Abstract
Poultries including chickens, ducks, geese, and pigeons are widely used in the biological and medical research in many aspects. The genetic quality of experimental poultries directly affects the results of the research. In this study, following electrophoresis analysis and short tandem repeat (STR) scanning, we screened out the microsatellite loci for determining the genetic characteristics of Chinese experimental chickens, ducks, geese, and pigeons. The panels of loci selected in our research provide a good choice for genetic monitoring of the population genetic diversity of Chinese native experimental chickens, ducks, geese, and ducks.
1. Introduction
Laboratory animals are important experimental materials for science research. They play key roles in the investigation of pathogenesis, diagnosis of diseases, pharmaceutical research, and other fields [1]. The genetic quality of laboratory animals directly affects the accuracy, repeatability, and scientificity of medical biological research results. Genetic monitoring is one of the effective methods to evaluate population's genetic diversity. Through genetic monitoring, whether genetic mutations and genetic pollution occurred can be analyzed.
Poultry, including chicken, duck, goose, and pigeon, has become commonly used laboratory animals [2]. They are easy to reproduce and hatch in vitro. Among them, chickens are the most widely used poultry in life science research [3, 4]. Ducks, geese, and pigeons also play important roles in the research of epidemiology, immunology, virology, and pharmacotoxicology [5–9]. There are many genetic analysis and quality control methods applied to chickens [10, 11]. However, at present, we find few reports about the genetic analysis systems and quality control methods of duck, goose, and pigeon populations, especially in the Chinese native groups.
Hence, in this study, we screened out the microsatellite loci with uniform distribution, stable amplification, and rich polymorphism in experimental chickens, ducks, geese, and pigeons with different genetic backgrounds [12]. We developed effective microsatellite marker systems to determine the genetic diversity of experimental chickens, ducks, geese, and pigeons, which will lay the foundation for the genetic quality control of them and promote the application of experimental poultry.
2. Materials and Methods
2.1. Animal Sample
Three outbred groups and three haplotype groups of experimental chicken were used in this research: outbred group BWEL-SPF chickens ((SCXK (black) 2017-005)), 40 samples, 37 weeks old, 6 males and 34 females, which has been closed for 20 generations; outbred group BM chicken (from BWEL chicken lineage (SCXK (black) 2017-005)), 40 samples, 14 weeks old, 6 males and 34 females; outbred group Beijing oil chickens, 46 samples. MHC haplotype chickens were bred from the 13th generation of BWEL chicken, the haplotype was continuously selected based on the MHC core genes, and the half-sibling or sibling mating method was used to breed to the 8th generation [13]. We selected 5 G1 haplotype chickens, 53 weeks old, 1 male and 4 females; 5 G2 haplotype chickens, 93 weeks, 1 male and 4 females; and 5 G7 haplotype chickens, 82 weeks, 1 male and 4 females. The Beijing oil chickens came from the Institute of Animal Science (IAS), Chinese Academy of Agricultural Sciences (CAAS). Other samples were from Harbin Veterinary Research Institute (HVRI), CAAS. All the samples were blood.
Two outbred groups and four haplotype groups of experimental duck (bred from Jinding (JD) duck lineage (SCXK (black) 2017-006)) were selected: outbred group 1, 40 samples, 37 weeks old, 6 males and 34 females; outbred group JD duck, 40 samples, 37 weeks old, 6 males and 34 females; 10 A haplotype ducks, 53 weeks old, 1 male and 4 females; 10 B haplotype ducks, 53 weeks old, 1 male and 4 females; 10 C haplotype ducks, 53 weeks old, 1 male and 4 females; 10 D haplotype ducks, 53 weeks old, 1 male and 4 females. All the samples are duck muscle tissue and were from HVRI, CAAS.
We collected two outbred groups of experimental geese: outbred group Guangdong Wuzong goose, 44 samples, 37 weeks old, 6 males and 34 females; outbred group Yangzhou goose, 44 samples, 37 weeks old, 6 males and 34 females. All the samples are goose liver tissue. Guangdong Wuzong geese were from Southern Medical University, and Yangzhou geese were from Yangzhou University.
Forty pigeons were randomly selected from two populations of white king pigeons and silver king pigeons, half male and half female, with no age limit. All the animals were from Liujinlong pigeon farms in Beijing. Their heart tissues were collected.
All breeding is carried out in accordance with Chinese agricultural standards NY/T 1901. What is more, all experiments followed the 3R principle.
2.2. Microsatellite Locus Selection
By searching PubMed and using SSR Hunter software to analyze animal gene information, we obtained microsatellite loci for further screening.
2.3. DNA Extraction
Phenol-chloroform extraction method was used to extract DNA from muscle, liver, and heart tissue. TIANamp Blood DNA Kits (Tiangen, Beijing, China) were used to extract DNA from chicken blood samples. All DNA concentrations were diluted to 50 ng/μL, stored in -20°C.
2.4. PCR Procedure and Agarose Gel Electrophoresis
The PCR was performed in a 20 μL reaction volume containing 10 μL Dream Taq Green PCR Master Mix (Thermo Fisher Scientific, Massachusetts, MA), 2 μL pure water (ddH2O), 10 pmol each primer, and 50 ng of the extracted DNA template. The PCR protocol was as follows: 94°C for 5 min, followed by 35 cycles of 94°C for 30 s, suitable temperature for 30 s, 72°C for 30 s, and a final extension at 72°C for 5 min. Amplified products were stored at -20°C for further analysis.
Amplified products were electrophoresed on a 2% agarose gel at 130 V, 90 min.
2.5. STR Scanning
We performed STR scanning on PCR amplification products of candidate loci. The forward primers of candidate microsatellite loci were fluorescent labelled with FAM, HEX, and TAMRA. The sample genome was amplified with fluorescent primers, and the amplified products were scanned by STR through 3730xl DNA Analyzer (Applied Biosystems, Thermo Fisher Scientific, Massachusetts, USA). All the STR scanning was performed by Beijing Tianyi Huiyuan Biotechnology Co., Ltd.
2.6. Data Analysis
GeneMarker V2.2.0 software was used to analyze the length of amplified fragments from different populations at each microsatellite locus. Popgene 3.2 software was used to analyze the observed number of alleles, effective number of alleles, Shannon's information index, and effective heterozygosity of microsatellite loci. The polymorphic information content of multiple sites was calculated using PIC calculation software (PIC_CALC.0.6).
3. Results
3.1. Microsatellite Locus Selection
3.1.1. Preliminary Screening of Microsatellite Loci by PCR
Firstly, we obtained the microsatellite locus information of experimental chickens, ducks, geese, and pigeons by searching previous reports on PubMed and using the SSR Hunter software to analyze the genetic information of different populations [14, 15]. We collected 72, 59, 57, and 61 microsatellite loci of experimental chicken, duck, goose, and pigeon, respectively.
In order to clarify the amplification conditions of the microsatellite loci and exclude the loci with poor specificity, we performed temperature gradient PCR and agarose gel electrophoresis of microsatellite loci. Then, we performed PCR amplification on the most suitable conditions and subjected the PCR products to agarose gel electrophoresis to screen out loci with suitable length, good polymorphism in outbred groups, good monomorphism in haplotypes, and high specificity. Taking the chicken GGNCAMZO locus and duck AY264 locus as example, the results are shown in Figure 1. GGNCAMZO locus is monomorphic in the haplotype chicken population, and AY264 locus is polymorphic in the outbred duck group.
Figure 1.
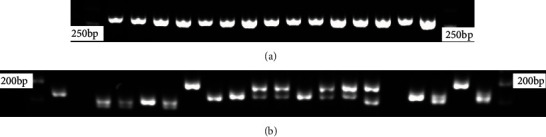
Results of agarose gel electrophoresis of microsatellite DNA locus GGNCAMZO in experimental chickens and locus AY264 in experimental ducks. (a) GGNCAMZO in haplotype chicken line G1. (b) AY264 in the outbred group of experimental ducks.
In summary, we selected 37 and 32 microsatellite loci with good polymorphism in the outbred groups and haplotypes of chicken, respectively [12, 16, 17]. In addition, 15 and 23 loci were screened in the outbred groups and haplotypes of duck, respectively [14, 18, 19]. In the outbred groups of goose and pigeon, 14 and 20 microsatellite loci were chosen [18, 20–23]. Loci in these panels would be candidate for the final microsatellite marker evaluation systems.
