Abstract
Context
Glycogen storage diseases are rare. Increased glycogen in the liver results in increased attenuation.
Objective
Investigate the association and function of a noncoding region associated with liver attenuation but not histologic nonalcoholic fatty liver disease.
Design
Genetics of Obesity-associated Liver Disease Consortium.
Setting
Population-based.
Main Outcome
Computed tomography measured liver attenuation.
Results
Carriers of rs4841132-A (frequency 2%-19%) do not show increased hepatic steatosis; they have increased liver attenuation indicative of increased glycogen deposition. rs4841132 falls in a noncoding RNA LOC157273 ~190 kb upstream of PPP1R3B. We demonstrate that rs4841132-A increases PPP1R3B through a cis genetic effect. Using CRISPR/Cas9 we engineered a 105-bp deletion including rs4841132-A in human hepatocarcinoma cells that increases PPP1R3B, decreases LOC157273, and increases glycogen perfectly mirroring the human disease. Overexpression of PPP1R3B or knockdown of LOC157273 increased glycogen but did not result in decreased LOC157273 or increased PPP1R3B, respectively, suggesting that the effects may not all occur via affecting RNA levels. Based on electronic health record (EHR) data, rs4841132-A associates with all components of the metabolic syndrome (MetS). However, rs4841132-A associated with decreased low-density lipoprotein (LDL) cholesterol and risk for myocardial infarction (MI). A metabolic signature for rs4841132-A includes increased glycine, lactate, triglycerides, and decreased acetoacetate and beta-hydroxybutyrate.
Conclusions
These results show that rs4841132-A promotes a hepatic glycogen storage disease by increasing PPP1R3B and decreasing LOC157273. rs4841132-A promotes glycogen accumulation and development of MetS but lowers LDL cholesterol and risk for MI. These results suggest that elevated hepatic glycogen is one cause of MetS that does not invariably promote MI.
Keywords: genetics, NALFD, glycogen, GWAS, triglyceride, metabolic syndrome
Glycogen storage diseases are caused by excess glycogen storage in tissues. Previously characterized glycogen storage diseases are rare, most being less than 1 in 20 000 live births (1). Increased glycogen accumulation in the liver results in increased liver attenuation. Genetic variants that increase liver attenuation in the population have not been fully explored.
We previously carried out a genome-wide association study (GWAS) of liver attenuation measured by computed tomography (CT) in individuals of European ancestry (2). The minor allele (A) of rs4240624 associated with increased liver attenuation, suggestive of increased liver glycogen. Importantly, rs4240624 did not significantly associate with histologic measures of nonalcoholic fatty liver disease (NAFLD), including steatosis, nonalcoholic steatohepatitis, and fibrosis in multiple studies (2, 3). We have since noted that the liver attenuation altering variants map to a long noncoding RNA LOC157273 ~190 kb upstream of a putative glycogen metabolism regulating gene, PPP1R3B. To investigate how these variants contribute to human disease, we carried out association analyses of the most significantly associated liver attenuation variant, rs4841132 (r2 = 0.99 with rs4240624) across multiple diseases and traits. We also examined how this variant affected gene expression in multiple tissues and datasets. We deleted the region containing this variant in a human liver hepatocarcinoma cell line and we examined how it affected PPP1R3B and LOC157273 RNA levels as well as glycogen.
Methods
Genetics of Obesity-associated Liver Disease Consortium
Eight cohorts (n = 16 234) with Illumina Exome array data were included (Supplementary Tables 1-5 (4)): AGES (5), FamHS (6), FHS (7), GENOA (8), IRASFS (9), JHS (10), MESA (11), and OOA (12, 13). All work was approved by local institutional review boards or equivalent committees and participants provided written informed consent.
Studies calculated a ratio of liver attenuation to external phantom (or spleen) for each individual to control for scan penetrance. Values were inverse normalized and regressed against variants for association using linear regression modeling (linear mixed modeling for related individuals) controlling for age, age2, sex, principal components, and study site (where appropriate). Variants with a minor allele count > 6, Hardy-Weinberg Equilibrium P > 1 × 10–6 and call rate > 98% from each study that did not have significant P values for heterogeneity were included. Variants in/near PPP1R3B were analyzed for association and results combined using a fixed effects meta-analysis in METAL (14).
