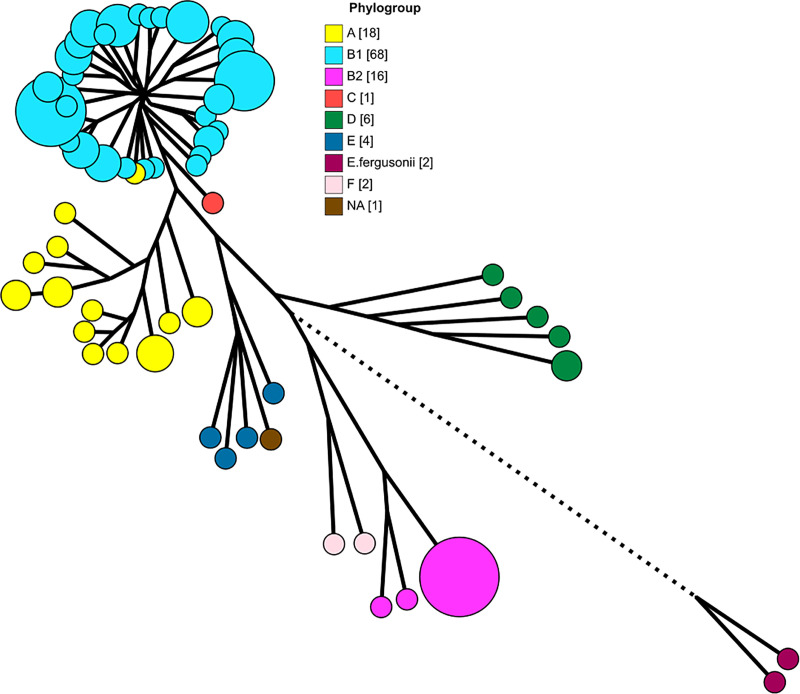
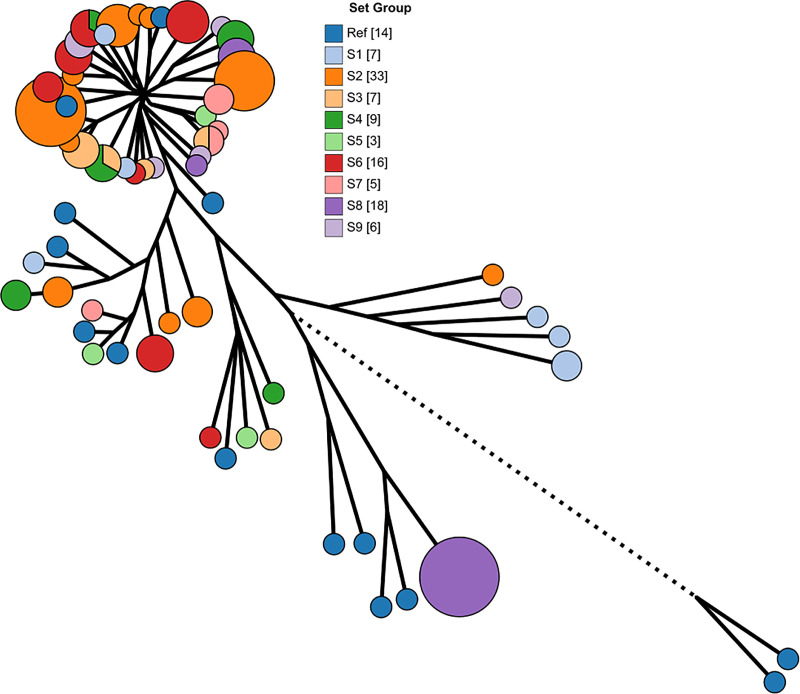
The pan-genome of our isolates and the 14 references strains that we included for comparison has a total of 25,526 genes, including 1794 core genes (in ≥ 99% of the genomes), 1201 soft core genes (in ≥ 95%), 2152 shell genes (in ≥ 15%), and 20,379 cloud genes (in < 15%). The phylogenetic tree generated from the core genes (Fig 2) reflects the ClermonTyping characterisation of our isolates except for isolate 2-8B (MLST 3519), which the in silico assays classified as phylogroup A but was closer to B1 isolates in the Mash estimation and the phylogenetic tree. We note that the MLST 3519 entries in the Warwick Enterobase database are also classified as phylogroup A with AxB1 lineage [22]. When the phylogenetic tree nodes are colour-coded by set (Fig 3), we observe that the evolutionary similarity of the isolates is not related to the water system that they came from.
Fig 2. Phylogenetic tree with nodes coloured by phylogroup as determined by ClermonTyping.
The bracketed numbers in the image keys indicate the number of isolates in each category. The tree is scaled to 125% to make visual parsing easier. Branches with length less than 0.0006 nucleotide substitutions per site were collapsed and the size of the nodes is scaled to indicate the number of isolates each one encompasses. The branch length for E. fergusonnii was shortened from 0.05 to 0.03 substitutions per site.
Fig 3. Phylogenetic tree with nodes coloured by sample set.
The ClermonTyping analysis classified 69 isolates from 21 sites as belonging to phylogenetic group B1, making B1 the most represented group in the sample set. The second-most prevalent is group A with 15 isolates from 8 sites, followed by group B2 with 14 isolates from 3 sites, group D with 6 isolates from 4 sites, and group E with 3 isolates from 3 sites. One isolate from S3C2 could not be classified by ClermonTyping, but the phylogenetic tree indicates that it belongs in phylogroup E (Fig 2). We did not identify cryptic clade E. coli, which are the group most strongly associated with environmental origins [13, 34].
The association between phylogroup and strain origin varies based on diet, hygiene, animal domestication status, and morphological and socioeconomic factors [8]. Some localised studies have found differences in the relative frequency of phylogroups by strain origin [57], but others have not [58]. A review of results from both higher and lower income countries in Europe, Africa, the Americas, Asia, and Australia [8] found that phylogroup B1 strains were dominant in animals (41%), followed by A (22%), B2 (21%), and D strains (16%); whereas A strains were most common in humans (40.5%), followed by B2 (25.5%) and then B1and D strains (17% each). Phylogroup B1 strains, which may be more common in animal faeces than in humans, comprised 78% of our PoC isolates and 50% of our PoU isolates. In contrast, A strains, which may be more common in humans, comprised 4% of our PoC isolates and 22% of our PoU isolates.
