This systematic review summarizes evidence comparing the sensitivities for detection of SARS-CoV-2 infection between nasopharyngeal swabs and saliva samples and estimates the incremental cost per additional SARS-CoV-2 infection detected with nasopharyngeal swabs.
Abstract
Background:
Nasopharyngeal swabs are the primary sampling method used for detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), but they require a trained health care professional and extensive personal protective equipment.
Purpose:
To determine the difference in sensitivity for SARS-CoV-2 detection between nasopharyngeal swabs and saliva and estimate the incremental cost per additional SARS-CoV-2 infection detected with nasopharyngeal swabs.
Data Sources:
Embase, Medline, medRxiv, and bioRxiv were searched from 1 January to 1 November 2020. Cost inputs were from nationally representative sources in Canada and were converted to 2020 U.S. dollars.
Study Selection:
Studies including at least 5 paired nasopharyngeal swab and saliva samples and reporting diagnostic accuracy for SARS-CoV-2 detection.
Data Extraction:
Data were independently extracted using standardized forms, and study quality was assessed using QUADAS-2 (Quality Assessment of Diagnostic Accuracy Studies 2).
Data Synthesis:
Thirty-seven studies with 7332 paired samples were included. Against a reference standard of a positive result on either sample, the sensitivity of saliva was 3.4 percentage points lower (95% CI, 9.9 percentage points lower to 3.1 percentage points higher) than that of nasopharyngeal swabs. Among persons with previously confirmed SARS-CoV-2 infection, saliva's sensitivity was 1.5 percentage points higher (CI, 7.3 percentage points lower to 10.3 percentage points higher) than that of nasopharyngeal swabs. Among persons without a previous SARS-CoV-2 diagnosis, saliva was 7.9 percentage points less (CI, 14.7 percentage points less to 0.8 percentage point more) sensitive. In this subgroup, if testing 100 000 persons with a SARS-CoV-2 prevalence of 1%, nasopharyngeal swabs would detect 79 more (95% uncertainty interval, 5 fewer to 166 more) persons with SARS-CoV-2 than saliva, but with an incremental cost per additional infection detected of $8093.
Limitation:
The reference standard was imperfect, and saliva collection procedures varied.
Conclusion:
Saliva sampling seems to be a similarly sensitive and less costly alternative that could replace nasopharyngeal swabs for collection of clinical samples for SARS-CoV-2 testing.
Primary Funding Source:
McGill Interdisciplinary Initiative in Infection and Immunity. (PROSPERO: CRD42020203415)
As of 16 December 2020, more than 74 million cases of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) have been diagnosed and more than 1.6 million persons have died of it (1). One of the most important components of public health strategies to contain SARS-CoV-2 is maintaining a high level of testing. Testing is prioritized for persons with symptoms of coronavirus disease 2019 (COVID-19) and contacts of those with confirmed SARS-CoV-2 infection (2), but it is often offered to persons at increased risk for exposure (such as health care workers). However, as economies and schools reopen, the pool of persons who may be at increased risk for SARS-CoV-2 exposure will grow (3), placing strain on testing systems.
Reverse transcriptase polymerase chain reaction (RT-PCR) on nasopharyngeal swabs is the reference method to detect SARS-CoV-2 (4). Yet, nasopharyngeal swabs present several barriers to reaching the level of testing required to meet demand and optimally control SARS-CoV-2. Their collection requires a trained health care professional (for example, a nurse), who must be in extensive personal protective equipment (5). Further, although more prominent early in the COVID-19 pandemic, supply chain issues (6) for nasopharyngeal swabs—and the transport media used during their transportation—still exist (7).
Saliva-based sampling for SARS-CoV-2 detection via RT-PCR has the potential to address many of the barriers associated with nasopharyngeal swab sampling (8). Saliva samples can be collected by the persons being tested themselves, with instruction from lower-cadre health care professionals or other personnel. This reduces exposure to health care workers and the need for personal protective equipment during collection. Saliva can be collected in sterile containers, removing the need for swabs. These practical advantages reduce human resource needs and could expand the number of persons who can be tested. However, the comparative sensitivity of saliva and nasopharyngeal swabs for SARS-CoV-2 detection is uncertain, which has impeded saliva's adoption.
We conducted a systematic review and meta-analysis to estimate the comparative sensitivity of saliva versus nasopharyngeal swabs for detection of SARS-CoV-2 and an economic evaluation to estimate the incremental cost per additional SARS-CoV-2 infection detected with nasopharyngeal swabs.
Methods
Systematic Review and Meta-analysis
This systematic review follows PRISMA (Preferred Reporting Items for Systematic reviews and Meta-Analyses) (9) and MOOSE (Meta-analysis of Observational Studies in Epidemiology) (10) guidelines, and its protocol is registered on PROSPERO (CRD42020203415).
Data Sources and Searches
We searched Medline and Embase from 1 January to 1 November 2020. We used a comprehensive search strategy (Table 1 of Supplement 1) with a combination of medical subject headings and free text containing concepts related to SARS-CoV-2, molecular diagnosis (such as RT-PCR), and respiratory specimens (such as nasopharyngeal swabs and saliva). No language restrictions were used. We additionally searched medRxiv and bioRxiv until 1 November 2020 for preprint literature; we used analytic code to screen for preprint manuscripts containing the words “COVID-19” or “SARS-CoV-2” and “saliva” in titles and abstracts before reviewer screening.
Study Selection
Eligible studies were randomized clinical trials, case series, cohort studies, case–control studies, and cross-sectional studies that reported accuracy of saliva-based sampling compared with nasopharyngeal swabs for SARS-CoV-2, reported at least 5 paired samples (that is, nasopharyngeal and saliva samples collected at the same time), and used the same method for detecting SARS-CoV-2 in nasopharyngeal and saliva samples. We excluded studies that did not assess diagnostic accuracy, as well as reviews, commentaries, editorials, case reports, mathematical modeling studies, economic analyses, and conference abstracts.
Three reviewers (M.L.B., S.P., and J.R.C.) independently screened all titles, abstracts, and full texts. At the full-text stage, reference lists were reviewed for relevant additional studies. Discordance on which studies to include was resolved by consensus.