3.1.2. STR Scanning Analysis
In order to further complete the microsatellite marker system, we performed STR scanning on the candidate microsatellite DNA loci matched microsatellite criteria and analyzed the length of the amplified product at the peak with GeneMarker software (V1.75). Taking the UU-CliμT47 locus as an example, it showed polymorphism in the outbred group of pigeon (Figure 2).
Figure 2.
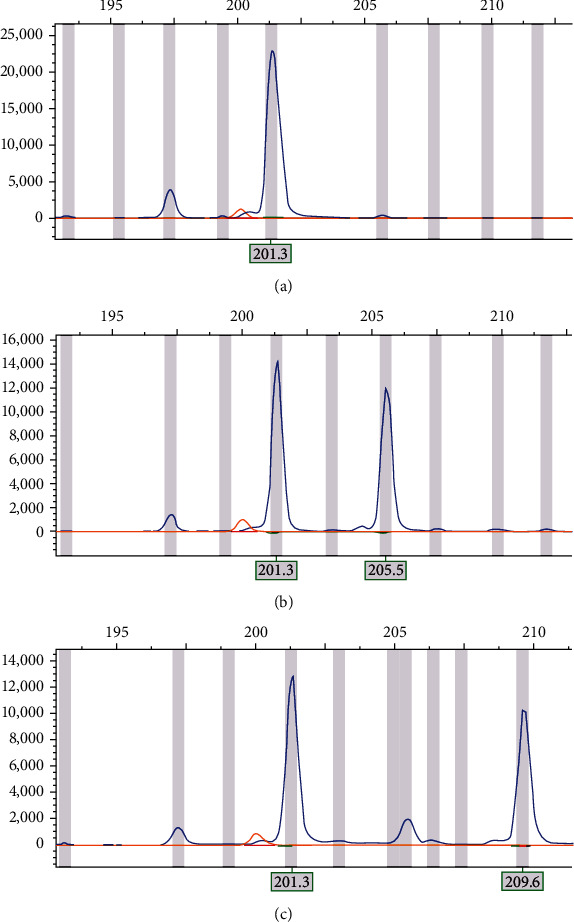
Results of UU-CliμT47 scan of the experimental pigeons. (a) The STR graph corresponding to the sample of haplotype under primer UU-CliμT47 shows homozygote with a wave peak of 201 bp. (b) The STR diagram corresponding to the sample of outbred groups under primer UU-CliμT47 shows heterozygote with two wave peaks of 201 bp and 205 bp, respectively. (c) The STR diagram corresponding to the sample of outbred groups under primer UU-CliμT47 shows heterozygote with two wave peaks of 201 bp and 209 bp, respectively.
We finally determined that in experimental chickens, 28 loci were selected for genetic monitoring in the outbred groups and 14 loci for haplotypes. All microsatellite DNA loci are shown in Table 1. There are 13 common loci.
Table 1.
Number of alleles, optimal amplification conditions, and fragment length of 29 alleles for the laboratory chickens.
| Loci | Primer sequence (5′-3′) | Temperature(°C) | Allele range | Applicable groups |
|---|---|---|---|---|
| MCW0029 | GTGGACACCCATTTGTACCCTATG | 63.8 | 139-188 | Outbred group |
| CATGCAATTCAGGACCGTGCA | ||||
| ADL0293 | GTAATCTAGAAACCCCATCT | 53.9 | 106-120 | Outbred group |
| ACATACCGCAGTCTTTGTTC | ||||
| ADL0317 | AGTTGGTTTCAGCCATCCAT | 58.5 | 177-219 | Outbred group |
| CCCAGAGCACACTGTCACTG | ||||
| GCT0016 | TCCAAGGTTCTCCAGTTC | 52.2 | 111-148 | Outbred group |
| GGCATAAGGATAGCAACAG | ||||
| ADL0304 | GGGGAGGAACTCTGGAAATG | 53.9 | 138-161 | Outbred group |
| CCTCATGCTTCGTGCTTTTT | ||||
| LEI0074 | GACCTGGTCCTGACATGGGTG | 58.5 | 221-243 | Outbred group |
| GTTTGCTGATTAGCCATCGCG | ||||
| ADL328 | CACCCATAGCTGTGACTTTG | 53.9 | 107-120 | Outbred group |
| AAAACCGGAATGTGTAACTG | ||||
| GGANTECl | GCGGGGCCGTTATCAGAGCA | 65.0 | 139-194 | Outbred group |
| AGTGCAGGGCGCTCCTGGT | ||||
| LEI094 | CAGGATGGCTGTTATGCTTCCA | 56.0 | 176-211 | Outbred group |
| CACAGTGCAGAGTGGTGCGA | ||||
| MCW0330 | TGGACCTCATCAGTCTGACAG | 58.5 | 217-287 | Outbred group |
| AATGTTCTCATAGAGTTCCTGC | ||||
| LEI0141 | CGCATTTGATGCATAACACATG | 52.2 | 221-245 | Outbred group |
| AAGGCAAACTCAGCTGGAACG | ||||
| MCW0087 | ATTTCTGCAGCCAACTTGGAG | 58.5 | 268-289 | Outbred group |
| CTCAGGCAGTTCTCAAGAACA | ||||
| MCW0347 | GCTTCCAGATGAGCTCCATGG | 52.0 | 121-149 | Outbred group |
| CACAGCGCTGCAGCAACTG | ||||
| ADL176 | TTGTGGATTCTGGTGGTAGC | 58.5 | 183-200 | Outbred group |
| TTCTCCCGTAACACTCGTCA | ||||
| ADL0201 | GCTGAGGATTCAGATAAGAC | 58.5 | 111-151 | Outbred group |
| AATGGCYGACGTTTCACAGC | ||||
| GGNCAMZO | GTCACTAGGTTAGCAGCATG | 56.0 | 234 | Outbred group |
| GCTGGATACAGACCTCGATT | Haplotype | |||
| GGAVIR | AGAGATGGTGCACGCAACCT | 60.7 | 86-89 | Outbred group |
| CGAGCACTTTCTGGCAGAGA | Haplotype | |||
| MCW0063 | GGCTCCAAAAGCTTGTTCTTAGCT | 53.9 | 116-146 | Outbred group |
| GAAAACCAGTAAAGCTTCTTAC | Haplotype | |||
| ADL185 | CATGGCAGCTGACTCCAGAT | 58.5 | 116-142 | Outbred group |
| AGCGTTACCTGTTCGTTTGC | Haplotype | |||
| GGMYC | CGAGGCGCTCTGCGAGTTTA | 62.4 | 139-151 | Outbred group |
| TGGGGACCTCTGGCTCTGAC | Haplotype | |||
| LEI0094 | GATCTCACCAGTATGAGCTGC | 53.9 | 250-283 | Outbred group |
| TCTCACACTGTAACACAGTGC | Haplotype | |||
| GGVITC | AGCCATCATTCAGGGCATCT | 58.5 | 86 | Outbred group |
| GATGTCCTGAGTGATGCTCA | Haplotype | |||
| ADL0292 | CCAAATCAGGCAAAACTTCT | 58.5 | 110-136 | Outbred group |
| AAATGGCCTAAGGATGAGGA | Haplotype | |||
| GGVITIIG | GGCAGGTTTCTAATGCCTGA | 56.0 | 186-189 | Outbred group |
| CCCATCGTTTCAACTGTATG | Haplotype | |||
| ADL166 | TGCCAGCCCGTAATCATAGG | 58.5 | 131-154 | Outbred group |
| AAGCACCACGACCCAATCTA | Haplotype | |||
| MCW0014 | AAAATATTGGCTCTAGGAACTGTC | 58.5 | 172-195 | Outbred group |
| ACCGGAAATGAAGGTAAGACTAGC | Haplotype | |||
| GGCYMA | AGCGAGGCGCTCTGCGAGTT | 64.6 | 140-153 | Outbred group |
| GGGCACCTCTGGCTCTGACC | Haplotype | |||
| MCW0402 | ACTGTGCCTAGGACTAGCTG | 56.0 | 141-229 | Outbred group |
| CCTAAGTCTGGGCTCTTCTG | Haplotype | |||
| STMSGGHU2-1A | CTTAATATGTGTGAGGTGGC | 53.9 | 235-238 | Haplotype |
| GTTCTCACAATTGCATTAGC |
In experimental duck populations, we chose 25 loci and 15 loci for genetic monitoring in the outbred duck groups and haplotype groups. There are 12 common loci. Microsatellite loci are shown in Table 2.