The dominance of phylogroup B1 and A strains in our samples does not necessarily indicate recent faecal contamination. Strains from B1 and A are better generalists and are more prevalent in freshwater samples than other strains [59]. B1 strains, especially, have been found to survive best in the environment [7, 60], and in freshwater specifically [9, 21, 61, 62], and phenotypes linked to environmental survival are relatively prevalent in B1 isolates [63, 64]. In contrast, B2 and D isolates do not survive well and are under-represented in freshwater [21, 59]. Less is known about phylogroup E, which is a small set of formerly unassigned strains that are relatively uncommon, historically difficult to cluster phylogenetically, and generally understudied [50, 65] (except for the O157:H7 serotype, which is clinically important but is excluded by typical culture methods that rely on β-glucuronidase activity).
Virulence genes
For the 103 non-chimeric isolates, a total of 184 virulence genes were identified with 100% identity and coverage. The complete list per isolate can be found in S2 Table. These genes represent 8 functional groups including secretion (67 genes), adherence (54 genes), iron uptake (46 genes), chemotaxis and motility (6 genes), invasion (5 genes), immune evasion (3 genes), autotransport (2 genes), and toxin production (1 gene). Most of the genes occur with low frequency across the isolates, and the number of genes possessed by any one isolate ranges from 34 to 94, with most having between 40 and 60 (S4 Fig). Isolates from phylogenetic group A have the least virulence genes (mean = 43, SD = 6.7), and isolates from groups B2 (63, 0.5), E (67, 1.7), and D (75, 9.2) have higher numbers than most B1 isolates (55, 10.1).
Due to the plasticity of the E. coli genome, it can be challenging to define and identify pathovars, but some combinations of virulence genes have been linked to different E. coli pathotypes [48, 66]. Genes for Shiga toxin production were not identified, ruling out presence of enterohemorrhagic E. coli (EHEC) or Shiga toxin-producing E. coli (STEC). Eight isolates from phylogroup B1 have multiple enteropathogenic E. coli (EPEC) associated virulence genes for adherence, autotransport, invasion, iron uptake, motility, secretion, and toxin production, but they lack key bundle-forming pili (bfp) and intimin (eae) genes, suggesting that they are neither typical nor atypical EPEC [67]. Similarly, all twenty isolates from phylogroups D or B2 have multiple genes that are associated with uropathogenic E. coli (UPEC) and / or neonatal meningitis-associated E. coli (NMEC), but all are missing key genes such as fdeC for adherence or cnf1 for toxin production.
None of our isolates were identified as complete pathovars, but the presence of virulence genes is informative, nonetheless, since virulence genes are more prevalent in strains isolated from humans and the trend is conserved within phylogroups [8, 21]. Four phylogroup B1 isolates have exceptionally high numbers of virulence genes (>80), suggesting that they are human derived. All four were isolated from PoU sites (S3C1U3, S6C1U1, S6C1U3). Furthermore, the phylogroup B2 and D isolates possessed more virulence genes than most of the A and B1 isolates. Taken together with evidence of their relatively poor survival in the environment [21, 59], this further supports that the PoC and PoU sites that had B2 or D isolates were subject to recent contamination. This applies to three PoC sites that were poorly protected from contamination (S1C1, an open reservoir; S8C1, an open dug well; S9C1, an unsealed concrete tank) and 4 household sites (S1C2U1, S2C1U2, S8C1U1, S8C1U2).
Antimicrobial resistance genes
For the 103 non-chimeric isolates, a total of 24 AMR genes were identified with 100% identity and coverage, with every isolate having at least one AMR gene associated with resistance to aminoglycosides (aph(6)-Id, aph(3'')-Ib, aadA1, aadA5, aac(3)-IId), beta-lactams (blaCTX-M-14, blaEC-5/8/13/15/18, blaTEM-1), erythromycin (mph(A)), quaternary ammonium compounds (qacEdelta1), sulphonamides (sul1/2), tetracycline (tet(A), tet(B)), or trimethoprim (dfrA1, dfrA5, dfrA14, dfrA17). Additionally, arsenite resistance gene arsB-mob, which codes for an arsenite efflux pump, was found in 82 isolates. Arsenic resistance can develop in response to use of arsenicals in antimicrobial drugs or in response to naturally occurring arsenic in the environment [68]. Concentrations of arsenic were low at the PoC sites, ranging from 0.04 to 0.95 ppb (measurement by inductively coupled plasma mass spectrometry; unpublished data from the monthly monitoring programme). As such, geogenic arsenic is unlikely to explain the prevalence of the arsB-mob gene. Furthermore, the absence of additional arsenic resistance genes suggests that the presence of arsB-mob may not be related to drug use or geogenic arsenic, rather research into broad arsenic resistance in prokaryotes points to ancestral gene clusters as a likely explanation [69].
Excluding arsB-mob, at least one AMR gene was found in each of the 103 isolates. Most of the genes occur with low frequency across the isolates (see S2 Table for a list of AMR genes per isolate), with 88 isolates having only 1 gene. AMR genes are transferrable between bacteria via plasmids and environmental reservoirs of resistance genes are widely recognised, including freshwater and drinking water systems [70, 71]. Nevertheless, multiple AMR genes may be more common in E. coli strains sourced from human as opposed to animal or naturalised populations [13, 72, 73]. Four PoU sites had isolates with multiple AMR genes including 3 phylogroup D isolates with 7 to 12 genes each (S1C2U1) and 6 phylogroup B1 isolates with 4 to 7 genes each (S2C2U1, S3C1U3, S7C1U1). Additionally, two PoC sites (S4C2 and S9C1, both unsealed concrete tanks) had phylogroup B1 isolates with 4 to 7 AMR genes each.
Multi-locus sequence types
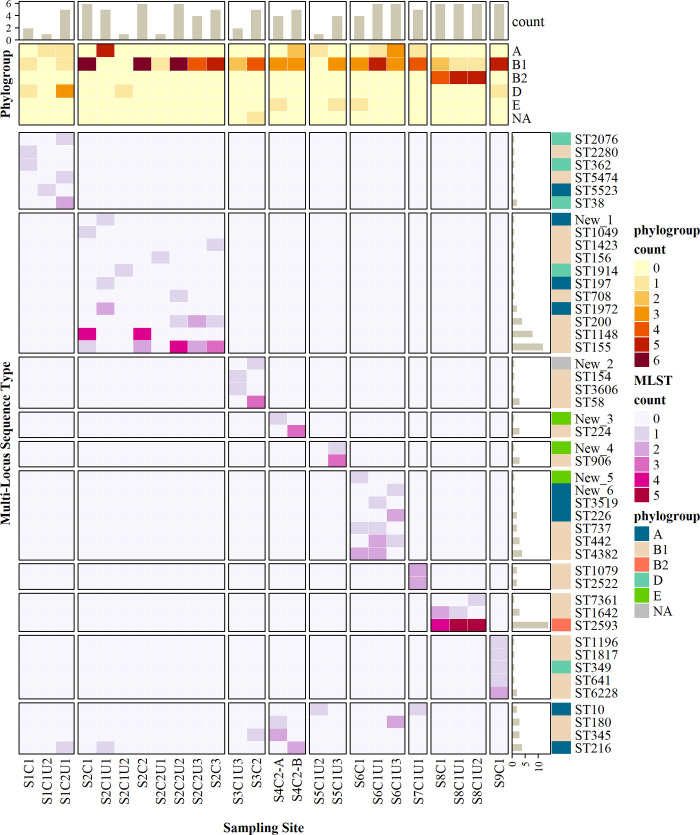
Using the Achtman MLST scheme, a total of 40 previously known sequence types were identified. As detailed in S3 Table, the number of entries for these MLSTs in the Warwick Enterobase database (as of May 2020) ranged from 1 to 7763; 10 of the MLSTs had not previously been identified in Kenya; and 6 of the isolates have MLST allele combinations that were not represented in the database. These new combinations are listed in S2 Table, we have labelled them New_1 to New_6.