Data Extraction and Quality Assessment
Two reviewers (M.L.B. and S.P.) independently extracted 25% of the data using a standardized form (fields are shown in Table 2 of Supplement 1); findings were checked for agreement. Concordance was high; thus, a single reviewer extracted the remaining data, and the other reviewer verified extractions. Extracted information included study design, location, enrollment dates, included population (persons presenting for SARS-CoV-2 testing or persons with confirmed SARS-CoV-2 infection), study setting (inpatient or outpatient), presence of symptoms when sampling was done, demographic information (age and sex), laboratory methods (analytic method used, primer, transport media, and cycle threshold values), and sampling method for saliva collection. We extracted the number of persons testing positive via nasopharyngeal swabs, saliva sampling, or on either sample. To complete missing data, we contacted 25 authors, of whom 18 (72%) replied.
Risk of bias and applicability concerns among included studies were assessed using an adapted QUADAS-2 (Quality Assessment of Diagnostic Accuracy Studies 2) (11) tool. The tool assessed the following domains: patient selection, performance of the index test, performance of the reference test, and flow and timing (Table 3 of Supplement 1). Two reviewers (M.L.B. and S.P.) independently assessed studies, and disagreement was resolved through consensus.
Data Synthesis and Analysis
The primary outcome of interest was the difference between saliva samples and nasopharyngeal swabs in sensitivity for SARS-CoV-2 detection; a positive result with either sample was considered the reference standard. The secondary outcome of interest was the sensitivity of saliva for SARS-CoV-2 detection, with a positive result with either saliva or nasopharyngeal swabs as the reference standard. Because we assumed that any positive result was a true positive, we could not estimate specificity.
In our primary analysis, we estimated the pooled difference in sensitivity between saliva samples and nasopharyngeal swabs for all included studies. To assess possible sources of heterogeneity, we did numerous stratified analyses. The a priori–specified analyses were on population sampled (persons presenting for SARS-CoV-2 testing or those with confirmed SARS-CoV-2 infection), age (adult or pediatric) and symptoms present at sampling (asymptomatic or symptomatic). The post hoc analyses were on study setting (outpatient or inpatient), method of saliva collection (general spitting technique, early-morning posterior oropharyngeal spitting technique, drooling technique, posterior pharyngeal spitting technique, or saliva collection device), use of transport media with the saliva sample (yes or no), analytic platform (laboratory-based RT-PCR or other), and number of risk of bias domains at low risk of bias (≥4 or <4). We pooled results only when at least 3 studies were included in stratified analyses; otherwise, we report only individual study estimates.
Meta-analyses were done with package meta, version 4.14 (12), and package metafor, version 2.4-0 (13), in R (R Foundation) (see Supplement 2 for additional methods and code). We estimated the difference and SE in sensitivity between saliva and nasopharyngeal swabs for each study, accounting for the paired nature of sample collection using the Wilson method (14), and calculated 95% CIs. We pooled estimates with random-effects meta-analysis using the inverse variance method and Sidik–Jonkman estimator with a Knapp–Hartung adjustment for heterogeneity (15–17). For the secondary analysis estimating the sensitivity of saliva sampling, we used random-effects meta-analysis with generalized linear mixed models (18); individual study estimates were logit-transformed for pooling, and pooled estimates were back-transformed. For all analyses, heterogeneity was reported using the I 2 statistic (19).
Economic Evaluation
Data Inputs
We collected costs associated with both nasopharyngeal swabs and saliva sampling using a microcosting approach. To arrive at costs per person sampled, we considered costs associated with materials (swabs, transport media, containers, and personal protective equipment) and personnel to collect the samples. On the basis of previous experience in Canada (3), we estimated that a nurse would take 6 minutes to conduct sampling with a nasopharyngeal swab (including changing of gloves and gown), whereas saliva-based sampling would be task-shifted to a lower-cadre health care professional or administrative personnel and would take 4 minutes (including changing of gloves). Transportation and laboratory costs for sample analysis were assumed to be identical. Cost estimates for materials and nurse salary (20) were from nationally representative sources in Canada in 2020 Canadian dollars. To enhance generalizability, we estimated the salary difference between a nurse and lower-cadre health care professional using data from an econometric analysis (21) for high-income countries; we assumed that administrative personnel would have the same salary. We converted cost estimates to U.S. dollars using exchange rates for materials costs (22) and purchasing power parity for personnel costs (23). Cost inputs are reported in Table 4 of Supplement 1.
Data Analysis
All analyses were conducted in R (see Supplement 2 for additional methods and code). The population of interest for the economic evaluation was persons presenting for SARS-CoV-2 testing (that is, without confirmed SARS-CoV-2 infection). We selected this group a priori because they were the most probable target population where saliva testing would be implemented. Using sensitivity difference estimates from our meta-analysis, we estimated the additional number of infections detected with nasopharyngeal swabs versus saliva sampling per 100 000 persons tested at population prevalence levels of 0.01%, 0.1%, 1%, and 10%. We fitted our estimate for the difference in sensitivity between nasopharyngeal swabs and saliva to a normal distribution and cost parameters to γ distributions (Table 5 of Supplement 1). We sampled from these distributions 1000 times and estimated the difference and 95% uncertainty interval (UI) in cost and number of persons correctly diagnosed with SARS-CoV-2 infection with nasopharyngeal swabs or saliva. Using these outputs, we calculated the proportion of samples where saliva was dominant to nasopharyngeal sampling (that is, lower cost and better sensitivity). We report point estimates of the incremental cost-effectiveness ratio and visualize uncertainty using a cost-effectiveness plane.
To cover a range of possible scenarios, we did post hoc secondary analyses to evaluate implications of sensitivity differences. We evaluated several scenarios where nasopharyngeal swabs were more sensitive than saliva (1, 2, 5, 10, and 20 percentage points more) at different SARS-CoV-2 prevalence estimates (0.01%, 0.1%, 1%, and 10%). For each analysis, we calculated the incremental cost per additional person with SARS-CoV-2 identified with nasopharyngeal swabs versus saliva.
Role of the Funding Source
This study was funded by the McGill Interdisciplinary Initiative in Infection and Immunity. The funders had no role in the design or conduct of the study; collection, management, analysis, or interpretation of the data; preparation, review, or approval of the manuscript; or decision to submit the manuscript for publication.
Results
Systematic Review and Meta-analysis
Characteristics of Included Studies
We identified 22 795 records for screening. After title and abstract screening, 127 studies entered full-text assessment. Overall, 37 studies (24–55-56–60) were included (Appendix Figure), comprising 7169 participants with 7332 paired saliva samples and nasopharyngeal swabs. Summary characteristics of included studies are reported in Table 1 and individual study characteristics in Tables 6 and 7 of Supplement 1. Of the 37 studies, 6 (16%) were at high or unclear risk of bias or applicability in 4 or more domains, 25 (68%) were at high or unclear risk of selection bias, and 32 (87%) were at high or unclear risk of bias due to blinding during sample analysis (Figure 1 of Supplement 1).
