Abstract
Background
Phylogenetic analysis can be used to assess human immunodeficiency virus (HIV) transmission in populations. We inferred the direction of HIV transmission using whole-genome HIV sequences from couples with known linked infection and known transmission direction.
Methods
Complete next-generation sequencing (NGS) data were obtained for 105 unique index–partner sample pairs from 32 couples enrolled in the HIV Prevention Trials Network (HPTN) 052 study (up to 2 samples/person). Index samples were obtained up to 5.5 years before partner infection; partner samples were obtained near the time of seroconversion. The bioinformatics method, phyloscanner, was used to infer transmission direction. Analyses were performed using samples from individual sample pairs, samples from all couples (1 sample/person; group analysis), and all available samples (multisample group analysis). Analysis was also performed using NGS data from defined regions of the HIV genome (gag, pol, env).
Results
Using whole-genome NGS data, transmission direction was inferred correctly (index to partner) for 98 of 105 (93.3%) of the individual sample pairs, 99 of 105 (94.3%) sample pairs using group analysis, and 31 of the 32 couples (96.9%) using multisample group analysis. There were no cases where the incorrect transmission direction (partner to index) was inferred. The accuracy of the method was higher with greater time between index and partner sample collection. Pol region sequences performed better than env or gag sequences for inferring transmission direction.
Conclusions
We demonstrate the potential of a phylogenetic method to infer the direction of HIV transmission between 2 individuals using whole-genome and pol NGS data.
Keywords: HIV, direction of transmission, phylogenetic analysis, next-generation sequencing, HPTN 052
We demonstrated that use of ultra-deep, whole-genome HIV sequence data for phyloscanner analysis improves accuracy for inferring the direction of HIV transmission. Pol region sequences performed better than env/gag sequences for inferring transmission direction.
(See the Editorial Commentary by Taylor and Sapién on pages 38–40.)
Significant advances have been made in identifying and implementing interventions for prevention of human immunodeficiency virus (HIV) infection, including early initiation of antiretroviral therapy [1, 2] and preexposure prophylaxis with antiretroviral drugs [3]. Further advances in HIV prevention may be facilitated by understanding the dynamics of HIV transmission at the individual and population levels. Because of the high levels of HIV genetic variation, phylogenetic methods can be used to identify genetically linked infections and infection clusters, providing insight into HIV transmission patterns [4–6].
Use of phylogenetic methods to identify “source” infections could help identify factors associated with HIV transmission [7], which could inform the design and implementation of HIV prevention interventions [8]. The potential to infer direction of transmission using phylogenetic methods has been explored using different sequencing and analysis methods [9–12]. Two recent studies used the bioinformatics method phyloscanner for this research [11, 13]. A study from our group used next-generation sequencing (NGS) data from the HIV env region to infer transmission direction [11]. That study analyzed samples from 33 index–partner pairs enrolled in the HIV Prevention Trials Network (HPTN) 052 study who had known linked infection and known transmission direction [2]. Transmission direction was inferred correctly in 55%–74% couples and incorrectly in 13%–21% couples, depending on analysis methods; no direction was inferred in the remaining cases [11]. In another study, phyloscanner was used to analyze whole-genome NGS data in a large sample set from Uganda; the false discovery rate for known transmission pairs (the proportion of sample pairs with incorrect transmission direction) was 16.3%, which may reflect relatively low sequencing depth and incomplete sequences in that data set [13].
In this report, we evaluated whether use of high-quality, ultra-deep whole-genome NGS data could improve accuracy for inferring HIV transmission direction. We used samples from index–partner pairs from HPTN 052. Most participants had samples available from 2 different study visits. This allowed us to assess whether inclusion of data from 2 samples per person improved the accuracy for inferring HIV transmission direction. We also compared results obtained using whole-genome data to results obtained using data from the gag, pol, or env genes.
METHODS
Study Cohort
Plasma samples were obtained from the HPTN 052 study, which evaluated the impact of early ART initiation on transmission from HIV-positive adults (“indexes”) to their HIV-negative sexual partners (“partners”) [2]. Samples were available from 32 couples who were previously confirmed to have genetically linked HIV infections [6, 14]. Four sample types were tested: index early samples (collected months or years before the partner’s seroconversion visit [n = 32]); index late samples (collected on a date close to the partner’s seroconversion visit [n = 31]); partner early samples (collected at the first HIV-positive visit [n = 32]); and partner late samples (collected weeks after the first HIV-positive visit [n = 29]).
