Abstract
Purpose
Deep learning can be used for improving the performance of computer-aided detection (CADe) in various medical imaging tasks. However, in computed tomographic (CT) colonography, the performance is limited by the relatively small size and the variety of the available training datasets. Our purpose in this study was to develop and evaluate a flow-based generative model for performing 3D data augmentation of colorectal polyps for effective training of deep learning in CADe for CT colonography.
Methods
We developed a 3D-convolutional neural network (3D CNN) based on a flow-based generative model (3D Glow) for generating synthetic volumes of interest (VOIs) that has characteristics similar to those of the VOIs of its training dataset. The 3D Glow was trained to generate synthetic VOIs of polyps by use of our clinical CT colonography case collection. The evaluation was performed by use of a human observer study with three observers and by use of a CADe-based polyp classification study with a 3D DenseNet.
Results
The area-under-the-curve values of the receiver operating characteristic analysis of the three observers were not statistically significantly different in distinguishing between real polyps and synthetic polyps. When trained with data augmentation by 3D Glow, the 3D DenseNet yielded a statistically significantly higher polyp classification performance than when it was trained with alternative augmentation methods.
Conclusion
The 3D Glow-generated synthetic polyps are visually indistinguishable from real colorectal polyps. Their application to data augmentation can substantially improve the performance of 3D CNNs in CADe for CT colonography. Thus, 3D Glow is a promising method for improving the performance of deep learning in CADe for CT colonography.
Keywords: Generative models, Data augmentation, Deep learning, Computer-aided detection, Virtual colonoscopy, Artificial intelligence
Introduction
Colorectal cancer (CRC) is the third most common cancer in terms of incidence and the second most common cancer in terms of mortality worldwide [1]. However, CRC can be prevented considerably by early detection and removal of its precursors, colorectal polyps [2]. Computed tomographic colonography (CTC) can be used both for detecting CRCs and for preventing CRCs by early detection of clinically significant polyps that could develop into cancers [3].
Deep learning based on convolutional neural networks (CNNs) has made it easy to obtain state-of-the-art results in various medical imaging tasks [4]. However, one of the limitations of deep learning is that the development of generalizable CNNs requires a large amount and great variety of training data [5]. In CTC, the available training datasets are relatively small and limited in numbers and variations.
The two principal approaches for addressing the issue of small training datasets in deep learning have included (1) transfer learning of CNNs and (2) data augmentation of training datasets. With the first approach, transfer learning, one starts with a CNN that has been pre-trained with a large number of images. The CNN is then adapted to the desired application by continuation of the training with the available domain-specific data. In CTC, transfer learning has been used successfully in training of CNNs for interpreting virtual endoluminal views [6], for detecting contrast-coated serrated polyps [7] and for performing electronic cleansing [8]. However, transfer learning has the limitations that most of the publicly available pre-trained CNNs have not been pre-trained with medical images and that most of these CNNs are 2D CNNs.
With the second approach, data augmentation, the training dataset is enhanced artificially so that the number and variety of training samples are increased. The most common method for implementing data augmentation has been the manipulation of the training samples by basic image processing operations [5]. In CTC, this approach has been used successfully for training of CNNs for detection of polyps [9] and masses [10], and for reduction in the number of false-positive (FP) detections in computer-aided detection (CADe) [11, 12]. However, the approach has the limitation that some of the image transforms that are commonly used with natural images are not appropriate for use with medical images. This can limit the number and variety of the obtainable training samples.
Recently, several studies have explored the possibility of performing data augmentation by use of generative adversarial networks (GANs) [13, 14]. In CTC, 3D GANs have been used for generation of synthetic polyps to improve the training of 3D CNNs in CADe [15]. However, the development of GANs that can generate realistic synthetic images at a high image resolution is known to suffer from various problems, such as non-convergence of the model parameters, mode collapse or training imbalance between the generator and the discriminator [5].
Flow-based generative models have several advantages over GANs [16]. Unlike GANs, they are designed to learn the distribution of the input data explicitly, thereby providing an exact latent-variable inference, log-likelihood evaluation and a meaningful latent space for performance of valid manipulations of the data. They are also highly parallelizable and can be implemented with a small memory footprint [16].
In this study, we investigated the feasibility of applying a flow-based generative model to 3D data augmentation of colorectal polyps in CTC. We hypothesized that (1) a 3D CNN that implements a state-of-the-art flow-based generative model known as Glow [16] (hereafter referred to as 3D Glow) can be trained to generate synthetic polyps that are visually indistinguishable from real colorectal polyps in CTC, and that (2) data augmentation by use of 3D Glow-generated synthetic polyps can be used for improving the performance of deep learning in differentiating between polyps and non-polyps in CADe for CTC. One could use successful development of 3D Glow to overcome the limitations of currently available CTC datasets, thereby enabling the training of generalizable high-performing 3D CNNs for CADe in CTC.