Table 2.
Number of alleles, optimal amplification conditions, and fragment length of 28 alleles for the laboratory ducks.
| Loci | Primer sequence(5′-3′) | Temperature (°C) | Allele range | Applicable groups |
|---|---|---|---|---|
| CAUD007 | ACTTCTCTTGTAGGCATGTCA | 60.8 | 100-190 | Outbred group |
| CACCTGTTGCTCCTGCTGT | ||||
| CAUD004 | TCCACTTGGTAGACCTTGAG | 60.8 | 234-385 | Outbred group |
| TGGGATTCAGTGAGAAGCCT | ||||
| CAUD023 | CACATTAACTACATTTCGGTCT | 51.4 | 163-234 | Outbred group |
| CAGCCAAAGAGTTCAACAGG | ||||
| CAUD027 | AGAAGGCAGGCAAATCAGAG | 66.0 | 70-180 | Outbred group |
| TCCACTCATAAAAACACCCACA | ||||
| CAUD001 | ACAGCTTCAGCAGACTTAGA | 55.5 | 150-247 | Outbred group |
| GCAGAAAGTGTATTAAGGAAG | ||||
| CAUD031 | AGCATCTGGACTTTTTCTGGA | 51.4 | 107-187 | Outbred group |
| CACCCCAGGCTCTGAGATAA | ||||
| CAUD032 | GAAACCAACTGAAAACGGGC | 58.1 | 96-206 | Outbred group |
| CCTCCTGCGTCCCAATAAG | ||||
| AY314 | CTCATTCCAATTCCTCTGTA | 50.3 | 112-329 | Outbred group |
| CAGCATTATTATTTCAGAAGG | ||||
| CMO211 | GGATGTTGCCCCACATATTT | 55.0 | 112-205 | Outbred group |
| TTGCCTTGTTTATGAGCCATT | ||||
| APH09 | GGATGTTGCCCCACATATTT | 58.0 | 134-190 | Outbred group |
| TTGCCTTGTTTATGAGCCATTA | ||||
| APH11 | GGACCTCAGGAAAATCAGTGTA | 58.5 | 183-185 | Outbred group |
| GCAGGCAGAGCAGGAAATA | ||||
| APL2 | GATTCAACCTTAGCTATCAGTCTCC | 58.5 | 115-125 | Outbred group |
| CGCTCTTGGCAAATGTCC | ||||
| CAUD011 | TGCTATCCACCCAATAAGTG | 50.3 | 145-223 | Outbred group |
| CAAAGTTAGCTGGTATCTGC | ||||
| CAUD006 | ATGGTTCTCTGTAGGCAATC | 63.5 | 183-290 | Outbred group |
| TTCTGCTTGGGCTCTTGGA | Haplotype | |||
| CAUD018 | TTAGACAAATGAGGAAATAGTA | 50.3 | 100-180 | Outbred group |
| GTCCAAACTAAATGCAGGC | Haplotype | |||
| CAUD010 | GGATGTGTTTTTCATTATTGAT | 50.3 | 138-200 | Outbred group |
| AGAGGCATAAATACTCAGTG | Haplotype | |||
| CAUD012 | ATTGCCTTTCAGTGGAGTTTC | 63.5 | 182-286 | Outbred group |
| CGGCTCTAAACACATGAATG | Haplotype | |||
| CAUD014 | CACAACTGACGGCACAAAGT | 58.1 | 136-200 | Outbred group |
| CTGAGTTTTTCCCGCCTCTA | Haplotype | |||
| CAUD034 | TACTGCATATCACTAGAGGA | 55.5 | 160-296 | Outbred group |
| TAGGCATACTCGGGTTTAG | Haplotype | |||
| CAUD035 | GTGCCTAACCCTGATGGATG | 63.5 | 174-282 | Outbred group |
| CTTATCAGATGGGGCTCGGA | Haplotype | |||
| APL579 | ATTAGAGCAGGAGTTAGGAGAC | 55.0 | 116-227 | Outbred group |
| GCAAGAAGTGGCTTTTTTC | Haplotype | |||
| AY258 | ATGTCTGAGTCCTCGGAGC | 58.1 | 89-162 | Outbred group |
| ACAATAGATTCCAGATGCTGAA | Haplotype | |||
| CMO212 | CTCCACTAGAACACAGACATT | 58.0 | 186-272 | Outbred group |
| CATCTTTGGCATTTTGAAG | Haplotype | |||
| CAUD028 | TACACCCAAGTTTATTCTGAG | 55.5 | 152-226 | Outbred group |
| ACTCTCCAGGGCACTAGG | Haplotype | |||
| CAUD026 | ACGTCACATCACCCCACAG | 60.8 | 134-196 | Outbred group |
| CTTTGCCTCTGGTGAGGTTC | Haplotype | |||
| APH18 | TTCTGGCCTGATAGGTATGAG | 58.0 | 178-325 | Haplotype |
| GAATTGGGTGGTTCATACTGT | ||||
| CAUD002 | CTTCGGTGCCTGTCTTAGC | 60.8 | 200-231 | Haplotype |
| AGCTGCCTGGAGAAGGTCT | ||||
| CAUD005 | CTGGGTTTGGTGGAGCATAA | 60.8 | 184-290 | Haplotype |
| TACTGGCTGCTTCATTGCTG |
14 microsatellite loci with good polymorphism were considered as microsatellite markers in the outbred group of goose. Table 3 demonstrates the number of alleles, optimal amplification conditions, and fragment length of 14 alleles for the outbred experiment geese.
Table 3.
Number of alleles, optimal amplification conditions, and fragment length of 14 alleles for the outbred colony laboratory geese.
| Loci | Primer sequence(5′-3′) | Temperature (°C) | Allele range |
|---|---|---|---|
| G-Ans17 | ACAAATAACTGGTTCTAAGCAC | 51.0 | 111–123 |
| AGAGGACTTCTATTCATAAATA | |||
| G-TTUCG1 | CCCTGCTGGTATACCTGA | 53.0 | 113-115 |
| GTGTCTACACAACAGC | |||
| G-APH13 | CAACGAGTGACAATGATAAAA | 53.0 | 163-165 |
| CAATGATCTCACTCCCAATAG | |||
| G-Ans02 | TTCTGTGCAGGGGCGAGTT | 58.0 | 202–230 |
| AGGGAACCGATCACGACATG | |||
| G-Ans07 | GACTGAGGAACTACAATTGACT | 62.0 | 236–246 |
| ACAAAGACTACTACTGCCAAG | |||
| G-Ans18 | GTGTTCTCTGTTTATGATATTAC | 56.0 | 229–237 |
| AACAGAATTTGCTTGAAACTGC | |||
| G-Ans25 | CACTTATTAATGGCACTTGAAA | 62.0 | 261–277 |
| GTTCTCTTGTCACAACTGGA | |||
| G-Hhiμ1b | ATCAAAGGCACAATGTGAAAT | 60.0 | 212–216 |
| AGTAAGGGGGCTTCCACC | |||
| G-CKW47 | AACTTCTGCACCTAAAAACTGTCA | 56.0 | 213-215 |
| TGCTGAGGTAACAGGAATTAAAA | |||
| G-Bcaμ5 | AGTGTTTCTTTCATCTCCACAAGC | 56.0 | 197-201 |
| AGACCACAATCGGACCACATATTC | |||
| G-Bcaμ7 | TAGTTTCTATTTGCACCCAATGGAG | 60.0 | 171-175 |
| CGGTCCTGTCCTTGTGCTGTAA | |||
| G-Bcaμ8 | CCCAAGACTCACAAAACCAGAAAT | 58.0 | 155-159 |
| ATGAAAGAAGAGTTAAACGTGTGCAA | |||
| G-CAUD006 | ATGGTTCTCTGTAGGCAATC | 56.0 | 170-210 |
| TTCTGCTTGGGCTCTTGGA | |||
| G-APH20 | ACCAGCCTAGCAAGCACTGT | 53.0 | 140-150 |
| GAGGCTTTAGGAGAGATTGAAAAA |
In the outbred group of pigeon, we finally screened out 16 microsatellite loci with good polymorphism, several alleles, and typical stutter peaks. All microsatellite locus information is shown in Table 4.
