SUMMARY
Dysregulation of the gut microbiome has been implicated in the progression of nonalcoholic fatty liver disease (NAFLD) to advanced fibrosis and cirrhosis. To determine the diagnostic capacity of this association, stool microbiomes were compared across 163 well-characterized participants encompassing non-NAFLD controls, NAFLD-cirrhosis patients and their first-degree relatives. Interrogation of shotgun metagenomic and untargeted metabolomic profiles using the Random Forest machine learning algorithm and differential abundance analysis identified discrete metagenomic and metabolomic signatures that were similarly effective in detecting cirrhosis (diagnostic accuracy 0.91, AUC). Combining the metagenomic signature with age and serum albumin levels accurately distinguished cirrhosis in etiologically and genetically distinct cohorts from geographically separated regions. Additional inclusion of serum aspartate aminotransferase levels, which are increased in cirrhosis patients, enabled discrimination of cirrhosis from earlier stages of fibrosis. These findings demonstrate that a core set of gut microbiome species may offer universal utility as a non-invasive diagnostic test for cirrhosis.
INTRODUCTION
Nonalcoholic fatty liver disease (NAFLD) is the leading cause of chronic liver disease worldwide and is estimated to affect 24% of the global population (Loomba and Sanyal, 2013; Younossi et al., 2018). NAFLD encompasses a spectrum of diseases prognostically sub-categorized into non-progressive nonalcoholic fatty liver (NAFL), progressive nonalcoholic steatohepatitis (NASH), and NAFLD-cirrhosis. NAFLD-cirrhosis, the most advanced form of the disease, is a major risk factor for hepatocellular carcinoma and the second leading indication for liver transplantation in the United States (Wong et al., 2014). Accurate, non-invasive tests to identify patients at greatest risk for advanced NAFLD are urgently needed.
A link between NAFL and the gut microbiome is supported by several studies (Da Silva et al., 2018; Hoyles et al., 2018; Raman et al., 2013; Sharpton et al., 2019). Consistent with this, we previously demonstrated that a gut microbiome-based metagenomic signature can differentiate between mild or moderate and advanced fibrosis in patients with biopsy-proven NAFLD, as well as defined a stool microbiome signature that accurately detected cirrhosis (UCSD twin and family cohort with and without NAFLD (Caussy et al., 2019b; Loomba et al., 2017)). However, given the influences of both host and environmental factors on the gut microbiome (Li et al., 2014), the universal applicability of microbiome-based diagnostic signatures was not known. Here, we define a gut microbiome signature for NAFLD-cirrhosis determined from metagenomic and metabolomic characterizations of stool microbiomes from a twin and family cohort comprised of NAFLD-cirrhosis probands and their first-degree relatives. Notably, this signature accurately identified patients with cirrhosis in 2 independent cohorts that included multiple cirrhotic etiologies: Chinese cohort (Qin et al., 2014) and Italian cohort (Iebba et al., 2018). The remarkable robustness of this signature in identifying cirrhosis across geographically and culturally distinct populations reinforces the intimate association of the gut and the liver, and demonstrates its potential utility as a diagnostic tool.
RESULTS
Characterization of the Study Population
The prospective cohort comprised 163 well-characterized participants with diagnoses that covered the spectrum of non-alcoholic fatty liver disease (NAFLD), including non-NAFLD controls, NAFLD-cirrhosis patients as well as their first-degree relatives. Detailed demographic, clinical, biochemical, and metabolic profiles of the entire cohort are provided in Figure S1A, and Tables 1 and S1.
Table 1:
Baseline characteristics between (i) non-NAFLD control group and (ii) NAFLD-cirrhosis group
| Characteristics | non-NAFLD group (N = 54) (i) (s.d.) | NAFLD-cirrhosis group (N = 27) (ii) (s.d.) | Wilcoxon p-value (i↔ii) |
|---|---|---|---|
| Demographics | |||
| Age (years) | 45.85 (19.86) | 64.74 (9.80) | <0.001 |
| Female (n%) | 39 (72.2%) | 22 (81.5%) | |
| White (n%) | 43 (79.6%) | 9 (33.3%) | |
| Hispanic (n%) | 8 (14.8%) | 15 (55.6%) | |
| BMI (kg/m2) | 26.07 (6.83) | 32.85 (10.06) | <0.001 |
| Clinical | |||
| Type 2 Diabetes (n%) | 1 (1.9% ) | 23 (85.2% ) | |
| Biological data | |||
| AST (U/L) | 22.31 (8.44) | 46.26 (23.11) | <0.001 |
| ALT (U/L) | 18.81 (8.84) | 44.3 (36.86) | <0.001 |
| Alk P (U/L) | 68.17 (19.04) | 112.19 (39.63) | <0.001 |
| GGT (Ui/L) | 17.63 (8.02) | 97.04 (57.71) | <0.001 |
| Total Bilirubin (mg/dL) | 0.51 (0.25) | 2.33 (6.63) | <0.01 |
| Direct Bilirubin (mg/dL) | 0.12 (0.04) | 1.36 (5.21) | <0.001 |
| Albumin (g/dL) | 4.53 (0.33) | 4.06 (0.61) | <0.001 |
| Glucose (mg/dl) | 86.09 (9.04) | 132.07 (61.85) | <0.001 |
| Hemoglobin A1c (%) | 5.65 (0.32) | 7.06 (1.88) | <0.001 |
| Insulin (U/ml) | 8.79 (6.35) | 43.52 (33.06) | <0.001 |
| Triglycerides (mg/dL) | 81.50 (44.73) | 147.42 (136.45) | <0.001 |
| Total cholesterol (mg/dL) | 188.57 (40.94) | 162.83 (64.23) | <0.05 |
| HDL-cholesterol (mg/dL) | 69.87 (19.89) | 49.17 (18.49) | <0.001 |
| LDL-cholesterol (mg/dL) | 102.43 (32.17) | 79.3 (32.42) | <0.01 |
| Platelet count (103/μL) | 246.94 (48.54) | 149.11 (72.73) | <0.001 |
| Prothrombin time | 10.56 (0.69) | 13.04 (3.66) | <0.001 |
| INR | 1.02 (0.07) | 1.22 (0.35) | <0.01 |
| Ferritin (ng/mL) | 103.89 (82.9) | 132.79 (123.19) | n.s. |
| Imaging data | |||
| MRI-PDFF (%) | 2.41 (0.86) | 5.16 (4.11) | <0.01 |
| MRE (kPa) | 2.13 (0.37) | 5.04 (2.68) | <0.001 |
The proband cohort (UCSD Cohort) comprised 27 prospectively recruited patients with NAFLD-cirrhosis (81.5% female; 16 with compensated cirrhosis, 11 with decompensated cirrhosis) and 54 non-NAFLD controls without liver disease (72.2% female). NAFLD diagnosis was based upon the American Association for the Study of Liver Diseases (AASLD) practice guidelines, and the presence of cirrhosis was confirmed by liver biopsy or imaging. The mean age and body-mass-index of the control group were lower than the NAFLD-cirrhosis group (45.9 ± 19.9 versus 64.7 ± 9.8 years, and 26.1 ± 6.8 versus 32.8 ± 10.1 kg/m2, respectively). Demographics, clinical history, and biochemical and imaging parameters are reported in Table 1.
Advanced magnetic resonance imaging (MRI) techniques were employed for liver phenotyping including MRI proton density fat fraction (MRI-PDFF) for fat content and magnetic resonance elastography (MRE) to measure liver stiffness as a correlative of fibrosis. Compared to the control group, the NAFLD-cirrhosis group had higher liver fat content (2.4 ± 0.9 % versus 5.2 ± 4.1%, p < 0.01; Table 1) and liver stiffness (2.13 ± 0.37 kPa versus 5.04 ± 2.68 kPa, p < 0.001; Table 1). Detailed associations between MRI-PDFF and MRE and demographics, metabolic risk factors, and biochemical and metabolic parameters are shown in Figure S1A–C.
Taxonomic Profiling
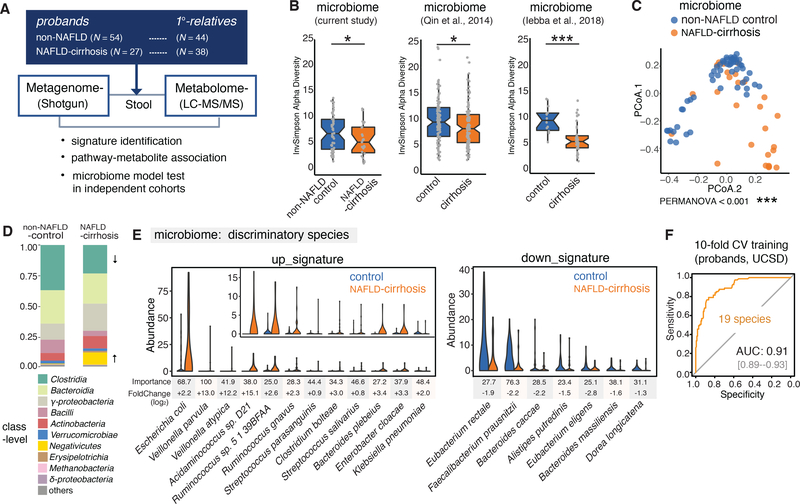
The compositions of the microbial communities and the abundance of microbial pathways were determined from shotgun sequencing of stool samples using Metagenomic Phylogenetic Analysis (MetaPhlAn2, (Segata et al., 2012) and HMP Unified Metabolic Analysis Network 2 (HUMAnN2, (Franzosa et al., 2018) respectively. In total, 1,216,335 gene-families and assembled sequences for 356 species were identified, and after removal of unannotated species, 310 microbial species representing 53 families and 115 genera were identified in the 163 patient samples (Figure 1A). Using the Inverse Simpson index to measure microbial richness, we found α-diversity to be decreased in the NAFLD-cirrhosis compared to the control group, consistent with a previous report (Figure 1B and S1D), (Caussy et al., 2019b). Similar enteric dysbiosis has been reported in patients with cirrhosis attributed to multiple etiologies (Figure 1B) (Iebba et al., 2018; Qin et al., 2014). Interestingly, the α-diversity in NAFLD-cirrhosis correlated with clinical parameters including levels of low-density lipoprotein (LDL), blood coagulation as measured by prothrombin time (PT) and international normalized ratio (INR), and glucose homeostasis reported by insulin levels (Figure S1E). In addition, a clear separation could be seen between NAFLD-cirrhosis and the non-NAFLD probands using a principal coordinate analysis (PCoA) coupled with weighted UniFrac, indicating altered β-diversity (Figure 1C; PERMANOVA < 0.001).