Flow-based generative model
Given an observed (complicated) probability distribution, a flow-based generative model provides a bijective mapping f between the observed distribution and a simple, well-understood target probability distribution, such as a standard Gaussian distribution. The desired computations can then be performed on the simple target distribution rather than on the observed distribution. Such a mapping that is constructed as a sequence of invertible bijective transforms is called the normalizing flow. Formally, let x denote an observation sampled from a training dataset D that has an unknown probability distribution p(x). The normalizing flow can be defined as
| 1 |
where z is a latent random variable of the desired target distribution. The corresponding flow-based generative model is constructed by optimizing the parameters of the component functions of Eq. (1), , by use of the maximum-likelihood method. The training loss is the negative log-likelihood over D, or
| 2 |
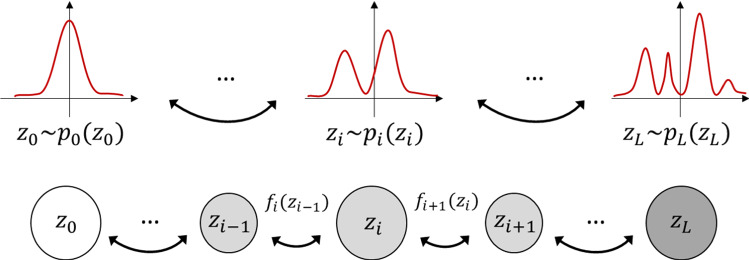
Figure 1 illustrates the construction of a normalizing flow. The path traversed by the random variables, , is a flow, and the full chain that is formed by the corresponding probability distributions is the normalizing flow.
Fig. 1.
Diagram of a normalizing flow between a simple Gaussian distribution and an observed distribution
3D Glow
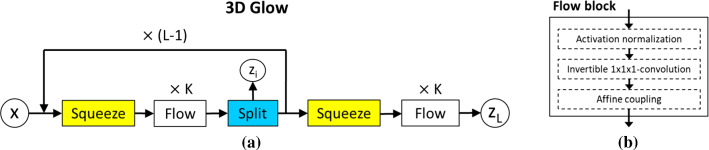
Figure 2a illustrates the overall architecture of our 3D Glow model. The model extends the originally proposed 2D Glow framework [16] into a 3D CNN for processing of volumetric CTC images.
Fig. 2.
a Illustration of the architecture of 3D Glow. The model implements K levels and L steps of flow [16]. b Design of a flow block
The model has three types of layer blocks. Squeeze transforms an input tensor, where is the input image size and c is the number of channels, into a tensor; thus, trading spatial size for number of channels [17]. Flow implements one step of the flow. Split splits the input tensor along the channel dimensions [16].
Figure 2b illustrates the design of a Flow block. There are three layers: an activation-normalization (actnorm) layer [16], an invertible -convolutional layer [16] and an affine-coupling layer [18]. The actnorm layer performs an affine transformation of the input tensor, where the trainable per-channel scale and bias parameters are initialized to yield a mean of zero and a standard deviation of one based on the first mini-batch [16]. The invertible -convolutional layer implements a trainable permutation that reverses the order of the input channels, where the weight matrix of the transform is initialized as a random rotation matrix [16]. The affine-coupling layer splits the input into two parts: the first d dimensions remain the same, whereas the latter dimensions, to D, undergo an affine transformation based on the scale and shift parameters as
| 3 |
| 4 |
where denotes an element-wise product and s(.) and t(.) are scale and translation functions that map .
It can be shown that the transformation above satisfies the basic properties required by efficient computation of the flow transformation [16]. The model is trained with the loss function of Eq. (2).
Construction of synthetic polyps
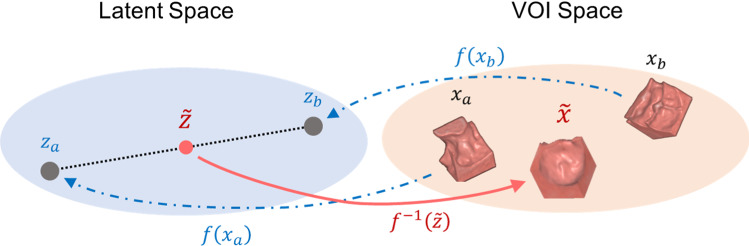
Figure 3 illustrates the principle of the 3D Glow-based data augmentation. The training of the model involves the learning of a bijective mapping from input polyp VOIs (volumes of interest, and ) to vectors of the latent space ( and ) by use of the latent variable z and the normalizing flow of Eq. (1).
