Abstract
The entrenched dogma of a sterile middle ear mucosa in health is incongruent with its periodic aeration and seeding with saliva aerosols. To test this, we sequenced 16S rRNA-V4 amplicons from otic secretions collected at the nasopharyngeal orifice of the tympanic tube and, as controls, oropharyngeal and buccal samples. The otic samples harbored a rich diversity of oral keystone genera and similar functional traits but were enriched in anaerobic genera in the Bacteroidetes (Prevotella and Alloprevotella), Fusobacteria (Fusobacterium and Leptotrichia) and Firmicutes (Veillonella) phyla. Facultative anaerobes in the Streptococcus genus were also abundant in the otic and oral samples but corresponded to distinct, and sometimes novel, cultivars, consistent with the ecological diversification of the oral migrants once in the middle ear microenvironment. Neutral community models also predicted a large contribution of oral dispersal to the otic communities and the positive selection of taxa better adapted to growth and reproduction under limited aeration. These results challenge the traditional view of a sterile middle ear in health and highlight hitherto unknown roles for oral dispersal and episodic ventilation in seeding and diversifying otic biofilms.
Keywords: Middle ear, Eustachian tube, Otic microbiome, Oral dispersal, Otic infections, Divers
Abbreviations: ET, Eustachian tube; OTUs, Operational taxonomic units; PCoA, Principle coordinates analysis; PCR, Polymerase chain reaction; rRNA, Ribosomal RNA
Highlights
-
•
The middle ear mucosa harbors a rich bacterial community in health.
-
•
Oral migration is the primary mechanism for seeding otic biofilms.
-
•
Periodic aeration of the middle ear enriches for anaerobic taxa and promotes the ecological diversification of oral migrants.
-
•
Our study challenges the entrenched dogma of a sterile middle ear in health.
Introduction
The notion that the healthy middle ear is sterile, like the entrenched dogma of lung sterility before [12], has been spread in the literature without strong scientific justification [42]. This tenet is also incongruent with the anatomy of the middle ear (Fig. 1a), which has adaptively evolved as an acoustic chamber (the tympanic cavity) while maintaining air exchange with the lower respiratory airways through its extended tube, the tympanic or Eustachian tube (ET). Indeed, the tympanic cavity houses the delicate chain of small bones (ossicles) that transmit sound-induced vibrations from the eardrum to the hearing organ (cochlea) in the inner ear [5]. A porous bone structure (mastoid antrum) with gas-filled, interconnected spaces communicates the cavity to the mastoid gas cell system to increase resonance and to provide a medium for acoustic insulation and sound dissipation [33]. Yet, the ET extension of the tympanic cavity connects the middle ear to the nasopharynx and, by extent, to the noisy background of the aerodigestive system. To prevent noise interference, the tube is passively closed at rest and opens when we swallow to aerate the otic tissues and relieve negative pressure building up in the tympanic cavity [2]. Yawning or inhaling deeply can exert positive pressure at the tube’s nasopharyngeal orifice, increasing the differential pressure with the tympanic cavity and forcing the opening of the tube [2]. ET aperture (patency) is also facilitated by the lubrication of the tubal walls with surfactants that reduce the surface tension of the mucoid layer [2]. Collectively, these physical and chemical mechanisms ensure that the tube dilates briefly (~400 ms) to minimize acoustic interference yet frequently (approximately every minute when we swallow) to provide adequate ventilation and pressure relief [2,5,48]. As a result, the middle ear mucosa (both the tympanic cavity and ET) is periodically seeded with microorganisms from the aerodigestive tract.
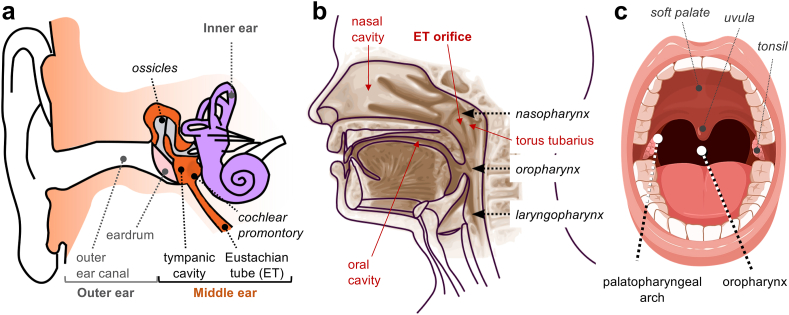
Fig. 1.
Anatomy of the ear, pharynx and oral cavity. (a) Anatomic structures in the outer, middle, and inner ear (illustration modified from Iain at the English Wikipedia, CC BY-SA 3.0). (b) Lateral cross section of the head showing the oral and nasal cavities, the three pharyngeal regions (naso-, oral-, and laryngo-) and the mucosal folds around the ET orifice and torus tubarius (illustration modified from Sémhur at Wikimedia Commons, CC BY-SA 3.0). (c) Frontal view of the oral cavity (licensed from Biorender and edited to add labels).
To minimize microbial infiltration, ET patency is timed to clear otic mucus and fluids that accumulate in the tympanic cavity as negative pressure builds. Mucociliary clearance facilitates the downward drainage of the secretions through the tube and, with them, foreign particles introduced with air [42]. Drainage is also facilitated by the pumping force exerted by the episodic contraction and extension of muscles around the tube while swallowing [26]. As in other mucosal tissues, the otic epithelium secretes to the mucosa glycoproteins (mucins) and proteins with antimicrobial activity or that bind viral or bacterial surface motifs to promote their recognition by macrophages and neutrophils [42], [82], [83]. The ET is narrower closer to the tympanic cavity, preventing the reflux of the mucus [2]. It also grows longer [67], curves slightly [2] and increases its tilt [57] into adulthood to facilitate drainage. To prevent mixing with nasal secretions, the ET orifice emerges into the nasopharynx as an elevated cartilage covered by mucosa (torus tubarius) (Fig. 1b). Additionally, this mucosal elevation is shaped like an inverted horseshoe that vertically drains the otic secretions behind the palatopharyngeal arch and prevents their re-aspiration (Fig. 1c).
The specialized anatomy of the middle ear and mechanisms for mucociliary and muscular clearance have been assumed to maintain the otic mucosa free of microbes in health [42]. Several studies [22,30,50,74] have attempted to confirm this, albeit with inconclusive results. All of these earlier studies collected samples from individuals undergoing transcanal surgical procedures designed to treat a number of otic conditions. These interventions reached the middle ear cavity through or around the eardrum [46], limiting sampling to small mucosal areas of the cochlear promontory that can be reached without perturbing the ossicular chain (Fig. 1a). Two independent pediatric studies that surgically collected biopsy mucosal specimens from this region confirmed the presence of bacterial microcolonies in patients with a history of otitis media [22,74] but only one detected microbial cells in samples from healthy individuals [74]. Non-invasive optical techniques also detected signals from bacterial biofilms in the middle ear of patients with a history of chronic ear infections but not in uninfected controls [54]. To improve the sensitivity of detection, Minami et al. [50] used transcanal surgery to collect middle ear mucosal swabs from pediatric and adult patients with or without a history of chronic ear infections. The detection in all the samples of a phylogenetically diverse pool of 16S rRNA amplicon sequences supported the conclusion that “the human middle ear is inhabited by more diverse microbial communities than was previously thought” [50]. However, the study did not include controls from the outer ear canal, which a more recent study showed to contaminate the swabs during sampling [30]. Contamination could explain why the otic samples were enriched (85%) in Proteobacteria and Actinobacteria [50], which are also the most abundant phyla in the outer ear canal [18,20]. Importantly, all of these earlier studies [22,30,50,74] considered as healthy controls, individuals undergoing surgery to treat otic conditions (e.g., otosclerosis, middle ear malformation, Bell’s palsy and deafness) that are associated with local inflammation and viral and bacterial infections [41,44,53,71]. Hence, the assumption that these individuals are healthy is questionable.
The uncertainty surrounding these earlier studies prompted us to design alternative approaches to investigate the microbiology of the middle ear. We reasoned that the growth of microcolonies in the middle ear mucosa in health would enrich for otic bacteria in secretions drained during the cycles of ET patency. To test this, we designed a pilot study and received institutional approval to non-invasively (through the mouth) collect otic secretions from the nasopharyngeal orifice of the ET. As a proof of concept, we collected otic, oropharyngeal and buccal samples from 23 healthy young adults for cultivation-independent analyses (16S–V4 amplicon sequences from 19 of the individuals) and recovery of otic and oral cultivars from the other 4 participants. The participants filled out a questionnaire and passed an onsite physical exam to establish health eligibility. The questionnaire also collected information about sports and recreational diving activities, which have been associated with increased pulmonary rates and, thus, more frequent middle ear ventilation [73]. The oral samples were collected from the oropharynx (the central hub for the distribution of saliva aerosols in the aerodigestive tract) and from the buccal mucosae (inner lining of the cheeks, upper gums and palate) that best represent the flexible and keratinized oral epithelia seeding the salivary microbiome [78]. Sequencing of 16S rRNA V4 region amplicons from the samples identified in the otic secretions a rich bacterial community of predominantly anaerobic taxa and revealed changes in community structure that correlated well with the frequency of otic aeration predicted for the participants. Furthermore, all of the subjects shared a core otic community taxonomically and metabolically similar to the oropharyngeal and buccal communities, albeit substantially different from the nasal microbiome. We also isolated from the three collection sites phylogenetically distinct species of facultative anaerobes, consistent with niche-specific adaptations. These results challenge the long-held view of a sterile otic mucosa and suggest instead that the middle ear is a dynamic ecosystem seeded by oral microbes and enriched in organisms better suited for growth and reproduction under episodic aeration.
Results
Sequencing of a diverse and robust microbiome in otic secretions collected from healthy, young adults
Using an institutionally approved protocol, we collected otic, oropharyngeal and buccal samples from 19 individuals (20 years old on average; Supplementary Table 1) for amplicon sequencing of the 16S rRNA variable 4 region (V4). All of the participants reported no recent history of respiratory infections and/or antibiotic treatment and passed an onsite physical exam that ruled out nasal congestion/dripping as well as inflammation and abnormalities of the nose, mouth, and ears. We also asked the participants to report recreational activities that could affect middle ear aeration and homeostasis such as swimming and diving. The survey identified 9 certified scuba divers who reported similar training in middle ear equalization techniques and diving experience at depths of >60 ft (Supplementary Table 2). Subjects who met the eligibility criteria and passed the onsite physical exam were asked to rinse their mouths with sterile saline to remove food debris and to performe a series of equalization exercises (deep inhaling, yawning and swallowing) that maximized otic drainage. The team’s physician used a tongue depressor to improve spatial access to the back of the mouth and introduced a sterile flocked swab behind the left and right palatopharyngeal arch to collect the otic secretions in the mucosal channel (torus tubarius) around the ET orifice (Fig. 1). The physician then used separate swabs to collect samples from the center of the oropharynx and from buccal mucosae (inner lining of the cheeks, upper gums and palate). Illumina sequencing of 16S–V4 amplicons from all the samples yielded a total of 12,219,721 reads, with an average number of 214,381 (±8132) reads for each region sampled per participant. To normalize differences in read number and therefore diversity among the samples, we rarified the sequences to a depth of 2,313 reads per sample prior to assigning operational taxonomic units (OTUs). We identified in the otic samples an average of 95 (±26) genus-level OTUs compared to 100 (±28) and 113 (±20) in the oropharyngeal and buccal samples, respectively. Thus, genus-level diversity in the otic secretions was within the levels obtained for the richly colonized mucosae of the oral cavity [78].
