ABSTRACT
Negative geotaxis (climbing) performance is a useful metric for quantifying Drosophila health. Manual methods to quantify climbing performance are tedious and often biased, while many available computational methods have challenging hardware or software requirements. We present an alternative: FreeClimber. This open source, Python-based platform subtracts a video's static background to improve detection for flies moving across heterogeneous backgrounds. FreeClimber calculates a cohort's velocity as the slope of the most linear portion of a mean vertical position versus time curve. It can run from a graphical user interface for optimization or a command line interface for high-throughput and automated batch processing, improving accessibility for users with different expertise. FreeClimber outputs calculated slopes, spot locations for follow-up analyses (e.g. tracking), and several visualizations and plots. We demonstrate FreeClimber's utility in a longitudinal study for endurance exercise performance in Drosophila mitonuclear genotypes using six distinct mitochondrial haplotypes paired with a common D. melanogaster nuclear background.
KEY WORDS: Automated behavioral analysis, Rapid iterative negative geotaxis assay, RING assay, Python, Spot detection, Insect locomotion, Longitudinal performance screen
Summary: FreeClimber quantifies the climbing velocity for a group of flies, eliminating systematic biases associated with traditional manual methods in a high throughput and automated (graphical and/or command line-based) platform.
INTRODUCTION
The Drosophila model system provides a rich set of genetic resources to explore the functional bases of traits at organismal, cellular and molecular levels (Bellen et al., 2011; Chow and Reiter, 2017; Lenz et al., 2013; Mackay et al., 2012). One of the most common Drosophila health metrics is locomotor capacity, easily measured using a negative geotaxis (climbing) assay (Gargano et al., 2005; Jones and Grotewiel, 2011). Here, flies are gently knocked to the bottom of a vial and are recorded by video as they instinctively climb upward (Ganetzky and Flanagan, 1978; Gargano et al., 2005). Climbing performance is often reported as some measure of the flies' position versus time, such as mean position at a time cut-off (Gargano et al., 2005; Lavoy et al., 2018) or time until a percentage of flies reach a set height (Ma et al., 2014; Podratz et al., 2013; Tsai et al., 2016; Xu et al., 2008).
The climbing assay's popularity is largely due to its accessibility. Experimental setups are easily engineered from common laboratory items, meaning they are relatively inexpensive to implement. Data collection is straightforward, only requiring simple image capture tools and basic software available on most computers. However, this assay's simplicity is beset by its tedious and time-consuming nature.
There is a rich history of using computers to automate the quantification of animal behavior studies (Hasegawa et al., 1988; Cole and Cheshire, 1996; Hoy et al., 1996; Ramazani et al., 2007; Geissmann et al., 2017). Several publications detail the automated conversion of visual media into data in the Drosophila climbing assay literature, though many are challenging to implement for the general community. Some of these platforms are detectors, while others are trackers. Detectors identify the x,y-coordinates of spots (flies) across frames, which can be evaluated as a function of position versus time [e.g. RflyDetection R module (Cao et al., 2017) and an ImageJ-based approach (Podratz et al., 2013)]. Trackers build on this with predictive linking to connect spots between frames based on their proximity and likelihood of being connected, e.g. the Hillary Climber tracks single flies in individual vials (Willenbrink et al., 2016), the iFly system tracks multiple flies in a single vial (Kohlhoff et al., 2011) and the DaRT system tracks multiple flies in multiple enclosures (Faville et al., 2015; Taylor and Tuxworth, 2019). Trackers are challenging to automate because they generally require supervision to discern erratic vertical motions (jumps and falls) or paths that laterally intersect with other flies (collision on the same plane or eclipse on separate planes) (Chenouard et al., 2014). Additionally, published methods for both detectors and trackers often require a homogeneous background, a custom setup, code from proprietary languages (MATLAB), and/or are only made available locally. Because of these and other factors, no platform is widely accepted by the Drosophila research community, despite the assay's ubiquity.
We created FreeClimber to address some of these major issues, correct for common biases in traditional manual approaches (such as irregular starting heights), and facilitate the generation of accurate and repeatable data that are more representative of the flies' motion. This Python 3-based platform can be run interactively, via a graphical user interface (GUI), or through a command line interface for automated and high-throughput batch processing. FreeClimber utilizes an efficient background subtraction step, so it performs respectably with heterogeneous backgrounds. Additionally, our detector implements a local linear regression for calculating a group's velocity (Olito et al., 2017), which captures an objective metric of fly climbing that may be challenging with traditional manual analyses (for a dynamic demonstration, see the first image in the FreeClimber description in GitHub: https://github.com/adamspierer/FreeClimber/#Overview). Finally, we demonstrate the utility of our platform for longitudinal Drosophila screens analyzing two original datasets of exercise-conditioned and unconditioned mitochondrial-nuclear (mitonuclear) introgression flies. We highlight FreeClimber's ability to quantify strong and subtle differences in phenotype across longitudinal and sample-rich studies, like those frequently conducted in Drosophila research.