Sample Preparation, Sequencing, and Data Processing
HIV RNA was extracted using the ViroSeq HIV-1 Genotyping System (Abbott Molecular). Viral RNA was reverse-transcribed using Superscript IV Reverse Transcriptase (Invitrogen) with 4 HIV-specific primers [15]. The complementary DNA was used to generate 4 overlapping amplicons (amplicon length: 1.9 kb, 3.6 kb, 3.0 kb, and 3.5 kb) [16]. A seminested polymerase chain reaction (PCR) assay was performed if the initial PCR failed [17]. Samples were not analyzed if amplification failed in any region. The 4 amplicons were pooled together for sequencing using 5 μL for the 1.9-kb amplicon and 10 μL for each of the other 3 amplicons. Primers are shown in Supplementary File 1. Samples were sequenced using MiSeq (2 × 250 cycles) with the MiSeq reagent kit version 3 (Illumina). NGS data were deposited in the National Center for Biotechnology Information’s sequence read archive (PRJNA588392).
For each sample, sequencing reads were first assembled into contigs using the iterative virus assembler method [18]. Contigs generated from raw reads were processed by SHIVER [19]. The resulting files of mapped reads were used for analysis. Consensus sequences from each sample were subtyped using the REGA subtyping tool (http://dbpartners.stanford.edu:8080/RegaSubtyping/stanford-hiv/typingtool).
Phylogenetic Analysis
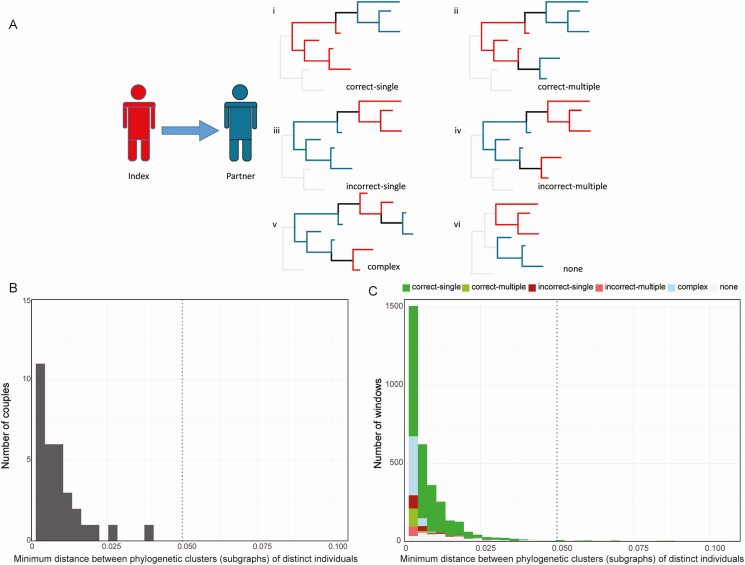
Phyloscanner (version 1.4.0) was used to analyze NGS data and infer the direction of transmission for each couple [12]. The command line specification is shown in Supplementary File 2. Phyloscanner infers phylogenies of the sampled reads and the ancestral host states in these phylogenies in discrete genomic windows [9]. Overlapping windows covered the whole HIV genome. In each window, all merged paired-end reads spanning the window were processed for tree construction. Transmission direction was inferred using windows of different widths (280–400 bp in increments of 20 bp). Six possible topological relationships were identified in each window for each sample pair: single ancestry with the correct transmission direction (correct-single); multiple ancestry with the correct transmission direction (correct-multiple); single ancestry with the incorrect transmission direction (incorrect-single); multiple ancestry with the incorrect transmission direction (incorrect-multiple); complicated relationship for which ancestry cannot be determined (complex); and no ancestry identified (none) (Figure 1A) [12]. An index–partner sample pair was classified as “linked” by this methodology if ≥ 50% of the windows had an ancestral or complex relationship with minimum subgraph distance < 0.05 substitutions per site. The inferred transmission direction was considered to be “correct” if the index and partner were “linked,” and if the correct ancestral relationship (index-to-partner) was present in ≥ 37.5% of windows with a minimum subgraph distance of < 0.05 substitutions per site. The inferred transmission direction was considered to be “incorrect” if the index and partner were “linked,” and if the incorrect ancestral relationship (partner-to-index) was present in ≥ 37.5% of windows with a minimum subgraph distance of < 0.05 substitutions per site. Sample pairs that did not meet these criteria were classified as “unknown.”
Figure 1.