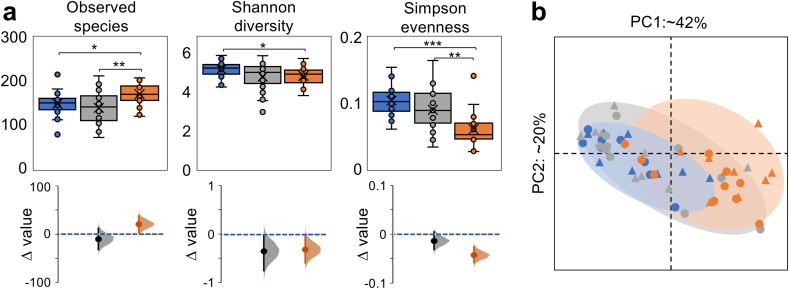
Alpha diversity analyses based on observed species, Shannon diversity and Simpson evenness provided evidence for the presence of a diverse and robust otic community (Fig. 2a). The number of observed otic species was within the ranges measured for the oropharyngeal and buccal microbiomes but Shannon and Simpson diversity tended to be higher in the otic than oral communities. The most notable differences were for Simpson’s evenness, which showed the greatest distribution of taxa in the otic communities compared to the oropharyngeal (p < 0.01) and buccal (p < 0.001) samples. Estimation plots of the standardized mean differences (Δ) among alpha diversity indices per site confirmed these differences (Fig. 2a). Notably, the distributions of Simpson’s evenness consistently followed the trend otic > oropharyngeal > buccal. Thus, the otic communities were more even than the oral (oropharyngeal and buccal) populations, a trait associated with microbiomes with the robustness and adaptability needed to function in a fluctuating environment [21]. Beta diversity analyses, on the other hand, highlighted similarities in the phylogenetic distribution of the otic and neighboring communities that agree well with a model of otic mucosal colonization by oral migrants (Fig. 2b). Principal Coordinates Analysis (PCoA) transformation plots of weighted UniFrac distances showed, for example, site-specific clustering of otic and buccal samples but some overlap in PCoA space along the variance obtained for the first axis. In contrast, the oropharyngeal sequences spread across the two axes to overlap with both the otic and buccal clusters. This is expected from the central role that the oropharynx is predicted to have in microbial immigration from the oral cavity to the middle ear. The periodic seeding of the oropharynx with saliva facilitates the aerial dispersal of aerosols with oral migrants, seeding the lower aerodigestive tract during inhalation and the middle ear cavity during exhalation. This shows as a substantial spatial overlap between the oropharyngeal and the otic and buccal communities in the PCoA ordination graphs (Fig. 2b).
Fig. 2.
Genus diversity in otic secretions.(a) Alpha diversity of the otic (blue), oropharyngeal (gray) and buccal (orange) communities based on richness (observed species), diversity (Shannon index) and evenness (Simpson index). Box plots show 50% of the diversity values in boxes, 25th and 75th percentiles as whiskers, median (line across the boxes), average (cross), outliers (circles outside the boxes) and confidence value from t-test comparisons (∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001; exact confidence values in Supplementary Table 4). Estimation graphics at the bottom show the mean (circle) diversity difference (Δ) of oropharyngeal or buccal samples versus the otic mean diversity (dashed blue line), the complete Δ distribution of values (shaded curve) and the 95% confidence interval of Δ (vertical line). (b) Principal Coordinates Analysis (PCoA) of weighted UniFrac distance in non-divers (circles) and divers (triangles) showing the spatial clustering of otic (blue) and buccal (orange) samples and overlap of these clusters with the central oropharyngeal samples (gray). Axes PC1 and PC2 show the proportion (%) of variance explained. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Otic community structure and evidence for adaptive responses to episodic aeration
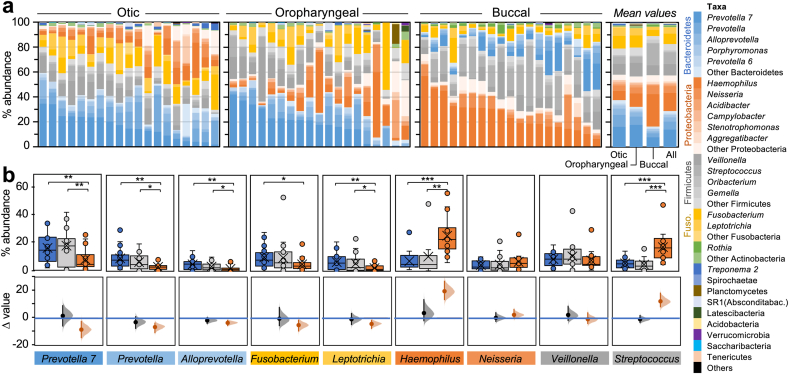
Taxonomic analyses of the OTUs assigned to the amplicon sequences identified the same dominant genera in all the samples but their abundance changed with the body site (Fig. 3a). Genera in the Bacteroidetes, Fusobacteria, Proteobacteria, and Firmicutes phyla collectively accounted for >95% of the OTUs identified in the otic, oropharyngeal and buccal communities. However, those in the obligate anaerobic phyla (Bacteroidetes and Fusobacteria) were more prevalent in the otic than in the oropharyngeal samples (55% and 48%, respectively) (Supplementary Table 3). These two phyla had the lowest representation in the more aerated buccal communities (24%), which enriched instead for facultative anaerobic and aerobic genera in the Proteobacteria and Firmicutes (72% of all the phylotypes). Similarly, all of the samples shared the same dominant genera, though their relative abundance was site-specific (Fig. 3b). These differences could not be attributed to the sex of the participants (Supplementary Fig. 1) but matched well with the frequency of aeration (therefore, oxygen availability) predicted at each body site (Fig. 3b). For example, the otic samples were dominated by strict anaerobes in the family Prevotellaceae (Prevotella and Alloprevotella), an oral group that can only proliferate in the low-oxygen communities of the subgingival plaque [19]. Obligate anaerobic genera in the Fusobacteria phylum (Fusobacterium and Leptotrichia) were also more highly represented in the otic than in the oropharyngeal and buccal microbiomes. Conversely, Haemophilus, a genus enriched in the aerated regions of the buccal mucosa [19], was less abundant in the otic samples than in the oropharynx and buccal microbiomes. Veillonella OTUs were also prevalent in all the samples (Fig. 3b). Despite their obligate anaerobic metabolism, Veillonella survive in the dental plaque as part of metabolically-dependent aggregates with lactate-producing species of Streptococcus [56,78]. The presence of both Veillonella and Streptococcus in the otic communities suggests similar metabolic associations in the middle ear mucosa. Yet, the facultatively anaerobic metabolism of Streptococcus allows them to also colonize aerated regions of the oral cavity, increasing their abundance in the buccal samples (Fig. 3b).
Fig. 3.
Genus-level structure of the otic communities in reference to oropharyngeal and buccal microbiomes. (a) Inter-individual differences in mean relative abundance (%) of genera (color-coded by phylum) at each collection site (b) Distribution of relative abundance values (top) and estimation plots (bottom) for dominant (>1%) otic genera. Data are color-coded for the otic (blue), oropharyngeal (gray) and buccal (orange) samples. Boxes in the bloxplots contain 50% of the values (horizontal line, median), whiskers the 25th and 75th percentiles, outliers (circles outsides the boxes) and t-test confidence values (∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001). Estimation plots showing the mean difference (Δ, solid circle) between otic (blue line at zero) and oropharyngeal (gray) or buccal (orange) samples, the complete Δ distribution (shaded curve), and 95% confidence interval of Δ (vertical line). Statistic values used to assess significance of the data are shown Supplementary Table 4. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The presence of 9 certified scuba divers in the cohort of participants prompted us to separately reconstruct the structure of their otic communities for comparisons with the non-divers group. Scuba divers are trained in equalization techniques that promote the frequent opening of the ET and the aeration of the middle ear cavity. Enhanced pulmonary functions in these individuals independently of diving habits have also been reported [73] that could impact the otic community structure. Indeed, despite the small number of participants in each subgroup, we observed significant changes in the representation of taxa based on their predicted metabolic response to aeration. In general, the otic communities of divers had less representation of anaerobic taxa that correlated well with increases in facultative anaerobic groups (Supplementary Fig. 2). For example, the mean relative abundance of the anaerobic Bacteroidetes and Fusobacteria phyla decreased (p = 0.02) from 62% in non-divers to 48% in divers (Supplementary Table 3). These decreases correlated well with changes in the relative abundance of two of the most abundant Bacteroidetes genera (Prevotella and Alloprevotella) and the fusobacterial genus Fusobacterium (Supplementary Fig. 2b). Divers also had a higher representation of oral genera such as Streptococcus and Veillonella (Supplementary Fig. 2b), whose metabolic dependence and co-aggregation in the oral cavity could facilitate their co-dispersal in aerosols.