MATERIALS AND METHODS
Drosophila husbandry and generation of lines
Six mitochondrial haplotypes (mtDNAs or mitotypes) were derived from four different Drosophila species: D. melanogaster [subtypes: OregonR (OreR) and Zimbabwe53 (Zim)], D. simulans (subtypes: siI and siII), D. mauritiana (subtype: maII) and D. yakuba (subtype: yakuba) (Ballard, 2000; Montooth et al., 2010; Mossman et al., 2016; Zhu et al., 2014). These mitotypes were each placed on a common, D. melanogaster w1118 nuclear background using balancer chromosome crosses and subsequent recurrent male backcrossing using w1118 males (Zhu et al., 2014). Drosophila mauritiana maII and D. yakuba lines were created by microinjection of cytoplasm donor into an egg (Ma et al., 2014).
Stocks were density controlled for two generations, whereby 20 females and 20 males were allowed to lay eggs for 3 days per brood. Fly cultures were held at 25°C on standard lab food (Mossman et al., 2016) and maintained on a 12 h:12 h light:dark schedule. Adult males were collected 3 days post-eclosion using light CO2 anesthetization and separated into vials of 20 flies. Flies were given 24 h to recover and transferred to new food every day. Testing day number in the longitudinal experiment does not include the 3 days post-eclosion before exercise conditioning began.
Experimental setup and video recording
Videos of hand-tapped vials are easily interpreted by FreeClimber, though more specialized setups are useful for standardizing the recording environment, reducing experimental variation and precisely timing video capture (Fig. S1). The main component of the setup is a custom polycarbonate climbing rig, composed of a fixed base with aluminium rails that a mobile chassis could slide along (Fig. S1B). This chassis held six evenly spaced, narrow glass vials that could be raised and subsequently dropped from a pre-designated height (7 cm) to control for the amount of force applied to all vials when beginning the assay. The base of the rig was mounted on a MakerBeam frame (MakerBeam, Utrecht, The Netherlands) and held in place with a setscrew. The frame also held a Huion L4S LED Light Pad (10.7 lumens/inch2; Fuyong, Bao'an District, Shenzhen, China) to backlight the flies. An 8 megapixel PiCamera (V2) attached to a Raspberry Pi 3 (Model B+; Raspberry Pi Foundation, Cambridge, UK) recorded videos from a fixed distance to standardize the video recording parameters.
We also wired a phototrigger to begin video recording as the climbing rig dropped and the assay began. The amount of light emitted from the LED light box was measured by a photoresistor (SEN-09088, Sparkfun Electronics, Niwot, CO, USA) and passed through an analog–digital converter (MCP3000, Adafruit Industries, New York, NY, USA). Both were wired to the Raspberry Pi through the general processing input output (GPIO) pins (Fig. S1C). The photoresistor was placed on a frame rail close to the LED light box, separated by an opaque tab attached to the climbing rig chassis (Fig. S1D) such that the light path was uninterrupted when the chassis was raised and blocked when it was lowered. After raising and dropping the climbing rig's chassis, a 5 s H264 video was recorded at 29 frames s−1. Flies were given 10 s from the end of the video recording to recover before they were tested again. Three technical replicates were recorded for each genotype–condition combination.
Overview of FreeClimber modes
The platform can be run in two modes: a GUI for optimizing detection parameters and running a single video, and a command line interface for high-throughput batch processing of many videos with pre-set detection parameters (Fig. 1A). It was designed with modules that are used across macOS, Windows and Linux operating systems, making it platform independent. Please refer to the FreeClimber description in GitHub for a complete guide on installation and usage, as well as tips and tricks for increasing data quality: https://github.com/adamspierer/FreeClimber/tree/JEB_release. The most current version is available at: https://github.com/adamspierer/FreeClimber/.
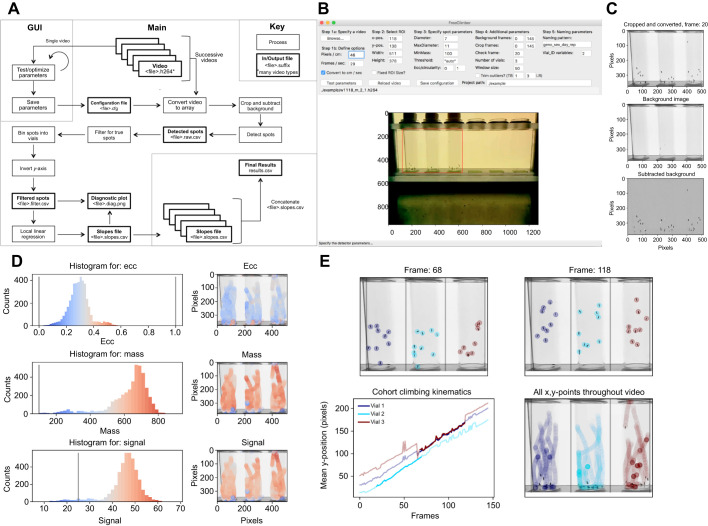
Fig. 1.
