Abstract
Purpose
Severity scoring is a key step in managing patients with COVID-19 pneumonia. However, manual quantitative analysis by radiologists is a time-consuming task, while qualitative evaluation may be fast but highly subjective. This study aims to develop artificial intelligence (AI)-based methods to quantify disease severity and predict COVID-19 patient outcome.
Methods
We develop an AI-based framework that employs deep neural networks to efficiently segment lung lobes and pulmonary opacities. The volume ratio of pulmonary opacities inside each lung lobe gives the severity scores of the lobes, which are then used to predict ICU admission and mortality with three different machine learning methods. The developed methods were evaluated on datasets from two hospitals (site A: Firoozgar Hospital, Iran, 105 patients; site B: Massachusetts General Hospital, USA, 88 patients).
Results
AI-based severity scores are strongly associated with those evaluated by radiologists (Spearman’s rank correlation 0.837, ). Using AI-based scores produced significantly higher () area under the ROC curve (AUC) values. The developed AI method achieved the best performance of AUC = 0.813 (95% CI [0.729, 0.886]) in predicting ICU admission and AUC = 0.741 (95% CI [0.640, 0.837]) in mortality estimation on the two datasets.
Conclusions
Accurate severity scores can be obtained using the developed AI methods over chest CT images. The computed severity scores achieved better performance than radiologists in predicting COVID-19 patient outcome by consistently quantifying image features. Such developed techniques of severity assessment may be extended to other lung diseases beyond the current pandemic.
Keywords: COVID-19, Chest CT, Patient outcome, Severity scoring, Artificial intelligence
Introduction
The SARS-CoV-2 (COVID-19) outbreak at the end of 2019, which results from contracting an extremely contagious beta-coronavirus, has spread worldwide and is responsible for the latest pandemic in human history. Prior studies report frequent use of chest computed tomography (CT) in patients suspicious of pneumonia, including COVID-19 [1, 4, 10, 14, 16]. Chest CT is often recommended to assess disease severity and monitor progression in patients with moderate to severe pneumonia as well as to assess suspected complications. In sites with limited availability of reverse transcription polymerase chain reaction (RT-PCR) and high disease prevalence, chest CT is also used in diagnosis for patients with suspected COVID-19 [1, 4, 10, 14, 16].
Recent clinical studies with chest CT have reported that the qualitative scoring of pulmonary opacities can help assess severe COVID-19 pneumonia. Examples include Yang et al. [16], summing up individual scores of 0–2 from 20 lung regions. Many clinical studies focus on qualitative assessment and grading of pulmonary involvement in each lung lobe to establish disease severity [7, 18]. For example, Li et al. [7] quantified the presence of consolidation from chest CT to derive a score between 0 and 5 for each of the five lobes. Summing all the scores together gives a total score in the range of [0, 25] indicating the severity of COVID-19 pneumonia. Their work [7] suggests that high CT severity scores (indicating extensive lobar involvement and consolidation) are associated with severe COVID-19 pneumonia.
In these clinical studies, radiologists visually assess the condition of each lobe and estimate the extent of opacities in each lung lobe to assign a severity score [7, 15, 18]. The scores are then added up to become the overall evaluation. However, such description is inconsistent, subjective, and suffers from intra- and inter-observer variations. To accurately quantify a patient’s condition and to relieve clinicians’ labor, an automated assessment of the extent of pulmonary parenchymal involvement and distribution of pulmonary opacities is useful since routine clinical interpretation of chest CT does not quantify the disease burden. To automate quantification of disease distribution and extent of pulmonary involvement, automatic segmentation of lung lobes, pulmonary opacities, and distribution of opacities within each lobe are desired. Tang et al. [11] employed threshold segmentation based on the Hounsfield units (HU) range of the ground glass opacity (GGO) to detect the severe disease on chest CT. He et al. [5] proposed a framework for joint lung lobe segmentation and severity assessment of COVID-19 in CT images. We hypothesized that AI-based automatic lung lobe segmentation and distribution of pulmonary opacities can help assess disease severity and outcomes in patients with COVID-19 pneumonia.
This work proposes an AI-assisted severity scoring method based on automated segmentation of the lung lobes and pulmonary opacities. We first analyze the correlation between scores obtained by AI and radiologists. We then show the statistics and data distribution of patients and derive the correlation between scores and patient outcomes. To quantify the correlation, we establish an evaluation scheme that uses three machine learning models for predicting the patient outcomes based on severity scores.
Data and annotation
Data sets
The deidentified data used in our work were acquired at two hospitals, i.e., Site A: Firoozgar Hospital (Tehran, Iran) and Site B: Massachusetts General Hospital (Boston, MA, USA). All the CT imaging data were from patients who underwent clinically indicated, standard-of-care, non-contrast chest CT.
Site A We reviewed medical records of adult patients admitted with known or suspected COVID-19 pneumonia from the site between February 23, 2020, and March 30, 2020. In the 117 patients with positive RT-PCR assay for COVID-19, three patients were excluded due to the presence of extensive motion artifacts on their chest CT. One patient was excluded due to the absence of ICU admission information. The radiologists annotated 105 CT volumes of the left 113 available scans excluding those with significant motion artifacts.