The oral cavity as a seeding source for the otic communities
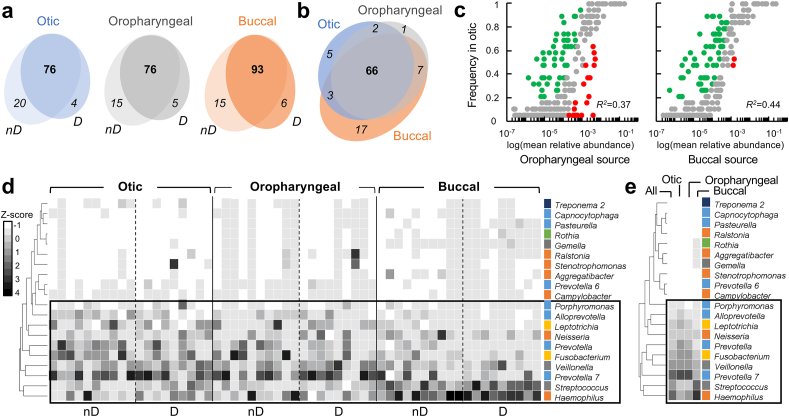
We gained insights into the contribution of oral bacteria to the seeding of the middle ear by defining the core microbiomes at each collection site and the degree of shared membership among the core communities (Fig. 4). We included in these analyses, genera represented in at least half of the participants within each group (non-divers and divers) and used this information to identify core members for each collection site (Fig. 4a). The core otic diversity comprised 76 genera and included 66 taxa from the oropharyngeal and buccal core communities (Fig. 4b). Thus, 87% of the otic genus diversity was shared with the neighboring oral communities. The remaining 13% consisted of low abundant OTUs (<0.1%) that were present in one or two sites only (Supplementary Table 5). Transience could explain the detection of some of these low abundant taxa in the otic samples. We detected, for example, Massilia, an aerobic genus that is ubiquitous in soils and enters and disperses through the aerodigestive tract via aerosols [60]. Also among the rare taxa was Peptococcus, a Firmicutes genus within the Gram-positive anaerobic cocci (GPAC) group that enters the aerodigestive tract via saliva or saliva aerosols [52]. Similarly, some of the otic OTUs were assigned to the Eubacterium brachy group, within the Firmicutes, and to uncultured Leptotrichiaceae within the Fusobacteria, which are groups of obligate anaerobes known to disperse through the aerodigestive tract as well [16,25].
Fig. 4.
Distribution and abundance of otic, oropharyngeal and buccal genera.(a–b) Core membership of genera shared by at least half of non-divers (nD) and divers (D) at each collection site (a) and among all microbiomes (b). (c) Neutral model fit of otic community assembly with the oropharyngeal or buccal communities as potential sources (R2, goodness of fit). Gray symbols represent neutrally distributed OTUs (within 95% confidence interval around the best-fit). Taxa above (green) or below (red) the confidence interval are more likely to be positively (over-represented) or negatively (under-represented) selected in the middle ear, respectively. (d–e) Heatmaps of individual (d) or average (e) Z-score transformed relative abundance (normalized z-score > −1.0) of the 20 dominant genera (color-coded by phylum: Bacteroidetes, blue; Fusobacteria, yellow; Firmicutes, gray; Proteobacteria, orange; Actinobacteria, green; Spirochaeta, dark blue; SR1, light blue; Planctomycetes, dark gold). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
We next implemented a neutral community assembly model, adapted from Sloan et al. [69] by Venkataraman et al. [76], to predict the contribution of unbiased (neutral) processes such as random dispersal and ecological drift (i.e., stochastic birth and death) to the otic core composition (Fig. 4c). A goodness-of-fit test (R2) determined how well (0, no fit; 1, perfect fit) these neutral processes explained the relative abundance of each otic OTU when the oropharyngeal or the buccal communities were considered as sources of migrants. We obtained similar R2 values when considering the oropharynx (R2 = 0.37) and buccal (R2 = 0.44) samples as source communities. This coefficient of determination is modest yet expected from inter-personal variation in microbial seeding, both in terms of the type and abundance of oral migrants (influenced by dietary preferences, among other factors) and the host physiology (e.g., frequency of aeration). Despite this uncertainty, the coefficient of determinations explained the presence of at least 70% of otic OTUs as the result of dispersal from the oral communities and ecological drift once in the middle ear environment. Most of the remaining OTUs fell above the confidence interval and, therefore, represented taxa over-represented in the otic communities compared to the two sources considered (green symbols in Fig. 4c). More than half of the over-represented OTUs (38 in all) deviated positively from the neutral model with both the oropharyngeal and buccal source communities (Supplementary Table 6). These taxa are predicted to have a competitive advantage in the middle ear, either because they are better fitted for growth in the otic mucosa and/or have a greater dispersal ability relative to other members of the source communities. The fact that most of these OTUs are rare oral taxa enriched in the otic samples (Supplementary Fig. 3) suggests, however, that they are not dispersal-limited but rather they have a growth advantage in the middle ear microenvironment.
The central location of the oropharynx in the aerodigestive system facilitates the seeding of its mucosa with saliva and the micro-aspiration of saliva aerosols into the middle ear. Therefore, we expected the oropharynx to contribute to the seeding of the middle ear more strongly than the buccal communities. To test this, we compared the mean relative abundance of the 66 genera that are shared by the three core communities (Supplementary Fig. 4). These analyses revealed a stronger positive association of otic taxa abundance with the oropharyngeal (Pearson’s correlation coefficient, R2 = 0.97) than with the buccal (R2 = 0.62) source communities, supporting the idea that the oropharynx serves as primary source for microbial immigration to the middle ear. The most significant deviations (at least two-fold increases or decreases in otic abundance compared to the oropharyngeal source) were for taxa with mean relative abundance <3% (Supplementary Fig. 4). Five of these OTUs (Treponema 2, Corynebacterium, Porphyromonas, Parvimonas, and Stenotrophomonas) were over-represented in the middle ear while the other 8 (Acidibacter, Corynebacterium 1, Ralstonia, Catonella, Rothia, Eubacterium yurii group, and other groups) were under-represented. Not surprisingly, these positive and negative deviations from the linear correlation of OTU abundance were exacerbated when comparing the otic to the buccal communities (Supplementary Fig. 4), which are more distant sources of microbial immigration.
Analyses of the abundance patterns among the dominant genera also provided evidence for the oropharynx serving as a central hub for the dispersal of oral migrants into the middle ear. For these analyses, we normalized taxa abundances per individual using the z-score method and generated a heatmap of the most abundant genera shared by the three core communities (Fig. 4d). We identified the same 20 dominant genera in all the samples, but the average abundance of each taxon changed with the collection site (Fig. 4e). The representation of the otic genera more closely mirrored the oropharyngeal than the buccal communities, further supporting the notion that the oropharynx is the primary seeding source. These analyses revealed abundance trends that supported, once again, the positive selection in the middle ear of obligate anaerobes such as those in the Bacteroidetes (family Prevotellaceae), Fusobacteria (Fusobacterium and Leptotrichia) and Firmicutes (Veillonella) phyla. By contrast, facultative anaerobic Proteobacteria (Haemophilus) and Firmicutes (Streptococcus) were most prevalent in the buccal samples. Neisseria OTUs provided an example of a genus that is similarly represented in the three locations and, therefore, neutrally distributed. This is a proteobacterial group of oropharyngeal and oral commensals that grows best aerobically [43] yet can disperse into the lower respiratory airways and lungs via saliva aerosols [1]. A similar mechanism of dispersal could explain the transience of these commensals in the middle ear.
Evidence for ecological diversification in the middle ear
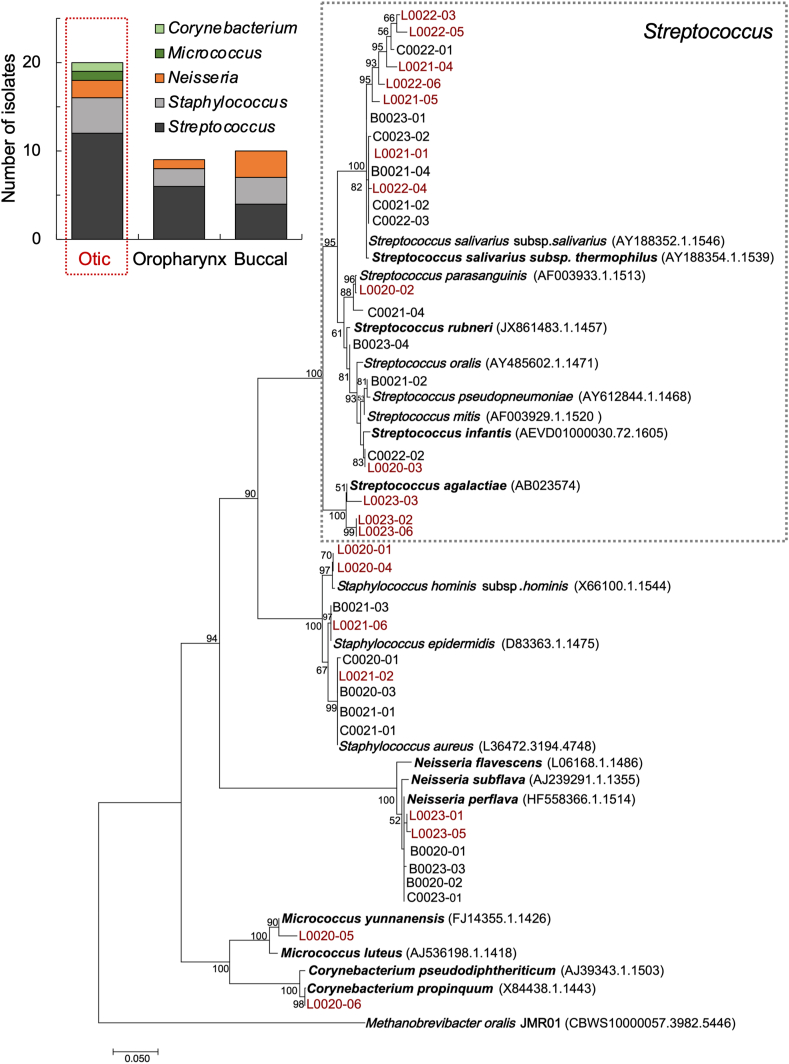
As part of the study, we recruited 4 additional participants and collected otic, oropharyngeal and buccal samples for cultivation experiments. These individuals went through the same eligibility criteria, physical exam and sample collection protocol as the rest of study subjects but the swabs were collected in a transport medium that preserved the viability of the cells for cultivation studies. We directly streaked freshly collected swabs onto plates of tryptic soy agar (TSA), a medium that supports the growth of Streptococcus and other heterotrophic bacteria dispersing through the respiratory airways [76]. After incubating the plates at 37 °C for 72 h, we visually inspected the colonies for morphology, color, shape and texture and ensured their purity through three passages on fresh TSA plates. This approach resulted in the recovery of 20 otic, 10 buccal and 9 oropharyngeal isolates. We next sequenced almost full-length amplicons of the 16S rRNA gene from each of the isolates for taxonomic classification at the genus and species levels using >95% and >98.7% identity cutoffs, respectively [70]. All of the isolates were closely related to species in the genera Streptococcus, Staphylococcus, Neisseria, Micrococcus and Corynebacterium. The majority of the isolates (79.5%) were Firmicutes in the Streptococcus genus (56.4%), consistent with this group’s abundance in the three microbiomes (Fig. 3) and growth advantage under the aerobic cultivation conditions used for their recovery. We also isolated species of Staphylococcus (23.1%), Neisseria (15.4%) and Actinobacteria (Micrococcus and Corynebacterium, 5.1%) (Fig. 5, inset). Phylogenetic analysis of partial 16S rRNA sequences obtained from the isolates and closest relatives showed a similar taxonomic distribution at the three collection sites but separation of some otic, oropharyngeal and buccal isolates that could have resulted from ecological diversification of oral migrants in the middle ear (Fig. 5).
