Highlights
-
•
Strain 135 is first dibenzothiophene-degrading Gordonia with genome fully assembled.
-
•
This is the first strain of Gordonia absorbing thiophene sulfur without dsz genes.
-
•
The strain utilized 45.26 % of dibenzothiophene within 150 h of growth at 26 °C.
Abbreviations: DBT, dibenzothiophene; SFM, sulfur-free mineral medium; HBPS, 2-hydroxybiphenyl 2-sulfinate; 2HВP, 2-hydroxybiphenyl
Keywords: Gordonia, Dibenzothiophene, Biodegradation, Genome assembly
Abstract
Sulfur is the third most abundant element in crude oil. Up to 70 % of sulfur in petroleum is found in the form of dibenzothiophene (DBT) and substituted DBTs. The aim of this work was to study the physiological, biochemical and genetical characteristics of Gordonia alkanivorans 135 capable of using DBT as the sole source of sulfur. The genome of G. alkanivorans 135 consists of a 5,039,827 bp chromosome and a 164,963 bp circular plasmid. We found the absence of dsz operon present in most DBT degrading bacteria, but discovered other genes that are presumably involved in DBT utilization by G. alkanivorans 135. The strain utilized 45.26 % of DBT within 150 h of growth at 26 °C. This is the first strain of Gordonia capable of absorbing thiophene sulfur without the aid of the dsz genes.
1. Introduction
Sulfur is one of the most commonly found elements in crude oil. All types of fossil fuel contain a variety of organic and inorganic sulfur compounds. Depending on the type of crude oil, the concentration of sulfur in the fraction used for the production of diesel fuel can vary from <500 to >5,000 mg/L [1]. Most of sulfur in crude oil is bound in the form of condensed thiophenes. Up to 70 % of sulfur in crude oil is in the form of dibenzothiophene (DBT) and its substituted derivatives (methylated and benzo-DBT) [2].
Heteroaromatic compounds containing sulfur in their structure are among the most environmentally stable components of crude oil [3]. The use of fuels containing sulfur compounds leads to emissions of sulfur oxides into the atmosphere and is one of the causes of acid rain. In addition, even low concentrations of sulfur oxides cause irritation of the human respiratory tract and damage to plants [4,5].
The problem of microbial conversion of DBT and its derivatives has been investigated in a number of works [1,5,6]. There are several known pathways of DBT catabolism by bacteria. The oxidative and carbon-carbon degradation of DBT known as the Kodama pathway consists of three main steps: lateral dioxygenation of one of the homolytic rings, ring disruption, and hydrolysis leading to hydroxyformyl benzothiophene as the final product. All steps of the Kodama pathway are executed by naphthalene degradation enzymes which utilize DBT as an alternative substrate. Although hydroxyformyl benzothiophene is considered a dead end product, other possible products and even complete mineralization of DBT by mixed bacterial cultures have been described [7]. The Kodama pathway does not involve the use of sulfur from DBT and therefore it is not a desulfurization pathway. Another pathway of ring disruption that results in mineralization of DBT has been described by Van Afferden et al. [8]. They have isolated Brevibacterium sp. DO capable of using DBT for growth as the sole source of carbon, sulfur and energy. During DBT mineralization, three metabolites have been identified: DBT sulfoxide (DBTO), DBT sulfone (DBTO2) and benzoate. This pathway resulted in the complete mineralization of DBT with the release of the sulfur atom in the form of sulfite which was subsequently oxidized to sulfate.
The ability of microorganisms to use DBT as the sole source of sulfur helps to remove excess sulfur from crude oil and products of its refining. The presence of DBT desulfurization pathways has been found in microorganisms of various genera [[9], [10], [11]]. The discovery of a highly specific 4S pathway (sulfur-specific pathway) in Rhodococcus erythropolis IGTS8 (ATCC 53968) isolated in 1990 at the Gas Technology Institute (USA) is of great importance for the study of microbial desulfurization of fossil fuels. Denom et al. [6] isolated from IGTS8 a large linear plasmid pSOX containing a fragment responsible for the oxidation of sulfur and the transfer of this trait to strains incapable of desulfurization. The soxABC (later renamed as dszABC) genes responsible for desulfurization comprise an operon-like structure. The dszC gene encodes a DBT monooxygenase which catalyzes the oxidation of DBT to DBT sulfoxide and then to DBT sulfone. The dszA gene encodes a DBT sulfone monooxygenase which catalyzes the oxidation of DBT sulfone to 2-hydroxybiphenyl 2-sulfinate (HBPS). Finally, the dszB gene encodes a HBPS sulfolyase (desulfinase) which catalyzes the conversion of HBPS to 2-hydroxybiphenyl and sulfite. Moreover, the 4S pathway requires an additional gene located on the chromosome – dszD. This gene encodes a NADH-FMN oxidoreductase which regenerates FMN H, a cofactor necessary for the first two reactions. The DszB desulfinase is present in the cytoplasm at a concentration several times lower than that of DszA and DszC, and it is the slowest of the three Dsz enzymes, which makes it the limiting stage of the desulfurization process. The three-dimensional structure of DszB indicates that this enzyme belongs to a new family of desulfinases which require cysteine for catalytic activity and the total amount of which is similar to the amount of periplasmic substrate-binding components of sulfur-regulated transporters of the ABC type [12].
The ability to use DBT as the sole source of sulfur is one of the distinctive features of bacteria of the genus Gordonia, which puts them, in terms of the diversity of metabolic capabilities, on a par with rhodococci, well-known destructors of various pollutants [6,13]. Strains of the genus Gordonia that catabolize DBT are often mentioned in the current literature [14,15]. In most cases, these strains have a classic set of genes that comprise the operon-like structure of dsz. The dszC gene encodes a DBT monooxygenase which catalyzes the oxidation of DBT to DBT sulfoxide and then to DBT sulfone. The dszA gene encodes a DBT sulfone monooxygenase which catalyzes the oxidation of DBT sulfone to 2-hydroxybiphenyl 2-sulfinate (HBPS). Finally, the dszB gene encodes a HBPS sulfolyase (desulfinase) which catalyzes the conversion of HBPS to 2-hydroxybiphenyl (2HВP) and sulfite. Moreover, the 4S pathway requires an additional dszD gene located on the chromosome. This gene encodes a NADH-FMN oxidoreductase which regenerates FMN H, a cofactor required for the first two reactions. Although there is no direct evidence that the dsz genes are inducible, they are inhibited by sulfate and sulfur-containing amino acids.
