Abstract
Although conformational dynamics of RNA molecules are potentially important in microRNA (miRNA) processing, the role of the protein binding partners in facilitating the requisite structural changes is not well understood. In previous work, we and others have demonstrated that nonduplex structural elements and the conformational flexibility they support are necessary for efficient RNA binding and cleavage by the proteins associated with the two major stages of miRNA processing. However, recent studies showed that the protein DGCR8 binds primary miRNA and duplex RNA with similar affinities. Here, we study RNA binding by a small recombinant construct of the DGCR8 protein and the RNA conformation changes that result. This construct, the DGCR8 core, contains two double-stranded RNA-binding domains (dsRBDs) and a C-terminal tail. To assess conformational changes resulting from binding, we applied small-angle x-ray scattering with contrast variation to detect conformational changes of primary-miR-16-1 in complex with the DGCR8 core. This method reports only on the RNA conformation within the complex and suggests that the protein bends the RNA upon binding. Supporting work using smFRET to study the conformation of RNA duplexes bound to the core also shows bending. Together, these studies elucidate the role of DGCR8 in interacting with RNA during the early stages of miRNA processing.
Significance
Regulation of messenger RNA lifetime and translation by microRNA (miRNA) is essential in multicellular organisms, yet the full mechanism whereby mature miRNA is generated from primary transcripts remains to be determined. Specifically, the molecular mechanism of precise cut placement by Drosha at the first stage of miRNA maturation remains in debate. Here, we apply contrast variation small-angle x-ray scattering to demonstrate that Drosha’s cofactor protein DGCR8 induces a bend in double-stranded RNA upon binding. This observation explains prior biochemical data on miRNA processing by Drosha while also providing the first demonstration of contrast variation small-angle x-ray scattering as a technique for monitoring RNA structural change in RNA-protein complexes.
Introduction
MicroRNAs (miRNAs) are short noncoding RNA molecules that facilitate post-transcriptional regulation of gene expression (1, 2, 3). The canonical processing of miRNA involves two major post-transcriptional stages (4). In the first stage, primary microRNA (pri-miRNA) is generated in the nucleus and is cleaved by the microprocessor complex, composed of the proteins Drosha and DGCR8, to form a hairpin-shaped precursor miRNA (pre-miRNA). In the second stage, the pre-miRNA is exported to the cytoplasm, wherein it is cleaved by Dicer to yield a mature miRNA (5). RNA structure and internal dynamics are known to play important roles in miRNA processing, but further investigation is needed to understand how they are coupled to biological function. For example, perturbed dynamics of pri-miRNA structures are hypothesized to contribute to cancer phenotypes associated with single-nucleotide polymorphisms in pri-miR-125a (6). In addition, structural mimicry by single-stranded RNA viruses exploits flexible regions that strongly resemble the terminal loops and internal bulges characteristic of pri-miRNA stem loops (7). The stem-loop precursor molecules of miRNA access diverse transient structures that ultimately affect their progression through the miRNA maturation pathway; however, detailed mechanisms that describe the roles of conformational dynamics and transitions in the RNA remain to be established. In an effort to refine the biological mechanism of miRNA processing, we and others have shown that the loops, bulges, and flanking RNA single strands in pri-miRNA—and the conformations associated with these structures—are important to RNA binding and cleavage by the microprocessor complex and Dicer (8, 9, 10). Interestingly, we also found that deformability in the cleavage site of pri-miRNA is not sensed by the microprocessor component DGCR8 and that DGCR8 binds pri-miRNA and duplex RNA with similar binding affinities (11). Although these studies provide growing biochemical evidence for the importance of RNA dynamics in miRNA processing, the paucity of available structures for pri-miRNA bound to the microprocessor complex leaves the role of DGCR8 in the first stage of miRNA processing unclear. Here, we explore the role of DGCR8 in modifying the average structure of the RNA upon complex formation by facilitating bending of pri-miRNA to kink the stem loop.
Just as the structures of folded RNA molecules are diverse (12), so are the mechanisms of protein partner recognition (13). In the context of pri-miRNA and pre-miRNA cleavage, virtually all of the proteins required for maturation include one or more double-stranded RNA binding domains (dsRBDs) (5). In partnership with catalytic centers formed from RNase III family enzymes, dsRBDs are ubiquitous among double-stranded RNA (dsRNA) processing proteins; however, substantial technical challenges have limited the number of high-resolution structures available that describe their interactions at the atomic scale. For the second stage of miRNA processing, in which Dicer cleaves the terminal loop off a pre-miRNA hairpin, this model suggests that the tandem dsRBDs from Dicer’s cofactor TRBP bind to a dsRNA duplex (14). This model revealed that the dsRBDs associate with the dsRNA in multiple orientations, which is consistent with prior in vitro biochemical results showing that TRBP binds dsRNA without sequence preference. Recently, a cryo-electron microscopy structure of Dicer bound to dsRNA revealed an intricate network of contacts between the enzyme and its substrate RNA (15). In the context of pri-miRNA cleavage by the microprocessor complex, minimally composed of Drosha and its cofactor DGCR8, gel filtration (16) and single-molecule FRET (smFRET) (17) studies established that the microprocessor is a 2:1:1 complex of DGCR8:Drosha:pri-miRNA. A recent structure of human Drosha implies two binding sites for DGCR8 (18). However, to date, no atomic-resolution model of the microprocessor complex involving all components (DGCR8, Drosha, and pri-miRNA) has been reported.
The dsRNA binding properties of DGCR8 have been extensively studied, both in the context of miRNA maturation and through other noncanonical roles in which DGCR8 must recognize dsRNA (19). Many noncanonical roles of DGCR8 are Drosha independent, suggesting that DGCR8 is often required to form productive complexes with dsRNA independent of the rest of the microprocessor complex (20). An early crystal structure of the DGCR8 dsRBDs led to the hypothesis that upon binding of RNA, DGCR8 would force bending of the pri-miRNA stem loop to engage both dsRBDs (21). Therefore, we have chosen to work with the same construct used in the crystal structure study, which is a minimal “DGCR8 core” composed of the two dsRBDs and a C-terminal tail (Fig. 1 A). We used this minimal construct to determine whether the dsRBDs in DGCR8 can bend RNA stem loops upon binding, which may be important for miRNA processing. Based on structural probing with the SHAPE method, we proposed two regions of inherent flexibility in pri-miRNA stem-loop structures (9); these regions are consistent with the high prevalence of non-Watson-Crick structural features observed in two similarly patterned clusters defined in bioinformatics studies (22). We hypothesized that DGCR8 function is dependent on the recognition of specific structural features in the miRNA precursor, such as loops, bulges, and single-stranded flanking segments. To our surprise, we found that the equilibrium constant for dsRNA binding by the DGCR8 core is insensitive to the presence of noncanonical structural elements within the stem (11). This study demonstrated that RNA deformability is not necessary for DGCR8 core binding, yet flexibility near the Drosha cleavage site is still required for efficient processing of pri-miR-16-1 (11). Thus, the function of the DGCR8 core might be to facilitate structural dynamics and may not require sequence specificity or noncanonical structural element recognition. Therefore, we designed our study to test the hypothesis that the DGCR8 core bends the pri-miRNA and duplex RNA upon binding.
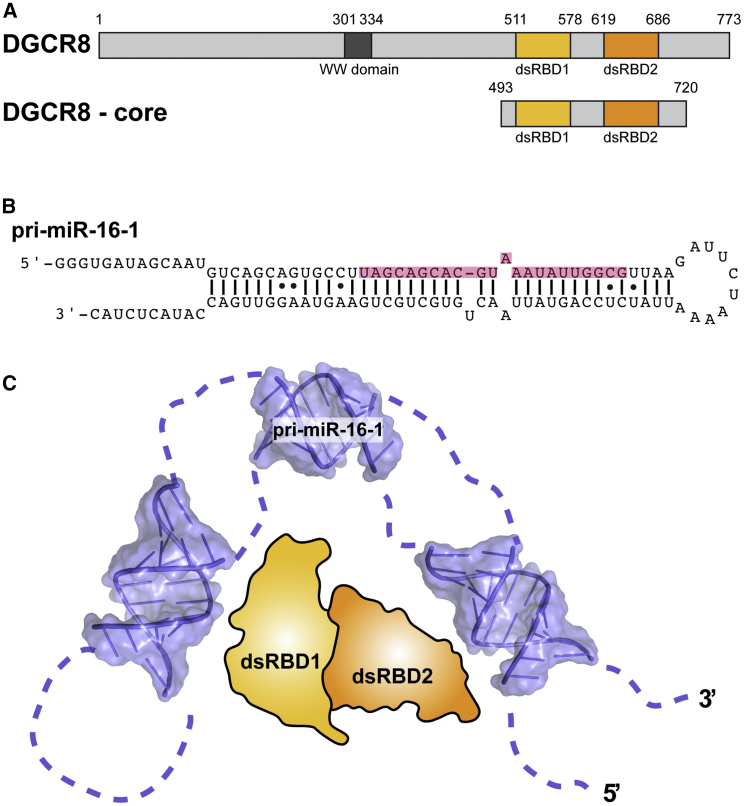
Figure 1.
