Abstract
T2-weighted (T2W) MRI provides high spatial resolution and tissue-specific contrast, but it is predominantly used for qualitative evaluation of prostate anatomy and anomalies. This retrospective multicenter study evaluated the potential of T2W image-derived textural features for quantitative assessment of peripheral zone prostate cancer (PCa) aggressiveness. A standardized preoperative multiparametric MRI was performed on 87 PCa patients across 6 institutions. T2W intensity and apparent diffusion coefficient (ADC) histogram, and T2W textural features were computed from tumor volumes annotated based on whole-mount histology. Spearman correlations were used to evaluate association between textural features and PCa grade groups (i.e. 1–5). Feature utility in differentiating and classifying low-(grade group 1) vs. intermediate/high-(grade group ≥ 2) aggressive cancers was evaluated using Mann–Whitney U-tests, and a support vector machine classifier employing “hold-one-institution-out” cross-validation scheme, respectively. Textural features indicating image homogeneity and disorder/complexity correlated significantly (p < 0.05) with PCa grade groups. In the intermediate/high-aggressive cancers, textural homogeneity and disorder/complexity were significantly lower and higher, respectively, compared to the low-aggressive cancers. The mean classification accuracy across the centers was highest for the combined ADC and T2W intensity-textural features (84%) compared to ADC histogram (75%), T2W histogram (72%), T2W textural (72%) features alone or T2W histogram and texture (77%), T2W and ADC histogram (79%) combined. Texture analysis of T2W images provides quantitative information or features that are associated with peripheral zone PCa aggressiveness and can augment their classification.
Subject terms: Cancer, Mathematics and computing
Introduction
Accurate assessment of localized prostate cancer aggressiveness is of utmost importance for determining patient treatment and follow-up strategies. Currently, this is determined based on Gleason and/or International Society of Urological Pathology (ISUP) grading1,2 of histological specimens, traditionally obtained by systematic transrectal ultrasound-guided biopsy sampling. Although the introduction of targeted approaches has improved biopsy sampling and cancer detection accuracies3, it is still limited to small portions of the prostate. With prostate cancer being a heterogeneous and multifocal disease, this can sometimes lead to inaccurate estimation of the disease extent, and thus undertreatment or overtreatment4,5. Moreover, biopsy sampling is invasive, and the risk of post-biopsy complications has become an increasing concern due to multidrug-resistance6. This makes repeated biopsy sampling unattractive in clinical practice, especially for active surveillance patients.
Multiparametric MRI (mpMRI) enables non-invasive acquisition of both anatomical [i.e. T2-weighted (T2W)] and functional [mainly diffusion-weighted (DW), and dynamic contrast-enhanced (DCE)] images of the entire prostate for cancer detection, staging, treatment planning, and response evaluation7. The introduction of mpMRI and the Prostate Imaging—Reporting and Data System (PI-RADS) guidelines8 have improved prostate cancer detection and accuracy9. DW and DCE MRI provide quantitative pathophysiological parameters such as apparent diffusion coefficient (ADC), and volume transfer constant (ktrans) and extravascular-extracellular volume fraction (Ve), which to some extent are capable of assessing prostate cancer aggressiveness10,11. Compared to DW and DCE, T2W MRI provides high spatial resolution and tissue-specific contrast, but currently, it is mainly used for qualitative evaluation of the prostate anatomy and anomalies.
Though important, qualitative assessment has several challenges and limitations, including dependency on subjective judgment of radiologists, which is prone to high inter-reader variability12 and the occurrence of equivocal findings in substantial number of cases13. Furthermore, with a multiparametric approach and the increasing availability of hybrid imaging modalities such as positron emission tomography/MRI14, the amount of data to be analyzed increases, making it also increasingly labor-intensive to manually collate all these images into meaningful information for clinical decision making.
Recently, radiomics, i.e. automatic high-throughput extraction of quantitative image features from radiological images and their subsequent analysis15,16, has gained attention with potential to overcome the above limitations and thus improve clinical decision making. Texture analysis constitutes a key methodology for extracting quantitative image features, particularly second- and high-order statistical image texture descriptors based on grey level co-occurrence matrix (GLCM)17 and grey level run length matrix (GLRLM)18, which examine spatial variations in pixel intensity distribution. Several interesting studies have reported the use of texture analysis in radiomics-based analysis of prostate cancer19–23, but have mostly been limited to single-center data. Previously, we24 showed that GLCM-based textural features derived from T2W images could potentially serve as non-invasive markers for assessing prostate cancer aggressiveness. Particularly, we found homogeneity and entropy features to correlate significantly with prostate cancer aggressiveness (i.e. grade groups 2 and 3) as defined on pathology, as well as with ADC and ktrans. Also, the augmentation of quantitative MRI parameters with T2W image textural features enabled better classification. However, these preliminary findings were based on a relatively small number of patients recruited from single center. The aim of this current work was to validate and extend these findings using a multicenter cohort, and to investigate their performance in the classification of biopsy-proven prostate cancers.
Materials and methods
Patient population and data collection
The patient cohort data for this retrospective multicenter study constitutes part of a prospectively collected (between June 2010 and August 2015) data for the Prostate Cancer localization with a Multiparametric MR Approach trial (PCa-MAP; ClinicalTrials.gov Identifier NCT01138527)25. Eligible patients (N = 128) from six institutions: Johns Hopkins University, Baltimore (n = 20); Norwegian University of Science and Technology, Trondheim (n = 22); Radboud University Medical Centre, Nijmegen (n = 30); University of California, Los Angeles (n = 20); University Health Network, Toronto (n = 10); and Medical University of Vienna (n = 26) were included in this study. All patients were diagnosed with primary prostate cancer and were scheduled to undergo preoperative mpMRI with subsequent radical prostatectomy. The Regional Committee for Medical and Health Ethics (Mid Norway), the PCa-MAP trial consortium review board, as well the review board of each participating institution (HIPAA-compliant for USA institutions) approved this study and waived the requirement for written informed consent. All methods were performed in accordance with local institutional, national and international guidelines and regulations.
MRI examination
The image acquisition protocol and/or settings were standardized across all centers, hence the term ‘single-arm’ in the title. All imaging was performed on 3T MRI systems (Siemens Healthineers) using standard vendor-supplied body and spine phased array coils for signal detection, without an endorectal coil. A minimum of four weeks was allowed between the last biopsy and MRI to avoid hemorrhage artifacts. The acquisition consisted of localizer scans, T2W, DW, DCE, and spectroscopic imaging. In this study, we utilized only the transverse T2W and DW images, which were acquired with a turbo spin-echo sequence (repetition/echo time (TR/TE): 4000/101 ms; field of view (FOV): 200 × 200 mm; matrix: 320 × 320; slice thickness: 3 mm; interslice gap: 0.6 mm), and a single-shot echo-planar sequence with four b-values: 0, 100, 400, and 800 s/mm2 (TR/TE: 3300/60 ms; FOV: 260 × 211 mm; matrix: 160 × 130; slice thickness: 3.6 mm), respectively. The images were oriented along the longest axis of the prostate, perpendicular to the urethra to best match routine histologic sectioning of the prostate. Pre-imaging preparations were performed in accordance with local institutional guidelines.