Fig. 3.
Illustration of 3D Glow-based data augmentation. Two training VOIs, and , are mapped to points and of the latent space, which are used for interpolating a new point . By mapping back to the VOI space, we generate a new synthetic VOI, , which is different from, yet similar to and
We interpolate the new samples in the latent space by use of linear interpolation between any two training samples. Given two vectors of the latent space, such as and , we calculate a new sample as , where is sampled randomly from the uniform distribution and scaled linearly to to avoid generating VOIs that would be nearly identical to the VOI of real polyps in the training dataset.
Materials
CTC cases
We constructed two independent CTC datasets for the experiments: a development dataset and a test dataset. The development dataset was constructed for providing samples of polyps for the training of 3D Glow for generating synthetic polyps and for providing samples of real polyps and normal anatomy for training of a 3D CNN for polyp detection. The independent test dataset was constructed for testing of the 3D CNN performance. The details of the training and testing of the polyp detection are explained in the section of “Polyp classification study” in “Evaluation methods” section.
We constructed a retrospective development dataset of 203 patients with 269 colonoscopy-confirmed polyps 6 mm in size from cases of various clinical CTC screening trials [19–22]. Most of the patients had been prepared with a cathartic bowel preparation and oral contrast (fecal tagging) by iodine and/or barium. The patients had been scanned in two (supine, prone and/or decubitus) positions by use of a total of 11 CT scanner models (Siemens, Philips, Toshiba, and GE Medical Systems) at 120 or 140 kVp, 0.54–0.97 mm pixel spacing, 0.75–5.00 mm slice thickness and 0.7–2.5 mm reconstruction interval. The spatial locations of the polyps on the CTC images were established by experienced radiologists.
We constructed an independent test dataset of 36 patients with 64 colonoscopy-confirmed polyps from two distinct subgroups of patients. The first subgroup of these patients (20 patients with 45 polyps) was prepared by use of a reduced cathartic bowel preparation with 18 g of magnesium citrate and fecal tagging by 50 ml of non-ionic iodine. The CTC acquisitions (SOMATOM Definition, Siemens Healthcare, Erlangen, Germany) were performed at 140 kVp with 640.6 mm collimation, pitch of 0.85, rotation time of 0.5 s, 1.0 mm slice thickness and a 0.6–0.7 mm reconstruction interval. The second subgroup (16 patients with 19 polyps) was prepared by use of the previously established protocol of polyethylene glycol solution plus contrast medium [23]. The CTC acquisitions (Acquilion, Canon Medical Systems, Tochigi, Japan) were performed at 120 kVp with 16 or 64 0.6 mm collimation, pitch of 0.81–0.94, rotation time of 0.5 s, 0.5 or 1.0 mm slice thickness and 0.8 or 1.0 mm reconstruction intervals.
Extraction of VOIs
For the experiments, we extracted three types of VOIs by use of the development dataset: 100 VOIs of real polyps, 100 VOIs of synthetic polyps and 100 VOIs of normal anatomy. The 100 VOIs of real polyps were those of randomly sampled real polyps. The VOIs of synthetic polyps were generated by 3D Glow after training with the VOIs of the 269 real polyps from the development dataset. The VOIs of normal anatomy were the VOIs of randomly sampled FP detections obtained at 100% polyp detection sensitivity by the 3D detection module of our previously developed CADe system [24–26].
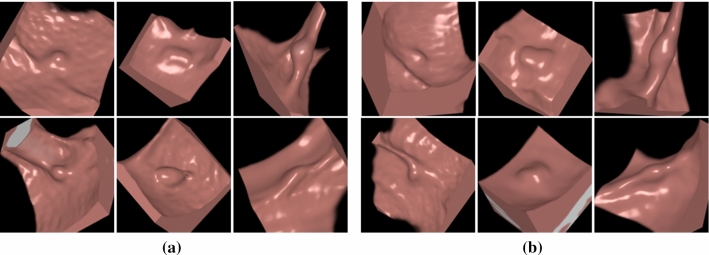
Figure 4 shows examples of the VOIs of the real polyps and 3D Glow-generated synthetic polyps that were included in our experiments. The synthetic polyps look visually indistinguishable from real polyps. Figure 5 shows a visual comparison of the radiomic values of the polyp regions of the real and synthetic polyps in terms of two radiomic features that we had previously identified as being most effective in the differentiation of true polyps from non-polyps in CADe [27]. The comparison indicates that the synthetic and real polyps are radiomically indistinguishable.
Fig. 4.
Virtual endoscopic views of the VOIs of a six real polyps and b six 3D Glow-generated synthetic polyps. Each VOI was voxels in size
Fig. 5.