Table 4.
Number of alleles, optimal amplification conditions, and fragment length of 16 alleles for the outbred colony laboratory pigeons.
| Loci | Primer sequence(5′-3′) | Temperature (°C) | Allele range |
|---|---|---|---|
| UU-Cli02 | TGGGCAAGGTACACTTTTAGGT | 61.0 | 158-170 |
| CTTTATGCTCCCCCTTGAGAT | |||
| UU-Cli06 | TTTGAAAAACATGGATTGTGC | 56.0 | 140-145 |
| AATTTGCAGAGGGTGAGTGG | |||
| PG5 | GTTCTTGGTGTTGCATGGATGC | 59.0 | 262-266 |
| AGTTACGAAATGATTGCCAGAAG | |||
| C26L9(1265223) | CAAAGCTGCTGACGTGAATCAA | 59.0 | 467-472 |
| AGAGACGCTCCATGCAAAAG | |||
| UU-Cli14 | CAGAACGTTTTGTTCTGTTTGG | 58.0 | 265-292 |
| TCTTGCTGCAGTCTTCATCC | |||
| C12L1(532572) | GTTGTTTGGCTGAGTGGACG | 62.0 | 126-136 |
| TCAACCAGGGGAATTGGCAG | |||
| C12L4(906353) | GCTGCTGTCTTCTTCATTGGG | 60.0 | 210-250 |
| TTAAAACCTCCCGTCTCCCTG | |||
| CliμD11 | CCAATCCCAAAGAGGATTAT | 58.0 | 78-98 |
| ACTGTCCTATGGCTGAAGTG | |||
| C26L10(1404758) | GCTGTCAGGTATCAGCCACAA | 59.0 | 211-226 |
| TCAGACCCACGAAAGCTGTAA | |||
| C26L4(568923) | CAACCCCATGTGGGTGAGAC | 63.0 | 357-432 |
| CACCACCACGTGGGACATC | |||
| PG4 | CCCATCTCCTGCCTGATGC | 64.0 | 136-170 |
| CACAGCAGGATGCTGCCTGC | |||
| UU-Cli12 | CGCCAGACTGTATTGTGAGC | 61.0 | 231-265 |
| AGCATGGCTGTTCTTTGAGG | |||
| CliμT47 | ATGTGTGTTTGTGCATGAAG | 56.0 | 183-214 |
| ATGAAAGCCTGTTAGTGGAA | |||
| CliμD32 | GAGCCATTTCAGTGAGTGACA | 60.0 | 136-158 |
| GTTTGCAGGAGCGTGTAGAGAAGT | |||
| UU-Cli07 | GCTGCCTGTTACTACCTGAGC | 61.0 | 277-310 |
| CTGGCCATGAAATGAACTCC | |||
| C26L1(20390) | AGGAGCCTACACTGGGTTTTC | 60.0 | 250-268 |
| TGTAGCTCTGCAATCAGCCT |
3.1.3. Analysis of Population Microsatellite Loci
We inputted the results of STR scanning into Popgene 3.2 to analyze experimental chicken in the outbred groups and the haplotypes at 29 loci. In the outbred groups, 28 microsatellite loci show a high degree of polymorphism, and the average number of observed alleles is 4.571. The average number of effective alleles is 3.270, and the average Shannon's information index is 1.198 (Table 5). Furthermore, the average effective heterozygosity is 0.492. The average polymorphism information content (PIC) is 0.610. All these data indicate a good genetic diversity of screening loci in the outbred groups and large heterozygosity difference among the laboratory experimental chicken populations.
Table 5.
Number of alleles, effective alleles, effective heterozygosity, PIC, and Shannon's index of the outbred colony chicken samples.
| Loci | Observed number of alleles | Effective number of alleles | Shannon's information index | Effective heterozygosity | PIC |
|---|---|---|---|---|---|
| MCW0029 | 4 | 2.931 | 1.209 | 0.579 | 0.603 |
| GGNCAMZO | 2 | 1.069 | 0.146 | 0.060 | 0.062 |
| ADL0293 | 5 | 3.200 | 1.311 | 0.573 | 0.634 |
| ADL0317 | 7 | 5.236 | 1.768 | 0.554 | 0.783 |
| GGAVIR | 3 | 1.916 | 0.796 | 0.456 | 0.408 |
| ADL0201 | 5 | 2.103 | 1.013 | 0.429 | 0.482 |
| GCT0016 | 5 | 3.042 | 1.274 | 0.337 | 0.618 |
| ADL0304 | 6 | 4.641 | 1.627 | 0.666 | 0.751 |
| MCW0402 | 8 | 6.042 | 1.881 | 0.702 | 0.813 |
| MCW0063 | 7 | 4.319 | 1.626 | 0.568 | 0.736 |
| ADL185 | 5 | 3.204 | 1.359 | 0.614 | 0.647 |
| GGMYC | 2 | 1.800 | 0.637 | 0.427 | 0.346 |
| LEI0094 | 6 | 3.674 | 1.468 | 0.562 | 0.683 |
| LEI0074 | 4 | 3.707 | 1.348 | 0.597 | 0.681 |
| ADL328 | 3 | 2.785 | 1.058 | 0.526 | 0.565 |
| GGVITC | 1 | 1.000 | 0.000 | 0.000 | 1.000 |
| GGANTECL | 3 | 2.897 | 1.080 | 0.600 | 0.580 |
| LEI094 | 6 | 4.444 | 1.579 | 0.690 | 0.738 |
| MCW0330 | 4 | 3.232 | 1.269 | 0.577 | 0.637 |
| LEI0141 | 4 | 3.162 | 1.229 | 0.341 | 0.623 |
| ADL0292 | 3 | 2.793 | 1.061 | 0.475 | 0.568 |
| GGVITIIG | 2 | 1.965 | 0.684 | 0.460 | 0.371 |
| MCW0087 | 8 | 5.930 | 1.898 | 0.544 | 0.810 |
| MCW0347 | 3 | 1.948 | 0.815 | 0.447 | 0.419 |
| ADL176 | 9 | 4.846 | 1.858 | 0.522 | 0.773 |
| ADL166 | 5 | 3.729 | 1.380 | 0.574 | 0.682 |
| MCW0014 | 5 | 4.342 | 1.543 | 0.592 | 0.735 |
| GGCYMA | 3 | 1.603 | 0.632 | 0.317 | 0.322 |
| Mean | 4.571 | 3.270 | 1.198 | 0.492 | 0.610 |
In the other 3 haplotype populations, 14 microsatellite loci showed monomorphism in each population but showed different lengths in different haplotype populations. The average number of observed alleles is 1.571. The average number of effective alleles, the average Shannon's information index, and the average effective heterozygosity are 1.433, 0.316, and 0.207, respectively (Table 6). The specific data of each haplotype population is shown in Supplementary Tables 1–3.
Table 6.