Histopathologic examination and tumor delineation
Patients underwent radical prostatectomy within 12 weeks after the MRI examination. The prostatectomy specimens were prepared locally according to histopathology protocols at each institution, which included fixation, serial sectioning (perpendicular to the urethra to facilitate spatial matching to MRI) into ~ 3–4 mm axial slices, and hematoxylin and eosin staining of microsections. An experienced local uro-pathologist examined the stained slides, outlined cancer foci, described cancer location, and graded them in accordance with the Gleason scoring system1,2.
The annotated whole-mount histology sections were visually matched to the T2W images based on anatomical landmarks such as urethra, ejaculatory ducts, size/shape of the peripheral zone and apex/base proximity. Moreover, descriptions from the pathology report were used as guidance. Tumor volumes of interest (VOIs) were then manually delineated (by Gabriel A. Nketiah, 5 years’ experience in prostate MRI; and guided by a radiologist Jurgen J. Fütterer with > 11 years’ experience) based on their location in histology and shape/appearance on the T2W images. The VOIs were subsequently transformed to the corresponding DW images via intensity-based rigid registration (Elastix toolbox26) using Mattes mutual information similarity metric (Fig. 1a). This was done by first co-registering the T2W images to the b = 0 s/mm2 images, and then applying the resulting transformation to the VOI masks. The co-registrations were visually verified, and manually corrected in case of mis-registration, for instance due to geometric distortion on the DW images. Each tumor was assigned a grade group (GG) according to the ISUP prostate cancer grading system2, and then dichotomized into low-(GG 1) and intermediate/high-(GG ≥ 2) aggressive cancers.
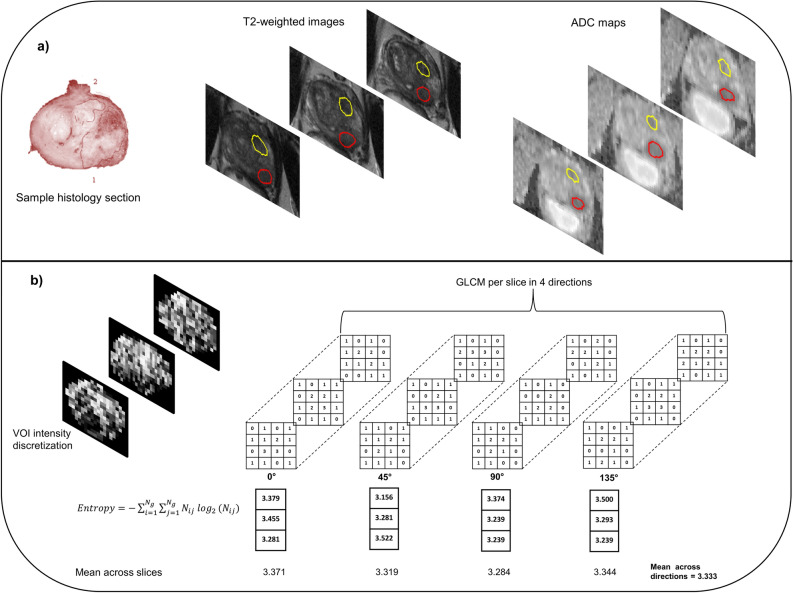
Figure 1.
Illustration of tumor volume-of-interest (VOI) delineation and grey level co-occurrence matric (GLCM) texture feature extraction. (a) Histopathology slides were visually matched to the T2-weighted (T2W) images using anatomical landmarks. Peripheral (red) and transition (yellow) zone tumors were manually outlined on the T2W images based on their location on histology and appearance on T2W images, and then transformed to corresponding apparent diffusion coefficient (ADC) maps via registration or manually if the registration was deemed suboptimal. (b) Textural feature extraction from T2W VOIs using 2D average approach. For illustration purposes, the number of grey level bins was limited to four in this schematic, instead of the 32 used in actual computations.
Post-processing and feature extraction
Two types of features were computed from the image VOIs: traditional intensity histogram features (number of features, nf = 11) from the T2W images and ADC maps, and second and high-order statistical image textural features (nf = 29) based on GLCM17 and GLRLM18 from the T2W images. The “2D average” approach27 was employed to compute textural features from the tumor VOIs (Fig. 1b). First, the intensities within each VOI were discretized into 32 grey levels via fixed bin number quantization. The GLCMs and GLRLMs were computed per slice at one-pixel distance (∂ = 1) in four symmetric directions, = 0°, 45°, 90° and 135°. Textural features were computed from each directional matrix, and the mean of each feature across the slices was obtained. Finally, the average of each feature across the four directions was calculated to eliminate potential differences in directionality. This 2D approach was preferred over 3D texture analysis due to the presence of interslice gaps in our data acquisition. ADC maps were calculated from the nonzero b-value DW image datasets (100, 400, and 800 s/mm2) by fitting a monoexponential decay model to the image intensities as a function of b-value in each voxel. ADC histogram features were then computed for each tumor VOI. The b = 0 s/mm2 image was excluded from ADC map computation to eliminate possible perfusion effects. The ADC was included as a benchmark metric for aggressiveness classification, since it has been previously shown to correlate with prostate cancer aggressiveness10.
Prior to feature extraction, the T2W images were corrected for intensity non-uniformity using the N4 bias field correction algorithm28, and subsequently normalized using the automated dual-reference tissue normalization approach29. Briefly, two aggregate feature channel object detectors were separately trained to detect fat and muscle tissue regions, from which reference intensity values (90th and 10th percentiles, respectively) were calculated, and then utilized to normalize the 3D image intensities to pseudo T2 values by linearly scaling the reference values to their corresponding T2 values at 3T from literature. Unlike T2W image intensities, the ADCs were not normalized because they are quantitative measurements in nature. Also, outlier voxels within each VOI, defined as intensities outside the range [µ − 3σ, µ + 3σ]30 were excluded; where μ and σ denote the mean and standard deviation of the intensities within each VOI. All features were computed in accordance with the image biomarker standardization initiative27.
Statistical analysis and classification modeling
Spearman correlation coefficients were calculated to investigate associations between the T2W image features and prostate cancer grade groups (i.e. 1–5). Differences in feature values between the two aggressiveness classes (i.e. low versus intermediate/high) were evaluated using two-tailed Mann–Whitney U-tests. p values were corrected for multiple testing using Benjamini and Hochberg’s approach31 at false discovery rate of 0.05, with values < 0.05 considered statistically significant.
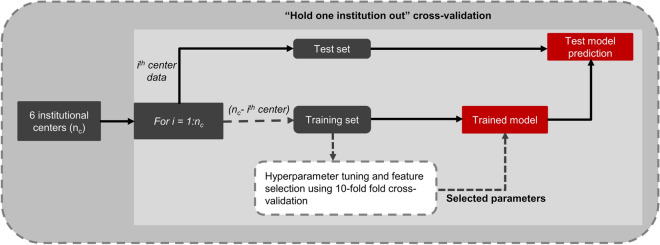
To evaluate the utility of the features in classifying the two cancer aggressiveness classes, a linear support vector machine (SVM) classifier was trained and tested separately for each feature set (i.e. ADC histogram, T2W histogram and T2W textural features), and the following combinations: T2W histogram + textural features, ADC histogram + T2W histogram, and ADC histogram + T2W histogram + textural features. In this analysis, we were particularly interested in how well cancer aggressiveness at one institution could be predicted by a model trained on data from the other institutions, using histogram features with and without textural features augmentation. For this, the classifier was iteratively (i = 1:number of institutions, ni) trained and tested, each time using data from ni − i institutions as training set, with the ith institution held out as an independent external test set (Fig. 2). The training employed stratified 10-fold cross-validation for hyperparameter tuning and feature selection. Hyperparameter (misclassification cost, C) tuning and feature selection during training were performed concurrently via grid search over seven logarithmically spaced values between − 1 and 1 inclusive, and using recursive feature elimination32, respectively. The hyperparameter and feature sets with the lowest mean misclassification error over all 10-folds were selected to build the model. The cross-validation partitioning of the data during training was done on patient level rather than on tumor level to ensure that multiple tumors from the same patient were all either in the training or in the validation subset. Predictions on the test set (i.e. data from the hold out institution) were however done on tumor-level.