DGCR8 core interacts with pri-miR-16-1 through two binding domains. (A) Domain architecture of the full-length DGCR8 protein and the protein construct used in this study (DGCR8 core) is shown. A WW domain responsible for DGCR8 assembly is annotated in gray and is absent in the DGCR8 core. The double-stranded RNA-binding domains (dsRBD1 and dsRBD2) are annotated in shades of orange. (B) Sequence and stem-loop structure of the primary miRNA pri-miR-16-1 that is the basis for the constructs used in the SAXS experiments are shown. The resulting mature miRNA is highlighted in pink. (C) Model for RNA binding and bending by the DGCR8 core is shown. To see this figure in color, go online.
Studying the structural changes of an RNA as it binds a protein partner requires the ability to distinguish signals from the RNA and protein in the complex. Small-angle x-ray scattering (SAXS) measurements provide global structural insights about freely diffusing macromolecules, including complexes, in solution. To address the challenge of distinguishing whether the SAXS signal originates from the protein, the RNA, or the whole complex and enable the study of conformational changes of the RNA, we applied contrast variation SAXS (CV-SAXS). CV-SAXS allows us to detect only the nucleic acid component of a protein-nucleic acid complex by masking the protein contribution to the scattering profile (23). We have previously demonstrated this technique in a study of DNA unwinding from histones in nucleosome core particles (24,25). Here, we describe the use of CV-SAXS to measure the conformations of pri-miR-16-1 in the presence and absence of the DGCR8 core.
CV-SAXS provides strong evidence that pri-miR-16-1 bends in the presence of the DGCR8 core. The ability of the protein to bind and bend RNAs with duplex elements is further supported by smFRET studies on a duplex RNA and DGCR8 core complex. Comparison of SAXS data of both canonical RNA duplexes and with duplexes that are interrupted by flexible regions near their centers provides additional insight into the conformations assumed by the protein-bound RNA. The tandem dsRBDs found in the DGCR8 core not only bend RNA stem loops upon binding but also bend canonical duplex RNAs that do not contain flexible loops or bulges. The indiscriminate bending of duplex RNA is consistent with our previous finding (11) that the binding affinities of duplex RNA and pri-miRNA for the DGCR8 core are very similar. Thus, we propose that both sequence-indiscriminate binding and bending are necessary for efficient processing of hundreds of structurally diverse primary miRNAs. We speculate that bending is a required part of a dynamic sequence of events involving catch and release of pri-miRNAs waiting to be processed. How bending and unbending of primary miRNA contribute to the energetics of the first stage of miRNA processing remains unclear. We suggest that the interactions of dsRBDs with RNA structures are important for better understanding miRNA processing in particular and duplex RNA-dsRBD interactions in general.
Materials and Methods
Expression and purification of proteins
The DGCR8 core construct (493–720) was prepared by amplifying DNA through PCR from the human DGCR8 gene, purchased from ATCC (Manassas, VA), which was then inserted into pET-47b(+) vector between the XmaI and EcoRI restriction sites, resulting in an in-frame 6x-His tag N-terminal to the DGCR8 coding sequence and followed by a 3C Protease recognition sequence. Treatment with 3C protease during purification results in a Gly-Pro-Gly cloning artifact N-terminal to the DGCR8 core. BL21 DE3 cells transformed with the DGCR8 core plasmid were grown in LB media at 37°C until the culture OD600 reached 0.8, at which point IPTG was added to a final concentration of 0.5 mM. Cultures were incubated for an additional 4 h at 37°C before harvesting. Cells were either stored at −80°C or lysed immediately. Cell pellets were resuspended in 50 mM sodium phosphate (pH 8.0), 50 mM sodium chloride, 20 mM imidazole, and 2 mM 2-mercaptoethanol supplemented with commercial protease inhibitor cocktail (Millipore Sigma, Burlington, MA) and 1 mM phenylmethanesulfonyl fluoride. Cells were lysed by sonication, and the lysate was clarified by centrifugation at 14,000 × g for 45 min at 4°C. Clarified lysate was applied to Ni-NTA resin equilibrated with lysis buffer, washed with lysis buffer supplemented with 0.1% Triton X-100, and eluted in 50 mM sodium phosphate (pH 8.0), 50 mM sodium chloride, 200 mM imidazole, and 2 mM 2-mercaptoethanol. Eluted protein treated with 3C protease was dialyzed against 2 L of lysis buffer overnight at 4°C. The dialysate was passed over a Ni-NTA column, then concentrated to <1 mL. Concentrated protein was further purified by FPLC on a Sephacryl S100 gel filtration column (Cytiva, Marlborough, MA) in 50 mM sodium phosphate (pH 8.0), 50 mM sodium chloride, 5 mM DTT. Fractions containing pure DGCR8 core by SDS-PAGE analysis were pooled and concentrated. Final concentration of the sample was determined via guanidine hydrochloride denaturation followed by UV absorption measurement, using ε = 544,800 (M ⋅ cm)−1 at 278 nm (11). DGCR8 core was buffer exchanged into 50 mM HEPES (pH 7.5) with 50 mM KCl, 5 mM DTT, and 1% glycerol before SAXS experiments.
Preparation of RNA molecules
To prepare pri-miR-16-1 for SAXS experiments, DNA purchased from Geneart (Thermo Fisher Scientific, Waltham, MA) containing a T7 promoter sequence at the 5′ end and an inverted BsaI cut site at the 3′ end was cloned into pUC19 (New England Biolabs, Ipswich, MA) and then transformed into DH5α-competent cells. These cells were grown overnight in LB media at 37°C until an effective OD of 3.75 was reached. The cells were then lysed, and the DNA was purified using a Plasmid Maxi Kit (Omega Bio-Tek, Norcross, GA). The recovered DNA was digested with BsaI overnight at 50°C. After digestion, calf intestinal alkaline phosphatase was added and incubated another 30 min at 37°C to prevent self-ligation. Postdigestion, the linearized DNA was extracted with phenolchloroform and precipitated with ethanol (26). The pri-miR-16-1 was transcribed overnight at 37°C by T7 polymerase using the T7 RiboMAX Express Large Scale RNA Production System (PROMEGA, Madison, WI) and subsequently buffer exchanged to a low-salt buffer (50 mM KCl, 50 mM HEPES (pH 7.5)). Formation of a homogenous stem-looped RNA molecule was confirmed by the presence of a single transition in UV-melting experiments (Cary 50; Agilent, Santa Clara, CA). The RNA was further purified using a Superdex 200 Increase 10 × 300 size-exclusion column (GE Healthcare, Chicago, IL). The final RNA concentration was determined by UV absorption at 260 nm, using ε = 1,125,400 (M ⋅ cm)−1. Pri-miR-16-1 RNA molecules were annealed at 85°C for 3 min, snap cooled to ice for 5 min, then brought to room temperature for an additional 5 min before storage at 4°C until needed for measurements.
The simpler RNA constructs for SAXS and FRET experiments were purchased desalted and HPLC purified from IDT (Coralville, IA). For SAXS, we used a canonical 25-basepair (bp) duplex RNA with sequence 5′-GCA UCU GGG CUA UAA AAG GGC GUC G and its complement. A flexible helix-junction-helix RNA molecule was made of two 12-bp helices joined by a single-stranded region of five uridine nucleotides: 5′- GCG AUU AGG AGG uuu uuG GGA GUA AAG GG. Complementary strands for the regions in capital letters were also purchased. Preparation of this flexible helix-junction-helix was described previously (27). The 29-bp RNA duplex samples for FRET were synthesized with an amino-modified dT at the desired site for internal labeling. RNA single strands were first labeled with the desired fluorophore (Alexa Fluor 488 TFP ester dye for donor and Cy5 NHS ester dye for acceptor; Thermo Fisher Scientific). The donor strand has a sequence of 5′-GCG A(dT-Alexa488)U AGG AGG CUG UUG GGA GUA AAG GG and the acceptor strand has a sequence of 3′-CGC UAA UCC UCC GAC AAC CCU CAU (dT-Cy5)UC CC. Excessive free dye in solution was removed by ethanol precipitation, and the two labeled strands were annealed together in a 1:2 donor/acceptor strand ratio at 92°C for 2 min to compensate for the lower-yield Cy5-labeled strand. The samples were slowly cooled to room temperature within an hour. Samples were then buffer exchanged to 30 mM KCl (for the duplex-junction-duplex) and 100 mM KCl (for the 25-bp duplex) in 50 mM potassium 3-(N-morpholino) propanesulfonic acid (K-MOPS) buffer.