Classes of topological relationships for index–partner couples and distance between phylogenetic clusters from different individuals and samples. A, Diagrams show examples of topological relationships that might be observed for sequences from index participants (red) and partners (blue); in the cases analyzed, human immunodeficiency virus was transmitted from the index to the partner. “Correct-single” indicates single ancestry with the correct direction of transmission (i); “correct-multiple” indicates multiple ancestry (transmission of multiple viral strains) with the correct transmission direction (ii); “incorrect-single” indicates single ancestry with the inverse (incorrect) transmission direction (iii); “incorrect-multiple” indicates multiple ancestry with the inverse (incorrect) transmission direction (iv); “complex” indicates that the relationship between the 2 hosts was too complex to predict transmission direction (v); “none” indicates no ancestry (vi). B, The histogram shows the distribution over the couples of the minimum subgraph distance obtained using phyloscanner; this analysis included data from 32 couples. All couples had mean minimum subgraph distance < 0.05 substitutions per site (indicated with a dotted line). C, The stacked bar graph shows the number of windows as a function of minimum subgraph distance; this analysis included data from 4607 windows from 105 index–partner sample pairs. The majority of windows (97.2%) had minimum subgraph distance < 0.05 substitutions per site (indicated with a dotted line). Colors indicate windows with the following results: single correct ancestry (correct-single), multiple correct ancestry (correct-multiple), single inverse ancestry (incorrect-single), multiple inverse ancestry (incorrect-multiple), complex (complex), and no relationship (none).
Transmission Direction Inferred Using Whole-genome NGS Data and Data From Defined Regions
Three approaches were used to analyze NGS data (Table 1). Sample pair analysis analyzed NGS data from a single sample from each index participant and a single sample from the corresponding partner; this approach requires knowledge of the relationships between study participants (ie, if 2 participants were a couple). Group analysis also included NGS data from a single sample for each participant, but combined data from all 32 couples; samples were selected based on the timing of sample collection and included 4 sets of samples: all index early samples plus all partner early samples (32 couples); all index early samples plus all partner late samples (24 couples); all index late samples plus all partner early samples (28 couples); and all index late samples plus all partner late samples (21 couples). Multisample group analysis was performed using the same approach as group analysis, but included all available NGS data from all 32 couples (data from 1 or 2 samples per person); this approach increased the number of NGS sequences analyzed for each individual.
Table 1.
Selection of Samples for phyloscanner Analysis
| Analysis Type | No. of Couples Included in Each phyloscanner Run | Samples Included in Each phyloscanner Run | Comments |
|---|---|---|---|
| Sample pair analysis | 1 couple | 1 sample per person | This approach required knowledge of the relationship between study participants (index–partner); a separate phyloscanner run was performed for each of the 105 possible sample pairs |
| Group analysis | All 32 couples | 1 sample per person | Samples were selected based on timing of sample collection; 4 phyloscanner runs were performed, with a separate run for each set of sample pairs (early/early, late/late, early/late, late/early); this approach provided data for all 105 possible sample pairs and did not require knowledge of the relationship between study participants |
| Multisample group analysis | All 32 couples | All available samples (1–2 samples/person) | This approach was the same as group analysis, but all data were included in a single phyloscanner run (data from 1–2 samples for each individual); this analysis did not provide data for individual sample pairs |
This table describes the approach used to select samples for each type of analysis (sample pair analysis; group analysis; multi-sample group analysis).
In a separate analysis, phyloscanner was also used to analyze NGS data from individual HIV genes (gag, pol, and env); this analysis was performed using a window width of 340 bp. The same threshold settings were used for analysis of whole-genome NGS data and NGS data from individual genes. The analysis was also performed using data from a short fragment of env gene (HXB2 coordinates: 7941–8264) that were used for analysis in a previous report [11].
Ethics Statement
The HPTN 052 study protocol was approved by the institutional review board or ethics committee at each study site, as well as by other local regulatory bodies. All study participants provided written informed consent.