Fig. 5.
Taxonomic and phylogenetic characterization of otic, oropharyngeal and buccal cultivars. The graph shows the number and genus assignment (based on 16S rRNA sequence) of otic, oropharyngeal and buccal isolates. The maximum-likelihood tree built with 16S rRNA sequences shows the phylogenetic placement of the otic (“L” designation, in maroon), oropharyngeal (“C”) and buccal (“B”) isolates and the closest strains (accession numbers, in parentheses). The scale bar indicates 5% divergence of 16S rRNA sequences filtered to a conservation threshold above 70% using the Living Tree Project (LTP) database [51,80]. The numbers at each node are bootstrap probabilities by 1000 replications above 50%. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The nearest neighbors to the Streptococcus sequences were species or subspecies within the Mitis group (Streptococcus mitis, Streptococcus infantis, Streptococcus oralis, Streptococcus pseudopneumoniae, Streptococcus parasaguinis and Streptococcus rubneri), the Salivarius group (Streptococcus salivarius), and the Lancefield’s group B streptococcus or GBS (Streptococcus agalactiae) [17,27]. Given the low discriminatory power of the 16S rRNA gene to classify human streptococcal isolates [64], we also screened the streptococcal isolates for hemolytic activity (Supplementary Table 7). All of the Streptococcus cultivars were α-hemolytic except for three γ-hemolytic isolates most closely related to S. oralis (Mitis group), S. salivarius (Salivarius group), and to S. agalactiae (GBS group). These results are consistent with the classification of streptococci within the Mitis and Salivarius groups as either α- or γ-hemolytic [17,65]. Furthermore, although nearly all strains of the GBS group are β-hemolytic [17], isolates of S. agalactiae have been recovered that are non-hemolytic (thus, they are classified as γ) [75].
The staphylococcal cultivars (20% otic, 22.2% oropharynx, 30% buccal) were all strains of Staphylococcus aureus, Staphylococcus epidermidis and Staphylococcus hominis (Fig. 5). Staphylococci are common residents of the nasal flora [3] and readily disperse into the neighboring oral cavity via the pharynx [55]. As a result, they are frequently isolated from oral and perioral regions [49,55]. The genus was however poorly represented in the otic, oropharyngeal or buccal 16S–V4 survey (Fig. 3), supporting the idea that they are transient members of these communities. We also recovered otic (10%), oropharyngeal (11%) and buccal (30%) strains closely related to Neisseria perflava (Fig. 5), an abundant member of the oropharyngeal flora [43] that disperses into the respiratory tract via saliva aerosols [1] and appeared transiently in the microbiome sequenced from otic secretions (Fig. 3b). Our cultivation approach also recovered in pure culture two otic Actinobacteria in the Micrococcus and Corynebacterium genera (Fig. 5). Actinobacteria is the most abundant nasal phylum [35] but only accounts for ~1% of the otic OTUs (Fig. 3). This is not unexpected considering that Micrococcus and Corynebacterium species are obligate or facultative aerobes and, therefore, they are more likely to be negatively selected in the less aerated environment of the middle ear.
Similar functional structure of the otic and oral microbiomes
Given the similarities in community membership among the three collection sites, we predicted a high degree of functional redundancy among their members as well. To test this, we used metabolic inference methods to predict the metabolic structure of the microbiome from otic secretions and describe relationships with the spatially close communities of the oropharynx and buccal mucosae. A heatmap of Z-score transformed relative proportion of functions represented at each collection site (1% cutoff) revealed a high degree of redundancy in core functions (Supplementary Fig. 5) and similar trends in non-divers and divers (Supplementary Fig. 6). All of the microbiomes had a high representation of membrane transport functions and modules associated with the metabolism of amino acid and carbohydrates (Supplementary Fig. 5). These metabolic functions often prevail in mucosal-associated communities, whose members support their growth using proteins and mucin glycoproteins secreted in the mucosa as sources of amino acids and carbohydrates [61]. Also with high representation in the three microbiomes were essential functions for genetic processing and information (replication and repair and, to a lesser extent, translation), which are critical to support cell growth (Supplementary Fig. 5). By contrast, cell motility functions were low. This is not unexpected given the primary contribution of passive mechanisms of dispersal (i.e., dispersal of saliva aerosols) to microbial immigration in these body sites.
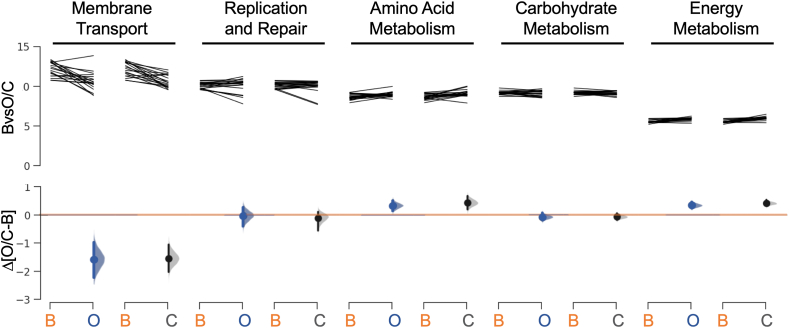
Despite the overall similarity in core functions at the three collection sites, we identified in pairwise comparisons some significant changes in the otic communities compared to the oropharyngeal and buccal sources (Fig. 6). The most notable differences were the lower representation of membrane transport functions yet higher relative proportion of amino acid and energy metabolism modules in the otic and oropharyngeal communities compared to the buccal communities. These differences likely reflect quantitative and qualitative changes in the nutrients that are available to support the growth of the resident microorganisms. Dietary substrates, which are abundant in the oral cavity, are less accessible in the oropharynx due to disturbance by the frequent cycles of air inhalation and exhalation. Dietary nutrients are also scarce in the middle ear due to the limited carriage of external nutrients in saliva aerosols. The oropharyngeal and, even more so, the otic communities are more likely to sustain their trophic webs with host-derived nutrients such as proteins and mucin glycoproteins secreted to the mucosa [61]. This helps explain why the relative proportion of membrane transport functions decreased while modules for amino acid and energy metabolism were more represented in the otic and oropharyngeal communities.
Fig. 6.
Taxonomic-based prediction of dominant metabolic functions. Pairwise comparisons of the abundance of the top 5 metabolic functions represented in the buccal (B) versus otic (O) or oropharyngeal (C; for center of the oropharynx) samples. Data from the statistical analyses is available in Supplementary Table 4.
Discussion
The identification of a diverse yet distinct microbiome in otic samples collected from healthy young adults challenges the entrenched view of a sterile middle ear mucosa and suggests instead that microorganisms colonize the otic mucosa and establish a site-specific community adapted to episodic ventilation. These results validate earlier studies, which reported the presence of bacterial microcolonies in otic mucosal samples collected from individuals that had no history of chronic otic infections [74]. We initially reasoned that these microcolonies could have resulted from the colonization and growth of oral bacteria introduced in the middle ear during the cycles of ventilation. As in other regions of the upper respiratory tract [28], mucociliary activity in the middle ear promotes the clearance of the otic mucus and, with it, bacteria residing in the mucosa. The periodic contraction and expansion of muscles around the ET also contributes to the clearance of mucus and fluids from the tympanic cavity and their drainage [26]. Non-invasive sampling of these secretions permitted the amplification of 16S–V4 rRNA bacterial sequences and the identification of an otic community with genus-level diversity comparable to the neighboring oral communities, which are among the richest and most diverse in the human body [78]. PCoA plots revealed the spatial clustering of the otic communities expected for site-specific populations yet overlap with the oropharyngeal and, to a lesser extent, buccal communities that we predicted to serve as seeding sources (Fig. 2b). Furthermore, the taxonomic composition of the otic communities was more similar to the oral microbiomes (Fig. 3) than to that reported for nasal microbiomes [35], despite the closer proximity of the ET orifice to the nasopharynx (Fig. 1b). Actinobacteria, for example, is the most abundant nasal phylum [35] but only accounted for ~1% of the otic OTUs (Supplementary Table 3). Instead, the otic secretions enriched (~38%) for genera in the Bacteroidetes, a strictly anaerobic phylum with lower representation (<10%) in the more aerated mucosae of the nasal cavity [35]. The relative abundance of Firmicutes in the otic samples (~21%) was within the ranges reported in the nasal microbiome [35] but while nasal Firmicutes are dominated by Staphylococcus [3], this genus was not significantly represented among the otic OTUs (Fig. 3). Rather, otic Firmicutes were dominated by Streptococcus and Veillonella, which are two of the most abundant genera in the oral cavity (Fig. 3). The otic communities also shared most of the keystone taxa of the core oral communities that we hypothesized would serve as sources of dispersal (Fig. 4). Indeed, the three mucosal sites sampled in this study harbored microbial communities dominated by the same 9 genera (Prevotella 7, Prevotella, Alloprovetella, Fusobacterium, Leptotrichia, Haemophilus, Veillonella, Streptococcus, and Neisseria) (Fig. 3). These genera represented four phyla (Bacteroidetes, Fusobacteria, Proteobacteria and Firmicutes) that collectively accounted for >90% of all the OTUs at each collection site. However, the relative abundance of anaerobic taxa (e.g., all of the Bacteroidetes and Fusobacterial genera) was higher in the otic secretions than in the neighboring oral mucosae, which selected instead for facultative aerobes in the Proteobacteria (Haemophilus) and Firmicutes (Streptococcus) (Fig. 5).
Alpha diversity analyses (Fig. 2a) provided additional evidence for the presence of an otic community as rich and diverse as the oral communities yet adapted to periodic aeration. The analyses revealed, for example, increases in species evenness in the otic communities that are often associated with microbiomes having the robustness and functional stability needed to adapt to environmental fluxes [21]. Redox fluctuations are expected in the middle ear, due to the intermittent pulses of air that enter the tympanic cavity when the ET opens. Episodic exposure of the otic mucosa to air provides a reasonable explanation for the enrichment in the otic communities of strict anaerobes, particularly those in the Bacteroidetes phylum (Fig. 3). The brief cycles of tubal dilation limit the volume of air entering the tympanic cavity to 4–5 μl [2]. Moreover, otic ventilation uses exhaled air [2], which has a lower concentration of oxygen than atmospheric air. Importantly, swallowing opens the ET once every minute during the wake hours but patency slows down to every 5 minutes during sleep [2]. This suggests that conditions of oxygen limitation prevail in the middle ear. Aerotolerant strains will have an adaptive advantage under these conditions and would be key to preserve community stability. The most prevalent otic Bacteroidetes were members of the family Prevotellaceae, which despite their strictly anaerobic metabolism can adaptively evolve oxygen tolerance under selective pressure [68]. The growth of these obligate anaerobic groups would, however, be sensitive to the frequency of aeration. This could explain why anaerobic genera such as Prevotella, Alloprevotella and Fusobacterium were less abundant in the otic secretions of divers (Supplementary Fig. 2), a group that has the equalization training associated with increased otic ventilation [2]. Furthermore, scuba diving, even if infrequent, can lead to subclinical changes in pulmonary functions [73] that could increase the rates of aerial dispersal into the middle ear and the abundance patterns of otic anaerobes.