Appendix Figure. Evidence search and selection.
Table 1. Summary Characteristics of Included Studies.
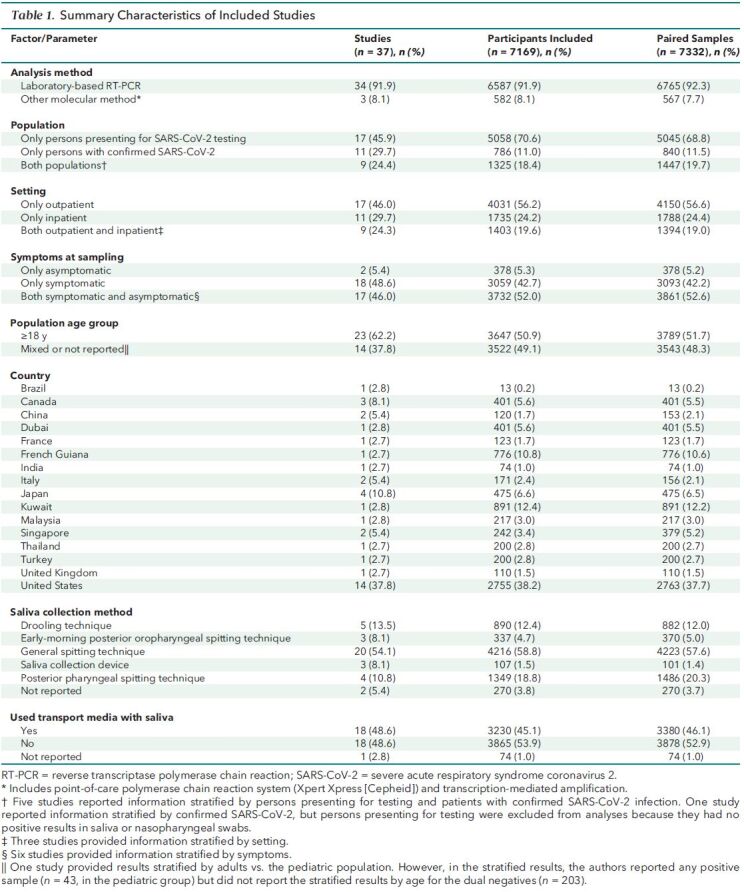
Of the 37 included studies, 34 (92%) used laboratory-based RT-PCR for SARS-CoV-2 detection and 3 (8%) used other methods (Table 8 of Supplement 1). Saliva was collected from participants using a general spitting technique in 20 studies (54%), drooling technique in 4 studies (11%), early-morning posterior oropharyngeal spitting technique in 4 studies (11%), posterior pharyngeal spitting technique in 4 studies (11%), and saliva collection device in 3 studies (8%) (Table 9 of Supplement 1 gives detailed collection descriptions). Transport media was used for saliva specimens in 18 studies (49%) (Table 10 of Supplement 1). Eighteen studies (49%) reported results for only symptomatic participants and 2 (5%) for only asymptomatic participants. The population of interest was only persons presenting for SARS-CoV-2 testing (that is, undiagnosed SARS-CoV-2 infection) in 17 studies (46%), whereas in 11 (30%) it was only persons with previously confirmed SARS-CoV-2 (by pharyngeal swab).
Primary Analysis
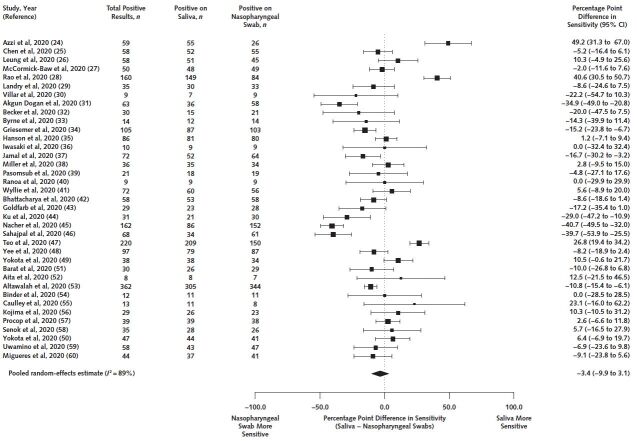
Among 7332 paired samples included, 2327 (32%) were positive on either nasopharyngeal swab or saliva. For our primary outcome, we estimated that saliva's sensitivity was 3.4 percentage points lower (95% CI, 9.9 percentage points lower to 3.1 percentage points higher) than that of nasopharyngeal swabs (Figure). Heterogeneity based on the I 2 statistic was 89%. For our secondary outcome, among the 2327 samples positive on either saliva or nasopharyngeal swabs, 1927 were positive with saliva, for a pooled sensitivity of 86.9% (CI, 82.3% to 90.4%).
Figure. Forest plot of all included studies in the primary analysis estimating the difference in sensitivity between saliva and nasopharyngeal swabs.
Stratified Analysis
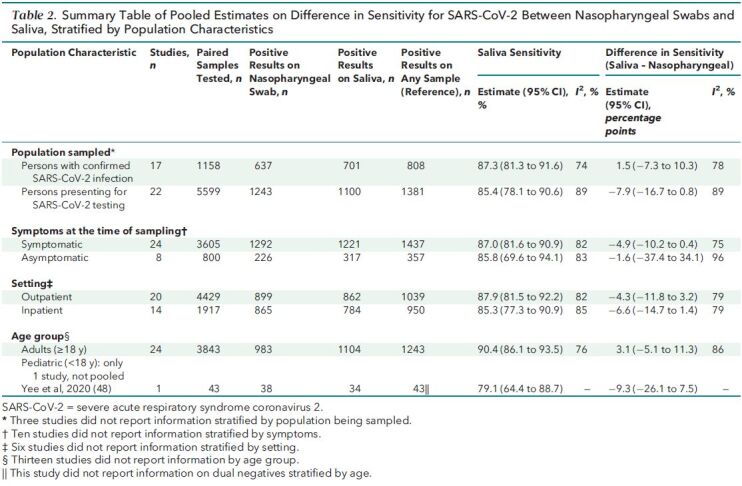
Table 2 shows results stratified by population characteristics, in which we observed no significant differences. Among 22 studies with data on persons presenting for SARS-CoV-2 testing consisting of 5599 paired samples, saliva was 7.9 percentage points less (CI, 16.7 percentage points less to 0.8 percentage point more) sensitive than nasopharyngeal swabs. Conversely, among 17 studies with data on persons with previously confirmed SARS-CoV-2 infection, saliva's sensitivity was 1.5 percentage points higher (CI, 7.3 percentage points lower to 10.3 percentage points higher) among 1158 paired samples (Table 11 of Supplement 1). Saliva was 4.9 percentage points less (CI, 10.2 percentage points less to 0.4 percentage point more) sensitive than nasopharyngeal swabs among symptomatic persons and 1.6 percentage points less (CI, 37.4 percentage points less to 34.1 percentage points more) sensitive than nasopharyngeal swabs among asymptomatic persons (Table 12 of Supplement 1). Differences in sensitivity did not differ between inpatients and outpatients (Table 13 of Supplement 1). In the only study with stratified data on pediatric participants, saliva was 9.3 percentage points less (CI, 26.1 percentage points less to 7.5 percentage points more) sensitive than nasopharyngeal swabs among 43 samples positive on either specimen (Table 14 of Supplement 1). Heterogeneity remained high (I 2 > = 75%) in stratified analyses.