Data collection for SAXS and CV-SAXS
SAXS data were collected at the Cornell High Energy Synchrotron Source (CHESS) G1 station with an x-ray energy of 9.95 keV. Profiles were recorded on a Pilatus 100 k detector (Dectris, Baden, Switzerland). The scattering angle was converted to momentum transfer (q = 4π sin θ/λ, where 2θ is the scattering angle and λ is the x-ray wavelength) based on scattering from a silver behenate calibration standard, analyzed via azimuthal integration using BioXtas RAW software (28,29). The distance from sample to detector was 1.517 m. SAXS samples, kept at ambient temperature (∼23°C), were loaded into a 2-mm quartz capillary sample flow cell, which allowed the sample to oscillate through the beam position to avoid radiation damage. 20 1-s exposures were recorded, and the images were normalized to x-ray intensity using the photon counts from the beamstop PIN diode. To control for radiation damage of the sample, exposures from the integrated individual frames were compared to ensure the absence of time-dependent changes. Data from statistically similar frames (with no time dependence) were averaged. To ensure good-quality SAXS data, scattering profiles of buffer were acquired before and after each sample measurement. These pre- and postsample buffer measurements were averaged and subtracted from the profile that contained the sample. Scattering profiles of the quartz capillary, both dry and filled with nanopure water, were acquired for use as an absolute calibration standard. The same quartz capillary was used for regular and CV-SAXS. Additional information about the SAXS samples, data collection, and analysis is shown in the Supporting Materials and Methods and summarized in Table S1, in compliance with the 2017 SAXS publication guidelines (30).
For regular SAXS measurements, SAXS data of pri-miR-16-1 alone (5, 10, and 20 μM), DGCR8 core alone (20 μM), and pri-miR-16-1-DGCR8 in complex at a 1:1 ratio (10 and 10 μM) were acquired in ordinary buffered conditions (50 mM HEPES (pH 7.5), 50 mM KCl, 5 mM DTT, and 1% glycerol). Guinier fits to display sample quality are shown as Figs. S1 and S2. SAXS curves from molecular controls, a 25-bp RNA duplex, and a 12-bp-junction-12-bp RNA construct (in 10 mM MOPS, 30–100 mM KCl (pH 7) buffer), were also measured. To perform SAXS with contrast variation, we used sucrose solutions as the contrast-matching agent. The theory and experimental considerations for CV-SAXS are reviewed by Tokuda et al. (23). To find the condition in which the electron density of the solution matches that of the DGCR8 protein, 80% (w/v) sucrose was prepared in the MOPS buffer described above. Scattering from DGCR8 protein mixed with the sucrose buffer at sucrose concentrations of 7.5, 15, 30, 47.5, and 50% was collected until the contrast match point (at 50% w/v sucrose), at which the buffer and protein sample are indistinguishable, was identified. More experimental details are presented in the Supporting Materials and Methods. Fig. S2 shows that the SAXS profile of the RNA and the measured Rg remain unchanged by the addition of 50% sucrose to the solvent, demonstrating that the RNA conformation is not altered by sucrose. However, the signal size is affected; the addition of 50% sucrose diminishes the strength of the RNA signal, reflecting the smaller electron density difference between solvent and solute. Fig. S3 demonstrates our empirical determination of the sucrose match point. The protein-RNA complex (20 μM, 1:1 ratio) and the RNA alone (20 μM) were prepared at this sucrose concentration for SAXS measurements. At least 15 min of equilibration was allowed for complex formation to occur for all buffer conditions.
Data analysis: absolute calibration and Rg measurements
After integration of the images from the detector, scattering curves were averaged and background subtracted using BioXtas RAW (28,29) and in-house-written MATLAB (The MathWorks, Natick, MA) scripts. Scattering intensity (I) is plotted against q. We used water as a calibrant to convert the y axis (I) to absolute units. To convert I(q) to absolute scattering units ([cm−1]), we use the equation
| (1) |
as well as the known scattering cross section of water, dΣ/dΩ = 0.0164 cm−1 at 23°C. I(q → 0)water was derived from the scattering of nanopure water minus scattering signal from an empty and dry quartz capillary, extrapolated to q = 0 (31,32). To show that the sample is monomeric, we calculate its molecular weight using the equation below and compare with the expected molecular weight of the molecule:
| (2) |
Here, Ms is the calculated molecular weight of the sample in g/mol, NA is Avogadro’s number, I(0)s is sample scattering intensity extrapolated to q = 0 (in cm−1) determined from either Guinier analysis or GNOM (33), c is the sample concentration in mg/mL and ΔρM is the excess scattering length per unit mass, 1.8 × 1010 cm/g for proteins and 3.6 × 1010 cm/g for RNA (34). Calculated radius of gyration (Rg) values reported here are from Guinier analysis (using either RAW or MATLAB), and the reported error was derived from standard deviation of multiple measurements. SAXS profiles are compared to those generated from available crystal structures using the program CRYSOL (35). The extracted molecular weights permitted determination of the stoichiometry of the protein-RNA complex formed.
Data analysis: molecular reconstructions and modeling
The program GNOM (33) was used to generate the associated pair-distance distribution functions, P(R), from the SAXS curves of sample with and without sucrose. GNOM outputs are the inputs to shape reconstruction algorithms using DAMMIF. Fifteen reconstructions were made in slow mode, averaged using DAMAVER, and compared with models by aligning the bead models to Protein Data Bank (PDB) files of molecular models using SUPCOMB. All programs were run through the SAXS analysis package ATSAS (36,37).
The model for pri-miR-16-1 was generated by running a short molecular dynamics (MD) simulation starting from the most probable SHAPE-constrained MC-Pipeline model reported in Quarles et al. (9) Simulations used a truncated construct in which the flanking tails and terminal loop were removed before simulation, leaving a 37-bp stem. Input files were generated with AmberTools 14.0 using ff14SB, which applies the ff99bsc0χOL3 force field to RNA molecules, 130,000 TIP3P waters in a truncated octahedron with a 12.0-Å distance between all RNA atoms and the nearest boundary, and 73 sodium ions for neutralization. Production MD simulations were run in AMBER 14 in the NPT ensemble with a pressure of 1.0 bar and a nonbonded cutoff of 12.0 Å, using a 2-fs timestep for 100 ns of total simulation time.
We used the fraction of common contact (FCC) algorithm (38) to determine angular differences between two models. From the SAXS reconstruction of the pri-miR-16-1 bound state, we determined a possible bent conformation of the pri-miR-16-1 using the FCC algorithm that we implemented in MATLAB. To measure the bend angle, we generated the most plausible bent conformation of pri-miR-16-1 by imposing conformational bending on the MD-simulated structure. With the MD structure as the starting point, we partitioned the structure into fixed and mobile parts using one basepair. A three-dimensional transformation matrix is applied to all the atomic coordinates of the mobile parts where θ is the polar (bending) angle with respect to the axis = (rx, ry, rz).
| (3) |
The axis was estimated to be the central axis of the pri-miR-16-1 RNA, and the FCC was defined as
| (4) |
where A is the pri-miR-16-1 atoms and B is the SAXS reconstruction. For each basepair as bending pivot, we searched for the bending angle θ that maximizes the FCC without steric clashes.
Data collection for single-molecule FRET
smFRET measurements were performed on freely diffusing labeled RNAs using a home-built microscope with confocal detection (39,40). Two hundred microliters of fluorescent RNA samples at nanomolar concentrations (with or without added DGCR8 core) were loaded into a chambered coverglass (Nunc; Thermo Fisher Scientific) and illuminated with a 488-nm laser through a 60× 1.2 NA objective. The fluorescence emission was collected through the same objective and split into donor and acceptor channels using a 550-nm longpass dichroic. The donor channel contained an additional 530/30 bandpass emission filter, whereas the acceptor channel contained a 630-nm longpass emission filter. Two 50-nm optical fibers provided confocal detection and photons were detected by two avalanche photodiodes (SPCM-AQR-14; Perkin Elmer, Santa Clara, CA). Data were acquired using a Flex-2kD correlator card (correlator.com) in photon counting mode. Photon arrival times were sampled in 25-ns intervals. For each data point, 20 sets of 30-s photon streams from donor and acceptor channels were collected. The data sets were processed in parallel by an all-photon burst search algorithm (41) with default parameters written in MATLAB. For each burst, the efficiency of energy transfer, EFRET, is
| (5) |
where IA and ID are the number of photons from the acceptor and donor channels respectively. The parameter γ accounts for the difference in quantum yields (Φ) and detection efficiencies (η) in both channels.
| (6) |
For our confocal setup, we found γ = 1.2. At least two independent measurements were taken to estimate the variance in the measurement of EFRET. The binding affinity of the labeled RNA to the DGCR8 core protein was measured on the same instrument by fitting diffusion parameters using fluorescence correlation spectroscopy (FCS) measurements as previously described (42,43).
