Abstract
Fusion of biological membranes, although mediated by divergent proteins, is believed to follow a common pathway. It proceeds through distinct steps, including docking, merger of proximal leaflets (stalk formation), and formation of a fusion pore. However, the structure of these intermediates is difficult to study because of their short lifetime. Previously, we observed a loosely and tightly docked state preceding leaflet merger using arresting point mutations in SNARE proteins, but the nature of these states remained elusive. Here, we used interferometric scattering (iSCAT) microscopy to monitor diffusion of single vesicles across the surface of giant unilamellar vesicles (GUVs). We observed that the diffusion coefficients of arrested vesicles decreased during progression through the intermediate states. Modeling allowed for predicting the number of tethering SNARE complexes upon loose docking and the size of the interacting membrane patches upon tight docking. These results shed new light on the nature of membrane-membrane interactions immediately before fusion.
Significance
Membrane merger through fusion is essential in a myriad of processes, including neurotransmission and viral infection. Despite the importance of understanding the process of membrane fusion, there are still many open questions about the exact mechanism. Here, we present a new technique that allows imaging the small vesicles while they are attached during fusion to another membrane with unprecedented temporal and spatial resolution. This approach in combination with modeling allowed us to extract novel features of fusion intermediates, which, until now, have remained elusive. In particular, we were able to dynamically distinguish two states that occur before fusion: a loosely docked and a tightly docked state. Characterizing these dynamic differences can help us to better understand membrane-membrane interactions during fusion.
Introduction
Soluble N-ethylmaleimide-sensitive factor activating protein receptor (SNARE) proteins mediate membrane fusion in the secretory pathway of eukaryotic cells and are widely used as model for studying fusion. They are small, mostly membrane-anchored proteins characterized by a conserved motif (SNARE-motif) that is usually located adjacent to the C-terminal membrane anchor (1). Upon contact between membranes destined to fuse, four complementary SNARE motifs assemble to form a trans-SNARE complex that cross-links the membranes. Assembly is initiated at the membrane-distal ends and then progresses toward the C-terminal membrane anchors (“zippering”), thus pulling the membranes tightly together (1, 2, 3). Zippering is highly exergonic and thus overcomes the energy barriers for membrane fusion (4, 5, 6, 7). After membrane merger is completed, all SNAREs of the complex are aligned in parallel in the same membrane (cis-SNARE complex).
Despite major progress, the structural transitions along the fusion pathway are far from clear. Fusion commences with membrane contact and then leads via nonbilayer transition states to the final opening of an aqueous fusion pore. Experimental evidence complemented with both continuum- and particle-based models have suggested tentative pathways involving tight membrane contact brought about by SNARE zippering, associated with local protrusions of high curvature in some models (8, 9, 10). This state may then lead to lipid tail splaying between the fusing bilayers (11), resulting in a merger of the proximal monolayers, yielding a structure that is also referred to as fusion stalk (see e.g., (12)). From the stalk intermediate, the system may progress to forming a hemifusion diaphragm (13) before the final opening of the fusion pore (8). For the transition from one intermediate to the next, energy barriers such as electrostatic repulsion or steric hydration forces need to be overcome (14, 15, 16).
Under physiological conditions, the lifetime of these intermediate states is very short. Indeed, only recently docking intermediates preceding hemifusion in SNARE-mediated fusion have been captured by cryoelectron microscopy (6,17,18). Deletion of a single amino acid in the SNARE synaptobrevin-2 (Δ84 syb) results in vesicles that are arrested in a tightly docked (but not hemifused) state in which the membranes are separated by less than 1 nm. In contrast, substitution of two amino acids more distal from the transmembrane domain (I45A, M46A; referred to as AA syb) uncovered a loosely docked state in which the membranes are separated by a larger gap only connected by reversible SNARE interactions (17). Finally, extended hemifusion diaphragms were observed when the wild-type (WT) syb protein was used (6).
In this study, we have used in vitro reconstitution to characterize the nature of the loose- and tight docking states. In particular, we have analyzed whether diffusion is affected by the nature of the docking intermediate. To this end, we have taken advantage of a previously established model system in which the SNARE-mediated docking and fusion of small vesicles with a single giant unilamellar vesicle (GUV) can be measured (19). In this work, we used the neuronal SNARE proteins syb, syntaxin-1, and SNAP-25 as models, with syb incorporated into small vesicles and an activated acceptor complex of syntaxin-1 and SNAP-25 (20) inserted into the GUV membrane. This system is ideally suited for studying particle diffusion on membranes because the membrane area is large but diffusion is not hindered by any type of surface contact as, for instance, in supported planar bilayers (21). We employed interferometric scattering (iSCAT) microscopy (22, 23, 24) to record three-dimensional (3D) trajectories of particle diffusion on the surface of GUVs. This technique has been shown to reach microsecond-temporal resolution and nanometer precision in detecting and tracking individual nanoparticles via the interference of their scattered light with a reference beam (see Materials and Methods). Using iSCAT, we could investigate the diffusion of unlabeled vesicles arrested at a loosely or tightly docked intermediate state using syb-point mutants as previously described (17). Modeling of the diffusion data allowed for approximating the number of SNARE complexes involved in vesicle docking (loose docking) as well as the size of the interaction interface (tight docking).
Materials and Methods
Protein expression and purification
Rattus norvegicus-derived SNARE proteins (syntaxin-1A (183–288) (25)), SNAP-25 (cysteine-free) (26), synaptobrevin-2 (WT (27), Δ84 (28), I45A, and M46A (AA) (17,29)), 1–96 (30), and synaptobrevin-2 fragment (49–96) (20) were expressed and purified as described before (17,31). In short, proteins were expressed in Escherichia coli strain BL21 (DE3) and purified via nickel-nitrilotriacetic acid affinity chromatography (Qiagen, Hilden, Germany) followed by ion exchange chromatography on an Äkta system (GE Healthcare, Chicago, IL). The preassembled so called ΔN complex (20) was used to mimic plasma membrane SNARE acceptor complex. It consists of syntaxin, SNAP-25, and syb fragment 49–96. The complex was obtained by mixing the monomers overnight at 4°C, followed by purification of the complex using ion exchange chromatography (MonoQ column) in a buffer containing CHAPS as described before (20,31).