Table 2. Summary Table of Pooled Estimates on Difference in Sensitivity for SARS-CoV-2 Between Nasopharyngeal Swabs and Saliva, Stratified by Population Characteristics.
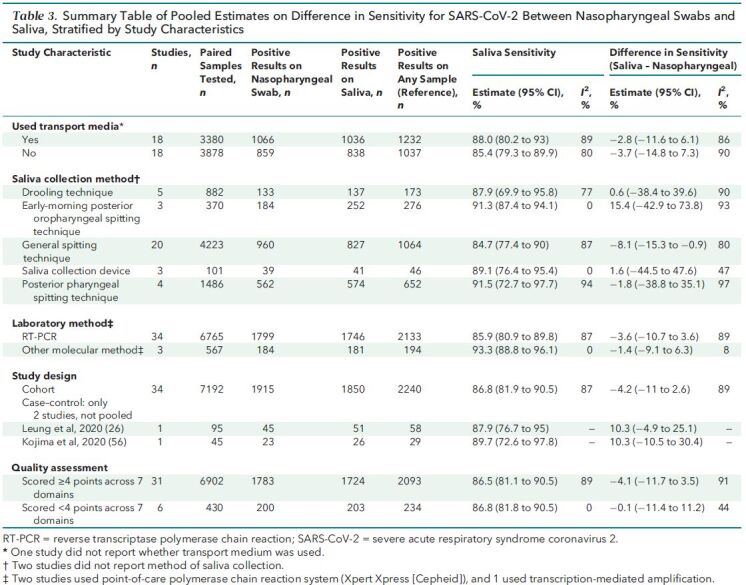
Results stratified by study-level characteristics are reported in Table 3; we found no significant differences in sensitivity except when considering the saliva collection method (Table 15 of Supplement 1). The sensitivity of saliva versus nasopharyngeal swabs was 15.4 percentage points higher (CI, 42.9 percentage points lower to 73.8 percentage points higher) with early-morning posterior oropharyngeal spitting technique (n = 370 paired samples), 1.6 percentage points higher (CI, 44.5 percentage points lower to 47.6 percentage points higher) with a saliva collection device (n = 101 paired samples), 0.6 percentage point higher (CI, 38.4 percentage points lower to 39.6 percentage points higher) with drooling technique (n = 882 paired samples), 1.8 percentage points lower (CI, 38.8 percentage points lower to 35.1 percentage points higher) with posterior pharyngeal spitting technique (n = 1486 paired samples), and 8.1 percentage points lower (CI, 15.3 to 0.9 percentage points lower) with a general spitting technique (n = 4223 paired samples). Transport media, analytic method, study design, and study quality (Tables 16 to 19, respectively, of Supplement 1) did not seem to affect sensitivity differences. Heterogeneity remained high (I 2 > = 75%) in most stratified analyses.
Table 3. Summary Table of Pooled Estimates on Difference in Sensitivity for SARS-CoV-2 Between Nasopharyngeal Swabs and Saliva, Stratified by Study Characteristics.
Economic Evaluation
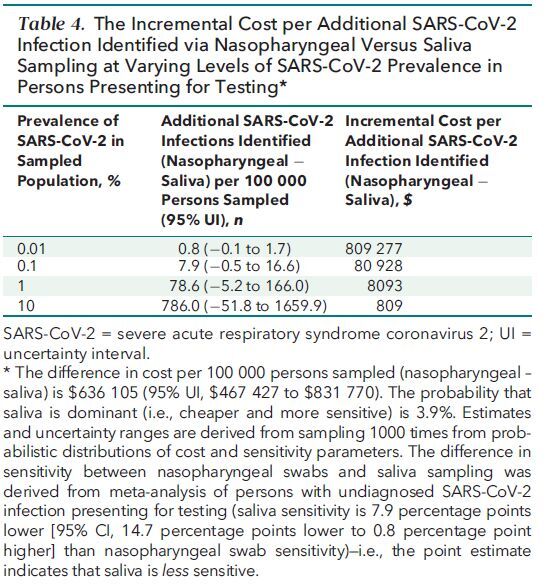
We estimated that the collection of specimens by saliva compared with nasopharyngeal swab would save $636 105 (95% UI, $467 427 to $831 770) per 100 000 persons sampled (Table 4). If the prevalence of SARS-CoV-2 is 1% among persons presenting for SARS-CoV-2 testing, then, on the basis of our estimates of the pooled difference in sensitivity in this population, we estimated that use of nasopharyngeal swabs would identify 79 more (95% UI, 5 fewer to 166 more) persons with SARS-CoV-2 infection per 100 000 persons sampled. This equated to a cost of $8093 per additional infection identified when using a nasopharyngeal swab. We estimated a 3.9% probability that saliva would both be cheaper and identify more persons with SARS-CoV-2 infection than nasopharyngeal swabs (Figure 2 of Supplement 1). In secondary analyses, the cost per additional person with SARS-CoV-2 identified varied proportionally with changes in prevalence and differences in sensitivity (Table 20 of Supplement 1).
Table 4. The Incremental Cost per Additional SARS-CoV-2 Infection Identified via Nasopharyngeal Versus Saliva Sampling at Varying Levels of SARS-CoV-2 Prevalence in Persons Presenting for Testing*.