Number of alleles, effective alleles, effective heterozygosity, and Shannon's index of the haplotype chicken samples.
| Loci | Observed number of alleles | Effective number of alleles | Shannon's information index | Effective heterozygosity |
|---|---|---|---|---|
| GGNCAMZO | 1 | 1.000 | 0.000 | 0.000 |
| GGAVIR | 2 | 1.923 | 0.673 | 0.480 |
| MCW0402 | 1 | 1.000 | 0.000 | 0.000 |
| MCW0063 | 1 | 1.000 | 0.000 | 0.000 |
| ADL185 | 3 | 2.174 | 0.898 | 0.540 |
| GGMYC | 1 | 1.000 | 0.000 | 0.000 |
| LEI0094 | 3 | 2.778 | 1.055 | 0.640 |
| GGVITC | 1 | 1.000 | 0.000 | 0.000 |
| ADL0292 | 2 | 1.471 | 0.500 | 0.320 |
| GGVITIIG | 2 | 2.000 | 0.693 | 0.500 |
| ADL166 | 1 | 1.000 | 0.000 | 0.000 |
| MCW0014 | 1 | 1.000 | 0.000 | 0.000 |
| GGCYMA | 1 | 1.000 | 0.000 | 0.000 |
| STMSGGHU2-1A | 2 | 1.724 | 0.611 | 0.420 |
| Mean | 1.571 | 1.434 | 0.316 | 0.207 |
In the outbred group of duck, 25 microsatellite loci show polymorphism. The average number of observed alleles is 7.520, and the average number of effective alleles in the population is 4.162. The average Shannon's information index is 1.574, and the average effective heterozygosity is 0.683. The average PIC is 0.698. These data showed that in the outbred groups, the genetic diversity of microsatellite DNA loci is better, and the genetic diversity of each locus is quite different. The specific results are shown in Table 7.
Table 7.
Number of alleles, effective alleles, effective heterozygosity, PIC, and Shannon's index of outbred colony duck samples.
| Loci | Observed number of alleles | Effective number of alleles | Shannon's information index | Effective heterozygosity | PIC |
|---|---|---|---|---|---|
| CMO211 | 8 | 4.628 | 1.698 | 0.764 | 0.752 |
| CAUD011 | 9 | 5.024 | 1.835 | 0.799 | 0.775 |
| CAUD027 | 9 | 3.698 | 1.588 | 0.654 | 0.696 |
| APH09 | 8 | 4.840 | 1.728 | 0.756 | 0.763 |
| AY314 | 12 | 7.285 | 2.165 | 0.806 | 0.848 |
| AY258 | 9 | 3.503 | 1.586 | 0.700 | 0.684 |
| CAUD018 | 4 | 2.941 | 1.194 | 0.640 | 0.596 |
| CAUD031 | 8 | 4.459 | 1.711 | 0.730 | 0.746 |
| CAUD026 | 7 | 4.674 | 1.697 | 0.750 | 0.757 |
| CAUD023 | 7 | 2.725 | 1.315 | 0.584 | 0.591 |
| CMO212 | 8 | 4.154 | 1.642 | 0.739 | 0.724 |
| CAUD006 | 4 | 3.333 | 1.280 | 0.440 | 0.645 |
| CAUD004 | 7 | 5.556 | 1.834 | 0.720 | 0.798 |
| CAUD001 | 6 | 5.000 | 1.696 | 0.600 | 0.772 |
| CAUD034 | 10 | 3.943 | 1.742 | 0.730 | 0.723 |
| CAUD007 | 8 | 3.894 | 1.639 | 0.714 | 0.713 |
| APL579 | 7 | 3.068 | 1.412 | 0.635 | 0.636 |
| CAUD010 | 6 | 4.655 | 1.630 | 0.768 | 0.753 |
| CAUD028 | 5 | 3.549 | 1.378 | 0.541 | 0.668 |
| CAUD012 | 7 | 3.122 | 1.354 | 0.652 | 0.630 |
| CAUD035 | 10 | 5.768 | 1.922 | 0.759 | 0.804 |
| CAUD014 | 9 | 3.600 | 1.448 | 0.696 | 0.672 |
| CAUD032 | 14 | 6.159 | 2.120 | 0.797 | 0.821 |
| APH11 | 2 | 1.923 | 0.673 | 0.479 | 0.365 |
| APL2 | 4 | 2.556 | 1.067 | 0.609 | 0.529 |
| Mean | 7.520 | 4.162 | 1.574 | 0.683 | 0.698 |
In 4 haplotype populations, 15 microsatellite loci show monomorphism in each population. The average number of observed alleles is 4.133, the average number of effective alleles is 2.863, and the average Shannon's information index is 1.153, indicating that the genetic diversity of the loci in these haplotype populations is poor; the average effective heterozygosity is 0.500, indicating that the heterozygosity difference is small and the genetic information of the selected loci is relatively single. See Table 8 for more detailed information, and the specific data in each haplotype population is shown in Supplementary Tables 4–7.
Table 8.
Number of alleles, effective alleles, effective heterozygosity, and Shannon's index of haplotype duck samples.
| Loci | Observed number of alleles | Effective number of alleles | Shannon's information index | Effective heterozygosity |
|---|---|---|---|---|
| CAUD002 | 3 | 2.020 | 0.857 | 0.360 |
| CAUD006 | 4 | 2.740 | 1.142 | 0.540 |
| CAUD018 | 3 | 1.802 | 0.746 | 0.400 |
| CAUD005 | 5 | 3.945 | 1.490 | 0.551 |
| APL579 | 5 | 2.632 | 1.205 | 0.500 |
| APH18 | 7 | 4.301 | 1.655 | 0.640 |
| CAUD010 | 3 | 2.597 | 1.010 | 0.420 |
| CAUD028 | 2 | 1.980 | 0.688 | 0.360 |
| CAUD012 | 3 | 2.597 | 1.010 | 0.420 |
| CAUD035 | 4 | 3.756 | 1.353 | 0.605 |
| CAUD014 | 4 | 3.509 | 1.306 | 0.580 |
| CAUD026 | 4 | 2.740 | 1.142 | 0.520 |
| CMO212 | 5 | 3.774 | 1.458 | 0.640 |
| AY258 | 4 | 2.353 | 1.063 | 0.500 |
| CAUD034 | 6 | 2.198 | 1.164 | 0.460 |
| Mean | 4.133 | 2.863 | 1.153 | 0.500 |
In the outbred colony of experimental goose, 14 loci were selected. The average number of observed alleles, the average number of effective alleles, the average Shannon's information index, the average effective heterozygosity, and the PIC are 4.714, 3.038, 1.195, 0.528, and 0.582, respectively. The microsatellite loci have large interindividual differences within the population, and the population has high gene stability (Table 9).
Table 9.
Number of alleles, effective alleles, effective heterozygosity, PIC, and Shannon's index of outbred colony goose samples.
| Loci | Observed number of alleles | Effective number of alleles | Shannon's information index | Effective heterozygosity | PIC |
|---|---|---|---|---|---|
| G-Ans17 | 4 | 1.843 | 0.775 | 0.441 | 0.388 |
| G-TTUCG1 | 3 | 2.255 | 0.943 | 0.381 | 0.494 |
| G-APH13 | 4 | 1.605 | 0.752 | 0.315 | 0.352 |
| G-Ans02 | 8 | 5.389 | 1.837 | 0.749 | 0.790 |
| G-Ans07 | 4 | 3.073 | 1.220 | 0.634 | 0.613 |
| G-Ans18 | 3 | 2.208 | 0.922 | 0.309 | 0.481 |
| G-Ans25 | 4 | 3.333 | 1.282 | 0.629 | 0.647 |
| G-Hhiμ1b | 4 | 2.965 | 1.147 | 0.471 | 0.594 |
| G-CKW47 | 4 | 3.143 | 1.238 | 0.573 | 0.623 |
| G-Bcaμ5 | 3 | 2.728 | 1.051 | 0.469 | 0.562 |
| G-Bcaμ7 | 6 | 2.731 | 1.158 | 0.455 | 0.562 |
| G-Bcaμ8 | 7 | 2.845 | 1.290 | 0.635 | 0.599 |
| G-CAUD006 | 4 | 3.704 | 1.344 | 0.602 | 0.680 |
| G-APH20 | 8 | 4.713 | 1.772 | 0.734 | 0.761 |
| Mean | 4.714 | 3.038 | 1.195 | 0.528 | 0.582 |
The selected microsatellite loci all show good polymorphism in the experimental outbred pigeon populations. A total of 16 loci were selected. The average number of observed alleles is 7.875. The average effective allele number is 4.554; the average Shannon's information index and the average effective heterozygosity are 1.559 and 0.649. The average PIC is 0.674 (Table 10).
Table 10.