When the ability of G. alkanivorans 135 to utilize thiophene sulfur was found, we suggested that the strain 135, like other DBT-desulfurizing Gordonia strains, carries dsz genes in the genome. But our study showed that the strain does not have the dsz genes. This is the first strain of Gordonia capable of absorbing thiophene sulfur without the aid of the dsz genes.
The aim of this work was to study the physiological, biochemical and genetical characteristics of the Gordonia alkanivorans 135 capable of using DBT as the sole source of sulfur.
2. Materials and methods
2.1. Chemicals
High purity grade (>98 %) phenanthrene, anthracene, naphthalene, hexadecane, eicosane, decane, toluene, benzol, dibenzothiophene, glucose, catechol were from Sigma–Aldrich (USA), Merck and Fluka (Germany). Dimethylformamide (DMFA) was from Riedel de Haen.
2.2. Microorganism and culture media
The microorganism used in this study was Gordonia alkanivorans 135 isolated in our laboratory [16].
As a culture medium, we used a desulfurized mineral medium (SFM) [17] containing: 1.22 g NH4Cl, 2.5 g KH2PO4, 2.5 g Na2HPO4·2H2O, 0.17 g MgCl2·6H2O, and Milli-Q water to 1 L. The SFM medium was supplemented with 0.5 mL of a solution of microelements without sulfur containing 25 g/L EDTA, 2.14 g/L ZnCl2, 2.5 g/L MnCl2·4H2O, 0.3 g/L CoCl2·6H2O, 0.2 g/L CuCl2·2H2O, 0.4 g/L NaMoO4·2H2O, 4.5 g/L CaCl2·2H2O, 2.9 g/L FeCl3·6H2O, 1.0 g/L H3BO3, and 0.1 g/L KI.
The medium was brought to pH 5.0, 7.0 or 9.0 with concentrated hydrochloric acid solution or 50 % sodium hydroxide solution.
As the only source of sulfur, we used a 0.1 M solution of DBT in DMFA; 200 μl of this solution was added per 100 mL of the SFM medium. As the only source of carbon we used a 40 % solution of glucose; 1 mL of this solution was added per 100 mL of the SFM medium.
2.3. Amplification of the genes of the dsz operon
DNA manipulations were carried out according to standard protocols [18]. The genes of the dsz operon (dszA, dszB) were searched for in the strain 135 using PCR with specific primers (Table 1). For the reactions, we used Q5 high fidelity polymerase; the temperatures and durations of the PCR steps were selected in accordance with the instruction for the enzyme. The annealing temperature was calculated using the NEB melting calculator [19]. Amplification was performed with a GeneAmp PCR System 2400. As a positive control, we used the DBT-desulfurizing strain Gordonia amicalis Ac-2795D [20]. Previously, the dsz genes have been found in the genome of the strain Gordonia amicalis Ac-2795D. As a negative control, we used the strain Gordonia sp. 1B from our laboratory collection. The strain 1B is not capable of growth in the SFM medium with DBT as the source of sulfur and/or carbon. The amplification products of appropriate size were observed in the positive control sample, whereas no signal was observed in the negative control samples.
Table 1.
Primers used in the work.
| Gene | Sequence | Annealing temperature, ºC | Source |
|---|---|---|---|
| dszA | for TGTTCCTGCCTGACGGATTG | 67 | This paper |
| rev TGAAGGTTGTCCTAACGGTCG | |||
| dszB | for ATCGAACTCGACGTCCTCAG | 67 | [20] |
| rev GGAACATCGACACCAGGACT |
2.4. Sequencing and analysis of the strain 135 genome
The genome of the strain was sequenced and assembled as described in [16]. The circular map of the chromosome was constructed using the DNAPlotter [21]. The COG analysis of the genes was carried out using the WebMGA [22]. Phylogenetic trees were constructed using the MEGA 7 software and REALPHY web service.
2.5. Calculation of position of ori in the plasmid
The starting point of replication on the plasmid was determined by calculating the GC skew using a python script [23]. The skew should reach a minimum at the position where the reverse half-strand ends and the forward half-strand begins, which is exactly the location of ori. Therefore, we used a new ori positioning algorithm: ori should be located where the skew is at a minimum. Accordingly, the script work results in the calculation of the minimum of the GC skew and the exact position in the nucleotide chain where the minimum is reached.
2.6. Strain cultivation
The seed culture was prepared by inoculating cells grown on plates into tubes containing 10 mL of the SFM medium (рH 7,0) supplemented with 2% (v/v) glucose and DBT, followed by incubation at 26 °C for 5 days with agitation in the orbital shaker at 180 rpm. The harvested cells were centrifuged at 10,000 rpm for 10 min at 4 °C and re-suspended in 10 mL of the SFM medium at the final concentration of 1 × 108 CFU/mL. The inoculate was introduced into test tubes (flasks) so that the initial concentration of cells in the medium was about 1 × 106 CFU/mL.
The ability of the microorganism to grow on various carbon sources was assessed in test tubes containing 10 mL of the SFM medium (pH 7.0) and DBT. The inoculum was added in an amount of 200 μl per tube. As carbon sources, we used phenanthrene, anthracene, naphthalene, hexadecane, eicosane, heptamethyl nonane, decan, toluene, benzene, glucose and catechol. The carbon sources were added to the medium at a concentration of 2% (v/v or w/v). The tubes with the microorganism were cultured at 26 °C for 5 days in the orbital shaker at 180 rpm.
The assessment of the dynamics of bacterial growth was carried out in 750-ml Erlenmeyer flasks containing 200 mL of SFM (at pH 5.0, 7.0 or 9.0) supplemented with 2% glucose as the sole source of carbon and energy and DBT as a source of sulfur. The inoculum was added into the flasks in an amount of 2 mL. The flasks were cultured at 26 °C for 7 days in the orbital shaker at 180 rpm.
The assessment of the reduction in dibenzothiophene was carried out in 750-ml Erlenmeyer flasks containing 200 mL of SFM (at pH 7.0) supplemented with 2% glucose as the sole source of carbon and energy and DBT as a source of sulfur. The inoculum was added into the flasks in an amount of 2 mL. The flasks were cultured at 26 °C for 7 days in the orbital shaker at 180 rpm.