Liposome preparation
Lipids (brain-derived phosphatidylcholine (PC), phosphatidylethanolamine (PE), and phosphatidylserine (PS) along with cholesterol (ovine wool)) were purchased from Avanti Polar Lipids (Alabaster, AL). For all liposome mixtures, lipids PC, PE, PS, and cholesterol were mixed in a ratio of 5:2:2:1, respectively. Liposomes were tested on regular basis for reconstitution efficiency of proteins as described before (6,31).
Large unilamellar vesicles (LUVs; diameter ∼100 nm) were prepared with a reverse phase evaporation method as previously described (6,19). Proteoliposomes were formed using a direct reconstitution method as previously described (6), with syb WT, Δ84, or AA reconstituted in a protein/lipid ratio of 1:500.
Small proteoliposomes (diameter ∼40 nm) were prepared as described before (19) by comicellization followed by size exclusion chromatography. During preparation, ΔN complex was added to yield a protein/lipid ratio of 1:1000.
GUV preparation
GUVs were prepared from small proteoliposomes containing ΔN complex by electroformation using in-house-built electroformation chambers containing platinum wires (32) using a protocol described previously (31).
iSCAT measurements
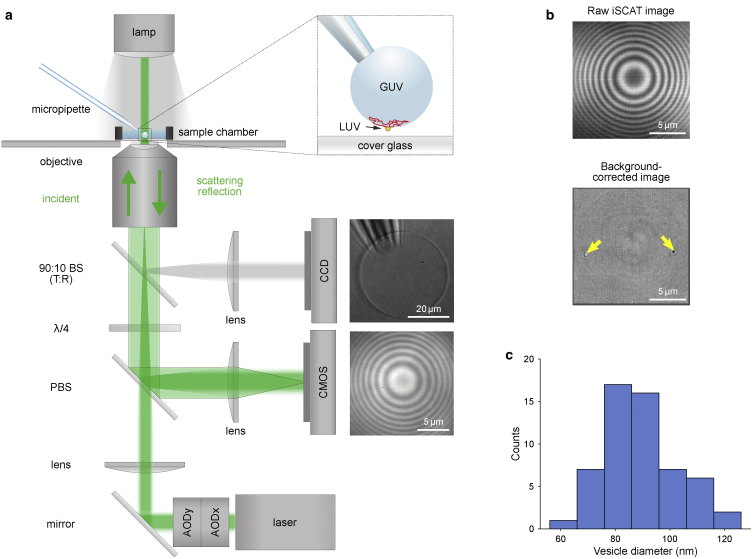
Liposomes were tracked using iSCAT microscopy (22, 23, 24). With iSCAT technology, we could reach microsecond-temporal resolution in 3D imaging by acquiring one image per time point instead of a z-stack from which data were extracted. Moreover, the interferometric nature of iSCAT provides a much higher axial resolution in comparison to confocal imaging (of the order of 1 nm) because the iSCAT contrast fully reverses within less than 100 nm of axial motion, providing a large gradient that reports on the axial location of the particle. The basic principle is schematically shown in Fig. 1 a (a more detailed description of the setup can be found in (33)). Briefly, the incoming light Einc of a 532-nm laser (Verdi 2G; Coherent, Santa Clara, CA) is scattered from the GUV and the liposomes (Escat) and partially reflected at the glass-sample interface (Eref). Escat and Eref are collected via an oil-immersion 60× objective (ApoN, NA = 1.49; Olympus, Shinjuku City, Tokyo, Japan) and imaged onto a CMOS camera (MV-D1024E-CL; Photonfocus, Lachen, Switzerland). The intensity at the camera is given by
| (1) |
and is thus dependent on the phase φ between the scatterer and the reflected light. Changes in the distance between the scatterer and the glass-sample interface result in a periodic modulation of the scatterer’s contrast. Scattering at the GUV surface therefore creates a ring pattern, as shown in Fig. 1 b. In the same fashion, the point spread function (PSF) of a liposome changes its amplitude when it travels along the GUV surface and thus appears either bright or dark on the GUV background.
Figure 1.
iSCAT detection of GUVs and liposomes. (a) Schematic illustration of the experimental optical setup allowing the observation of GUVs in bright field (on the CCD camera) and iSCAT (on the CMOS camera) as well as micropipette manipulation of GUVs (see inset). BS, beam splitter; λ/4, quarter wave plate; PBS, polarizing beam splitter; AOD, acousto-optical deflector. (b) iSCAT GUV image before (upper image) and after (lower image) background-correction procedure. In the background-corrected image, two docked LUVs become visible (marked with yellow arrows) with the PSF shape of the wide-field iSCAT microscope encoding the particular height of the particles. (c) A histogram of the average LUV size determined based on their contrast in iSCAT (NLUV = 56).
To hold the GUVs in place above the field of view, we used micropipette aspiration (34) using glass pipettes with an ∼5-μm opening connected to a height-adjustable water tank and a micromanipulator (Sensapex, Oulu, Finland). Using a 90:10 (T:R) beam splitter, a part of the collected light was sent onto a CCD camera (SensiCam; PCO, Kelheim, Germany) for monitoring the GUV aspiration process in bright field.
Videos of liposomes bound to GUVs were recorded typically for 5 s at a 1-kHz frame rate. For liposome tracking, we performed background correction based on image registration (details of the procedure can be found in the Supporting Materials and Methods). The iSCAT contrast of the liposomes typically amounts to 2–3%. In the background-corrected images, we fit a two-dimensional (2D) Gaussian function to each PSF for determining the 2D projected position. The particle height zi in each frame is calculated based on the GUV center (xGUV, yGUV), the GUV radius rGUV (which we extract from the bright field image), and the 2D particle localization (xi, yi):
| (2) |
We determined the center of the GUV using two consecutive routines adopted from (35), which are described in the Supporting Materials and Methods.