Site B We reviewed medical records of adult patients admitted with COVID-19 symptom between March 24 and May 9, 2020. 125 RT-PCR positive admitted patients underwent unenhanced chest CT are selected to form this dataset. The radiologists labeled 88 patients with both ICU admission and mortality risk.
Demographic statistics Tables 1and 2 summarize the demographic data from Sites A and B, respectively. Several data variables (age, gender, white blood cell (WBC) count, lymphocyte count (Lym), lymphocyte-to-WBC ratio (Lym_r)) have significantly different values between patients with and without ICU admission.
Table 1.
Demographic statistics (mean ± SD, except for gender) for Site A dataset
| ICU admission | Not admitted | ICU admitted |
|---|---|---|
| Gender (M:F) | 43:28 | 27:12 |
| Age (year) | ||
| Lym_r (%) | 8.3 | |
| WBC | ||
| Lym |
Table 2.
Demographic statistics (mean ± SD, except for gender) for Site B dataset
| ICU admission | Not admitted | ICU admitted |
|---|---|---|
| Gender (M:F) | 18:21 | 24:25 |
| Age (year) | ||
| Lym_r (%) | ||
| WBC | ||
| Lym |
Annotation of severity
Two thoracic subspecialty radiologists (one with 16 years of experience and the other with 14 years of experience) reviewed all CT images without the knowledge of clinical features, laboratory data, and patient outcomes. Chest CT images were reviewed on DICOM image viewer (MicroDicom DICOM Viewer, Sofia, Bulgaria) in lung windows (window width 1500 HU, window level-600 HU). The radiologists recorded the type of pulmonary opacities (ground glass opacities, mixed ground glass and consolidative opacities, consolidation, organizing pneumonia, nodular pattern, and ground glass opacities with septal thickening (crazy-paving pattern)). Extent of involvement of each lobe (right upper, right middle, right lower, left upper, and left lower lobes) by the pulmonary opacities was assessed using a previously described scale (0: 0% lobar involvement; 1: < 5% involvement of lobar volume; 2: 5–25% involvement of lobe; 3: 26–50% lobar involvement; 4: 51–75% lobar involvement; 5: >75% lobar involvement) [10]. The two thoracic subspecialty radiologists reviewed all CT images independently. Any discordance between them was resolved by consensus readout of cases. Total extent of pulmonary opacities was estimated by adding the scores of all lobes (lowest score 0, highest possible score 25) [8, 10, 13].
AI-assisted severity assessment
Since pulmonary opacity (PO) is an important criterion in terms of the patient severity assessment, we further evaluated our AI-based quantification scores against radiologists’ manually graded scores in patients’ outcome prediction. In order to obtain explainable severity scores, we divide the AI-assisted procedure into two steps as same as radiologists. In the first step, we use deep learning-based method to automatically segment the lung lobes and pulmonary opacities. Then, severity scores of each patient are computed from the sizes of the lobes and pulmonary opacities. In this way, the severity scores are not dependent on the radiologists’ annotation. Furthermore, we use the association of severity scores with patient outcome, ICU admission, and mortality risk of patients with COVID-19 pneumonia, as the criterion to evaluate the severity assessment between AI against radiologists.
Deep learning-based image segmentation
This work employs deep neural networks to segment both lungs, five lung lobes (left upper lobe, left lower lobe, right upper lobe, right middle lobe, right lower lobe) and pulmonary opacity regions of infection from non-contrast chest CT examinations. For network training, we semiautomatically labeled all five pulmonary lobes in 71 CT volumes from Site A using chest imaging platform [17]. A radiologist (M.K.K.) annotated the lung opacities slice by slice in 105 CT volumes from Site A. For lung lobe segmentation, we adopted the automated lung segmentation method proposed by Hofmanninger et al. [6]. Their work provides a trained U-net model for lung segmentation. The U-Net consists of an encoder with regular convolutions and max pooling layers, and a decoder that applies transposed convolutions along with regular convolutions. The network consists of 19 convolutional layers. Each convolutional block in the encoder and decoder uses two 33 convolutional layers. Finally, a 11 convolutional layer is applied to squeeze the number of feature map channels into 2. To improve the model performance, residual connections between the encoder and decoder were employed. Same as their pre-processing step, the intensity HU range is cropped into the window of [− 1024, 600] and then normalized into [0, 1]. The pre-trained model1 was fine-tuned with a learning rate of using our annotated data. During tuning, each slice was randomly cropped into patches with size . The tuned model was then applied to segment all the chest CT volumes.
Segmentation of pulmonary opacities was completed by our previously proposed method, Pyramid Input Pyramid Output Feature Abstraction Network (PIPO-FAN) [3] with publicly released source code.2 The network integrates image pyramid and multi-scale feature analysis into one single end-to-end framework. It applies spatial pyramid pooling on one 2D slice to generate pyramid input and hierarchically fuses semantically similar features after convolutional blocks. Pyramid features were then adaptively fused via attention module to obtain the lesion map of the slice. In the pre-processing step, all slices were resampled into a fixed resolution of pixels, and to improve the contrast of pulmonary opacity, the intensity HU range was cropped into a window of . In training, the learning rate was set to be 0.002. Softmax activation function with threshold 0.5 was used to obtain the infection area. Morphological operation was applied to refine the segmentation of lung lobes and pulmonary opacities. Using a 33 kernel which only connects the nearest neighbors to the center, we first perform the opening operation to remove small noisy segmentation and then apply the closing operation using the same kernel to generate a smooth segmentation.