RESULTS
Samples Used in Analysis
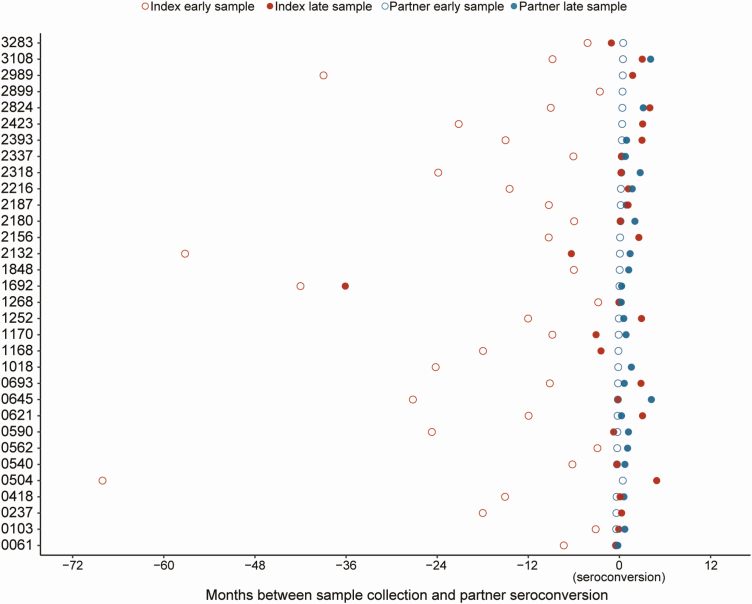
Amplification and sequencing were successful for 116 of the 124 (93.5%) samples from 32 couples (Figure 2). The data set included NGS data from 105 unique index–partner sample pairs (1 index sample plus 1 partner sample). The viral loads of the 116 samples ranged from 691 to > 750 000 copies/mL (median, 70 526 copies/mL). HIV subtypes were the same for each index–partner pair and were consistent with previously reported results [14]; the subtypes included C (80.2%), B (10.3%), A1 (6.0%), and D (3.4%). The timing of sample collection for each couple is shown in Figure 2. The median times between collection of paired samples were as follows: (1) early index samples and early partner samples, 360 days (range, 84–2055 days); (2) early index samples and late partner samples, 378 days (range, 91–1758 days); (3) late index samples and early partner samples, 38.5 days (range, 0–1083 days); (4) late index samples and late partner samples, 28.5 days (range, 1–126 days).
Figure 2.
The plot shows the timing of sample collection for 32 couples. Index participants were human immunodeficiency virus (HIV) positive at study enrollment; partners were HIV negative at study enrollment. Next-generation sequencing data were obtained for 105 unique sample pairs (32 index early/partner early; 24 index early/partner late; 28 index late/partner early; 21 index late/partner late). Data from each couple are shown in 1 row; couple identifiers are shown on the y-axis. The x-axis shows the number of months between sample collection and partner seroconversion; negative numbers indicate that samples were collected before partner seroconversion. Partner samples used for these analyses were collected at the first HIV-positive visit (early) and at a subsequent study visit (late) at/after time of seroconversion. Index samples used for these analyses were collected at an earlier study visit (early) or near the time of partner seroconversion (late). Index samples are shown in red; partner samples are shown in blue. Early samples are shown with open circles; late samples are shown with filled circles.
Topological Relationships
A high average read depth was obtained for most of the samples (Supplementary File 3). The average depth in the region used for phylogenetic analysis among all 116 samples (HXB2 coordinates: 520–9480) was 34 758X. Phyloscanner was then used to determine the topological relationship between paired individuals for each genomic window (Figure 1A). The “minimum subgraph distance” between individuals was determined for each window based on the ancestral state reconstruction of hosts on the phylogeny. Subgraphs are defined as the sets of tips and internal nodes of a phylogeny that are reconstructed as belonging to the same individual [12]. Distance is defined as the shortest patristic distance between 2 nodes, 1 from each individual in a potential transmission pair [12]. For each couple, we determined the mean minimum subgraph distance across all of the windows spanning the HIV genome (Figure 1B); this value reflects the genetic relationship between the HIV strains from the 2 individuals analyzed. The mean subgraph distance for all 32 couples was < 0.05 substitutions per site, indicating close relatedness of sequences from all index–partner pairs. Figure 1C also shows the distribution of minimum subgraph distances for all sample pairs, grouped by the topological relationship of each couple. Most windows (97.2%) had a minimum subgraph distance of < 0.05; 51.4% of those windows had single correct ancestry (51.4%, Figure 1C). Minimum subgraph distances < 0.05 were also observed for pairs with complex relationships (18.4%), multiple correct ancestry (15.4%), and no relationship determined (none, 8.2%). Both single correct ancestry and multiple correct ancestry were considered to be evidence of a correct ancestral topological relationship for a given window (index to partner transmission). Only 6.7% of all windows had an incorrect ancestry inferred (partner-to-index transmission, with single or multiple incorrect ancestry in 3.6% and 3.1% of cases, respectively). The distribution of topologies obtained for each sample is shown in Supplementary File 4.