Figure 1. Major alterations in the gut microbiome profiles of the NAFLD-cirrhosis group compared to the non-NAFLD control group.
(A) Study overview depicting the study cohort, sample collection, and stool metagenomic and metabolomic analyses.
(B) Inverse Simpson α-diversity scores highlighted significant decreases in the richness of gut microbiota from the NAFLD-cirrhosis group (N = 27) compared to the non-NAFLD control group (N = 54; P < 0.05). Significant α-diversity differences were also observed between non-NAFLD control group and NAFLD-cirrhosis patients in cohorts from China and Italy (P < 0.05 and P < 0.001, respectively). Gray dots represent values for individual participants. Boxes represent the interquartile range (IQR) between the first and third quartiles. Median values are represented by horizontal lines within the boxes. Notches represent 95% confidence intervals for the medians. Whiskers indicate the range from minimum (first quartile – 1.5*IQR) to maximum (third quartiles + 1.5*IQR). The estimate_richness function of phyloseq was utilized for this analysis. *P < 0.05, ***P < 0.001. T-test was used to determine significance.
(C) Principal coordinate analysis demonstrating significant separation between stool samples from NAFLD-cirrhosis and non-NAFLD control groups, using weighted-UniFrac distances. Blue dots represent individual non-NAFLD control participants. Orange dots represent individual NAFLD-cirrhosis patients. PERMANOVA was performed to determine significance (P < 0.001).
(D) Stacked bar plots depicting class-level differences in gut microbiome composition between the NAFLD-cirrhosis and non-NAFLD control groups. The “Other” subcategory included viruses, fungi and rare species (< 1%).
(E) Relative abundances of top discriminatory microbial species for the prediction of NAFLD-cirrhosis. Violin plots depict the relative abundances of the top 19 discriminatory species identified by Random Forest (RF) machine learning in the NAFLD-cirrhosis and non-NAFLD control groups. Species were chosen from the highest scores of Mean Decrease in Gini using RF feature selection. Importance scores in the RF classification model and fold-change levels in log2 scale are noted below the plot for each species.
(F) Receiver operating characteristic (ROC) curve of the RF model using 19 discriminatory species in the UCSD proband cohort including 27 NAFLD-cirrhosis and 54 non-NAFLD control stool samples. Random Forest (RF) method was used with train function of R’s caret package. For training set, 10-fold cross-validation (CV) was applied with trainControl function. To compute and visualize AUC from ROC outcome, the pROC package was utilized.
Major differences in the gut microbial communities between the NAFLD-cirrhosis and control probands were observed at taxonomical levels from phylum down to family (Figure S1F–H). Notable compositional shifts in NAFLD-cirrhosis probands included an enrichment in Negativicutes and a reduction in Clostridia classes (Figure 1D). These class level shifts are consistent with previous studies of microbial populations in patients with advanced fibrosis due to NAFLD or cirrhosis of multiple etiologies (Iebba et al., 2018; Loomba et al., 2017; Qin et al., 2014).
A Metagenomic-derived Gut Microbiome Signature for NAFLD-cirrhosis
Notable correlations were observed between microbial species and clinical metadata associated with NAFLD-cirrhosis (Figure S2). Of the 310 microbial species identified, 108 showed significant correlation with metadata in the proband cohort. For example, the levels of Veillonella parvula inversely correlated with albumin and platelet counts, parameters decreased in cirrhosis (Figure S2). In contrast, species enriched in the control group including Eubacterium eligens showed a positive correlation with albumin and platelet counts (Figure S2). The sharp dichotomy observed in associations between certain species altered in diseased probands and clinical metadata led to the hypothesis that a selection of gut microbial organisms may be indicative of the disease state. To explore this notion, Random Forest (RF) machine learning was employed to identify key discriminatory species of NAFLD-cirrhosis. Specifically, differentially abundant microbial species were detected using the DESeq2 tool, and a NAFLD-cirrhosis signature identified using the RF classifier for feature selection. Remarkably, this approach identified a gut microbiome signature comprised of 19 discriminatory species that accurately detected NAFLD-cirrhosis in the proband cohort (area under the receiver operating curve [AUC] of 0.91, Figure 1E and 1F).
The identified disease signature included increases in the levels of Veillonella parvula, Veillonella atypica, Ruminococcus gnavus, Clostridium bolteae and Acidaminococcus sp. D21 accompanied by decreases in the abundances of Eubacterium eligens, Eubacterium rectale and Faecalibacterium prausnitzii (Figure 1E). Notably, despite its low abundance, Veillonella parvula was the most discriminatory adverse species for the association with NAFLD-cirrhosis. A second member of the class Negativicutes, Acidaminococcus sp. D21 levels were 15-fold higher in the NAFLD-cirrhosis group. In contrast, Eubacterium rectale and Faecalibacterium prausnitzii are members of the class Clostridia which decreased with NAFLD severity, and have previously been associated with NAFLD and cirrhosis (Loomba et al., 2017; Qin et al., 2014). Faecalibacterium prausnitzii is a beneficial commensal species with anti-inflammatory functions that are decreased in several intestinal and metabolic disorders including inflammatory bowel disease (IBD), colorectal cancer (CRC), obesity, celiac disease as well as cirrhosis from multiple etiologies (Olsson et al., 2019; Qin et al., 2014; Zaarour et al., 2019). In our dataset, it is the most critical beneficial species for discriminating between the NAFLD-cirrhosis and control groups. Consistent with the RF model at the species level, the genera with the highest discriminatory values included Veillonella and Faecalibacterium in the up- and down-signatures, respectively (Figure S3A).
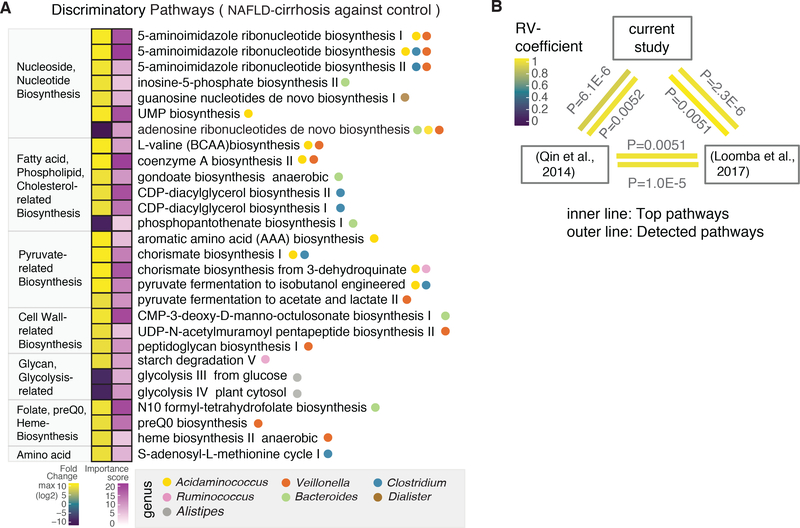
Metagenomic sequencing facilitated the identification of 188 individual strains, of which 53 showed differential abundance between groups, as well as correlations with clinically important parameters (Figure S3B and 3C). Dorea longicatena (GCF_000154065), reduced 1.3-fold in NAFLD-cirrhosis, and Acidaminococcus sp. D21 (GCF_000174215), enriched 15-fold, were the most important reduced and enriched strains for predicting disease status (Figure S3C). Of note, Veillonella dispar (GCF_000160015) levels were 8.6-fold higher in the NAFLD-cirrhosis compared to the control probands (adjusted P = 4.79 E-43). In addition, 7,355 functional pathways were identified, of which 39 differed between the NAFLD-cirrhosis and control probands including those involved in the biosynthesis of aromatic amino acids (AAA), branched-chain amino acids (BCAA), fatty acids, and nucleotides (Figure 2A). An association of dysregulated pathways with specific microbial species and genera highlighted Acidaminococcus and Veillonella, both of which have been associated with AAA and BCAA synthesis. Importantly, these discriminatory pathways correlated with clinical metadata, and are consistently dysregulated in independent cohorts from China and San Diego (Figure 2B and S4). These findings suggest that dysregulation of essential microbial metabolic processes may contribute to disease progression in NAFLD-cirrhosis.
Figure 2. Microbial functional pathways altered in cirrhosis.
(A) Microbial pathways associated with NAFLD-cirrhosis and non-NAFLD control groups were identified using the HUMAnN2 tool, which includes an analysis of microbial gene-families. Discriminatory pathways were selected using feature selection by Random Forest (RF) and differential abundance analyses through the computation of mean decrease in Gini and fold-change. Left column denotes fold-change, based on log2 scale. In the fold-change color scale, yellow represents microbial pathways that were increased in the NAFLD-cirrhosis group compared to the non-NAFLD control group. Purple represents pathways that were decreased in the NAFLD-cirrhosis group compared to the non-NAFLD control group. Right column denotes importance of the pathway in the RF model, based on Mean Decrease in Gini score. In the associated color scale, pathway importance is represented by a gradient going from light purple to dark purple. Circles denote specific genera that significantly associate with discriminatory functional pathways.
(B) A multivariate RV-coefficient analysis was performed with HUMAnN2 pathway outcomes from three different datasets including the current study, Chinese cohort and our previous study. The statistic tool, the FactorMineR package, was utilized to calculate RV-coefficient scores among cohorts. The color of the inner line represents the significant of top discriminatory pathways shown in Figure 2A. Outer line represents the significance of pattern alteration from all detected pathways.