Results
SAXS with absolute calibration confirms protein-RNA complex formation
To test the hypothesis that DGCR8 can bend primary miRNA, we used a minimal construct of DGCR8 (DGCR8 core) that consists of the two dsRBDs and a C-terminal tail (Fig. 1 A). We used the pri-miR-16-1 sequence to assess binding to the DGCR8 core; this RNA contains the loops, bulges, and flanking single-stranded RNA found in many primary miRNAs (Fig. 1 B). Binding of dsRBDs to duplex RNA is not sequence specific (14,44). In our previous work, we showed that the DGCR8 core and pri-miR-16-1 bind with a KD of ∼1 μM (11). Using a combination of CV-SAXS and smFRET, we present evidence to support a model in which pri-miR-16-1 bends upon binding to the DGCR8 core (modeled in Fig. 1 C).
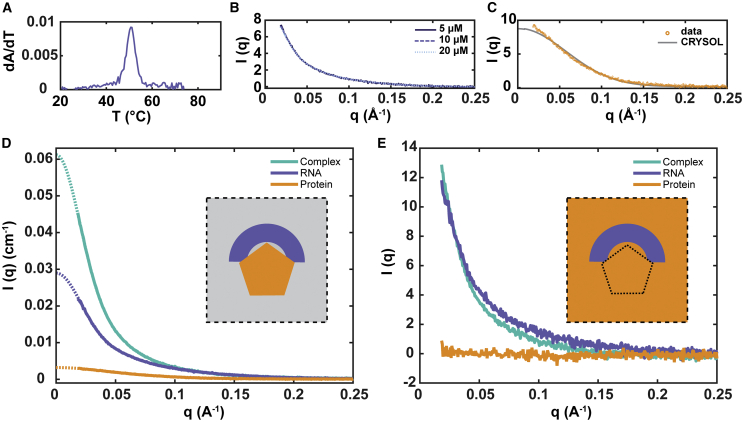
To study the DGCR8-pri-miR-16-1 complex using SAXS, we first established scattering profiles for monomeric DGCR8 core and pri-mi-16-1 in isolation from each other and subsequently demonstrated that the protein-RNA pair forms a 1:1 complex. Fig. 2 A shows the UV-melting profile of pri-miR-16-1, which indicates a single transition at 51°C that reflects melting of the stem loop to single strands. Note that a mixture of monomers and dimers (monomers that hybridized to form duplex RNA) would have two melting transitions. Fig. 2 B shows SAXS profiles of the RNA alone at different pri-miR-16-1 concentrations: 5, 10, and 20 μM. Because RNA molecules are negatively charged, it is important that the SAXS curves fall on top of each other within error to demonstrate the absence of concentration-dependent structural changes resulting from intermolecular repulsive interactions at the buffer conditions and concentrations used. This control also confirms the absence of aberrant assembly or aggregation at higher RNA concentrations (45). Using Guinier analysis, we established that the Rg from three independent measurements of the pri-miR-16-1 is 44.0 ± 1.4 Å. Fig. 2 C shows SAXS profiles of the DGCR8 core at a concentration of 20 μM compared with the simulated profile generated from a CRYSOL (35) calculation from the crystal coordinates (PDB: 2YT4) (21). The measured Rg of the DGCR8 core is 23.7 ± 1.1 Å, which is close to the CRYSOL-predicted value of 20.7 Å. This close (but imperfect) match between crystal and solution structures likely reflects the exploration of conformational space in solution. Absolute calibration with water as a standard, as described in Materials and Methods, allows conversion of SAXS intensity profiles to absolute units and confirms that the protein and RNA are monomeric when alone and form a 1:1 protein-RNA complex when together. The Guinier plots are shown in Fig. S1. Fig. 2 D shows profiles of the protein alone, RNA alone, and the protein-RNA complex in absolute scattering units.
Figure 2.
RNA pri-miR-16-1 and the protein DGCR8 core are monomers in solution and form a 1:1 complex in the bound state. (A) UV-melting profile, dA/dT, of 0.5 μM pri-miR-16-1 is given, showing a single melting transition at 51°C. (B) SAXS profiles of pri-miR-16-1 at 5, 10, and 15 μM show no concentration dependence of the scattering. (C) Experimental and predicted SAXS profiles of the protein DGCR8 core are shown. (D) SAXS profiles, in absolute units, of DGCR8 core protein alone (orange), pri-miR-16-1 RNA alone (blue), and the complex formed (cyan) by binding of pri-miR-16-1 RNA to the DGCR8 core are shown. The dotted lines in the figure denote application of GNOM to extrapolate the scattering curves to I(q = 0). (E) SAXS profiles of protein alone (orange), RNA alone (blue), and complex (cyan) in contrast-matched buffer containing 50% sucrose are given. Insets in (D) and (E) are illustrations of the contrast-matching method. Under aqueous buffer conditions, without sucrose (inset of D), signals from both protein (orange) and nucleic acid (blue) are reported. Under contrast-matching conditions (inset of E) when molecules are in a buffer with sucrose concentration that matches the electron density of the protein (orange), only signal from the nucleic acid component (blue) of an RNA-protein complex contributes to the measured SAXS curve. To see this figure in color, go online.
To understand how complex formation is determined in absolute units, recall that the SAXS measurements report the scattering intensity, I(q). I(q) is the product of the scattering amplitude and its complex conjugate (23,46). The amplitude, A(q), can be described as the product of this scattering factor, f, which is dependent on the solvent-corrected number of electrons in the macromolecule and an angle (q)-dependent form factor, F(q), that reflects the arrangement of electrons in the macromolecule. For a single-component system, the scattering amplitude is given by A(q) = f1F1(q), and the measured scattering intensity can be written as
| (7) |
where P1(q) is the partial scattering form factor of this molecule, given by F1(q)F1∗(q), integrated over all space. P1(q) reflects the shape of the macromolecule, and its value is unity at q = 0.
For a two-component system, the total scattering amplitude reflects the contribution from each component: A = f1F1(q) + f2F2(q). The resulting expression for scattering intensity contains cross-terms in proportion to various products of the form factors, F1(q) and F2(q). For this system, the scattering intensity I(q) is given by
| (8) |
where P1(q) and P2(q) are the partial scattering form factors for each of the two components and reflect their individual contributions. The cross-term, P12(q), contains information about the relative distributions of electrons between the two components (23).
Fig. 2 D shows the SAXS profiles of DGCR8 protein alone (in orange), pri-miR-16-1 RNA alone (in blue), and the DGCR8-RNA complex (in cyan) mixed at equimolar concentrations of 10 μM each. The scattering signal of the complex is greater than what is expected if the protein and RNA solutions are mixed but do not associate. This signal increase arises from the cross-term, 2f1f2P12(q), and is a clear indication that a complex has formed. We note that the absolute scattering intensity of the protein is much less than the RNA 1) because of the relative sizes of each molecule and 2) because proteins are less electron dense than nucleic acids.
We also used absolute calibration to compute molecular weights of the DGCR8 core and the pri-mirR16 as described in the Materials and Methods. The calculated molecular weight of the DGCR8 core is 21,400 Da (within 18% of the 26,000-Da expected value), and the calculated molecular weight of pri-miR-16-1 is 35,970 Da (within 1% of the 36,170-Da expected value). These numbers are in good agreement considering the other sources of error in using SAXS for molecular weight estimates (e.g., estimates of the scattering length per unit mass, estimates of sample concentration, etc.). Overall, they provide confidence that the samples are monodisperse.
The absolute scattering at q = 0 can also determine whether the complex is formed in a 1:1 ratio. Fig. 2 D shows the application of GNOM (33) to extrapolate the scattering curves to I(q = 0). We used the extrapolated values derived from the RNA- and protein-alone samples to estimate the I(0) of the protein-RNA complex using
| (9) |
From this equation, we predict that the I(0) of the complex should be 0.067 cm−1, which is within 9% of the measured value of 0.061 cm−1, supporting the formation of a 1:1 complex between DGCR8 core protein and pri-miR-16-1RNA. The generated Rg from the complex is 50.2 ± 0.8 Å, but it is difficult to distinguish the contribution of the different molecules from the scattering profile of the complex, so we next turned to contrast variation to exclusively observe scattering from the RNA molecules by screening out the protein signal.