Inferred Transmission Direction Using Whole-genome NGS Data
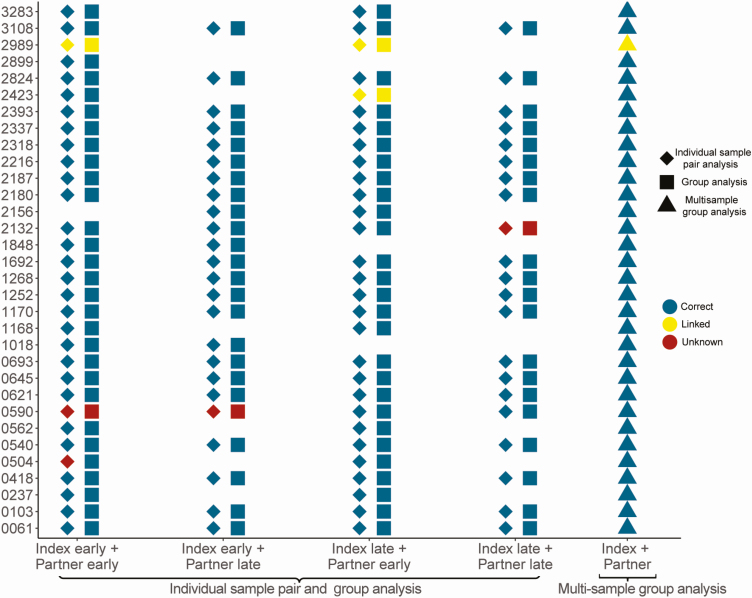
We first analyzed individual samples from known couples using whole-genome NGS data for the 105 unique sample pairs. Each analysis included a single index sample and a single partner sample, to prevent inference from sequences from other individuals who may have been part of the same transmission network. Overall, correct transmission direction was inferred using this method in at least 95 of the 105 sample pairs using any window width from 300 bp to 360 bp (Supplementary File 5A). The fraction of pairs with the correct transmission direction was 93.3% (98/105 sample pairs) using a window width of 320 bp (Figure 3; Supplementary File 5A). Four sample pairs had unknown status; the remaining 3 sample pairs were classified as linked, with no transmission direction inferred. In 1 pair of samples (#0504, index early and partner early), the correct transmission direction was inferred using 280-bp windows, but this pair had unknown status with wider windows.
Figure 3.
Inferred direction of transmission using different sample sets. The plot shows the inferred relationship between sequences from each couple based on analysis of data from different sample sets using a window width of 320 bp (individual sample pair analysis and group) and 380 bp (multisample group analysis). Each symbol (diamond, square, or triangle) represents the inferred relationship based on a single analysis; data from each couple are shown in 1 row, with participant identifier numbers shown on the y-axis. Diamonds show results obtained using data from individual sample pairs (1 sample per participant; 1 couple per analysis); squares show results obtained using group analysis (samples selected based on the timing of sample collection; all couples combined); triangles show results obtained using multisample group analysis (up to 2 samples per participant, all couples combined). Colors of symbols correspond to the inferred transmission direction: blue symbols indicate the correct inferred direction; yellow symbols indicate pairs that were classified as linked with no direction inferred; red symbols indicate that the analysis did not yield a result for linkage or transmission direction. Blank spaces indicate that the specific combination of samples (eg, index early+partner late) was not available for analysis.
We also performed group analysis, which included data from all couples with 1 sample per person in a single run. Using window widths of 320 bp or 340 bp, the correct transmission direction was inferred in 99 of the 105 (94.3%) sample pairs (Figure 3; Supplementary File 5B). The results from the remaining 6 pairs matched those from the individual pair analysis (3 status unknown; 3 linked with no direction inferred).
As a final step, we performed multisample group analysis, which combined NGS data from up to 2 samples per person; this approach provided more sequence data for each participant. Using 380-bp and 400-bp window widths, transmission direction was correctly inferred for 31 of the 32 (96.9%) couples (Figure 3; Supplementary File 5B). In 1 couple (#2989), transmission direction was not inferred using any of the 3 approaches, even though the viral sequences from this couple had been confirmed to be linked in a previous study [11]. In 3 couples (#2423, #2132, and #0590), individual sample pair analysis and group analysis did not infer transmission direction, while multisample group analysis established the correct direction.