Discussion
In this meta-analysis of 37 studies comprising 7169 participants providing 7332 paired saliva samples and nasopharyngeal swabs, we found no statistically significant difference in sensitivity between these specimens for SARS-CoV-2 detection. In the economic evaluation of the subgroup of undiagnosed persons presenting for SARS-CoV-2 testing, in whom nasopharyngeal swabs were nonsignificantly more sensitive, the incremental cost per additional SARS-CoV-2 infection detected with nasopharyngeal swabs versus saliva was $8093 if the prevalence was 1%, although UIs were wide. These data suggest that saliva sampling could be an important alternative to nasopharyngeal swabs.
We found indications that the method of saliva collection might affect sensitivity. Studies using a general spitting technique for saliva collection showed a significantly lower sensitivity for saliva than for nasopharyngeal swabs. These data suggest use of other saliva collection techniques (such as early-morning posterior oropharyngeal spitting or drooling) when possible. Saliva sensitivity was not significantly different from nasopharyngeal swab sensitivity among asymptomatic persons and outpatients (suggesting milder disease). These results suggest that saliva may be a particularly useful method of sample collection in community settings.
Previous systematic reviews and meta-analyses have compared nasopharyngeal swabs with saliva sampling (61, 62). These reviews were done earlier in the pandemic and were limited by few studies (≤8) and the participant populations (majority symptomatic) examined at the time. Our meta-analysis builds on this literature with more diverse participant populations, settings, and saliva collection methods. An important additional advantage is our paired economic evaluation, making explicit the potential tradeoffs with moving to saliva sampling. At a prevalence of 1%, our analysis suggests that the added cost ($8093) of detecting an additional SARS-CoV-2 infection with nasopharyngeal swabs could be used to collect more than 3900 saliva samples.
Saliva sampling is an immediate way to expand testing access, while freeing up much-needed health care resources. Saliva sampling has already launched in some jurisdictions (63–65), and a laboratory protocol has received emergency use authorization from the U.S. Food and Drug Administration (66, 67). Although laboratories analyzing saliva will need to validate analytic methods, this can be done and implemented much more quickly than approving, producing, and distributing new tests, such as those intended to be used daily or at the point of care (68).
Maintaining a high level of testing has been repeatedly shown to be an important part of public health strategies to contain SARS-CoV-2 (2, 69, 70). Although laboratory reagents (such as primers and extraction reagents) are an ongoing bottleneck, so too is access to nasopharyngeal swabs (and viral transport media) and trained health care professionals to administer them (71–73). Such methods as pooling samples may overcome some reagent shortages when SARS-CoV-2 prevalence is low (74), but no such methods are available for swabs. Even if a minority of persons may not be able to produce adequate amounts of saliva—and thus would require a nasopharyngeal swab—nasopharyngeal swabs are an uncomfortable method of specimen collection (75) that also carries risk for occupational exposure to the health care workers collecting the samples. We expect that a less invasive and cheaper approach with similar sensitivity may allow a rapid increase in testing, while freeing up much-needed health care professionals for forthcoming vaccinations.
The most important strength of this study is the large number of studies included in the meta-analysis, with participants from many settings with diverse clinical characteristics. These qualities permitted extensive stratified analyses to examine potentially important sources of variability, such as saliva collection method, study setting, testing purpose, and presence of symptoms. We found consistent results in nearly all stratified analyses, enhancing generalizability. In addition, pairing the meta-analysis with an economic evaluation provides data to policymakers about cost and feasibility should they consider adopting saliva sampling. The probabilistic nature of the analysis makes explicit the uncertainty in our estimates, allowing informed decision making in various realistic prevalence scenarios.
These analyses also have limitations. We used an imperfect reference standard assuming that tests would not result in false positives. This precludes estimation of specificity and results in the sampling method with the most positive results being the most sensitive method. Contamination is the most likely source of false positives (76). However, we did not observe systematic trends across included studies, which may suggest that contamination with 1 sampling method was driving results. Different methods of saliva collection and transport media were used, although in most cases these did not seem to affect results. In the subgroup of participants with paired samples who already had confirmed SARS-CoV-2 infection, the method of initial diagnosis was by pharyngeal swab in all studies. This might be expected to bias estimates in favor of nasopharyngeal swabs, but sensitivity differences were nonsignificant. Many studies had risk of bias due to lack of blinding during analysis. Although samples are unlikely to be easily identified after processing in the laboratory, it is uncertain what effect this might have had on outcomes. We assumed that laboratory costs for saliva and nasopharyngeal samples were identical; some samples may require additional processing, but this is unlikely to significantly affect our findings. Further, we did not consider potential downstream costs and impacts. However, we judge it unlikely that these would be affected by method of sampling. The economic and public health implications of missing SARS-CoV-2 infections are important, but no sample collection method, including nasopharyngeal swabs, is 100% sensitive. Missed infections already occur, particularly if persons at risk are not tested at all. Finally, we identified only 1 study that stratified pediatric results. Caution is required in generalizing findings to pediatrics, although saliva sample collection in children may be preferable given the difficulties of nasopharyngeal swabbing in this population (63).
In this study, saliva sampling had similar yield to and lower costs than nasopharyngeal swabs for detecting SARS-CoV-2. Given these findings, plus the advantages of reduced invasiveness, reduced need for trained health care professionals, lower risk for occupational exposure, and reduced need for specialized supplies, we suggest that saliva sampling should replace nasopharyngeal swabs in most populations being tested for SARS-CoV-2.
Supplementary Material
Footnotes
This article was published at Annals.org on 12 January 2021.
References
- 1. Johns Hopkins Coronavirus Resource Center. COVID-19 map. Accessed at https://coronavirus.jhu.edu/map.html on 16 December 2020.
- 2. Aleta A , Martín-Corral D , Pastore y Piontti A , et al. Modelling the impact of testing, contact tracing and household quarantine on second waves of COVID-19. Nat Hum Behav. 2020;4:964-971. [PMID: ] doi: 10.1038/s41562-020-0931-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Campbell JR , Uppal A , Oxlade O , et al. Active testing of groups at increased risk of acquiring SARS-CoV-2 in Canada: costs and human resource needs. CMAJ. 2020;192:E1146-E1155. [PMID: ] doi: 10.1503/cmaj.201128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sethuraman N , Jeremiah SS , Ryo A . Interpreting diagnostic tests for SARS-CoV-2. JAMA. 2020;323:2249-2251. [PMID: ] doi: 10.1001/jama.2020.8259 [DOI] [PubMed] [Google Scholar]
- 5. Alberta Health Services. IPC recommendations PPE table for assessment centres during COVID-19. Updated 18 December 2020. Accessed at www.albertahealthservices.ca/assets/healthinfo/ipc/hi-ipc-assmt-cntrs-covid-ppe-matrx-res-topics-z0-emerging-issues.pdf on 17 September 2020.
