Abstract
Background: Lung adenocarcinoma (LUAD) is the most frequent histological type of lung cancer, and its incidence has displayed an upward trend in recent years. Nevertheless, little is known regarding effective biomarkers for LUAD.
Methods: The robust rank aggregation method was used to mine differentially expressed genes (DEGs) from the gene expression omnibus (GEO) datasets. The Search Tool for the Retrieval of Interacting Genes (STRING) database was used to extract hub genes from the protein–protein interaction (PPI) network. The expression of the hub genes was validated using expression profiles from TCGA and Oncomine databases and was verified by real-time quantitative PCR (qRT-PCR). The module and survival analyses of the hub genes were determined using Cytoscape and Kaplan–Meier curves. The function of KIF4A as a hub gene was investigated in LUAD cell lines.
Results: The PPI analysis identified seven DEGs including BIRC5, DLGAP5, CENPF, KIF4A, TOP2A, AURKA, and CCNA2, which were significantly upregulated in Oncomine and TCGA LUAD datasets, and were verified by qRT-PCR in our clinical samples. We determined the overall and disease-free survival analysis of the seven hub genes using GEPIA. We further found that CENPF, DLGAP5, and KIF4A expressions were positively correlated with clinical stage. In LUAD cell lines, proliferation and migration were inhibited and apoptosis was promoted by knocking down KIF4A expression.
Conclusion: We have identified new DEGs and functional pathways involved in LUAD. KIF4A, as a hub gene, promoted the progression of LUAD and might represent a potential therapeutic target for molecular cancer therapy.
Keywords: Bioinformatical analysis, Biomarker, Differentially expressed genes, KIF4A, Lung adenocarcinoma
Background
Lung cancer is one of the most frequent malignant tumors in clinical practice [1]. Its morbidity and mortality have ranked first among all types of malignant tumors, and it has been recognized as the most widespread cancer in the world [2]. More than 80% of lung cancers are non-small cell lung cancers (NSCLC), and lung adenocarcinoma (LUAD) accounts for over 70% of NSCLC [3,4]. In recent years, the treatment of LUAD has continuously improved, and a series of new diagnosis and treatment technologies have been applied in clinical practice, which can improve the treatment outcomes of LUAD patients to some extent [5–7]. Nevertheless, the 5-year survival rate is still very low, and outcomes are still unsatisfactory [8]. At present, early diagnosis and treatment can greatly improve prognosis and survival rates of LUAD patients [9,10]. Therefore, it is crucial to fully elucidate the molecular mechanism involved in LUAD.
In recent years, with the development of microarrays and high-throughput sequencing technologies allowing large open data resources, such as The Cancer Genome Atlas (TCGA) database and Gene Expression Omnibus (GEO) datasets, large amounts of genetic data have been generated [10–12]. Researchers can effectively mine big data to identify novel cancer-related genes [13–15]. At present, how to extract valuable information from this huge database has become a new research direction, and bioinformatics has effectively solved this problem. Bioinformatics can screen for availably and mine the microarray data, and ultimately applies relevant software to graph the results, so as to reveal genes potentially implicated in oncogenesis at the molecular level [16,17]. Consequently, bioinformatics can provide new diagnostic markers and novel ideas for the early diagnosis and treatment of LUAD.
The purpose of the present study was to identify the potential key genes and pathways associated with the carcinogenesis of LUAD. We first compared the gene expression profiles of LUAD and adjacent normal lung tissues utilizing the data in the GEO public database. Next, we systematically analyzed the expression profiles of microarray data and screened DEGs. Meanwhile, we determined the Gene ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and protein–protein interactions (PPIs) network analyses of the robust DEGs in LUAD. Further, we also obtained and analyzed the expression and prognosis of the related hub genes using the Oncomine database and the TCGA and GTEx datasets in GEPIA. We validated the mRNA expression of the seven hub genes in clinical samples of LUAD and in paired normal tissues. Furthermore, we investigated the potential functional mechanism of KIF4A in regulating LUAD cell proliferation, migration, and apoptosis.
Materials and methods
Data acquisition and preprocessing
Processed microarray data were searched from the websites of GEO (available: http://www.ncbi.nlm.nih.gov/geo/) using “LUAD” as the keyword. Of these, we selected and downloaded the GSE85716, GSE32863, and GSE116959 datasets (Supplementary Table S1). DEGs were identified by the Bioconductor limma package and the robust rank aggregation (RRA) method was used to integrate and rank all of the DEGs from three GEO datasets. In addition, the edgeR package was used to screen DEGs with thresholds of |log2fold change(FC)|>1 and the thresholds of the adjusted p-value false discovery rate (FDR)<0.05.
Hierarchical clustering analysis
The top 20 robust DEGs were determined using the robust RRA method. As in a previous report [18], the R language package was applied for hierarchical clustering analysis based on the TPM values of the genes in each group. In the analysis, TPM values represented the expression levels of genes, and the gene expression patterns were further clustered into the same or similar genes. In the clustergram, differences in subregional information were represented using different colors, and the different experimental conditions were determined using the clustering pattern control model.
GO analysis
Direct annotation of gene function can obtain multiple and complicated functional nodes, which can result in the redundancy of results. Thus, we applied the GO analysis method to extract and integrate data and further increase the reliability of the gene analysis. GO analysis mainly includes cellular components (CC), molecular functions (MF), and biological processes (BP). In the present study, GO enrichment analysis of DEGs was also analyzed using the Bioinformatics Tool (DAVID, version 6.8, https://david.ncifcrf.gov/) [19,20].
KEGG analysis
KEGG analysis, created by the Kanehisa Laboratories, is a collection of databases that can integrate genomes, biological pathways, diseases, drugs, and chemicals [21]. KOBAS 2.0 software (http://kobas.cbi.pku.edu.cn/) was used to carry out KEGG pathway enrichment analysis for the DEGs in LUAD [22]. A P-value <0.05 was used as the screening threshold to explore the biological signaling pathways involved in DEGs in LUAD.
PPI analysis
The STRING (http://string-db.org/) is a database that can search for and predict the interactions between known proteins [23]. The online software string (comprehensive score>0.9) was adopted to analyze the PPIs of DEGs, and then a PPI diagram including the DEGs was drawn based on the analysis. Meanwhile, a number of nodes in the network was obtained to denote genetic relationships through network structure analysis, and the core genes were defined by the number of nodes in the network. Cytoscape software was used to generate the PPI network diagram. The software includes two algorithms: MCC and DMNC, which can calculate the top 10 genes and then cross-cut their intersections. The hub genes were obtained from the PPI network and were then subjected to verification of their expression, survival analysis, and other analyses.
Application of oncomine database
The Oncomine database (http://www.oncomine.org) was utilized to obtain the expression of seven hub genes in LUAD and in adjacent normal lung tissues. The screening conditions were as follows: (1) Genes: BIRC5, DLGAP5, CENPF, KIF4A, TOP2A, AURKA, or CCNA2; (2) Analysis Type: LUAD versus Normal Analysis; (3) Threshold using P-value<0.05; FC: All; Gene Rank: All.