Figure 2.
Schematic of the cross-validation scheme employed in support vector machine classifier training and testing across six institutional centers. At each iteration, data from one institution was held out for testing, and data from the remaining 5 institutions used for training.
Receiver operating characteristic (ROC) curves were computed for each test set, from which the area under the curve (AUC) and 95% confidence intervals (95CI), accuracy, sensitivity and specificity were calculated to evaluate the performances of the classifiers (i.e. feature sets) across the centers. The optimal threshold for calculating the accuracy, sensitivity and specificity was determined from the training based on the Youden index33, and then applied to the test set.
The added value of T2W textural features was evaluated using two approaches. First, at each institution, the AUCs before (i.e. ADC histogram, T2W histogram, and ADC + T2W histogram) and after augmentation with T2W textural features (i.e. T2W histogram + textural features, and ADC histogram + T2W histogram + textural features) were compared using Delong’s nonparametric approach for comparing the areas under two or more correlated receiver operating characteristic curves34. Secondly, the differences in performances across the institutions before and after augmentation with T2W textural features were compared using paired student t-test.
Prior to the classification modelling, two-way ANOVA was performed to evaluate potential effects of data origin (i.e. institution) and cancer aggressiveness on the features. Features for which the interaction between institution and cancer aggressiveness or main effect of institution were significant were excluded from the model. Each feature was log transformed to meet normality assumption requirement of ANOVA. The SVM classification modelling was performed with scikit-learn library35 in python (version 3.7, www.python.org), and other statistical analyses were performed in MATLAB R2019a (Mathworks).
Results
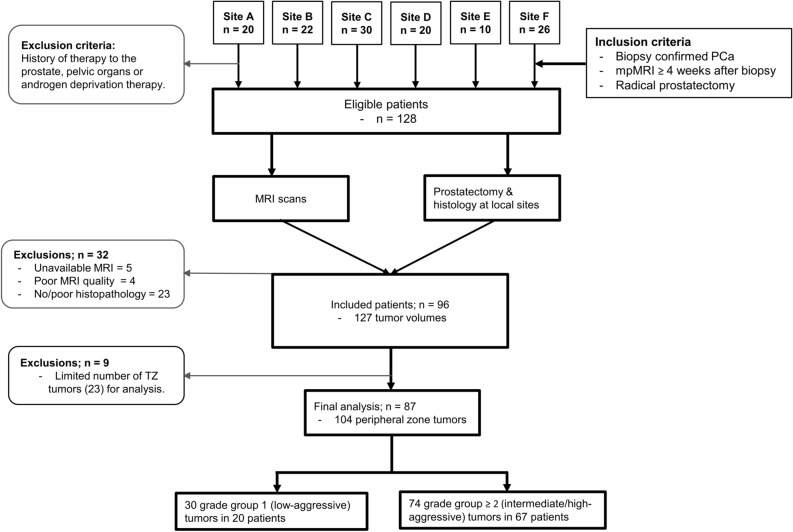
Out of the 128 eligible patients, 32 patients were excluded due to unavailable MRI (n = 5), MRI artifacts and/or distortion (n = 4), no pathology report/grading (n = 15), unsatisfactory matching between histopathology and MRI (n = 8). Data of 96 patients (mean age = 61.3 ± 6.1 years) for whom good quality MRI and post-surgical histopathology data were available were included in this study. In these patients, 127 tumor volumes (mean [range] = 469 [101–1397] voxels) were identified, of which 104 were in the peripheral zone and 23 were in the transition zone. Figure 3 shows the flowchart of patient inclusion and exclusion. Due to the limited number of transition zone tumors, only the peripheral zone tumors (in 87 patients) were analyzed, of which 30 were stratified as low-aggressive, and 74 as intermediate/high-aggressive cancers. The overview of the characteristics of patients and tumors is given in Table 1.
Figure 3.
Flow chart of patients and datasets included in study. mpMRI multiparametric MRI, PCa prostate cancer, TZ transition zone.
Table 1.
Characteristics of patients and tumors.
| Institutions | |||||||
|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | Overall | |
| Number of patients | 17 | 15 | 21 | 16 | 8 | 10 | 87 |
| Age, years [range] | 56 [46–70] | 61 [52–73] | 62 [51–69] | 60 [55–69] | 64 [50–71] | 64 [52–76] | 62 [46–76] |
| PSA ng/mL [range] | 5.5 [1.7–18] | 7.3 [1.9–11.2] | 5.4 [2.5–20.8] | 6.1 [3.6–12.5] | 5.9 [3.1–17.3] | 6.4 [5.2–31] | 6.1 [1.7–31] |
| Tumor grade group (aggressiveness class) | |||||||
| 1 (L) | 5 | 1 | 10 | 3 | 6 | 5 | 30 |
| 2 (I) | 8 | 9 | 11 | 7 | 1 | – | 36 |
| 3 (H) | 4 | 7 | 1 | 9 | 1 | 4 | 26 |
| 4 (H) | 1 | – | 4 | - | 1 | – | 6 |
| 5 (H) | 1 | – | 1 | 1 | 1 | 2 | 6 |
| Total | 19 | 17 | 27 | 20 | 10 | 11 | 104 |
PSA prostate specific antigen, L low, I intermediate, H high.
Feature association with prostate cancer grade group
The Spearman correlation between the image features and cancer grade groups was significant (p < 0.05) for eight T2W intensity histogram and nine textural features (Table 2). Differences in features between the two cancer aggressiveness classes were significant for seven intensity histogram and 16 textural features. Generally, the textural features reflected higher disorder/complexity (i.e. high entropy or lower homogeneity) in intermediate/high-aggressive tumors, and vice versa in low-aggressive tumors (Fig. 4). As expected, the majority of these features (7 intensity and 9 texture features) were found to be common between the two statistical tests.
Table 2.