Metabolite Abundances Associated with NAFLD-cirrhosis
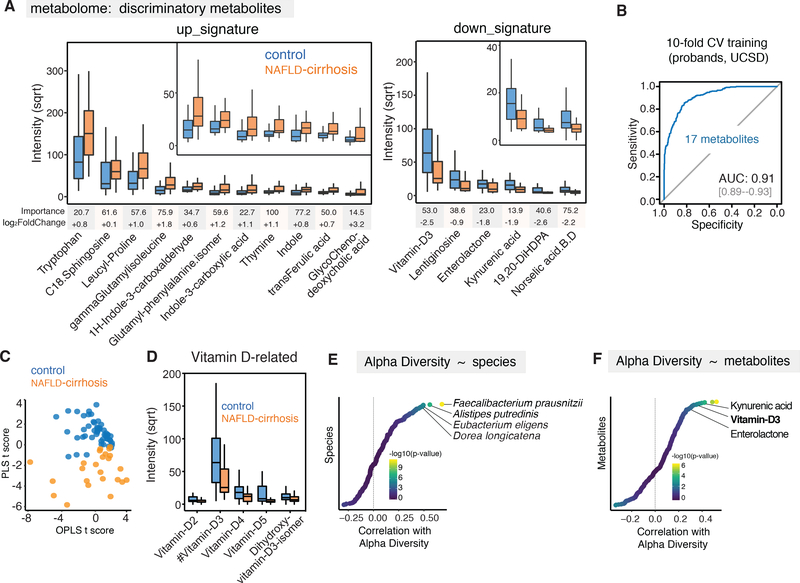
To further refine the disease signature, we quantified the abundances of 435 distinct stool metabolites in NAFLD-cirrhosis and control probands, of which 75 associated with clinically significant metadata in NAFLD-cirrhosis (Figure S5) (Lai et al., 2018; Tsugawa et al., 2015). Feature selection using RF machine learning combined with differential expression/abundance revealed 17 metabolites that, in combination, were able to accurately detect NAFLD-cirrhosis (AUC; 0.91, Figure 3A and 3B). Consistent with this, the levels of these 17 metabolites were sufficient to discriminate NAFLD-cirrhosis from control probands in a principal component analysis (Figure 3C). Metabolites with the greatest predictive power for identifying NAFLD-cirrhosis included those involved in metabolism of aromatic amino acids (AAA), branched chain amino acids (BCAA), bile acids, and Vitamin D. Tryptophan and related metabolites such as indole and kynurenic acid also featured prominently in the metabolomic signature. Products of microbial dissimilation of tryptophan (indole and indole-related compounds) were increased, while the metabolite kynurenic acid was reduced in NAFLD-cirrhosis samples, resulting in an overall increase in tryptophan levels (Figure 3A). NAFLD-cirrhosis samples were enriched in leucine and phenylalanine derivatives such as leucyl-proline, γ-glutamyl-isoleucine, and glutamyl-phenylalanine isomer (Figure 3A). Glutamyl-phenylalanine isomer, the leading discriminatory metabolite, was previously reported to be elevated in serum samples from non-alcoholic steatohepatitis (NASH) patients (Kalhan et al., 2011). These observations are intriguing given past studies that have shown gut microbial sources of aromatic and branched chain amino acid derivatives influence intestinal permeability, systemic immunity, and liver steatosis (Dodd et al., 2017; Hoyles et al., 2018).
Figure 3. Identification of discriminatory metabolites for cirrhosis.
(A) 17 discriminatory metabolites were identified in NAFLD-cirrhosis group versus non-NAFLD control group. Metabolites were chosen from the highest scores of Mean Decrease in Gini using Random Forest (RF) feature selection. Importance scores in the RF classification model and fold-change levels in square root (sqrt) scale are presented below the plot for each metabolite. Boxes represent the interquartile range (IQR) between the first and third quartiles. Median values are represented by horizontal lines within the boxes. Notches represent 95% confidence intervals for the medians. Whiskers indicate the range from minimum (first quartile - 1.5*IQR) to maximum (third quartiles + 1.5*IQR).
(B) ROC curve of the RF model using 17 discriminatory metabolites in the UCSD proband cohort. RF method was used with train function of R’s caret package. For training set, 10-fold cross-validation (CV) was applied with trainControl function.
(C) Discriminative power of significant metabolites using a multivariate model, Orthogonal PLS (OPLS). R’s ropls package was utilized to compute Partial Least Squares (PLS) and OPLS scores. The scatter plot was generated with the ggplot2.
(D) Decreased abundance of vitamin D derivatives in NAFLD-cirrhosis group relative to non-NAFLD control group. Decreased intensity of vitamin D2–5 and dihydroxy-vitamin D3 isomer in square root scale were shown using the boxplots.
(E) Correlation analysis between relative abundance of species and α-diversity using Spearman’s test. Statistic outcome was shown with gradient colors in -log10(P-value) scale. Correlation r scores were presented in x-axis.
(F) Correlation analysis between metabolites and α-diversity using Spearman’s test. Statistic outcome was shown with gradient colors in -log10(P-value) scale. Correlation r scores were presented in x-axis.
C18-Sphingosine and the bile acid glycochenodeoxycholic acid (GCDCA) were also enriched in NAFLD-cirrhosis samples (Figure 3A). Interestingly, GCDCA is a ligand for the farnesoid X receptor (FXR), an important target in experimental NASH therapeutics. In addition, vitamin D and its derivatives were reduced in NAFLD-cirrhosis stools (Figure 3F). Given that vitamin D deficiency has been linked to many cancers including hepatocellular carcinoma (HCC), and the high risk for HCC among patients with cirrhosis, this finding warrants further exploration of a possible causative association.
It is worth noting that several microbial species (e.g., Faecalibacterium prausnitzii, Alistipes putredinis, Eubacterium eligens, Dorea longicatena) and key metabolites including kynurenic acid, vitamin D3, and enterolactone strongly correlate with α-microbial diversity, suggesting a potential role for these discriminatory species and metabolites in maintaining microbiome richness (Figure 3D and 3E).
Association of Microbes and Metabolites with NAFLD-cirrhosis
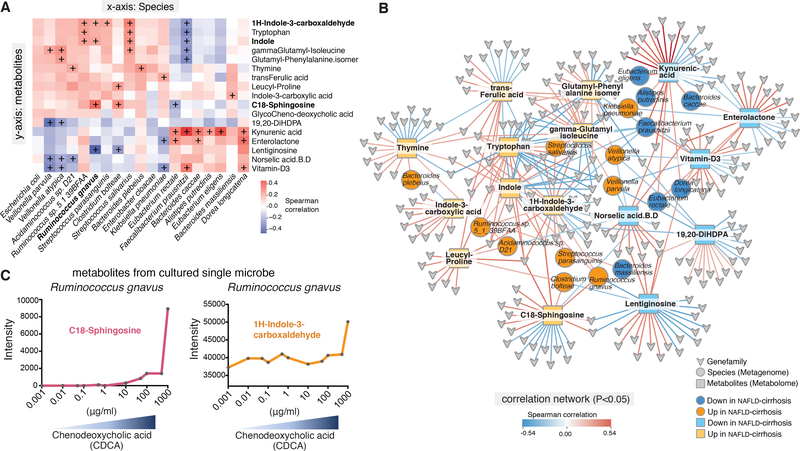
Microbial metabolites have been tightly linked to pathophysiological processes contributing to NAFLD, IBD and colon cancer (Perry et al., 2016). In study probands, correlated abundances between key discriminatory microbial species and metabolites were observed (Figure 4A). For example, levels of C18-Sphingosine correlated with increased abundances of Ruminococcus gnavus. In contrast, levels of kynurenic acid positively correlated with Faecalibacterium prausnitzii (Figure S6A and S6B). To further examine extended network links, abundances of microbial gene-families from discriminatory species were evaluated for associations with key metabolite levels. The resulting network contained 33 nodes and 338 edges representing significant microbial gene-families, metagenome-derived species, and metabolites (Figure 4B).
Figure 4. Interaction network of metagenomic and metabolomic features for cirrhosis.
(A) Key discriminatory microbial species show significant associations with specific stool metabolites. The heatmap depicts correlative relationships between 19 discriminatory microbial species and 17 metabolites identified in the NAFLD-cirrhosis and non-NAFLD control groups. Color scale represents Spearman correlation coefficients. Red denotes strong positive correlations. Blue denotes strong negative correlations. Correlation r scores were visualized with gradient colors using the ggplot2 package of R. + denotes p < 0.05.
(B) Network map depicting relationships between species, metabolites, and microbial gene-families that are significantly associated with either the up- or down-signature in the NAFLD-cirrhosis group (P < 0.05). Color scale represents the Spearman correlation coefficient. Red denotes positive associations between components of the network; blue denotes negative associations. Species nodes are represented as circles. Orange denotes species that are significantly associated with the up-signature in the NAFLD-cirrhosis group. Dark blue denotes species that are significantly associated with the down-signature in the NAFLD-cirrhosis group. Metabolite nodes are represented as squares. Yellow denotes metabolites that are significantly associated with the up-signature in the NAFLD-cirrhosis group. Light blue denotes metabolites that are significantly associated with the down-signature in the NAFLD-cirrhosis group. Microbial gene-family nodes are represented as arrowheads. Interactions were visualized using Cytoscape.
(C) Anaerobic culturing of Ruminococcus gnavus with increased chenodeoxycholic acid (CDCA) demonstrates increased production of discriminatory metabolites (e.g., C18-Sphingosine and 1H-Indole-3-carboxaldehyde).
To in part validate these associations, we sought to confirm the link between increased C18-Spinghosine and Ruminococcus gnavus (Figure 4A and B). Ruminococcus gnavus was cultured under anaerobic conditions in the presence of increasing concentrations of chenodeoxycholic acid (CDCA) to approximate the increased bile acid levels seen in NAFLD-cirrhosis patients (Acharya and Bajaj, 2019), and the levels of metabolites including aromatic and branched-chain amino acids determined by LC-MS/MS. Consistently, the levels of C18-Sphingosine and 1H-Indole-3-carboxaldehyde produced by Ruminococcus gnavus increased with CDCA concentrations (Figure 4C). The data in Figures 4A and B support altered metabolite production as a plausible mechanism by which changes in the gut microbiome are communicated to the liver. An important direction for future studies is to define the relationships between liver disease and key microbial species and metabolites.