The diffusion coefficient was calculated for each trajectory from the first two points of the mean-squared displacement (MSD) with time lag of and ( = 1 ms):
| (3) |
Modeling of loose docking
Liposomes loosely docked on the GUV surface were assumed to be tethered by different numbers of SNARE complexes. Their diffusion was simulated using random walk (36) of individual SNARE complexes in a 2D membrane (similar to (37)). The constraining circular area (50 nm in radius) was introduced to reflect the maximal surface of the liposome-GUV-docking interface with the size resembling maximal LUV membrane deformation during docking as seen in (6,17,38). Other radii (5–100 nm) of constraint areas, including an adaptive one (constraint radius increasing with increasing number of tethering SNARE complexes), were also tested; however, they did not yield substantially different results (data not shown). The effect of solvent friction on vesicle’s diffusion in this case was assumed to be negligible (see Fig. S6 and corresponding Supporting Materials and Methods). It also seems unlikely that membrane deformation generated here by diffusing SNARE complexes would result in major decrease of the diffusion coefficient. Therefore, we did not introduce such energy potential in our model.
Initially, a various number of tethering SNARE complexes are randomly distributed within the constraining area. Each SNARE complex displacement dx was determined in every time step dt (usually 100 μs):
| (4) |
with DSNARE being the diffusion coefficient of the assembled SNARE complex of 3.8 μm2/s as determined by means of fluorescence correlation spectroscopy with fluorescently labeled versions of neuronal SNAREs incorporated into GUVs in (39). The absolute position of a vesicle after each displacement step was then calculated as center of mass of all simulated proteins. Each 5-s-long simulation was performed 20 times to determine average MSD and diffusion coefficient of the vesicle. Further, we have investigated how clustering of SNARE complexes influences diffusion of loosely docked vesicles, but we did not observe any large effects (Fig. S7). The source code used for simulations in Octave (40) and MATLAB (MathWorks, Natick, MA) are available at Zenodo (41).
Modeling of tight docking
The diffusion speed of tightly docked vesicles was modeled with the use of an analytical expression (42) for the model developed earlier by Hughes et al. (43) to predict diffusion of membrane-embedded objects (proteins) based on the hydrodynamic radius of the membrane-embedded part. The version of the approximation used in this study was presented by (44):
| (5) |
with the parameter ε being dependent on the inclusion radius R:
| (6) |
Explanation of used symbols can be found in Table 1. Values used in the model are given in Table 2.
Table 1.
| Symbol | Definition |
|---|---|
| D | diffusion coefficient |
| kB | Boltzmann’s constant |
| T | temperature (in K) |
| μm | membrane viscosity (in Pa · s) |
| h | bilayer thickness |
| γ | Euler’s constant |
| ε | parameter derived from Eq. 6 |
| a1, b1, a2, b2 | equation scaling coefficients (45) |
| R | membrane inclusion radius |
| μs | viscosity of surrounding medium (in Pa · s) |
Table 2.
| Parameter | Value | Comment |
|---|---|---|
| kB | physical constant | |
| T | 293 K | ∼20°C |
| μm | 111 mPa · s | calculated from diffusion coefficient 3.9 μm2/s determined by FCS in (39) |
| h | 4.6 nm | (37) |
| γ | 0.5772 | physical constant |
| a1 | 0.433274 | (45) |
| b1 | 2.74819 | (45) |
| a2 | 0.670045 | (45) |
| b2 | 0.614465 | (45) |
| R | 0.6 nm | for single SNARE complex transmembrane domain (46) |
| μs | 0.96 mPa · s | (44) |
Simulated example trajectories of tightly docked vesicles were generated as random walk (Eq. 4) of objects of a certain radius characterized by the diffusion coefficient obtained from Eq. 5.
Results
To study loosely and tightly docked intermediates, a GUV containing SNARE acceptor complexes was immobilized with a micropipette (see Fig. 1 a) and brought in close proximity to the cover glass. Next, we added LUVs (80–100 nm in diameter) containing syb and used iSCAT microscopy to track LUVs on the lower hemisphere of individual GUVs from a single focus position (47). The LUVs readily bound to the GUV and either fused with it (as for WT syb (19)) or they remained docked on its surface and diffused (Videos S1, S2, and S3). Detection of the LUV-scattering signal was greatly improved with a background-correction procedure (compare upper and lower image in Fig. 1 b) described in more detail in Supporting Materials and Methods. The iSCAT contrast of a nano-object is directly proportional to its polarizability, which is, in turn, linked to the particle size (24). The size of the LUVs was estimated from the observed iSCAT contrast to be ∼90 nm (see Fig. 1 c). Thus, an LUV appears as the iSCAT PSF of microscope, which consists of few lobes encoding the 3D position of the scatterer (48). This PSF could be fitted with a physical model to extract the third dimension (49); however, here, we fitted the main lobe for the lateral localization and used the change of the central lobe contrast (50,51) as well as the geometry of the GUVs (47) to extract the axial position of the vesicles (Fig. 1 b).
Frame rate = 1 kHz, recording time = 1s, videos slowed-down 20×, frame size = 16.28 μm × 16.28 μm.
Frame rate = 1 kHz, recording time = 1s, videos slowed-down 20×, frame size = 16.28 μm × 16.28 μm.
Frame rate = 1 kHz, recording time = 1s, videos slowed-down 20×, frame size = 16.28 μm × 16.28 μm.