Nutritional variables cannot be excluded as selective forces in the middle ear either. Oral Bacteroidetes, for example, also disperse into the digestive tract and proliferate in the strictly anaerobic environment of the colon [13]. Yet, we sequenced a higher proportion of Bacteroidetes phylotypes in the otic samples than typically detected in the colon (~16%) [13]. This is because gut Bacteroidetes play a key role in the degradation of complex dietary carbohydrates, a physiological activity that slows down their growth in the colon [13]. Otic Bacteroidetes however are more likely to specialize at the degradation of host-derived nutrients such as lipids, proteins and mucin glycoproteins secreted by the mucosal epithelium. Bacteroidetes are well equipped to break down mucin [72], an abundant source of protein and carbohydrates in the otic mucosa [61]. Mucin-derived sugars could support the growth of fermentative bacteria and the formation of syntrophic microcolonies in the middle ear mucosa. These syntrophic interactions could be similar to those established during the degradation of mucin and fiber by gut Bacteroidetes, which provide fermentable sugars to support the growth of strictly anaerobic Firmicutes in the class Clostridia [13].. This class was under-represented (~4%) in the otic communities. Instead, otic Firmicutes were represented by Streptococcus and Veillonella, abundant oral taxa that form metabolically linked co-aggregates during the primary colonization of the tooth surface [56]. The metabolic co-dependence of these two bacteria is established with the fermentation of sugars to lactate by the Streptococcus partner and the fermentation of lactate by Veillonella to produce propionate, acetate, CO2 and H2 [8,9]. A syntrophic consortium between otic Bacteroidetes, Streptococcus and Veillonela could promote the degradation and fermentation of mucin sugars into short chain fatty acids critical to mucosal health. The lactate dependency of Veillonella may also permit direct syntrophic interactions with Bacteroidetes partners that ferment simple sugars into lactate [63]. The high representation of pathways for carbohydrate and amino acid metabolism and reduced membrane transport predicted for the otic phylotypes (Fig. 6) does indeed support a trophic web in the otic mucosa driven by the metabolism of mucins and relying on lactate cross-feeding.
The similarities between the otic and gut trophic webs are not unexpected given the fact that both mucosae are seeded with oral microbes and both sites enrich for anaerobic migrants. A wide range of microbes enter the mouth with air, food, and by host-to-host contact before dispersing into the aerodigestive tract via air and/or saliva [58]. The oral cavity provides a heterogenous landscape (tooth surfaces, tongue, gingival crevices, palate, etc.) that locally selects for the growth of specific taxa, increasing their representation in saliva and their dispersal potential to other parts of the gastrointestinal tract and the lower respiratory airways [1]. Neutral models of community assembly suggest that the majority of microbial sequences recovered from the upper gastrointestinal tract and the lungs in healthy individuals disperse from oral reservoirs [76]. Similarly, oral dispersal had a profound influence in shaping the composition of the communities sequenced from otic secretions. Indeed, we identified in the oropharyngeal and buccal communities most of the core otic taxa (87% of the otic OTUs) (Fig. 4b), including many taxa known to disperse through the aerodigestive tract with air and/or saliva. Further, most (~70%) of the shared OTUs were neutrally distributed in a neutral model fit when considering the oropharynx or buccal communities as potential sources of immigration (Fig. 4c). ET patency only lasts about 400 miliseconds but occurs frequently (about 1,000 times a day just by swallowing) [6], providing a constant source of oral migrants. The process is analogous to the subclinical micro-aspiration of saliva aerosols that seeds the lower airways and the healthy lungs with oral microbes [28]. But unlike the seeding of the lungs with saliva aerosols carried during inhalation, microbial immigration into the middle ear is via exhaled air drawn from the lungs during ET patency. Not surprisingly, oral microbes commonly carried into the lower respiratory tract via aerosols such as Prevotella, Veillonella, Streptococcus, and Fusobacterium [28] were also among the most abundant in the otic secretions (Fig. 4). The adapted insular model of the lung microbiome [11] based on island biogeography theory [47] explains the constant presence of these microbes and their abundance in the lower airways as an equilibrium between the rates of microbial immigration (primarily by micro-aspiration during inhalation) and extinction (exhalation, coughing, etc.) with little contribution from the local growth of its members [11]. A similar model could explain the colonization of the middle ear with oral seeds, though with some notable differences. The middle ear is but a few centimeters away from the richly colonized mucosa of the oropharynx (the lungs are about half a meter), which could significantly increase the rates of microbial immigration. Spatial proximity could also permit the active migration of oropharyngeal bacteria such as via swarming, a mode of flagellar motility that is stimulated by mucosal surfactants and mucin [79,81]. Extinction rates in the middle ear would be determined by the efficiency of mucociliary and muscular clearance [26,66]. The enrichment of specific groups in the otic samples suggests that these mechanisms for mucus clearance cannot prevent the colonization of the otic mucosa by oral migrants. Furthermore, most of the otic OTUs that deviated from the neutral model were over-represented in the otic communities compared to the oropharyngeal or buccal sources (Fig. 4c), suggesting a process of positive selection. Among the oral sequences enriched in the otic environment were strict anaerobes within the Bacteroidetes, Fusobacteria and Firmicutes. Also over-represented were many Proteobacterial sequences assigned to groups of facultative anaerobes, which could have a competitive advantage for growth with intermittent aeration. Indeed, otic Proteobacteria represented 16% of the sequences identified in the non-divers otic samples and had an even higher representation (~25%) in the more aerated middle ear mucosa of divers. By contrast, many oral Proteobacteria are negatively selected under the strictly anaerobic conditions of the colon, reducing their representation to ~0.1% of the gut phylotypes [13]. Dispersal ability could have also contributed to the over-representation of some oral taxa in the otic communities. More frequent otic ventilation, as predicted for divers, could also increase the rates of micro-aspiration of saliva aerosols from the oropharynx and the dispersal of oral migrants. This could explain the higher representation in divers of facultative anaerobic taxa such as Streptococcus, a genus that is most abundant in the buccal communities (Fig. 4).
The correlations between otic community structure and aeration cannot rule out the influence of other host variables. Scuba diving can cause small changes in pulmonary function that could increase the rates of aerial dispersal into the middle ear [73]. Anatomy and even body position and posture are known to influence ET function [2]. Thus, ET opening time is shorter when lying down because the increased blood flow to the head and neck causes venous engorgement around the tympanic tube [31,39]. As a result, the mean volume of air passing through the tube’s lumen is two thirds lower when in the supine than the prone position [29]. Thus, increased otic ventilation in divers could have resulted from higher levels of physical activity in this group. Further, subtle differences in the anatomy of the ET are not uncommon [2] and could have affected the rates of microbial immigration over extinction and, by extent, the structure of the otic community. Importantly, we selected a homogenous cohort of young adults among the college population but we cannot exclude that dietary preferences contributed to intra- or inter-group differences. Future investigations could address these variables in larger surveys that select participants based on their dietary and exercise routines. Also important for future studies are insights into the intrinsic and extrinsic factors that lead to the positive or negative selection of taxa in the middle ear, particularly otopathogens. Our study highlighted correlations that point at a critical role for aeration and host-derived nutrients in the assembly of otic biofilms, but other variables such as pressure fluctuations and sound-induced vibrations could exert selective pressure and permit the diversification of oral taxa into lineages better suited for growth and reproduction in the middle ear mucosa. In support of this, we recovered from the otic samples cultivars that were closely related yet phylogenetically distinct to oral isolates (Fig. 5). These otic lineages could, in turn, influence the functionality of the otic community, its interactions with the host mucosa and the outcome of infections.
Methods
Study design. We recruited 23 healthy adult individuals by advertisement at the Michigan State University campus and diving shops in the state of Michigan from September to November of 2017. The study was approved by Michigan State University’s Institutional Review Board (IRB #17-502) and all subjects gave signed, informed consent to participate. The participants were scheduled for a physical exam and sample collection on November 18th of 2018 in a clinic room at the Clinical and Translational Sciences Institute on the Michigan State University campus. Each volunteer filled out a questionnaire to provide relevant information (age, sex, race, medical history with a focus on ear, nose, and mouth conditions and past antibiotic use; recreational/professional exercise and aquatic activities). Exclusion criteria were pregnancy, chronic health problems, asthma, persistent allergies, and recent (less than 3 months) antibiotic treatment or conditions affecting the ear, nose and throat (e.g., bronchitis, strep throat, ear infections, hearing loss, problems with balance, etc.). Participants that showed proof of current scuba diving certification (n=9) were asked to provide information about their diving training (Supplementary Table 2). A physician specifically trained for the task interviewed each participant in the presence of the team leader and prior to sample collection to ensure they understood the study protocol and rule out mental incapacity and/or cognitive impairment for consent. The physician then performed a general otolaryngologic examination (otoscopic examination of the ear, nose, and mouth) to rule out inflammation or other conditions affecting participant’s eligibility. The results of the otoscopic exam were recorded in an examination sheet.
Eligible participants (n = 23) were asked to rinse their mouth with a sterile saline solution to remove food debris and to follow a series of deep inhalation and yawn cycles that forced the opening the tympanic tube (they had to hear a “pop”) and the drainage of the middle ear fluids. They were then asked to swallow to naturally open the ET one more time and to open their mouth to initiate sample collection. We used a sterile tongue depressor to improve access to the back of the mouth and prevent oral and/or tonsil contamination. Sample collection was with FLOQSwabs™ (Copan) and storage was in collection tubes filled with eNAT™ (DNA sequencing; 19 participants) or ESwab™ (cultivation; 4 participants) collection tubes (Copan). We first collected in a single swab the left and right otic secretions, using an ascending motion to swab the mucosal channels that laterally drain the otic fluids behind the palatopharyngeal arch. The physician then used separate swabs to collect control samples from the central region of the oropharynx and from the inner lining of the cheeks and upper gingiva and palate (buccal samples). To preserve the anonymity of the participants, we barcoded the swab samples and all the forms and questionnaire collected for each individual. All samples were stored in the collection tubes at 4 °C for 8–24 h before transport to the lab for immediate processing.