Application of contrast variation SAXS to visualize RNA in protein-RNA complexes
To visualize the RNA component of the protein-RNA complex, we can change the contrast of the solvent to equal that of the protein. Under this condition, any remaining signal (above background) should arise solely from the RNA. As discussed in the Materials and Methods, CV-SAXS exploits the fact that the SAXS signal arises from the electron density difference between the macromolecule and the solvent. The scattering form factor of a molecule, described above, is given by fM = ΔρMVM, where VM indicates the volume of the macromolecule. In an environment in which there is no solvent (i.e., in vacuum), fM is essentially equal to the number of electrons within the macromolecule. When the macromolecule is in a buffered saline solution, fM reflects the number of excess electrons above that contained by the solvent in the same volume: ΔρM = ρM − ρM,solv. We explained this concept in detail in a 2016 review article (23).
Given a two-component system made of an RNA molecule (component 1) and a protein molecule (component 2), the SAXS scattering intensity is described by Eq. 8. If the solvent electron density (ρsolv) is increased to equal that of the protein molecule (ρ2), then f2 = 0 and the second and third terms of Eq. 8 vanish. For the pri-miR-16-1 and DGCR8 core system, f2 = 0 when the solvent is made up of 50% (w/v) sucrose. The scattering signal from the protein component of the complex disappears as shown in Fig. 2 E; under 50% sucrose conditions, the retained scattering signal of the complex is exclusively from the RNA. Based on the association constant of the protein and RNA, a very small fraction of free protein and free RNA may remain in solution. The signal from the free protein is blanked out by the addition of sucrose. The small signal from the free RNA may add some heterogeneity to the scattering profile of the RNA in the complex. In Fig. 2 E, we find that the SAXS profile of the pri-miR-16-1 RNA by itself (in the absence of its protein partner) is distinct from the RNA profile in complex with the DGCR8 core. Because SAXS reports the overall shape of the molecule, we deduced that the conformation of pri-miR-16-1 changes upon protein binding.
Contrast variation SAXS shows bending of miR16 in DGCR8-mir16 complex
CV-SAXS at the correct match point nullifies the signal from the protein and still reports on RNA conformation in the presence of the protein. We applied the program GNOM to generate the real-space pair-distance distribution function (P(R)) of pri-miR-16-1 without and with the DGCR8 core, using the relevant I vs. q scattering curves as input. Note that with the DGCR8 core, the RNA remains bound to the protein in the presence of sucrose, and the contrast agent simply makes the protein invisible, as illustrated in the insets in Fig. 2, D and E.
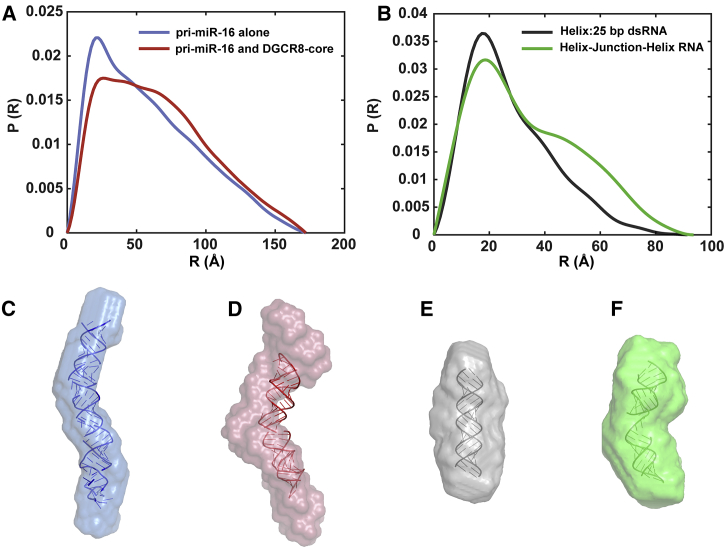
The P(R) curves for the pri-miR-16-1 (alone and complexed with the DGCR8 core) are shown in Fig. 3 A. The RNA-alone curve, in blue, displays a pair-distance distribution characteristic of a dsRNA duplex; a peak close to 20 Å signifies the helix diameter, and a subsequent linearly decaying profile reflects its roughly cylindrical global structure. The value of R at which P(R) drops to zero indicates the maximal dimension of the molecule, DMAX. The RNA in complex is in red, and the protein is invisible because of contrast matching. The DMAX-value is comparable between the RNA alone (blue) and RNA in complex (red), but there are notable changes in the P(R) curve that suggest a bent structure. Upon protein binding, the peak at 20 Å is less pronounced, and there is a higher density of pair-distribution distances on length scales between 50 and 100 Å not seen in the RNA by itself. These features suggest a deviation from a rigid cylindrical rod profile and that distant parts of the molecule can come closer together. Similar changes are detected when comparing the different scattering profiles and P(R)-values of rigid 25-bp RNA duplexes and flexible 12-bp-5U-nucleotide-12-bp (helix-junction-helix) RNA constructs. The pair-distance distribution functions of these constructs (computed using the same GNOM analysis) are shown in Fig. 3 B. The control molecules are shorter than the pri-miR-16-1 and display smaller DMAX-values, but the overall features are comparable. The P(R) curve for the pri-miR-16-1 by itself closely resembles the rigid dsRNA duplex, whereas the pri-miR-16-1 in complex has features that reflect the added flexibility in the helix-junction-helix construct. This latter molecule has been extensively studied by our group (27,39). Overall, this simple comparison already suggests that, in complex, the protein bends the RNA.
Figure 3.
Comparisons of pair-distance distribution functions and solution SAXS reconstructions show notable bend of pri-miR16-1 upon binding the DGCR8 core. (A) P(R) of pri-miR-16-1 alone (blue) and pri-miR-16-1 bound to the DGCR8 core (red) with the protein signal blanked out by the contrast-matching agent (50% sucrose) is shown. (B) P(R) of a rigid model system duplex RNA (gray) and a flexible helix-junction-helix RNA molecule (green) is shown. For ease of comparison, P(R) curves are normalized by dividing each curve by the area under the curve. Shape envelopes from SAXS reconstructions: (C) pri-miR-16-1 alone (blue), (D) pri-miR-16-1 bound to DGCR8 core (red) superimposed to PDB models described in the text, (E) duplex RNA (gray), and (F) helix-junction-helix RNA (green) also superimposed to PDB models are shown. The PDB models in (C) and (F) are were determined from MD simulations. The PDB model in (D) was determined by imposing a bend in the PDB model from (C) such that the value of the FCC is 1 when compared to the SAXS reconstruction of pri-miR-16-1 bound to the DGCR8 core. The PDB model shown in (E) is from a Nucleic Acid Builder canonical A-form RNA helix. To see this figure in color, go online.
We visualized the shape of the molecules through the application of reconstruction algorithms (see Materials and Methods) to determine low-resolution three-dimensional solution structures from SAXS scattering profiles. The averaged dummy atom model from reconstructions are shown in Fig. 3, C–F as semitransparent shape envelopes. The individual reconstructions for pri-miR16-1 (alone and in complex with the DGCR8 core) from DAMMIF are shown in Figs. S4 and S5. The data and models have been assigned ID numbers SASBDB: SASDJV7 and SASBDB: SASDJW7 by the Small Angle Scattering Biological Data Bank (47). In Fig. 3 C, the reconstructed pri-miR-16-1 alone, shown as a light blue shape envelope, aligns well with the truncated pri-miR-16-1 model PDB structure in dark blue. The PDB model was generated from an MD simulation that was run to equilibrate the most probable pri-miR-16-1 structure predicted by MC-Pipeline (9). The transparent red solution structure depicts RNA in complex with protein (Fig. 3 D). Here, the shape reconstructions of the RNA in complex show a clear signature of bending that is absent in the pri-miR-16-1 alone, suggesting that the DGCR8 core bends the primary miRNA. To measure the bend angle, we generated the most plausible bent conformation of pri-miR-16-1 by imposing conformational bending on the dark blue MD-simulated structure using the FCC algorithm (38) and requiring that it follow the shape of the light red shape reconstruction. This procedure is described in the Materials and Methods. The resulting bent model that gives a maximal FCC-value of 1 is colored dark red and superimposed on the light red solution structure in Fig. 3 D. The final estimate of the bending angle is 39.8°, with the pivot point located near the center of the molecule.
To further illustrate the extent of differences between a straight conformation and the bent molecular model consistent with our CV-SAXS data, we applied the SAXS reconstruction algorithms to the 25-bp canonical RNA duplex and the 12-bp-5 nt-12-bp helix-junction-helix. The shape reconstruction and superimposed PDB models are shown in Fig. 3, E and F, respectively. The PDB model for the 25-bp canonical duplex RNA was made using Nucleic Acid Builder (48). The PDB model for the helix-junction-helix at the low-salt conditions shown here (30 mM KCl) was derived from MD simulations described previously (27). At higher salt concentrations, the helix-junction-helix molecule can access more conformations, and the helices can come closer together. Because the pri-miR-16-1 is kept at low salt (50 mM KCl), with and without the DGCR8 core, we chose to compare it with the low-salt state of the helix-junction-helix construct. The conformation of the duplex RNA does not depend on salt conditions at this resolution.