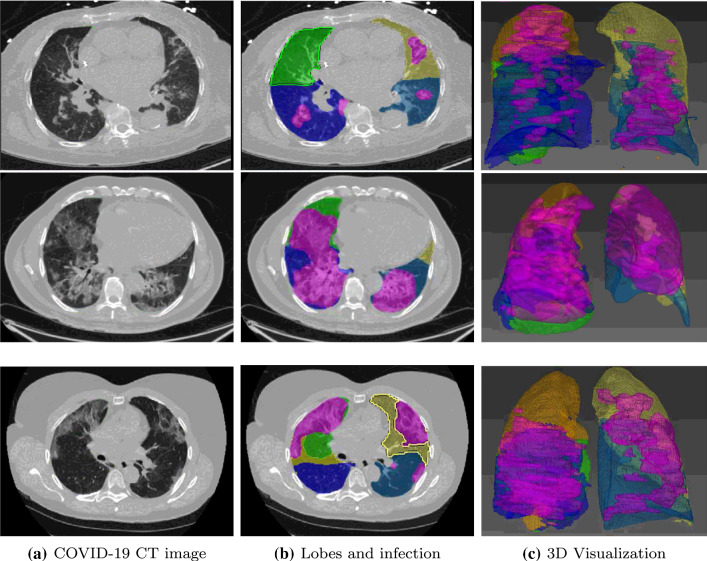
Figure 1 shows the segmentation results of lung lobes and pulmonary opacities. From axial and 3D view, we can see that the segmentation model can smoothly and accurately predict isolated regions with pulmonary opacities. Based on the area of pulmonary opacities, we then computed the ratio of opacity volume over lobes, which is a widely used measurement to describe the severity [11, 20]. The dice similarity coefficient (DSC) was used to quantitatively evaluate the segmentation accuracy. For Site A dataset, we obtained DSC scores of 82.5% and 90.0% on lung opacity segmentation and lobe segmentation, respectively.
Fig. 1.
Segmentation results of lung lobes and pulmonary opacities. Areas colored in magenta indicate segmented lesions. Other colored areas represent the segmented lobes, with orange: right upper, green: right middle, navy: right lower, yellow: left upper and ocean: left lower
Severity scoring
Based on the segmentation process in the section “Deep learning-based image segmentation,” the AI-based quantification scores are obtained based on the segmentation results with similar procedure, followed by radiologists (see section “Annotation of severity” for more details). First, RPO (ratio of pulmonary opacities) over lobe was calculated for each lobe and graded into 6 levels (0–5). Then, the final score for a patient was the sum of the scores of the 5 lobes which ranges from 0 to 25.
To show relationship between the severity scores and patient outcome, patients were divided into 4 groups (Group I, II, III and IV) based on their final outcome from mild to severe. Groups I and II included recovered patients without admission to ICU. Group I consisted of patients discharged from hospital within 7 days. Group II included patients with more than 7 days of hospital stay. Group III patients had ICU admission and recovered, whereas Group IV patients succumbed to COVID-19 pneumonia. We then divided the severity scores into four buckets. The patients were first divided into two groups using the mean severity score 15 assigned by the radiologists. Each group is then further divided by halving the buckets. We display the statistic of severity groups in different buckets and evaluate the correlation between severity buckets and patient outcome using mean absolute error (MAE).
Severity analysis
This work establishes an evaluation scheme to quantify the correlation between severity scores and patient outcome. The severity scores are used as an input to different machine learning models for predicting patient outcome. The AUC for this task indicates the correlation between scores and patient outcomes. Considering the variations introduced by different prediction targets, models and datasets, to obtain a more objective evaluation, we conducted experiments for both ICU admission and mortality prediction with 3 different models (i.e., support vector machine (SVM), random forest (RF) and logistic regression (LR)) on the two datasets. Radial basis function kernels were used to construct SVM models. Squared L2 norm is used as a regularization term for the SVM and LR models. The RF model has 300 trees, and the Gini index is used as the criterion for calculating information gain. We bootstrap AUCs with 1000 entries to obtain the 95% confidence interval.
In addition to the scores estimated by radiologists and out AI methods, we also include another two groups of scores, i.e., threshold-based scores, and the mixture of radiologists’ scores and the AI-based scores. As HU corresponds to GGO regions [2, 11], the method regards voxels within this threshold as pulmonary opacities and further calculates the severity scores. To investigate whether the scores of our AI method are complementary with those of the radiologists on patient outcome prediction, we merged their scores (denoted as AI + Radiologists). Specifically, while all other methods use 5 scores of 5 lobes as inputs of the prediction models, AI + Radiologists uses all 10 scores obtained by both the AI methods and the radiologists as inputs. The average results of the three models is used to compute AUC of each scoring method. We conducted one-tailed z-test [19] on ROC curves using scores from radiologists and other severity scoring methods.
Experimental results
This section presents the results of the developed techniques. We show the effectiveness of our proposed segmentation-based severity scoring on the two datasets separately through comparison with different severity scoring methods.