Inferred Transmission Direction Using NGS Data From Individual Gene Segments
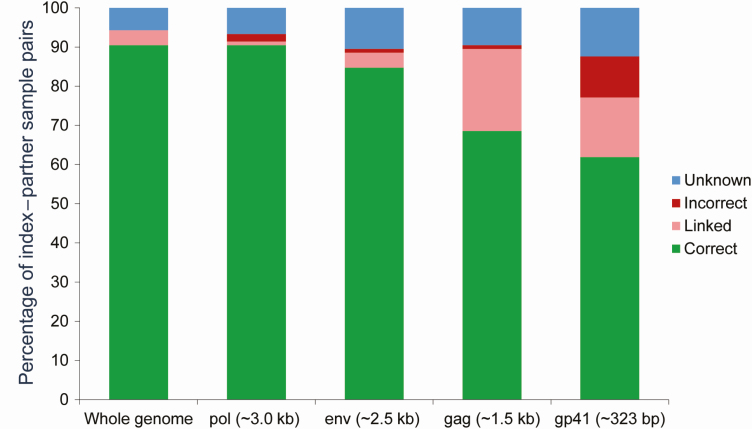
In addition, we evaluated the accuracy of the inferred transmission direction using NGS sequence data from specific genomic regions (gag, pol, or env gene, or short fragment of env) [11]. Those results were compared to results from whole-genome NGS (Figure 4). The same criteria were used to infer transmission direction in these analysis that were used for whole-genome analysis (see Methods). Using the window width of 340 bp, the correct transmission was inferred in 90.5% of the sample pairs using pol sequences, 88.4% using env sequences, 68.6% using gag sequences, and 61.9% using sequences from the short env fragment. One or 2 sample pairs had incorrect transmission direction inferred using NGS sequencing from individual genes; this was not observed using whole-genome NGS data. Using the short env fragment, > 10% of sample pairs had the incorrect transmission direction inferred, which was similar to the percentage of pairs with incorrect transmission direction inferred in a prior study (range, 13%–21%), that used the same env fragment with an overlapping sample set and independent sequence data set [11].
Figure 4.
Inferred direction of transmission using whole-genome sequences compared to gag, pol, and env sequences. A comparison of phyloscanner results using next-generation sequencing (NGS) data from single genes (gag, pol, or env) or NGS data from the whole human immunodeficiency virus genome. The plot shows the percentage of sample pairs with the correct/incorrect inferred transmission direction (green bars/red bars). Peach bars indicate pairs that were classified as linked with no direction determined. Blue bars indicate that the analysis did not yield a result for linkage or transmission direction. The analysis was performed using a window width of 340 bp with sequences from 105 sample pairs; the same transmission inference criteria were used for the whole genome and for single genes.
DISCUSSION
Previous studies have used whole-genome NGS data and env NGS data to infer transmission direction; those studies correctly inferred transmission direction in most cases (55%–74% and 83.7% of couples [11, 13]). However, in both studies, the incorrect transmission direction was inferred in several cases (13%–21% and 16.3% [11, 13]). In contrast, our analysis, which used ultra-deep whole-genome NGS data, inferred the correct transmission direction in 31 of 32 (96.9%) couples with no incorrect results. The methods used for this analysis did not require use of clinical or epidemiology data or assumptions about index or partner characteristics [20–22].
An important variable in the analysis was the width of the genomic windows used to determine topological relationships between index and partner sequences. Shorter window widths allow inclusion of more sequence data, but may not include enough information about within-host diversity to infer transmission direction. In contrast, longer window widths may limit the number of sequencing reads available for analysis, since sequence reads from a sample pair must fully span a window in the phyloscanner analysis; loss of data increases the likelihood that no direction of transmission will be inferred [11]. Because high average read depth and sequencing coverage were achieved for most samples, a wider window width was used in this study compared to previous work (250 bp) [13]; this could explain the higher accuracy for inferring transmission direction. We also observed that sample pairs with longest time intervals had the highest percentage of windows with correct ancestry at any window width (Supplementary File 6). This trend may not be observed in other settings and populations. As viral populations evolve over time in individuals with source and recipient infections, the distance between subgraphs may increase, impacting the ability to accurately infer transmission direction.
Phyloscanner can use information from phylogenies across the whole genome, which decreases the impact of random error and increases accuracy. The accuracy of the inferred transmission direction was higher using whole-genome sequences (93.3%) or pol sequences (90.5%), compared to env sequences (88.4%), gag sequences (68.6%), or a short env fragment (61.9%). The accuracy using the short env fragment was lower in this study than in a previous study that included 24 of the same couples (74%) [11]. This may reflect use of different sequencing methods (amplicon sequencing [11] vs shotgun sequencing in this report). It may also reflect the inclusion of different samples and/or couples in the 2 studies.