- 6. Webber L, Jewett C. Testing swabs run in short supply as makers try to speed up production. NPR. 18 March 2020. Accessed at www.npr.org/sections/health-shots/2020/03/18/817801222/testing-swabs-run-in-short-supply-as-makers-try-to-speed-up-production on 17 September 2020.
- 7. Norman H. COVID testing choke points. Kaiser Health News. 19 August 2020. Accessed at https://khn.org/news/covid-testing-choke-points on 17 September 2020.
- 8. Canadian Agency for Drugs and Technologies in Health. Saliva-based tests to detect active severe acute respiratory syndrome coronavirus 2 infection. Updated 3 June 2020. Accessed at https://cadth.ca/saliva-based-tests-detect-active-severe-acute-respiratory-syndrome-coronavirus-2-infection on 17 September 2020.
- 9. Liberati A , Altman DG , Tetzlaff J , et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. Ann Intern Med. 2009;151:W65-94 [DOI] [PubMed] [Google Scholar]
- 10. Stroup DF , Berlin JA , Morton SC , et al; Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. Meta-analysis of observational studies in epidemiology: a proposal for reporting. JAMA. 2000;283:2008-12. [PMID: ] doi: 10.1001/jama.283.15.2008 [DOI] [PubMed] [Google Scholar]
- 11. Whiting PF , Rutjes AW , Westwood ME , et al; QUADAS-2 Group. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155:529-36. doi: 10.7326/0003-4819-155-8-201110180-00009 [DOI] [PubMed] [Google Scholar]
- 12. Balduzzi S , Rücker G , Schwarzer G . How to perform a meta-analysis with R: a practical tutorial. Evid Based Ment Health. 2019;22:153-160. [PMID: ] doi: 10.1136/ebmental-2019-300117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Viechtbauer W. Conducting meta-analyses in R with the metafor package. J Stat Softw. 2010;36. doi:10.18637/jss.v036.i03
- 14. Newcombe RG . Improved confidence intervals for the difference between binomial proportions based on paired data. Stat Med. 1998;17:2635-50. [PMID: ] [PubMed] [Google Scholar]
- 15. Veroniki AA , Jackson D , Viechtbauer W , et al. Methods to estimate the between-study variance and its uncertainty in meta-analysis. Res Synth Methods. 2016;7:55-79. [PMID: ] doi: 10.1002/jrsm.1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Veroniki AA , Jackson D , Bender R , et al. Methods to calculate uncertainty in the estimated overall effect size from a random-effects meta-analysis. Res Synth Methods. 2019;10:23-43. [PMID: ] doi: 10.1002/jrsm.1319 [DOI] [PubMed] [Google Scholar]
- 17. DerSimonian R , Kacker R . Random-effects model for meta-analysis of clinical trials: an update. Contemp Clin Trials. 2007;28:105-14. [PMID: ] [DOI] [PubMed] [Google Scholar]
- 18. Hamza TH , van Houwelingen HC , Stijnen T . The binomial distribution of meta-analysis was preferred to model within-study variability. J Clin Epidemiol. 2008;61:41-51. [PMID: ] [DOI] [PubMed] [Google Scholar]
- 19. Higgins JP , Thompson SG , Deeks JJ , et al. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557-60. [PMID: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Government of Canada. Job bank: compare wages. 2020. Accessed at www.jobbank.gc.ca/trend-analysis/search-wages on 14 May 2020.
- 21. Serje J , Bertram MY , Brindley C , et al. Global health worker salary estimates: an econometric analysis of global earnings data. Cost Eff Resour Alloc. 2018;16:10. [PMID: ] doi: 10.1186/s12962-018-0093-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. World Bank. Official exchange rate (LCU per US$, period average) - Canada, United States. 2020. Accessed at https://data.worldbank.org/indicator/PA.NUS.FCRF?locations=CA-US on 17 September 2020.
- 23. World Bank. PPP conversion factor, GDP (LCU per international $) - Canada, United States. 2020. Accessed at https://data.worldbank.org/indicator/PA.NUS.PPP?locations=CA-US on 17 September 2020.
- 24. Azzi L , Baj A , Alberio T , et al; ASST dei Sette Laghi Rapid Salivary Test Nurse staff Research Group. Rapid salivary test suitable for a mass screening program to detect SARS-CoV-2: a diagnostic accuracy study [Letter]. J Infect. 2020;81:e75-e78. [PMID: ] doi: 10.1016/j.jinf.2020.06.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen JH , Yip CC , Poon RW , et al. Evaluating the use of posterior oropharyngeal saliva in a point-of-care assay for the detection of SARS-CoV-2. Emerg Microbes Infect. 2020;9:1356-1359. [PMID: ] doi: 10.1080/22221751.2020.1775133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Leung EC , Chow VC , Lee MK , et al. Deep throat saliva as an alternative diagnostic specimen type for the detection of SARS-CoV-2. J Med Virol. 2020. [PMID: ] doi: 10.1002/jmv.26258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McCormick-Baw C , Morgan K , Gaffney D , et al. Saliva as an alternate specimen source for detection of SARS-CoV-2 in symptomatic patients using Cepheid Xpert Xpress SARS-CoV-2 [Letter]. J Clin Microbiol. 2020;58. [PMID: ] doi: 10.1128/JCM.01109-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rao M , Rashid FA , Sabri FSAH , et al. Comparing nasopharyngeal swab and early morning saliva for the identification of SARS-CoV-2. Clin Infect Dis. 2020. [PMID: ] doi: 10.1093/cid/ciaa1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Landry ML , Criscuolo J , Peaper DR . Challenges in use of saliva for detection of SARS CoV-2 RNA in symptomatic outpatients. J Clin Virol. 2020;130:104567. [PMID: ] doi: 10.1016/j.jcv.2020.104567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Villar LM , da Costa VD , Marques BCL , et al. Usefulness of saliva samples for detecting SARS-CoV-2 RNA among liver disease patients [Letter]. J Infect. 2020. [PMID: ] doi: 10.1016/j.jinf.2020.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Akgun Dogan O, Kose B, Agaoglu NB, et al. Does sampling saliva increase detection of SARS-CoV-2 by RT-PCR? Comparing saliva with oro-nasopharyngeal swabs. medRxiv. Preprint posted online 28 July 2020. doi:10.1101/2020.07.26.20158618 [DOI] [PMC free article] [PubMed]