We refer to a system and its associated PoCs and PoUs as a set. The main discriminating factor between the sequence types is the set that the isolate came from (Fig 4). The sequence types generally do not overlap between sets, except in the cases of ST10, SW180, ST345, and ST216. We are also interested in the overlap in sequence types between matched PoC and PoU sites. There was no overlap for set 1, but sets 2, 6, and 8 did have overlap. For sets 3 and 5, no E. coli was isolated from the PoCs (except S3C2, which is an unsealed concrete water tank at a school that is unrepresentative of the wider distribution system). For sets 4, 7, and 9, E. coli was only isolated from one site each.
Fig 4. Heatmap of phylogenetic groups and MLSTs identified for each sampling site.
Colour gradients indicate number of isolates. Bar chart annotations show total counts of isolates per MLST (right) and per site (top). The heatmap was created with R package ‘ComplexHeatmap’ [74].
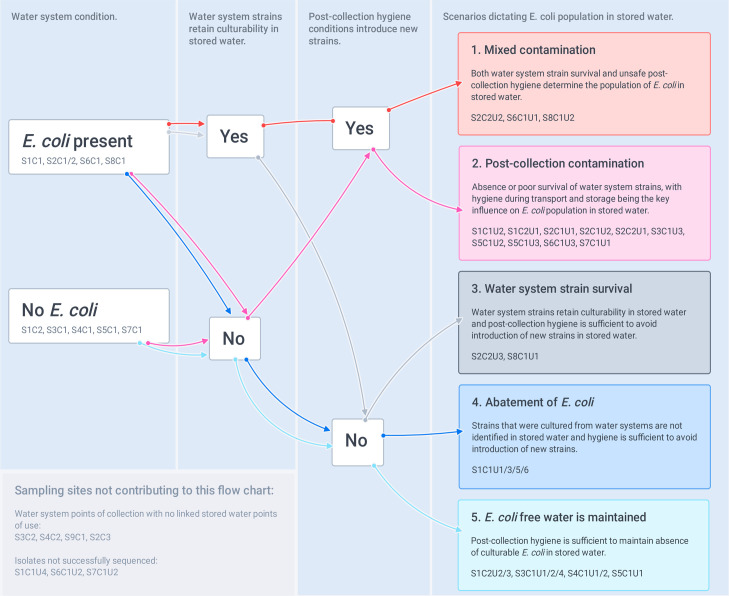
Logically, five scenarios may dictate the population of E. coli in PoU water, these are permutations of three factors: presence or absence of E. coli in the PoC water, whether PoC strains retain culturability in PoU water, and whether post-collection hygiene conditions introduce new strains. Although the isolates analysed in our study allow only a partial view of the diversity of E. coli strains at each sampling site, comparison of the MLSTs isolated from matched PoC and PoU sites suggests that multiple PoUs exemplified each of the five scenarios (Fig 5).
Fig 5. Five scenarios theorised to explain the population of E. coli in stored water at point of use.
Scenarios are based on sustained presence of strains that were isolated from the water supply system and addition of new strains during post-collection transport and storage.
In scenarios 3, 4, and 5 no additional health hazard is introduced at the household level: E. coli in PoU water is determined by PoC water quality and persistence or abatement of strains in storage containers. Abatement of E. coli does not equate to abatement of pathogens (decreased health risk), so PoC results should be prioritised in these scenarios. In scenarios 1 and 2, inadequate hygiene at the household level does influence PoU E. coli population, either as the exclusive driver (scenario 2) or in combination with persistence of PoC E. coli strains (scenario 1). Only five of the households that we sampled had overlap in MLSTs between PoC and PoU (scenarios 1 and 3), this points to strain persistence but is not evidence of regrowth. Nevertheless, the results are interesting in light of research that a) found increase in E. coli between PoC and PoU was unrelated to household-level sanitary or hygiene factors [31] and b] demonstrated regrowth in household storage containers within 48 hours [75]. Conversely, Scenario 2, post-collection contamination, appears to be the most common for the households that we sampled, which is consistent with studies that relate PoU water quality deterioration to unsafe storage [28, 76].
The health implications of post-collection contamination are debated. In households like those in our study area, with low levels of access to sanitation and hygiene facilities, E. coli and other faecal indicators and pathogens are widespread on surfaces and in food produce [77] and it is likely that strains circulate between humans, animals, and the domestic environment [8, 78]. Large-scale randomised control trials investigating the impact of water, sanitation, and hygiene interventions on health outcomes for communities in Kenya, Bangladesh and elsewhere have demonstrated the importance of considering multiple faecal exposure pathways [79]. In a household setting, where there are multiple pathways for exposure to pathogens that are circulating in the household environment, focussing on only one of these pathways (stored water) is unlikely to reduce the burden of enteric disease in the household [80, 81]. In contrast, a contaminated water distribution system may pose a unique threat as a pathway for spreading disease between households and, in some cases, between communities.
Allelic diversity
In addition to directly comparing MLSTs, we queried the differences between sites further by using the MLST genes to analyse differences in the diversity of the isolates from the lower (<50 CFU/100mL) and higher (>50 CFU/100mL) concentration PoC and PoU sites. We found all loci are polymorphic in all groupings (Table 3) and the diversity of alleles (hj) ranges from 0.32 to 0.91. A permutation test comparing all four of the sub-groups (PoU-H, PoU-L, PoC-H, and PoC-L) found that the grouping has an effect on diversity (z = 2.47, p = 0.0499); more specifically, pairwise tests found that the PoC-L group is less diverse than both the PoC-H group (z = 2.12, p = 0.03) and the PoU-H group (z = 2.29, p = 0.02). No other pairwise differences are significant, including comparisons of the PoU sites versus the PoC sites (z = 1.13, p = 0.26), the high concentration sites versus the low concentration sites (z = 1.82, p = 0.07), or the PoU-L sites versus the PoU-H sites (z = 1.4, p = 0.17).