All results are derived from three independent replicates.
2.7. Assessment of growth dynamics of the strain in flasks
The abundance of microorganisms was determined by the method of serial dilutions followed by plating on Petri dishes with the Luria Bertani agar. The dishes were incubated for 3 days at a temperature of 26 °C, and then the number of grown colonies was determined. Based on the results obtained, a microorganism growth curve was constructed. All results are derived from three independent replicates.
2.8. Determination of degree of DBT degradation
The degree of degradation (DD) of DBT was determined by the difference in its residual concentration in the medium with microorganisms compared to the control (medium without microorganisms):
| DD = (Xc − Xi)/Xc × 100 % |
where Xc is the DBT concentration (mg/L) in the medium without microorganisms, and Xi is the DBT concentration in the inoculated flask (mg/L).
The residual concentration of dibenzotiophene was analyzed by liquid chromatography (Agilent 1100-DAD) using a silica gel-packed column (Alltech) with the length of 250 mm, diameter of 4.6 mm, grain size of 5 μm grafted with octadecyl groups. The used wavelength was 280 nm. The mobile phase was (A) water; (B) acetonitrile. The gradient was as follows: 0 min – 50 % eluent B, 5 min – 80 % eluent B, 7−12 min – 95 % eluent B. The eluent flow rate was 1 mL/min, injection volume — 5 mkl. The analysis duration was 12 min; system stabilization took 5 min [24]. The aliquote of culture broth (5 mL) was brought to pH 2.2 with H2S04 (conc.) and extracted twice with 5 mL of dichloromethane. The extract was evaporated under vacuum and dissolved in 10 mL of acetonitrile. A blank sample without bacterial extract was also analyzed by HPLC. All results are derived from three independent replicates.
Metabolites of DBT-degradation were analyzed by GC–MS (Agilent 5975C Series GC / MSD) using a capillary column VF-5 ms (Varian) with the length of 15 m, diameter of 0.25 mm, 0.50 μm film thickness. We used helium as a carrier gas; temperature of injector was 270 °C. The temperature gradient from 40 °C to 300 °C (15 °C/min) followed by 10 min of isotherm was used. Velocity of carrier gas was 1 mL/min, injection volume — 1 μL. The analysis duration was 30 min. Energy of ionization was 70 eV, temperature of source - 230 °C, full-scan mode, 1 scan/sec.
3. Results
3.1. The ability of the strain to grow on various carbon sources
Colonies of the strain 135 are 2−4 mm in diameter, round, smooth, with folded edges. As the sole carbon sources, we used either acetate, benzoate, citrate, formate, succinate and pyruvate, but not oxalate. Previously, we established that the strain is capable of growth in mineral media with various hydrocarbons as the sole carbon source in the temperature range of 20−40 °C; the optimal cultivation temperature was determined to be 26 °C [25].
The strain G. alkanivorans 135 is not capable of growth in mineral media with DBT as the sole source of carbon and energy. The strain Gordonia alkanivorans 135 was found to be capable of growth in a sulfur-free mineral medium with the only source of sulfur (DBT) on the following carbon sources: hexadecane, eicosane, n-decane and catechol (the final cell concentration exceeded than 1 × 108 CFU / ml). It should be noted that the strain was not capable of growth in a medium containing naphthalene, phenanthrene, anthracene, toluene, and benzene (the final cell concentration was less than 1 × 104 CFU / ml).
3.2. Amplification of genes of the dsz operon
Amplification did not find genes of the dsz operon in the strain 135.
3.3. Sequencing and genome analysis of the Gordonia alkanivorans 135
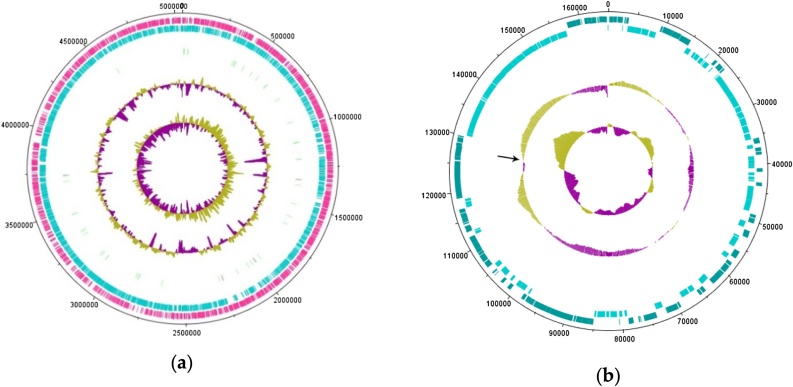
The genome of the strain was sequenced and completely assembled [16]. The Gordonia alkanivorans135 genome consists of a 5,039,827 bp circular chromosome (GC content of 67.44 %, NZ_CP046257) and a 164,963 bp circular plasmid, namely, pG135 (GC content of 64.58 %, NZ_CP046258). The chromosome contains 4,492 coding sequences, 4 rRNA clusters (5S, 16S, and 23S), and 51 tRNAs (Fig. 1a and b).
Fig. 1.
Circular maps of Gordonia alkanivorans 135 chromosome (a) and plasmid pG135 (b). The outer tracks represent the predicted CDSs of the forward and reverse strands. The red tracks represent tRNAs; the violet strings, rRNA clusters. Two inner tracks display the GC plot and the GC skew. The expected starting point of replication is indicated by an arrow.
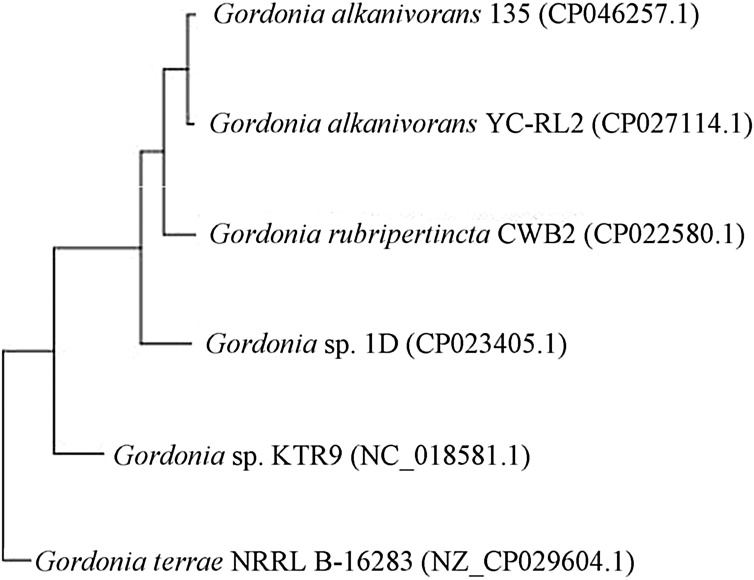
To determine the taxonomic position of the G. alkanivorans 135 within the Gordonia genus, a comparison of the strain 135 genome with the genomes of other Gordoniae was carried out (Fig. 2).