The results are summarized in three parts. In the first part, we computed the correlation coefficient between severity scores assigned by radiologists and computed by our deep learning-based segmentation to demonstrate a good consistency between AI and radiologists. In the second part, we display the statistics of number and proportion of patients in severity groups on different buckets based on section “Severity scoring.” The number of patients in each group is overlaid on the corresponding segment. We use the mean absolute error (MAE) between severity score and patient severity group to evaluate the consistency between severity scores and patient outcome.
Finally, to further evaluate the association between the scores and patient outcome, we use the different groups of scores to do ICU admission and mortality prediction, respectively. We computed the AUC for mortality and ICU admission prediction using the severity scores to evaluate the association between scores and patient outcomes. Cross-dataset validation (training on Site A and testing on Site B; training on Site B and testing on Site A) was performed for each model on each group of severity scores to compute the AUC.
Correlation between AI and radiologists
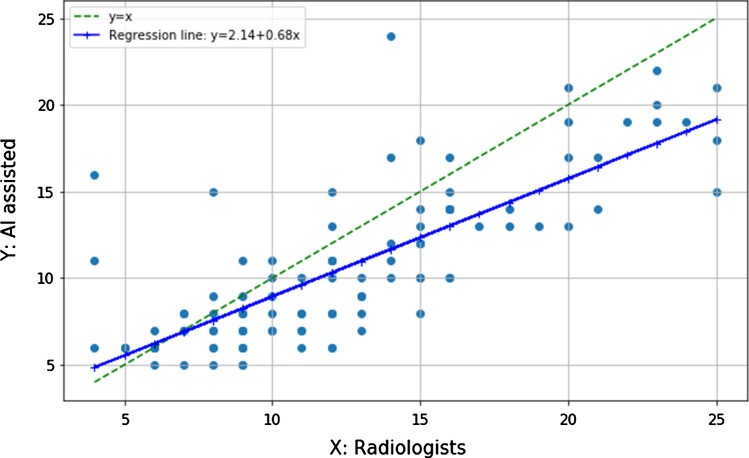
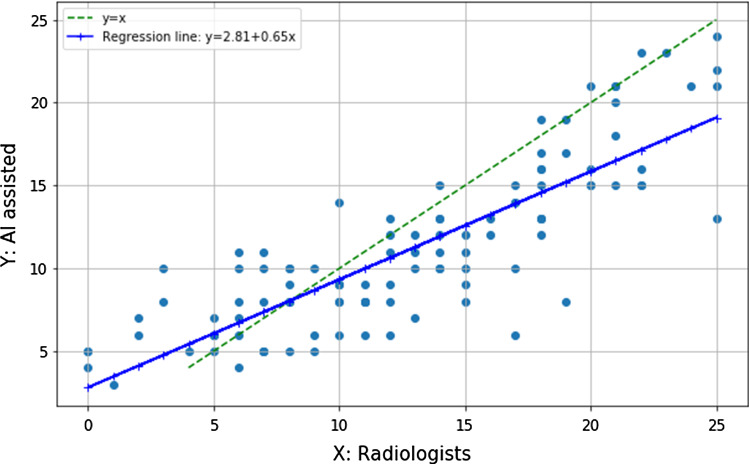
The score for a patient is the sum of the scores of the 5 lobes which ranges from 0 to 25. Similar to the work of Li et al. [9], we evaluated Spearman’s rank correlation and associated p-value to determine the strength of the correlation between scores computed by AI-assisted method and assigned by radiologists on Site A and Site B dataset. Figure 2 shows the correlation between the two types of severity scores. We obtained a considerable positive Spearman’s rank correlation of 0.770 on Site A dataset (). Figure 3 shows the correlation between the two types of severity scores of 88 patients. We obtained a considerable positive Spearman’s rank correlation of 0.837 on Site B dataset (), indicating that the AI-assisted prediction can obtain consistent results with radiologists.
Fig. 2.
Correlation between the severity scores assigned by radiologists and computed by our deep learning-based segmentation on Site A dataset
Fig. 3.
Correlation between the severity scores assigned by radiologists and computed by our deep learning-based segmentation on Site B dataset
Severity scores and patient outcome
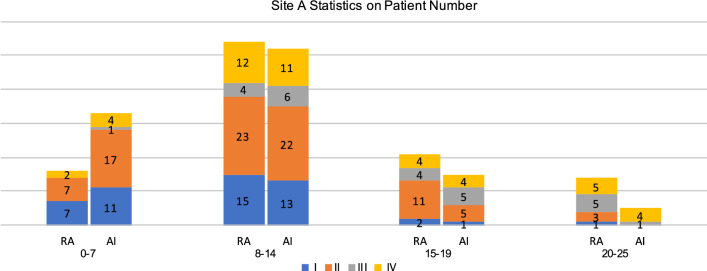
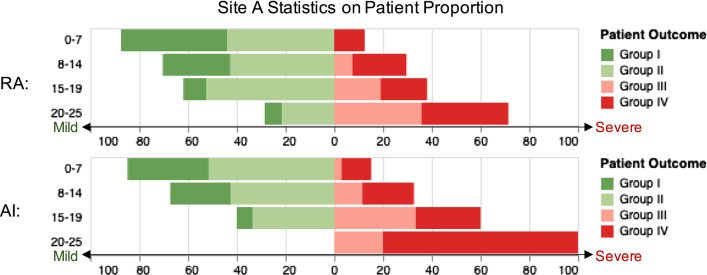
Figures 4 and 5 display the statistics on number and proportion of 105 patients in Site A dataset falling into the four score buckets based on the computed severity scores, assigned by radiologists and our AI algorithm, respectively. As one can see from the figure, patient severity groups are positively correlated with the severity scores buckets. In Fig. 5, we can see that for our AI method, no patients in Group III and Group IV are in the bucket [20,25]. For quantitative evaluation, we then computed the MAE between severity score buckets and patient severity groups. Each bucket of severity score is paired with one severity group. The MAE for radiologists and AI quantification computed using the number is 88 and 84, respectively. The MAE computed using the proportion is 3.51 and 2.65. Our AI method obtained comparable MAE when compared to the radiologists, i.e., the difference in the MAE scores is not statistically different.