- 32. Becker D, Sandoval E, Amin A, et al. Saliva is less sensitive than nasopharyngeal swabs for COVID-19 detection in the community setting. medRxiv. Preprint posted online 17 May 2020. doi:10.1101/2020.05.11.20092338
- 33. Byrne RL , Kay GA , Kontogianni K , et al. Saliva alternative to upper respiratory swabs for SARS-CoV-2 diagnosis. Emerg Infect Dis. 2020;26:2770-2771. [PMID: ] doi: 10.3201/eid2611.203283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Griesemer SB, Van Slyke G, Ehrbar D, et al. Evaluation of specimen types and saliva stabilization solutions for SARS-CoV-2 testing. medRxiv. Preprint posted online 18 June 2020. doi:10.1101/2020.06.16.20133041 [DOI] [PMC free article] [PubMed]
- 35. Hanson KE , Barker AP , Hillyard DR , et al. Self-collected anterior nasal and saliva specimens versus health care worker-collected nasopharyngeal swabs for the molecular detection of SARS-CoV-2. J Clin Microbiol. 2020;58. [PMID: ] doi: 10.1128/JCM.01824-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Iwasaki S , Fujisawa S , Nakakubo S , et al. Comparison of SARS-CoV-2 detection in nasopharyngeal swab and saliva [Letter]. J Infect. 2020;81:e145-e147. [PMID: ] doi: 10.1016/j.jinf.2020.05.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jamal AJ , Mozafarihashjin M , Coomes E , et al; Toronto Invasive Bacterial Diseases Network COVID-19 Investigators. Sensitivity of nasopharyngeal swabs and saliva for the detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Clin Infect Dis. 2020. [PMID: ] doi: 10.1093/cid/ciaa848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Miller M, Jansen M, Bisignano A, et al. Validation of a self-administrable, saliva-based RT-qPCR test detecting SARS-CoV-2. medRxiv. Preprint posted online 9 June 2020. doi:10.1101/2020.06.05.20122721
- 39. Pasomsub E , Watcharananan SP , Boonyawat K , et al. Saliva sample as a non-invasive specimen for the diagnosis of coronavirus disease 2019: a cross-sectional study. Clin Microbiol Infect. 2020. [PMID: ] doi: 10.1016/j.cmi.2020.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ranoa DRE, Holland RL, Alnaji FG, et al. Saliva-based molecular testing for SARS-CoV-2 that bypasses RNA extraction. bioRxiv. Preprint posted online 18 June 2020. doi:10.1101/2020.06.18.159434
- 41. Wyllie AL , Fournier J , Casanovas-Massana A , et al. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2 [Letter]. N Engl J Med. 2020;383:1283-1286. [PMID: ] doi: 10.1056/NEJMc2016359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bhattacharya D, Parai D, Rout UK, et al. Saliva as a potential clinical specimen for diagnosis of SARS-CoV-2. medRxiv. Preprint posted online 11 September 2020. doi:10.1101/2020.09.11.20192591
- 43. Goldfarb DM, Tilley P, Al-Rawahi GN, et al. Self-collected saline gargle samples as an alternative to healthcare worker collected nasopharyngeal swabs for COVID-19 diagnosis in outpatients. medRxiv. Preprint posted online 14 September 2020. doi:10.1101/2020.09.13.20188334 [DOI] [PMC free article] [PubMed]
- 44. Ku CW, Shivani D, Kwan JQT, et al. Validation of self-collected buccal swab and saliva as a diagnostic tool for COVID-19. medRxiv. Preprint posted online 5 October 2020. doi:10.1101/2020.10.03.20205278 [DOI] [PMC free article] [PubMed]
- 45. Nacher M, Mergeay-Fabre M, Blanchet D, et al. Prospective comparison of saliva and nasopharyngeal swab sampling for mass screening for COVID-19. medRxiv. Preprint posted online 24 September 2020. doi:10.1101/2020.09.23.20150961 [DOI] [PMC free article] [PubMed]
- 46. Sahajpal NS, Mondal AK, Ananth S, et al. SalivaAll: clinical validation of a sensitive test for saliva collected in healthcare and community settings with pooling utility for SARS-CoV-2 mass surveillance. medRxiv. Preprint posted online 1 September 2020. doi:10.1101/2020.08.26.20182816 [DOI] [PMC free article] [PubMed]
- 47. Teo AKJ, Choudhury Y, Tan IB, et al. Validation of saliva and self-administered nasal swabs for COVID-19 testing. medRxiv. Preprint posted online 14 August 2020. doi:10.1101/2020.08.13.20173807
- 48. Yee R, Truong T, Pannaraj PS, et al. Saliva is a promising alternative specimen for the detection of SARS-CoV-2 in children and adults. medRxiv. Preprint posted online 27 October 2020. doi:10.1101/2020.10.25.20219055 [DOI] [PMC free article] [PubMed]
- 49. Yokota I, Hattori T, Shane PY, et al. Equivalent SARS-CoV-2 viral loads between nasopharyngeal swab and saliva in symptomatic patients. medRxiv. Preprint posted online 3 September 2020. doi:10.1101/2020.09.01.20186254 [DOI] [PMC free article] [PubMed]
- 50. Yokota I , Shane PY , Okada K , et al. Mass screening of asymptomatic persons for SARS-CoV-2 using saliva. Clin Infect Dis. 2020. [PMID: ] doi: 10.1093/cid/ciaa1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Barat B, Das S, De Giorgi V, et al. Pooled saliva specimens for SARS-CoV-2 testing. medRxiv. Preprint posted online 5 October 2020. doi:10.1101/2020.10.02.20204859