Validation of Microbiome Cirrhosis Signature
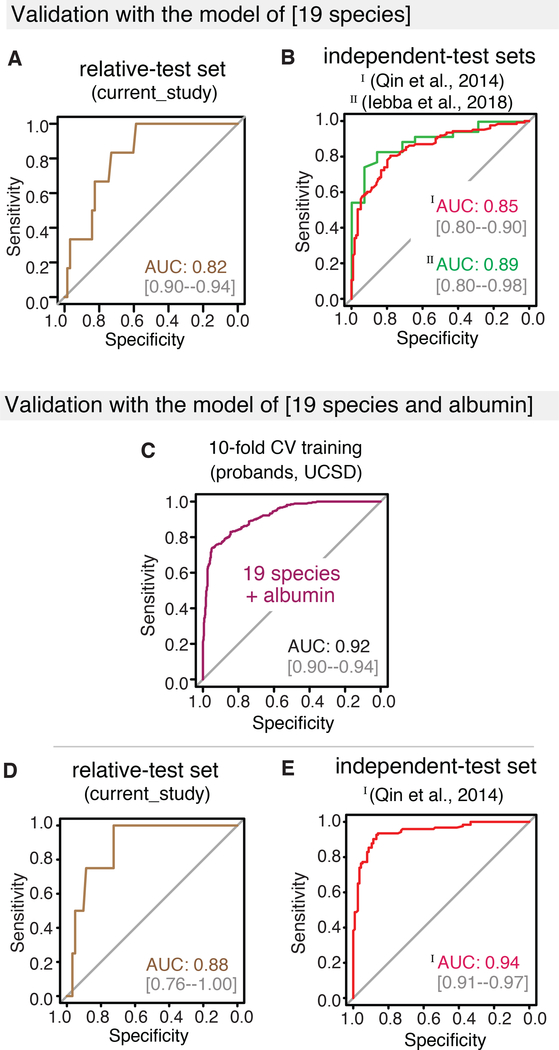
To evaluate the utility of the metagenomic gut microbiome signature for the detection of cirrhosis, we tested its diagnostic accuracy in the first-degree relative cohort comprised of 51 non-NAFLD, 21 NAFLD without advanced fibrosis, 3 NAFLD-related fibrosis, and 7 NAFLD-cirrhosis participants (Table S1). In this cohort, the combination of our microbiome signature with proband age (AUC 0.91 in training cohort, Figure S6C) identified cirrhosis with a diagnostic accuracy of AUC 0.88 (Figure 5A). Subsequently, we determined its utility in published studies that included diverse disease etiologies (alcoholic liver disease, hepatitis B, hepatitis C, and NAFL) (Qin, 2014, Iebba, 2018). Interrogation of the data from an Italian cohort (16S rRNA sequencing data from 46 cirrhotic patients and 14 healthy age-matched controls) using the trained model identified cirrhosis with a diagnostic accuracy of AUC 0.89 (Figure 5B) (Iebba, 2018). Similarly, reanalysis of a quantitative metagenomic study conducted in a Han Chinese population (123 diseased patients and 114 healthy controls) identified cirrhosis with comparable accuracy to that reported with their “15 gene marker” signature (AUC 0.86 and 0.84, respectively; Figure 5B). Given the array of clinical data normally available for patients presenting with possible cirrhosis, we explored including BMI and serum markers such as albumin, alanine aminotransferase (ALT), aspartate aminotransferase (AST), bilirubin, cholesterol, gamma-glutamyltransferase (GGT), glucose, insulin, and platelet count to improve the diagnostic accuracy of the microbiome and age signature. In sensitivity analysis, several clinical parameters greatly improved the diagnostic accuracy of the signature, thereby identifying cirrhosis (Figure S6D). For example, combining the 19 discriminatory species and age with serum albumin levels, available in both current and Chinese studies, improved the diagnostic accuracy in the proband cohort to AUC 0.92 (Figure S6E). Notably, this combination signature showed improved diagnostic accuracy in both our relative and the Chinese cohorts (AUC 0.91 and 0.95, respectively, Figure 5C and 5D).
Figure 5. Validation of the machine learning RF model from stool metagenome for cirrhosis.
(A) Validation of the RF model using 19 discriminatory species and age in the UCSD relatives cohort for NAFLD-cirrhosis. ROC curve shows the diagnostic accuracy of the RF model in identifying cirrhosis in the UCSD relatives cohort. The 19-discriminatory species and age identified NAFLD-cirrhosis with a robust accuracy of AUC 0.88. RF modeling was implemented using the train function in R’s caret package. To compute and visualize AUC from ROC outcome, the pROC package was utilized.
(B) External validation of the RF model in geographically independent cohorts of patients with cirrhosis. ROC curves show the diagnostic accuracy of the RF model in identifying cirrhosis in cohorts from China (123 cirrhosis patients and 114 controls; total N=237) and Italy (35 cirrhosis and 14 controls; total N = 49). The 19-discriminatory species and age identified cirrhosis of multiple etiologies with robust accuracy. For Chinese cohort (red line) the AUC was 0.86. Validation in Italian dataset was performed with 19 species, due to limitation of demographic data. For the Italian cohort (green line), the AUC was 0.89.
(C-D) Validation of the RF model using 19 discriminatory species, age and serum albumin in the UCSD relative and Chinese cohorts. ROC curves show the diagnostic accuracy of a 22-feature RF model combining 19-microbial species, age, and serum albumin scores in identifying cirrhosis in geographically independent cohorts--the UCSD relative and Chinese cohorts. The addition of serum albumin levels to the 19 discriminatory species improved the model’s diagnostic accuracy in the (C) UCSD relative cohort (AUC 0.91) and (D) Chinese cohort (AUC 0.95).
(E-F) Accuracy of microbiome-based signature to differentiate cirrhosis from fibrosis in a previously-described mixed fibrosis cohort. ROC curves show the diagnostic accuracy of an RF model combining 19-microbial species and age in identifying cirrhosis in a cohort of NAFLD patients with mixed fibrosis stages (stage 0 [NAFL, N = 36]; stages 1–3 [mid fibrosis, N = 41]; stage 4 [cirrhosis, N = 9]).
(G-H) Effect of including serum liver damage marker AST on the accuracy of discriminating cirrhosis from fibrosis in the mixed cohort described in (E-F).
(I-J) Relative abundances of signature microbial species with disease progression. Data from the current NAFLD-cirrhosis study (27 cirrhosis) was combined with the mixed fibrosis cohort described in (E-F) and analyzed using Kruskal-Wallis ANOVA test, fast zero-inflated negative binomial mixed model (FZINBMM) and DESeq2. * p < 0.05, ** p < 0.01, *** p < 0.001.
Model Validation in the Mixed Fibrosis Cohort
Given its clinical importance, we next ran a sensitivity analysis to determine whether our combination signature could discriminate cirrhosis from fibrosis. To explore this possibility, we interrogated a previously described cohort of biopsy-proven NAFLD patients that included 36 patients with fibrosis stage 0 (NAFL), 41 patients with fibrosis stages 1–3 (mild or moderate fibrosis) and 9 patients with stage 4 fibrosis (cirrhosis) (Loomba et al., 2017). Encouragingly, the model was able to distinguish cirrhosis from NAFL and mild or moderate fibrosis (AUC 0.85 and AUC 0.84, respectively; Figure 5E and 5F). Furthermore, incorporation of serum AST levels, which are increased in cirrhosis patients, both improved the signature’s accuracy in the UCSD cohort (Figure S6F) and markedly improved its ability to distinguish cirrhosis from NAFL and fibrosis (AUC 0.94 and 0.91, respectively; Figure 5G and 5H). To examine the pattern of specific microbial abundance, we further interrogated the previous cohort of fibrosis stage 0–4 by adding 27 cirrhosis of the current study. Significant changes in the relative abundance of individual signature species were seen across NAFL, mild or moderate fibrosis and cirrhosis groups (Kruskal-Wallis ANOVA test, fast zero inflated negative binomial mixed model [FZINBMM] and DESeq2; Figure 5I and 5J). The robustness of this gut microbiome based signature across geographically and culturally distinct populations, as well as in mixed fibrosis patients, attests to its potential utility as a diagnostic approach for cirrhosis detection.
DISCUSSION
Broadly applicable, non-invasive methods for diagnosing cirrhosis are currently not available. Here we identify a gut microbiome-derived signature that, when combined with the patient age, accurately detected cirrhosis in a well-characterized NAFLD-cirrhosis population. Importantly, this microbiome-based signature achieved similar diagnostic accuracies in Chinese and Italian cohorts, where the underlying causes of cirrhosis included viral and alcohol-induced liver damage. Furthermore, combining serum albumin levels with the signature microbial species and patient age in our RF model improved the diagnostic accuracy in both our NAFLD-cirrhosis and the Chinese cohort (AUC of 0.91 and 0.95, respectively). Moreover, this signature was sufficient to distinguish earlier stage fibrosis from cirrhosis.
We demonstrate here that a core gut microbiome signature can identify cirrhosis across geographically separated cohorts, independent of disease etiology and the effects of host genetics and environmental factors on the gut microbiome. The apparent universal nature of our microbiome-derived signature suggests that key microbial species within the signature may play causal roles in the pathophysiology of cirrhosis. Indeed, the abundances of several Veillonella species were commonly altered in patients in all 3 cirrhosis studies. In further support of a potential causal association, a metabolite-derived signature was able to achieve similar diagnostic accuracy, and significant correlations were found between this and the microbiome-derived signature. Moreover, levels of several signature metabolites including tryptophan, leucine derivatives, and kynurenic acid are also altered in a rat model of cirrhosis, suggesting that dysregulation of microbial AAA and BCAA synthesis may affect liver biology (Chang et al., 2017). Epidemiologic principles indicate that the greater the heterogeneity in the validation cohort (i.e., the etiology of cirrhosis, diet, ethnicity) the more generalizable the finding of a universal cirrhosis signature (Guyatt et al., 2000a; Guyatt et al., 2000b). Thus, the finding that our microbiome-derived signature accurately distinguishes liver cirrhosis independent of the aforementioned confounding factors speaks to the universality of this signature. Our multi-omic analyses provide a roadmap for future investigations into causal associations between key microbial species and their metabolites in the development and progression of cirrhosis.