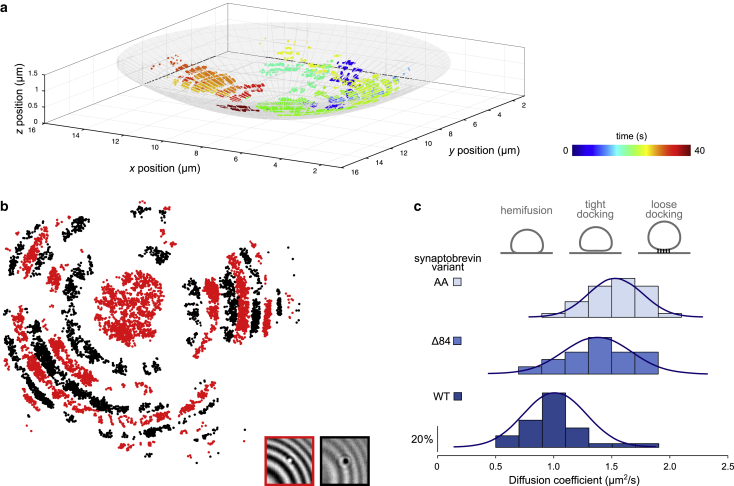
Lack of photobleaching in iSCAT allowed for monitoring the 3D motion of docked vesicles over prolonged periods (as shown exemplarily in Fig. 2 a; Fig. S2) while maintaining nanometer spatial precision (see Fig. S1 and corresponding Supporting Materials and Methods). Docked vesicles closely follow the topography of the GUV surface, thus displaying a characteristic periodic ring-shaped contrast pattern (see Fig. 2 b), similar to the Newton rings encountered in Fig. 1 b (also see inset in Fig. 2 b). We then compared surface diffusion of docked vesicles containing either WT syb or the docking mutants Δ84 syb and AA syb (described above). As expected, all three vesicle populations dock to GUVs that contain acceptor SNARE complexes. However, when we determined their diffusion coefficients from the measured 3D trajectories (Fig. 2 a), significant differences were observed between the three populations (Fig. 2 c). Vesicles carrying the AA syb mutants showed the fastest diffusion, followed by those containing the Δ84 mutant, whereas vesicles with the WT protein were the slowest. These data show that the different states uncovered previously by electron microscopy (17) indeed represent physical differences in the nature of membrane attachment between the two vesicles.
Figure 2.
Tracking of docked LUVs on a GUV surface. (a) 3D-reconstructed trajectory of a single liposome docked on the GUV surface tracked for 40 s. The color bar shows the time evolution. The sphere represents a GUV surface with a radius of 18 μm. (b) xy projection of the docked liposome’s trajectory from (a). The point color depends on the LUV iSCAT signal intensity reflecting fluctuations in the LUV z-position relative to the GUV membrane. Regions marked in black and red correspond to the regions of dark and bright iSCAT particle contrasts, respectively (see lower insets). The gaps in the trajectory between regions with positive and negative contrast result from the fact that the vesicle is barely visible when undergoing a contrast switch. (c) Histograms presenting diffusion coefficients of LUVs docked on the GUV surface depending on the syb variant (AA, Δ84, or WT). Bin width is 0.2 μm2/s. Blue lines represent a Gaussian fit. NAA = 32, NΔ84 = 17, NWT = 32. Illustrations above the graph depict possible docking states of the vesicles.
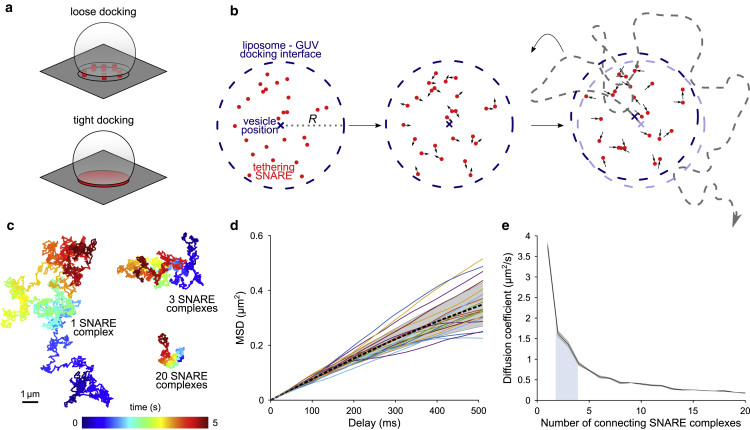
As discussed above, the loosely docked state is probably governed exclusively by trans interactions between individual SNAREs whereas in the tightly docked state the membranes are firmly attached, forming a disk that is probably rigid (6,17). Using the experimentally obtained diffusion data, rather than extracting data directly from recorded trajectories (as it was demonstrated before for virion tracking in total internal reflection fluorescence microscopy (52)), we have therefore carried out random walk simulations to evaluate whether the data are compatible with these models. First, we modeled the diffusion of vesicles connected by multiple (1–20) SNARE complexes (Fig. 3 a upper panel). To gain insight into the diffusion of a liposome docked on the GUV surface while being tethered to different numbers of SNARE complexes we simulated random walks of individual SNARE complexes within the constraining circular area to reflect the maximal docking interface area (Fig. 3 b, for details see Materials and Methods). Vesicle position was repeatedly determined after each displacement step as the center of mass of all simulated proteins (Fig. 3 b) to form a 5-s-long trajectory (see Fig. 3 c). Next, MSD curves were generated from the trajectories (Fig. 3 d), and diffusion coefficients for a varying number of tethering SNARE complexes were calculated (Fig. 3 e). The outcome of simulations confirms that the diffusion coefficient of a vesicle decreases with an increasing number of tethering SNARE complexes (Fig. 3, c and e). Diffusion speeds obtained in simulations and experiments let us conclude that loosely docked vesicles (mainly AA syb and Δ84 syb) observed with iSCAT are attached to the GUV with maximally 3–4 assembled SNARE complexes. Docked vesicles moving at the maximal speed, on the other hand, are found to be attached via two SNARE complexes. Vesicles attached with one SNARE complex were probably rejected from our analyses because of large fluctuations in the position perpendicular to the GUV surface and the lack of a ring-shaped amplitude pattern as seen in Fig. 2 b.