These findings demonstrate that whole-genome NGS data can accurately infer the direction of transmission in couples with recent transmission. Of note, the first stages of the analysis in this report were performed without input from the authors who developed the SHIVER and phyloscanner methods (C. W., M. H. and C. F.); this reduced the investigator bias.
There were several limitations in this study. First, the number of couples analyzed was relatively small. Second, all of the partners analyzed in this report were likely to have acquired HIV through sexual transmission; further research is needed to determine whether these methods are accurate for inferring transmission direction in other risk groups [23], since transmission mode may influence the diversity of HIV in newly infected individuals [24]. Third, our results were only obtained from couples with recent partner infections; in HPTN 052, partners were not followed after seroconversion was confirmed. Fourth, the HPTN 052 study enrolled serodiscordant couples in stable sexual partnerships; we cannot exclude the possibility that unsampled individuals were the source of the partner’s infection. Fifth, most of the couples in this study had subtype C HIV infection; further research is needed to determine if our findings are related to viral subtypes. Finally, most current molecular HIV surveillance systems use data from HIV drug resistance testing, which is usually obtained using population/bulk sequencing; those data cannot be used for the type of analysis described in this report. Large, comprehensive sets of ultra-deep NGS data would be needed to use these methods for molecular surveillance.
While use of phylogenetic methods to infer transmission direction has important research applications, it raises serious ethical concerns at both the group and individual level [25]. Activities that could expose other individuals to HIV risk are criminalized in several states in the United States [26, 27] and in other countries. Care is needed to avoid releasing data on HIV transmission direction in settings where results could be linked to individuals, as this could result in social harm, stigma, and legal penalties, including incarceration. For these reasons, anonymization of samples and sequence data is recommended when using methods to infer transmission direction. More extensive evaluation of these methods is also needed before results from this type of analysis are used in legal settings [28].
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Author contributions. Y. Z. and S. H. E. designed the study, analyzed the data, and prepared the manuscript. Y. Z. performed laboratory testing. C. W. and M. H. provided advice on use of phyloscanner software. C. F. provided advice on phylogenetic analysis. O. L. and M. K. G. provided input into data presentation. S. H. assisted with sample selection. E. P. M. is the HPTN Laboratory Center quality assurance/quality control representative for HPTN 052. M. M. and T. G. are HPTN 052 study managers. M. C. H., N. K., J. G. H., J. K., V. A., B. R. S., B. G., J. H. P., S. C., and J. M. are principal investigators for different HPTN 052 sites. Y. Q. C. is the statistician for HPTN 052. M. S. C. is the HPTN 052 protocol chair. All of the authors contributed to manuscript preparation and reviewed the manuscript before publication.
Acknowledgments. The authors thank the HIV Prevention Trials Network (HPTN) 052 study team and participants for providing the samples and data used in this study; Ethan Wilson and Victor Akelo for reviewing the paper and providing comments; and the laboratory staff who helped with sample management and testing.
Disclaimer. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Financial support. This work was supported by the Division of AIDS of the US National Institute of Allergy and Infectious Diseases (NIAID); and by the Office of AIDS Research of the US National Institutes of Health (grant numbers UM1-AI068613 to S. H. E., UM1-AI068617 to Deborah J. Donnell, and UM1-AI068619 to Wafaa M. El-Sadr). C. F. has received a research grant for the Phylogenetics and Networks for Generalized HIV Epidemics in Africa consortium from the Bill & Melinda Gates Foundation. Additional support was provided by the Division of Intramural Research, NIAID.