Gene expression profiling interactive analysis (GEPIA) database
GEPIA is a public database established for expression profiling analysis of cancer and normal genes [24]. GEPIA analysis contains the expression analysis of RNA sequencing data from 9736 tumors and 8587 normal samples in TCGA (http://cancergenome.nih.gov/) [25] and Genotype-Tissue Expression (GTEx, http://commonfund.nih.gov/GTEx/) projects [26,27]. In our study, the expression of the seven hub genes was extracted from the TCGA database. In addition, the survival and disease-free survival analyses of the seven hub genes were also obtained based on TCGA and the GTEx data in GEPIA. Moreover, we also obtained the correlation of CENPF, DLGAP5, KIF4A, and the clinical stage of LUAD.
Total RNA isolation and real-time quantitative PCR (qRT-PCR)
In total, samples from 10 LUAD patients (along with paired normal lung samples) that had undergone surgical treatment in our hospital, were available for examination in our study. All collected tissues were confirmed histologically by two independent pathologists. Written consent was obtained from all patients who were recruited. This research was approved by the Ethics Committee of the First Hospital of Changsha.
Total RNA from LUAD samples (n=10) and paired normal samples (n=10) was isolated using the RNeasy Mini Kit (Cat.74101, Qiagen, Germany) according to the manufacturer's instructions. The synthesis of cDNA used for genes was performed using the Bestar™ qPCR RT kit (DBI; #DBI-0) using 2 μg RNA. The relative mRNA levels of BIRC5, DLGAP5, CENPF, KIF4A, TOP2A, AURKA, and CCNA2 were determined by real-time quantitative PCR (qRT-PCR) using a 20 μl reaction system. The PCR process was performed on an ABI PRISM 7500 real-time PCR system (Applied Biosystems, Carlsbad, CA, U.S.A.) using the following settings: 95°C for 2 min, followed by 40 cycle of 94°C for 20 s, 58°C for 20 s, and 72°C for 20s. GAPDH was used as the internal normalized reference gene. The fold change was determined as: 2−ΔΔCt (ΔΔCt = (ΔCt of genes of interest) − (ΔCt of GAPDH). The primer sequences are listed in Supplementary Table S2.
Cell culture and reagents
The human LUAD cell lines HCC827 and A549 were purchased from AbZyme Biotechnology Inc (Jiangsu, China). The human bronchial epithelial cells Beas2B were purchased from Zhong Qiao Xin Zhou Biotechnology Inc (Shanghai, China). Beas2B cells were grown in DMEM (Invitrogen Life Technologies, Carlsbad, CA, U.S.A.) supplemented with 5% fetal bovine serum (FBS, Gibco BRL, Gaithersburg, MD, U.S.A.). HCC827 and A549 cells were cultured in RPMI-1640 (Hyclone, Logan, UT, U.S.A.) supplemented with 10% FBS. Rabbit monoclonal antibodies against KIF4A were obtained from Abcam (#ab124903, Abcam, Cambridge, MA, U.S.A.).
Western blot analysis
After treatment, the cells were collected, washed and lysed with ice-cold RIPA lysis buffer (Beyotime Inst. Biotech) with 1 mmol/l PMSF. Protein concentrations were calculated using BCA assay kits (Beyotime Inst. Biotech). Total cellular protein (20 μg) was subjected to 12% SDS-PAGE and transferred to PVDF membranes (Millipore). The membranes were blocked with 5% nonfat milk powder at room temperature for 2 h, followed by immunoblotting with primary antibodies at 4°C overnight and immunoblotting with HRP-conjugated secondary antibody at room temperature for 1 h. Following each step, the membranes were washed three times with PBST for 5 min. Finally, the blots were developed using an enhanced chemiluminescence system (Pierce). Actin was used as a loading control.
Colony formation assay
Five hundred exponentially growing cells were plated in six-well cell culture plates in a total volume of 2 ml of medium and incubated for 6 h. Next, 2 mM of KIF4A-siRNA was added to the medium, and incubation continued for 6 h. Following replacement with fresh medium, colony formation was monitored. Ten days later, the colonies were fixed with 75% ethanol and stained with methylene blue. The optical density (OD) values were detected using a microplate reader.
Flow cytometry for quantitative analysis of apoptosis
After treatment, 3 × 105 cells were collected used for each sample. For apoptosis detection, cells were stained using the Annexin V-FITC Apoptosis Detection Kit I (BD Biosciences, San Diego, CA) according to the manufacturer’s recommendation. The stained cells were determined by flow cytometry (BD FACS Canto) and analyzed by the FCS Express v2.0 software, as in our previous studies [28].
Transwell assay
Cells were seeded in the chamber with 5 × 104 cells in 100 μl FBS-free culture medium. A 600 μl volume of culture medium (10% FBS) was added to the 24-well plate. After a 24 h incubation, the suspended cells were washed and removed, then fixed with methylalcohol for 30 min. Subsequently, the non-migrated cells were removed and the migrated cells were stained with 1% Crystal Violet. The OD values were detected using a microplate reader as described in our previous studies [29].
RNA interference to inhibit KIF4A expression
A mixture of 200 nM KIF4A-siRNA (KIF4A-siRNA-1: GGAACAGGGCAACAACTCT; KIF4A-siRNA-2: TAAGGATACCCTTCTATCT; KIF4A-siRNA-3:TGCTGTTTGAGGAACGAAA) (HonorGene, Changsha, China) and control siRNA were added to each well of a six-well plate for 6 h, after which fresh RPMI media containing 10% serum was added. Cells were harvested 48 h later.
Statistical analysis
All statistical analyses were performed using SPSS 19.0 (SPSS Inc., Chicago, IL, U.S.A.). All of the data are presented as mean ± standard deviation (SD). Statistical significance comparing two groups was evaluated by the Student’s t test. A P-value<0.05 was considered statistically significant.
Results
Identification of DEGs from GSE85716, GSE32863, and GSE116959 datasets
To identify genes that might be involved in the tumorigenesis of LUAD, we analyzed the DEGs in the GSE85716, GSE32863, and GSE116959 datasets. We used a Volcano plot to present DEGs in LUAD and nontumor lung tissues based on the data from these three databases; the red dots indicated genes that were up-regulated in LUAD, and the green dots indicated genes that were down-regulated in LUAD (Figure 1A–C). Next, we screened the top 20 up-regulated and down-regulated genes in the GSE85716, GSE32863, and GSE116959 datasets, respectively, and the differential distribution of DEGs between adjacent normal lung and LUAD tissues was displayed using the hierarchical clustering (Figure 1D). The up- and downregulated DEGs are listed in Supplementary Tables S3 and 4.
Figure 1. Identification of DEGs from GSE85716, GSE32863, and GSE116959 datasets.
(A–C) Volcano plot showing the DEGs of the GSE85716, GSE32863, and GSE116959 datasets. (D) Hierarchical clustering comparing the top 20 up-regulated and down-regulated genes in adjacent normal lung tissues and LUAD tissues.