Fig. 4.
For different buckets divided by severity scores obtained by AI and radiologists, values of number of patients from different patient groups (I, II, III, IV) are presented on Site A dataset. RA: Results of radiologists’ annotation. AI: Results of the AI-based method. Heights of the bars represent the number
Fig. 5.
For each severity score bucket shown in vertical, horizontal stacked bars present the proportion of patient number of one severity group to patient number of all severity groups of Site A in the bucket. Widths of the bars represent such proportion (mild patient: green for group I and light green for group II; severe patient: light red for group III and red for group IV)
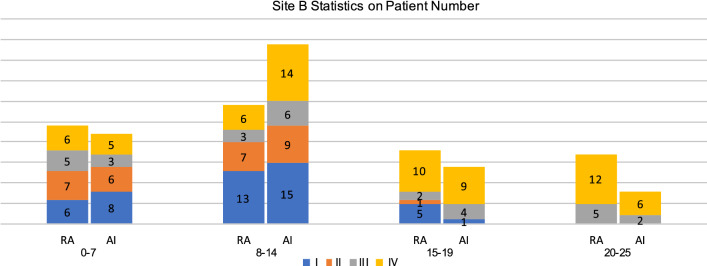
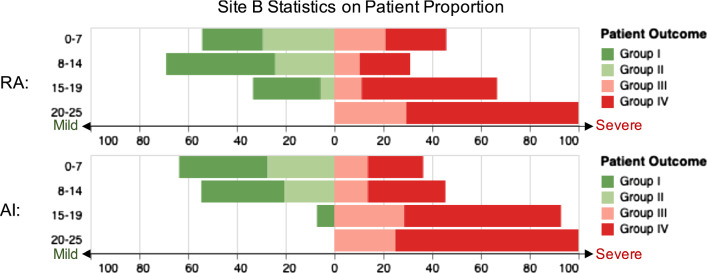
Figures 6 and 7 display the statistics on number and proportion of 88 patients in Site B dataset falling into different categories based on the computed severity scores, given by radiologists and our AI algorithm, respectively. The MAE for radiologists and AI quantification computed using the number is 89 and 89, respectively. The MAE computed by the proportion is 3.88 and 3.38 for the radiologists and the AI, respectively. That means the AI-assisted method achieved comparable consistency with patient severity groups than radiologists. In Fig. 7, we can see that for our AI method, the proportion of the severe patients (Group III and Group IV) monotonically increases with the raising of the severity score, while the result of the radiologists has a drawback on the second score bucket.
Fig. 6.
For different buckets divided by severity scores obtained by AI and radiologists, values of number of patients from different patient groups (I, II, III, IV) are presented on Site B dataset. RA: Results of radiologists’ annotation. AI: Results of the AI-based method. Heights of the bars represent the number
Fig. 7.
For each severity score bucket shown in vertical, horizontal stacked bars present the proportion of patient number of one severity group to patient number of all severity groups of Site B in the bucket. Widths of the bars represent such proportion (mild patient: green for group I and light green for group II; severe patient: light red for group III and red for group IV)
Patient outcome prediction
Results on Site A
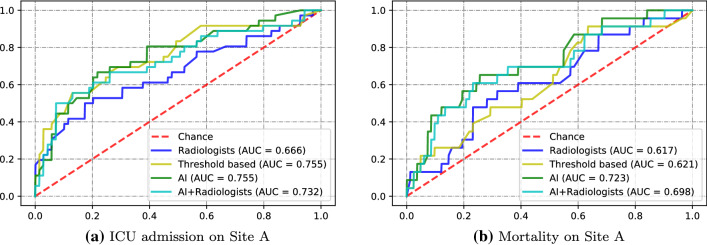
Table 3 summarizes the AUCs and 95% confidence intervals of three machine learning models on ICU admission and mortality. The three machine learning models were trained on Site B and tested on Site A. The bold values represent the best AUCs. We can see that AI achieved higher AUC than radiologists under all models and tasks. We further use the mean of the scores from the three models as a simple ensemble strategy to compute the AUC value for each severity scoring method. ROC curves on ICU admission and mortality prediction are shown in Fig. 8a, b. AI obtains best AUC 0.755 and 0.723 on ICU admission and mortality, respectively. One-tailed z-test is used to evaluate the statistical significance between radiologists and other scoring methods. Threshold-based method outperforms radiologists in ICU admission with although performance in mortality prediction was not significantly different (). AI significantly outperforms radiologists with in ICU admission and on mortality.
Table 3.