Potential conflicts of interest. M. C. reports travel and advisory board fees from Gilead and Merck, honoraria for editorial work from UpToDate, and honoraria for educational material development from MedScape, outside the submitted work. All other authors report no potential conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Cohen MS, Chen YQ, McCauley M, et al. HPTN 052 Study Team Antiretroviral therapy for the prevention of HIV-1 transmission. N Engl J Med 2016; 375:830–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cohen MS, Chen YQ, McCauley M, et al. HPTN 052 Study Team Prevention of HIV-1 infection with early antiretroviral therapy. N Engl J Med 2011; 365:493–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Desai M, Field N, Grant R, McCormack S. Recent advances in pre-exposure prophylaxis for HIV. BMJ 2017; 359:j5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hassan AS, Pybus OG, Sanders EJ, Albert J, Esbjörnsson J. Defining HIV-1 transmission clusters based on sequence data. AIDS 2017; 31:1211–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hué S, Clewley JP, Cane PA, Pillay D. HIV-1 pol gene variation is sufficient for reconstruction of transmissions in the era of antiretroviral therapy. AIDS 2004; 18:719–28. [DOI] [PubMed] [Google Scholar]
- 6. Eshleman SH, Hudelson SE, Redd AD, et al. Treatment as prevention: characterization of partner infections in the HIV Prevention Trials Network 052 trial. J Acquir Immune Defic Syndr 2017; 74:112–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dennis AM, Herbeck JT, Brown AL, et al. Phylogenetic studies of transmission dynamics in generalized HIV epidemics: an essential tool where the burden is greatest? J Acquir Immune Defic Syndr 2014; 67:181–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pasquale DK, Doherty IA, Sampson LA, et al. Leveraging phylogenetics to understand HIV transmission and partner notification networks. J Acquir Immune Defic Syndr 2018; 78:367–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Romero-Severson EO, Bulla I, Leitner T. Phylogenetically resolving epidemiologic linkage. Proc Natl Acad Sci U S A 2016; 113:2690–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Leitner T, Romero-Severson E. Phylogenetic patterns recover known HIV epidemiological relationships and reveal common transmission of multiple variants. Nat Microbiol 2018; 3:983–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rose R, Hall M, Redd AD, et al. Phylogenetic methods inconsistently predict direction of HIV transmission among heterosexual pairs in the HPTN052 cohort. J Infect Dis 2019; 220:1406–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wymant C, Hall M, Ratmann O, et al. Phyloscanner: inferring transmission from within- and between-host pathogen genetic diversity. Mol Biol Evol 2017; 35:719–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ratmann O, Grabowski MK, Hall M, et al. PANGEA Consortium and Rakai Health Sciences Program Inferring HIV-1 transmission networks and sources of epidemic spread in Africa with deep-sequence phylogenetic analysis. Nat Commun 2019; 10:1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Eshleman SH, Hudelson SE, Redd AD, et al. Analysis of genetic linkage of HIV from couples enrolled in the HIV Prevention Trials Network 052 trial. J Infect Dis 2011; 204:1918–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Berg MG, Yamaguchi J, Alessandri-Gradt E, Tell RW, Plantier JC, Brennan CA. A pan-HIV strategy for complete genome sequencing. J Clin Microbiol 2016; 54:868–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gall A, Ferns B, Morris C, et al. Universal amplification, next-generation sequencing, and assembly of HIV-1 genomes. J Clin Microbiol 2012; 50:3838–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cousins MM, Donnell D, Eshleman SH. Impact of mutation type and amplicon characteristics on genetic diversity measures generated using a high-resolution melting diversity assay. J Mol Diagn 2013; 15:130–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hunt M, Gall A, Ong SH, et al. IVA: accurate de novo assembly of RNA virus genomes. Bioinformatics 2015; 31:2374–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wymant C, Blanquart F, Golubchik T, et al. Easy and accurate reconstruction of whole HIV genomes from short-read sequence data with SHIVER. Virus Evol 2018; 4:vey007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. de Oliveira T, Kharsany AB, Gräf T, et al. Transmission networks and risk of HIV infection in KwaZulu-Natal, South Africa: a community-wide phylogenetic study. Lancet HIV 2017; 4:e41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kenah E, Britton T, Halloran ME, Longini IM Jr. Molecular infectious disease epidemiology: survival analysis and algorithms linking phylogenies to transmission trees. PLoS Comput Biol 2016; 12:e1004869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ratmann O, van Sighem A, Bezemer D, et al. Sources of HIV infection among men having sex with men and implications for prevention. Sci Transl Med 2016; 8:320ra2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Leitner T. Phylogenetics in HIV transmission: taking within-host diversity into account. Curr Opin HIV AIDS 2019; 14:181–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bar KJ, Li H, Chamberland A, et al. Wide variation in the multiplicity of HIV-1 infection among injection drug users. J Virol 2010; 84:6241–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Coltart CEM, Hoppe A, Parker M, et al. Ethics in HIV Phylogenetics Working Group Ethical considerations in global HIV phylogenetic research. Lancet HIV 2018; 5:e656–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cann D, Harrison SE, Qiao S. Historical and current trends in HIV criminalization in South Carolina: implications for the southern HIV epidemic. AIDS Behav 2019; 23:233–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lehman JS, Carr MH, Nichol AJ, et al. Prevalence and public health implications of state laws that criminalize potential HIV exposure in the United States. AIDS Behav 2014; 18:997–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Abecasis AB, Pingarilho M, Vandamme AM. Phylogenetic analysis as a forensic tool in HIV transmission investigations. AIDS 2018; 32:543–54. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.