Overview of FreeClimber platform. (A) Flow diagram of FreeClimber's processes. (B) The graphical user interface (GUI) is designed for parameter optimization with the region of interest (ROI) outlined in red. (C) Visualization of background subtraction shows the original image recolored and cropped (top), the static background matrix (middle) and the final subtracted image used for spot detection (bottom). (D) Optimization plots visualize the distribution and location of each spot and its respective metric: ecc (eccentricity, roundness), mass and signal. (E) Visualization of spot locations in the first and last frames of the most linear segment of all flies climbing (top), the most linear portion (darker shade) of the mean-vertical position versus the frame curve plotted over all frames (lighter shade) for an indicated vial (lower-left), and all x,y-coordinates throughout the video (lower-right). Note: with the exception of A, all figure panels are generated as outputs or from screenshots of FreeClimber.
Video preprocessing and background subtraction
Videos of all common formats (.avi, .h264, .mov, .mp4, etc.) can be read into integer-based n-dimensional arrays (nd-array) using the FFmpeg-python package (v.4.0.4; https://github.com/kkroening/ffmpeg-python). This package requires FFmpeg (v.4.3.1; https://ffmpeg.org/), which must be installed prior to use. Following user-defined parameters, videos are cropped for the appropriate frame range and positional region of interest (ROI) (Fig. 1B) before being converted to grayscale. A matrix representing the static background is calculated from the median pixel intensity of each x,y-coordinate across a user-defined number of frames (default is all frames). This background matrix is subtracted from each individual frame's pixel intensity matrix, resulting in a new nd-array corresponding with only regions of movement (flies) in the video (Fig. 1C).
Detector optimization and spot detection
The background-subtracted frames are passed to a Python implementation of the Crocker and Weeks particle-tracking algorithm trackpy (v.0.4.2; Crocker and Grier, 1996) for spot detection. Spots are identified from clusters of pixels that meet user-defined parameters for the expected spot diameter (diameter) and maximum diameter (maxsize). These values are saved as a csv (comma separated values) file and can be used as input with trackpy to track spots, separately (see ‘Step 3: Link features into particle trajectories’ in trackpy: http://soft-matter.github.io/trackpy/dev/tutorial/walkthrough.html).
Spots are filtered and must exceed thresholds for spot roundness (ecc; eccentricity), minimum integrated brightness (minmass) and signal threshold (threshold) to be considered a ‘true’ spot. The signal threshold may be provided by the user, or FreeClimber will calculate one using the SciPy (v.1.4.1) functions: peak_prominences and find_peaks. This method finds the local minimum between two signal peaks in a histogram of spot signal values, or is assigned to be one half of the value of the global maximum if there is only one peak. These three spot parameters can be visualized for all spots in a video to assist in the optimization process (Fig. 1D).
True spots are transformed and filtered, then binned into vials based on their x-position relative to equally spaced bins (user-defined number of vials) between the minimum and maximum x-values (Fig. 1E). True spots and their respective vial assignments are saved in a separate csv file for users to explore.
Calculating climbing velocity, via local linear regression
The mean y-position for all spots in a vial is calculated for each frame. A sliding window is applied to the mean y-position versus time (velocity) curve to calculate the most linear (greatest regression coefficient) segment of the curve, via local linear regression. The slope of this segment is considered to be the vial's velocity (Olito et al., 2017) (Fig. 1E). In videos where the P-value for the regression is not significant (P≥0.05), the slope is set to 0. If specified by the user, slopes can be converted from pixels per frame to centimeters per second so results can be compared across studies and filming setups.
Detection parameters are saved as a text-based configuration (.cfg suffix) file that contains all the detection presets so others can replicate results. Ideally, this file is published as a supplemental file with future studies.
Automated, high-throughput detection of climbing velocity across many videos
Once the detector is optimized, it can be run from the command line on many files of the same type. Using the configuration file created in the GUI, the same settings can be applied over all the videos with the specified video type nested in the ‘path_project’ path. The command line interface has several optional arguments for processing subsets of videos (all, unprocessed or a custom list), generating optimization plots, and preventing the final concatenation of all results files. For more information, please consult the tutorial (https://github.com/adamspierer/FreeClimber/blob/master/TUTORIAL.md/).