List of computed features from T2-weighted images and ADC maps, and their association with prostate cancer aggressiveness.
| Feature type | List of computed features | Correlation (ρ) with cancer grade group | Differentiation of low versus I/H aggressive cancers: p values | ||
|---|---|---|---|---|---|
| ADC | T2W | ADC | T2W | ||
| Intensity histogram | Kurtosis | − 0.15 | 0.14 | 0.133 | 0.283 |
| Maximum | − 0.37* | − 0.23* | 0.015 | 0.283 | |
| Mean | − 0.45* | − 0.34* | 0.001 | 0.016 | |
| Median | − 0.44* | − 0.34* | 0.002 | 0.015 | |
| Minimum | − 0.50* | − 0.46* | < 0.0001 | < 0.0001 | |
| 10th percentile | − 0.51* | − 0.39* | < 0.0001 | 0.002 | |
| 25th percentile | − 0.47* | − 0.35* | 0.001 | 0.006 | |
| 75th percentile | − 0.41* | − 0.33* | 0.005 | 0.027 | |
| 90th percentile | − 0.40* | − 0.31* | 0.006 | 0.048 | |
| Skewness | − 0.06 | 0.08 | 0.597 | 0.135 | |
| Standard deviation | 0.14 | − 0.04 | 0.152 | 0.422 | |
| Textural (GLCM & GLRLM) features | ASM | − 0.34* | 0.043 | ||
| Autocorrelation | − 0.02 | 0.597 | |||
| Cluster Prominence | − 0.19 | 0.048 | |||
| Cluster shade | 0.02 | 0.422 | |||
| Contrast | − 0.18 | 0.019 | |||
| Correlation | 0.03 | 0.597 | |||
| Difference entropy | − 0.09 | 0.084 | |||
| Difference variance | − 0.19 | 0.019 | |||
| Dissimilarity | − 0.17 | 0.027 | |||
| Entropy | 0.38* | 0.028 | |||
| IMC 1 | 0.51* | < 0.0001 | |||
| IMC 2 | − 0.48* | < 0.0001 | |||
| IDM | 0.13 | 0.048 | |||
| Maximum probability | − 0.32* | 0.048 | |||
| Sum average | − 0.02 | 0.495 | |||
| Sum entropy | 0.11 | 0.940 | |||
| Sum variance | − 0.23* | 0.027 | |||
| Variance | − 0.25* | 0.012 | |||
| Grey level non-uniformity | 0.52* | < 0.0001 | |||
| HGLRE | − 0.06 | 0.364 | |||
| LGLRE | − 0.18 | 0.345 | |||
| Long run emphasis | 0.17 | 0.046 | |||
| Long run high GLE | − 0.02 | 0.651 | |||
| Long run low GLE | − 0.14 | 0.557 | |||
| RLNU | 0.50* | 0.001 | |||
| Run percentage | − 0.17 | 0.046 | |||
| Short run emphasis | − 0.15 | 0.051 | |||
| Short run high GLE | − 0.07 | 0.293 | |||
| Short run low GLE | − 0.18 | 0.330 | |||
Bolded p values indicate features differed significantly between low- (grade group 1) and I/H-aggressive (grade group ≥ 2) prostate cancers.
ASM angular second moment, IMC information measure of correlation, IDM inverse difference moment, HGLRE high grey level run emphasis, LGLRE Low grey level run emphasis, GLE grey level emphasis, RLNU run length non-uniformity, I/H intermediate/high, ρ spearman correlation coefficient between features and cancer grade groups.
*Indicates feature correlated significantly with cancer grade group. The correlation coefficients were calculated for the entire grade groups (i.e. 1–5) and not for the dichotomized aggressive classes (i.e. grade groups 1 and ≥ 2).
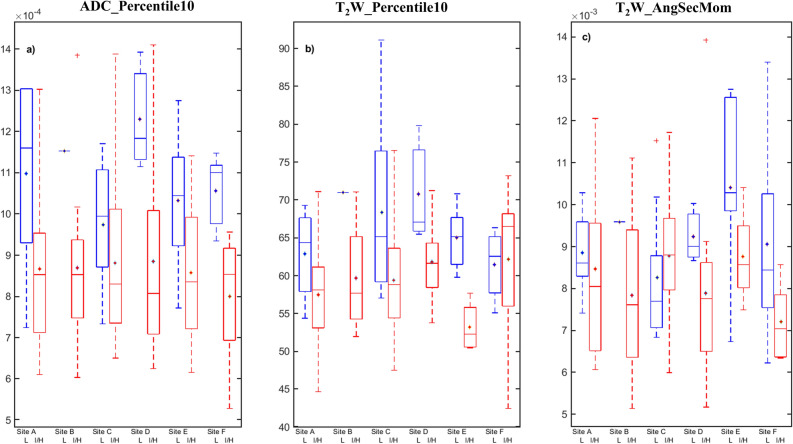
Figure 4.
Box plots showing the distributions of apparent diffusion coefficient (ADC) and T2-weighted (T2W) histogram (10th percentile), and T2W textural (angular second moment) features between low-aggressive (L) and intermediate/high-aggressive (I/H) prostate cancers at the participating institutions or sites. Notice that, where the T2W histogram feature (middle; Site F) overlap between the two cancer groups, an improved separation is observed for the textural feature (right; Site F), and vice-versa, indicating that the feature sets contain complementary information. *Indicates mean value.
The two-way ANOVA showed no statistically significant interaction between the effects of data origin (institution) and cancer aggressiveness on any feature. Similarly, the main effect of institution was not significant.
Classification of low versus intermediate/high-aggressive cancers
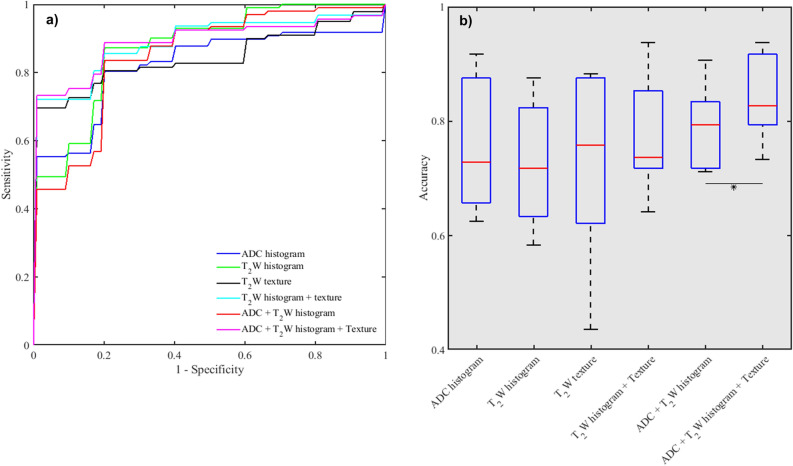
Table 3 and Fig. 5 show comparisons of the performance of the feature sets in SVM classification of the two aggressiveness classes at the various institutions. The added value of T2W textural features varied generally between the sites. At the individual centers, the augmentation of ADC and T2W histogram features with T2W textural features resulted in improvement in AUCs at four centers, though not statistically significant (p > 0.05). When considering the overall performance across the centers, the differences in AUCs before and after augmentation with textural features were not significant. However, the difference in accuracy was significant (p = 0.0218) for ADC + T2W histogram versus ADC histogram + T2W histogram + T2W texture features (Fig. 5). In terms of feature importance within the classifier, a similar trend as in the feature association with cancer grade group was observed. Textural features relating to similarity (grey level non-uniformity), maximum probability, and textural complexity (information measure of correlation) were the most frequently selected, in addition to minimum and 10th percentile from intensity histogram (Table 4).
Table 3.