Number of alleles, effective alleles, effective heterozygosity, PIC, and Shannon's index of outbred colony pigeon samples.
| Loci | Observed number of alleles | Effective number of alleles | Shannon's information index | Effective heterozygosity | PIC |
|---|---|---|---|---|---|
| UU-Cli02 | 5 | 3.613 | 1.374 | 0.694 | 0.672 |
| UU-Cli06 | 4 | 2.921 | 1.163 | 0.383 | 0.593 |
| PG5 | 2 | 1.681 | 0.595 | 0.397 | 0.323 |
| C26L9(1265223) | 4 | 2.576 | 1.076 | 0.602 | 0.533 |
| UU-Cli14 | 10 | 5.144 | 1.923 | 0.787 | 0.784 |
| C12L1(532572) | 4 | 2.810 | 1.118 | 0.487 | 0.575 |
| C12L4(906353) | 11 | 6.375 | 2.052 | 0.766 | 0.825 |
| CliμD11 | 7 | 4.541 | 1.682 | 0.734 | 0.750 |
| C26L10(1404758) | 11 | 9.118 | 2.281 | 0.860 | 0.880 |
| C26L4(568923) | 13 | 5.854 | 2.062 | 0.807 | 0.812 |
| PG4 | 10 | 6.847 | 2.017 | 0.767 | 0.836 |
| UU-Cli12 | 8 | 2.825 | 1.364 | 0.623 | 0.599 |
| CliμT47 | 7 | 3.492 | 1.413 | 0.658 | 0.666 |
| CliμD32 | 9 | 6.695 | 1.991 | 0.807 | 0.833 |
| UU-Cli07 | 5 | 1.352 | 0.592 | 0.252 | 0.251 |
| C26L1(20390) | 16 | 7.014 | 2.244 | 0.759 | 0.844 |
| Mean | 7.875 | 4.554 | 1.559 | 0.649 | 0.674 |
3.1.4. Population Genetic Structure Analysis
Among the three outbred chicken groups, the mean number of observed alleles, the mean number of effective alleles, the mean Shannon's information index, and the mean effective heterozygosity are shown in Table 11. All these data are the highest in the Beijing oil chicken, indicating the best gene diversity.
Table 11.
Comparison of mean observed allele number, mean effective allele number, mean Shannon's index, and mean effective heterozygosity among the outbred colonies of chickens.
| Colonies | Mean observed number of alleles | Mean effective number of alleles | Mean Shannon's information index | Mean effective heterozygosity |
|---|---|---|---|---|
| BWEL | 2.857 | 2.024 | 0.730 | 0.424 |
| BM | 2.857 | 2.132 | 0.802 | 0.485 |
| Beijing oil chicken | 4.464 | 2.821 | 1.088 | 0.569 |
In the haplotype chicken populations, the highest mean observed number of alleles is observed in G7groups. Haplotype G7 has the highest mean effective allele number and the highest mean Shannon's information index. The mean effective heterozygosity of haplotype G7 is 0.364. The genetic heterozygosity of the 3 populations is very low, and the consistency is good (Table 12).
Table 12.
Comparison of mean observed allele number, mean effective allele number, mean Shannon's index, and mean effective heterozygosity among the haplotype chickens.
| Colonies | Mean observed number of alleles | Mean effective number of alleles | Mean Shannon's information index | Mean effective heterozygosity |
|---|---|---|---|---|
| G1 | 1.571 | 1.434 | 0.316 | 0.207 |
| G2 | 1.643 | 1.409 | 0.335 | 0.224 |
| G7 | 2.000 | 1.626 | 0.548 | 0.364 |
In the two outbred groups of duck, the mean number of observed alleles, the mean effective number of alleles, the mean Shannon's index, and the mean effective heterozygosity of outbred group 1 are higher than those of outbred group JD, indicating that outbred group 1 had better diversity. The results are shown in Table 13. Among the 4 haplotype populations, the highest mean number of alleles is observed in haplotype A. Haplotype A has the highest mean Shannon's information index. The highest mean effective heterozygosity in the duck groups is 0.489 in haplotype A (Table 14). The genetic heterozygosity of 4 populations is in good agreement.
Table 13.
Comparison of mean observed allele number, mean effective allele number, mean Shannon's index, and mean effective heterozygosity among the outbred colonies of ducks.
| Colonies | Mean observed number of alleles | Mean effective number of alleles | Mean Shannon's information index | Mean effective heterozygosity |
|---|---|---|---|---|
| 1 | 6.320 | 3.518 | 1.410 | 0.685 |
| JD | 5.280 | 3.466 | 1.335 | 0.680 |
Table 14.
Comparison of mean observed allele number, mean effective allele number, mean Shannon's index, and mean effective heterozygosity among the haplotype ducks.
| Colonies | Mean observed number of alleles | Mean effective number of alleles | Mean Shannon's information index | Mean effective heterozygosity |
|---|---|---|---|---|
| A | 2.400 | 2.022 | 0.760 | 0.489 |
| B | 2.333 | 2.029 | 0.745 | 0.484 |
| C | 2.400 | 1.912 | 0.726 | 0.459 |
| D | 2.333 | 1.944 | 0.701 | 0.442 |
In the two outbred groups of goose, the mean number of observed alleles, the mean effective number of alleles, and the mean Shannon's index of Guangdong Wuzong goose are higher than those of Yangzhou goose, indicating that Guangdong Wuzong goose has a better diversity (Table 15).
Table 15.
Comparison of mean observed allele number, mean effective allele number, mean Shannon's index, and mean effective heterozygosity among the outbred colonies of geese.
| Colonies | Mean observed number of alleles | Mean effective number of alleles | Mean Shannon's information index | Mean effective heterozygosity |
|---|---|---|---|---|
| Guangdong Wuzong | 4.000 | 2.769 | 1.112 | 0.618 |
| Yangzhou | 3.714 | 2.155 | 0.802 | 0.439 |
The analysis of the two main experimental pigeon populations used for scientific research shows that the mean effective heterozygosity of two populations is 0.647 and 0.651, respectively. The mean number of observed alleles, the mean effective number of alleles, and the mean Shannon's index are higher in white king pigeons than in silver king pigeons. The comparison of the data is shown in Table 16.
Table 16.
Comparison of mean observed allele number, mean effective allele number, mean Shannon's index, and mean effective heterozygosity among the outbred colonies of pigeons.
| Colonies | Mean observed number of alleles | Mean effective number of alleles | Mean Shannon's information index | Mean effective heterozygosity |
|---|---|---|---|---|
| Silver king | 6.125 | 3.260 | 1.307 | 0.647 |
| White king | 7.375 | 4.247 | 1.435 | 0.651 |
4. Discussion
Poultries are widely used and are indispensable supporting conditions for the life sciences and biomedicine industries. Specific pathogen-free (SPF) chicken embryos are used in the manufacture and quality control of biological product [4]; ducks play an important role in the research of avian influenza, fatty liver, duck hepatitis A, and duck hepatitis B [5–7]; goose blood contains a higher concentration of immunoglobulin, which is often used in pharmacology and toxicology research [8]; pigeons belong to the class of birds and are considered as important animal model in avian influenza research [9]. With the increasing demand for experiment poultry, people are paying more attention to the genetic structure analysis and genetic quality control. However, the current methods of genetic structure analysis and genetic quality control for experimental poultry animals are insufficient.
Coat colour gene testing method, biochemical marker gene testing method, immune marker gene testing method, and DNA molecular marker method are popular methods for genetic monitoring. Microsatellite DNA, mitochondrial DNA (mtDNA), restriction fragment length polymorphism (PCR-RFLP), single-stranded conformation polymorphism (PCR-SSCP), and specific gene polymorphisms are commonly used DNA molecular marker methods [24–27]. Among them, microsatellite DNA has become valuable tools for evaluating population genetic diversity due to their unique virtue.
Microsatellite DNA is characterized by short tandem repeats (STRs) of 1 to 6 nucleotides in eukaryotic genome with a random manner [28]. It has rich polymorphism and large genetic information. Microsatellite can be used to distinguish heterozygous from homozygous because of their codominant inheritance feature [29]. In previous studies, microsatellites have been used as biomarkers for monitoring rodent genetic traits [30, 31]. With the deep understanding of microsatellites, it plays a more important role in genetic monitoring for being simple, clear, and stable in operation. In this research, we screened out microsatellite loci with suitable length and high specificity as candidate loci by gel electrophoresis firstly. Then, we performed STR scanning on these candidate loci. Microsatellite loci with good polymorphism, abundant alleles in the outbred groups, and good monomorphism in the haplotype populations were selected to form the microsatellite marker system. We analyzed the average effective allele number, average Shannon's index, average effective heterozygosity, and other analytical indices to estimate genetic variation in different groups.