DNA extraction and 16S rRNA amplicon sequencing. We vortexed the transport tubes at medium speed for 1 min to detach the specimens and used a 400-μl aliquot of the cell suspension for DNA extraction with the FastDNA™ Spin kit (MP Biomedicals). The protocol for DNA extraction followed manufacturer’s recommendations, except that cell lysis used 800 μl of cell lysis buffer instead of the recommended 1 ml. Sample homogenization used a Mini-Beadbeater (BioSpec Products) operated at maximum speed for 40 s and followed three cycles of mechanical homogenization interspersed with sample cooling on ice for 1 min. DNA quantification in the samples was with a NanoDop™ (Thermo Fisher Scientific) and a Qubit™ dsDNA HS assay (Thermo Fisher Scientific). The quality of the extracted DNA for amplification of the V4 region of the 16S rRNA genes was tested using dual indexed Illumina compatible primers (515f [GTGCCAGCMGCCGCGGTAA] and 806r [GGACTACHVGGGTWTCTAAT]) [36] in a 25-μl PCR reaction containing 12.5 μl 2xGoTaq® Green Master Mix (Promega), 0.1 μM of each primer, 0.2–20 ng of DNA template, and nuclease-free water. The PCR amplification also included controls that replace the DNA template with the elution buffer used in DNA extraction, sterilized water and eNAT transport medium. Visualization of PCR products was on 1.2% agarose gels. Library preparation and amplicon sequencing were performed by the Genomics Core staff at Michigan State University’s Research Technology Support Facility (RTSF) and followed standard protocols for PCR-amplification of the V4 hypervariable region of the 16S rRNA gene using the 515f/806r primer pair, normalization of the PCR products in SequalPrep DNA Normalization plates (Thermo Fisher Scientific), and cleaning of the pooled samples with AMPureXP magnetic SPRI beads (Beckman Coulter). Quality control and DNA amplicon quantification used the Qubit™ dsDNA HS assay, LabChip® GX DNA HS assay (PerkinElmer), and Kapa Library Quantification kit for Illumina platforms (Kapa Biosystems). Sequencing of the pooled amplicons was on an Illumina MiSeq v2 standard flow cell using a 500 cycle v2 reagent cartridge for 250-bp paired-end reads and used standard Illumina quality control steps, including base calling by Illumina Real Time Analysis (RTA) v1.18.54, demultiplexing, adaptor and barcode removal and RTA output conversion to FastQ format with Illumina Bcl2fastq v2.19.1.
Sequence data processing and analysis. We used USEARCH (v10.0.240) [15] to process the paired-end reads (FASTQ files) and merge paired-end sequences, quality-filter, dereplicate, remove singletons, pick OTUs and match them against the Silva database. Briefly, processing of raw reads and quality filtering used the UPARSE pipeline [14] and clustered into operational taxonomic units (OTUs) at 97% identity against the Silva Database (v. 1.19) [59] using previously outlined protocols [40,62]. Reads without a database match were clustered in de novo mode at 97% identity [59]. Taxonomic assignment and diversity analyses used the QIIME workflow [4]. A summary of the script used in this study is available on Github (https://github.com/mutantjoo0/RegueraLab_ONR).
Intra-group (alpha) and inter-group (beta) diversity analyses used the functions available in the QIIME pipeline [59]. Alpha diversity analyses measured in each sample the species richness (number of observed species), their abundance and evenness (Shannon diversity index, H) and evenness (Simpson). Beta diversity analyses applied the weighted UniFrac metrics [45] to calculate pairwise phylogenetic distances between sets of sequences and generate a distance matrix for Principal Coordinates Analysis (PCoA). MS Excel 2016 was then used to visualize the inter-group relatedness in PCoA plots and calculate the statistical significance of the Unifrac distance between the otic and control samples with the t-test (∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001). InteractiVenn [23] was used to generate Venn diagrams and identify the core microbiome. When indicated, we applied estimation statistics (www.estimationstats.com) to assess the effect of size distribution and generate two-group estimation plots, as described elsewhere [7,24]. The estimation plots show the mean difference (Δ), the bootstrapped distribution of the mean difference and the bootstrapped 95% confidence interval of mean difference.
We analyzed the 16S–V4 phylogenetic data with the PICRUSt software package [38] to predict functional traits based on the Kyoto Encyclopedia of Genes and Genomes Orthology (KO) classification [32] on the Nephele cloud platform [77]. Visualization of the taxonomic and functional profiles for each sample used heatmaps generated with the HemI toolkit [10]. To illustrate the normalized distribution across the values, we used row Z-score normalized relative abundances and relative proportions or applied the average linkage clustering method and Euclidean distance metric to calculate pair wise distance and similarity for hierarchical clustering in heatmaps. The neutral community model applied in this study was adapted from Sloan et al. [69] using custom R scripts developed by Venkataraman et al. [76]. Briefly, the analyses calculated OTU abundance in the source (number of OTU sequences with OTU in source community/total no. of sequences in source community) and the frequency of detection of each OTU in the otic communities (no. of participants with that OTU detected in the otic sample/total no. of individuals surveyed). A beta probability distribution was then used to predict the frequency of detection of each OTU in the otic samples as a result of neutral processes (dispersal and ecological drift) [69]. After optimizing the fitting parameter using a least-squares approach, we calculated the 95% binomial proportion confidence intervals for the neutral model and the goodness-of-fit coefficient of determination (R2), which ranges from 0 (no fit) to 1 (perfect fit). OTUs that fall outside the upper or lower confidence intervals are not considered to be neutrally distributed and identify OTUs likely to undergo positive or negative selection in the target community, respectively.
Cultivation and isolation procedures. We vortexed the ESwab™ collection tubes for 30 s to detach the cells from the flocked swabs and transferred 200 μl aliquots of the cell suspension to a sterile cryovial containing an equal volume of cryogenic preservation medium (filter-sterilized Lysogenic Broth [LB] with 50% glycerol) and stored at −80 °C for long-term preservation. Using the sample collection swab, we streaked the remaining cell suspension onto solidified TSA medium (30g/L Tryptic Soy Broth (Sigma Aldrich) and 15g/L Bacto Agar (BD)) and incubated at 37 °C for 48–72 h until colonies were visible. Single colonies showing different morphological traits (size, color, shape, texture) were re-streaked up to three times to ensure purity and reproducibility of the colony phenotype. The isolates were then grown overnight in liquid Tryptic Soy Broth medium (TSB; Sigma Aldrich) at 37 °C with gentle agitation. A 200-μl aliquot of the culture was transferred to a sterile cryovial, mixed with an equal volume of the cryogenic preservation medium (LB-50% glycerol) and stored at – 80 °C.
Phylogenetic analyses. DNA extraction was from cells harvested by centrifugation from 2 ml TSB cultures grown at 37 °C for 24 h and used the FastDNA™ Spin kit (MP Biomedicals) following manufacturer’s recommendations. The DNA served as template for PCR-amplification of 16S rDNA fragments using bacterial universal primers (27F: AGAGTTTGATCMTGGCTCAG; 1492R: TACGGYTACCTTGTTACGACTT) [37] and GoTaq® Green Master Mix (Promega). After purification, we concentrated the amplified DNA fragments with the DNA Clean & ConcentratorTM-5 kit (Zymo Research) prior to Sanger sequencing with the forward or reverse primers using ABI Prism BigDyeTM Terminator v3.1 Cycle Sequencing Kit on an ABI 3730xl DNA Analyzer (Applied Biosystems) at Michigan State’s Research Technology Support Facility Genomic Core. To minimize sequencing errors, we trimmed the 3′ end of the amplicons after the first 900-bp sequence. We also removed 50 nucleotides at the 5′ end to produce a high-quality 850-bp amplicon sequence for homology searches in the GenBank database. For ambiguous matches, we aligned the forward and reverse contigs and repeated the search. Taxonomic assignation used a species cutoff value of 98.65% and a genus cutoff of 97% [34]. We used the Living Tree Project (LTP) database (https://www.arb-silva.de) [51], [80] to identify the type strain for each of the closest species relatives and used their 16S rRNA genes as reference sequences for phylogenetic analysis with the 850-bp forward amplicons. Using the MUSCLE tool [84], [85] in the MEGA X software [86], we aligned the sequences before building a maximum likelihood tree and calculating the bootstrap confidence values at each node from 1000 replications. The tree shows bootstrap values above the 50% confidence threshold.
Hemolysis assays. The hemolytic activity of the cultivars was tested in microcolonies grown on blood agar plates prepared with 30 g/L of TSB (Sigma Aldrich), 15 g/L of Bacto Agar (BD) and 50 ml/L of sheep’s blood (Sigma Aldrich). The bacterial strains were first grown overnight in TSB at 37 °C with gentle agitation before spot-plating 5 μl of the culture on the blood agar plates. After plating them, we allowed the culture drops to dry at room temperature for 30 min before incubation at 37 °C for 24 h in a CO2 incubator. The hemolytic activity was characterized as either alpha (media discoloration around the area of growth), beta (clearing around the area of growth) or gamma (no hemolysis).
Availability of data and materials
Raw sequence reads have been deposited in the NCBI Sequence Read Archive under BioProject ID: PRJNA473788.
Ethics approval and consent to participate
The research proposed in this study was approved on May 17 of 2017 by the Institutional Review Board (IRB) at Michigan State University, East Lansing, Michigan, United States of America. The committee found the research project to be appropriate in design, to protect the rights and welfare of human subjects, and to meet the requirements of Michigan State University’s Federal Wide Assurance and the Federal Guidelines (45 CFR 46 and 21 CFR Part 50).
Consent for publication
Not applicable.
Funding
This work was supported by award N00014-17-2678 from the United States Office of Naval Research to G.R. and K.K.
Author information
Affiliations
Department of Microbiology and Molecular Genetics, Michigan State University, 567 Wilson Rd., Rm 6190, Biomedical and Physical Sciences building; East Lansing, MI 48824, USA.
CRediT authorship contribution statement
Joo-Young Lee: Data curation, Formal analysis, processed samples for Illumina sequencing and analyzed the 16S–V4 data, participated in the collection of the samples, All authors read and approved the final version of the manuscript. Kristin M. Jacob: Formal analysis, processed samples for cultivation experiments, J-YL amplified and sequenced the 16S rRNA sequences, and performed the phylogenetic analyses and hemolysis assays, participated in the collection of the samples, All authors read and approved the final version of the manuscript. Kazem Kashefi: participated in the collection of the samples, All authors read and approved the final version of the manuscript. Gemma Reguera: conceived the study and planned the sample collection protocol with contributions from KK, Writing - original draft, wrote the first draft of the manuscript and incorporated contributions from the other authors, participated in the collection of the samples, All authors read and approved the final version of the manuscript.