GO and KEGG analyses of robust DEGs in LUAD
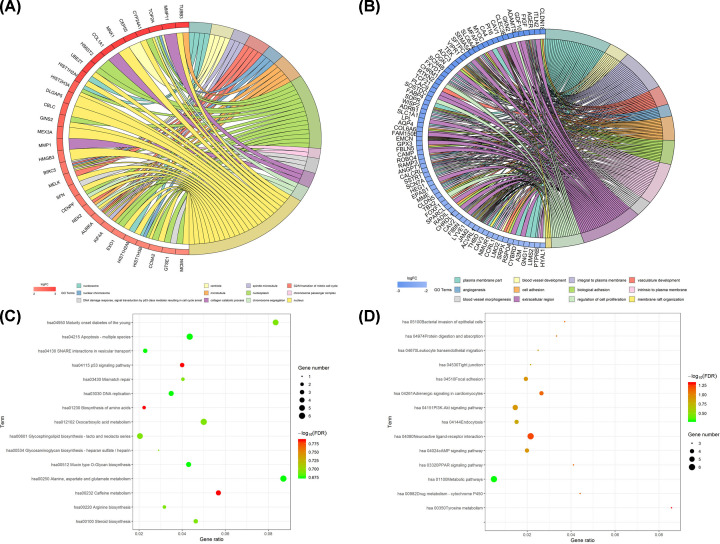
Based on the expression profile of DEGs between adjacent normal lung and LUAD tissues, GO and KEGG analyses for DEGs were performed. The results of GO analysis revealed that the up-regulated DEGs were enriched in CC terms (nucleosome, centriole, spindle microtubule, nuclear chromosome, microtubule, nucleoplasm, chromosome passenger complex, and nucleus) and in BP terms (G2/M transition of mitotic cell cycle, DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest, collagen catabolic process, and chromosome segregation) (Figure 2A). The down-regulated DEGs were enriched in CC terms (plasma membrane part, integral to plasma membrane, intrinsic to plasma membrane, and extracellular region) and BP terms (blood vessel development, vasculature development, angiogenesis, cell adhesion, biological, and blood vessel morphogenesis) (Figure 2B). The results of the GO function annotation for the up-regulated and down-regulated DEGs in LUAD are shown in Tables 1 and 2. Furthermore, KEGG pathway analysis disclosed that the up-regulated DEGs mainly participated in regulating glycosaminoglycan biosynthesis-heparan sulfate/heparin, mismatch repair, and biosynthesis of amino acids (Figure 2C); the down-regulated DEGs mainly participated in regulating neuroactive ligand–receptor interactions, metabolic pathways, and the PI3K-Akt signaling pathway (Figure 2D). The detailed KEGG pathways for the up-regulated and down-regulated DEGs in LUAD are shown in Tables 3 and 4.
Figure 2. Functional characteristics analyses of robust DEGs.
GO enrichment analyses of (A) the up-regulated DEGs and (B) the down-regulated DEGs. KEGG pathway enrichment analyses of (C) the up-regulated DEGs and (D) the down-regulated DEGs.
Table 1. The GO analysis of the up-regulated DEGs in LUAD.
| Category | ID | Term | Genes | adj_pval |
|---|---|---|---|---|
| CC | GO:0000786 | Nucleosome | HIST2H3A, HIST1H3B, HIST1H2AI, HIST1H2AM | 0.0020621 |
| CC | GO:0005814 | Centriole | BIRC5, AURKA, CEP55, TOP2A | 0.00347557 |
| CC | GO:0005876 | Spindle microtubule | KIF4A, BIRC5, AURKA | 0.00623428 |
| BP | GO:0000086 | G2/M transition of mitotic cell cycle | NEK2, BIRC5, AURKA, MELK | 0.00702927 |
| CC | GO:0000228 | Nuclear chromosome | HIST1H3B, BIRC5, TOP2A | 0.00894277 |
| CC | GO:0005874 | Microtubule | KIF4A, NEK2, BIRC5, AURKA, TUBB3 | 0.00964457 |
| CC | GO:0005654 | Nucleoplasm | EXO1, HIST2H3A, GINS2, CYP24A1, KIF4A, CENPF, AURKA, BIRC5, MCM4, GTSE1, HIST1H3B, HS6ST2, CCNA2, TOP2A, UBE2T | 0.01268473 |
| CC | GO:0032133 | Chromosome passenger complex | BIRC5, AURKA | 0.01337317 |
| BP | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest | AURKA, SFN, GTSE1 | 0.01356678 |
| BP | GO:0030574 | Collagen catabolic process | COL1A1, MMP1, MMP11 | 0.01441139 |
| BP | GO:0007059 | Chromosome segregation | NEK2, CENPF, TOP2A | 0.01616706 |
| CC | GO:0005634 | Nucleus | EXO1, HIST2H3A, CYP24A1, GINS2, HMGB3, NEK2, DLGAP5, MEX3A, CENPF, BIRC5, AURKA, SFN, MCM4, CBLC, MNX1, HIST1H2AI, HIST1H3B, HIST1H2AM, TOP2A, CCNA2, UBE2T, TUBB3, MELK | 0.01726561 |
Table 2. The GO analysis of the down-regulated DEGs in LUAD.
| Category | ID | Term | Genes | adj_pval |
|---|---|---|---|---|
| CC | GO:0044459 | Plasma membrane part | FXYD1, CAV3, CAV2, EMCN, CAV1, CLDN18, ACVRL1, LIMS2, CLDN5, SLC6A4, MME, AQP4, GNG11, VIPR1, NMUR1, SDPR, TEK, CALCRL, SCN7A, SLC1A1, CCRL1, PTPRB, RAMP3, AGER, LYVE1, THBD, ADRB1, SSTR1, CHRM1, CYBRD1, CA4, SCN4B, JAM2 | 0.00108239 |
| BP | GO:0001568 | Blood vessel development | SEMA5A, CAV1, EMCN, ACVRL1, LMO2, EPAS1, FOXF1, ROBO4, ANGPT1, FIGF | 0.00230295 |
| CC | GO:0005887 | Integral to plasma membrane | FXYD1, PTPRB, RAMP3, CAV2, CAV1, ACVRL1, SLC6A4, AQP4, MME, VIPR1, AGER, LYVE1, ADRB1, THBD, SSTR1, NMUR1, CHRM1, TEK, SCN4B, SCN7A, CALCRL, JAM2, SLC1A1, CCRL1 | 0.00271098 |
| BP | GO:0001944 | Vasculature development | SEMA5A, TCF21, CAV1, EMCN, ACVRL1, LMO2, EPAS1, FOXF1, ROBO4, ANGPT1, FIGF | 0.00271098 |
| BP | GO:0001525 | Angiogenesis | SEMA5A, EMCN, ACVRL1, EPAS1, ROBO4, ANGPT1, FIGF | 0.00337538 |
| BP | GO:0007155 | Cell adhesion | CLDN18, EMCN, CLDN5, RADIL, SEMA5A, LYVE1, WISP2, SRPX, COL6A6, FBLN5, TEK, JAM2, MFAP4, AOC3 | 0.00362546 |
| BP | GO:0022610 | Biological adhesion | CLDN18, EMCN, CLDN5, RADIL, SEMA5A, LYVE1, WISP2, SRPX, COL6A6, FBLN5, TEK, JAM2, MFAP4, AOC3 | 0.00494697 |
| CC | GO:0031226 | Intrinsic to plasma membrane | FXYD1, PTPRB, RAMP3, CAV2, CAV1, ACVRL1, SLC6A4, AQP4, MME, VIPR1, AGER, LYVE1, ADRB1, THBD, SSTR1, NMUR1, CHRM1, TEK, SCN4B, SCN7A, CALCRL, JAM2, SLC1A1, CCRL1 | 0.00522618 |
| BP | GO:0048514 | Blood vessel morphogenesis | SEMA5A, CAV1, EMCN, ACVRL1, EPAS1, FOXF1, ROBO4, ANGPT1, FIGF | 0.00561097 |
| CC | GO:0005576 | Extracellular region | A2M, EMCN, ITLN2, OGN, RSPO4, WISP2, COL6A6, SOSTDC1, GPX3, HEG1, SFTPC, ANGPT1, FIBIN, FAM150B, PI16, FIGF, MYOC, LPL, HYAL1, SPARCL1, CAMP, AGER, PLAC9, C2ORF40, GKN2, CHRDL1, ADAMTS8, THBD, CLEC3B, FBLN5, GDF10, MFAP4 | 0.00599981 |
| BP | GO:0042127 | Regulation of cell proliferation | HYAL1, CAV2, CAV1, ACVRL1, TBX2, VIPR1, AGER, WISP2, ADAMTS8, SSTR1, CHRM1, TEK, RTKN2, FABP4, CALCRL, FIGF | 0.00656309 |
| BP | GO:0031579 | Membrane raft organization | CAV3, CAV2, CAV1 | 0.00679421 |
Table 3. The KEGG analysis of the up-regulated DEGs in LUAD.