The mean effective number of alleles is an indicator of genetic variation and mutation drift balance. In our study, Beijing oil chicken has the highest mean effective allele number of three outbred chicken populations; outbred duck group 1 has higher mean effective allele number than outbred duck group JD. The outbred goose group Guangdong Wuzong and outbred pigeon group white king have the highest mean number of effective alleles in outbred goose populations and outbred pigeon populations, respectively. The higher mean effective number of alleles indicates that the population can maintain the original gene and avoid new variations under the pressures from genetic drift and artificial selection. The results show that Beijing oil chicken, outbred duck group 1, Guangdong Wuzong goose, and white king pigeon are the most stable strains in the outbred group of experiment chicken, duck, goose, and pigeon groups in this research, respectively.
The mean effective heterozygosity of a population is an important indicator of population genetic diversity and can reflect the richness of the detected genes. It is generally believed that when the mean effective heterozygosity of the population is less than 0.5, it indicates that the individual differences in the population are small and the genetic heterozygosity is low, which does not conform to the genetic characteristics of an outbred group animal. When the mean effective heterozygosity of the population is higher than 0.7, its genetic diversity is high [32].
Hence, we found that the mean effective heterozygosity of BWEL, BM, and Beijing oil chicken groups is all greater than 0.5, which conforms to the characteristics of the outbred group. The mean effective heterozygosity of BWEL and BM chicken groups is nearly 0.5. The average effective heterozygosity of G1, G2, and G7 groups is all less than 0.5. It is also consistent with the background that BWEL, BM, and Beijing oil chickens are outbred colonies; Beijing oil chicken has abundant genetic diversity and high selection potential for it has the highest mean effective heterozygosity among the outbred chicken groups in this study. This may be due to the large population. Duck group 1 and JD duck all have a mean effective heterozygosity greater than 0.680 which indicates a high genetic diversity. The mean effective heterozygosity of Guangdong Wuzong goose group, silver king pigeon group, and white king pigeon group is all greater than 0.5 which reflects abundant genetic diversity. The mean effective heterozygosity of three haplotype chicken groups and four haplotype duck groups is 0.207 and 0.500, respectively. The result indicates a good consistency in haplotype chickens and ducks. This may be the result of long-term full-sib and half-sib reproduction. Chickens and ducks are more widely used in biological research, and the breeding standards are stricter, while geese and pigeons are more useful in agriculture. Haplotype chickens have lower mean effective heterozygosity than haplotype duck populations, which is consistent with a longer history of breeding in experimental chickens.
When measuring the degree of gene variation, PIC is often used as a variation index. It is generally believed that when PIC is between 0.25 and 0.5, it is moderately polymorphic, and <0.25 shows a low level of polymorphism, when PIC is greater than 0.5, it means a high level of polymorphism [33]. In our microsatellite marker system, most of the microsatellite sites have a PIC greater than 0.5 that show high polymorphism. All these data prove that our microsatellite marker system provides rich genetic information, which can be used as effective genetic markers. In our study, highly polymorphic microsatellite marker systems showed powerful markers for quantifying genetic variations within and between poultry populations. We will collect more samples to make a more accurate description of genetic structure of the Chinese experimental chickens, ducks, geese, and pigeons in the future [34].
5. Conclusions
In conclusion, we identified appropriate microsatellite marker systems for native experimental chickens, ducks, geese, and pigeons in China. The combination of loci selected in our research provides a good choice for genetic monitoring of the quality and the population genetic diversity of poultry stocks.
Acknowledgments
We are very grateful to the Institute of Animal Science, Chinese Academy of Agricultural Sciences, Harbin Veterinary Research Institute, Southern Medical University and Yangzhou University for providing animal samples for this study. This work was supported by the Beijing Municipal Science and Technology Projects (No. D181100000518002), Support Project of High-level Teachers in Beijing Municipal Universities in the Period of 13th Five-year Plan (Grant Number IDHT20170516), and the National Key Research and Development Plan of China (No. 2017YFD0501602).
Contributor Information
Changlong Li, Email: licl@ccmu.edu.cn.
Zhenwen Chen, Email: czwen@ccmu.edu.cn.
Data Availability
All data, models, and code generated or used during the study appear in the submitted article.
Conflicts of Interest
The authors declare that there is no conflict of interest regarding the publication of this paper.
Supplementary Materials
Supplementary Table 1: number of alleles, effective alleles, effective heterozygosity, and Shannon's index of the G1 haplotype chicken population. Supplementary Table 2: number of alleles, effective alleles, effective heterozygosity, and Shannon's index of the G2 haplotype chicken population. Supplementary Table 3: number of alleles, effective alleles, effective heterozygosity, and Shannon's index of the G7 haplotype chicken population. Supplementary Table 4: number of alleles, effective alleles, effective heterozygosity, and Shannon's index of the A haplotype duck population. Supplementary Table 5: number of alleles, effective alleles, effective heterozygosity, and Shannon's index of the B haplotype duck population. Supplementary Table 6: number of alleles, effective alleles, effective heterozygosity, and Shannon's index of the C haplotype duck population. Supplementary Table 7: number of alleles, effective alleles, effective heterozygosity, and Shannon's index of the D haplotype duck population.
References
- 1.Neff E. P. What is a lab animal? Lab Anim (NY) 2018;47(9):223–227. doi: 10.1038/s41684-018-0135-3. [DOI] [PubMed] [Google Scholar]
- 2.Bello M. B., Yusoff K., Ideris A., Hair-Bejo M., Peeters B. P. H., Omar A. R. Diagnostic and vaccination approaches for Newcastle disease virus in poultry: the current and emerging perspectives. BioMed Research International. 2018;2018:18. doi: 10.1155/2018/7278459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dequéant M. L., Pourquié O. Chicken genome: new tools and concepts. Developmental dynamics : an official publication of the American Association of Anatomists. 2005;232(4):883–886. doi: 10.1002/dvdy.20266. [DOI] [PubMed] [Google Scholar]
- 4.Yin H. B., Chen C. H., Darre M. J., Donoghue A. M., Donoghue D. J., Venkitanarayanan K. Phytochemicals reduce aflatoxin-induced toxicity in chicken embryos. Poultry Science. 2017;96(10):3725–3732. doi: 10.3382/ps/pex190. [DOI] [PubMed] [Google Scholar]
- 5.Ramey A. M., Reeves A. B., Drexler J. Z., et al. Influenza A viruses remain infectious for more than seven months in northern wetlands of North America. Proceedings of the Royal Society B: Biological Sciences. 2020;287(1934):p. 20201680. doi: 10.1098/rspb.2020.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fu Y., Pan M., Wang X., Xu Y., Yang H., Zhang D. Molecular detection and typing of duck hepatitis A virus directly from clinical specimens. Veterinary Microbiology. 2008;131(3-4):247–257. doi: 10.1016/j.vetmic.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 7.Pan Q., Liu L., Wang Y., et al. The first whole genome sequence and pathogenicity characterization of a fowl adenovirus 4 isolated from ducks associated with inclusion body hepatitis and hydropericardium syndrome. Avian Pathology. 2017;46(5):571–578. doi: 10.1080/03079457.2017.1311006. [DOI] [PubMed] [Google Scholar]
- 8.Huang T., Wu K., Yuan X., et al. Molecular analysis of the immunoglobulin genes in goose. Developmental and Comparative Immunology. 2016;60:160–166. doi: 10.1016/j.dci.2016.02.024. [DOI] [PubMed] [Google Scholar]