Fig. 2.
Neighbor‐joining phylogenetic tree of Gordoniae based on whole‐genome alignment.
The phylogenetic tree confirms that the closest relative of the strain 135 is Gordonia alkanivorans YC-RL2 (CP027114) (ANI value of 97.45 %)
The plasmid pG135 is related to pKB1 from G. westfalica Kb1 (NC_005307) (91.31 %) and pYYC01 from G. alkanivorans YC-RL2 (CP027115) (92.08 %). The starting point of replication on the plasmid pG135 is expected to be located in the region with the minimum GC skew value (indicated by the arrow).
3.4. Analysis of clusters of orthologous genes (COGs) in the strain 135
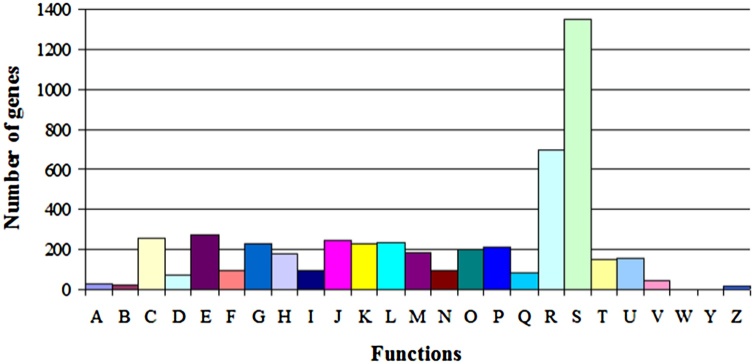
Of the 4,492 CDSs found, 4,091 may be assigned to 25 different categories of clusters of orthologous groups (COGs). These results suggest that the DBT-degrading strain 135 possesses efficient lipid, carbohydrate, amino acid transport, and metabolism (Fig. 3).
Fig. 3.
COG function classification of Gordonia alkanivorans 135. (A) RNA processing and modification. (B) Chromatin structure and dynamics. (C) Energy production and conversion. (D) Cell cycle control, cell division, chromosome partitioning. (E) Aminoacid transport and metabolism. (F) Nucleotide transport and metabolism. (G) Carbohydrate transport and metabolism. (H) Coenzyme transport and metabolism. (I) Lipid transport and metabolism. (J) Translation, ribosomal structure, and biogenesis. (K) Transcription.(L) Replication, recombination, and repair, (M) Cell wall/membrane/envelope biogenesis. (N) Cell motility. (O) Posttranslational modification, protein turnover, chaperones. (P) Inorganic ion transport and metabolism. (Q) Secondary metabolites biosynthesis, transport, and catabolism. (R) General function prediction only. (S) Function unknown. (T) Signal transduction mechanisms. (U) Intracellular trafficking, secretion, and vesicular transport. (V) Defense mechanisms. (Z) Cytoskeleton.
3.5. Synteny analysis of genomes of strain 135 and closest related strains
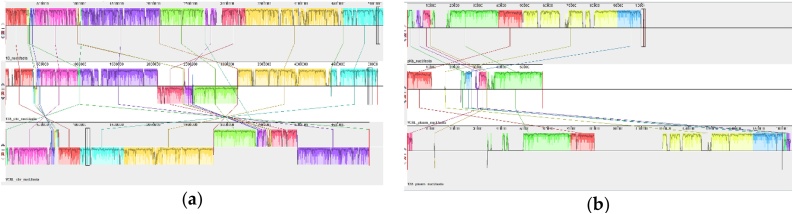
A Mauve-based comparison was performed between the complete genomes of the strain 135, its closest relative Gordonia alkanivorans YC-RL2 and the strain Gordonia sp. 1D. The strain Gordonia sp. 1D, like the strain 135, utilizes DBT. The diagram shows the homology regions of the three genomes (Fig. 4a).
Fig. 4.
Mauve visualization of locally collinear blocks identified between (a) chromosomes of Gordonia alkanivorans 135, Gordonia sp. 1D, and G. alkanivorans YC-RL2, (b) plasmids pG135 from G. alkanivorans 135, pKb1 from G. westfalica Kb1 (NC_005307), and pYYC01 from G. alkanivorans YC-RL2 (CP027115). Vertical bars demarcate interchromosomal boundaries.
The results obtained are characteristic of the genomes of Gordonia strains known for their instability and variability. The plasmid pG135 contains 181 open reading frames. The predicted products of 104 of them are similar to proteins with a known function and 77 are hypothetical. The plasmid pG135 is related to the plasmid pKB1 from G. westfalica Kb1 (NC_005307) (ANI value of 91.31 %) and pYYC01 from G. alkanivorans YC-RL2 (CP027115) (92.08 %) (Fig. 4b). The plasmid pG135 contains two conservative regions. The first one is located between 49,379 and 80,956 bp and the second one between 109,021 and 164,963 bp. These sequence regions were also found on related plasmids pKB1 and pYYC01.
In addition, the plasmid contains regions that are not present in related plasmids of Gordonia strains. The plasmid contains 8 transposase genes and no tra genes that encode proteins required for conjugation. The functions of the plasmid pG135 are still unclear. We found no catabolic genes responsible for the degradation of pollutants, with the exception of several genes involved in heavy metals’ transport. One copy of Type I restriction-modification (RM) gene system (M, S, R subunits) was found. The genes of Type III RM system found in the sequence of the plasmid pG135 were presumably transferred from Mycobacteria.
We found no clusters of secondary metabolite production on the plasmid pG135. And it is curious that the megaplasmid of 164 kbp without widespread catabolic genes is stable in the strain. The functions of the plasmid will be the subject of further research.