- 52. Aita A , Basso D , Cattelan AM , et al. SARS-CoV-2 identification and IgA antibodies in saliva: one sample two tests approach for diagnosis. Clin Chim Acta. 2020;510:717-722. [PMID: ] doi: 10.1016/j.cca.2020.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Altawalah H , AlHuraish F , Alkandari WA , et al. Saliva specimens for detection of severe acute respiratory syndrome coronavirus 2 in Kuwait: a cross-sectional study. J Clin Virol. 2020;132:104652. [PMID: ] doi: 10.1016/j.jcv.2020.104652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Binder RA , Alarja NA , Robie ER , et al. Environmental and aerosolized severe acute respiratory syndrome coronavirus 2 among hospitalized coronavirus disease 2019 patients. J Infect Dis. 2020;222:1798-1806. [PMID: ] doi: 10.1093/infdis/jiaa575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Caulley L , Corsten M , Eapen L , et al. Salivary detection of COVID-19 [Letter]. Ann Intern Med. 2020. doi: 10.7326/M20-4738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kojima N , Turner F , Slepnev V , et al. Self-collected oral fluid and nasal swab specimens demonstrate comparable sensitivity to clinician-collected nasopharyngeal swab specimens for the detection of SARS-CoV-2. Clin Infect Dis. 2020. [PMID: ] doi: 10.1093/cid/ciaa1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Procop GW , Shrestha NK , Vogel S , et al. A direct comparison of enhanced saliva to nasopharyngeal swab for the detection of SARS-CoV-2 in symptomatic patients. J Clin Microbiol. 2020;58. [PMID: ] doi: 10.1128/JCM.01946-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Senok A , Alsuwaidi H , Atrah Y , et al. Saliva as an alternative specimen for molecular COVID-19 testing in community settings and population-based screening. Infect Drug Resist. 2020;13:3393-3399. [PMID: ] doi: 10.2147/IDR.S275152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Uwamino Y , Nagata M , Aoki W , et al. Accuracy and stability of saliva as a sample for reverse transcription PCR detection of SARS-CoV-2 [Letter]. J Clin Pathol. 2021;74:67-68. [PMID: ] doi: 10.1136/jclinpath-2020-206972 [DOI] [PubMed] [Google Scholar]
- 60. Migueres M , Mengelle C , Dimeglio C , et al. Saliva sampling for diagnosing SARS-CoV-2 infections in symptomatic patients and asymptomatic carriers [Letter]. J Clin Virol. 2020;130:104580. [PMID: ] doi: 10.1016/j.jcv.2020.104580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Peeters E, Kaur Dhillon Ajit Singh S, Vandesompele J, et al. Rapid systematic review of the sensitivity of SARS-CoV-2 molecular testing on saliva compared to nasopharyngeal swabs. medRxiv. Preprint posted online 6 August 2020. doi:10.1101/2020.08.05.20168716
- 62. Czumbel LM , Kiss S , Farkas N , et al. Saliva as a candidate for COVID-19 diagnostic testing: a meta-analysis. Front Med (Lausanne). 2020;7:465. [PMID: ] doi: 10.3389/fmed.2020.00465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Weichel A. Goodbye, nasal swabs: B.C. announces non-invasive COVID-19 test for students. CTV News. 17 September 2020. Accessed at https://bc.ctvnews.ca/goodbye-nasal-swabs-b-c-announces-non-invasive-covid-19-test-for-students-1.5109803 on 20 September 2020.
- 64. Sataline S. Hong Kong's key to keeping COVID out is in its airport. Intelligencer. 24 August 2020. Accessed at https://nymag.com/intelligencer/2020/08/hong-kongs-key-to-keeping-covid-out-is-in-its-airport.html on 20 September 2020.
- 65. University of Chicago. On-campus COVID-19 testing dashboard. Accessed at https://splunk-public.machinedata.illinois.edu/en-US/app/uofi_shield_public_APP/home on 20 September 2020.
- 66. Vogels CBF, Watkins AE, Harden CA, et al. SalivaDirect: a simplified and flexible platform to enhance SARS-CoV-2 testing capacity. medRxiv. Preprint posted online 28 September 2020. doi:10.1101/2020.08.03.20167791 [DOI] [PMC free article] [PubMed]
- 67. U.S. Food and Drug Administration. Coronavirus (COVID-19) update: FDA issues Emergency Use Authorization to Yale School of Public Health for SalivaDirect, which uses a new method of saliva sample processing. 15 August 2020. Accessed at www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-issues-emergency-use-authorization-yale-school-public-health on 20 September 2020.
- 68. Larremore DB, Wilder B, Lester E, et al. Test sensitivity is secondary to frequency and turnaround time for COVID-19 surveillance. medRxiv. Preprint posted online 8 September 2020. doi:10.1101/2020.06.22.20136309
- 69. Tuite AR , Fisman DN , Greer AL . Mathematical modelling of COVID-19 transmission and mitigation strategies in the population of Ontario, Canada. CMAJ. 2020;192:E497-E505. [PMID: ] doi: 10.1503/cmaj.200476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Grassly NC , Pons-Salort M , Parker EPK , et al; Imperial College COVID-19 Response Team. Comparison of molecular testing strategies for COVID-19 control: a mathematical modelling study. Lancet Infect Dis. 2020;20:1381-1389. [PMID: ] doi: 10.1016/S1473-3099(20)30630-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Tong S. Testing, testing: labs, supply chain overwhelmed by coronavirus surge and diagnostic demand. Marketplace. 7 July 2020. Accessed at www.marketplace.org/2020/07/07/covid19-labs-supply-chain-overwhelmed-virus-surge-testing-demand on 20 September 2020.
- 72. Grant K, Gray J. As demand for virus testing spikes, officials look for new methods. The Globe and Mail. 17 September 2020. Accessed at www.theglobeandmail.com/canada/article-bc-offers-covid-19-saliva-tests-for-children-demand-spikes-for on 20 September 2020.
- 73. Watson E. Coronavirus testing: nationwide shortage of reagents limiting Hampton Roads hospitals, labs. 13News Now. 11 August 2020. Accessed at www.13newsnow.com/article/news/health/coronavirus/coronavirus-testing-nationwide-shortage-of-reagents-limiting-hampton-roads-hospitals-labs/291-0f649f23-ac14-41b9-8e91-51e2e6a23d03 on 20 September 2020.
- 74. U.S. Food and Drug Administration. Pooled sample testing and screening testing for COVID-19. Accessed at www.fda.gov/medical-devices/coronavirus-covid-19-and-medical-devices/pooled-sample-testing-and-screening-testing-covid-19 on 20 September 2020.
- 75. Frazee BW , Rodríguez-Hoces de la Guardia A , Alter H , et al. Accuracy and discomfort of different types of intranasal specimen collection methods for molecular influenza testing in emergency department patients. Ann Emerg Med. 2018;71:509-517.e1. [PMID: ] doi: 10.1016/j.annemergmed.2017.09.010 [DOI] [PubMed] [Google Scholar]
- 76. Surkova E , Nikolayevskyy V , Drobniewski F . False-positive COVID-19 results: hidden problems and costs. Lancet Respir Med. 2020;8:1167-1168. [PMID: ] doi: 10.1016/S2213-2600(20)30453-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.