Table 3. Genetic diversity of alleles (hj) and average genetic diversity (H) for groupings based on source type (PoC vs PoU) and E. coli concentration level (higher or lower than 50 MPN/100mL).
| Groups | H | Diversity of Alleles (hj) | ||||||
|---|---|---|---|---|---|---|---|---|
| adk | fumC | gyrB | icd | mdh | purA | recA | ||
| All | 0.80 | 0.63 | 0.83 | 0.90 | 0.87 | 0.85 | 0.70 | 0.81 |
| PoU | 0.82 | 0.72 | 0.88 | 0.88 | 0.89 | 0.86 | 0.70 | 0.82 |
| PoC | 0.76 | 0.51 | 0.76 | 0.90 | 0.84 | 0.81 | 0.71 | 0.76 |
| Higher | 0.82 | 0.74 | 0.84 | 0.89 | 0.87 | 0.84 | 0.77 | 0.80 |
| Lower | 0.70 | 0.44 | 0.78 | 0.84 | 0.82 | 0.80 | 0.54 | 0.64 |
| PoU-H | 0.81 | 0.79 | 0.84 | 0.85 | 0.85 | 0.82 | 0.80 | 0.74 |
| PoU-L | 0.72 | 0.52 | 0.85 | 0.81 | 0.87 | 0.78 | 0.44 | 0.76 |
| PoC-H | 0.81 | 0.65 | 0.80 | 0.91 | 0.89 | 0.86 | 0.73 | 0.81 |
| PoC-L | 0.62 | 0.32 | 0.67 | 0.76 | 0.77 | 0.75 | 0.62 | 0.45 |
That the allelic diversity of E. coli isolates from low concentration PoCs is significantly less than from high concentration sites is interesting in light of studies that found lower allelic diversity and greater genome similarity in samples from environmental sources compared to samples from faecal sources [13, 82, 83]. The comparison suggests that PoC sites with low concentration of E. coli may be more associated with naturalised E. coli. Although not significantly different from the high concentration sites, the low concentration PoU sites also had lower allelic diversity and, therefore, may have been less affected by recent contamination. This interpretation of allelic diversity supports use of E. coli risk categories [26] and aligns with research findings that indicate a threshold effect, with significant increase in diarrhoeal disease burden only associated with high concentrations (>1000/100 mL) of E. coli [84].
Interpretations of E. coli results, however, should not rely exclusively on concentration: the health implications of differences in E. coli concentrations are context dependent and naturalisation is not the only process that confuses the relationship between E. coli and health hazard. For example, the water for set 1 is sourced from an open reservoir system, which has animal and human activity in the catchment area and does not include treatment. Thus, the low concentration of E. coli in S1C1, and absence in S1C2, may be better interpreted as indicating poor survival of E. coli in the reservoir–likely due to a combination of predation, competition, UV radiation, and absence of surfaces for biofilm formation [9, 10]–rather than absence of faecal contamination. Furthermore, one of the isolates from S1C1 was from phylogroup D with numerous virulence genes, a likely indicator of recent human faecal contamination. Additionally, for sets 3 and 5 the water is chlorinated in the distribution system, which gives more assurance of safety, but some pathogens are more resistant to chlorine than E. coli [26]. Generally, concentrations of E. coli do not correlate with concentrations of pathogens: the transport and survival patterns of E. coli vary considerably from those of faecal pathogens, particularly viruses and protozoa which tend to be more robust [25]–so the likelihood that water has been contaminated with faecal matter must be prioritised over E. coli sampling results.
Summary and recommendations
Although definitive attribution is not possible, the strains that most likely originated from human and/or recent faeces were found in poorly protected PoC water (4 sites including an unfenced open reservoir, unfenced open dug well, and two unsealed concrete tanks) or PoU water (12 out of 30 PoU sites). These were the 34 isolates from phylogroups A, B2, and D, and the 16 from phylogroup B1 with >80 virulence genes or multiple AMR genes. The other B1 isolates with fewer virulence genes account for almost half of our sample (48%), likely because B1 strains are generally better adapted to the freshwater environment. Allelic diversity comparisons suggest that naturalised E. coli may be particularly relevant at PoC sites with lower E. coli concentrations (<50 / 100mL). And for PoU sites, analysis based on five theorised PoU E. coli population scenarios underscores the difficulty of interpreting health risk from grab samples.
Placing our findings in relation to the literature, we develop two main recommendations. Firstly, we emphasise the inadequacy of judging hazard based on single E. coli samples at either PoC or PoU. Tracking sanitary conditions and E. coli concentrations over time can inform a more reliable understanding of hazard. In addition to E. coli sampling, rapid, in situ measurements such as turbidity or tryptophan-like fluorescence may be useful for high frequency tracking of water quality variability; although they have their own limitations, these measures can indicate process changes in water systems including, for example, response to rainfall events [85, 86], which may help differentiate between naturalised E. coli and contamination events. And regardless of water quality measures, sanitary inspection is needed to confirm the current and prospective safety of a system. Studies have found weak or no correlations between sanitary inspection scores and microbial water quality as measured by faecal indicator bacteria (FIB) [87], but this does not diminish the importance of the inspections given what we know of FIB results having multiple possible explanations.
Secondly, we recommend that PoC and PoU E. coli samples should not be compared directly in terms of their health hazard implications. E. coli in PoC water (especially when concentration is high) should be prioritised for interventions with a focus on water safety management to provide safe water at the PoC. On the other hand, PoU samples are more difficult to interpret because uncertainty is introduced by variability in: PoC quality, persistence of strains, and post-collection hygiene. Positive E. coli samples at household level could indicate no additional health hazard but, conservatively, they should be interpreted as indicating a hazardous household environment, generally. Effective intervention at the household-level requires a multi-pathway approach that goes beyond water treatment and safe storage.