3.6. Search for genes presumably involved in DBT degradation
It has previously been shown that the 4S pathway of DBT utilization by actinobacteria, including Gordonia, is the most common way for these bacteria to assimilate sulfur from condensed thiophenes without destroying carbon-carbon bonds [2]. Despite the ability of the strain 135 to utilize DBT as a source of sulfur, the genes of the complete dsz operon were not identified in the strain. Therefore, a search was carried out for related genes that can presumably be involved in the process of DBT catabolism. The search performed using the Prokka, RAST and GenBank NCBI services identified two such genes in the chromosome of the G. alkanivorans 135 (Table 2).
Table 2.
Related genes possibly involved in DBT catabolism.
| Gene | Related gene from DBT-desulfurizing Gordonia | Designation |
|---|---|---|
| SfnB family sulfur acquisition oxidoreductase | dszC Dibenzothiophene desulfurization enzyme C | QGP87489.1 |
| acyl-CoA dehydrogenase | dszC Dibenzothiophene desulfurization enzyme C | QGP87979.1 |
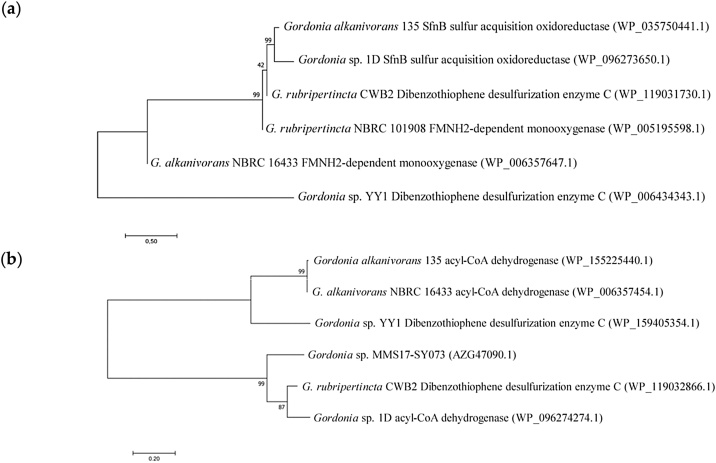
The sfnB genes (Fig. 5a) are common in both gram-negative (Pseudomonas, Acinetobacter) and gram-positive bacteria, including actinobacteria of various genera (Gordonia, Rhodococcus, Nocardia).
Fig. 5.
Neighbor‐joining phylogenetic tree of G. alkanivorans 135 and related strains based on sequences of genes presumably involved in DBT degradation. (a) SfnB family sulfur acquisition oxidoreductase, QGP87489.1, (b) acyl-CoA dehydrogenase, QGP87979.1.
There are data that the sfnB genes and the dszC dibenzothiophene desulfurization gene are related [26]. An analysis of the genome region containing this gene showed that it is part of a group of genes involved in sulfur catabolism. The group is conservative; the gene surroundings do not change in related strains (G. alkanivorans YC-RL2 and Gordonia sp. 1D).
The sequence of acyl-CoA dehydrogenase (Fig. 5b) of the Gordonia alkanivorans strain 135 is related to the sequence of the dszC gene of the Gordonia sp. MMS17-SY073 (AZG47090.1), as well as the sequence of the soxC5 gene (alternative name of dsz) of the G. rubripertincta strain CWB2.
The genes dszA, dszB, and dszD were not identified in the genome of the strain 135.
3.7. Dynamics of strain growth in a mineral medium with DBT and glucose
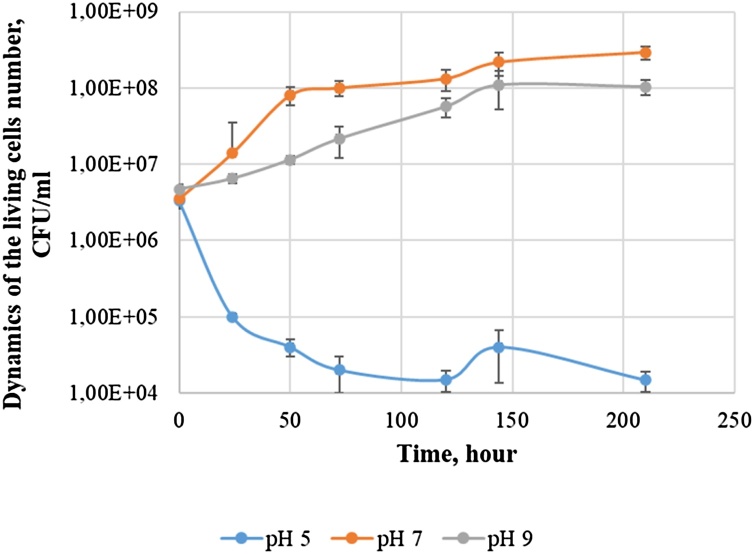
An analysis of the dynamics of the abundance of microorganisms cultivated in the SFM medium with DBT and glucose as sources of sulfur and carbon at temperatures 26 °C and pH (5, 7 and 9) was performed (Fig. 6).
Fig. 6.
Dynamics of the living cells number of Gordonia alkanivorans 135 during growth in a mineral medium with DBT and glucose at temperatures of 26 °C.
The maximum cell count was achieved when the culture grew at a temperature of 26 °C and pH of 7.0. At pH 7.0, the midpoint of the exponential growth phase was at 24 h of growth. The exponential phase ended after 50 h. A slight increase in the cell count was also observed in the period from 55 to 144 h.
At pH 9.0, the midpoint of the exponential growth phase was at 75 h of growth. The exponential phase ended after 144 h. pH 9.0 slightly inhibited the growth of the strain, which was manifested in a decrease in the maximum cell count by 0.5 orders of magnitude compared to pH 7.0. At pH 5.0, we observed a toxic effect on the culture growth – the number of viable cells was drastically decreasing.