Performance of ADC, T2W histogram and T2W textural features in SVM classification of low-(grade group 1) vs. intermediate/high-aggressive (grade group ≥ 2) prostate cancers at the various institutions.
| Feature type | Center for Testing: AUC [95CI] % | Mean [RD] | |||||
|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | ||
| ADC histogram | 71 [37–1] | 100 [*Nan] | 66 [44–88] | 88 [73–100] | 67 [20–100] | 100 [*Nan] | 82 [17] |
| T2W histogram | 83 [60–100] | 100 [*Nan] | 86 [70–100] | 94 [81–100] | 88 [61–100] | 73 [39–100] | 87 [7] |
| T2W texture | 77 [55–99] | 100[*Nan] | 51[28–74] | 96[87–100] | 96[84–100] | 87[64–100] | 84 [16] |
| T2W histogram + Texture | 80 [60–100] | 94 [*Nan] | 76 [57–95] | 100 [*Nan] | 92 [73–100] | 93 [78–100] | 89 [8] |
| ADC + T2W histogram | 74 [42–100] | 94 [*Nan] | 78 [57–98] | 96 [87–100] | 88 [65–100] | 80 [47–100] | 85 [9] |
| ADC + T2W + Texture | 77 [56–99] | 94 [*Nan] | 75[54–95] | 100[*Nan] | 96[84–100] | 93[78–100] | 89 [10] |
| Accuracy % | |||||||
| ADC histogram | 66 | 88 | 66 | 79 | 63 | 92 | 75 [14] |
| T2W histogram | 79 | 88 | 64 | 82 | 58 | 63 | 72 [15] |
| T2W texture | 62 | 78 | 44 | 88 | 88 | 73 | 72 [18] |
| T2W histogram + Texture | 72 | 94 | 64 | 85 | 75 | 72 | 77 [11] |
| ADC + T2W histogram | 79 | 91 | 71 | 79 | 83 | 72 | 79 [7] |
| ADC + T2W + Texture | 79 | 94 | 73 | 85 | 92 | 80 | 84 [8] |
| Sensitivity/specificity % | |||||||
| ADC histogram | 71/60 | 75/100 | 82/50 | 59/100 | 75/50 | 83/100 | 74/77 [8/30] |
| T2W histogram | 79/80 | 75/100 | 88/40 | 65/100 | 100/17 | 67/60 | 79/66 [13/41] |
| T2W texture | 64/60 | 56/100 | 47/40 | 76/100 | 75/100 | 76/80 | 64/80 [13/25] |
| T2W histogram + Texture | 64/80 | 88/100 | 88/40 | 71/100 | 100/50 | 83/60 | 82/72 [12//30] |
| ADC + T2W histogram | 79/80 | 81/100 | 82/60 | 59/100 | 100/67 | 83/60 | 81/78 [10/20] |
| ADC + T2W + Texture | 79/80 | 88/100 | 76/70 | 71/100 | 100/83 | 100/60 | 86/82[12/15] |
An SVM classifier was trained and tested for each individual institution, where the test data were from the institution being evaluated, and the training data were obtained from the remaining 5 institutions.
ADC apparent diffusion coefficient, T2W T2-weighted, CI confidence interval, SVM support vector machine, RD relative deviation.
*Nan indicates there is no interval.
Figure 5.
Performance of apparent diffusion coefficient (ADC), T2-weighted (T2W) histogram and T2W textual features in SVM classification of low-aggressive (grade group 1) versus intermediate/high-aggressive (grade group ≥ 2) prostate cancers. (a) Receiver operating characteristic curves showing the added value of T2W image textural features over traditional intensity histogram features alone. (b) Box plot comparing classification accuracies. * indicates significant difference.
Table 4.
Frequency of feature selection for SVM classification of low-(grade group 1) vs. intermediate/high-(grade group ≥ 2) aggressive prostate cancers across six institutions.
| Features | Classification model | |||||
|---|---|---|---|---|---|---|
| ADC | T2W Histogram | T2W Texture | T2W + Texture | ADC + T2W | ADC + T2W + Texture | |
| ADC kurtosis | 5 | 5 | 2 | |||
| ADC maximum | 2 | 1 | 0 | |||
| ADC mean | 4 | 4 | 1 | |||
| ADC median | 2 | 3 | 0 | |||
| ADC minimum | 6 | 5 | 3 | |||
| ADC 10th percentile | 4 | 5 | 6 | |||
| ADC 25th percentile | 2 | 5 | 3 | |||
| ADC 75th percentile | 0 | 3 | 1 | |||
| ADC 90th percentile | 3 | 2 | 0 | |||
| ADC Skewness | 2 | 3 | 0 | |||
| ADC standard deviation | 4 | 4 | 2 | |||
| T2W kurtosis | 4 | 1 | 1 | 0 | ||
| T2W maximum | 6 | 4 | 5 | 0 | ||
| T2W mean | 1 | 1 | 1 | 0 | ||
| T2W median | 3 | 1 | 1 | 1 | ||
| T2W minimum | 6 | 5 | 6 | 6 | ||
| T2W 10th percentile | 4 | 3 | 4 | 4 | ||
| T2W 25th percentile | 2 | 1 | 4 | 1 | ||
| T2W 75th percentile | 3 | 4 | 2 | 0 | ||
| T2W 90th percentile | 1 | 1 | 0 | 0 | ||
| T2W skewness | 2 | 2 | 4 | 1 | ||
| T2W standard deviation | 4 | 5 | 4 | 5 | ||
| T2W ASM | 1 | 3 | 1 | |||
| T2W Autocorrelation | 1 | 2 | 0 | |||
| T2W cluster prominence | 0 | 1 | 0 | |||
| T2W cluster shade | 5 | 2 | 1 | |||
| T2W contrast | 2 | 2 | 2 | |||
| T2W correlation | 1 | 1 | 0 | |||
| T2W difference entropy | 2 | 1 | 0 | |||
| T2W difference variance | 2 | 3 | 1 | |||
| T2W dissimilarity | 0 | 2 | 0 | |||
| T2W entropy | 1 | 1 | 0 | |||
| T2W IMC 1 | 6 | 3 | 4 | |||
| T2W IMC2 | 3 | 3 | 3 | |||
| T2W IDM | 0 | 1 | 0 | |||
| T2W maximum probability | 2 | 5 | 4 | |||
| T2W sum average | 0 | 2 | 0 | |||
| T2W sum entropy | 2 | 4 | 1 | |||
| T2W sum variance | 0 | 2 | 0 | |||
| T2W variance | 1 | 3 | 0 | |||
| T2W GLNU | 5 | 4 | 4 | |||
| T2W HGLRE | 0 | 1 | 0 | |||
| T2W LGLRE | 0 | 2 | 0 | |||
| T2W long run emphasis | 0 | 2 | 0 | |||
| T2W long run high GLE | 2 | 3 | 0 | |||
| T2W long run low GLE | 1 | 3 | 1 | |||
| T2W RLNU | 1 | 1 | 1 | |||
| T2W run percentage | 0 | 0 | 0 | |||
| T2W short run emphasis | 0 | 1 | 0 | |||
| T2W short run high GLE | 0 | 0 | 0 | |||
| T2W short run low GLE | 1 | 1 | 0 | |||
SVM support vector machine, ADC apparent diffusion coefficient, T2W T2-weighted, ASM angular second moment, IMC information measure of correlation, IDM inverse difference moment, GLNU grey level non-uniformity, HGLRE high grey level run emphasis, LGLRE low grey level run emphasis, GLE grey level emphasis, RLNU run length non-uniformity.