NAFLD is considered a hepatic manifestation of metabolic syndrome and is closely associated with obesity, insulin resistance and type 2 diabetes. Distinct from viral or alcoholic fatty liver driven cirrhosis, NAFLD-cirrhosis is a consequence of accumulated metabolic damage rather than external insults. The finding of a core set of microbial species that are broadly relevant to cirrhosis indicates previously unappreciated commonalities in the hepatic damage response. Alternatively, the severity of liver damage in cirrhosis may result in a convergence of dysbiosis. Further studies in cohorts of single etiologies will be necessary to distinguish these possibilities.
Our finding of a universal microbiome-derived signature lays the foundations for a stool-based diagnostic test for advanced fibrosis or cirrhosis among high-risk individuals. This represents an important advance for the field, because the diagnosis of cirrhosis continues to be challenging due to the limited accuracy of serum markers and the expense and restricted availability of MRE technologies. Rapid, inexpensive methods to identify patients with cirrhosis is important due to their heightened risks for hepatocellular carcinoma and liver failure. Our discovery of a universal gut microbiome-derived signature that accurately identifies cirrhosis, regardless of etiology, has immense potential to improve disease diagnosis, especially in resource-limited settings where hepatologists and/or imaging may not be available. Similar to screening for colon cancer, we believe a diagnostic panel could be developed to detect cirrhosis in high-risk individuals such as patients with type 2 diabetes. In addition, the identification of pathogenic and beneficial microbial species may lead to novel therapies for severe forms of NAFLD.
Limitations of Study
Limitations of this study include the relatively small sizes of the experimental cohorts, the lack of differentiation between compensated and decompensated cases, and limited access to clinical parameters in the Italian and Chinese validation cohorts. In addition, the association between microbial species identified in our study and cirrhosis does not demonstrate causality. Multi-center studies of well-phenotyped patients with mixed stages of hepatic fibrosis will be needed to further validate study findings.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for reagents should be directed and will be fulfilled by the Lead Contact, (Rohit Loomba; roloomba@ucsd.edu)
Materials availability
All reagents generated in this study are available from the Evans lab or various core services. All human data are available from the Loomba laboratory.
Data and Code Availability
The accession numbers for shotgun sequencing datasets reported in this paper are publicly available at the European Genome-Phenome Archive (EGAS000010046). The study was conducted for identifying NAFLD-related biomarkers.
Bioinformatics workflows for metagenomics and data are available at: https://bitbucket.org/biobakery/ and http://bioconductor.org/packages/release/data/experiment/html/curatedMetagenomicData.html.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human Subjects
Study Participants
This study included participants from the Familial Cirrhosis cohort and Twins and Family cohort who were prospectively recruited between December 2011 and December 2017 at the University of California, San Diego (UCSD) NAFLD Research Center. All subjects underwent an exhaustive standardized clinical research visit that included a detailed medical history, physical examination, and testing to rule out other causes of chronic liver diseases (see inclusion and exclusion criteria), and fasting laboratory tests (Caussy et al., 2019a; Caussy et al., 2017; Caussy et al., 2019b; Loomba et al., 2015). All study subjects had also participated in a biobank initiative (Caussy et al., 2019b). At the time of each research visit, subjects provided stool and fasting serum samples, which were collected and immediately stored in a −80°C freezer.
Definition of NAFLD
Participants were considered to have NAFLD if they had hepatic steatosis (MRI-PDFF ≥5%) and no secondary causes of hepatic steatosis due to use of steatogenic medications, other liver diseases, or significant alcohol intake (see Exclusion Criteria). Definition for NAFLD was based upon the American Association for the Study of Liver Study (AASLD) Practice Guidelines (Chalasani et al., 2012).
Definition of cirrhosis and advanced fibrosis
Participants met the criteria for NAFLD-related cirrhosis if they had NAFLD according to the definition above and biopsy-proven cirrhosis (histologic stage 4 fibrosis). For the diagnosis of advanced fibrosis, MRE is the most accurate non-invasive test to date (Dulai et al., 2016; Hsu et al., 2019; Loomba et al., 2014; Park et al., 2016; Sun et al., 2016). We have previously validated that a liver stiffness cut point of > 3.63 kPa on MRE provides an accuracy of 0.92 for the detection of advanced fibrosis. Therefore, advanced fibrosis among first-degree relatives was determined by imaging evidence of nodularity and presence of intra-abdominal varices, other imaging evidence of portal hypertension, or through liver stiffness assessment by MRE with a threshold ≥ 3.63 kPa (Cui et al., 2015; Hsu et al., 2019; Loomba et al., 2014). The presence of cirrhosis was determined by a liver stiffness assessment by MRE with a threshold ≥ 4.69 kPa Hsu et al. If MRE was not performed due to contraindications, then a transient elastography assessment with a VCTE threshold ≥ 11.8 kPa was used as criteria for advanced fibrosis (Hsu et al., 2019).
NAFLD-cirrhosis probands and their first-degree relatives
This study included 27 NAFLD-cirrhosis probands and 38 of their first-degree relatives from the Familial Cirrhosis cohort prospectively recruited at the UCSD NAFLD Research Center (Caussy et al., 2019a; Caussy et al., 2017; Caussy et al., 2019b). NAFLD-cirrhosis probands had documented evidence of NAFLD (see definition of NAFLD) as well as cirrhosis (see definition of cirrhosis), proven by either biopsy or by meeting imaging criteria.
Inclusion criteria for NAFLD-cirrhosis probands and their first-degree relatives
Probands and first-degree relatives were required to be at least 18 years old. Probands were required to have a documented diagnosis of NAFLD-cirrhosis either by liver biopsy or by imaging evidence, as defined by criteria in the protocol. First-degree relatives (sibling, child, or parent) who did not meet any exclusion criteria were also included in the study, if they gave written informed consent. Subjects were included if they were twin, sibling, or parent-offspring pairs, at least 18 years old, and willing and able to complete all research procedures and observations (Cui et al., 2016; Loomba et al., 2015).
Exclusion criteria for NAFLD-cirrhosis probands and their first-degree relatives
Participants were excluded from the study if they met any of the following criteria: significant alcohol intake (>10 g/day in females or >20 g/day in males) for at least 3 consecutive months over the previous 12 months or if the quantity of alcohol consumed could not be reliably ascertained; clinical or biochemical evidence of liver diseases other than NAFLD (e.g., viral hepatitis, HIV, coeliac disease, cystic fibrosis, autoimmune hepatitis); metabolic and/or genetic liver disease (e.g., Wilson’s disease, haemochromatosis, polycystic liver disease, alpha-1-antitrypsin deficiency, dysbetalipoproteinaemia); clinical or laboratory evidence of systemic infection or any other clinical evidence of liver disease associated with hepatic steatosis; use of drugs known to cause hepatic steatosis (e.g., amiodarone, glucocorticoids, methotrexate, L-asparaginase and valproic acid) for at least 3 months in the last past 6 months; history of bariatric surgery; presence of systemic infectious illnesses; females who were pregnant or nursing at the time of the study; contraindications to MRI (e.g., metal implants, severe claustrophobia, body circumference greater than the imaging chamber); any other condition(s) which, based on the principal investigator’s opinion, may significantly affect the participant’s compliance, competence, or ability to complete the study.
Non-NAFLD control probands and their first-degree relatives
The study included 54 non-NAFLD control probands and 44 of their community-dwelling first-degree relatives (i.e., twins, parents, offspring, or siblings) from the previous Twin and Family study (Caussy et al., 2019a; Caussy et al., 2017; Caussy et al., 2019b; Cui et al., 2016; Zarrinpar et al., 2016); 7 NAFLD patients with advanced fibrosis in the Twin and Family study were included into relative cohorts. Twins with no evidence of either NAFLD (MRI-PDFF < 5%) or advanced fibrosis (MRE < 3.63 kPa) were assigned to the non-NAFLD controls subgroup.
Justification for not performing a liver biopsy
Liver biopsy was not used for hepatic fat content and fibrosis assessment of non-NAFLD controls and first-degree relatives. As such individuals were asymptomatic with no suspected liver disease, performing a liver biopsy would have been unethical. A non-invasive, accurate quantitative imaging method was used instead to estimate liver fat and fibrosis. We have previously shown that MRI-PDFF is a highly precise, accurate, and reproducible non-invasive biomarker for the quantification of liver fat content (Noureddin et al., 2013; Reeder, 2013). In addition, MRE is the most accurate, currently available, non-invasive quantitative biomarker of liver fibrosis (Cui et al., 2015; Hsu et al., 2019; Loomba et al., 2014). MRE has been shown to be have excellent diagnostic accuracy in differentiating between normal liver and mild fibrosis (stage 0–2) and between non-advanced fibrosis and advanced fibrosis (stage 3–4) (Hsu et al., 2019; Kim et al., 2013; Yin et al., 2016).
Sample size and power estimation
In our previous study, we identified significant differences between two groups (AUC 0.88) that comprised 16 individuals with NASH-cirrhosis/advanced fibrosis and 33 controls (Loomba et al., 2017). Therefore, the current study, which includes 27 participants with NAFLD-cirrhosis and 54 controls, would be adequate to detect clinically meaningful differences between the sub-groups with a power of at least 80% with an error rate, 0.01.
Patient consent
All subjects provided a written informed consent, and the study protocol was approved by the UCSD Institutional Review Board (approval numbers: UCSD IRB #140084 and #111282). The study complies with all relevant ethical regulations for research with human subjects.