Files containing regression results (including slopes) for each vial in a video are saved with the ‘slopes.csv’ suffix. These files are all concatenated into a single ‘results.csv’ that resides in the ‘path_project’ folder for separate statistical analysis.
Power Tower: the Drosophila treadmill
The Power Tower automates the process of repeatedly eliciting the negative geotaxis (climbing) startle response, effectively acting as a treadmill (Sujkowski et al., 2018; Tinkerhess et al., 2012). Experimental and control flies on the Power Tower were set up in glass vials with food. Flies allowed to ‘exercise’ were placed in vials with the foam stopper at the top to allow climbing, while their ‘unexercised’ control siblings were placed in vials with the foam stopper 1 cm from the food to limit mobility. Flies were knocked down once every 15 s while on the Power Tower.
Longitudinal exercise conditioning and testing
A longitudinal study over the course of 3 weeks was conducted with male flies from six mitochondrial haplotypes listed above. Male flies, aged 3 days post-eclosion, were divided into two groups of 12 vials containing 20 flies under light-CO2 anesthesia. Flies were conditioned on the Power Tower each weekday for 2 h the first week, 2.5 h the second week and 3 h the third week (Piazza et al., 2009). Fly climbing performance was determined using the RING assay (rapid iterative negative geotaxis assay; Gargano et al., 2005) in the hour preceding each weekdays' conditioning program. Of the 1010 videos recorded, only three were discarded as a result of experimenter error during data acquisition.
Endurance exercise fatigue testing
A separate cohort of male flies, aged and collected similarly to the longitudinal cohort, was used to study the mitotypes' ability to resist endurance climbing fatigue. In this treatment, four vials containing 25 flies were set up on the Power Tower (similar to above) and either allowed to exercise (fatigued) or not allowed to exercise (rested). Flies' initial climbing performance was assayed before they were placed on the Power Tower for six consecutive hours and then assayed hourly during the Power Tower treatment. In total, 297 videos were analyzed.
Statistical analysis on longitudinal data
ANOVA of repeated measures was conducted using the statsmodels (v.0.10.0) module in Python. The ANOVA was used to quantify significant differences between mitochondrial haplotypes, exercise conditions and the interaction between the two. This test was conducted using absolute velocity and the normalized climbing index, which represents the climbing velocity normalized by the average velocity from each genotype's initial time point.
RESULTS AND DISCUSSION
Local linear regression outperforms a time-based cutoff for climbing velocity
The mean vertical position versus time curve is generally concave (Fig. 2A–C) with progress occasionally lagging in the first several frames as flies react to the stimulus, and plateauing at the end as flies reach the top. Traditional manual metrics quantifying the mean vertical position at 2 s (or any time point) overestimates the cohorts' velocity because it assumes flies increase their vertical position linearly. Flies do not necessarily climb in a straight line, and flies can also have a delayed reaction to the stimulus. This analytical method also assumes flies start at the bottom of the vial. Some flies jump when startled and/or begin at a non-zero starting height, which can create biological noise that is amplified if only a single frame is considered for a time- or position-based cutoff. Furthermore, reducing a 3D object to a 2D image causes issues as depth is translated into height. Flies starting at the bottom-front of the vial may have a different starting height from those starting at the bottom-back.
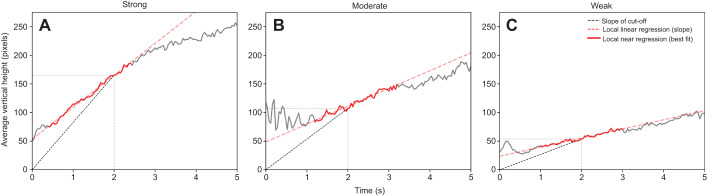
Fig. 2.
Local linear regression provides an objective measure of climbing performance. Mean vertical position versus time (velocity, solid gray line) plots for a cohort of flies measured at (A) 5 days, (B) 10 days and (C) 20 days post-eclosion. The slope of the standard, time-based cut-off at 2 s (black dashed line) is steeper than the line of best fit (red dashed line) during the most linear 2 s of a 5 s climb (red solid line). Note: these figure panels were not generated by FreeClimber.
A local linear regression is an appropriate workaround to many of these issues. Rather than using a static snapshot to measure performance, like most traditional manual approaches, this method considers all flies across all frames as it calculates the slope of the segment with the greatest regression coefficient over n-consecutive frames of the mean vertical position versus time curve. Accordingly, the starting position of flies in the vial and potential for unpredictable climbing behaviors (jumping, falling, eclipsed by other flies, hiding/undetected, etc.) have reduced impact on the metric.