Declaration of competing interest
The authors declare that they have no competing interests.
Acknowledgements
The authors thank the study participants who kindly provided the swab samples and team’s physician, Dr. Lauryn Przeslawski, for assistance with sample collection. We are also grateful to Dr. Emily Chrisovan at Michigan State’s Research Technology Support Facility for advice with amplicon sequencing, to Hugh McCullough for assistance with 16S rRNA sequencing of cultivars, and to Drs. Ashley Shade, Patrick Kearns, and Nejc Stopnisek at Michigan State and Dr. Arvind Venkataraman at Procter & Gamble for bioinformatics support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioflm.2020.100041.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Bassis C.M., Erb-Downward J.R., Dickson R.P., Freeman C.M., Schmidt T.M., Young V.B., Beck J.M., Curtis J.L., Huffnagle G.B. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. mBio. 2015;6 doi: 10.1128/mBio.00037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bluestone C.D. BC Decker Inc; Hamilton, Ontario: 2005. The Eustachian Tube: structure, function, and role in the middle ear. [Google Scholar]
- 3.Bomar L., Brugger S.D., Lemon K.P. Bacterial microbiota of the nasal passages across the span of human life. Curr Opin Microbiol. 2018;41:8–14. doi: 10.1016/j.mib.2017.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Pena A.G., Goodrich J.K., Gordon J.I., Huttley G.A., Kelley S.T., Knights D., Koenig J.E., Ley R.E., Lozupone C.A., McDonald D., Muegge B.D., Pirrung M., Reeder J., Sevinsky J.R., Turnbaugh P.J., Walters W.A., Widmann J., Yatsunenko T., Zaneveld J., Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christensen-Dalsgaard J., Carr C.E. Evolution of a sensory novelty: tympanic ears and the associated neural processing. Brain Res Bull. 2008;75:365–370. doi: 10.1016/j.brainresbull.2007.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cinamon U. Passive and dynamic properties of the eustachian tube: quantitative studies in a model. Otol Neurotol. 2004;25:1031–1033. doi: 10.1097/00129492-200411000-00030. [DOI] [PubMed] [Google Scholar]
- 7.Claridge-Chang A., Assam P.N. Estimation statistics should replace significance testing. Nat Methods. 2016;13:108–109. doi: 10.1038/nmeth.3729. [DOI] [PubMed] [Google Scholar]
- 8.Cotter P.D., Hill C. Surviving the acid test: responses of gram-positive bacteria to low pH. Microbiol Mol Biol Rev. 2003;67:429–453. doi: 10.1128/mmbr.67.3.429-453.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delwiche E.A., Pestka J.J., Tortorello M.L. The Veillonellae: gram-negative cocci with a unique physiology. Annu Rev Microbiol. 1985;39:175–193. doi: 10.1146/annurev.mi.39.100185.001135. [DOI] [PubMed] [Google Scholar]
- 10.Deng W., Wang Y., Liu Z., Cheng H., Xue Y. HemI: a toolkit for illustrating heatmaps. PloS One. 2014;9 doi: 10.1371/journal.pone.0111988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dickson R.P., Erb-Downward J.R., Huffnagle G.B. Towards an ecology of the lung: new conceptual models of pulmonary microbiology and pneumonia pathogenesis. Lancet Respir Med. 2014;2:238–246. doi: 10.1016/S2213-2600(14)70028-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dickson R.P., Erb-Downward J.R., Martinez F.J., Huffnagle G.B. The microbiome and the respiratory tract. Annu Rev Physiol. 2016;78:481–504. doi: 10.1146/annurev-physiol-021115-105238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eckburg P.B., Bik E.M., Bernstein C.N., Purdom E., Dethlefsen L., Sargent M., Gill S.R., Nelson K.E., Relman D.A. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edgar R.C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 15.Edgar R.C. Updating the 97% identity threshold for 16S ribosomal RNA OTUs. Bioinformatics. 2018 doi: 10.1093/bioinformatics/bty113. [DOI] [PubMed] [Google Scholar]
- 16.Eisenberg T., Fawzy A., Nicklas W., Semmler T., Ewers C. Phylogenetic and comparative genomics of the family Leptotrichiaceae and introduction of a novel fingerprinting MLVA for Streptobacillus moniliformis. BMC Genom. 2016;17:864. doi: 10.1186/s12864-016-3206-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Facklam R. What happened to the streptococci: overview of taxonomic and nomenclature changes. Clin Microbiol Rev. 2002;15:613. doi: 10.1128/Cmr.15.4.613-630.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frank D.N., Spiegelman G.B., Davis W., Wagner E., Lyons E., Pace N.R. Culture-independent molecular analysis of microbial constituents of the healthy human outer ear. J Clin Microbiol. 2003;41:295–303. doi: 10.1128/jcm.41.1.295-303.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao L., Xu T., Huang G., Jiang S., Gu Y., Chen F. Oral microbiomes: more and more importance in oral cavity and whole body. Protein Cell. 2018;9:488–500. doi: 10.1007/s13238-018-0548-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grice E.A., Kong H.H., Conlan S., Deming C.B., Davis J., Young A.C., Program N.C.S., Bouffard G.G., Blakesley R.W., Murray P.R., Green E.D., Turner M.L., Segre J.A. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324:1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haig S.J., Quince C., Davies R.L., Dorea C.C., Collins G. The relationship between microbial community evenness and function in slow sand filters. mBio. 2015;6 doi: 10.1128/mBio.00729-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hall-Stoodley L., Hu F.Z., Gieseke A., Nistico L., Nguyen D., Hayes J., Forbes M., Greenberg D.P., Dice B., Burrows A., Wackym P.A., Stoodley P., Post J.C., Ehrlich G.D., Kerschner J.E. Direct detection of bacterial biofilms on the middle-ear mucosa of children with chronic otitis media. J Am Med Assoc. 2006;296:202–211. doi: 10.1001/jama.296.2.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heberle H., Meirelles G.V., da Silva F.R., Telles G.P., Minghim R. InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinf. 2015;16:169. doi: 10.1186/s12859-015-0611-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ho J., Tumkaya T., Aryal S., Choi H., Claridge-Chang A. Moving beyond P values: data analysis with estimation graphics. Nat Methods. 2019;16:565–566. doi: 10.1038/s41592-019-0470-3. [DOI] [PubMed] [Google Scholar]
- 25.Holdeman L.V., Cato E.P., Burmeister J.A., Moore W.E.C. Descriptions of Eubacterium timidum sp. nov., Eubacterium brachy sp. nov., and Eubacterium nodatum sp. nov. isolated from human periodontitis. Int J Syst Evol Microbiol. 1980;30:163–169. doi: 10.1099/00207713-30-1-163. [DOI] [Google Scholar]
- 26.Honjo I., Hayashi M., Ito S., Takahashi H. Pumping and clearance function of the eustachian tube. Am J Otolaryngol. 1985;6:241–244. doi: 10.1016/S0196-0709(85)80095-8. [DOI] [PubMed] [Google Scholar]
- 27.Huch M., De Bruyne K., Cleenwerck I., Bub A., Cho G.S., Watzl B., Snauwaert I., Franz C.M., Vandamme P. Streptococcus rubneri sp. nov., isolated from the human throat. Int J Syst Evol Microbiol. 2013;63:4026–4032. doi: 10.1099/ijs.0.048538-0. [DOI] [PubMed] [Google Scholar]
- 28.Huffnagle G.B., Dickson R.P., Lukacs N.W. The respiratory tract microbiome and lung inflammation: a two-way street. Mucosal Immunol. 2017;10:299–306. doi: 10.1038/mi.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ingelstedt S., Ivarsson, Jonson B. Mechanics of the human middle ear: pressure regulation in aviation and diving, a non-traumatic method. Acta Otolaryngol. 1967;228:1–58. [PubMed] [Google Scholar]
- 30.Jervis-Bardy J., Leong L.E.X., Papanicolas L.E., Ivey K.L., Chawla S., Woods C.M., Frauenfelder C., Ooi E.H., Rogers G.B. Examining the evidence for an adult healthy middle ear microbiome. mSphere. 2019;4 doi: 10.1128/mSphere.00456-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jonson B., Rundcrantz H. Posture and pressure within the internal jugular vein. Acta Otolaryngol. 1969;68:271–275. doi: 10.3109/00016486909121565. [DOI] [PubMed] [Google Scholar]
- 32.Kanehisa M., Goto S., Sato Y., Furumichi M., Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2011;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keefe D.H. Acoustical transmission-line model of the middle-ear cavities and mastoid air cells. J Acoust Soc Am. 2015;137:1877–1887. doi: 10.1121/1.4916200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim M., Oh H.S., Park S.C., Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014;64:346–351. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]
- 35.Koskinen K., Reichert J.L., Hoier S., Schachenreiter J., Duller S., Moissl-Eichinger C., Schopf V. The nasal microbiome mirrors and potentially shapes olfactory function. Sci Rep. 2018;8:1296. doi: 10.1038/s41598-018-19438-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kozich J.J., Westcott S.L., Baxter N.T., Highlander S.K., Schloss P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lane D.J. 16S/23S rRNA sequencing. In: Stackebrandt E., Goodfellow M., editors. Nucleic acid techniques in bacterial systematics. John Wiley & Sons; New York: 1991. pp. 115–175. [DOI] [Google Scholar]
- 38.Langille M.G.I., Zaneveld J., Caporaso J.G., McDonald D., Knights D., Reyes J.A., Clemente J.C., Burkepile D.E., Vega Thurber R.L., Knight R., Beiko R.G., Huttenhower C. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leclerc J.E., Doyle W.J., Karnavas W. Physiological modulation of eustachian tube function. Acta Otolaryngol. 1987;104:500–510. doi: 10.3109/00016488709128281. [DOI] [PubMed] [Google Scholar]
- 40.Lee S.H., Sorensen J.W., Grady K.L., Tobin T.C., Shade A. Divergent extremes but convergent recovery of bacterial and archaeal soil communities to an ongoing subterranean coal mine fire. ISME J. 2017;11:1447–1459. doi: 10.1038/ismej.2017.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liktor B., Szekanecz Z., Batta T.J., Sziklai I., Karosi T. Perspectives of pharmacological treatment in otosclerosis. Eur Arch Oto-Rhino-Laryngol. 2013;270:793–804. doi: 10.1007/s00405-012-2126-0. [DOI] [PubMed] [Google Scholar]