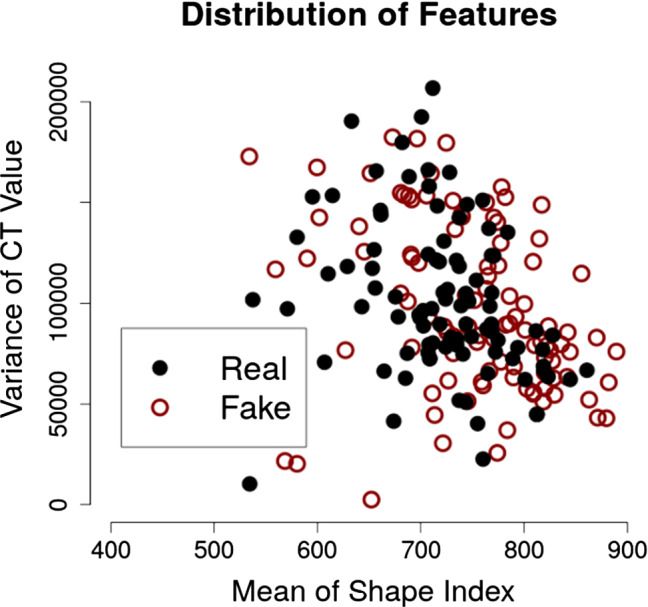
Visual comparison of the volumetric shape and texture features of the 100 real polyps (“Real”) and 100 3D Glow-generated synthetic polyps (“Fake”)
For independent testing, we acquired all of the VOIs of the polyp candidates that were detected from the test dataset (see the subsection of “CTC cases”) by the detection module of our previously developed CADe system. There were 92 true-positive (TP) detections 6 mm in their largest diameter (6–9 mm: 61 TPs) and 6519 FP detections. To calculate meaningful area-under-the-curve (AUC) values for receiver operating characteristic (ROC) analysis, we balanced the number of TP and FP detections in the test dataset by data augmentation of the TP VOIs with 3D rotations. This yielded a total of 12,407 VOIs for independent testing.
Evaluation methods
Observer study
Three experienced observers (two physicians and one physicist, each with experience of reading > 500 CTC studies) attempted to differentiate the VOIs of the 100 real polyps from those of the 100 3D Glow-generated synthetic polyps. The VOIs were loaded to a commercial CTC reading workstation (AZE Virtual Place Raijin, Canon Inc., Tokyo, Japan), where they were presented to the readers in random order. The readers were instructed to evaluate the VOIs interactively by use of the standard 2D and 3D CTC reading modes. For each VOI, the reader recorded his/her confidence that the VOI contained a real polyp, on a scale from 0 (synthetic polyp) to 10 (real polyp). The reading time was unlimited.
The discrimination performance was evaluated by use of ROC analysis, where the confidence levels recorded by each reader were analyzed by use of the pROC package (version 1.16.2) [28] in R (version 3.6.3) [29]. The ROC curves were generated by use of binomial fitting. The 95% confidence intervals were computed by use of bootstrapping with 1000 replicates. The difference of the AUC value from 50% (the level of not being able to tell the difference between a real polyp and a synthetic polyp) was tested by use of a bootstrap test with 1000 replicates, where indicated a statistically significant difference.
Polyp classification study
We trained our previously developed 3D DenseNet CNN [30] to detect polyps with five data augmentation approaches: baseline augmentation, nonlinear augmentation, 3D GAN with baseline augmentation, 3D Glow with baseline augmentation and 3D Glow with nonlinear augmentation. The baseline augmentation included random flipping and/or 1–3-times zooming of the VOIs. The nonlinear augmentation included the baseline augmentation plus 3D shifting, 3D rotation and/or application of Gaussian noise to the CT values of the VOIs. The GAN-based augmentation was based on our previously developed 3D self-attention GAN method for generating synthetic 3D polyp VOIs [15].
For the training of the 3D DenseNet, 200 VOIs of real polyps and normal anatomy were extracted from the development dataset as described in section “Extraction of VOIs”. In the baseline and nonlinear augmentation approaches, the 3D DenseNet was trained with the 200 VOIs augmented by each approach. In the Glow- and GAN-based augmentations, the training dataset also included 100 synthetic polyp VOIs generated by each method.
After training with data augmentation, the 3D DenseNet was tested with the 12,407 VOIs extracted from the independent test dataset. The likelihoods for the presence of a polyp in a VOI as estimated by the 3D DenseNet were analyzed by use of the pROC package (version 1.16.2) [28] in R (version 3.6.3) [29]. The classification performance was evaluated for the clinically significant polyp size categories of 6 mm (all polyps) and 6–9 mm (small polyps) by use of the AUC as the performance metric. Bootstrapping with 1000 replicates was used for calculation of the 95% confidence intervals of the AUC values as well as for testing of the difference of the AUC values, where indicated a statistically significant difference.