| Term | Count | Ratio | FDR |
|---|---|---|---|
| Hsa00534 Glycosaminoglycan biosynthesis-heparan sulfate/heparin | 1 | 0.02912621 | 0.20276459 |
| Hsa03430 Mismatch repair | 2 | 0.04032258 | 0.20276459 |
| Hsa01230 Biosynthesis of amino acids | 2 | 0.02234637 | 0.16376332 |
| Hsa00100 Steroid biosynthesis | 3 | 0.0462963 | 0.20276459 |
| Hsa04115 p53 signaling pathway | 3 | 0.04 | 0.16376332 |
| Hsa04130 SNARE interactions in vesicular transport | 3 | 0.02285714 | 0.21164483 |
| Hsa00512 Mucin type O-Glycan biosynthesis | 4 | 0.04301075 | 0.21164483 |
| Hsa00601 Glycosphingolipid biosynthesis-lacto and neolacto series | 5 | 0.02040816 | 0.20276459 |
| Hsa04950 Maturity onset diabetes of the young | 5 | 0.08333333 | 0.20276459 |
| Hsa012102 Oxocarboxylic acid metabolism | 6 | 0.05 | 0.20276459 |
| Hsa00220 Arginine biosynthesis | 2 | 0.03174603 | 0.20276459 |
| Hsa04215 Apoptosis-multiple species | 6 | 0.04347826 | 0.21164483 |
| Hsa00250 Alanine, aspartate and glutamate metabolism | 6 | 0.08695652 | 0.21164483 |
| Hsa00232 Caffeine metabolism | 4 | 0.05681818 | 0.16376332 |
| Hsa03030 DNA replication | 4 | 0.03488372 | 0.21164483 |
Table 4. The KEGG analysis of the down-regulated DEGs in LUAD.
| Term | Count | Ratio | FDR |
|---|---|---|---|
| Hsa04080 Neuroactive ligand-receptor interaction | 6 | 0.02158273 | 0.06041271 |
| Hsa01100 Metabolic pathways | 6 | 0.00483871 | 0.50410934 |
| Hsa04151 PI3K-Akt signaling pathway | 5 | 0.01457726 | 0.12284827 |
| Hsa04261 Adrenergic signaling in cardiomyocytes | 4 | 0.02649007 | 0.07636768 |
| Hsa04024 cAMP signaling pathway | 4 | 0.0199005 | 0.12086446 |
| Hsa04510 Focal adhesion | 4 | 0.01941748 | 0.12086446 |
| Hsa04144 Endocytosis | 4 | 0.01515152 | 0.1506064 |
| Hsa00350 Tyrosine metabolism | 3 | 0.08571429 | 0.04917388 |
| Hsa00982 Drug metabolism - cytochrome P450 | 3 | 0.04411765 | 0.07636768 |
| Hsa03320 PPAR signaling pathway | 3 | 0.04109589 | 0.07636768 |
| Hsa05100 Bacterial invasion of epithelial cells | 3 | 0.03703704 | 0.08027977 |
| Hsa04974 Protein digestion and absorption | 3 | 0.03333333 | 0.09115089 |
| Hsa04670 Leukocyte transendothelial migration | 3 | 0.025 | 0.12284827 |
| Hsa04530 Tight junction | 3 | 0.02158273 | 0.1506064 |
Establishment of the PPI network and module analyses of genes in LUAD
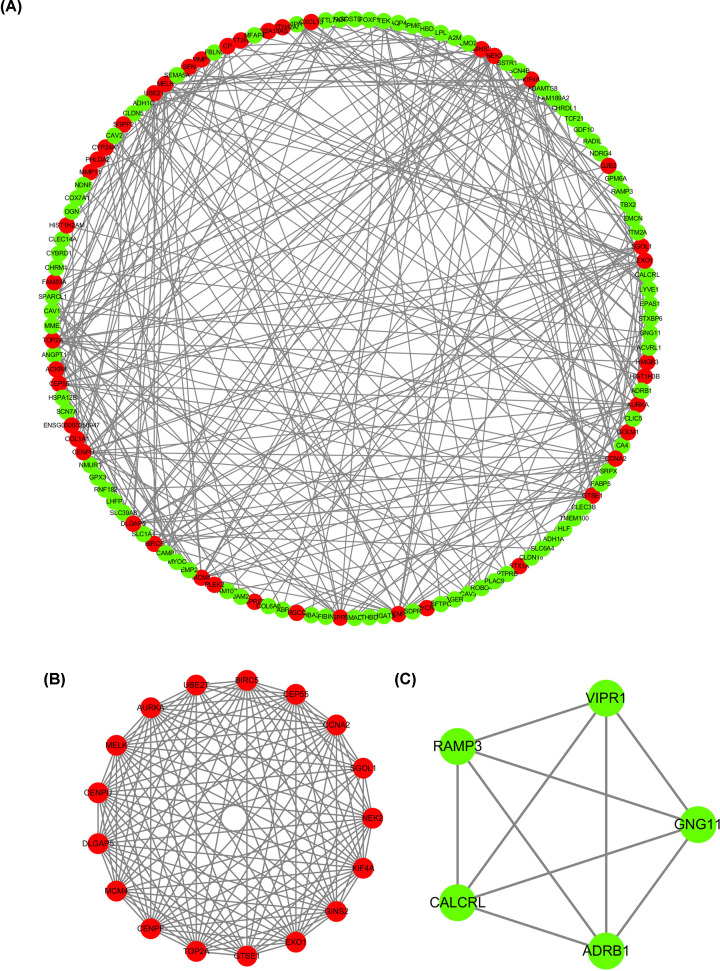
To further predict the underlying functions of DEGs, we analyzed any potential co-expressed mRNAs of these DEGs and determined their associations by PPIs. As presented in Figure 3A, the entire PPI network revealed potential interrelationships between a number of genes, which might provide a foundation for the pathway research in LUAD. Moreover, we also analyzed two of the modules, and the relative networks are displayed in Figure 3B,C. Genes with the most highly connected cluster were extracted using the MCODE plug-in in Cytoscape after PPI networks were analyzed. The DMNC and MCC algorithms of Cytoscape were used to identify hub genes. The top ten hub genes based on the two methods were screened, and seven mutual hub genes were isolated, including BIRC5, DLGAP5, CENPF, KIF4A, TOP2A, AURKA, and CCNA2. These seven genes were all significantly up-regulated in LUAD samples compared with normal samples and were used for further analysis.