FRET confirms SAXS measurements
The above studies present evidence suggesting that pri-miR-16-1 bends in the presence of the DGCR8 core. To further explore this model, we performed smFRET experiments on a complex consisting of the DGCR8 core and duplex RNA. We first attempted to construct a FRET-labeled hybridized molecule from two single-stranded fluorescently labeled RNA strands with the bulges and mismatches found in pri-mirR16-1; however, the resulting constructs were unstable. We therefore selected a 29-bp RNA duplex as a FRET construct. This duplex has a similar length to the stem of pri-miR-16-1 and stays hybridized under our smFRET conditions. Its selection is consistent with our previous work in which we showed that the DGCR8 core binds short canonical duplex RNA with a KD of ∼3–4 μM (11), similar to that of the pri-miR16-1 (albeit slightly weaker). These prior results suggested that it would be feasible to test whether the bending of the duplex RNA occurs upon binding the DGCR8 core through changes in FRET efficiency.
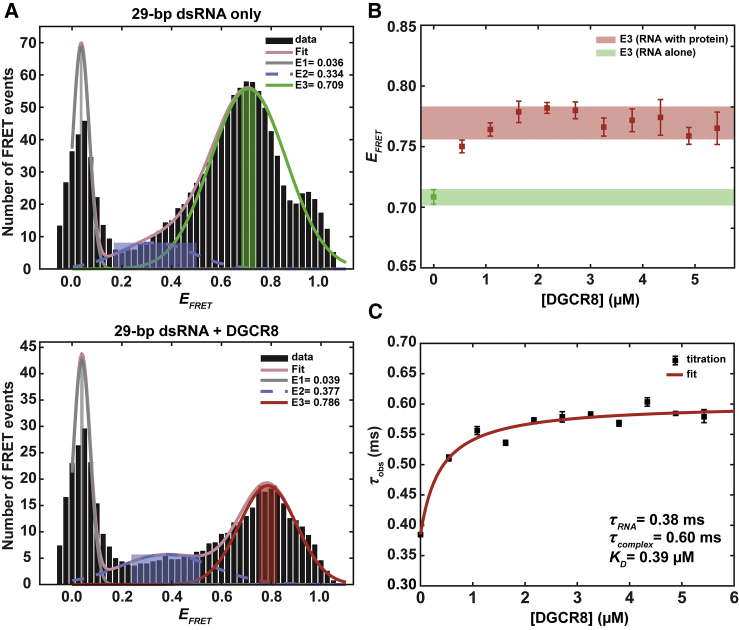
Using the smFRET experimental setup and the 29-bp labeled RNA construct described in Materials and Methods, we measured energy transfer between dyes on the RNA both in the presence and absence of protein. Fig. 4 A shows representative smFRET histograms of the labeled RNA by itself (Fig. 4 A, top) and labeled RNA with DGCR8 core (Fig. 4 A, bottom). We fit the FRET histogram to a model of three Gaussians to determine the relevant FRET signals, as described previously (39). The first two peaks, E1 and E2, do not change appreciably between free and protein-bound conditions; we associate the FRET signal due to the distance between the two dyes to the third FRET peak, E3. The shift in the E3 peak position to a higher value (in the bottom histogram) reflects the decrease in separation distance between the two dyes. Without the protein, this peak is centered near EFRET = 0.7 (green peak, top histogram), and it shifts closer to EFRET = 0.8 (red peak, bottom histogram) after the addition of the DGCR8 core, reflecting increased FRET efficiency. Fig. 4 B shows EFRET-values for E3 as a function of DGCR8 concentration and confirms that the E3 FRET efficiency shifts (from the green bar to the red bar) upon binding of the DGCR8 core.
Figure 4.
Single-molecule FRET experiments show a conformational change in a 29-bp duplex RNA upon binding of DGCR8 core. (A) FRET histograms of the RNA alone (top) and the RNA with the DGCR8 core (bottom) are given. The FRET signal resulting from energy transfer from donor dye to acceptor dye is assumed to be the third FRET peak, E3 (green peak in the top histogram and red peak in the bottom), because of the distance it represents. The peak position of E3 shifts to a higher EFRET-value upon protein binding. The zero-FRET peak (E1, gray) and low peak next to it (E2, blue) are included in a three-Gaussian fitting to precisely identify the position of E3. E1, the zero-FRET peak, can be attributed to the presence of donor-only RNA strands and/or formation of a dark triplet state of the acceptor dye (49). E2 arises from noise in the histograms because of its wide distribution across all FRET values. (B) EFRET-values for E3 increase upon titration of DGCR8 core to the labeled RNA. The colored bars display the error range of the FRET efficiencies across multiple titrations. The error bars for each data point indicate the 95% confidence intervals of the mean of the peak. The lower EFRET-value near 0.7 for RNA alone (green bar) reflect the separation distance between the two dyes for a rigid duplex, and the higher EFRET-values for the protein-bound state (red bar) show that the dyes moved closer together upon protein binding. (C) FCS titration curve showing the increase in observed diffusion time when a mixture of the bound protein-RNA complex and unbound RNA are present is given. The solid line is the fit from Eq. 10, which allows us to determine the binding constant. To see this figure in color, go online.
To measure the binding affinity for the DGCR8 core and the labeled RNA construct, we used the same smFRET setup and correlator card for FCS measurements, as described previously (42). We observed a shift from a single-component diffusion (RNA alone) to a two-component system when the DGCR8 core was incubated with the RNA. The protein-bound RNA diffuses more slowly than the RNA alone (43). Fig. 4 C shows the effective change in diffusion time, τd, when there are two components present in the solution: the RNA alone versus the RNA in complex with the DGCR8 core. The protein is unlabeled and does not emit a fluorescent signal. The diffusion time is represented by the variable τd, and the fit to τd determines the binding affinity:
| (10) |
Here, M denotes the DGCR8 core concentration and KA = 1/KD. The KD from the fit was 0.39 ± 0.14 μM, where the error represents the 68% confidence bounds associated with the fit. We therefore verified that the DGCR8 core binds 29-bp labeled RNA with a similar KD to that specified in Quarles et al. (11). Our FCS-based measurements show slightly tighter binding than measured previously.
We used the following well-known equation to convert the transfer efficiency to average dye-dye distance, R, given the Forster distance, R0, between Alexa 488 and Cy5 is 52 Å.
| (11) |
Using this equation and the average E3-values in Fig. 4 B, we found a decrease in separation distance between the two dyes from 48 to 43 Å upon binding the DGCR8 core. The distance of 48 Å is consistent with separation distance of the Alexa488 and Cy5 conjugated along the 29-bp dsRNA. Because duplex RNA is fairly rigid, the observation of a decrease in separation distance between the two dyes provides more support for the conclusion derived from the SAXS experiments that the DGCR8 core bends the RNA upon binding.
Discussion
DGCR8 and Drosha are the minimal components of the protein complex called the microprocessor, which is a critical component to the first stage of the miRNA maturation process. In the microprocessor, Drosha functions as a ribonuclease and determines the cleavage sites in the primary miRNA (16). The DGCR8 protein has multiple functions, but it primarily recognizes the substrate pri-miRNA to be cleaved by Drosha (20,50). The mechanism by which DGCR8 works with Drosha to direct pri-miRNA cleavage is not widely understood. A recent crystal structure (18) of a large fragment of human Drosha with a fragment of the DGCR8 C-terminal tail provided substantial insight into how Drosha facilitates the first stage of miRNA processing but also led to new questions about the structural dynamics of the process.
Comprehensive structural mechanistic studies of DGCR8-pri-miRNA complexes have proven challenging because of the dynamics of the interaction and the lack of RNA sequence specificity of DGCR8. Here, our implementation of both SAXS and smFRET show that, together with indiscriminate binding, indiscriminate bending of RNA also occurs. The use of absolute calibration and CV-SAXS confirmed the 1:1 binding of the complex and, in conjunction with modeling, revealed that the protein induces a notable bend in the RNA. smFRET experiments carried out on duplexes (not on the pri-miR-16-1 RNA, which proved technically challenging) suggest that the DGCR8 core bends basepaired molecules, as well as molecules with more design flexibility. Taken together, these results suggest that the DGCR8 core indiscriminately bends both canonical duplex RNA and pri-miRNA, consistent with a model first proposed by Sohn et al. (21) based on the crystal structure of the DGCR8 core and ensemble FRET measurements. They suggested that, based on the positioning of the two dsRBDs within the crystal structure, a long duplex RNA needs to be substantially kinked for two regions of the molecule to be bound to each dsRBD. We depict this model in Fig. 1 C. From our findings, we can support this model for DGCR8-pri-miRNA interaction in which a single RNA molecule is bent upon binding to DGCR8. Finally, these observations are consistent with our previous work, showing that the binding affinity of DGCR8 to both duplex RNA and pri-miRNA is fairly similar and that flexibility or lack thereof in the potential Drosha cleavage site is not sensed by the dsRBDs of the DGCR8 core (11). We suggested that this indiscriminate binding is likely a feature of the RNA binding domains in DGCR8 that facilitates processing of hundreds of structurally diverse primary miRNAs (11).