Discussion
T2W MRI provides high spatial resolution and tissue-specific contrast compared to DW and DCE imaging, but it is predominantly limited to qualitative radiological evaluation of the prostate. In a preliminary study using single-center data24, we showed that quantitatively derived T2W image textural features have the potential to serve as non-invasive markers for assessing aggressiveness. In this work, we extended and confirmed these findings in a multicenter cohort. T2W image textural features, particularly those reflecting homogeneity/similarity (angular second moment, run length non-uniformity, grey level non-uniformity), disorder (entropy) and textural complexity (information measure of correlation) correlated significantly with prostate cancer aggressiveness; and differed significantly between low- and intermediate/high-aggressive prostate cancers as defined by histopathology. Compared to the classifier based on the commonly used histogram metrics from ADC and T2W images, the classifier utilizing histogram features augmented with T2W textural features performed better, an indication that quantitative texture analysis of anatomical images has the potential to reveal additional morphological and pathophysiological information for radiomics-based assessment of prostate cancer aggressiveness.
The usefulness of entropy/complexity and homogeneity associated textural features in prostate cancer aggressiveness assessment and classification was shown in our previous study24, and has also been reported by others23,36,37. Histologically, aggressive prostate cancers are characterized by poor differentiation, glandular structure deformation, and loss of cellular integrity of the prostate gland. This disrupts the tissue cyto-architectural patterns, potentially leading to decreased homogeneity and high disorder. Correlations between textural features and prognostic factors and clinical outcome have also been reported36,38. If validated, these quantitatively derived T2W image features could be combined with other MRI parameters as evidence-based markers for prostate cancer. In the context of this study setup, the findings could particularly be useful in active surveillance situations to follow-up on low-risk cancer patients thereby limiting the need for repeated biopsies.
Although a number of promising studies have reported the utility of MRI texture analysis in prostate cancer19–23,36–38, very few are based on multicenter cohorts22 or focused on aggressiveness prediction/classification19,21,23,36. Multicenter data sharing is important to fulfill the high data demand for training radiomics-based decision support systems. Furthermore, multicenter studies are necessary to ascertain the applicability and robustness of texture analysis and radiomics, and to facilitate their clinical transition across centers. Texture analysis, which considers spatial relationships between pixels rather than individual pixel intensities as in a histogram, could possibly contribute to overcome the inter-institution and scanner variability challenges associated with multicenter data. Compared to DW and DCE imaging, T2W imaging is generally regarded as the most stable sequence in terms of imperviousness to scanner variations and gradient artifacts, and tolerance in patients (i.e. contrast agent-free). Although these factors add to its importance, T2W imaging is not currently used for quantitative assessment of prostate cancer aggressiveness mainly due to the non-quantitative nature of its signal intensities.
We found the classification performances of the individual feature sets (ADC histogram, T2W histogram and T2W texture) across the sites to be complementary and in some cases comparable, while the best performance was achieved when the feature sets were combined. The latter observation confirms the preliminary findings from our single-center study24. The improved classification performance of T2W intensity histogram features compared to some reported studies19,37 may be attributable to the post-processing (i.e. intensity non-uniformity correction and standardization) of the T2W images, which ensured that the intensities were comparable and had consistent quantitative interpretation across the patients and institutions. Our results are consistent with the findings of previous single-center studies by Fehr et al.37 and Chen et al.19 who also reported improved characterization of prostate cancer aggressiveness when using combined textural features from ADC and T2W images, compared to only mean ADC values or the individual feature sets respectively. On the contrary, Bonekamp et al.20 found the performance of mean ADC values to be comparable to that of combined radiomics features from ADC and T2W images when classifying benign versus malignant prostate lesions. One possible explanation for this observation is that their study was focused on cancer detection, which could be regarded as a relatively simpler task than aggressiveness classification. Similarly, the aggressiveness dichotomization in our study [i.e. low (grade group 1) vs. intermediate/high (grade group 2–5)] could be considered as a relatively simple classification task compared to for instance grade group 3 versus grade group 4, or the classification of the five distinct groups as in the PROSTATEx-2 Challenge39. Surprisingly, in some instances the classifier performance was reduced when textural features were added. Whereas this observation is worth further investigation, we suppose it could be due to potential inefficiencies in the feature selection process.
The goal of this study was to investigate the added value of T2W image textural features in prostate cancer aggressiveness assessment with respect to traditional histogram features. Hence, we opted to limit our texture feature extraction to this image sequence even though textural features can also be extracted from ADC maps. Similarly, a wide variety of other radiomic feature types exist that can be extracted including shape, Gabor, wavelet, grey level size zone matrix, neighbouring grey tone difference matrix, etc., features40,41. Our choice of using only GLCM and GLRLM features is not based on preference, but, these features have been extensively studied in different tissues and image modalities, and are generally regarded as intuitive. Nevertheless, including additional features may further enhance the classification of prostate cancer aggressiveness.
Feature repeatability and reproducibility are important aspects of radiomics pipeline, especially in a multicenter setting as in this study. Ideally, one would expect a good radiomic feature to be reproducible or stable across centers under the same conditions. A number of studies have recently investigated the repeatability of radiomic features mostly via test–retest analysis39,42–44. However, there is no consensus on which feature sets are most repeatable, mainly because the features are heavily influenced by pre-processing configurations during extraction. Most studies43,44 show that shape features are more repeatable. In our analysis we found neither interaction between the effects of data origin and aggressiveness nor the main effect of data origin to be significant on any feature, an indication that features were reproducible or stable across the institutions. Despite this, the classification accuracies varied generally within mean relative deviation [7–18%] across the institutions, which may suggest cross-site inconsistency in feature performance. Even though the two statistics (ANOVA & classification) are not directly comparable, a plausible explanation for this latter observation could be due to the highly imbalanced distribution of the data (i.e. classification classes) across the institutions. Even if significant effects were found in the former, it could be due to other potential confounding factors such as inherent differences in the cohort tumor heterogeneity across centers or possible inter-observer differences in the histopathologic grading that was used as ground truth, rather than the features. An alternative and possibly a more robust method of evaluating cross-site reproducibility of radiomics features could be through bootstrapping as employed in the studies by Chirra et al.39 and Leo et al. studies45, but this method requires a relative higher number of patients per-center.
Our study data and hence methods had some limitations. Despite being a multicenter study, our cohort size is relatively small and the distribution of patients or cancer aggressiveness classes across the centers is imbalanced (Table 1). This could affect our study results, especially the significance of T2W textural features in the classification performance. For instance, in a similar classification task performed by Fehr et al.37, the added value of textural features was found to be significant only after high imbalances in data was corrected for via sample augmentation. Clinically, prostate cancers are usually categorized into three aggressiveness classes (low, intermediate and high) or more46. Due to our study cohort size, however, tumor aggressiveness was stratified into low and intermediate/high. Although this stratification is inadequate for prostate cancer management, it serves as an important basis in the disease management pathway and could also be used as a benchmark for further clinical evaluation in larger multicenter studies. For instance, it is important to rule out low-risk cancers not needing active treatment, which otherwise can be overtreated with associated side effects. The inadequate number of transition zone tumors for analysis limits the applicability of our results to only the peripheral zone. Cancers originating from the transition zone have different radiomic features22, they tend to be elusive and are primarily assessed based on their appearance on T2W imaging8. Hence, the utility of quantitatively derived T2W image textural features for transition zone tumor characterization would be of great interest. Machine learning classifier performances are typically evaluated based on their classification accuracy. In this study for instance, the added of value T2W textural features can be better appreciated when looking at accuracy scores. Imbalances in datasets that are unrepresentative of the overall population can therefore lead to inflated assessment of the classifier accuracy. However, this was taken into account during the training and prediction (i.e. balanced accuracy) by adjusting the sample weights to be inversely proportional to the class frequencies. Feature repeatability is an important aspect of radiomics pipeline, hence, the lack of this tests in our study is another limitation.