Figure 3.
Simulation of diffusion of loosely docked vesicles tethered by multiple SNARE complexes. (a) Schematic illustration of a vesicle tethered by multiple SNARE complexes to GUV membrane (loose docking, SNARE complexes in red) and vesicle that is tightly docked on a GUV surface with interacting docking membrane interfaces marked in red. (b) Schematic illustration of the random walk diffusion simulation of SNARE proteins (red points) with liposome position determination (cross) and with a diffusional constraint R for SNARE proteins representing maximal contact area between LUV and GUV (dashed blue line). Liposome trajectory after multiple displacement steps is schematically shown with a gray dashed line. (c) Representative trajectories obtained in simulations with 1, 3, or 20 tethering SNARE complexes. (d) MSD obtained from 20 simulation runs with 20 tethering SNARE complexes. The mean is represented by the dashed black line and the standard deviation of the mean is shown by the shaded area. (e) Dependence of LUV’s diffusion coefficient on the number of tethering complexes. The mean is represented by the black line and the standard deviation of the mean is shown by the shaded area around it. Each diffusion coefficient was determined in 20 independent simulation runs. The blue area below the graph shows the range of experimentally measured diffusion values for syb Δ84 and AA.
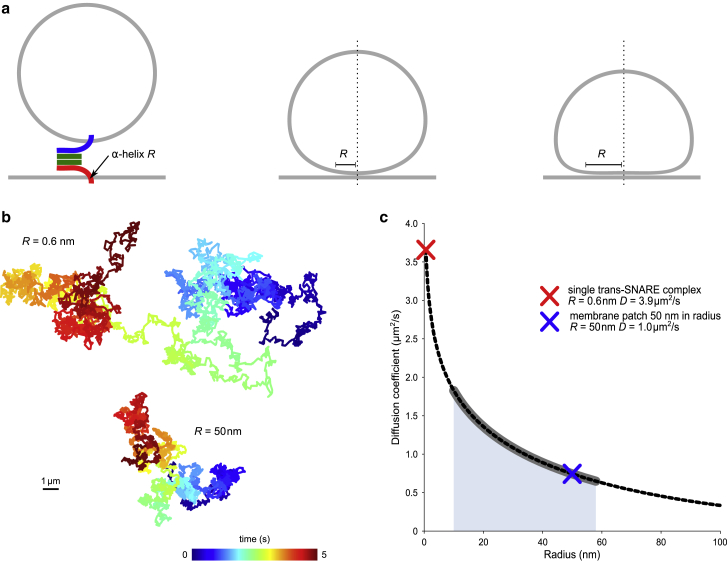
Next, we modeled the diffusion of vesicles that were attached to the GUV through tight docking interfaces by assuming a disk-like-docking interface that diffuses as a whole (see Materials and Methods). Here, we have used an analytical approximation of the translational diffusion in the membrane (42,43) to calculate the theoretical diffusion coefficient of a tightly docked vesicle with a given docking interface radius (Fig. 4 a, see Materials and Methods for details). The resulting diffusion coefficients (Fig. 4, b and c) decrease with increasing interface size at a slower pace than in the case of a loose docking model (Fig. 3 e). Remarkably, diffusion coefficient values of the slowest vesicles containing syb mutants (i.e., potentially not hemifused, Fig. 2 c) correspond to the modeled interface radius of around 50 nm, which, in turn, correlates with maximal feasible LUV deformations based on electron microscopy studies (6,17).
Figure 4.
Modeling the diffusion of tightly docked vesicles. (a) Schematic illustration displaying different docking stages that can be captured by the model: point attachment by a single tethering SNARE complex, intermediate, and extended docking interfaces with varying radii R of the membrane interaction surface. (b) Example trajectories of vesicles over 5 s, moving with diffusion speeds obtained from the model (see (c)) corresponding to point attachment (R = 0.6 nm) and extended docking interface (R = 50 nm). (c) Analytical solution of the tight docking model (dashed line) with parameters (Table 2) corresponding to the experimental conditions in this work. Thickened gray line and blue area show the range of experimentally measured diffusion values for syb Δ84 and AA. Diffusion coefficient values of the trajectories shown in (b) are marked with crosses.
Discussion
Here, we utilized in vitro reconstitution using previously described SNARE mutants (6,17) to characterize recently discovered early membrane fusion intermediates (6,17). Employing iSCAT microscopy in this work allowed, for the first time, to our knowledge, for extended 3D-tracking of vesicles’ dynamics on large, free-standing membranes with very low curvature as typical for cell membranes thus avoiding disturbance by surface contact or edge effects that may occur in supported or pore-spanning membranes (33). With this approach, we were able to observe that docked vesicle’s arrest at subsequent membrane fusion intermediate states (loose docking, tight docking, and hemifusion) is reflected in decreasing diffusion coefficients.
The loosely docked state, stabilized here by point mutations in the SNARE synaptobrevin, may serve as a model for partially assembled SNARE complexes in which progression of zippering is thought to be arrested by regulatory proteins (1,53). In our experimental system, we find that such loosely docked vesicles, on average, diffuse faster than other docked liposomes, which indicates less interactions between docked membranes. Moreover, the number of connecting SNARE complexes predicted by our model (up to 3–4) is in good agreement with current, albeit rather indirect, estimates for the number of SNARE complexes required for fusion (38,54,55).