Although the interactions between dsRNA and single dsRBDs are well understood (44), we are still learning how proteins with two or more dsRBDs function. Existing models for dsRBD interaction with dsRNA derived from x-ray crystallography, electron microscopy, NMR spectroscopy, and mass spectrometry yield important insights into the structure of dsRBD-RNA complexes (14,15,21). Binding of the DGCR8 core to RNA molecules happens through the two dsRBDs. These dsRBDs have an αβββα structure and bind duplex RNA through sequence-independent nonspecific interactions (44,51). Examples from the literature for other proteins, such as the protein kinase PKR (52,53) or the Dicer binding partner TRBP (14), can help address the general question of how proteins with multiple dsRBDs bind RNA. Like the DGCR8 core, PKR and TRBP have multiple dsRBDs; the difference is that in PKR and TRBP, the dsRBDs are allowed to have independent motions from one another in the absence of RNA. The dsRBDs in DGCR8 are in stable contact with one another in the absence of RNA, and these rigid contacts probably dictate the bending of duplex RNA that we observed here.
Conclusion
Our observation that a pri-miRNA bends upon binding to the DGCR8 dsRBDs contributes to the global understanding of RNA-protein systems and RNA-protein interactions in general and how the microprocessor complex efficiently processes pri-miRNA in particular. Hypothetically, RNA bending by DGCR8 could facilitate cleavage by Drosha because the physical bend can relax the strain in the rigid duplex, expose the cleavage site, and stimulate cleavage and the subsequent release of the cleaved pri-miRNA. We can also imagine a dynamic picture of a cluster of primary miRNA within a single transcript or the accumulation of pri-miRNA within transcription-associated subnuclear compartments waiting to be cleaved by the Drosha-DGCR8 complex and how bending contributes to the efficiency and energetics of the process. Bending could be a necessary component of a dynamic sequence of events involving catch and release of pri-miRNAs waiting to be processed. In this hypothetical picture, it is interesting to infer how the proposed 2:1 DGCR8:Drosha stoichiometry (16,17) plays a role in final cut-site energetics and dynamics, and we look forward to further studies to visualize how this could play out. From previous studies (8,9), we understand that a sequential model of DGCR8 recruitment before Drosha cleavage is unlikely. Bending of RNA by DGCR8 supports the notion that DGCR8 and Drosha work together for efficient and effective processing. Although DGCR8-induced RNA bending could facilitate Drosha binding, without sequence specificity it is not sufficient to establish the cleavage site with the required single-nucleotide precision. Mechanisms to establish the necessary specificity, such as through the presence of mismatched GHG motifs (54), offer tantalizing insights into how RNA bending by DGCR8 and the presence of sites with high SHAPE reactivity near the Drosha cut site may synergistically lead to both fast and precise cleavage. Finally, our findings here highlight how protein binding can contribute to RNA structural changes. In a recent study, we showed that proteins partners can help stabilize RNA structure (55). Work by other groups also showed the generality of mechanisms in which proteins help fold RNA. Williamson and co-workers recently showed that transient protein-RNA interactions guide ribosomal RNA folding (56), and Walter and co-workers have shown proteins can facilitate RNA unfolding by untying an RNA pseudoknot (57). We are now beginning to understand the intricate and dynamic relationship between RNA and proteins. Future work should address the kinetics and energetics of RNA binding and conformational change and how these events drive biological function.
Author Contributions
S.A.P. designed SAXS research, performed SAXS research, and analyzed data. Y.-L.C. designed FRET research, performed FRET research, and analyzed data. E.T.U. performed SAXS research and analyzed data. E.C.C. performed SAXS research. L.P. designed research and oversaw the project. S.A.S. designed research and oversaw the project. All authors contributed to writing the manuscript.
Acknowledgments
We thank Richard Gillilan of the CHESS and the Pollack and Showalter group members for experimental assistance.
This work is based upon research conducted at the Center for High Energy X-ray Sciences, which is supported by the National Science Foundation under award DMR-1829070, and the Macromolecular Diffraction at CHESS (MacCHESS) facility, which is supported by award 1-P30-GM124166-01A1 from the National Institute of General Medical Sciences, National Institutes of Health, and by New York State’s Empire State Development Corporation. L.P. is supported by National Institutes of Health R35-GM122514. S.A.S. is supported by National Institutes of Health R01-GM098451.
Editor: Mark Williams.
Footnotes
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2020.10.038.
Contributor Information
Lois Pollack, Email: lp26@cornell.edu.
Scott A. Showalter, Email: sas76@psu.edu.
Supporting Material
References
- 1.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Kim V.N. MicroRNA biogenesis: coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 3.Ameres S.L., Zamore P.D. Diversifying microRNA sequence and function. Nat. Rev. Mol. Cell Biol. 2013;14:475–488. doi: 10.1038/nrm3611. [DOI] [PubMed] [Google Scholar]
- 4.Michlewski G., Cáceres J.F. Post-transcriptional control of miRNA biogenesis. RNA. 2019;25:1–16. doi: 10.1261/rna.068692.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ha M., Kim V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 6.Dallaire P., Tan H., Major F. Structural dynamics control the MicroRNA maturation pathway. Nucleic Acids Res. 2016;44:9956–9964. doi: 10.1093/nar/gkw793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burke J.M., Kelenis D.P., Sullivan C.S. A central role for the primary microRNA stem in guiding the position and efficiency of Drosha processing of a viral pri-miRNA. RNA. 2014;20:1068–1077. doi: 10.1261/rna.044537.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roth B.M., Ishimaru D., Hennig M. The core microprocessor component DiGeorge syndrome critical region 8 (DGCR8) is a nonspecific RNA-binding protein. J. Biol. Chem. 2013;288:26785–26799. doi: 10.1074/jbc.M112.446880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quarles K.A., Sahu D., Showalter S.A. Ensemble analysis of primary microRNA structure reveals an extensive capacity to deform near the Drosha cleavage site. Biochemistry. 2013;52:795–807. doi: 10.1021/bi301452a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Acevedo R., Orench-Rivera N., Showalter S.A. Helical defects in microRNA influence protein binding by TAR RNA binding protein. PLoS One. 2015;10:e0116749. doi: 10.1371/journal.pone.0116749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quarles K.A., Chadalavada D., Showalter S.A. Deformability in the cleavage site of primary microRNA is not sensed by the double-stranded RNA binding domains in the microprocessor component DGCR8. Proteins. 2015;83:1165–1179. doi: 10.1002/prot.24810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bevilacqua P.C., Ritchey L.E., Assmann S.M. Genome-wide analysis of RNA secondary structure. Annu. Rev. Genet. 2016;50:235–266. doi: 10.1146/annurev-genet-120215-035034. [DOI] [PubMed] [Google Scholar]
- 13.Hentze M.W., Castello A., Preiss T. A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell Biol. 2018;19:327–341. doi: 10.1038/nrm.2017.130. [DOI] [PubMed] [Google Scholar]
- 14.Masliah G., Maris C., Allain F.H. Structural basis of siRNA recognition by TRBP double-stranded RNA binding domains. EMBO J. 2018;37:e97089. doi: 10.15252/embj.201797089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Z., Wang J., Wang H.W. Cryo-EM structure of human dicer and its complexes with a pre-miRNA substrate. Cell. 2018;173:1191–1203.e12. doi: 10.1016/j.cell.2018.03.080. [DOI] [PubMed] [Google Scholar]
- 16.Nguyen T.A., Jo M.H., Woo J.S. Functional anatomy of the human microprocessor. Cell. 2015;161:1374–1387. doi: 10.1016/j.cell.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 17.Herbert K.M., Sarkar S.K., Steitz J.A. A heterotrimer model of the complete microprocessor complex revealed by single-molecule subunit counting. RNA. 2016;22:175–183. doi: 10.1261/rna.054684.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwon S.C., Nguyen T.A., Woo J.S. Structure of human DROSHA. Cell. 2016;164:81–90. doi: 10.1016/j.cell.2015.12.019. [DOI] [PubMed] [Google Scholar]