3.8. Assessment of the ability of strain 135 to utilize DBT
The ability of strain 135 to utilize DBT was evaluated at pH 7.0 and 26 °C after 144 h of growth. The residual content of DBT was assessed by liquid chromatography. The degree of degradation was assessed in comparison with a control system without microorganisms. We found that the strain is capable of degrading 45.26 ± 6.24 % of DBT. To confirm our result, we analyzed concentrated extracts with DBT which was dissolved in 0.5 mL of acetonitrile. We also analyzed extracts using an Agilent 6890 gas chromatograph (Agilent, USA) with a flame-ionization detector equipped with a DB-1 (100 % dimethylpolysiloxane) fused capillary column (30 m ×0.25 mm). For this, the extract was dissolved in 0.5 mL of dichloromethane. We examined 10 extracts from the culture broth of both types (with or without microorganisms). The reliability of the DBT analysis was confirmed by the standard addition method. The extraction efficiency was found to be 95 %. The significance of differences in the DBT concentration between variants (with or without microorganisms) was confirmed by ANOVA (p ≤ 0.05).
Moreover, concentrated extracts were analyzed using an Agilent 5975C Series GC / MSD equipped with a DB-1 column (30 m ×0.25 mm). Degradation products of DBT, including 2-hydroxybiphenyl and biphenyl, were not detected.
4. Discussion
The ability of Gordonia strains to utilize sulfur-containing hydrocarbon derivatives is widely known. This ability was found in representatives of various species: G. aichiensis 51 [27], Gordonia sp. strain CYSK1 [28], G. amicalis DSM 44461 T [29] utilized dibenzothiophene, G. desulfuricans strain 213E utilized benzothiophene. The G. alkanivorans strain 1B is also known [17]; according to the authors, the strain utilized DBT, BT, DBT-sulfone and alkylated compounds of the thiophene series.
We found that the strain Gordonia alkanivorans 135 is capable of degradation of various carbon substrates (glucose, hexadecane, eicosane, n-decane and catechol) in a mineral medium supplemented with sulfate. In addition, it is capable of growth in the SFM medium with the above carbon substrates and DBT as the sole source of sulfur. At the same time, the strain is not capable of growth in the SFM medium with DBT as the sole source of both sulfur and carbon. No naphthalene degradation genes were found in the strain 135, which was confirmed by its inability to grow in a medium with naphthalene. There are no data in the literature on the use of the Kodama pathway for DBT catabolism by Gordoniae, which is also confirmed by the results presented here.
We assumed that initially the strain Gordonia alkanivorans 135 is not capable of utilizing DBT as a carbon source, but it can use DBT as the only source of sulfur necessary for the degradation of the carbon substrate (in our study of glucose) when cultured in the SFM medium. Similar processes have been described in numerous studies of DBT catabolism by Gordoniae [17,[27], [28], [29], [30], [31]].
Fig. 6 shows the growth curves of the strain under various conditions. At pH 9.0, the culture entered the stationary growth phase after 144 h, at pH 7.0 – after 50 h. μmax of the strain 135 at pH 7.0 was 0.062 h−1, which was 3 times higher than μmax at pH 9.0. This is comparable to the value of μmax (0.0575 h−1) obtained by Alves L. et al. [30]. In the work of Shavandi [15], the strain G. alkanivorans RIPI90A reached the stationary growth phase under similar cultivation conditions after 50 h. Aminsefat A. et al. [2] demonstrated that the Gordonia sp. AHV-01 enters the stationary growth phase after 96 h. In the work of Akhtar N. et al. [31], the strain Gordonia sp. 4 N reached the stationary growth phase after 84 h.
The degree of DBT degradation by Gordonia alkanivorans 135 after 144 h of growth was 45.26 %. Table 3 presents data on DBT degradation by different Gordonia strains.
Table 3.
The comparison of desulfurizing efficiency of the Gordonia strains.
| Reported Strains | Desulfurizing Efficiency | Days of Incubation | Concentration of DBT | References |
|---|---|---|---|---|
| Gordonia alkanivorans 1B | 77 % | 7 | 0.5 mM | [32] |
| Gordonia alkanivorans RIPI90A | 90 % | 10 | 0,5 mM | [1] |
| Gordonia rubropertincta (MTCC 289) | 99 % | 10 | 0,5 mM | [33] |
| Gordonia sp. HS126-4 N | 95 % | 2 | 0,2 mM | [13] |
| Gordonia alkanivorans 135 | 45 % | 6 | 0,2 mM | This study |
| Gordonia sp. ZD-7 | 44 % | 1 | 0.2 mM | [34] |
| Gordonia amicalis 6-1 | 98 % | 15 | 0,1 mM | [19] |
The Gordonia strains are known to be capable of desulfurization of dibenzothiophene (DBT) with the formation of 2-hydroxybiphenyl (2-HBP), the final product of the 4S pathway [[32], [33], [34], [35],14]. In the works presented in Table 3, the authors observed the maximum accumulation of this metabolite by the time of complete depletion of DBT in the system and entering the stationary growth phase by the culture. The degree of DBT degradation by Gordonia alkanivorans 135 after 144 h of growth was 45.26 %, which is lower than that for other known Gordoniae (Table 3). In our work, the degradation products of DBT, including 2-hydroxybiphenyl and biphenyl, were not detected by GC—MS analysis. We assume that this may be due to the degradation of the DBT metabolites in the culture broth. However, in similar experiments using GC—MS analysis under the same conditions, a number of research groups have earlier demonstrated the formation of the above metabolites [14,[29], [30], [31], [32], [33], [34]].
Usually, Gordonia strains capable of DBT desulfurization carry the dsz operon on the chromosome [24,17,35,36]. Preliminary amplification did not find the dsz operon genes in the strain 135, which is consistent with the data on the absence of accumulation of 2HBP. There is no information in the literature on DBT-catabolizing Gordonia strains that do not have genes of the dsz operon. Gordonia strains with such properties (the ability to actively utilize DBT in the absence of the dsz operon genes) have not been previously reported; therefore, we sequenced, assembled and analyzed the genome of the strain 135. This is the first dibenzothiophene-degrading Gordonia strain, the genome of which was completely assembled. In the genome of the strain, we performed a search for genes potentially involved in the DBT catabolism. The annotation performed using KEGG showed that sulfur metabolism in the strain 135 is controlled by 22 genes, including a SfnB family sulfur acquisition oxidoreductase and an acyl CoA dehydrogenase. As a result of the genome analysis of the strain, we found the genes catA encoding a catechol-1,2-dioxygenase and catC encoding a muconolactone D-isomerase that allow to perform the degradation of catechol. These genes are likely to be capable of contributing to the degradation of metabolites formed after the extraction of sulfur from DBT. Taking into account the observed genetic, physiological and biochemical parameters, the DBT degradation by the Gordonia alkanivorans 135 is likely to proceed via an alternative pathway that has not yet been described in the literature. The obtained results require further detailed genetic studies.