Figure 3. Establishment of the PPI network and modules analyses of genes in LUAD.
(A) The overall PPI network in LUAD. (B) The PPI network of module 1 in LUAD. (C) The PPI network of module 2 in LUAD.
Validation of seven hub genes expression in LUAD samples based on TCGA database
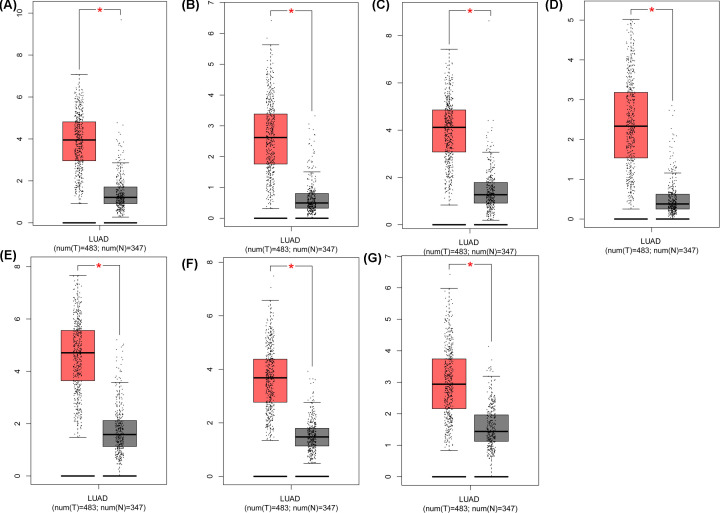
We utilized the Oncomine database to further reveal the expression of these seven hub genes in LUAD. The expression of BIRC5, DLGAP5, CENPF, KIF4A, TOP2A, AURKA, and CCNA2 (Figure 4) was significantly higher in LUAD tissue samples than that in nontumor lung tissue samples. Meanwhile, TCGA database was used to further identify the levels of the seven hub genes in LUAD. The results from the TCGA database displayed the same expression trends as in the Oncomine database (Figure 5). Therefore, these seven hub genes were significantly highly expressed in LUAD.
Figure 4. Validation of the expression of seven hub genes in LUAD samples based on the Oncomine database.
The expression levels of (A) BIRC5, (B) DLGAP5, (C) CENPF, (D) KIF4A, (E) TOP2A, (F) AURKA, and (G) CCNA2 in LUAD were obtained from the Oncomine database.
Figure 5. Identification of the expression of the seven hub genes in LUAD samples in the TCGA database.
Box plots show the expression of (A) BIRC5, (B) DLGAP5, (C) CENPF, (D) KIF4A, (E) TOP2A, (F) AURKA, and (G) CCNA2 in LUAD from the TCGA database. *P<0.05.
Overall survival and disease-free survival analyses of seven hub genes in LUAD
To further assess the prognostic values of the seven hub genes in LUAD, TCGA, and GTEx datasets were obtained from GEPIA. The results of the overall survival analyses revealed that the high expression of BIRC5, DLGAP5, CENPF, KIF4A, TOP2A, AURKA, and CCNA2 (Figure 6) was associated with shorter overall survival than in patients with low expression. Simultaneously, the data also indicated that high expression of the seven hub genes could shorten the disease-free survival in LUAD (Figure 7). Therefore, the seven hub genes were associated with poor prognosis of LUAD.
Figure 6. Overall survival analyses of the seven hub genes in LUAD on the basis of the TCGA and GTEx data in GEPIA.
Statistical graphs indicate the results of overall survival analyses of (A) BIRC5, (B) DLGAP5, (C) CENPF, (D) KIF4A, (E) TOP2A, (F) AURKA, and (G) CCNA2 in LUAD. Red lines represent the high expression of genes and the blue lines represent low expression of genes.
Figure 7. Disease-free survival analyses of the seven hub genes in LUAD based on TCGA and GTEx data in GEPIA.
In accordance with the TCGA and GTEx data in GEPIA, the disease-free survival of the seven hub genes are shown: (A) BIRC5, (B) DLGAP5, (C) CENPF, (D) KIF4A, (E) TOP2A, (F) AURKA, and (G) CCNA2.
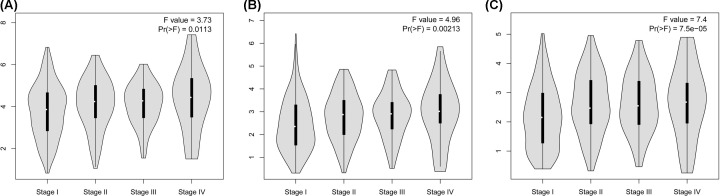
CENPF, DLGAP5, and KIF4A expression was positively correlated with the clinical stage of LUAD
We screened the genes (CENPF, DLGAP5, and KIF4A), which were most likely to be associated with LUAD and analyzed any possible relationships with the clinical stage of LUAD. As displayed in Figure 8, the expression levels of CENPF, DLGAP5, and KIF4A were dramatically increased in the different stages of LUAD, and the expression of CENPF, DLGAP5, and KIF4A were gradually increased as the LUAD stages increased. Thus, we suggested that high expression of CENPF, DLGAP5, and KIF4A might be associated with higher stages of LUAD. Meanwhile, the seven hub genes including KIF4A, CENPF, and DLGAP5 contributed mostly to the GO analysis results (Figure 2A, Table 1).
Figure 8. CENPF, DLGAP5, and KIF4A expression was positively correlated with the clinical stage of LUAD.
The levels of CENPF, DLGAP5, and KIF4A expression were analyzed in different stages of LUAD.
Expression validation of the seven hub genes by qRT-PCR
We compared the mRNA expression levels of the seven hub genes in LUAD tissues and paired normal tissues. As shown in Supplementary Figure S1, compared with their expression in paired normal samples, the expression of BIRC5, DLGAP5, CENPF, KIF4A, TOP2A, AURKA, and CCNA2 was significantly increased in LUAD samples (P<0.01).
KIF4A was highly expressed in LUAD cells
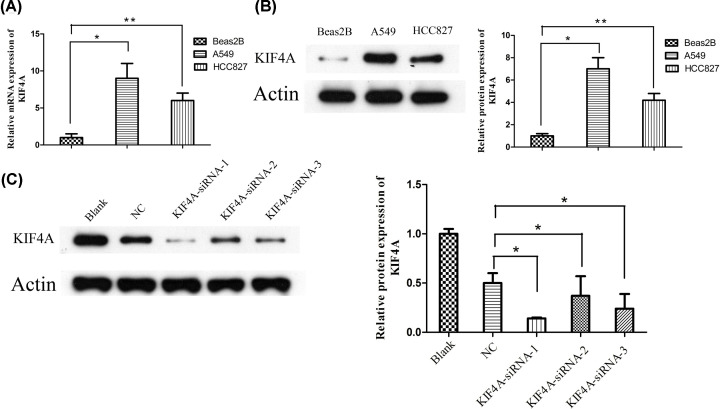
After identifying the seven hub genes and given the limited number of studies focusing on the relationship between KIF4A and the progression of LUAD, we further studied the expression and function of KIF4A in vitro. As shown in Figure 9A,B, the mRNA and protein expression levels of KIF4A were upregulated in LUAD cells (HCC827 and A549) compared with that in normal bronchial epithelial cells Beas2B.