- 9.Kwon J. H., Noh Y. K., Lee D. H., et al. Experimental infection with highly pathogenic H5N8 avian influenza viruses in the mandarin duck (Aix galericulata) and domestic pigeon (Columba livia domestica) Veterinary Microbiology. 2017;203:95–102. doi: 10.1016/j.vetmic.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 10.Nunome M., Kinoshita K., Ishishita S., Ohmori Y., Murai A., Matsuda Y. Genetic diversity of 21 experimental chicken lines with diverse origins and genetic backgrounds. Experimental Animals. 2019;68(2):177–193. doi: 10.1538/expanim.18-0139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strillacci M. G., Cozzi M. C., Gorla E., et al. Genomic and genetic variability of six chicken populations using single nucleotide polymorphism and copy number variants as markers. Animal : an international journal of animal bioscience. 2017;11(5):737–745. doi: 10.1017/s1751731116002135. [DOI] [PubMed] [Google Scholar]
- 12.Khatib H., Genislav E., Crittenden L. B., Bumstead N., Soller M. Sequence-tagged microsatellite sites as markers in chicken reference and resource populations. Animal Genetics. 1993;24(5):355–362. doi: 10.1111/j.1365-2052.1993.tb00340.x. [DOI] [PubMed] [Google Scholar]
- 13.Gao C., Han L., Han J., et al. Establishment of six homozygous MHC- _B_ haplotype populations associated with susceptibility to Marek 's disease in Chinese specific pathogen-free BWEL chickens. Infection, Genetics and Evolution. 2015;29:15–25. doi: 10.1016/j.meegid.2014.10.031. [DOI] [PubMed] [Google Scholar]
- 14.Seo D., Bhuiyan M. S., Sultana H., Heo J. M., Lee J. H. Genetic diversity analysis of south and east Asian duck populations using highly polymorphic microsatellite markers. Asian-Australasian Journal of Animal Sciences. 2016;29(4):471–478. doi: 10.5713/ajas.15.0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stringham S. A., Mulroy E. E., Xing J., et al. Divergence, convergence, and the ancestry of feral populations in the domestic rock pigeon. Current biology : CB. 2012;22(4):302–308. doi: 10.1016/j.cub.2011.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seo J. H., Lee J. H., Kong H. S. Assessment of genetic diversity and phylogenetic relationships of Korean native chicken breeds using microsatellite markers. Asian-Australasian Journal of Animal Sciences. 2017;30(10):1365–1371. doi: 10.5713/ajas.16.0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rudresh B. H., Murthy H. N. N., Jayashankar M. R., Nagaraj C. S., Kotresh A. M., Byregowda S. M. Microsatellite based genetic diversity study in indigenous chicken ecotypes of Karnataka. Veterinary world. 2015;8(8):970–976. doi: 10.14202/vetworld.2015.970-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang Y., Tu J., Cheng X., et al. Characterization of 35 novel microsatellite DNA markers from the duck (Anas platyrhynchos) genome and cross-amplification in other birds. Genetics, selection, evolution : GSE. 2005;37(5):455–472. doi: 10.1186/1297-9686-37-5-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su Y., Long R., Chen G., Wu X., Xie K., Wan J. Genetic analysis of six endangered local duck populations in China based on microsatellite markers. Journal of Genetics and Genomics. 2007;34:1010–1018. doi: 10.1016/s1673-8527(07)60114-3. [DOI] [PubMed] [Google Scholar]
- 20.Lee J. C.-I., Tsai L.-C., Kuan Y.-Y., et al. Racing pigeon identification using STR and chromo-helicase DNA binding gene markers. Electrophoresis. 2007;28(23):4274–4281. doi: 10.1002/elps.200700063. [DOI] [PubMed] [Google Scholar]
- 21.Andres K., Kapkowska E. Applicability of anatid and galliform microsatellite markers to the genetic diversity studies of domestic geese (Anser anser domesticus) through the genotyping of the endangered zatorska breed. BMC Research Notes. 2011;4(1):1–10. doi: 10.1186/1756-0500-4-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parada R., Ksiazkiewicz J., Kawka M., Jaszczak K. Studies on resources of genetic diversity in conservative flocks of geese using microsatellite DNA polymorphic markers. Molecular Biology Reports. 2012;39(5):5291–5297. doi: 10.1007/s11033-011-1327-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lebas N. R., Spencer P. B. Polymorphic microsatellite markers in the ornate dragon lizard, Ctenophorus ornatus. Molecular ecology. 2000;9(3):365–366. doi: 10.1046/j.1365-294x.2000.00874.x. [DOI] [PubMed] [Google Scholar]
- 24.Barbaric I., Stewart M., Wells S., Dear T. N. A new coat color mouse line for testing germline transmission of embryonic stem cells while retaining an inbred genetic background. Journal of the American Association for Laboratory Animal Science. 2007;46(3):37–40. [PubMed] [Google Scholar]
- 25.Weitzmann M. N., Woodford K. J., Usdin K. The mouse Ms6-hm hypervariable microsatellite forms a hairpin and two unusual tetraplexes. The Journal of Biological Chemistry. 1998;273(46):30742–30749. doi: 10.1074/jbc.273.46.30742. [DOI] [PubMed] [Google Scholar]
- 26.Bulfield G., Bantin G. Genetic monitoring of inbred strains of mice using electrophoresis and electrofocusing. Laboratory Animals. 1981;15(2):147–149. doi: 10.1258/002367781780958973. [DOI] [PubMed] [Google Scholar]
- 27.Fahey J. R., Katoh H., Malcolm R., Perez A. V. The case for genetic monitoring of mice and rats used in biomedical research. Mammalian genome : official journal of the International Mammalian Genome Society. 2013;24(3-4):89–94. doi: 10.1007/s00335-012-9444-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y. C., Korol A. B., Fahima T., Beiles A., Nevo E. Microsatellites: genomic distribution, putative functions and mutational mechanisms: a review. Molecular Ecology. 2002;11(12):2453–2465. doi: 10.1046/j.1365-294x.2002.01643.x. [DOI] [PubMed] [Google Scholar]
- 29.Grover A., Sharma P. C. Development and use of molecular markers: past and present. Critical Reviews in Biotechnology. 2014;36(2):290–302. doi: 10.3109/07388551.2014.959891. [DOI] [PubMed] [Google Scholar]
- 30.Liu X., Yu X., Xu Y., et al. Development of an effective microsatellite marker system to determine the genetic structure of Meriones meridianus populations. Experimental Animals. 2020;69(2):224–232. doi: 10.1538/expanim.19-0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Du X. Y., Li W., Sa X. Y., et al. Selection of an effective microsatellite marker system for genetic control and analysis of gerbil populations in China. Genetics and Molecular Research. 2015;14(3):11030–11042. doi: 10.4238/2015.September.21.16. [DOI] [PubMed] [Google Scholar]
- 32.Chakraborty R., Zhong Y., de Andrade M., Clemens P. R., Fenwick R. G., Caskey C. T. Linkage disequilibria among (CA)n polymorphisms in the human dystrophin gene and their implications in carrier detection and prenatal diagnosis in Duchenne and Becker muscular dystrophies. Genomics. 1994;21(3):567–570. doi: 10.1006/geno.1994.1315. [DOI] [PubMed] [Google Scholar]
- 33.Vanhala T., Tuiskula-Haavisto M., Elo K., Vilkki J., Mäki-Tanila A. Evaluation of genetic variability and genetic distances between eight chicken lines using microsatellite markers. Poultry Science. 1998;77(6):783–790. doi: 10.1093/ps/77.6.783. [DOI] [PubMed] [Google Scholar]
- 34.Pritchard J. K., Stephens M., Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: number of alleles, effective alleles, effective heterozygosity, and Shannon's index of the G1 haplotype chicken population. Supplementary Table 2: number of alleles, effective alleles, effective heterozygosity, and Shannon's index of the G2 haplotype chicken population. Supplementary Table 3: number of alleles, effective alleles, effective heterozygosity, and Shannon's index of the G7 haplotype chicken population. Supplementary Table 4: number of alleles, effective alleles, effective heterozygosity, and Shannon's index of the A haplotype duck population. Supplementary Table 5: number of alleles, effective alleles, effective heterozygosity, and Shannon's index of the B haplotype duck population. Supplementary Table 6: number of alleles, effective alleles, effective heterozygosity, and Shannon's index of the C haplotype duck population. Supplementary Table 7: number of alleles, effective alleles, effective heterozygosity, and Shannon's index of the D haplotype duck population.
Data Availability Statement
All data, models, and code generated or used during the study appear in the submitted article.