Our study also lacks comparison with clinical readings involving PI-RADS scores, which were not available for this study. However, previous studies19,47 have shown radiomics-based models from combined ADC and T2W to outperform PI-RADS scores in prostate cancer detection and aggressiveness assessment. Our study setup was based on biopsy proven cases. The performance of textural features in prostate cancer detection and characterization remains to be evaluated prospectively without knowledge of lesion presence and location, especially as textural properties could be affected by inflammation or prostatitis. Also, in future multicenter studies, it would be interesting to perform PI-RADS scoring and VOI segmentation locally at each institution similarly to the histopathology evaluation. Ultimately, further studies in a much larger multicenter cohort are worth investigating.
Conclusion
This multicenter study confirms that texture analysis of T2W images provides quantitative information for assessment of peripheral zone prostate cancer aggressiveness. T2W MRI-derived textural features correlated significantly with pathological findings (cancer grade group) from multiple institutions and were sensitive to underlying pathological differences between low- and intermediate/high-grade prostate cancers in the peripheral zone. Although, we found the added value of T2W textural features in the classification of these cancer aggressiveness classes to be moderate, our study suggests that T2W textural features may have the potential to improve prostate cancer classification in multicenter settings. With a wide array of proposed radiomics features, further studies in larger multicenter cohort would be needed to ascertain their added value and robustness.
Author contributions
G.A.N., M.E., T.F.B. and K.M.S. conceived and designed the study. T.W.S., M.C.M., T.F.B. and K.M.S. performed data acquisition and provided administrative and technical support. G.A.N. performed data/statistical analysis and drafted and/or prepared the manuscript including figures and tables. G.A.N., M.E., T.W.S., M.C.M., T.F.B. and K.M.S. were involved in data interpretation and critical revision of the manuscript for intellectual content. The PCa-MAP Consortium organized the clinical trial (ClinicalTrials.gov Identifier NCT01138527) and collected data, part of which was used in this study.
Funding
The Norwegian Cancer Society (100792–2013); and the liaison Committee between the Central Norway Regional Health Authority and the Norwegian University of Science and Technology (90368401 and 90265300).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
A comprehensive list of consortium members appears at the end of the paper.
Contributor Information
Gabriel A. Nketiah, Email: gabriel.a.nketiah@ntnu.no
The PCa-MAP Consortium:
Ulrike I. Attenberger, Pascal A. T. Baltzer, Tone F. Bathen, Jurgen J. Fütterer, Masoom A. Haider, Thomas H. Helbich, Berthold Kiefer, Marnix C. Maas, Katarzyna J. Macura, Daniel J. A. Margolis, Anwar R. Padhani, Stephen H. Polanec, Marleen Praet, Tom W. Scheenen, Stefan O. Schoenberg, Kirsten M. Selnæs, Theodorus H. van der Kwast, Geert M. Villeirs, Trond Viset, and Heninrich von Busch
References
- 1.Gleason DF, Mellinger GT. Prediction of prognosis for prostatic adenocarcinoma by combined histological grading and clinical staging. J. Urol. 1974;111:58–64. doi: 10.1016/S0022-5347(17)59889-4. [DOI] [PubMed] [Google Scholar]
- 2.Epstein JI, et al. A contemporary prostate cancer grading system: a validated alternative to the Gleason score. Eur. Urol. 2015;69:428–435. doi: 10.1016/j.eururo.2015.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van der Leest M, et al. Head-to-head comparison of transrectal ultrasound-guided prostate biopsy versus multiparametric prostate resonance imaging with subsequent magnetic resonance-guided biopsy in biopsy-naïve men with elevated prostate-specific antigen: a large prospective multicenter clinical study. Eur. Urol. 2019;75:570–578. doi: 10.1016/j.eururo.2018.11.023. [DOI] [PubMed] [Google Scholar]
- 4.Corcoran NM, et al. Upgrade in Gleason score between prostate biopsies and pathology following radical prostatectomy significantly impacts upon the risk of biochemical recurrence. BJU Int. 2011;108:E202–E210. doi: 10.1111/j.1464-410X.2011.10119.x. [DOI] [PubMed] [Google Scholar]
- 5.Epstein JI, Feng Z, Trock BJ, Pierorazio PM. Upgrading and downgrading of prostate cancer from biopsy to radical prostatectomy: incidence and predictive factors using the modified Gleason grading system and factoring in tertiary grades. Eur. Urol. 2012;61:1019–1024. doi: 10.1016/j.eururo.2012.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liss MA, et al. Fluoroquinolone resistant rectal colonization predicts risk of infectious complications after transrectal prostate biopsy. J. Urol. 2014;192:1673–1678. doi: 10.1016/j.juro.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 7.Scheenen TWJ, Rosenkrantz AB, Haider MA, Fütterer JJ. Multiparametric magnetic resonance imaging in prostate cancer management: current status and future perspectives. Investig. Radiol. 2015;50:594–600. doi: 10.1097/RLI.0000000000000163. [DOI] [PubMed] [Google Scholar]
- 8.Turkbey B, et al. Prostate imaging reporting and data system version 2.1: 2019 update of prostate imaging reporting and data system version 2. Eur. Urol. 2019;76:340–351. doi: 10.1016/j.eururo.2019.02.033. [DOI] [PubMed] [Google Scholar]
- 9.Fütterer JJ, et al. Can clinically significant prostate cancer be detected with multiparametric magnetic resonance imaging? A systematic review of the literature. Eur. Urol. 2015;68:1045–1053. doi: 10.1016/j.eururo.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 10.Hambrock T, et al. Relationship between apparent diffusion coefficients at 3.0-T MR imaging and Gleason grade in peripheral zone prostate cancer. Radiology. 2011;259:453–461. doi: 10.1148/radiol.11091409. [DOI] [PubMed] [Google Scholar]
- 11.Vos EK, et al. Assessment of prostate cancer aggressiveness using dynamic contrast-enhanced magnetic resonance imaging at 3 T. Eur. Urol. 2013;64:448–455. doi: 10.1016/j.eururo.2013.05.045. [DOI] [PubMed] [Google Scholar]
- 12.Muller BG, et al. Prostate cancer: interobserver agreement and accuracy with the revised prostate imaging reporting and data system at multiparametric MR imaging. Radiology. 2015;277:741–750. doi: 10.1148/radiol.2015142818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ullrich T, et al. Risk stratification of equivocal lesions on multiparametric magnetic resonance imaging of the prostate. J. Urol. 2018;199:691–698. doi: 10.1016/j.juro.2017.09.074. [DOI] [PubMed] [Google Scholar]
- 14.Jena A, et al. Improving Diagnosis of primary prostate cancer with combined 68Ga–prostate-specific membrane antigen–HBED-CC simultaneous PET and multiparametric MRI and clinical parameters. Am. J. Roentgenol. 2018 doi: 10.2214/AJR.18.19585. [DOI] [PubMed] [Google Scholar]