Limitations and further research
The growth of E. coli is influenced by physicochemical characteristics of water such as nutrient levels, salinity, and temperature, as well as microbiome characteristics such as competition and predation [9, 10]. But given the sample size of our study, we are not able to query the impact of these factors on the balance between growth and die-off of E. coli strains. Similarly, our study did not focus on temporal change in E. coli populations. Only one site was sampled twice: S4C2 had no overlap in MLSTs between the two samples taken two weeks apart. This suggests that continual contamination is driving the population dynamics at this site rather than persistent dominance of strains in biofilms or otherwise. The site is a poorly protected concrete tank with multiple openings situated in a market square. Furthermore, the water at the site is saline (median 4.1 mS/cm), and salinity is known to inhibit E. coli survival in water [88]. Thus, the conditions at this site seem to enable ongoing input of new E. coli whilst discouraging E. coli survival and growth, which could explain the lack of overlap in the time-separated samples. A larger study incorporating a temporal dimension would improve insight into E. coli population dynamics in water systems over time–including the impact of sanitary conditions and physiochemical and microbiome characteristics. Additionally, a larger study would enable better characterisation of strain diversity within samples if more isolates per sample were analysed (S3 Fig).
Another avenue for further work is prompted by the prevalence of phylogroup B1 isolates in our samples, given their association with animal faeces [8]. Multiple studies have now emphasised the importance of animal management as a key sanitary factor influencing drinking water safety [89–91]; however, the importance of zoonotic transmission is not well established in the water, sanitation and hygiene (WASH) literature and recent models relating health outcomes to WASH factors have excluded zoonotic pathways in part due “to data scarcity on animal faeces and animal presence” [92 p279]. Further work in this space would be valuable.
Finally, the limitations of genomic characterisation for informing on strain origin is a key constraint of this study–we are able to comment on the likelihood of isolates being naturalised or recently sourced from faeces but cannot definitively identify them as such. To-date there have been few studies focussing on genomic characterisation of E. coli from drinking water supplies, but as the collective dataset grows it will enable metanalyses and more robust statistics that will improve our ability to distinguish naturalised strains and better understand the origins, diversity, and dynamics of E. coli populations in water supplies.
Supporting information
The boxes show median values and span lower to upper quartiles, the whiskers show the lowest and highest datums within 1.5 times the interquartile range.
(TIFF)
The boxes show median values and span lower to upper quartiles, the whiskers show the lowest and highest datums within 1.5 times the interquartile range.
(TIFF)
(TIFF)
(TIFF)
Includes points of collection (PoC) and points of use (PoU) with median E. coli, pH and conductivity (EC) from monitoring programme results.
(XLSX)
Includes Sequencing, Assembly, Phylogroup, MLST, Virulome, and Resistome Results.
(XLSX)
(XLSX)
Acknowledgments
The authors thank Cliff Nyaga, Musenya Sammy, Mbogo Mwaniki and the rest of the Rural Focus Limited and REACH Kenya teams for their support in making the fieldwork possible.
Data Availability
The raw sequence read files have been deposited in the European Nucleotide Archive (ENA) as study accession PRJEB40218 (http://www.ebi.ac.uk/ena/data/view/PRJEB40218).
Funding Statement
S.N. and K.C. are supported by the REACH programme funded by UK Aid from the UK Foreign, Commonwealth and Development Office (FCDO | https://www.gov.uk/government/organisations/foreign-commonwealth-development-office) for the benefit of developing countries (Programme Code 201880). S.N. is further supported by the Commonwealth Scholarship Commission in the UK (https://www.gov.uk/government/organisations/commonwealth-scholarship-commission-in-the-uk). G.G. is funded by the National Institute for Health Research (project reference 17/63/82) funded by UK Aid (FCDO) to support global health research. However, the views expressed and information contained in this article are not necessarily those of or endorsed by the FCDO, which can accept no responsibility for such views or information, or for any reliance placed on them. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Chaudhuri RR, Henderson IR. The evolution of the Escherichia coli phylogeny. Infect Genet Evol. 2012;12(2):214–26. 10.1016/j.meegid.2012.01.005 [DOI] [PubMed] [Google Scholar]
- 2.Leclerc H, Mossel D a, Edberg SC, Struijk CB. Advances in the bacteriology of the coliform group: their suitability as markers of microbial water safety. Annu Rev Microbiol. 2001;55:201–34. 10.1146/annurev.micro.55.1.201 [DOI] [PubMed] [Google Scholar]
- 3.Alm EW, Walk ST, Gordon DM. The Niche of Escherichia coli. In: Walk ST, Feng PCH, Whittam TS, editors. Population Genetics of Bacteria: a Tribute to Thomas S Whittam. Washington, D.C.: ASM Press; 2011. p. 69–89. [Google Scholar]
- 4.Savageau MA. Escherichia coli Habitats, Cell Types, and Molecular Mechanisms of Gene Control. Am Nat. 1983;122(6):732–44. [Google Scholar]
- 5.Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, Bidet P, et al. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 2009;5(1). 10.1371/journal.pgen.1000344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rasko DA, Rosovitz MJ, Myers GSA, Mongodin EF, Fricke WF, Gajer P, et al. The pangenome structure of Escherichia coli: Comparative genomic analysis of E. coli commensal and pathogenic isolates. J Bacteriol. 2008;190(20):6881–93. 10.1128/JB.00619-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergholz PW, Noar JD, Buckley DH. Environmental patterns are imposed on the population structure of Escherichia coli after fecal deposition. Appl Environ Microbiol. 2011;77(1):211–9. 10.1128/AEM.01880-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tenaillon O, Skurnik D, Picard B, Denamur E. The population genetics of commensal Escherichia coli. Nat Rev Microbiol. 2010;8(3):207–17. 10.1038/nrmicro2298 [DOI] [PubMed] [Google Scholar]