Climbing performance is easily quantified for short- and long-term longitudinal studies
Once detection parameters are optimized, FreeClimber can batch process videos. Previous studies demonstrate climbing performance can be affected by genotype (Gargano et al., 2005; Holmbeck et al., 2015; Lavoy et al., 2018), environment (Piazza et al., 2009; Tinkerhess et al., 2012), and genotype×environment effects (Holmbeck et al., 2015; Sujkowski et al., 2018). To demonstrate the utility of the FreeClimber software, we tested a set of six, phylogenetically diverse (Ballard, 2000; Montooth et al., 2009), mitochondrial-nuclear (mitonuclear) introgression flies (mitotypes; Fig. 3A). These mitotypes were derived from four different Drosophila species: D. melanogaster [subtypes: OregonR (OreR) and Zimbabwe53 (Zim)], D. simulans [subtypes: siI and siII (sm21)], D. mauritiana (maII) and D. yakuba (yak). All were paired with a common D. melanogaster (w1118) nuclear background (genotype notation: mito;nuclear). Four of these lines (OreR;w1118, siI;w1118, sm21;w1118 and Zim;w1118) were previously shown to have weak to moderate climbing performance ability (Sujkowski et al., 2018), while two (yak;w1118 and maII;w1118) were previously untested. Under the disrupted co-adaptation hypothesis (Montooth et al., 2010; Rand et al., 2004), we would expect to see a negative relationship between the divergence between a mitonuclear pairing and climbing performance. More distantly related pairings have greater opportunity to accumulate mitonuclear incompatibilities, which would hinder performance. A previous study observed that D. yakuba mtDNA paired with D. melanogaster nuclear DNA (yak;w1118) was longer lived compared with its native mitochondrial-nuclear pairing (D. melanogaster; D. melanogaster) (Ma and O'Farrell, 2016). Our motivation was to use FreeClimber as an independent test of this observation that a yak;w1118 was not a ‘disrupted’ genotype.
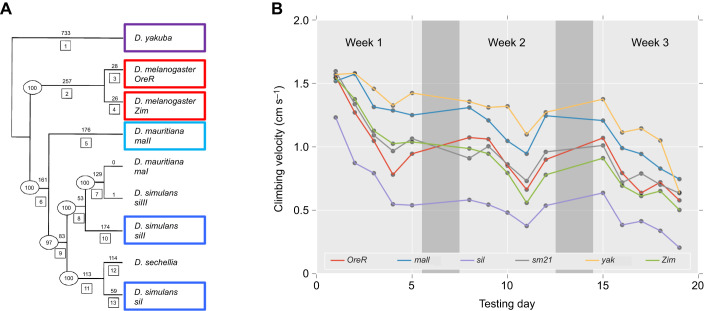
Fig. 3.
Mitochondrial haplotypes show a differential response to endurance exercise conditioning and resistance to endurance fatigue. (A) Phylogenetically distinct mitochondrial haplotypes were derived from four clades (D. melanogaster, red; D. simulans, blue; D. mauritiana, cyan; and D. yakuba, purple). Phylogenetic tree modified from Ballard (2000) with sm21 as a subgroup of siII. (B) These mitochondrial haplotypes, on a common nuclear background (D. melanogaster w1118), were subjected to a 3 week endurance exercise conditioning program. Flies were tested on weekdays (light gray) before exercise conditioning on a Power Tower, and allowed to rest on weekends (dark gray). Points represent the mean velocity for all flies of a genotype across replicates (n=1007 videos total). As there was no significant conditioning effect (P=0.83; Table S1), conditions were averaged together. Most mitotypes began at roughly the same starting velocity, though differed in their age-associated performance decline; yak;w1118 (yak, yellow) and maII;w1118 (maII, blue) were the strongest overall, while siI;w1118 (siI, purple) was the weakest. Note: the color codes differ between A and B, and these figure panels were not generated by FreeClimber.
We conducted a longer-term longitudinal experiment asking whether mitochondrial haplotypes respond differently to a 3 week exercise-conditioning program. We conditioned 12 cohorts (6 mitotypes×2 conditioning conditions) of 20 male flies following a prescribed conditioning protocol (Sujkowski et al., 2018; Tinkerhess et al., 2012), comparing the daily climbing performance of conditioned cohorts against that of unconditioned controls. Flies experienced age-associated declines in climbing performance (Fig. 3B) that were significant by mitotype (P<0.0001), but not for conditioning (P=0.83), or mitotype×conditioning effects (P=0.26) (Fig. S2A, Table S1). When normalizing performance by their respective mean first day performance (normalized climbing index), mitotype (P<0.0005) and mitotype×conditioning (P<0.0001) were both significant. While there was no significant exercise conditioning effect, the unconditioned flies generally outperformed their conditioned counterparts. This would suggest exercise conditioning on the Power Tower may not always be beneficial for the flies. We previously demonstrated mitotypes on the w1118 nuclear background are not sensitive to exercise conditioning effects (Sujkowski et al., 2018), which our results support.