- 19.Pong S.K., Gullerova M. Noncanonical functions of microRNA pathway enzymes - Drosha, DGCR8, Dicer and Ago proteins. FEBS Lett. 2018;592:2973–2986. doi: 10.1002/1873-3468.13196. [DOI] [PubMed] [Google Scholar]
- 20.Macias S., Cordiner R.A., Cáceres J.F. DGCR8 acts as an adaptor for the exosome complex to degrade double-stranded structured RNAs. Mol. Cell. 2015;60:873–885. doi: 10.1016/j.molcel.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sohn S.Y., Bae W.J., Cho Y. Crystal structure of human DGCR8 core. Nat. Struct. Mol. Biol. 2007;14:847–853. doi: 10.1038/nsmb1294. [DOI] [PubMed] [Google Scholar]
- 22.Warf M.B., Johnson W.E., Bass B.L. Improved annotation of C. elegans microRNAs by deep sequencing reveals structures associated with processing by Drosha and Dicer. RNA. 2011;17:563–577. doi: 10.1261/rna.2432311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tokuda J.M., Pabit S.A., Pollack L. Protein-DNA and ion-DNA interactions revealed through contrast variation SAXS. Biophys. Rev. 2016;8:139–149. doi: 10.1007/s12551-016-0196-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Y., Tokuda J.M., Pollack L. Revealing transient structures of nucleosomes as DNA unwinds. Nucleic Acids Res. 2014;42:8767–8776. doi: 10.1093/nar/gku562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Y., Tokuda J.M., Pollack L. Asymmetric unwrapping of nucleosomal DNA propagates asymmetric opening and dissociation of the histone core. Proc. Natl. Acad. Sci. USA. 2017;114:334–339. doi: 10.1073/pnas.1611118114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wostenberg C., Quarles K.A., Showalter S.A. Dynamic origins of differential RNA binding function in two dsRBDs from the miRNA “microprocessor” complex. Biochemistry. 2010;49:10728–10736. doi: 10.1021/bi1015716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y.L., Lee T., Pollack L. Conformations of an RNA helix-junction-helix construct revealed by SAXS refinement of MD simulations. Biophys. J. 2019;116:19–30. doi: 10.1016/j.bpj.2018.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nielsen S.S. BioXTAS RAW, a software program for high-throughput automated small-angle X-ray scattering data reduction and preliminary analysis. J. Appl. Cryst. 2009;42:959–964. [Google Scholar]
- 29.Hopkins J.B., Gillilan R.E., Skou S. BioXTAS RAW: improvements to a free open-source program for small-angle X-ray scattering data reduction and analysis. J. Appl. Cryst. 2017;50:1545–1553. doi: 10.1107/S1600576717011438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trewhella J., Duff A.P., Whitten A.E. 2017 publication guidelines for structural modelling of small-angle scattering data from biomolecules in solution: an update. Acta Crystallogr. D Struct. Biol. 2017;73:710–728. doi: 10.1107/S2059798317011597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orthaber D., Bergmann A., Glatter O. SAXS experiments on absolute scale with Kratky systems using water as a secondary standard. J. Appl. Cryst. 2000;33:218–225. [Google Scholar]
- 32.Meisburger S.P., Pabit S.A., Pollack L. Determining the locations of ions and water around DNA from X-ray scattering measurements. Biophys. J. 2015;108:2886–2895. doi: 10.1016/j.bpj.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Svergun D.I. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Cryst. 1992;25:495–503. [Google Scholar]
- 34.Mylonas E., Svergun D.I. Accuracy of molecular mass determination of proteins in solution by small-angle X-ray scattering. J. Appl. Cryst. 2007;40:245–249. [Google Scholar]
- 35.Svergun D., Barberato C., Koch M.H. CRYSOL - a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Cryst. 1995;28:768–773. [Google Scholar]
- 36.Franke D., Svergun D.I. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Cryst. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Franke D., Petoukhov M.V., Svergun D.I. ATSAS 2.8: a comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J. Appl. Cryst. 2017;50:1212–1225. doi: 10.1107/S1600576717007786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodrigues J.P.G.L.M., Trellet M., Bonvin A.M.J.J. Clustering biomolecular complexes by residue contacts similarity. Proteins. 2012;80:1810–1817. doi: 10.1002/prot.24078. [DOI] [PubMed] [Google Scholar]
- 39.Sutton J.L., Pollack L. Tuning RNA flexibility with helix length and junction sequence. Biophys. J. 2015;109:2644–2653. doi: 10.1016/j.bpj.2015.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roy R., Hohng S., Ha T. A practical guide to single-molecule FRET. Nat. Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nir E., Michalet X., Weiss S. Shot-noise limited single-molecule FRET histograms: comparison between theory and experiments. J. Phys. Chem. B. 2006;110:22103–22124. doi: 10.1021/jp063483n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pabit S.A., Sutton J.L., Pollack L. Role of ion valence in the submillisecond collapse and folding of a small RNA domain. Biochemistry. 2013;52:1539–1546. doi: 10.1021/bi3016636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kelly S.M., Pabit S.A., Berland K.M. Recognition of polyadenosine RNA by zinc finger proteins. Proc. Natl. Acad. Sci. USA. 2007;104:12306–12311. doi: 10.1073/pnas.0701244104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tian B., Bevilacqua P.C., Mathews M.B. The double-stranded-RNA-binding motif: interference and much more. Nat. Rev. Mol. Cell Biol. 2004;5:1013–1023. doi: 10.1038/nrm1528. [DOI] [PubMed] [Google Scholar]
- 45.Pabit S.A., Qiu X., Pollack L. Both helix topology and counterion distribution contribute to the more effective charge screening in dsRNA compared with dsDNA. Nucleic Acids Res. 2009;37:3887–3896. doi: 10.1093/nar/gkp257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Svergun D.I., Koch M.H.J. Small-angle scattering studies of biological macromolecules in solution. Rep. Prog. Phys. 2003;66:1735–1782. [Google Scholar]
- 47.Kikhney A.G., Borges C.R., Svergun D.I. SASBDB: towards an automatically curated and validated repository for biological scattering data. Protein Sci. 2020;29:66–75. doi: 10.1002/pro.3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Macke T.J., Case D.A. Modeling unusual nucleic acid structures. In: Leontis N.B., SantaLucia J., editors. Vol. 682. American Chemical Society; 1997. pp. 379–393. (Molecular Modeling of Nucleic Acids, ACS Symposium Series). [Google Scholar]
- 49.Dey S.K., Pettersson J.R., Peteanu L.A. Eliminating spurious zero-efficiency FRET states in diffusion-based single-molecule confocal microscopy. J. Phys. Chem. Lett. 2018;9:2259–2265. doi: 10.1021/acs.jpclett.8b00362. [DOI] [PubMed] [Google Scholar]
- 50.Calses P.C., Dhillon K.K., Taniguchi T. DGCR8 mediates repair of UV-induced DNA damage independently of RNA processing. Cell Rep. 2017;19:162–174. doi: 10.1016/j.celrep.2017.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chang K.-Y., Ramos A. The double-stranded RNA-binding motif, a versatile macromolecular docking platform. FEBS J. 2005;272:2109–2117. doi: 10.1111/j.1742-4658.2005.04652.x. [DOI] [PubMed] [Google Scholar]
- 52.Zheng X., Bevilacqua P.C. Activation of the protein kinase PKR by short double-stranded RNAs with single-stranded tails. RNA. 2004;10:1934–1945. doi: 10.1261/rna.7150804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Patel S., Blose J.M., Bevilacqua P.C. Specificity of the double-stranded RNA-binding domain from the RNA-activated protein kinase PKR for double-stranded RNA: insights from thermodynamics and small-angle X-ray scattering. Biochemistry. 2012;51:9312–9322. doi: 10.1021/bi300935p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kwon S.C., Baek S.C., Kim V.N. Molecular basis for the single-nucleotide precision of primary microRNA processing. Mol. Cell. 2019;73:505–518.e5. doi: 10.1016/j.molcel.2018.11.005. [DOI] [PubMed] [Google Scholar]
- 55.Welty R., Rau M., Hall K.B. Ribosomal protein L11 selectively stabilizes a tertiary structure of the GTPase center rRNA domain. J. Mol. Biol. 2020;432:991–1007. doi: 10.1016/j.jmb.2019.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Duss O., Stepanyuk G.A., Williamson J.R. Transient protein-RNA interactions guide nascent ribosomal RNA folding. Cell. 2019;179:1357–1369.e16. doi: 10.1016/j.cell.2019.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lund P.E., Chatterjee S., Walter N.G. Protein unties the pseudoknot: S1-mediated unfolding of RNA higher order structure. Nucleic Acids Res. 2020;48:2107–2125. doi: 10.1093/nar/gkz1166. Published online December 13, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.