- 9.Berthe T, Ratajczak M, Clermont O, Denamur E, Petit F. Evidence for coexistence of distinct Escherichia coli populations in various aquatic environments and their survival in estuary water. Appl Environ Microbiol. 2013;79(15):4684–93. 10.1128/AEM.00698-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ishii S, Sadowsky MJ. Escherichia coli in the environment: Implications for water quality and human health. Microbes Environ. 2008;23(2):101–8. 10.1264/jsme2.23.101 [DOI] [PubMed] [Google Scholar]
- 11.Luby SP, Halder AK, Huda TM, Unicomb L, Islam MS, Arnold BF, et al. Microbiological contamination of drinking water associated with subsequent child diarrhea. Am J Trop Med Hyg. 2015;93(5):904–11. 10.4269/ajtmh.15-0274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferguson AS, Mailloux BJ, Ahmed KM, Van Geen A, McKay LD, Culligan PJ. Hand-pumps as reservoirs for microbial contamination of well water. J Water Health. 2011;9(4):708–17. 10.2166/wh.2011.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Devane ML, Moriarty E, Weaver L, Cookson A, Gilpin B. Fecal indicator bacteria from environmental sources; strategies for identification to improve water quality monitoring. Water Res. 2020;185 10.1016/j.watres.2020.116204 [DOI] [PubMed] [Google Scholar]
- 14.Power ML, Littlefield-Wyer J, Gordon DM, Veal DA, Slade MB. Phenotypic and genotypic characterization of encapsulated Escherichia coli isolated from blooms in two Australian lakes. Environ Microbiol. 2005;7(5):631–40. 10.1111/j.1462-2920.2005.00729.x [DOI] [PubMed] [Google Scholar]
- 15.Byappanahalli MN, Whitman RL, Shively DA, Sadowsky MJ, Ishii S. Population structure, persistence, and seasonality of autochthonous Escherichia coli in temperate, coastal forest soil from a Great Lakes watershed. Environ Microbiol. 2006;8(3):504–13. 10.1111/j.1462-2920.2005.00916.x [DOI] [PubMed] [Google Scholar]
- 16.Texier S, Prigent-Combaret C, Gourdon MH, Poirier MA, Faivre P, Dorioz JM, et al. Persistence of Culturable Escherichia coli Fecal Contaminants in Dairy Alpine Grassland Soils. J Environ Qual. 2008;37(6):2299–310. 10.2134/jeq2008.0028 [DOI] [PubMed] [Google Scholar]
- 17.Gordon DM, Bauer S, Johnson JR. The genetic structure of Escherichia coli populations in primary and secondary habitats. Microbiology. 2002;148(5):1513–22. 10.1099/00221287-148-5-1513 [DOI] [PubMed] [Google Scholar]
- 18.Nandakafle G, Huegen T, Potgieter SC, Steenkamp E, Venter SN, Brözel VS. Niche preference of Escherichia coli in a peri-urban pond ecosystem. bioRxiv [Preprint] 2020. [cited 2020 December 15]. Available from: https://www.biorxiv.org/content/10.1101/2020.01.30.926667v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whittam TS. Clonal dynamics of Escherichia coli in its natural habitat. Antonie Van Leeuwenhoek. 1989;55(1):23–32. 10.1007/BF02309616 [DOI] [PubMed] [Google Scholar]
- 20.Zhi S, Banting G, Stothard P, Ashbolt NJ, Checkley S, Meyer K, et al. Evidence for the evolution, clonal expansion and global dissemination of water treatment-resistant naturalized strains of Escherichia coli in wastewater. Water Res. 2019;156:208–22. 10.1016/j.watres.2019.03.024 [DOI] [PubMed] [Google Scholar]
- 21.Touchon M, Perrin A, de Sousa JAM, Vangchhia B, Burn S, O’Brien CL, et al. Phylogenetic background and habitat drive the genetic diversification of Escherichia coli. PLoS Genet. 2020;16(6). 10.1371/journal.pgen.1008866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou Z, Alikhan NF, Mohamed K, Fan Y, Achtman M. The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res [Internet]. 2020;30(1):138–52. Available from: http://enterobase.warwick.ac.uk/ 10.1101/gr.251678.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blyton MDJ, Gordon DM. Genetic attributes of E. coli isolates from chlorinated drinking water. PLoS One. 2017;12(1):1–14. 10.1371/journal.pone.0169445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jang J, Hur HG, Sadowsky MJ, Byappanahalli MN, Yan T, Ishii S. Environmental Escherichia coli: ecology and public health implications—a review. J Appl Microbiol. 2017;123(3):570–81. 10.1111/jam.13468 [DOI] [PubMed] [Google Scholar]
- 25.Charles KJ, Nowicki S, Bartram JK. A framework for monitoring the safety of water services: from measurements to security. npj Clean Water. 2020;3(36):1–6. [Google Scholar]
- 26.WHO. Guidelines for drinking-water quality: fourth edition incorporating the first addendum [Internet]. 4th ed. Geneva, Switzerland: WHO Press; 2017. Available from: http://apps.who.int/iris/bitstream/handle/10665/254637/9789241549950-eng.pdf;jsessionid=2E1B7E42B16F03AB88E253B119976D63?sequence=1 [PubMed]
- 27.JMP. Water quality monitoring [Internet]. UNICEF WHO Joint Monitoring Programme; 2020. Available from: https://washdata.org/monitoring/drinking-water/water-quality-monitoring
- 28.Sobsey MD. Managing Water in the Home: Accelerated Health Gains from Improved Water Supply [Internet]. World Health Organization; 2002. Available from: http://www.bvsde.paho.org/bvsacd/who/sobs.pdf [Google Scholar]
- 29.Wright J, Gundry S, Conroy R. Household drinking water in developing countries: A systematic review of microbiological contamination between source and point-of-use. Trop Med Int Heal. 2004;9(1):106–17. 10.1046/j.1365-3156.2003.01160.x [DOI] [PubMed] [Google Scholar]
- 30.Shields KF, Bain RES, Cronk R, Wright JA, Bartram J. Association of supply type with fecal contamination of source water and household stored drinking water in developing countries: A bivariate meta-analysis. Environ Health Perspect. 2015;123(12):1222–31. 10.1289/ehp.1409002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trevett AF, Carter RC, Tyrrel SF. Water quality deterioration: A study of household drinking water quality in rural Honduras. Int J Environ Health Res. 2004;14(4):273–83. 10.1080/09603120410001725612 [DOI] [PubMed] [Google Scholar]
- 32.Trevett AF, Carter RC, Tyrrel SF. The importance of domestic water quality management in the context of faecal-oral disease transmission. J Water Health. 2005;3(3):259–70. 10.2166/wh.2005.037 [DOI] [PubMed] [Google Scholar]
- 33.Burke-Gaffney H. The Classification of The Colon-Aerogenes Group of Bacteria in Relation to their Habitat and its Application to the Sanitary Examination of Water Supplies in the Tropics and in Temperate Climates: A Comparative Study of 2500 Cultures. J Hyg (Lond). 1932;32(1):85–131. 10.1017/s002217240001785x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ingle DJ, Clermont O, Skurnik D, Denamur E, Walk ST, Gordon DM. Biofilm Formation by and Thermal Niche and Virulence Characteristics of Escherichia spp. Appl Environ Microbiol. 2011;77(8):2695–700. 10.1128/AEM.02401-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matthews RL, Tung R. Broader incubation temperature tolerances for microbial drinking water testing with enzyme substrate tests. J Water Health. 2014;12(1):113–21. 10.2166/wh.2013.076 [DOI] [PubMed] [Google Scholar]