We found D. melanogaster pairings (OreR;w1118 and Zim;w1118) were intermediate performers and two divergent lines performed the same (sm21;w1118) or much worse (siI;w1118), supporting the disrupted co-adaptation hypothesis. However, the two most divergent pairings (maII;w1118 and yak;w1118) performed the best. We note that D. melanogaster and D. yakuba species are reproductively incompatible and separated by 7–10 million years of divergence (Consortium, 2007). This finding provides independent support that this yak;w1118 genotype is not dysfunctional, representing a challenge to the disrupted co-adaptation hypothesis.
Finally, we tested ability of a separate cohort of the same mitotypes to resist fatigue in a shorter-term, 6 h fatigue assay. We followed a similar Power Tower protocol as for the longitudinal study, but instead used four cohorts of 25 flies and flies were on the Power Tower for one 6 h stretch. We measured initial climbing performance and performance after each hour. Previous studies have shown this fatigue assay is distinct from a longitudinal aging assay (Sujkowski and Wessells, 2018). We observed significant mitotype, fatigue and mitotype×fatigue effects for the absolute velocities, but only a significant fatigue effect after normalizing the slopes to the initial time point (Fig. S2B, Table S1). This fatigue resistance test demonstrates that the Power Tower effectively elicits a consistent climbing phenotype that can slowly fatigue flies over a 6 h window. Interestingly, rested yak;w1118 were strong performers, though their fatigued counterparts had the greatest variation between time points. It is possible that yak;w1118 are strong climbers when undisturbed, but more variable in the climbing performance when stressed.
Conclusion
FreeClimber is a free and easy-to-use platform for quantifying the climbing velocity for cohorts of flies. It automates the tedious process of detecting flies, and reliably quantifies an objective measure of climbing performance. We applied our platform to measure the longitudinal climbing performance during an exercise-conditioning program and during a resistance to endurance fatigue assay across six mitonuclear introgression lines. We demonstrate our platform's ability to identify both strong and subtle differences between genotypes, and its ability to work with longitudinal datasets.
Supplementary Material
Acknowledgements
We acknowledge Leann Biancani for her assistance with Drosophila husbandry, John Murphy in the Brown University BioMed Machine Shop and Benjamin Wilks for their technical expertise with the climbing apparatus, and Kathryn Russo for her time during testing. We thank the Brown University Computational Biology Core (Ashok Ragavendran, August Guang and Joselynn Wallace) for their technical assistance, and Tom Roberts for providing instruction and materials in the circuit assembly, and for critical feedback on this manuscript. We also thank the three anonymous reviewers for critical feedback.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: A.N.S., D.M.R.; Methodology: A.N.S., C.-T.Z.; Software: A.N.S., C.-T.Z.; Validation: A.N.S.; Formal analysis: A.N.S., D.Y.; Investigation: A.N.S., D.Y.; Resources: D.M.R.; Writing - original draft: A.N.S., D.M.R.; Writing - review & editing: A.N.S., D.Y., D.M.R.; Visualization: A.N.S.; Supervision: D.M.R.; Funding acquisition: D.M.R.
Funding
A.N.S., C.T.Z. and this work were supported by the National Institutes of Health [R01 GM067862 and 1R01AG027849 to D.M.R.]. D.Y. was supported by a Brown University Undergraduate Research and Teaching Award (UTRA). The Power Tower was supported by a Sigma Xi Grants-in-Aid of Research (to A.N.S.). This research was partially supported by Institutional Development Award Number P20GM109035 from the National Institute of General Medical Sciences of the National Institutes of Health, which funds COBRE Center for Computational Biology of Human Disease. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Deposited in PMC for release after 12 months.