Figure 9. KIF4A was significantly up-regulated in LUAD cells.
(A) mRNA expression levels of KIF4A in the LUAD cells were higher than those in normal bronchial epithelial cells. (B) Expression of KIF4A protein in the LUAD cell lines was higher than in normal bronchial epithelial cells. (C) The protein expression levels of KIF4A targeted by siRNA-1, siRNA-2, and siRNA-3. *, P-value<0.05; **, P-value<0.01.
Knockdown of KIF4A decreased the migration in A549 cell lines
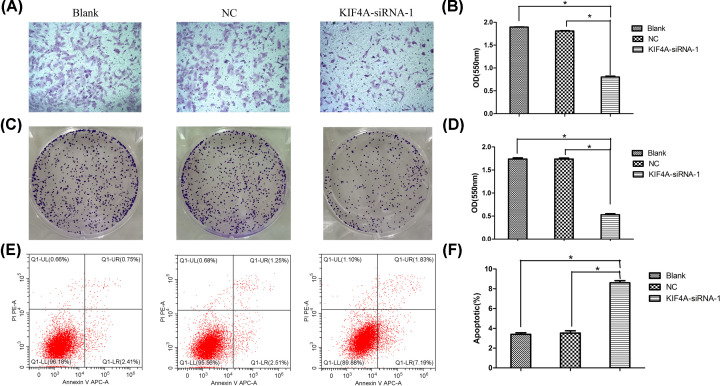
To further confirm the role of KIF4A, siRNA experiments were performed in A549 cells. The efficiency of the knockdown of KIF4A expression was examined by Western blotting, as shown in Figure 9C. To investigate the relationship between KIF4A and migration, we conducted the Transwell assay. The results revealed that the OD value of migrated KIF4A-siRNA-1 cells was decreased significantly, compared with that in the blank and the negative control groups (P<0.05) (Figure 10A,B).
Figure 10. Silencing KIF4A inhibited proliferation, migration, and promoted apoptosis in the A549 cell line.
(A and B) Silencing KIF4A suppressed the migration of A549 cells. (C and D) KIF4A knockdown decreased cell proliferation capacity of A549 cells. (E and F) KIF4A knockdown promoted the apoptotic rate of A549 cells; *, P-value<0.05.
Effect of KIF4A on the proliferation and apoptosis of A549 cell lines
The colony formation assay showed that the OD values of the clone numbers decreased significantly in the KIF4A-siRNA-1 cells compared with that of the blank group and the negative control group (P<0.05) (Figure 10C,D). In addition, the apoptosis rate of the KIF4A-siRNA-1 cells increased significantly compared with that in control group as determined by the flow cytometry assay (P<0.05) (Figure 10E,F). Together, these results indicated that KIF4A suppression decreased cell proliferation and increased apoptosis abilities in A549 cell lines.
Discussion
In recent years, with the development of high-throughput sequencing and bioinformatics, a novel research model was formed [30,31]. Massive quantities of research data generated by microarrays have been uploaded to the GEO database for sharing, and scientific researchers classified and downloaded the relevant data in accordance with their area of interest [32]. Meanwhile, researchers have further integrated and compared similar experimental data carried out by different laboratories, so as to unearth potentially useful data.
LUAD is a type of lung cancer with a high degree of malignancy, poor prognosis, and characterized by the increased likelihood of developing chest, liver, bone, and even intracranial metastasis [33,34]. At present, the majority of LUAD patients are diagnosed in the advanced stages and cannot be treated with surgery due to the presence of distant metastasis [35]. Although chemoradiotherapy can improve symptoms, the adverse reactions are significant [36]. Therefore, attempts at improving the survival rate of patients have become the main purpose of LUAD treatment. High-throughput genetic sequencing and bioinformatics might provide a new approach for the study of LUAD pathogenesis.
In our study, we selected ‘LUAD’ as the keyword to search for related datasets from the GEO datasets, and then performed a detailed biological analysis of these DEGs. Subsequently, in order to better understand the DEGs and their associated gene function, related pathways, protein interaction in LUAD, we conducted bioinformatics studies on LUAD using GO functional annotation, KEGG pathway analysis, and PPIs analysis. Overall, our results may provide a theoretical basis for exploring the pathogenesis of LUAD and further studies.
The results of the GO function annotation revealed that the upregulated DEGs in LUAD were mainly concentrated in the nucleosome, centriole, and spindle microtubule, while the down-regulated DEGs in LUAD were mainly enriched in the plasma membrane, in blood vessel development, and in functions integral to the plasma membrane. Meanwhile, KEGG pathway analysis revealed that the up-regulated DEGs in LUAD were mainly enriched in alanine, aspartate, and glutamate metabolism, and oxocarboxylic acid metabolism, while the downregulated DEGs in LUAD were mainly enriched in neuroactive ligand–receptor interactions and in tyrosine metabolism. Moreover, we obtained the key DEGs in LUAD using PPI network analysis.
With the intensive study of the PPI network, the topological structure of the PPI network obtained is frequently associated with disease-related proteins [37,38]. Previous studies have documented that the proteins in the same disease are more likely to interact and be co-expressed in the network [39], while the functional changes of neighboring node proteins associated with a disease are more likely to cause the same or similar diseases [40,41]. Meanwhile, the smaller nodes that interact with numerous proteins in the protein network are called hubs [42]. Hub genes may have more crucial biological functions in disease development. In biological networks, the change in expression of hub proteins will generate serious destruction of the connected network and may seriously affect the biological functions of cells [43]. Currently, a large number of key hub genes have been identified in a variety of cancers, such as hepatocellular carcinoma [44], bladder cancer [45], renal cell carcinoma [46], and osteosarcoma [47]. However, a number of hub genes in LUAD remain undetermined. In our study, we have filtered seven key hub genes including BIRC5, DLGAP5, CENPF, KIF4A, TOP2A, AURKA, and CCNA2 in LUAD. Simultaneously, we found that these seven hub genes were significantly up-regulated in LUAD using Oncomine data and TCGA datasets, and up-regulation of these seven hub genes were associated with a poor prognosis of LUAD. In addition, we determined that the expression of CENFP, KIF4A, and DLGAP5 might be associated with the metastasis of LUAD. Moreover, we further confirmed the increased mRNA expression level of these seven key hub genes in primary LUAD samples compared with paired normal samples.
In the present study, we provided evidence that KIF4A was significantly up-regulated in LUAD cells. To date, only one study has reported that the knockdown KIF4A might suppress the growth of NSCLC cell lines [48]. As for other tumor types, KIF4A has been reported to enhance carcinoma development by promoting cell cycle progression in vitro and in vivo in colorectal cancer and hepatocellular carcinoma [49,50]. Our study revealed that silencing KIF4A expression inhibited proliferation, migration, and promoted apoptosis in A549 cell lines, which was in agreement with the previous studies indicated above.