Results
The observer study
Figure 6 shows the ROC curves of the performance of the three observers in the differentiation of the 3D Glow-generated synthetic polyps from real polyps. The ROC curves indicate that the observers were unable to differentiate between the polyps. Table 1 shows an analysis of the AUC values of the ROC curves and their statistical significance with respect to difference from 50%. None of the AUC values of the ROC curves were statistically significantly different from 50% ().
Fig. 6.

ROC curves of the three observers for the differentiation between synthetic polyps and real polyps
Table 1.
The AUC values, 95% confidence intervals (CIs) and p values of the three ROC curves in Fig. 6
| AUC (%) (95% CI) | p value | |
|---|---|---|
| Observer 1 | 52.7 [40.7, 62.5] | 0.65 |
| Observer 2 | 51.9 [43.6, 60.3] | 0.81 |
| Observer 3 | 50.3 [42.8, 59.1] | 0.84 |
The polyp classification study
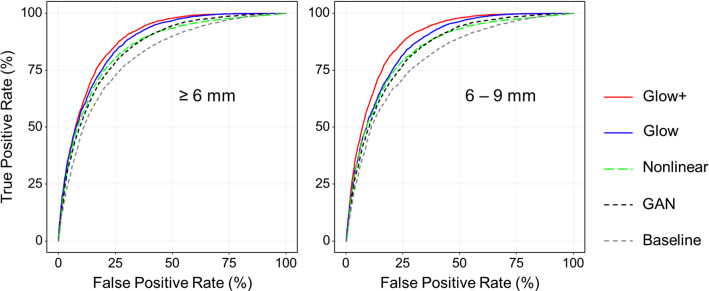
Figure 7 shows the ROC curves representing the classification performance of the 3D DenseNet between polyps and non-polyps based on the training with the different augmentation approaches. The ROC curves on the left show the classification performance for all polyps in our test dataset, whereas the ROC curves on the right show the performance for the clinically significant small polyps (6–9 mm in size), the detection of which is generally challenging. In both cases, the use of the 3D Glow-based augmentation approaches, Glow+ (red curves) and Glow (blue curves) yielded a higher performance than did any of the other augmentation approaches, and Glow+ yielded the highest performance.
Fig. 7.
ROC curves representing the performance of the 3D DenseNet in the classification of polyp candidates with respect to the five different augmentation approaches used for the training step. The left plot shows the ROC curves for all polyps in our test dataset, whereas the right plot shows the ROC curves for clinically significant small polyps. Abbreviations Glow+ = 3D Glow with nonlinear augmentation; Glow = 3D Glow with baseline augmentation; Nonlinear = nonlinear augmentation only; Baseline = baseline augmentation only; GAN = 3D GAN with baseline augmentation
Table 2 lists the AUC values obtained from the ROC curves in Fig. 7, as well as their pairwise comparison results. The upper table shows the results for all polyps, whereas the lower table shows the results for the clinically significant small polyps (6–9 mm in size). In both cases, the 3D Glow-based augmentation approaches (Glow+ and Glow) yielded statistically significantly higher AUC values than did the other augmentation approaches, and Glow+ yielded the highest AUC values.
Table 2.
AUC values obtained from the ROC curves in Fig. 7, and their pairwise comparison results
| Glow+ | Glow | Nonlinear | GAN | Baseline | |
|---|---|---|---|---|---|
| 6 mm | |||||
| Glow+ | 88.1 [87.5, 88.7] | 0.00013 | < 0.0001 | < 0.0001 | < 0.0001 |
| Glow | 0.9 | 87.2 [86.5, 87.8] | < 0.0001 | < 0.0001 | < 0.0001 |
| Nonlinear | 3.4 | 2.5 | 84.7 [84.1, 85.4] | 0.49 | < 0.0001 |
| GAN | 3.5 | 2.6 | 0.1 | 84.6 [83.9, 85.3] | < 0.0001 |
| Baseline | 7.0 | 6.1 | 3.6 | 3.5 | 81.1 [80.3, 81.8] |
| 6–9 mm | |||||
| Glow+ | 88.7 [88.1, 89.2] | < 0.0001 | < 0.0001 | < 0.0001 | < 0.0001 |
| Glow | 2.6 | 86.1 [85.6, 86.7] | < 0.0001 | < 0.0001 | < 0.0001 |
| Nonlinear | 4.6 | 2.0 | 84.1 [83.4, 84.8] | 0.51 | < 0.0001 |
| GAN | 4.8 | 2.2 | 0.2 | 83.9 [83.3, 84.6] | < 0.0001 |
| Baseline | 8.3 | 5.7 | 3.7 | 3.5 | 80.4 [79.6, 81.4] |
The diagonal components show the AUC values in percentage with the 95% confidence interval in square brackets. The upper triangle shows the p values obtained from a bootstrap test (1000 replicates) for the pairwise comparison of the AUC values. The lower triangle shows the pairwise differences in the AUC values
Glow+ = 3D Glow with nonlinear augmentation; glow = 3D Glow with baseline augmentation; nonlinear = nonlinear augmentation only; baseline = baseline augmentation only; GAN = 3D GAN with baseline augmentation
Discussion
Although deep learning has made it relatively easy to obtain state-of-the-art results of various medical imaging tasks, in CTC, the available training datasets are relatively small, and the numbers of clinically significant polyps are even smaller. In this study, we investigated the feasibility of developing a flow-based generative model for simulating colorectal polyps to improve the training of 3D CNNs in CADe for CTC. The results of our experiments indicate that this is indeed feasible.