Comparison of AI with radiologists in predicting ICU admission and mortality prediction on Site A dataset
| Outcome | Severity scoring | SVM | RF | LR | |||
|---|---|---|---|---|---|---|---|
| Mean | 95% CI | Mean | 95% CI | Mean | 95% CI | ||
| ICU admission | Radiologists | 0.689 | (0.594, 0.778) | 0.663 | (0.562, 0.759) | 0.642 | (0.535, 0.739) |
| Threshold [11] | (0.666, 0.830) | 0.706 | (0.607,0.796) | (0.713, 0.854) | |||
| AI segmentation (AI) | 0.733 | (0.641, 0.816) | (0.698, 0.848) | 0.723 | (0.623, 0.810) | ||
| AI + radiologists | 0.726 | (0.639, 0.804) | 0.759 | (0.638, 0.836) | 0.681 | (0.575, 0.778) | |
| Mortality | Radiologists | 0.639 | (0.527, 0.743) | 0.644 | (0.531, 0.738) | 0.566 | (0.441, 0.674) |
| Threshold [11] | 0.605 | (0.479, 0.718) | 0.593 | (0.474, 0.702) | 0.614 | (0.480, 0.741) | |
| AI segmentation (AI) | (0.626, 0.812) | (0.623, 0.820) | (0.545, 0.777) | ||||
| AI + radiologists | 0.693 | (0.589, 0.788) | 0.719 | (0.618, 0.807) | 0.638 | (0.505, 0.758) | |
Fig. 8.
Comparison of severity scoring methods under ROC curves on Site A
Results on site B dataset
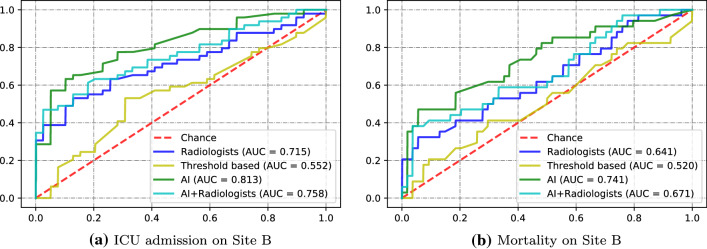
The same set of experiments were repeated on the Site B dataset. The three machine learning models are trained on Site A and tested in Site B. Table 4 summarizes the AUCs of different models with 95% confidence interval indicated by ICU admission and mortality on Site B dataset. The bold values represent the best AUCs. We can see that AI achieved best AUC under all models and tasks. ROC curves on ICU admission and mortality prediction are shown in Fig. 9a, b. AI obtains best AUC 0.813 and 0.741 on ICU admission and mortality, respectively. One-tailed z-test is used to evaluate the statistical significance between radiologists and other scoring methods. Radiologists outperform threshold-based method in ICU admission with ; both methods had similar performance for mortality prediction (). AI significantly outperforms radiologists with in ICU admission and on mortality.
Table 4.
Comparison of AI with radiologists in predicting ICU admission and mortality prediction on Site B dataset
| Outcome | Severity scoring | SVM | RF | LR | |||
|---|---|---|---|---|---|---|---|
| Mean | 95% CI | Mean | 95% CI | Mean | 95% CI | ||
| ICU admission | Radiologists | 0.680 | (0.581, 0.773) | 0.695 | (0.600, 0.782) | 0.705 | (0.608, 0.794) |
| Threshold [11] | 0.719 | (0.624, 0.808) | 0.703 | (0.599, 0.796) | 0.701 | (0.602, 0.795) | |
| AI segmentation (AI) | (0.675, 0.851) | (0.665, 0.840) | (0.675, 0.851) | ||||
| AI + radiologists | 0.725 | (0.634, 0.811) | 0.741 | (0.657, 0.821) | 0.725 | (0.634, 0.811) | |
| Mortality | Radiologists | 0.566 | (0.447, 0.679) | 0.616 | (0.504, 0.727) | 0.670 | (0.602, 0.795) |
| Threshold [11] | 0.584 | (0.481, 0.685) | 0.579 | (0.470,0.679) | 0.605 | (0.505, 0.701) | |
| AI segmentation (AI) | (0.545, 0.766) | (0.578, 0.779) | (0.642, 0.828) | ||||
| AI + radiologists | 0.603 | (0.496, 0.719) | 0.631 | (0.530, 0.735) | 0.716 | (0.623, 0.810) | |
Fig. 9.
Comparison of severity scoring methods under ROC curves on Site B
Discussion
In the current clinical radiology practice, radiologists from hospitals do not perform a quantitative or semiquantitative assessment of disease severity or distribution of pulmonary opacities. This lack of quantification is related to the fact that they are not trained and required to assign severity scores in patients with pneumonia. While in patients with cancer and focal lesions, radiologists measure and compare single or volumetric dimension of focal lesions, in patients with diffuse and ill-defined disease patterns found in pneumonia such measurements are not feasible and practical. As a result, radiology reports in patients with COVID-19 pneumonia are limited to semantic description of extent (such as diffuse, multifocal, or localized) and type of opacities rather than an assigned severity score. However, prior studies with subjective severity scores from both chest radiography and CT report on their ability to predict disease severity and patient outcome [12, 16]. Threshold segmentation can detect coarse opacities with some degree, but it is known to be less accurate than AI-based methods.