METHOD DETAILS
Clinical assessments and laboratory tests
All study participants underwent a standardized clinical research visit at the UCSD NAFLD Research Center. A detailed medical history was obtained from all participants. A physical exam, which included vital signs, height, weight, and anthropometric measurements, was performed by a trained clinical investigator. Body mass index was defined as the body weight (in kilograms) divided by height (in meters) squared. Alcohol consumption was documented outside clinical visits and confirmed in the research clinic using the Alcohol Use Disorders Identifications Test and the Skinner questionnaire. A detailed history of medications was obtained; no patient took medications known or suspected to cause steatosis or steatohepatitis. Other causes of liver disease and secondary causes of hepatic steatosis were systemically ruled out using detailed history and laboratory data. After completion of the history and physical examination, participants underwent comprehensive fasting laboratory tests including metabolic and liver assessments (Caussy et al., 2019a; Caussy et al., 2018b; Caussy et al., 2017; Caussy et al., 2019b; Cui et al., 2016; Loomba et al., 2015; Zarrinpar et al., 2016). At the time of each research visit, patients provided stool samples, which were collected and immediately stored in a −80°C freezer.
MRI-PDFF assessment
MRI was performed at the UCSD MR3T Research Laboratory using the 3T research scanner (GE Signa EXCITE HDxt; GE Healthcare, Waukesha, WI) with all participants in the supine position. MRI-PDFF was used to measure hepatic fat content, and MRE was used to measure liver fibrosis. For MRI-PDFF, multiple echo sequences are acquired at different times when fat and water signals are nominally in phase or out of phase with each other. Data from each echo time are passed into an algorithm that estimates and corrects T2* effects, models the fat signal as a superposition of multiple frequency components, and estimates fat and water proton densities from which the fat content is calculated. A magnitude-based technique was applied to echo sequences to avoid phase errors, which can adversely affect fat quantification. This algorithm is applied to source images using custom analysis software developed at the UCSD Liver Imaging Group to generate a PDFF parametric map depicting fat quantity and distribution throughout the liver (Patel et al., 2013; Permutt et al., 2012). Image analysts were blinded to all clinical and biochemical data as well as the participants’ study group.
MRE assessment
For MRE, a standard 60-Hz shear-wave was generated by an acoustic passive driver attached to the body wall anterior to the liver and coupled to an acoustic active driver outside the MR examination room. Shear waves were imaged using a two-dimensional gradient-recalled echo MRE pulse sequence with oscillating motion-sensitizing gradients synchronized to the shear wave frequency. Four noncontiguous axial slices (10 mm thick with 10 mm inter-slice gap) were acquired, each during a 16-second breath-hold, through the widest transverse dimension of the liver with short recovery times in between. The acquisition parameters were as follows: repetition time, 50 milliseconds; echo time, 20.2 milliseconds; flip angle, 30 degrees; matrix, 256 × 64; field of view, 48 × 48 cm; one-signal average; receiver bandwidth ± 33 kHz; and parallel imaging accelerating factor, 2. Total acquisition time was approximately 2 minutes. Wave images from each slice location were automatically processed on the scanner computer, using an inversion algorithm, to generate axial liver stiffness maps called elastograms. Elastograms were transferred and analyzed offline by a trained image analyst with at least 6 months of MRE image analysis experience who was blinded to clinical and histologic data.
Ultrasound-based VCTE assessment
Participants from the Familial Cirrhosis cohort also underwent an ultrasound-based vibration controlled transient elastography (VCTE) assessment using a FibroScan. VCTE was performed by a trained technician, using the FibroScan® 502 Touch model (M Probe; XL Probe; Echosens, Paris, France). VCTE measurements were obtained while subjects held a supine position with the right arm fully adducted. The area of abdomen at the location of the right liver lobe was scanned during a 10-second breath hold. Participants were asked to fast at least 3 hours prior to the exam. A minimum of 10 valid measurements were used to determine the median liver stiffness measurement (LSM) in kilopascals (kPa) and the interquartile range (IQR). In accordance with the manufacturer’s protocol, all patients were first scanned using the M probe (3.5 MHz). If indicated by the equipment upon initial assessment, patients were re-scanned using the XL probe (2.5 MHz) (Caussy et al., 2018a; Hsu et al., 2019). The threshold used for the classification of cirrhosis (stage 4 fibrosis) was VCTE > 11.8 kPa as previously determined in reference (Hsu et al., 2019). Eleven first-degree relatives of NAFLD-cirrhosis probands did not have an MRE assessment due to contraindications. For those individuals, the presence of cirrhosis (fibrosis stage 4) was determined using a VCTE threshold > 11.8 kPa as previously described (Hsu et al., 2019).
Human stool sample preparation
Human stool samples were collected from participants in the Familial Cirrhosis and Twins and Family cohorts according to approved HRPP protocols (UCSD IRB #140084 and #111282). All samples had been previously processed for quantitative metagenomic sequencing and analysis. The unprocessed swabs from original double-headed swabs used to collect the samples were placed in a deep-well 2-mL polypropylene 96-well microtiter plate according to the plate maps. The material was then extracted in 300 μL 50% MeOH overnight, concentrated in centrifugal evaporator and redissolved in 100 μL of 50% MeOH with internal standards.
Microbiome profiling
DNA extraction from human stool samples and shotgun sequencing were performed by the Center for Microbiome Innovation (CMI) at University of California, San Diego (UCSD). DNA sequencing libraries were prepared using Nextera Library Prep Kits (Illumina). Shotgun DNA sequencing was performed on the Illumina HiSeq4000 platform.
Metagenomic analysis
Raw fastq reads were quality-checked. Briefly, skewer (version 0.2.2), an accurate adapter trimmer, was utilized with the paired-end mode. Unwanted human reads were identified and removed by Bowtie2 mapping against the human genome reference (hg19), followed by bam2fastq with --unaligned --no-aligned --force options. After trimming and filtering, metagenomic read data was processed with the HUMAnN2 pipeline (version 0.11.2). For taxonomic profiling, MetaPhlAn2 (version 2.7.7) embedded in HUMAnN2 was utilized with the default setting. For functional analysis, HUMAnN2 was conducted with the UniRef90 database. Initially, Bowtie2 mapped reads were analyzed with the functionally pre-annotated pan-genome of ChocoPhlAn. Unmapped reads were then aligned to UniRef90 with the translated search tool, DIAMOND. Utilizing the MetaCyc pathway database, HUMAnN2 algorithm generated gene-family abundance, pathway abundance and pathway coverage. Moreover, abundance outputs were normalized with the humann2_renorm_table command. Metagenomic profiling outcomes were transformed into the R object using the curatedMetagenomicData package based on ExperimentHub platform.
Shotgun functional profiling
To identify altered microbial pathways between NAFLD-cirrhosis and non-NAFLD control groups, the HUMAnN2 tool including the analysis of microbial gene-families was utilized. HUMAnN2 outcome was transformed into curatedMetagenomicData format and loaded into R object. Top discriminatory pathways were selected using feature selection by Random Forest (RF) and differential abundance analyses from DESeq2 through the computation of mean decrease in Gini and fold-change.
Microbiome data extraction from external cohorts
The curatedMetagenomicData tool was used to access the metagenomic data from the Chinese cohort (Qin et al., 2014) and our previous study (Loomba et al., 2017), and taxonomy table and metadata including albumin were extracted using the query function. For cirrhosis data from the Italian cohort, raw fastq reads were curated from PRJNA471972. Using the package, dada2, reads were pre-processed with default setting. Taxonomy profiling on the RDP reference was conducted with assignTaxonomy and addSpecies commands. Assigned taxa were transformed into phyloseq format.
Metabolite profiling
Metabolite extraction from human stool samples and the subsequent LC-MS/MS was performed by CMI at the University of California, San Diego (Wang et al., 2020).
Data conversion
Metabolomic data was processed using MS-DIAL. In brief, LC-MS/MS raw data was converted into .abf format using Abf (Analysis Base File) Converter. The converted files were then imported to MS-DIAL (version 3.66) for metabolic feature extraction, alignment, and putative annotation. Detailed parameters were as follows:Tolerances of MS1 0.01 Da and MS2 0.025 Da were employed. For isotope recognition: maximum charged number: 2; for metabolic feature picking: minimum peak height: 1000, mass slice width 0.1; smoothing method: linear weighted moving average; smoothing level 3, minimum peak width: 5 scans. For alignment, retention time tolerance was set to 0.2 min and MS1 tolerance was 0.015 Da. After data processing in MS-DIAL, manual data checking was performed to visually check and exclude false positive metabolic features. The refined metabolic feature table was exported from MS-DIAL and used for downstream statistical analysis. Metabolic features with low MS signals that were not picked up by MS-DIAL were later manually checked in the raw LC-MS data.
Metabolite annotations
Metabolite annotations were manually performed using MS-Finder (version 3.20). Each annotated metabolite was assigned a mass error and MS2 matching score, based upon the MS2 spectrum similarity to matches in the default MS2 spectral library in MS-Finder. Identified metabolites with the smallest mass error and highest MS2 matching score were selected and added to the list of 159 unknown features.The threshold for confident identification was set to be mass error ≤ 10 ppm and MS2 matching score ≥ 8 (out of 10). However, metabolites that did not meet these thresholds were not always false positive identifications. Therefore, metabolites of interest with MS2 matching scores between approximately 7.5 to 8.0 were manually identified.
Validation of metabolites synthesized by single microbe cultured with bile acid
Bacterial pellets from Ruminococcus gnavus strain VPI C7–9 (#29149) were purchased from ATCC and rehydrated in 0.5ml ATCC 260 broth medium (Tryptic Soy Broth (BD 211825) 30g, Sheep Blood 50ml, DI water 950ml) under anaerobic conditions. 50μl of resuspended culture was then plated on ATCC 260 Medium (Tryptic Soy Agar (BD 236950) 40g with 5% Sheep Blood (defibrinated) 50 ml, DI Water 950 ml). The remaining rehydrated bacterial culture was transferred to 5ml ATCC 260 broth medium. All cultures were incubated in an anaerobic atmosphere containing a gas mix of 5% hydrogen and 95% nitrogen at 37°C for 24–48 hours. Ruminococcus gnavus cultures were treated with 12 concentrations of CDCA (final concentrations ranging from 0.001 μg/ml to 1000 μg/ml) in ATCC 260 medium and cultured in total anaerobic conditions. After culturing for 48hrs, supernatants were collected for untargeted metabolomics. To extract metabolites, 200 μL supernatants was first mixed with 600 μL LC-MS grade ice-cold methanol. The solution was placed in a −20°C freezer for 2 hours to denature and precipitate the proteins. Further centrifugation (17,530 g, 4°C, 15 min) removed the precipitated proteins and the supernatant was carefully transferred to a new vial. The solvent was evaporated in a Speedvac at 4°C. The dried sample was reconstituted in 200 μL solvent (ACN:H2O=1:3, v:v). The reconstituted sample was centrifuged (17,530 g, 4°C, 15 min) again to remove any insoluble particles. The final solution was transferred into the LC glass insert for untargeted LC-MS/MS analysis in data-dependent acquisition mode on a Bruker Impact II Ultra-High Resolution Qq-Time-Of-Flight Mass Spectrometer (UHR-QqTOF-MS) coupled with an Agilent 1290 Infinity II Ultra High-Performance Liquid Chromatography (UHPLC) system.