Conclusions
We have identified new DEGs and pathways active in LUAD, and more specifically, KIF4A as a hub gene with the capacity to promote the progression of LUAD. Our findings provide researchers with a potential therapeutic target for molecular cancer therapy.
Supplementary Material
Acknowledgements
We acknowledge and appreciate our colleagues for their valuable efforts and comments relative to this paper.
Abbreviations
- DEG
differentially expressed gene
- GEO
gene expression omnibus
- LUAD
lung adenocarcinoma
- PPI
protein–protein interaction
- qRT-PCR
real-time quantitative PCR
- STRING
Search Tool for the Retrieval of Interacting Genes
Data Availability
The datasets used or analyzed during the current study are available from the GEO (https://www.ncbi.nlm.nih.gov/geo/), Oncomine (https://www.oncomine.org/resource/login.html), GEPIA (http://gepia.cancer-pku.cn/), STRING (https://string-db.org/), KOBAS (http://kobas.cbi.pku.edu.cn/), DAVID (https://david.ncifcrf.gov/), and TCGA (https://cancergenome.nih.gov/) databases.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of Hunan [grant number 2020JJ5866], grants from Science and Technology Project of Changsha [grant number kq1907013], and grants from Xiangya Hospital Central South University postdoctoral foundation.
Author Contribution
Yexun Song conceived the project and designed the experiments. Wenfang Tang analyzed the data and performed most of the experiments. Hui Li carried out the bioinformatics analysis, wrote the manuscript, and provided technical support.
References
- 1.Gazdar A.F. and Zhou C. (2018) Lung cancer in never-smokers: a different disease. IASLC Thoracic Oncology, pp. 23.e3–29.e3, Elsevier [Google Scholar]
- 2.Torre L.A., Siegel R.L. and Jemal A. (2016) Lung cancer statistics. Lung Cancer and Personalized Medicine, pp. 1–19, Springer; 10.1007/978-3-319-24223-1_1 [DOI] [PubMed] [Google Scholar]
- 3.Oser M.G., Niederst M.J., Sequist L.V. and Engelman J.A. (2015) Transformation from non-small-cell lung cancer to small-cell lung cancer: molecular drivers and cells of origin. Lancet Oncol. 16, e165–e172 10.1016/S1470-2045(14)71180-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hirsch F.R., Suda K., Wiens J. and Bunn P.A. Jr (2016) New and emerging targeted treatments in advanced non-small-cell lung cancer. Lancet North Am. Ed. 388, 1012–1024 10.1016/S0140-6736(16)31473-8 [DOI] [PubMed] [Google Scholar]
- 5.Liu Z., Liu B. and Li C. (2019) A case report of metastatic bilateral ovarian cancer due to non-small cell lung cancer with ALK gene rearrangement. Eur. J. Gynaecol. Oncol. 40, 151–153 [Google Scholar]
- 6.Asiedu M.K., Reed N.R., Aubry M.C., Roden A.C. and Wigle D.A. (2019) Genomic Alterations in Lung Adenocarcinoma Precursor Lesions, AACR [Google Scholar]
- 7.Coroller T.P., Grossmann P., Hou Y., Velazquez E.R., Leijenaar R.T., Hermann G. et al. (2015) CT-based radiomic signature predicts distant metastasis in lung adenocarcinoma. Radiother. Oncol. 114, 345–350 10.1016/j.radonc.2015.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin J.J., Cardarella S., Lydon C.A., Dahlberg S.E., Jackman D.M., Jänne P.A. et al. (2016) Five-year survival in EGFR-mutant metastatic lung adenocarcinoma treated with EGFR-TKIs. J. Thoracic Oncol. 11, 556–565 10.1016/j.jtho.2015.12.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inamura K. and Ishikawa Y. (2016) MicroRNA in lung cancer: novel biomarkers and potential tools for treatment. J. Clin. Med. 5, 36 10.3390/jcm5030036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Postmus P., Kerr K., Oudkerk M., Senan S., Waller D., Vansteenkiste J. et al. (2017) Early and locally advanced non-small-cell lung cancer (NSCLC): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 28, iv1–iv21 10.1093/annonc/mdx222 [DOI] [PubMed] [Google Scholar]
- 11.Vinner L., Mourier T., Friis-Nielsen J., Gniadecki R., Dybkaer K., Rosenberg J. et al. (2015) Investigation of human cancers for retrovirus by low-stringency target enrichment and high-throughput sequencing. Sci. Rep. 5, 13201 10.1038/srep13201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen L., Sun F., Yang X., Jin Y., Shi M., Wang L. et al. (2017) Correlation between RNA-Seq and microarrays results using TCGA data. Gene 628, 200–204 10.1016/j.gene.2017.07.056 [DOI] [PubMed] [Google Scholar]
- 13.Yuan C., Xiang L., Bai R., Cao K., Gao Y., Jiang X. et al. (2019) MiR-195 restrains lung adenocarcinoma by regulating CD4+ T cell activation via the CCDC88C/Wnt signaling pathway: a study based on the Cancer Genome Atlas (TCGA), Gene Expression Omnibus (GEO) and bioinformatic analysis. Ann. Transl. Med. 7, 263 10.21037/atm.2019.05.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu Q., Sun Y., Zhou Q., He Q. and Qian H. (2018) Identification of key genes and pathways by bioinformatics analysis with TCGA RNA sequencing data in hepatocellular carcinoma. Mol. Clin. Oncol. 9, 597–606 10.3892/mco.2018.1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee E. and Moon A. (2016) Identification of biomarkers for breast cancer using databases. J. Cancer Prevent. 21, 235 10.15430/JCP.2016.21.4.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu W., Sun Y., Zheng X., Ma J., Hu X.-Y., Gao T. et al. (2018) Identification of gastric cancer-related circular RNA through microarray analysis and bioinformatics analysis. BioMed Res. Int. 2018, 2381680 10.1155/2018/2381680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu K., Fu Q., Liu Y. and Wang C. (2019) An integrative bioinformatics analysis of microarray data for identifying hub genes as diagnostic biomarkers of preeclampsia. Biosci. Rep. 39, BSR20190187 10.1042/BSR20190187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang S., Wang L., Li S., Zhang W., Ma X., Cheng G. et al. (2018) Identification of Potential Key Genes Associated with Adipogenesis through Integrated Analysis of Five Mouse Transcriptome Datasets. Int. J. Mol. Sci. 19, 3557 10.3390/ijms19113557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen S., Kong J., Qiu Y., Yang X., Wang W. and Yan L. (2019) Identification of core genes and outcomes in hepatocellular carcinoma by bioinformatics analysis. J. Cell. Biochem. 120, 10069–10081 10.1002/jcb.28290 [DOI] [PubMed] [Google Scholar]