Data availability
FreeClimber source code, tutorial, examples and result files used for statistical analysis are available online from the GitHub repository (https://github.com/adamspierer/FreeClimber/tree/JEB_release). The most updated version of FreeClimber can be found at: https://github.com/adamspierer/FreeClimber/
Supplementary information
Supplementary information available online at https://jeb.biologists.org/lookup/doi/10.1242/jeb.229377.supplemental
References
- Ballard J. W. O. (2000). Comparative genomics of mitochondrial DNA in members of the Drosophila melanogaster subgroup. J. Mol. Evol. 51, 48-63. 10.1007/s002390010066 [DOI] [PubMed] [Google Scholar]
- Bellen H. J., Levis R. W., He Y. C., Carlson J. W., Evans-Holm M., Bae E., Kim J., Metaxakis A., Savakis C., Schulze K. L. et al. (2011). The Drosophila gene disruption project: progress using transposons with distinctive site specificities. Genetics 188, U731-U341 10.1534/genetics.111.126995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W. Z., Song L., Cheng J. J., Yi N., Cai L. Y., Huang F. D. and Ho M (2017). An automated rapid iterative negative geotaxis assay for analyzing adult climbing behavior in a Drosophila Model of neurodegeneration. J. Vis. Exp. 127, 56507 10.3791/56507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenouard N., Smal I., De Chaumont F., Maška M., Sbalzarini I. F., Gong Y., Cardinale J., Carthel C., Coraluppi S. and Winter M (2014). Objective comparison of particle tracking methods. Nat. Methods 11, 281 10.1038/nmeth.2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow C. Y. and Reiter L. T (2017). Etiology of human genetic disease on the fly. Trends Genet. 33, 391-398. 10.1016/j.tig.2017.03.007 [DOI] [PubMed] [Google Scholar]
- Cole B. J. and Cheshire D (1996). Mobile cellular automata models of ant behavior: movement activity of Leptothorax allardycei. Am. Nat. 148, 1-15. 10.1086/285908 [DOI] [Google Scholar]
- Consortium D. G. (2007). Evolution of genes and genomes on the Drosophila phylogeny. Nature 450, 203 10.1038/nature06341 [DOI] [PubMed] [Google Scholar]
- Crocker J. C. and Grier D. G (1996). Methods of digital video microscopy for colloidal studies. J. Colloid Interface Sci. 179, 298-310. 10.1006/jcis.1996.0217 [DOI] [Google Scholar]
- Faville R., Kottler B., Goodhill G. J., Shaw P. J. and van Swinderen B (2015). How deeply does your mutant sleep? Probing arousal to better understand sleep defects in Drosophila. Sci. Rep. 5 10.1038/srep08454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganetzky B. and Flanagan J. R (1978). Relationship between senescence and age-related-changes in 2 wild-type strains of Drosophila-melanogaster. Exp. Gerontol. 13, 189-196. 10.1016/0531-5565(78)90012-8 [DOI] [PubMed] [Google Scholar]
- Gargano J. W., Martin I., Bhandari P. and Grotewiel M. S (2005). Rapid iterative negative geotaxis (RING): a new method for assessing age-related locomotor decline in Drosophila. Exp. Gerontol. 40, 386-395. 10.1016/j.exger.2005.02.005 [DOI] [PubMed] [Google Scholar]
- Geissmann Q., Garcia Rodriguez L., Beckwith E. J., French A. S., Jamasb A. R. and Gilestro G. F (2017). Ethoscopes: an open platform for high-throughput ethomics. PLoS Biol. 15, e2003026 10.1371/journal.pbio.2003026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa K., Tanakadate A. and Ishikawa H (1988). A method for tracking the locomotion of an isolated microorganism in real time. Physiol. Behav. 42, 397-400. 10.1016/0031-9384(88)90282-X [DOI] [PubMed] [Google Scholar]
- Holmbeck M. A., Donner J. R., Villa-Cuesta E. and Rand D. M. (2015). A Drosophila model for mito-nuclear diseases generated by an incompatible interaction between tRNA and tRNA synthetase. Dis. Model. Mech. 8, 843-854. 10.1242/dmm.019323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoy J. B., Koehler P. G. and Patterson R. S (1996). A microcomputer-based system for real-time analysis of animal movement. J. Neurosci. Methods 64, 157-161. 10.1016/0165-0270(95)00121-2 [DOI] [PubMed] [Google Scholar]
- Jones M. A. and Grotewiel M (2011). Drosophila as a model for age-related impairment in locomotor and other behaviors. Exp. Gerontol. 46, 320-325. 10.1016/j.exger.2010.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlhoff K. J., Jahn T. R., Lomas D. A., Dobson C. M., Crowther D. C. and Vendruscolo M (2011). The iFly tracking system for an automated locomotor and behavioural analysis of Drosophila melanogaster. Integrative Biology 3, 755-760. 10.1039/c0ib00149j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoy S., Chittoor-Vinod V. G., Chow C. Y. and Martin I (2018). Genetic modifiers of neurodegeneration in a drosophila model of Parkinson's disease. Genetics 209, 1345-1356. 10.1534/genetics.118.301119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz S., Karsten P., Schulz J. B. and Voigt A (2013). Drosophila as a screening tool to study human neurodegenerative diseases. J. Neurochem. 127, 453-460. 10.1111/jnc.12446 [DOI] [PubMed] [Google Scholar]