- 15.Stoyanova R, et al. Prostate cancer radiomics and the promise of radiogenomics. Transl. Cancer Res. 2016;5:432–447. doi: 10.21037/tcr.2016.06.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278:563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haralick RM, Shanmugam K, Dinstein I. Textural features for image classification. IEEE Trans. Syst. Man Cybern. 1973;3:610–621. doi: 10.1109/TSMC.1973.4309314. [DOI] [Google Scholar]
- 18.Galloway M. Texture analysis using gray level run lengths. Comput. Graph. Image Process. 1975;4:172–179. doi: 10.1016/S0146-664X(75)80008-6. [DOI] [Google Scholar]
- 19.Chen T, et al. Prostate cancer differentiation and aggressiveness: assessment with a radiomic-based model vs. PI-RADS v2. J. Magn. Reson. Imaging. 2019;49:875–884. doi: 10.1002/jmri.26243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonekamp D, et al. Radiomic machine learning for characterization of prostate lesions with MRI: comparison to ADC values. Radiology. 2018;289:128–137. doi: 10.1148/radiol.2018173064. [DOI] [PubMed] [Google Scholar]
- 21.Toivonen J, et al. Radiomics and machine learning of multisequence multiparametric prostate MRI: towards improved non-invasive prostate cancer characterization. PLoS ONE. 2019;14:e0217702. doi: 10.1371/journal.pone.0217702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ginsburg SB, et al. Radiomic features for prostate cancer detection on MRI differ between the transition and peripheral zones: preliminary findings from a multi-institutional study. J. Magn. Reson. Imaging. 2017;46:184–193. doi: 10.1002/jmri.25562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fehr D, et al. Automatic classification of prostate cancer Gleason scores from multiparametric magnetic resonance images. Proc. Natl. Acad. Sci. 2015;112:E6265–E6273. doi: 10.1073/pnas.1505935112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nketiah G, et al. T2-weighted MRI-derived textural features reflect prostate cancer aggressiveness: preliminary results. Eur. Radiol. 2016 doi: 10.1007/s00330-016-4663-1. [DOI] [PubMed] [Google Scholar]
- 25.Maas MC, et al. A single-arm, multicenter validation study of prostate cancer localization and aggressiveness with a quantitative multiparametric magnetic resonance imaging approach. Investig. Radiol. 2019;54:437–447. doi: 10.1097/RLI.0000000000000558. [DOI] [PubMed] [Google Scholar]
- 26.Klein S, Staring M, Murphy K, Viergever MA, Pluim JPW. Elastix: a toolbox for intensity-based medical image registration. IEEE Trans. Med. Imaging. 2010;29:196–205. doi: 10.1109/TMI.2009.2035616. [DOI] [PubMed] [Google Scholar]
- 27.Zwanenburg A, Leger S, Vallières M, Löck S. Image biomarker standardisation initiative. Radiology. 2019 doi: 10.17195/candat.2016.08.1. [DOI] [Google Scholar]
- 28.Tustison NJ, et al. N4ITK: improved N3 bias correction. IEEE Trans. Med. Imaging. 2010;29:1310–1320. doi: 10.1109/TMI.2010.2046908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sunoqrot MRS, Nketiah GA, Selnæs KM, Bathen TF, Elschot M. Automated reference tissue normalization of T2-weighted MR images of the prostate using object recognition. Magn. Reson. Mater. Phys. Biol. Med. 2020 doi: 10.1007/s10334-020-00871-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Collewet G, Strzelecki M, Mariette F. Influence of MRI acquisition protocols and image intensity normalization methods on texture classification. Magn. Reson. Imaging. 2004;22:81–91. doi: 10.1016/j.mri.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 31.Benjamini Y, Hochberg Y. On the adaptive control of the false discovery rate in multiple testing with independent statistics. J. Educ. Behav. Stat. 2000;25:60–83. doi: 10.3102/10769986025001060. [DOI] [Google Scholar]
- 32.Guyon I, Weston J, Barnhill S, Vapnik V. Gene selection for cancer classification using support vector machines. Mach. Learn. 2002;46:389–422. doi: 10.1023/A:1012487302797. [DOI] [Google Scholar]
- 33.Youden WJ. Index for rating diagnostic tests. Cancer. 1950;3:32–35. doi: 10.1002/1097-0142(1950)3:1<32::AID-CNCR2820030106>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 34.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. doi: 10.2307/2531595. [DOI] [PubMed] [Google Scholar]
- 35.Pedregosa F, et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- 36.Vignati A, et al. Texture features on T2-weighted magnetic resonance imaging: new potential biomarkers for prostate cancer aggressiveness. Phys. Med. Biol. 2015;60:2685–2701. doi: 10.1088/0031-9155/60/7/2685. [DOI] [PubMed] [Google Scholar]
- 37.Wibmer A, et al. Haralick texture analysis of prostate MRI: utility for differentiating non-cancerous prostate from prostate cancer and differentiating prostate cancers with different Gleason scores. Eur. Radiol. 2015;25:2840–2850. doi: 10.1007/s00330-015-3701-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gnep K, et al. Haralick textural features on T2-weighted MRI are associated with biochemical recurrence following radiotherapy for peripheral zone prostate cancer. J. Magn. Reson. Imaging. 2017;45:103–117. doi: 10.1002/jmri.25335. [DOI] [PubMed] [Google Scholar]
- 39.Chirra P, et al. Multisite evaluation of radiomic feature reproducibility and discriminability for identifying peripheral zone prostate tumors on MRI. J. Med. Imaging. 2019;6:1. doi: 10.1117/1.JMI.6.2.024502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Castellano G, Bonilha L, Li LM, Cendes F. Texture analysis of medical images. Clin. Radiol. 2004;59:1061–1069. doi: 10.1016/j.crad.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 41.Zwanenburg A, et al. The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology. 2020;295:328–338. doi: 10.1148/radiol.2020191145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwier M, et al. Repeatability of multiparametric prostate MRI radiomics features. OPEN. 2019;1:1. doi: 10.1038/s41598-019-45766-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peerlings J, et al. Stability of radiomics features in apparent diffusion coefficient maps from a multi-centre test-retest trial. Sci. Rep. 2019;9:1–10. doi: 10.1038/s41598-019-41344-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fiset S, et al. Repeatability and reproducibility of MRI-based radiomic features in cervical cancer. Radiother. Oncol. 2019;135:107–114. doi: 10.1016/j.radonc.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 45.Leo P, et al. Evaluating stability of histomorphometric features across scanner and staining variations: prostate cancer diagnosis from whole slide images. J. Med. Imaging. 2016;3:047502. doi: 10.1117/1.JMI.3.4.047502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mottet N, et al. EAU-ESTRO-SIOG guidelines on prostate cancer. Part 1: screening, diagnosis, and local treatment with curative intent. Eur. Urol. 2017;71:618–629. doi: 10.1016/j.eururo.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 47.Wang J, et al. Machine learning-based analysis of MR radiomics can help to improve the diagnostic performance of PI-RADS v2 in clinically relevant prostate cancer. Eur. Radiol. 2017;27:4082–4090. doi: 10.1007/s00330-017-4800-5. [DOI] [PubMed] [Google Scholar]