Tightly docked membranes have only recently been observed in fusion reactions mediated by SNARE proteins (6), viral fusion proteins (56), as well as mitochondrial fusion proteins (57). Additionally, this state appears to be also independent of the continued presence of the tethering proteins because docking becomes irreversible upon SNARE complex disassembly (17). Moreover, development of tight membrane-membrane contact patch was also inferred from protein-free coarse-grain simulations (58), providing further evidence that it constitutes a true fusion intermediate. Taken together, it seems that tight docking is independent of the nature of the fusion protein and is thus an intrinsic feature of the bilayer. In this work, we detected a high proportion of vesicles with relatively large tight attachment interfaces, exceeding 10 nm in radius. Together with previous electron microscopy observations of large, flattened, interacting membrane patches (6,56,57), it suggests that once formed, expansion of such an adhesion interface can progress up to the point when further membrane deformation is not possible or when there is steric hindrance coming from protein tethers. It remains to be established how the repulsive forces of the negatively charged membranes are overcome at close distance and also whether, and to which extent, diffusion of membrane constituents is hindered in the disk-like contact zone.
Do tightly docked intermediates represent fusion intermediates in living cells? Extended disk-shaped contacts were frequently observed in classical electron microscopy studies of secretory vesicles (59), but they were later attributed to fixation artifacts (60). However, more recently docked vesicles with subnanometer distance from the plasma membrane were seen in electron microscopy images of synaptic vesicles in neurons obtained with high-pressure freezing (61,62). Understanding the physical parameters governing this state such as adhesive and repulsive forces at the contact zone, the lateral mobility of molecules within and between the membranes, and the energy barriers leading from this state to nonbilayer intermediates, such as fusion stalks, will be instrumental for unraveling the mechanism of membrane fusion. The methodology applied here may be useful for analyzing the intermediate states of other fusion proteins such as those mediating mitochondrial or viral fusion.
Author Contributions
A.W., R.J., and V.S. designed the study. A.W. prepared all samples, performed confocal imaging, wrote code and performed numerical simulations, and analyzed and evaluated all data. S.S. prepared GUVs and performed and evaluated iSCAT experiments. R.G.M. contributed new analytic tools for iSCAT and evaluated iSCAT experiments. A.W., S.S., R.J., and V.S. wrote the manuscript.
Acknowledgments
We thank Martin Kaller for support with iSCAT experiments.
This work was supported by funds from the Max Planck Society (to R.J. and V.S.), United States National Institutes of Health Grant No. 2 P01 GM072694 (to R.J.), and an Alexander von Humboldt Professorship (to V.S.).
Editor: Rumiana Dimova.
Footnotes
Agata Witkowska’s present address is Department of Molecular Pharmacology and Cell Biology, Leibniz-Forschungsinstitut für Molekulare Pharmakologie (FMP), Berlin, Germany.
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2020.10.033.
Contributor Information
Agata Witkowska, Email: agata.witkowska@wp.eu.
Vahid Sandoghdar, Email: vahid.sandoghdar@mpl.mpg.de.
Reinhard Jahn, Email: rjahn@gwdg.de.
Supporting Citations
References (63,64) appear in the Supporting Material.
Supporting Material
References
- 1.Jahn R., Fasshauer D. Molecular machines governing exocytosis of synaptic vesicles. Nature. 2012;490:201–207. doi: 10.1038/nature11320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker R.W., Hughson F.M. Chaperoning SNARE assembly and disassembly. Nat. Rev. Mol. Cell Biol. 2016;17:465–479. doi: 10.1038/nrm.2016.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rizo J., Xu J. The synaptic vesicle release machinery. Annu. Rev. Biophys. 2015;44:339–367. doi: 10.1146/annurev-biophys-060414-034057. [DOI] [PubMed] [Google Scholar]
- 4.Fasshauer D., Antonin W., Jahn R. SNARE assembly and disassembly exhibit a pronounced hysteresis. Nat. Struct. Biol. 2002;9:144–151. doi: 10.1038/nsb750. [DOI] [PubMed] [Google Scholar]
- 5.Gao Y., Zorman S., Zhang Y. Single reconstituted neuronal SNARE complexes zipper in three distinct stages. Science. 2012;337:1340–1343. doi: 10.1126/science.1224492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hernandez J.M., Stein A., Jahn R. Membrane fusion intermediates via directional and full assembly of the SNARE complex. Science. 2012;336:1581–1584. doi: 10.1126/science.1221976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Südhof T.C., Rothman J.E. Membrane fusion: grappling with SNARE and SM proteins. Science. 2009;323:474–477. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chernomordik L.V., Kozlov M.M. Mechanics of membrane fusion. Nat. Struct. Mol. Biol. 2008;15:675–683. doi: 10.1038/nsmb.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Efrat A., Chernomordik L.V., Kozlov M.M. Point-like protrusion as a prestalk intermediate in membrane fusion pathway. Biophys. J. 2007;92:L61–L63. doi: 10.1529/biophysj.106.103341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuzmin P.I., Zimmerberg J., Cohen F.S. A quantitative model for membrane fusion based on low-energy intermediates. Proc. Natl. Acad. Sci. USA. 2001;98:7235–7240. doi: 10.1073/pnas.121191898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Risselada H.J., Kutzner C., Grubmüller H. Caught in the act: visualization of SNARE-mediated fusion events in molecular detail. ChemBioChem. 2011;12:1049–1055. doi: 10.1002/cbic.201100020. [DOI] [PubMed] [Google Scholar]