- 42.Lim D.J., Chun Y.M., Lee H.Y., Moon S.K., Chang K.H., Li J.D., Andalibi A. Cell biology of tubotympanum in relation to pathogenesis of otitis media - a review. Vaccine. 2001;19:S17–S25. doi: 10.1016/s0264-410x(00)00273-5. [DOI] [PubMed] [Google Scholar]
- 43.Liu G., Tang C.M., Exley R.M. Non-pathogenic Neisseria: members of an abundant, multi-habitat, diverse genus. Microbiology. 2015;161:1297–1312. doi: 10.1099/mic.0.000086. [DOI] [PubMed] [Google Scholar]
- 44.Liu J., Li Y., Yuan X., Lin Z. Bell’s palsy may have relations to bacterial infection. Med Hypotheses. 2009;72:169–170. doi: 10.1016/j.mehy.2008.09.023. [DOI] [PubMed] [Google Scholar]
- 45.Lozupone C., Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luers J.C., Huttenbrink K.B. Surgical anatomy and pathology of the middle ear. J Anat. 2016;228:338–353. doi: 10.1111/joa.12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.MacArthur R.H., Wilson E.O. An equilibrium theory of insular zoogeography. Evolution. 1963;17:373–387. doi: 10.2307/2407089. [DOI] [Google Scholar]
- 48.Manley G.A. An evolutionary perspective on middle ears. Hear Res. 2010;263:3–8. doi: 10.1016/j.heares.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 49.McCormack M.G., Smith A.J., Akram A.N., Jackson M., Robertson D., Edwards G. Staphylococcus aureus and the oral cavity: an overlooked source of carriage and infection? Am J Infect Contr. 2015;43:35–37. doi: 10.1016/j.ajic.2014.09.015. [DOI] [PubMed] [Google Scholar]
- 50.Minami S.B., Mutai H., Suzuki T., Horii A., Oishi N., Wasano K., Katsura M., Tanaka F., Takiguchi T., Fujii M., Kaga K. Microbiomes of the normal middle ear and ears with chronic otitis media. Laryngoscope. 2017;127:E371–E377. doi: 10.1002/lary.26579. [DOI] [PubMed] [Google Scholar]
- 51.Munoz R., Yarza P., Ludwig W., Euzeby J., Amann R., Schleifer K.H., Glockner F.O., Rossello-Mora R. Release LTPs104 of the all-species living tree. Syst Appl Microbiol. 2011;34:169–170. doi: 10.1016/j.syapm.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 52.Murphy E.C., Frick I.M. Gram-positive anaerobic cocci--commensals and opportunistic pathogens. FEMS Microbiol Rev. 2013;37:520–553. doi: 10.1111/1574-6976.12005. [DOI] [PubMed] [Google Scholar]
- 53.Namin A.W., Czerny M.S., Antisdel J.L. Facial paresis secondary to an aggressive acute bacterial rhinosinusitis. JAMA Otolaryngol Head Neck Surg. 2015;141:856–857. doi: 10.1001/jamaoto.2015.1086. [DOI] [PubMed] [Google Scholar]
- 54.Nguyen C.T., Jung W., Kim J., Chaney E.J., Novak M., Stewart C.N., Boppart S.A. Noninvasive in vivo optical detection of biofilm in the human middle ear. Proc Natl Acad Sci USA. 2012;109:9529–9534. doi: 10.1073/pnas.1201592109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ohara-Nemoto Y., Haraga H., Kimura S., Nemoto T.K. Occurrence of staphylococci in the oral cavities of healthy adults and nasal oral trafficking of the bacteria. J Med Microbiol. 2008;57:95–99. doi: 10.1099/jmm.0.47561-0. [DOI] [PubMed] [Google Scholar]
- 56.Palmer R.J., Diaz P.I., Kolenbrander P.E. Rapid succession within the Veillonella population of developing human oral biofilm in situ. J Bacteriol. 2006;188:4117. doi: 10.1128/JB.01958-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Proctor B. Anatomy of the eustachian tube. Arch Otolaryngol. 1973;97:2–8. doi: 10.1001/archotol.1973.00780010006002. [DOI] [PubMed] [Google Scholar]
- 58.Proctor D.M., Shelef K.M., Gonzalez A., Davis C.L., Dethlefsen L., Burns A.R., Loomer P.M., Armitage G.C., Ryder M.I., Millman M.E., Knight R., Holmes S.P., Relman D.A. Microbial biogeography and ecology of the mouth and implications for periodontal diseases. Periodontol. 2020;82:26–41. doi: 10.1111/prd.12268. 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glockner F.O. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ravva S.V., Sarreal C.Z., Mandrell R.E. Bacterial communities in aerosols and manure samples from two different dairies in central and Sonoma valleys of California. PloS One. 2011;6 doi: 10.1371/journal.pone.0017281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reddy M.S., Murphy T.F., Faden H.S., Bernstein J.M. Middle ear mucin glycoprotein: purification and interaction with nontypable Haemophilus influenzae and Moraxella catarrhalis. Otolaryngol Head Neck Surg. 1997;116:175–180. doi: 10.1016/S0194-59989770321-8. [DOI] [PubMed] [Google Scholar]
- 62.Rideout J.R., He Y., Navas-Molina J.A., Walters W.A., Ursell L.K., Gibbons S.M., Chase J., McDonald D., Gonzalez A., Robbins-Pianka A., Clemente J.C., Gilbert J.A., Huse S.M., Zhou H.W., Knight R., Caporaso J.G. Subsampled open-reference clustering creates consistent, comprehensive OTU definitions and scales to billions of sequences. PeerJ. 2014;2:e545. doi: 10.7717/peerj.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rios-Covian D., Salazar N., Gueimonde M., de los Reyes-Gavilan C.G. Shaping the metabolism of intestinal Bacteroides population through diet to improve human health. Front Microbiol. 2017;8 doi: 10.3389/fmicb.2017.00376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rossi-Tamisier M., Benamar S., Raoult D., Fournier P.E. Cautionary tale of using 16S rRNA gene sequence similarity values in identification of human-associated bacterial species. Int J Syst Evol Microbiol. 2015;65:1929–1934. doi: 10.1099/ijs.0.000161. [DOI] [PubMed] [Google Scholar]
- 65.Ruoff K.L. Miscellaneous catalase-negative, gram-positive cocci: emerging opportunists. J Clin Microbiol. 2002;40:1129–1133. doi: 10.1128/jcm.40.4.1129-1133.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sade J. Mucociliary flow in the middle ear. Ann Otol Rhinol Laryngol. 1971;80:336–341. doi: 10.1177/000348947108000306. [DOI] [PubMed] [Google Scholar]
- 67.Sadler-Kimes D., Siegel M.I., Todhunter J.S. Age-related morphologic differences in the components of the eustachian tube/middle ear system. Ann Otol Rhinol Laryngol. 1989;98:854–858. doi: 10.1177/000348948909801104. [DOI] [PubMed] [Google Scholar]
- 68.Silva V.L., Carvalho M.A., Nicoli J.R., Farias L.M. Aerotolerance of human clinical isolates of Prevotella spp. J Appl Microbiol. 2003;94:701–707. doi: 10.1046/j.1365-2672.2003.01902.x. [DOI] [PubMed] [Google Scholar]
- 69.Sloan W.T., Lunn M., Woodcock S., Head I.M., Nee S., Curtis T.P. Quantifying the roles of immigration and chance in shaping prokaryote community structure. Environ Microbiol. 2006;8:732–740. doi: 10.1111/j.1462-2920.2005.00956.x. [DOI] [PubMed] [Google Scholar]
- 70.Stackebrandt E., Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today. 2006;33:152–155. [Google Scholar]
- 71.Sziklai I., Batta T.J., Karosi T. Otosclerosis: an organ-specific inflammatory disease with sensorineural hearing loss. Eur Arch Oto-Rhino-Laryngol. 2009;266:1711–1718. doi: 10.1007/s00405-009-0967-y. [DOI] [PubMed] [Google Scholar]
- 72.Tailford L.E., Crost E.H., Kavanaugh D., Juge N. Mucin glycan foraging in the human gut microbiome. Front Genet. 2015;6:81. doi: 10.3389/fgene.2015.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tetzlaff K., Thomas P.S. Short- and long-term effects of diving on pulmonary function. Eur Respir Rev. 2017;26 doi: 10.1183/16000617.0097-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tonnaer E.L., Mylanus E.A., Mulder J.J., Curfs J.H. Detection of bacteria in healthy middle ears during cochlear implantation. Arch Otolaryngol Head Neck Surg. 2009;135:232–237. doi: 10.1001/archoto.2008.556. [DOI] [PubMed] [Google Scholar]
- 75.Tsao R.Y., Lutwick L. Non-hemolytic group B streptococcus as a cause of chemotherapy port infection. IDCases. 2017;10:53–54. doi: 10.1016/j.idcr.2017.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Venkataraman A., Bassis C.M., Beck J.M., Young V.B., Curtis J.L., Huffnagle G.B., Schmidt T.M. Application of a neutral community model to assess structuring of the human lung microbiome. mBio. 2015;6 doi: 10.1128/mBio.02284-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Weber N., Liou D., Dommer J., MacMenamin P., Quiñones M., Misner I., Oler A.J., Wan J., Kim L., Coakley McCarthy M., Ezeji S., Noble K., Hurt D.E. Nephele: a cloud platform for simplified, standardized and reproducible microbiome data analysis. Bioinformatics. 2017;34:1411–1413. doi: 10.1093/bioinformatics/btx617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Welch J.L.M., Dewhirst F.E., Borisy G.G. Biogeography of the oral microbiome: the site-specialist hypothesis. Annu Rev Microbiol. 2019 doi: 10.1146/annurev-micro-090817-062503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xu J., Platt T.G., Fuqua C. Regulatory linkages between flagella and surfactant during swarming behavior: lubricating the flagellar propeller? J Bacteriol. 2012;194:1283–1286. doi: 10.1128/JB.00019-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yarza P., Richter M., Peplies J., Euzeby J., Amann R., Schleifer K.H., Ludwig W., Glockner F.O., Rossello-Mora R. The All-Species Living Tree project: a 16S rRNA-based phylogenetic tree of all sequenced type strains. Syst Appl Microbiol. 2008;31:241–250. doi: 10.1016/j.syapm.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 81.Yeung A.T., Parayno A., Hancock R.E. Mucin promotes rapid surface motility in Pseudomonas aeruginosa. mBio. 2012;3 doi: 10.1128/mBio.00073-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McGuire Surfactant in the middle ear and eustachian tube: a review. Int J Pediatr Otorhinolaryngol. 2002;66:1–15. doi: 10.1016/S0165-5876(02)00203-3. [DOI] [PubMed] [Google Scholar]
- 83.Wu Surfactant proteins A and D inhibit the growth of Gram-negative bacteria by increasing membrane permeability. J Clin Invest. 2003;111:1589–1602. doi: 10.1172/JCI16889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Edgar MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinf. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Edgar MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kumar Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequence reads have been deposited in the NCBI Sequence Read Archive under BioProject ID: PRJNA473788.