Results analysis
The study confirms that AI-based severity scoring method yields AUCs in ICU admission and mortality prediction, on Site A and B datasets, that exceed those of radiologists. Considering the large differences in patient demographic statistics and clinical protocols at the two participating sites, the results highlight the robustness of the AI-based method and their generalization ability of the extracted scores.
The statistical test of differences between ROC curves using scores from AI and radiologists suggests that AI significantly outperforms radiologists (A B: on ICU admission, on mortality; B A: on ICU admission, on mortality). The results outline that the correlation between AI-assisted severity score and prognosis of patients has stronger correlation with patient outcomes than radiologists, which could effectively improve the accuracy of severity scoring system.
We explored the potential of combining the scores of AI and radiologists to further improve the prognosis of COVID-19 pneumonia. Both score concatenation and sum were tested. However, compared with AI-based method, the combination of scores of AI and radiologists did not present a significant improvement. We also used investigated other machine learning models, decision tree (DT) and multilayer perceptron (MLP), to predict patient outcomes. The two models did not perform as well as the three models reported in this paper. Besides the choice of classification model, the segmentation quality of pulmonary lobes and opacities may be an additional factor that limits the performance of outcome prediction. More accurate image segmentation will give a more precise lung opacity quantification, which, in turn, may help to improve the performance of our AI method further. Also, ancillary imaging findings not assessed with our segmentation tool such as coronary artery disease, cardiac enlargement, pleural effusions, co-existing bronchial wall thickening or emphysema, and mediastinal lymphadenopathy, may have affected the disease outcome, and thus decreased the performance of our models.
The AUC obtained for mortality prediction is lower than in ICU admission prediction. Since mortality is influenced by several factors including comorbidities, local treatment condition and disease exacerbation, mortality prediction is more challenging than ICU admission. However, the AI method can still persistently improve the AUC over radiologists for this task by 10%, which further demonstrate the effectiveness of scores extracted from AI.
Future directions
Although scores from radiologists may not be as accurate as AI, they may convey overall condition of severe patients. For example, although not included in our study, radiologists assessed findings beyond pulmonary opacities such as architectural distortion in lungs, pleural effusions, mediastinal/hilar lymphadenopathy, cardiac enlargement, and increased subcutaneous fat stranding or attenuation suggestive of anasarca and underlying fluid overload. Li et al. [9] showed that such findings can be learned by AI algorithms and help longitudinal disease evaluation on COVID-19 pneumonia.
For future work, we will continue to explore developing AI algorithms, which may efficiently incorporate the knowledge of radiologists with an overall evaluation of patient condition. It is also worth noting that the developed techniques of severity assessment may extend to other lung diseases beyond the current pandemic. As the world awaits the introduction of disease prevention and specific treatment for those infected by COVID-19 pneumonia, it is important that future versions of AI-based time-to-event measures assess the extent of chronic changes from the substantial patient population who had initial recovery but might have long lasting sequelae from their infection.
Conclusions
This paper has proposed an AI-assisted severity scoring method based on automatic segmentation of lung lobes and pulmonary opacities. The severity scores obtained by AI have shown to be consistent with those obtained by radiologists in the two datasets. We have further quantitatively evaluated the scores based on patient outcomes and demonstrated that the AI segmentation-based method was significantly more accurate than current severity scoring by radiologists only. The results suggest that AI can significantly improve the patient outcome prediction for patients with severe COVID-19 pneumonia. We believe such techniques have the potential to numerous clinical applications involving COVID-19 pneumonia.
Acknowledgements
This work was partially supported by National Institute of Biomedical Imaging and Bioengineering (NIBIB) under award R21EB028001 and National Heart, Lung, and Blood Institute (NHLBI) under Award R56HL145172.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
The study received IRB approval at both participating sites. Need for informed consent was waived due to the retrospective nature of the study.
Informed consent
There is no informed consent required for the work reported in this manuscript.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mannudeep K. Kalra, Email: mkalra@mgh.harvard.edu
Pingkun Yan, Email: yanp2@rpi.edu.