- 12.Aeffner S., Reusch T., Salditt T. Energetics of stalk intermediates in membrane fusion are controlled by lipid composition. Proc. Natl. Acad. Sci. USA. 2012;109:E1609–E1618. doi: 10.1073/pnas.1119442109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang L., Huang H.W. Observation of a membrane fusion intermediate structure. Science. 2002;297:1877–1879. doi: 10.1126/science.1074354. [DOI] [PubMed] [Google Scholar]
- 14.Israelachvili J.N. Interactions of biological membranes and structures. In: Israelachvili J.N., editor. Intermolecular and Surface Forces. Third Edition. Academic Press; 2011. pp. 577–616. [Google Scholar]
- 15.Schneck E., Sedlmeier F., Netz R.R. Hydration repulsion between biomembranes results from an interplay of dehydration and depolarization. Proc. Natl. Acad. Sci. USA. 2012;109:14405–14409. doi: 10.1073/pnas.1205811109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hernández J.M., Podbilewicz B. The hallmarks of cell-cell fusion. Development. 2017;144:4481–4495. doi: 10.1242/dev.155523. [DOI] [PubMed] [Google Scholar]
- 17.Yavuz H., Kattan I., Jahn R. Arrest of trans-SNARE zippering uncovers loosely and tightly docked intermediates in membrane fusion. J. Biol. Chem. 2018;293:8645–8655. doi: 10.1074/jbc.RA118.003313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prinslow E.A., Stepien K.P., Rizo J. Multiple factors maintain assembled trans-SNARE complexes in the presence of NSF and αSNAP. eLife. 2019;8:e38880. doi: 10.7554/eLife.38880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Witkowska A., Jahn R. Rapid SNARE-mediated fusion of liposomes and chromaffin granules with giant unilamellar vesicles. Biophys. J. 2017;113:1251–1259. doi: 10.1016/j.bpj.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pobbati A.V., Stein A., Fasshauer D. N- to C-terminal SNARE complex assembly promotes rapid membrane fusion. Science. 2006;313:673–676. doi: 10.1126/science.1129486. [DOI] [PubMed] [Google Scholar]
- 21.Domanska M.K., Kiessling V., Tamm L.K. Single vesicle millisecond fusion kinetics reveals number of SNARE complexes optimal for fast SNARE-mediated membrane fusion. J. Biol. Chem. 2009;284:32158–32166. doi: 10.1074/jbc.M109.047381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindfors K., Kalkbrenner T., Sandoghdar V. Detection and spectroscopy of gold nanoparticles using supercontinuum white light confocal microscopy. Phys. Rev. Lett. 2004;93:037401. doi: 10.1103/PhysRevLett.93.037401. [DOI] [PubMed] [Google Scholar]
- 23.Kukura P., Ewers H., Sandoghdar V. High-speed nanoscopic tracking of the position and orientation of a single virus. Nat. Methods. 2009;6:923–927. doi: 10.1038/nmeth.1395. [DOI] [PubMed] [Google Scholar]
- 24.Taylor R.W., Sandoghdar V. Interferometric scattering microscopy: seeing single nanoparticles and molecules via Rayleigh scattering. Nano Lett. 2019;19:4827–4835. doi: 10.1021/acs.nanolett.9b01822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schuette C.G., Hatsuzawa K., Jahn R. Determinants of liposome fusion mediated by synaptic SNARE proteins. Proc. Natl. Acad. Sci. USA. 2004;101:2858–2863. doi: 10.1073/pnas.0400044101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fasshauer D., Antonin W., Jahn R. Mixed and non-cognate SNARE complexes. Characterization of assembly and biophysical properties. J. Biol. Chem. 1999;274:15440–15446. doi: 10.1074/jbc.274.22.15440. [DOI] [PubMed] [Google Scholar]
- 27.Stein A., Weber G., Jahn R. Helical extension of the neuronal SNARE complex into the membrane. Nature. 2009;460:525–528. doi: 10.1038/nature08156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siddiqui T.J., Vites O., Fasshauer D. Determinants of synaptobrevin regulation in membranes. Mol. Biol. Cell. 2007;18:2037–2046. doi: 10.1091/mbc.E07-01-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wiederhold K., Kloepper T.H., Fasshauer D. A coiled coil trigger site is essential for rapid binding of synaptobrevin to the SNARE acceptor complex. J. Biol. Chem. 2010;285:21549–21559. doi: 10.1074/jbc.M110.105148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Margittai M., Fasshauer D., Langen R. Homo- and heterooligomeric SNARE complexes studied by site-directed spin labeling. J. Biol. Chem. 2001;276:13169–13177. doi: 10.1074/jbc.M010653200. [DOI] [PubMed] [Google Scholar]
- 31.Witkowska A., Jablonski L., Jahn R. A convenient protocol for generating giant unilamellar vesicles containing SNARE proteins using electroformation. Sci. Rep. 2018;8:9422. doi: 10.1038/s41598-018-27456-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stein H., Spindler S., Sandoghdar V. Production of isolated giant unilamellar vesicles under high salt concentrations. Front. Physiol. 2017;8:63. doi: 10.3389/fphys.2017.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spindler S., Sibold J., Sandoghdar V. High-speed microscopy of diffusion in pore-spanning lipid membranes. Nano Lett. 2018;18:5262–5271. doi: 10.1021/acs.nanolett.8b02240. [DOI] [PubMed] [Google Scholar]
- 34.Mitchison J.M., Swann M.M. The mechanical properties of the cell surface: I. The cell elastimeter. J. Exp. Biol. 1954;31:443–460. [Google Scholar]
- 35.Dobroiu A., Alexandrescu A., Damian V.S. Centering and profiling algorithm for processing Newton’s rings fringe patterns. Opt. Eng. 2000;39:3201–3207. [Google Scholar]
- 36.Einstein A. Über die von der molekularkinetischen Theorie der Wärme geforderte Bewegung von in ruhenden Flüssigkeiten suspendierten Teilchen. Ann. Phys. 1905;322:549–560. [Google Scholar]
- 37.Witkowska A. 2017. 2D dye diffusion model. Zenodo. [Google Scholar]
- 38.Hernandez J.M., Kreutzberger A.J.B., Jahn R. Variable cooperativity in SNARE-mediated membrane fusion. Proc. Natl. Acad. Sci. USA. 2014;111:12037–12042. doi: 10.1073/pnas.1407435111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bacia K., Schuette C.G., Schwille P. SNAREs prefer liquid-disordered over “raft” (liquid-ordered) domains when reconstituted into giant unilamellar vesicles. J. Biol. Chem. 2004;279:37951–37955. doi: 10.1074/jbc.M407020200. [DOI] [PubMed] [Google Scholar]