In the first experiment, we performed an observer study to compare the visually observed characteristics of 3D Glow-generated synthetic polyps with those of real polyps. To the best of our knowledge, this is the first study to show that it is possible to generate VOIs of synthetic polyps that are visually equivalent to VOIs of real polyps in CTC.
In the second experiment, we investigated the effect of the training of deep learning (3D DenseNet) with synthetic polyps generated by 3D Glow on the differentiation of polyps from non-polyps in CTC. To the best of our knowledge, this is the first study to show that data augmentation by synthetic polyps can be used to yield a statistically significant performance improvement in deep learning for CTC. We demonstrated that the use of synthetic polyps generated by 3D Glow outperformed the use of traditional nonlinear and GAN-based augmentations, and that the classification performance was highest when 3D Glow was used in combination with a nonlinear augmentation.
The observed improvement in the polyp classification performance by use of the 3D Glow was high for small polyps (6–9 mm in size). This result is important, because it is the detection of small polyps that remains challenging in CTC, whereas many clinical studies have demonstrated that existing CADe systems can detect large polyps in CTC at a high accuracy [31, 32]. Therefore, 3D Glow could have a meaningful impact in improving the detection of small polyps.
We performed the polyp detection by use of volumetric analysis of 3D CT colonography data, because this enables deep learning to review the complete region of the colon regardless of obstructions such as collapsed or poorly distended colon segments. Although other methods such as virtual endoscopic views have been used previously for polyp detection by deep learning [6], such approaches are limited to well-distended regions of the colon, whereas obstructed regions that can hide clinically significant polyps or masses would need to be identified and reviewed by other means. Also, the visual appearance of a virtual endoscopic view depends on the rendering algorithm and its visualization parameters, whereas 3D CT colonography data are readily standardized by Hounsfield units. Thus, the volumetric analysis provides the most uniform approach for the application of deep learning to polyp detection in CT colonography.
This study had several limitations. First, the number of datasets was necessarily relatively small. Second, there are various other data augmentation methods besides the nonlinear image transforms and the GAN-based method that we used as reference augmentation methods in this study. However, a thorough systematic assessment of their comparative performance against 3D Glow will require a separate study. Third, the study could be expanded by consideration of the unique demands of CTC, such as various bowel preparations or polyps that tend to be underrepresented in CTC datasets. Addressing such issues provides topics for future studies.
Conclusion
We developed a 3D CNN based on a flow-based generative model for generating synthetic colorectal polyps in CTC. Our results indicate that the generated synthetic polyps are visually indistinguishable from real polyps and that data augmentation by such polyps can significantly improve the performance of deep learning in CADe for CTC.
Acknowledgements
We thank the Enterprise Research Infrastructure and Services at Mass General Brigham (Boston, MA) for their provision of high-performance computing services.
Funding
This study was supported partly by the National Institutes of Health (NIH) under Award Nos. R01CA212382 and R01EB023942. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. J.J.N. and H.Y. are co-inventors of electronic cleansing and computer-aided detection software patents.
Compliance with ethical standards
Conflict of interest
The other authors declare that they have no conflict of interest.
Ethical standards
This retrospective single-center study was approved by the Mass General Brigham (MGB) institutional review board (IRB).
Human and animal rights statement
All procedures involving human participants were performed in accordance with the ethical standards of the IRB and with the 1964 Declaration of Helsinki and its later amendments.