5. Conclusion
For the first time, we assembled the complete genome of a dibenzothiophene-desulfurizing strain Gordonia alkanivorans 135. We found genes presumably involved in the process of DBT degradation. These genes are distantly related to the dszC gene, but other genes of the dsz operon that are usually present in DBT-desulfurizing Gordoniae were not found in the genome of the strain 135. Gordonia strains capable of DBT desulfurization in the absence of the dsz operon genes have not been previously reported. Taking into account the observed physiological and biochemical parameters, the degradation of DBT by the Gordonia alkanivorans 135 is likely to proceed via an alternative pathway that has not been previously described in the literature.
Funding
This work was financially supported by the Russian Science Foundation (grant No. 19-74-00097).
CRediT authorship contribution statement
Yanina Delegan: Conceptualization, Supervision, Funding acquisition, Writing - original draft, Writing - review & editing. Yulia Kocharovskaya: Methodology, Investigation, Visualization. Ekaterina Frantsuzova: Methodology, Investigation, Visualization. Rostislav Streletskii: Methodology, Investigation, Validation. Anna Vetrova: Project administration, Data curation, Writing - original draft, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Mohebali G., Ball A., Kaytash A. Biodesulfurization potential of a newly isolated bacterium Gordonia alkanivorans RIPI90A. Enzyme Microb. Technol. 2007;40:578–584. [Google Scholar]
- 2.Aminsefat A., Rasekh B., Ardakani M.R. Biodesulfurization of dibenzothiophene by Gordonia sp. AHV-01 and optimization by using of response surface design procedure. Mikrobiologiia. 2012;81:171–176. [PubMed] [Google Scholar]
- 3.Pokethitiyook P., Tangaromsuka J., Kruatrachue M., Kalambahetib C., Borole A. Biological removal of organic sulphur by bacterial strains isolated in Thailand. Science Asia. 2008;34:361–366. doi: 10.2306/scienceasia1513-1874.2008.34.361. [DOI] [Google Scholar]
- 4.Li F., Zhang Z., Feng J., Cai X., Xu P. Biodesulfurization of DBT in tetradecane and crude oil by a facultative thermophilic bacterium Mycobacterium goodii X7B. J. Biotechnol. 2007;127:222–228. doi: 10.1016/j.jbiotec.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 5.Tao F., Yu B., Xu P., Ma C.Q. Biodesulfurization in biphasic systems containing organic solvents. Appl. Environ. Microbiol. 2006;72:4604–4609. doi: 10.1128/AEM.00081-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denome S.A., Olson E.S., Young K.D. Identification and cloning of genes involved in specific desulfurization of dibenzothiophene by Rhodococcus sp. strain IGTS8. Appl. Environ. Microbiol. 1993;59:2837–2843. doi: 10.1128/aem.59.9.2837-2843.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta N., Roychoudhury P.K., Deb J.K. Biotechnology of desulfurization of diesel: prospects and challenges. Appl. Microbiol. Biotechnol. 2005;66:356–366. doi: 10.1007/s00253-004-1755-7. [DOI] [PubMed] [Google Scholar]
- 8.van Afferden M., Tappe D., Beyer M., Trüper H.G., Klein J. Biochemical mechanisms for the desulfurization of coal-relevant organic sulfur compounds. Fuel. 1993;72(12):1635–1643. doi: 10.1016/0016-2361(93)90348-6. [DOI] [Google Scholar]
- 9.Bahuguna A., Lily M.K., Munjal A., Singh R.N., K D. Desulfurization of dibenzothiophene (DBT) by a novel strain Lysinibacillus sphaericus DMT-7 isolated from diesel contaminated soil. J. Environ. Sci. (China) 2011;23:975–982. doi: 10.1016/s1001-0742(10)60504-9. [DOI] [PubMed] [Google Scholar]
- 10.Buzanello E.B., Rezende R.P., Sousa F.M., Marques Ede L., Loguercio L.L. A novel Bacillus pumilus-related strain from tropical landfarm soil is capable of rapid dibenzothiophene degradation and biodesulfurization. BMC Microbiol. 2014;14:257. doi: 10.1186/s12866-014-0257-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Araujo H.W.C., de Freitas Siva M.C., Lins C.I.M., do Nascimento A.E., da Silva C.A.A., Campos-Takaki G.M. Oxidation of dibenzothiophene (DBT) by Serratia marcescens UCP 1549 formed biphenyl as final product. Biotechnol. Biofuels. 2012;(5):33. doi: 10.1186/1754-6834-5-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee W.C., Ohshiro T., Matsubara T., Izumi Y., Tanokura M. Crystal structure and desulfurization mechanism of 2’-hydroxybiphenyl-2-sulfinic acid desulfinase. J. Biol. Chem. 2006;281:32534–32539. doi: 10.1074/jbc.M602974200. [DOI] [PubMed] [Google Scholar]
- 13.Davoodi-Dehaghani F., Vosoughi M., Ziaee A.A. Biodesulfurization of dibenzothiophene by a newly isolated Rhodococcus erythropolis strain. Bioresour. Technol. 2010;101:1102–1105. doi: 10.1016/j.biortech.2009.08.058. [DOI] [PubMed] [Google Scholar]
- 14.Akhtar N., Akhtar K., Ghauri M.A. Biodesulfurization of thiophenic compounds by a 2-Hydroxybiphenyl-Resistant Gordonia sp. HS126-4N carrying dszABC genes. Curr. Microbiol. 2018;75:597–603. doi: 10.1007/s00284-017-1422-8. [DOI] [PubMed] [Google Scholar]
- 15.Shavandi M., Sadeghizadeh M., Khajeh K., Mohebali G., Zomorodipour A. Genomic structure and promoter analysis of the dsz operon for dibenzothiophene biodesulfurization from Gordonia alkanivorans RIPI90A. Appl. Microbiol. Biotechnol. 2010;87:1455–1461. doi: 10.1007/s00253-010-2605-4. [DOI] [PubMed] [Google Scholar]
- 16.Delegan Y., Valentovich L., Vetrova A., Frantsuzova E., Kocharovskaya Y. Complete genome sequence of Gordonia alkanivorans 135, a promising dibenzothiophene- and hydrocarbon-degrading strain. Microbiol. Resour. Announc. 2020:9. doi: 10.1128/MRA.01450-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alves L., Salgueiro R., Rodrigues C., Mesquita E., Matos J., Gírio F.M. Desulfurization of dibenzothiophene, benzothiophene, and other thiophene analogs by a newly isolated bacterium, Gordonia alkanivorans strain 1B. Appl. Biochem. Biotechnol. 2005;120:199–208. doi: 10.1385/abab:120:3:199. [DOI] [PubMed] [Google Scholar]
- 18.Sambrook J., Russell D.W. 3rd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. Molecular Cloning, a Laboratory Manual. [Google Scholar]
- 19.NEB. NEB Tm Calculator to estimate an appropriate annealing temperature when using NEB PCR products. Availabe online: https://tmcalculator.neb.com (accessed on 2011).