- 40.Eaton J.W., Bateman D., Wehbring R. 2018. GNU Octave version 4.4.0 manual: a high-level interactive language for numerical computations. Free Software Foundation, Inc. [Google Scholar]
- 41.Witkowska A. Simulation of diffusion of a multitethered vesicle. Zenodo. 2020 doi: 10.5281/zenodo.3337618. [DOI] [Google Scholar]
- 42.Petrov E.P., Schwille P. Translational diffusion in lipid membranes beyond the Saffman-Delbruck approximation. Biophys. J. 2008;94:L41–L43. doi: 10.1529/biophysj.107.126565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hughes B.D., Pailthorpe B.A., White L.R. The translational and rotational drag on a cylinder moving in a membrane. J. Fluid Mech. 1981;110:349–372. [Google Scholar]
- 44.Weiß K., Neef A., Enderlein J. Quantifying the diffusion of membrane proteins and peptides in black lipid membranes with 2-focus fluorescence correlation spectroscopy. Biophys. J. 2013;105:455–462. doi: 10.1016/j.bpj.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guigas G., Weiss M. Influence of hydrophobic mismatching on membrane protein diffusion. Biophys. J. 2008;95:L25–L27. doi: 10.1529/biophysj.108.136069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sutton R.B., Fasshauer D., Brunger A.T. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 47.Spindler S., Ehrig J., Sandoghdar V. Visualization of lipids and proteins at high spatial and temporal resolution via interferometric scattering (iSCAT) microscopy. J. Phys. D Appl. Phys. 2016;49:274002. [Google Scholar]
- 48.Taylor R.W., Gholami Mahmoodabadi R., Sandoghdar V. Interferometric scattering microscopy reveals microsecond nanoscopic protein motion on a live cell membrane. Nat. Photonics. 2019;13:480–487. [Google Scholar]
- 49.Gholami Mahmoodabadi R., Taylor R.W., Sandoghdar V. Point spread function in interferometric scattering microscopy (iSCAT). Part I: aberrations in defocusing and axial localization. Opt. Express. 2020;28:25969–25988. doi: 10.1364/OE.401374. [DOI] [PubMed] [Google Scholar]
- 50.Krishnan M., Mojarad N., Sandoghdar V. Geometry-induced electrostatic trapping of nanometric objects in a fluid. Nature. 2010;467:692–695. doi: 10.1038/nature09404. [DOI] [PubMed] [Google Scholar]
- 51.de Wit G., Albrecht D., Kukura P. Revealing compartmentalized diffusion in living cells with interferometric scattering microscopy. Biophys. J. 2018;114:2945–2950. doi: 10.1016/j.bpj.2018.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Block S., Zhdanov V.P., Höök F. Quantification of multivalent interactions by tracking single biological nanoparticle mobility on a lipid membrane. Nano Lett. 2016;16:4382–4390. doi: 10.1021/acs.nanolett.6b01511. [DOI] [PubMed] [Google Scholar]
- 53.Gipson P., Fukuda Y., Brunger A.T. Morphologies of synaptic protein membrane fusion interfaces. Proc. Natl. Acad. Sci. USA. 2017;114:9110–9115. doi: 10.1073/pnas.1708492114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mohrmann R., de Wit H., Sørensen J.B. Fast vesicle fusion in living cells requires at least three SNARE complexes. Science. 2010;330:502–505. doi: 10.1126/science.1193134. [DOI] [PubMed] [Google Scholar]
- 55.Sinha R., Ahmed S., Klingauf J. Two synaptobrevin molecules are sufficient for vesicle fusion in central nervous system synapses. Proc. Natl. Acad. Sci. USA. 2011;108:14318–14323. doi: 10.1073/pnas.1101818108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gui L., Ebner J.L., Lee K.K. Visualization and sequencing of membrane remodeling leading to influenza virus fusion. J. Virol. 2016;90:6948–6962. doi: 10.1128/JVI.00240-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brandt T., Cavellini L., Cohen M.M. A mitofusin-dependent docking ring complex triggers mitochondrial fusion in vitro. eLife. 2016;5:e14618. doi: 10.7554/eLife.14618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kasson P.M., Lindahl E., Pande V.S. Atomic-resolution simulations predict a transition state for vesicle fusion defined by contact of a few lipid tails. PLoS Comput. Biol. 2010;6:e1000829. doi: 10.1371/journal.pcbi.1000829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Palade G.E., Bruns R.R. Structural modulations of plasmalemmal vesicles. J. Cell Biol. 1968;37:633–649. doi: 10.1083/jcb.37.3.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rash J.E. The rapid-freeze technique in neurobiology. Trends Neurosci. 1983;6:208–212. [Google Scholar]
- 61.Imig C., Min S.-W., Cooper B.H. The morphological and molecular nature of synaptic vesicle priming at presynaptic active zones. Neuron. 2014;84:416–431. doi: 10.1016/j.neuron.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 62.Maus L., Lee C., Cooper B.H. Ultrastructural correlates of presynaptic functional heterogeneity in hippocampal synapses. Cell Rep. 2020;30:3632–3643.e8. doi: 10.1016/j.celrep.2020.02.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goldman A.J., Cox R.G., Brenner H. Slow viscous motion of a sphere parallel to a plane wall—I Motion through a quiescent fluid. Chemical Engineering Science. 1967;22:637–651. [Google Scholar]
- 64.Kyoung M., Sheets E.D. Vesicle diffusion close to a membrane: intermembrane interactions measured with fluorescence correlation spectroscopy. Biophysical Journal. 2008;95:5789–5797. doi: 10.1529/biophysj.108.128934. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Frame rate = 1 kHz, recording time = 1s, videos slowed-down 20×, frame size = 16.28 μm × 16.28 μm.
Frame rate = 1 kHz, recording time = 1s, videos slowed-down 20×, frame size = 16.28 μm × 16.28 μm.
Frame rate = 1 kHz, recording time = 1s, videos slowed-down 20×, frame size = 16.28 μm × 16.28 μm.