- Ma H. S. and O'Farrell P. H (2016). Selfish drive can trump function when animal mitochondrial genomes compete. Nat. Genet. 48, 798.-+ 10.1038/ng.3587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H. S., Xu H. and O'Farrell P. H (2014). Transmission of mitochondrial mutations and action of purifying selection in Drosophila melanogaster. Nat. Genet. 46, 393.-+ 10.1038/ng.2919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T. F. C., Richards S., Stone E. A., Barbadilla A., Ayroles J. F., Zhu D. H., Casillas S., Han Y., Magwire M. M., Cridland J. M. et al. (2012). The Drosophila melanogaster genetic reference panel. Nature 482, 173-178. 10.1038/nature10811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montooth K. L., Abt D. N., Hofmann J. W. and Rand D. M (2009). Comparative genomics of Drosophila mtDNA: novel features of conservation and change across functional domains and lineages. J. Mol. Evol. 69, 94-114. 10.1007/s00239-009-9255-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montooth K. L., Meiklejohn C. D., Abt D. N. and Rand D. M (2010). Mitochondrial–nuclear epistasis affects fitness within species but does not contribute to fixed incompatibilities between species of Drosophila. Evolution 64, 3364-3379. 10.1111/j.1558-5646.2010.01077.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossman J. A., Biancani L. M., Zhu C. T. and Rand D. M (2016). Mitonuclear epistasis for development time and its modification by diet in Drosophila. Genetics 203, 463 10.1534/genetics.116.187286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olito C., White C. R., Marshall D. J. and Barneche D. R (2017). Estimating monotonic rates from biological data using local linear regression. J. Exp. Biol. 220, 759-764. 10.1242/jeb.148775 [DOI] [PubMed] [Google Scholar]
- Piazza N., Gosangi B., Devilla S., Arking R. and Wessells R (2009). Exercise-training in young drosophila melanogaster reduces age-related decline in mobility and cardiac performance. PLoS ONE 4, e5886 10.1371/journal.pone.0005886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podratz J. L., Staff N. P., Boesche J. B., Giorno N. J., Hainy M. E., Herring S. A., Klennert M. T., Milaster C., Nowakowski S. E., Krug R. G. et al. (2013). An automated climbing apparatus to measure chemotherapy-induced neurotoxicity in Drosophila melanogaster. Fly 7, 187-192. 10.4161/fly.24789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramazani R. B., Krishnan H. R., Bergeson S. E. and Atkinson N. S (2007). Computer automated movement detection for the analysis of behavior. J. Neurosci. Methods. 162, 171-179. 10.1016/j.jneumeth.2007.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand D. M., Haney R. A. and Fry A. J (2004). Cytonuclear coevolution: the genomics of cooperation. Trends Ecol. Evol. 19, 645-653. 10.1016/j.tree.2004.10.003 [DOI] [PubMed] [Google Scholar]
- Sujkowski A. and Wessells R (2018). Using Drosophila to understand biochemical and behavioral responses to exercise. Exerc. Sport Sci. Rev. 46, 112-120. 10.1249/JES.0000000000000139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sujkowski A., Spierer A. N., Rajagopalan T., Bazzell B., Safdar M., Imsirovic D., Arking R., Rand D. M. and Wessells R (2018). Mito-nuclear interactions modify Drosophila exercise performance. Mitochondrion. 47: 188-205. 10.1016/j.mito.2018.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M. J. and Tuxworth R. I (2019). Continuous tracking of startled Drosophila as an alternative to the negative geotaxis climbing assay. J. Neurogenet. 33, 190-198. 10.1080/01677063.2019.1634065 [DOI] [PubMed] [Google Scholar]
- Tinkerhess M. J., Ginzberg S., Piazza N. and Wessells R. J (2012). Endurance training protocol and longitudinal performance assays for Drosophila melanogaster. J. Vis. Exp. 61, 3786 10.3791/3786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai H. Z., Lin R. K. and Hsieh T. S (2016). Drosophila mitochondrial topoisomerase III alpha affects the aging process via maintenance of mitochondrial function and genome integrity. J. Biomed. Sci. 23, 38 10.1186/s12929-016-0255-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willenbrink A. M., Gronauer M. K., Toebben L. F., Kick D. R., Wells M. and Zhang B (2016). The Hillary Climber trumps manual testing: an automatic system for studying Drosophila climbing. J. Neurogenet. 30, 205-211. 10.1080/01677063.2016.1255211 [DOI] [PubMed] [Google Scholar]
- Xu H., DeLuca S. Z. and O'farrell P. H (2008). Manipulating the metazoan mitochondrial genome with targeted restriction enzymes. Science 321, 575-577. 10.1126/science.1160226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C. T., Ingelmo P. and Rand D. M (2014). GxGxE for lifespan in Drosophila: mitochondrial, nuclear, and dietary interactions that modify longevity. PLoS Genet. 10, e1004354 10.1371/journal.pgen.1004354 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.