- 36.Seemann T, Goncalves da Silva A, Bulach D, Schultz M, Kwong J, Howden B. Nullarbor [Internet]. Github. [cited 2020 Sep 15]. Available from: https://github.com/tseemann/nullarbor
- 37.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–20. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wood DE, Salzberg SL. Kraken: Ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15(3). 10.1186/gb-2014-15-3-r46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–77. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beghain J, Bridier-Nahmias A, Le Nagard H, Denamur E, Clermont O. ClermonTyping: An easy-to-use and accurate in silico method for Escherichia genus strain phylotyping. Microb Genomics. 2018;4(7):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clermont O, Dixit OVA, Vangchhia B, Condamine B, Dion S, Bridier-Nahmias A, et al. Characterization and rapid identification of phylogroup G in Escherichia coli, a lineage with high virulence and antibiotic resistance potential. Environ Microbiol. 2019;21(8):3107–17. 10.1111/1462-2920.14713 [DOI] [PubMed] [Google Scholar]
- 42.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva E V., Zdobnov EM. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31(19):3210–2. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- 43.Waterhouse RM, Tegenfeldt F, Li J, Zdobnov EM, Kriventseva E V. OrthoDB: A hierarchical catalog of animal, fungal and bacterial orthologs. Nucleic Acids Res. 2013;41(D1):358–65. 10.1093/nar/gks1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, et al. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31(22):3691–3. 10.1093/bioinformatics/btv421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Price MN, Dehal PS, Arkin AP. FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5(3). 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou Z, Alikhan NF, Sergeant MJ, Luhmann N, Vaz C, Francisco AP, et al. Grapetree: Visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res. 2018;28(9):1395–404. 10.1101/gr.232397.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seemann T. Abricate [Internet]. Github. [cited 2020 Sep 15]. Available from: https://github.com/tseemann/abricate
- 48.Chen L, Zheng D, Liu B, Yang J, Jin Q. VFDB 2016: Hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res. 2016;44(D1):D694–7. 10.1093/nar/gkv1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feldgarden M, Brover V, Haft DH, Prasad AB, Slotta DJ, Tolstoy I, et al. Validating the AMRFinder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob Agents Chemother. 2019;63(11):1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, et al. Sex and virulence in Escherichia coli: An evolutionary perspective. Mol Microbiol. 2006;60(5):1136–51. 10.1111/j.1365-2958.2006.05172.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sahl JW, Matalka MN, Rasko DA. Phylomark, a tool to identify conserved phylogenetic markers from whole-genome alignments. Appl Environ Microbiol. 2012;78(14):4884–92. 10.1128/AEM.00929-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Larsen M V., Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, et al. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol. 2012;50(4):1355–61. 10.1128/JCM.06094-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nei M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics. 1978;89(3):583–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Strasser H, Weber C. On the Asymptotic Theory of Permutation Statistics. Math Methods Stat. 1999;8(2):220–50. [Google Scholar]
- 55.Hothorn T, Hornik K, van de Wiel MA, Zeileis A. Implementing a class of permutation tests: The coin package. J Stat Softw [Internet]. 2008;28(8). Available from: https://www.jstatsoft.org/article/view/v028i08 [Google Scholar]
- 56.Genz A, Bretz F. Computation of Multivariate Normal and t Probabilities. Heidelberg, Germany: Springer-Verlag; 2009. [Google Scholar]
- 57.Smati M, Clermont O, Bleibtreu A, Fourreau F, David A, Daubié AS, et al. Quantitative analysis of commensal Escherichia coli populations reveals host-specific enterotypes at the intra-species level. Microbiologyopen. 2015;4(4):604–15. 10.1002/mbo3.266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Julian TR, Islam MA, Pickering AJ, Roy S, Fuhrmeister ER, Ercumen A, et al. Genotypic and phenotypic characterization of Escherichia coli isolates from feces, hands, and soils in rural Bangladesh via the Colilert Quanti-Tray System. Appl Environ Microbiol. 2015;81(5):1735–43. 10.1128/AEM.03214-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Donnenberg MS. Escherichia coli: Pathotypes and Principles of Pathogenesis. 2nd ed Donnenberg MS, editor. Academic Press; 2013. [Google Scholar]
- 60.Walk ST, Alm EW, Calhoun LM, Mladonicky JM, Whittam TS. Genetic diversity and population structure of Escherichia coli isolated from freshwater beaches. Environ Microbiol. 2007;9(9):2274–88. 10.1111/j.1462-2920.2007.01341.x [DOI] [PubMed] [Google Scholar]
- 61.Ratajczak M, Laroche E, Berthe T, Clermont O, Pawlak B, Denamur E, et al. Influence of hydrological conditions on the Escherichia coli population structure in the water of a creek on a rural watershed. BMC Microbiol. 2010;10 10.1186/1471-2180-10-222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Orsi RH, Stoppe NC, Sato MIZ, Ottoboni LMM. Identification of Escherichia coli from groups A, B1, B2 and D in drinking water in Brazil. J Water Health. 2007;5(2):323–7. 10.2166/wh.2007.028 [DOI] [PubMed] [Google Scholar]
- 63.White AP, Sibley KA, Sibley CD, Wasmuth JD, Schaefer R, Surette MG, et al. Intergenic sequence comparison of Escherichia coli isolates reveals lifestyle adaptations but not host specificity. Appl Environ Microbiol. 2011;77(21):7620–32. 10.1128/AEM.05909-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Méric G, Kemsley EK, Falush D, Saggers EJ, Lucchini S. Phylogenetic distribution of traits associated with plant colonization in Escherichia coli. Environ Microbiol. 2013;15(2):487–501. 10.1111/j.1462-2920.2012.02852.x [DOI] [PubMed] [Google Scholar]
- 65.Clermont O, Christenson JK, Denamur E, Gordon DM. The Clermont Escherichia coli phylo-typing method revisited: Improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep. 2013;5(1):58–65. 10.1111/1758-2229.12019 [DOI] [PubMed] [Google Scholar]
- 66.Kaper JB, Nataro JP, Mobley HLT. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2(2):123–40. 10.1038/nrmicro818 [DOI] [PubMed] [Google Scholar]