Informed consent
Informed consent was waived for this study by the MGB IRB.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Tomoki Uemura and Janne J. Näppi contributed equally to this work.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel R, Torre L, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Winawer S, Fletcher R, Miller L, Godlee F, Stolar M, Mulrow C, Woolf S, Glick S, Ganiats T, Bond J, Rosen L, Zapka J, Olsen S, Giardiello F, Sisk J, Van Antwerp R, Brown-Davis C, Marciniak D, Mayer R. Colorectal cancer screening: clinical guidelines and rationale. Gastroenterology. 1997;112:594–642. doi: 10.1053/gast.1997.v112.agast970594. [DOI] [PubMed] [Google Scholar]
- 3.U.S. Preventive Services Task Force: Screening for colorectal cancer: U.S. Preventive Services Task Force Recommendation Statement. JAMA 315:2564–2575 (2016) [DOI] [PubMed]
- 4.Litjens G, Kooi T, Bejnordi B, Setio A, Ciompi F, Ghafoorian M, van der Laark J, van Ginneken B, Sánchez C. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 5.Shorten C, Khoshgoftaar T (2019) A survey on image data augmentation for deep learning. J Big Data 6:60 [DOI] [PMC free article] [PubMed]
- 6.Näppi J, Hironaka T, Regge D, Yoshida H (2016) Deep transfer learning of virtual endoluminal views for the detection of polyps in CT colonography. In: Proceedings of SPIE medical imaging: computer-aided diagnosis. SPIE Press, vol 97852B, pp V1–8
- 7.Näppi J, Pickhardt P, Kim D, Hironaka T, Yoshida H (2017) Computer-aided detection of serrated polyps with a deep convolutional neural network in CT colonography. Int J CARS 12S1: 144–146
- 8.Tachibana R, Näppi J, Ota J, Kohlhase N, Hironaka T, Kim S, Regge D, Yoshida H. Deep learning electronic cleansing for single- and dual-energy CT colonography. Radiographics. 2018;38:2034–2050. doi: 10.1148/rg.2018170173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Y, Ren Y, Fu L, Xiong J, Larsson R, Xu X, Sun J, Zhao J (2018) A 3D convolutional neural network framework for polyp candidates detection on the limited dataset of CT colonography. In: Conf Proc IEEE Eng Med Biol Soc. Honolulu, HI, USA, pp 678–681 [DOI] [PubMed]
- 10.Näppi J, Hironaka T, Yoshida H (2017) Detection of colorectal masses in CT colonography: application of deep residual networks for differentiating masses from normal colon anatomy. In: SPIE medical imaging: computer-aided diagnosis. Houston, Texas, United States, vol 10575, p 1057518
- 11.Roth H, Lu L, Liu J, Yao J, Seff A, Cherry K, Kim L, Summers R. Improving computer-aided detection using convolutional neural networks and random view aggregation. IEEE Trans Med Imaging. 2016;35:1170–1181. doi: 10.1109/TMI.2015.2482920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uemura T, Lu H, Kim H, Tachibana R, Hironaka T, Näppi J, Yoshida H (2018) 3D deep residual convolutional networks for computer-aided detection of polyps in CT colonography. Int J CARS 13S1:94–95
- 13.Yi X, Walia E, Babyn P. Generative adversarial network in medical imaging: a review. Med Image Anal. 2019;58:101552. doi: 10.1016/j.media.2019.101552. [DOI] [PubMed] [Google Scholar]
- 14.Shin HC, Tenenholtz NA, Rogers JK, Schwarz CG, Senjem ML, Gunter JL, Andriole KP, Michalski M. Medical image synthesis for data augmentation and anonymization using generative adversarial networks. In: Gooya A, Goksel O, Oguz I, Burgos N, editors. Simulation and synthesis in medical imaging. Cham: Springer; 2018. pp. 1–11. [Google Scholar]
- 15.Uemura T, Näppi J, Lu H, Tachibana R, Hironaka T, Yoshida H (2019) Ensemble 3D residual network (E3D-ResNet) for reduction of false-positive polyp detections in CT colonography. In: Proc SPIE medical imaging 2019: computer-aided diagnosis, p 1095038
- 16.Kingma D, Prafulla D (2018) Glow: generative flow with invertible convolutions. In: Bengio S, Wallach H, Larochelle H, Grauman K, Cesa-Blanchl N, Garnett R (eds) 32nd conference on neural information processing systems (NIPS), Montreal, Canada, pp 10235–10244
- 17.Dinh L, Sohl-Dickstein J, Bengio Y (2017) Density estimation using real NVP. In: 5th International conference on learning representations (ICLR)