References
- 1.Ai T, Yang Z, Hou H, Zhan C, Chen C, Lv W, Tao Q, Sun Z, Xia L. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020 doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chao H, Fang X, Zhang J, Homayounieh F, Arru CD, Digumarthy SR, Babaei R, Mobin HK, Mohseni I, Saba L, Carriero A, Falaschi Z, Pasche A, Wang G, Kalra MK, Yan P. Integrative analysis for COVID-19 patient outcome prediction. Med Image Anal. 2021;67:101844. doi: 10.1016/j.media.2020.101844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fang X, Yan P. Multi-organ segmentation over partially labeled datasets with multi-scale feature abstraction. IEEE Trans Med Imaging. 2020;39(11):3619–3629. doi: 10.1109/TMI.2020.3001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harmon SA, Sanford TH, Xu S, Turkbey EB, Roth H, Xu Z, Yang D, Myronenko A, Anderson V, Amalou A, Blain M, Kassin M, Long D, Varble N, Walker SM, Bagci U, Ierardi AM, Stellato E, Plensich GG, Franceschelli G, Girlando C, Irmici G, Labella D, Hammoud D, Malayeri A, Jones E, Summers RM, Choyke PL, Xu D, Flores M, Tamura K, Obinata H, Mori H, Patella F, Cariati M, Carrafiello G, An P, Wood BJ, Turkbey B. Artificial intelligence for the detection of COVID-19 pneumonia on Chest CT using multinational datasets. Nat Commun. 2020;11(1):4080. doi: 10.1038/s41467-020-17971-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He K, Zhao W, Xie X, Ji W, Liu M, Tang Z, Shi F, Gao Y, Liu J, Zhang J, Shen D (2020) Synergistic learning of lung lobe segmentation and hierarchical multi-instance classification for automated severity assessment of COVID-19 in CT images. arXiv preprint arXiv:2005.03832 [DOI] [PMC free article] [PubMed]
- 6.Hofmanninger J, Prayer F, Pan J, Rohrich S, Prosch H, Langs G. Automatic lung segmentation in routine imaging is primarily a data diversity problem, not a methodology problem. Eur Radiol Exp. 2020 doi: 10.1186/s41747-020-00173-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li K, Wu J, Wu F, Guo D, Chen L, Fang Z, Li C. The clinical and chest CT features associated with severe and critical COVID-19 pneumonia. Investig Radiol. 2020;55(6):327–331. doi: 10.1097/RLI.0000000000000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li L, Qin L, Xu Z, Yin Y, Wang X, Kong B, Bai J, Lu Y, Fang Z, Song Q, Cao K, Liu D, Wang G, Xu Q, Fang X, Zhang S, Xia J, Xia J. Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT. Radiology. 2020 doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li MD, Arun NT, Gidwani M, Chang K, Deng F, Little BP, Mendoza DP, Lang M, Lee SI, O’Shea A, Parakh A, Singh P, Kalpathy-Cramer J. Automated assessment and tracking of covid-19 pulmonary disease severity on chest radiographs using convolutional siamese neural networks. Radiol Artif Intell. 2020;2(4):e200079. doi: 10.1148/ryai.2020200079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shan F, Gao Y, Wang J, Shi W, Shi N, Han M, Xue Z, Shi Y (2020) Lung infection quantification of COVID-19 in CT images with deep learning. arXiv preprint arXiv:2003.04655
- 11.Tang Z, Zhao W, Xie X, Zhong Z, Shi F, Liu J, Shen D. Severity assessment of coronavirus disease 2019 COVID-19 using quantitative features from Chest CT images. Phys Med Biol. 2020 doi: 10.1088/1361-6560/abbf9e. [DOI] [PubMed] [Google Scholar]
- 12.Toussie D, Voutsinas N, Finkelstein M, Cedillo MA, Manna S, Maron SZ, Jacobi A, Chung M, Bernheim A, Eber C, Concepcion J, Fayad Z, Gupta YS. Clinical and chest radiography features determine patient outcomes in young and middle age adults with COVID-19. Radiology. 2020;297:201754. doi: 10.1148/radiol.2020201754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong HYF, Lam HYS, Fong AHT, Leung ST, Chin TWY, Lo CSY, Lui MMS, Lee JCY, Chiu KWH, Chung T, Lee EYP, Wan EYF, Hung FNI, Lam TPW, Kuo M, Ng MY. Frequency and distribution of chest radiographic findings in COVID-19 positive patients. Radiology. 2020 doi: 10.1148/radiol.2020201160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie X, Zhong Z, Zhao W, Zheng C, Wang F, Liu J. Chest CT for typical 2019-nCoV pneumonia: relationship to negative RT-PCR testing. Radiology. 2020;296:200343. doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiong Y, Sun D, Liu Y, Fan Y, Zhao L, Li X, Zhu W. Clinical and high-resolution CT features of the COVID-19 infection: comparison of the initial and follow-up changes. Investig Radiol. 2020;55(6):332–339. doi: 10.1097/RLI.0000000000000674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang R, Li X, Liu H, Zhen Y, Zhang X, Xiong Q, Luo Y, Gao C, Zeng W. Chest CT severity score: an imaging tool for assessing severe COVID-19. Radiol Cardiothorac Imaging. 2020;2(2):e200047. doi: 10.1148/ryct.2020200047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yip SS, Parmar C, Blezek D, Estepar RSJ, Pieper S, Kim J, Aerts HJ. Application of the 3d slicer chest imaging platform segmentation algorithm for large lung nodule delineation. PLoS ONE. 2017;12(6):e0178944. doi: 10.1371/journal.pone.0178944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao W, Zhong Z, Xie X, Yu Q, Liu J. Relation between chest CT findings and clinical conditions of coronavirus disease (COVID-19) pneumonia: a multicenter study. Am J Roentgenol. 2020;214(5):1072–1077. doi: 10.2214/AJR.20.22976. [DOI] [PubMed] [Google Scholar]
- 19.Zhou XH, McClish DK, Obuchowski NA. Statistical methods in diagnostic medicine. New York: Wiley; 2009. [Google Scholar]
- 20.Zhu X, Song B, Shi F, Chen Y, Hu R, Gan J, Zhang W, Li M, Wang L, Gao Y, Shan F, Shen D. Joint prediction and time estimation of COVID-19 developing severe symptoms using chest CT scan. Med Image Anal. 2021;67:101824. doi: 10.1016/j.media.2020.101824. [DOI] [PMC free article] [PubMed] [Google Scholar]