- 20.Klopfenstein D., Zhang L., Pedersen B.S., Ramírez F., Vesztrocy A.W., Naldi A. et al. (2018) GOATOOLS: A Python library for Gene Ontology analyses. Sci. Rep. 8, 10872 10.1038/s41598-018-28948-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanehisa M., Goto S., Kawashima S., Okuno Y. and Hattori M. (2004) The KEGG resource for deciphering the genome. Nucleic Acids Res. 32, D277–D280 10.1093/nar/gkh063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Z., Shang P., Li Q., Wang L., Chamba Y., Zhang B. et al. (2017) iTRAQ-based proteomic analysis reveals key proteins affecting muscle growth and lipid deposition in pigs. Sci. Rep. 7, 46717 10.1038/srep46717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mering C.V., Huynen M., Jaeggi D., Schmidt S., Bork P. and Snel B. (2003) STRING: a database of predicted functional associations between proteins. Nucleic Acids Res. 31, 258–261 10.1093/nar/gkg034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang Z., Li C., Kang B., Gao G., Li C. and Zhang Z. (2017) GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 45, W98–W102 10.1093/nar/gkx247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tomczak K., Czerwińska P. and Wiznerowicz M. (2015) The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemporary Oncol. 19, A68 10.5114/wo.2014.47136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lonsdale J., Thomas J., Salvatore M., Phillips R., Lo E., Shad S. et al. (2013) The genotype-tissue expression (GTEx) project. Nat. Genet. 45, 580 10.1038/ng.2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Consortium G. (2015) The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660 10.1126/science.1262110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song Y.X., Li W., Peng X.W., Xie J., Li H.Q. and Tan G.L. (2017) Inhibition of autophagy results in a reversal of taxol resistance in nasopharyngeal carcinoma by enhancing taxol-induced caspase-dependent apoptosis. Am. J. Transl. Res. 9, 1934–1942 [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X.Y., Wang K., Xiao J., Song Y.X. and Li H.Q. (2020) Neferine sensitized Taxol-resistant nasopharygeal carcinoma to Taxol by inhibiting EMT via downregulating miR-130b-5p. Biochem. Bioph. Res. Co. 531, 573–580 10.1016/j.bbrc.2020.08.008 [DOI] [PubMed] [Google Scholar]
- 30.Fonseca N.A., Rung J., Brazma A. and Marioni J.C. (2012) Tools for mapping high-throughput sequencing data. Bioinformatics 28, 3169–3177 10.1093/bioinformatics/bts605 [DOI] [PubMed] [Google Scholar]
- 31.Nakazato T., Ohta T. and Bono H. (2013) Experimental design-based functional mining and characterization of high-throughput sequencing data in the sequence read archive. PLoS ONE 8, e77910 10.1371/journal.pone.0077910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M. et al. (2012) NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 41, D991–D995 10.1093/nar/gks1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Network CGAR (2014) Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543 10.1038/nature13385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bandoh N., Ichikawa H., Asahi A., Kono M., Harabuchi S., Sato R. et al. (2019) Lung Adenocarcinoma with Cheek Dysesthesia as an Initial Symptom: A Case Report and Literature Review. Case Rep. Oncol. 12, 650–656 10.1159/000502053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Popper H.H. (2016) Progression and metastasis of lung cancer. Cancer Metastasis Rev. 35, 75–91 10.1007/s10555-016-9618-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hsia T.C., Liang J.A., Li C.C. and Chien C.R. (2018) Comparative effectiveness of concurrent chemoradiotherapy versus EGFR‐tyrosine kinase inhibitors for the treatment of clinical stage IIIb lung adenocarcinoma patients with mutant EGFR. Thoracic. Cancer 9, 1398–1405 10.1111/1759-7714.12847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Francois M., Donovan P. and Fontaine F. (2018) Modulating transcription factor activity: Interfering with protein-protein interaction networks. Seminars Cell Development. Biol. 99, 12–19Elsevier [DOI] [PubMed] [Google Scholar]
- 38.Zhao B., Wang J., Li M., Li X., Li Y., Wu F.-X. et al. (2016) A new method for predicting protein functions from dynamic weighted interactome networks. IEEE Trans. Nanobioscience 15, 131–139 10.1109/TNB.2016.2536161 [DOI] [PubMed] [Google Scholar]
- 39.Goh K.-I., Cusick M.E., Valle D., Childs B., Vidal M. and Barabási A.-L. (2007) The human disease network. Proc. Natl. Acad. Sci. 104, 8685–8690 10.1073/pnas.0701361104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oti M., Snel B., Huynen M.A. and Brunner H.G. (2006) Predicting disease genes using protein–protein interactions. J. Med. Genet. 43, 691–698 10.1136/jmg.2006.041376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karni S., Soreq H. and Sharan R. (2009) A network-based method for predicting disease-causing genes. J. Comput. Biol. 16, 181–189 10.1089/cmb.2008.05TT [DOI] [PubMed] [Google Scholar]
- 42.Yu D., Lim J., Wang X., Liang F. and Xiao G. (2017) Enhanced construction of gene regulatory networks using hub gene information. BMC Bioinformatics 18, 186 10.1186/s12859-017-1576-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Batada N.N., Hurst L.D. and Tyers M. (2006) Evolutionary and physiological importance of hub proteins. PLoS Comput. Biol. 2, e88 10.1371/journal.pcbi.0020088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen P., Wang F., Feng J., Zhou R. and Zhao Q. (2017) Co-expression network analysis identified six hub genes in association with metastasis risk and prognosis in hepatocellular carcinoma. Oncotarget 8, 48948–48958 10.18632/oncotarget.16896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang D.Q., Zhou C.K., Chen S.Z., Yue Y. and Shi B.K. (2017) Identification of hub genes and pathways associated with bladder cancer based on co-expression network analysis. Oncol. Lett. 14, 1115 10.3892/ol.2017.6267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y., Chen L., Wang G., Cheng S. and Wang X. (2018) Fifteen hub genes associated with progression and prognosis of clear cell renal cell carcinoma identified by coexpression analysis: WANG et al. J. Cell. Physiol. 234, 10225–10237 [DOI] [PubMed] [Google Scholar]
- 47.Wu Y.S., Liu J.J. and Zheng C.J. (2017) The mAP-KL algorithm combined with mutual information network used to screen hub genes in osteosarcoma. Current Bioinformatics 11, 1 10.2174/1574893611666160726170147 [DOI] [Google Scholar]
- 48.Taniwaki M., Takano A., Ishikawa N., Yasui W., Inai K., Nishimura H. et al. (2007) Activation of KIF4A as a prognostic biomarker and therapeutic target for lung cancer. Clin. Cancer Res. 13, 6624–6631 10.1158/1078-0432.CCR-07-1328 [DOI] [PubMed] [Google Scholar]
- 49.Hou P.-F., Jiang T., Chen F., Shi P.-C., Li H.-Q., Bai J. et al. (2018) KIF4A facilitates cell proliferation via induction of p21-mediated cell cycle progression and promotes metastasis in colorectal cancer. Cell Death Dis. 9, 477 10.1038/s41419-018-0550-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bai L., Ren Y. and Cui T. (2020) Overexpression of CDCA5, KIF4A, TPX2, and FOXM1 Coregulated Cell Cycle and Promoted Hepatocellular Carcinoma Development. J. Comput. Biol. 27, 965–974 10.1089/cmb.2019.0254 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used or analyzed during the current study are available from the GEO (https://www.ncbi.nlm.nih.gov/geo/), Oncomine (https://www.oncomine.org/resource/login.html), GEPIA (http://gepia.cancer-pku.cn/), STRING (https://string-db.org/), KOBAS (http://kobas.cbi.pku.edu.cn/), DAVID (https://david.ncifcrf.gov/), and TCGA (https://cancergenome.nih.gov/) databases.