QUANTIFICATION AND STATISTICAL ANALYSIS
Development of a model utilizing stool-derived signatures
To develop models capable of distinguishing NAFLD-cirrhosis from non-NAFLD control samples, we utilized a supervised learning algorithm available in the Classification And Regression Training (caret) package in R to independently identify signatures from metagenomic and metabolomic datasets. A priori relative abundance taxonomic tables were pre-processed using preProcess and predict functions with the zv, scale and center methods. Briefly, the model was trained using the Caret parameter as follows; 10 fold cross-validation, smote sampling mode and 10 repeats in trainControl. To determine the best hyperparameter setting, mtry value for the number of variables randomly sampled as candidates at each split was set at the range (0, 10, 20, 30, 40, 50) or (−1, 0 and +1 of the square root of the feature number) on grid tuning. The ntree setting for the number of trees to grow was 501 with the ROC metric option. For feature selection, features (species or metabolites with patient blood data) from the feature importance scores of the RF outcome were examined to find a set of features that trains a forest with the highest overall accuracy of sample classification. To do so, importance scores of features were extracted using varImp function and tested iteratively by ordering from highest to lowest Mean Decrease in Gini index. Subsequently, different combinations of features from the top rank were compared by calculating AUC. Feature selection was utilized to optimize the numbers of the signature sets, with the top-performing models utilizing 19 microbiome species in combination with participant age, and 17 metabolites, respectively. Further optimization of the microbiome+age signature was performed by incorporating serum albumin levels, which were available in the metadata from both the UCSD and Chinese cohorts. The model training set included 81 stool samples from non-NAFLD controls and NAFLD-cirrhosis (fibrosis stage 4) probands. Subsequently, the model was validated in the UCSD relatives, Chinese and Italian cohorts. To calculate AUC, RF outcome was matched to disease status using auc function of the pROC package. To evaluate the synergistic effect between species and metabolites, a combination of features was utilized to distinguish NAFLD-cirrhosis in the training set (from the UCSD cohort).
Model Validation
In order to test and validate the RF model from the UCSD training set in external datasets, we curated and established metagenomic data and 16S taxonomy in the Chinese (total 237 samples) and Italian cohorts (49 samples) as described above. Machine learning models were trained with features, including discriminatory species, as described above. Sequentially, we tested models in the Chinese and Italian datasets. To do so, we extracted 19 species from all datasets. If the species was absent, abundance was regarded as zero. We then tested models in the Chinese and Italian datasets using predict function with the option of type prob. For AUC score, roc function was utilized using the setting of boot.n; 100 and ci.alpha; 0.9. Since the Chinese dataset included albumin scores, we further tested our model in the Chinese cohort. For testing the model in the mixed fibrosis cohort, the previous dataset was downloaded and established. The 86 samples were split into two validation sets: 1) NAFL only vs cirrhosis (stage 4) or 2) mild fibrosis (stage 1–3) vs cirrhosis (stage 4). The previously retained RF model was tested in these two sets to evaluate whether it could accurately identify cirrhosis cases in a cohort of NAFLD patients with a wide range of fibrosis levels. To screen for other clinical features that could improve diagnostic accuracy, clinical metadata (e.g., AST) was added to the taxa table, and RF training and testing were conducted. To visualize AUC outcome, plot function was utilized in R.
Statistical analysis
In order to quantify within-sample diversity, we computed alpha-diversity scores (Inverse Simpson) using the phyloseq package and calculated P-values with t-test. For between-sample diversity, we utilized the ordinate function in phyloseq with the top significant microbiomes. PCoA plot was generated with weighted-Unifrac distances. To examine the significant alteration of ordination, we performed permutational analysis of variance (PERMANOVA) using the adonis function from the R package, vegan. To identify important features including microbial data and metabolites, we used the DESeq2 pipeline and the feature selection of Random Forest. To compare altered pathways in different cirrhosis datasets, we performed the RV-correlation analysis using the coeffRV of the FactorMineR package. First, we compared the matrices of the top significant pathways from the current study to other datasets. Subsequently, we conducted RV-correlation analysis with all detected and common pathways between datasets. To measure multivariate analysis with significant metabolites, we used R’s ropls package and computed Partial Least Squares (PLS) and Orthogonal PLS (OPLS) scores. For the statistic reported in figure 5I and 5J, we utilized Kruskal-Wallis ANOVA test, fast zero-inflated negative binomial mixed model (FZINBMM) and DESeq2 using custom R commands. The most significant outcomes were indicated using asterisks.
Supplementary Material
KEY RESOURCES TABLE.
ACKNOWLEDGMENTS
We thank Leyna Nguyen for her assistance with data verification. This work was funded by grants from the National Institute of Environmental Health Sciences of the National Institutes of Health under Award Number P42ES010337 and NIH (HL088093). R.M.E is an investigator of the Howard Hughes Medical Institute and March of Dimes Chair in Molecular and Developmental Biology at the Salk Institute and is supported by the NIH (DK057978, HL105278, ES010337), the Cancer Center (CA014195), a NOMIS Foundation Distinguished Scientist and Scholar Award, and a grant from the Fondation Leducq. R.L. receives funding support from NIEHS (5P42ES010337), NCATS (5UL1TR001442), NIDDK (U01DK061734, R01DK106419, P30DK120515, R01DK121378), NHLBI (P01HL147835), and DOD PRCRP (W81XWH-18-2-0026).
Footnotes
DECLARATION OF INTERESTS
The authors declare no conflicts of interest.
ACCESSION CODES
Microbiome sequencing data are accessible through accession number EGAS000010046.
SUPPLEMENTAL INFORMATION
Supplemental Information includes one table and six figures, and can be found with this article online.
REFERENCES
- Acharya C, and Bajaj JS (2019). Altered Microbiome in Patients With Cirrhosis and Complications. Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association 17, 307–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caussy C, Alquiraish MH, Nguyen P, Hernandez C, Cepin S, Fortney LE, Ajmera V, Bettencourt R, Collier S, Hooker J, et al. (2018a). Optimal threshold of controlled attenuation parameter with MRI-PDFF as the gold standard for the detection of hepatic steatosis. Hepatology (Baltimore, Md) 67, 1348–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caussy C, Bhargava M, Villesen IF, Gudmann NS, Leeming DJ, Karsdal MA, Faulkner C, Bao D, Liu A, Lo MT, et al. (2019a). Collagen formation assessed by PRO-C3 is an heritable trait and is associated with liver fibrosis assessed by MRE. Hepatology (Baltimore, Md). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caussy C, Hsu C, Lo MT, Liu A, Bettencourt R, Ajmera VH, Bassirian S, Hooker J, Sy E, Richards L, et al. (2018b). Link between gut-microbiome derived metabolite and shared gene-effects with hepatic steatosis and fibrosis in NAFLD. Hepatology (Baltimore, Md). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caussy C, Soni M, Cui J, Bettencourt R, Schork N, Chen CH, Ikhwan MA, Bassirian S, Cepin S, Gonzalez MP, et al. (2017). Nonalcoholic fatty liver disease with cirrhosis increases familial risk for advanced fibrosis. J Clin Invest 127, 2697–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caussy C, Tripathi A, Humphrey G, Bassirian S, Singh S, Faulkner C, Bettencourt R, Rizo E, Richards L, Xu ZZ, et al. (2019b). A gut microbiome signature for cirrhosis due to nonalcoholic fatty liver disease. Nature communications 10, 1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalasani N, Younossi Z, Lavine JE, Diehl AM, Brunt EM, Cusi K, Charlton M, and Sanyal AJ (2012). The diagnosis and management of non-alcoholic fatty liver disease: practice Guideline by the American Association for the Study of Liver Diseases, American College of Gastroenterology, and the American Gastroenterological Association. Hepatology (Baltimore, Md) 55, 2005–2023. [DOI] [PubMed] [Google Scholar]
- Chang H, Meng HY, Liu SM, Wang Y, Yang XX, Lu F, and Wang HY (2017). Identification of key metabolic changes during liver fibrosis progression in rats using a urine and serum metabolomics approach. Scientific reports 7, 11433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Ang B, Haufe W, Hernandez C, Verna EC, Sirlin CB, and Loomba R (2015). Comparative diagnostic accuracy of magnetic resonance elastography vs. eight clinical prediction rules for non-invasive diagnosis of advanced fibrosis in biopsy-proven non-alcoholic fatty liver disease: a prospective study. Alimentary pharmacology & therapeutics 41, 1271–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Chen CH, Lo MT, Schork N, Bettencourt R, Gonzalez MP, Bhatt A, Hooker J, Shaffer K, Nelson KE, et al. (2016). Shared genetic effects between hepatic steatosis and fibrosis: A prospective twin study. Hepatology (Baltimore, Md) 64, 1547–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Silva HE, Teterina A, Comelli EM, Taibi A, Arendt BM, Fischer SE, Lou W, and Allard JP (2018). Nonalcoholic fatty liver disease is associated with dysbiosis independent of body mass index and insulin resistance. Scientific reports 8, 1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd D, Spitzer MH, Van Treuren W, Merrill BD, Hryckowian AJ, Higginbottom SK, Le A, Cowan TM, Nolan GP, Fischbach MA, et al. (2017). A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 551, 648–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulai PS, Sirlin CB, and Loomba R (2016). MRI and MRE for non-invasive quantitative assessment of hepatic steatosis and fibrosis in NAFLD and NASH: Clinical trials to clinical practice. Journal of hepatology 65, 1006–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzosa EA, McIver LJ, Rahnavard G, Thompson LR, Schirmer M, Weingart G, Lipson KS, Knight R, Caporaso JG, Segata N, et al. (2018). Species-level functional profiling of metagenomes and metatranscriptomes. Nature methods 15, 962–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyatt GH, Haynes RB, Jaeschke RZ, Cook DJ, Green L, Naylor CD, Wilson MC, and Richardson WS (2000a). Users’ Guides to the Medical Literature: XXV. Evidence-based medicine: principles for applying the Users’ Guides to patient care. Evidence-Based Medicine Working Group. Jama 284, 1290–1296. [DOI] [PubMed] [Google Scholar]