- 20.Borzenkov I.A., Sokolova D.S., Nazina T.N., Babich T.L., Semenova E.M., Ershov A.P., Khisametdinov M.R. 2018. Gordonia AMICALIS Strain with Ability of Generation Directly in Oil Reservoir of OIL-DISPLACING Agent - bioPAV and Decreasing Content of Organosulfur Compounds of Oil. RU2673747C1, 16.05. [Google Scholar]
- 21.Carver T., Thomson N., Bleasby A., Berriman M., Parkhill J. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics. 2009;25:119–120. doi: 10.1093/bioinformatics/btn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu S., Zhu Z., Fu L., Niu B., Li W. WebMGA: a customizable web server for fast metagenomic sequence analysis. BMC Genomics. 2011;12:444. doi: 10.1186/1471-2164-12-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delegan Y. 2020. GC-skew Calculation Using a Python Script. Availabe online: https://gist.github.com/Melania2006/10dad87338f5fbee547a855636298ce3 (Accessed on 11.01.2020) [Google Scholar]
- 24.Bhanjadeo M.M., Rath K., Gupta D., Pradhan N., Biswal S.K., Mishra B.K., Subudhi U. Differential desulfurization of dibenzothiophene by newly identified MTCC strains: influence of Operon Array. PLoS One. 2018;13 doi: 10.1371/journal.pone.0192536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delegan Y.A., Frantsuzova E.E., Vetrova A.A. The strain Gordonia alkanivorans 135 is a promising destructor of dibenzothiophene. Biotekhnologiya. 2020;36(4):121–125. doi: 10.21519/0234-2758-2020-36-4-121-125. [DOI] [Google Scholar]
- 26.Habe H., Kouzuma A., Endoh T., Omori T., Yamane H., Nojiri H. Transcriptional regulation of the sulfate-starvation-induced gene sfnA by a sigma54-dependent activator of Pseudomonas putida. Microbiology. 2007;153:3091–3098. doi: 10.1099/mic.0.2007/008151-0. [DOI] [PubMed] [Google Scholar]
- 27.Finkel’shtein Z.I., Baskunov B.P., Golovlev E.L., Golovleva L.A. Desulfurization of 4,6-dimethyldibenzothiophene and dibenzothiophene by Gordona aichiensis 51. Chemistry. 1999 [Google Scholar]
- 28.Rhee S.K., Chang J.H., Chang Y.K., Chang H.N. Desulfurization of dibenzothiophene and diesel oils by a newly isolated Gordona strain CYKS1. Appl. Environ. Microbiol. 1998;64:2327–2332. doi: 10.1128/aem.64.6.2327-2331.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim S.B., Brown R., Oldfield C., Gilbert S.C., Iliarionov S., Goodfellow M. Gordonia amicalis sp. nov., a novel dibenzothiophene-desulphurizing actinomycete. Int. J. Syst. Evol. Microbiol. 2000;50(Pt 6):2031–2036. doi: 10.1099/00207713-50-6-2031. [DOI] [PubMed] [Google Scholar]
- 30.Alves L., Paixão S.M. Enhancement of dibenzothiophene desulfurization by Gordonia alkanivorans strain 1B using sugar beet molasses as alternative carbon source. Appl. Biochem. Biotechnol. 2014;172(March (6)):3297–3305. doi: 10.1007/s12010-014-0763-z. [DOI] [PubMed] [Google Scholar]
- 31.Akhtar N., Ghauri M.A., Akhtar K. Dibenzothiophene desulfurization capability and evolutionary divergence of newly isolated bacteria. Arch. Microbiol. 2016;198(August (6):509–519. doi: 10.1007/s00203-016-1209-5. [DOI] [PubMed] [Google Scholar]
- 32.Alves L., Matos J., Tenreiro R., Girio F.M. Evidence for the role of zinc on the performance of dibenzothiophene desulfurization by Gordonia alkanivorans strain 1B. J. Ind. Microbiol. Biotechnol. 2008;35:69–73. doi: 10.1007/s10295-007-0278-5. [DOI] [PubMed] [Google Scholar]
- 33.Mohebali G., Ballb A.S., Rasekha B., Kaytasha A. Biodesulfurization potential of a newly isolated bacterium Gordonia alkanivorans RIPI90A. Enzyme Microb. Technol. 2008;40:578–584. doi: 10.1099/mic.0.2008/017608-0. [DOI] [Google Scholar]
- 34.Bhanjadeo M.M., Rath K., Gupta D., Pradhan N., Biswal S.K., Mishra B.K., Subudhi U. Differential desulfurization of dibenzothiophene by newly identified MTCC strains: influence of Operon Array. PLoS One. 2018;13(March (3)) doi: 10.1371/journal.pone.0192536. e0192536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li W., Wang M.D., Chen H., Chen J.M., Shi Y. Biodesulfurization of dibenzothiophene by growing cells of Gordonia sp. in batch cultures. Biotechnol. Lett. 2006;28(August (15)):1175–1179. doi: 10.1007/s10529-006-9070-2. Epub 2006 Jun 27. PMID: 16802103. [DOI] [PubMed] [Google Scholar]
- 36.Bhanjadeo M.M., Rath K., Gupta D., Pradhan N., Biswal S.K., Mishra B.K., Subudhi U. Differential desulfurization of dibenzothiophene by newly identified MTCC strains: Influence of Operon Array. PLoS One. 2018;13(March (3)) doi: 10.1371/journal.pone.0192536. e0192536. [DOI] [PMC free article] [PubMed] [Google Scholar]