- 18.Dinh L, Krueger D, Bengio Y (2014) NICE: Non-linear independent components estimation. Technical report
- 19.Pickhardt P, Choi J, Hwang I, Butler J, Puckett M, Hildebrandt H, Wong R, Nugent P, Mysliwiec P, Schindler W. Computed tomographic virtual colonoscopy to screen for colorectal neoplasia in asymptomatic adults. N Engl J Med. 2003;349:2191–2200. doi: 10.1056/NEJMoa031618. [DOI] [PubMed] [Google Scholar]
- 20.Rockey DC, Paulson E, Niedzwiecki D, Davis W, Bosworth HB, Sanders L, Yee J, Henderson J, Hatten P, Burdick S, Sanyal A, Rubin DT, Sterling M, Akerkar G, Bhutani MS, Binmoeller K, Garvie J, Bini EJ, McQuaid K, Foster WL, Thompson WM, Dachman A, Halvorsen R. Analysis of air contrast barium enema, computed tomographic colonography, and colonoscopy: prospective comparison. Lancet. 2005;365(9456):305–311. doi: 10.1016/S0140-6736(05)17784-8. [DOI] [PubMed] [Google Scholar]
- 21.Johnson CD, Chen MH, Toledano AY, Heiken JP, Dachman A, Kuo MD, Menias CO, Siewert B, Cheema JI, Obregon RG, Fidler JL, Zimmerman P, Horton KM, Coakley K, Iyer RB, Hara AK, Halvorsen RA, Casola G, Yee J, Herman BA, Burgart LJ, Limburg PJ. Accuracy of CT colonography for detection of large adenomas and cancers. N Engl J Med. 2008;359(12):1207–1217. doi: 10.1056/NEJMoa0800996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Regge D, Laudi C, Galatola G, Della Monica P, Bonelli L, Angelelli G, Asnaghi R, Barbaro B, Bartolozzi C, Bielen D, Boni L, Borghi C, Bruzzi P, Cassinis MC, Galia M, Gallo TM, Grasso A, Hassan C, Laghi A, Martina MC, Neri E, Senore C, Simonetti G, Venturini S, Gandini G. Diagnostic accuracy of computed tomographic colonography for the detection of advanced neoplasia in individuals at increased risk of colorectal cancer. JAMA. 2009;301(23):2453–2461. doi: 10.1001/jama.2009.832. [DOI] [PubMed] [Google Scholar]
- 23.Nagata K, Okawa T, Honma A, Endo S, Kudo SE, Yoshida H. Full-laxative versus minimum-laxative fecal-tagging CT colonography using 64-detector row CT: prospective blinded comparison of diagnostic performance, tagging quality, and patient acceptance. Acad Radiol. 2009;16(7):780–789. doi: 10.1016/j.acra.2008.12.027. [DOI] [PubMed] [Google Scholar]
- 24.Näppi J, Yoshida H. Fully automated three-dimensional detection of polyps in fecal-tagging CT colonography. Acad Radiol. 2007;25:287–300. doi: 10.1016/j.acra.2006.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshida H, Näppi J. Three-dimensional computer-aided diagnosis scheme for detection of colonic polyps. IEEE Trans Med Imaging. 2001;20:1261–1274. doi: 10.1109/42.974921. [DOI] [PubMed] [Google Scholar]
- 26.Näppi J, Yoshida H. Adaptive correction of the pseudo-enhancement of CT attenuation for fecal-tagging CT colonography. Med Image Anal. 2008;12:413–426. doi: 10.1016/j.media.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Näppi J, Yoshida H. Automated detection of polyps in CT colonography: evaluation of volumetric features for reduction of false positives. Acad Radiol. 2002;9:386–397. doi: 10.1016/S1076-6332(03)80184-8. [DOI] [PubMed] [Google Scholar]
- 28.Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, Mller M. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.R Core Team: R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria (2020) https://www.R-project.org/
- 30.Uemura T, Näppi J, Hironaka T, Hyougseop K, Yoshida H (2020) Comparative performance of 3D-DenseNet, 3D-ResNet, and 3D-VGG models in polyp detection for CT colonography. In: Proceedings of SPIE medical imaging 2020: computer-aided diagnosis, p 1131435. 10.1117/12.2549103
- 31.Regge D, Halligan S. CAD: How it works, how to use it, performance. Eur J Radiol. 2013;82:1171–1176. doi: 10.1016/j.ejrad.2012.04.022. [DOI] [PubMed] [Google Scholar]
- 32.Näppi J, Uemura T, Kim S, Kim H, Yoshida H (2020) Comparative performance of 3D machine-learning and deep-learning models in the detection of small polyps in dual-energy CT colonography. In: Proceedings of SPIE medical imaging 2020: computer-aided diagnosis, p 113143C. 10.1117/12.2549793