- Guyatt GH, Naylor D, Richardson WS, Green L, Haynes RB, Wilson MC, Cook DJ, and Jaeschke RZ (2000b). What is the best evidence for making clinical decisions? Jama 284, 3127–3128. [PubMed] [Google Scholar]
- Hoyles L, Fernandez-Real JM, Federici M, Serino M, Abbott J, Charpentier J, Heymes C, Luque JL, Anthony E, Barton RH, et al. (2018). Molecular phenomics and metagenomics of hepatic steatosis in non-diabetic obese women. Nature medicine 24, 1070–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu C, Caussy C, Imajo K, Chen J, Singh S, Kaulback K, Le MD, Hooker J, Tu X, Bettencourt R, et al. (2019). Magnetic Resonance vs Transient Elastography Analysis of Patients With Nonalcoholic Fatty Liver Disease: A Systematic Review and Pooled Analysis of Individual Participants. Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association 17, 630–637.e638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iebba V, Guerrieri F, Di Gregorio V, Levrero M, Gagliardi A, Santangelo F, Sobolev AP, Circi S, Giannelli V, Mannina L, et al. (2018). Combining amplicon sequencing and metabolomics in cirrhotic patients highlights distinctive microbiota features involved in bacterial translocation, systemic inflammation and hepatic encephalopathy. Scientific reports 8, 8210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalhan SC, Guo L, Edmison J, Dasarathy S, McCullough AJ, Hanson RW, and Milburn M (2011). Plasma metabolomic profile in nonalcoholic fatty liver disease. Metabolism: clinical and experimental 60, 404–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Kim WR, Talwalkar JA, Kim HJ, and Ehman RL (2013). Advanced fibrosis in nonalcoholic fatty liver disease: noninvasive assessment with MR elastography. Radiology 268, 411–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z, Tsugawa H, Wohlgemuth G, Mehta S, Mueller M, Zheng Y, Ogiwara A, Meissen J, Showalter M, Takeuchi K, et al. (2018). Identifying metabolites by integrating metabolome databases with mass spectrometry cheminformatics. Nature methods 15, 53–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Jia H, Cai X, Zhong H, Feng Q, Sunagawa S, Arumugam M, Kultima JR, Prifti E, Nielsen T, et al. (2014). An integrated catalog of reference genes in the human gut microbiome. Nature biotechnology 32, 834–841. [DOI] [PubMed] [Google Scholar]
- Loomba R, and Sanyal AJ (2013). The global NAFLD epidemic. Nature reviews Gastroenterology & hepatology 10, 686–690. [DOI] [PubMed] [Google Scholar]
- Loomba R, Schork N, Chen CH, Bettencourt R, Bhatt A, Ang B, Nguyen P, Hernandez C, Richards L, Salotti J, et al. (2015). Heritability of Hepatic Fibrosis and Steatosis Based on a Prospective Twin Study. Gastroenterology 149, 1784–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomba R, Seguritan V, Li W, Long T, Klitgord N, Bhatt A, Dulai PS, Caussy C, Bettencourt R, Highlander SK, et al. (2017). Gut Microbiome-Based Metagenomic Signature for Non-invasive Detection of Advanced Fibrosis in Human Nonalcoholic Fatty Liver Disease. Cell metabolism 25, 1054–1062.e1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomba R, Wolfson T, Ang B, Hooker J, Behling C, Peterson M, Valasek M, Lin G, Brenner D, Gamst A, et al. (2014). Magnetic resonance elastography predicts advanced fibrosis in patients with nonalcoholic fatty liver disease: a prospective study. Hepatology (Baltimore, Md) 60, 1920–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noureddin M, Lam J, Peterson MR, Middleton M, Hamilton G, Le TA, Bettencourt R, Changchien C, Brenner DA, Sirlin C, et al. (2013). Utility of magnetic resonance imaging versus histology for quantifying changes in liver fat in nonalcoholic fatty liver disease trials. Hepatology (Baltimore, Md) 58, 1930–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson LM, Poitou C, Tremaroli V, Coupaye M, Aron-Wisnewsky J, Backhed F, Clement K, and Caesar R (2019). Gut microbiota of obese subjects with Prader-Willi syndrome is linked to metabolic health. Gut. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CC, Nguyen P, Hernandez C, Bettencourt R, Ramirez K, Fortney L, Hooker J, Sy E, Alquiraish MH, Valasek MA, et al. (2016). Magnetic Resonance Elastography vs Transient Elastography in Detection of Fibrosis and Noninvasive Measurement of Steatosis in Patients with Biopsy-proven Nonalcoholic Fatty Liver Disease. Gastroenterology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel NS, Peterson MR, Brenner DA, Heba E, Sirlin C, and Loomba R (2013). Association between novel MRI-estimated pancreatic fat and liver histology-determined steatosis and fibrosis in non-alcoholic fatty liver disease. Alimentary pharmacology & therapeutics 37, 630–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Permutt Z, Le TA, Peterson MR, Seki E, Brenner DA, Sirlin C, and Loomba R (2012). Correlation between liver histology and novel magnetic resonance imaging in adult patients with non-alcoholic fatty liver disease - MRI accurately quantifies hepatic steatosis in NAFLD. Alimentary pharmacology & therapeutics 36, 22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry RJ, Peng L, Barry NA, Cline GW, Zhang D, Cardone RL, Petersen KF, Kibbey RG, Goodman AL, and Shulman GI (2016). Acetate mediates a microbiome-brain-beta-cell axis to promote metabolic syndrome. Nature 534, 213–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin N, Yang F, Li A, Prifti E, Chen Y, Shao L, Guo J, Le Chatelier E, Yao J, Wu L, et al. (2014). Alterations of the human gut microbiome in liver cirrhosis. Nature 513, 59–64. [DOI] [PubMed] [Google Scholar]
- Raman M, Ahmed I, Gillevet PM, Probert CS, Ratcliffe NM, Smith S, Greenwood R, Sikaroodi M, Lam V, Crotty P, et al. (2013). Fecal microbiome and volatile organic compound metabolome in obese humans with nonalcoholic fatty liver disease. Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association 11, 868–875.e861–863. [DOI] [PubMed] [Google Scholar]
- Reeder SB (2013). Emerging quantitative magnetic resonance imaging biomarkers of hepatic steatosis. Hepatology (Baltimore, Md) 58, 1877–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Waldron L, Ballarini A, Narasimhan V, Jousson O, and Huttenhower C (2012). Metagenomic microbial community profiling using unique clade-specific marker genes. Nature methods 9, 811–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpton SR, Ajmera V, and Loomba R (2019). Emerging Role of the Gut Microbiome in Nonalcoholic Fatty Liver Disease: From Composition to Function. Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association 17, 296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Cui H, Li N, Wei Y, Lai S, Yang Y, Yin X, and Chen DF (2016). Comparison of FIB-4 index, NAFLD fibrosis score and BARD score for prediction of advanced fibrosis in adult patients with non-alcoholic fatty liver disease: A meta-analysis study. Hepatol Res 46, 862–870. [DOI] [PubMed] [Google Scholar]
- Tsugawa H, Cajka T, Kind T, Ma Y, Higgins B, Ikeda K, Kanazawa M, VanderGheynst J, Fiehn O, and Arita M (2015). MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nature methods 12, 523–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Jarmusch AK, Vargas F, Aksenov AA, Gauglitz JM, Weldon K, Petras D, da Silva R, Quinn R, Melnik AV, et al. (2020). Mass spectrometry searches using MASST. Nature biotechnology 38, 23–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong RJ, Cheung R, and Ahmed A (2014). Nonalcoholic steatohepatitis is the most rapidly growing indication for liver transplantation in patients with hepatocellular carcinoma in the U.S. Hepatology (Baltimore, Md) 59, 2188–2195. [DOI] [PubMed] [Google Scholar]
- Yin M, Glaser KJ, Talwalkar JA, Chen J, Manduca A, and Ehman RL (2016). Hepatic MR Elastography: Clinical Performance in a Series of 1377 Consecutive Examinations. Radiology 278, 114–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younossi Z, Anstee QM, Marietti M, Hardy T, Henry L, Eslam M, George J, and Bugianesi E (2018). Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nature reviews Gastroenterology & hepatology 15, 11–20. [DOI] [PubMed] [Google Scholar]
- Zaarour M, Zaharia R, Bretault M, Jublanc C, De Marcellus C, Bouillot JL, Lefebvre H, Oppert JM, Aron-Wisnewsky J, and Raffin-Sanson ML (2019). Laparoscopic Revision of Bariatric Surgeries in Two Patients with Severe Resistant Hypocalcemia After Endocrine Cervical Surgery. Obesity surgery. [DOI] [PubMed] [Google Scholar]
- Zarrinpar A, Gupta S, Maurya MR, Subramaniam S, and Loomba R (2016). Serum microRNAs explain discordance of non-alcoholic fatty liver disease in monozygotic and dizygotic twins: a prospective study. Gut 65, 1546–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession numbers for shotgun sequencing datasets reported in this paper are publicly available at the European Genome-Phenome Archive (EGAS000010046). The study was conducted for identifying NAFLD-related biomarkers.
Bioinformatics workflows for metagenomics and data are available at: https://bitbucket.org/biobakery/ and http://bioconductor.org/packages/release/data/experiment/html/curatedMetagenomicData.html.