- 67.Robins-Browne RM, Bordun AM, Tauschek M, Bennett-Wood VR, Russell J, Oppedisano F, et al. Escherichia coli and community-acquired gastroenteritis, Melbourne, Australia. Emerg Infect Dis. 2004;10(10):1797–805. 10.3201/eid1010.031086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ferrie JE. Arsenic, antibiotics and interventions. Int J Epidemiol. 2014;43(4):977–82. 10.1093/ije/dyu152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fekih I Ben, Zhang C, Li YP, Zhao Y, Alwathnani HA, Saquib Q, et al. Distribution of arsenic resistance genes in prokaryotes. Front Microbiol. 2018;9(OCT):1–11. 10.3389/fmicb.2018.02473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010;74(3):417–33. 10.1128/MMBR.00016-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Haberecht HB, Nealon NJ, Gilliland JR, Holder A V., Runyan C, Oppel RC, et al. Antimicrobial-Resistant Escherichia coli from Environmental Waters in Northern Colorado. J Environ Public Health. 2019. 10.1155/2019/3862949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Iramiot JS, Kajumbula H, Bazira J, De Villiers EP, Asiimwe BB. Whole genome sequences of multi-drug resistant Escherichia coli isolated in a Pastoralist Community of Western Uganda: Phylogenomic changes, virulence and resistant genes. PLoS One. 2020;15(5). 10.1371/journal.pone.0231852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mainda G, Lupolova N, Sikakwa L, Richardson E, Bessell PR, Malama SK, et al. Whole genome sequence analysis reveals lower diversity and frequency of acquired antimicrobial resistance (AMR) genes in E. coli from dairy herds compared with human isolates from the same region of central Zambia. Front Microbiol. 2019;10(MAY). 10.3389/fmicb.2019.01114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32(18):2847–9. 10.1093/bioinformatics/btw313 [DOI] [PubMed] [Google Scholar]
- 75.Momba MNB, Kaleni P. Regrowth and survival of indicator microorganisms on the surfaces of household containers used for the storage of drinking water in rural communities of South Africa. Water Res. 2002;36(12):3023–8. 10.1016/s0043-1354(02)00011-8 [DOI] [PubMed] [Google Scholar]
- 76.Levy K, Nelson KL, Hubbard A, Eisenberg JNS. Following the water: A controlled study of drinking water storage in Northern Coastal Ecuador. Environ Health Perspect. 2008;116(11):1533–40. 10.1289/ehp.11296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pickering AJ, Julian TR, Marks SJ, Mattioli MC, Boehm AB, Schwab KJ, et al. Fecal contamination and diarrheal pathogens on surfaces and in soils among Tanzanian households with and without improved sanitation. Environ Sci Technol. 2012;46(11):5736–43. 10.1021/es300022c [DOI] [PubMed] [Google Scholar]
- 78.Montealegre MC, Talavera Rodríguez A, Roy S, Hossain MI, Islam MA, Lanza VF, et al. High Genomic Diversity and Heterogenous Origins of Pathogenic and Antibiotic-Resistant Escherichia coli in Household Settings Represent a Challenge to Reducing Transmission in Low-Income Settings. mSphere. 2020;5(1). 10.1128/mSphere.00704-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Arnold BF, Null C, Luby SP, Colford JM. Implications of WASH Benefits trials for water and sanitation–Authors’ reply. Lancet Glob Heal [Internet]. 2018;6(6):e616–7. Available from: 10.1016/S2214-109X(18)30229-8 [DOI] [PubMed] [Google Scholar]
- 80.Feachem RG, Burns E, Cairncross S, Cronin A, Cross P, Curtis D, et al. Water, health and development: an interdisciplinary evaluation. London, UK: Tri Med Books Ltd.; 1978. [Google Scholar]
- 81.Robb K, Null C, Teunis P, Yakubu H, Armah G, Moe CL. Assessment of fecal exposure pathways in low-income urban neighborhoods in Accra, Ghana: rationale, design, methods, and key findings of the Sanipath study. Am J Trop Med Hyg. 2017;97(4):1020–32. 10.4269/ajtmh.16-0508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McLellan SL. Genetic diversity of Escherichia coli isolated from urban rivers and beach water. Appl Environ Microbiol. 2004;70(8):4658–65. 10.1128/AEM.70.8.4658-4665.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Perchec-Merien AM, Lewis GD. Naturalized Escherichia coli from New Zealand wetland and stream environments. FEMS Microbiol Ecol. 2013;83(2):494–503. 10.1111/1574-6941.12010 [DOI] [PubMed] [Google Scholar]
- 84.Moe CL, Sobsey MD, Samsa GP, Mesolo V. Bacterial indicators of risk of diarrhoeal disease from drinking-water in the Philippines. Bull World Health Organ. 1991;69(3):305–17. [PMC free article] [PubMed] [Google Scholar]
- 85.Nowicki S, Lapworth DJ, Ward JST, Thomson P, Charles K. Tryptophan-like fluorescence as a measure of microbial contamination risk in groundwater. Sci Total Environ. 2019;646:782–91. 10.1016/j.scitotenv.2018.07.274 [DOI] [PubMed] [Google Scholar]
- 86.Carstea EM, Popa CL, Baker A, Bridgeman J. In situ fluorescence measurements of dissolved organic matter: A review. Sci Total Environ. 2020;699:134361 10.1016/j.scitotenv.2019.134361 [DOI] [PubMed] [Google Scholar]
- 87.Kelly ER, Cronk R, Kumpel E, Howard G, Bartram J. How we assess water safety: A critical review of sanitary inspection and water quality analysis. Sci Total Environ. 2020;718:137237 10.1016/j.scitotenv.2020.137237 [DOI] [PubMed] [Google Scholar]
- 88.Rozen Y, Belkin S. Survival of enteric bacteria in seawater: FEMS Microbiol Rev. 2001;25:513–29. 10.1111/j.1574-6976.2001.tb00589.x [DOI] [PubMed] [Google Scholar]
- 89.Daniel D, Diener A, Van De Vossenberg J, Bhatta M, Marks SJ. Assessing drinking water quality at the point of collection and within household storage containers in the hilly rural areas of mid and far-western Nepal. Int J Environ Res Public Health. 2020;17(7). 10.3390/ijerph17072172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dufour A, Bartram J, Bos R, Gannon V. Animal Waste, Water Quality and Human Health. London, UK: World Health Organization; 2012. [Google Scholar]
- 91.Hamzah L, Boehm AB, Davis J, Pickering AJ, Wolfe M, Mureithi M, et al. Ruminant Fecal Contamination of Drinking Water Introduced Post-Collection in Rural Kenyan Households. Int J Environ Res Public Health. 2020;17(2):1–23. 10.3390/ijerph17020608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wolf J, Johnston R, Hunter PR, Gordon B, Medlicott K, Prüss-Ustün A. A Faecal Contamination Index for interpreting heterogeneous diarrhoea impacts of water, sanitation and hygiene interventions and overall, regional and country estimates of community sanitation coverage with a focus on low- and middle-income countries. Int J Hyg Environ Health. 2019;222(2):270–82. 10.1016/j.ijheh.2018.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]