UK Biobank Liver Fat Analyses
UK Biobank (UKBB) liver fat analyses were performed under Resource Project #18120. Briefly, UKBB is a prospective epidemiological study of phenotyped individuals aged 40 to 69 at the time of recruitment from the United Kingdom who have been genotyped (15). Genotypes of the UKBB participants were assayed using either of two genotyping arrays, the Affymetrix UK BiLEVE Axiom array or Affymetrix UKBiobank Axiom array. These arrays were augmented by imputation of ~96 million genetic variants from the Haplotype Reference Consortium (http://www.haplotype-reference-consortium.org/), 1000 Genomes (https://www.internationalgenome.org/), and the UK 10K (https://www.uk10k.org/) projects. Individuals were excluded if they were designated by the UKBB as outliers based on either genotyping missingness rate or heterogeneity, whose sex inferred from the genotypes did not match their self-reported sex and who were not of white British ancestry. Finally, individuals were removed if they had missingness > 5% across variants which passed quality control procedures. Characteristics of the participants (N = 408 961) are shown in Supplementary Table 6 (4). Association analyses were carried out for inverse normally transformed liver magnetic resonance imaging proton density fat fraction in UKBB using linear mixed modeling controlling for sex, array batch, UKBB Assessment Center, age, age2, and the first 10 genomic principal components using SAIGE (16). Imputation quality scores (IQS) for rs4841132 (IQS = 1), rs738409 (IQS = 1), rs58542926 (IQS = 1), rs378140 (IQS = 0.99), and rs61756425 (IQS = 1) were excellent.
Michigan Genomics Initiative Cohort
University of Michigan Health System patients were recruited on the day of their elective procedure using an opt-in written informed consent for broad long-term use of their electronic health information and genetic data (17). Laboratory values, diagnoses, demographics, and vital signs were extracted for patients seen between January 2012 and December 31, 2015. All available laboratory values were extracted and the mean ± SD for each trait and each individual calculated with exclusion of measures that were > 1 SD to decrease entry errors. Continuous traits were inverse normally transformed. NAFLD was identified by International Classification of Diseases, 9th edition (ICD-9; before 2015) or ICD-10 (after 2015) diagnoses: 571.8 and K76.0. Cirrhosis was identified using ICD-10 diagnosis: K70.2, K70.3, K70.4, K71.7, and K.74*. Characteristics of the participants are shown in Supplementary Table 7 (4). Genotyping was performed on the Illumina HumanCoreExome array. IQS for rs4841132 (IQS = 1), rs738409 (IQS = 1), rs58542926 (IQS = 1), rs378140 (IQS = 0.88), and rs61756425 (IQS = 0.99) were excellent.
Massachusetts General Hospital Biopsies
Liver (n = 566), subcutaneous (N = 608), and omental fat (n = 741) were obtained from patients undergoing bariatric surgery. RNA expression was assessed using a custom Agilent microarray targeting 34 694 known/predicted genes. Genotyping was performed on the Illumina 650Y array (18). eQTLs were called using Matrix EQTL (https://cran.r-project.org/web/packages/MatrixEQTL/), running a linear model with a window of ±1 Mb for cis eQTLs. Conditional analysis was run using stepwise linear regression.
STARNET Liver Biopsies
Liver tissue from 600 coronary artery disease patients in the Stockholm-Tartu Atherosclerosis Reverse Networks Engineering Task (STARNET) study were used (19). Samples were genotyped using the Illumina OmniExpressExome-8v1 array. RNA sequencing was carried out using the Illumina TruSeq stranded mRNA kit followed by mapping reads with STAR v.2.3.0e (20) (Genome Reference Consortium GRCh37). Gene expression was determined using DESeq2 v.1.8.1 (21) adjusted for age, sex, protocol, and laboratory covariates. Conditional analyses were carried out as in the Massachusetts General Hospital (MGH) sample.
Allelic expression imbalance was fit to 1 of 2 models in R. For rs4841132, a binomial model was used to test for imbalance between expression of the A/G alleles in heterozygotes (confirmed by genotyping) only. For PPP1R3B, rs330915 in the 3′ UTR was used to identify allele-specific expression and the rank of the absolute level of imbalance was regressed in a generalized linear model against whether the sample was heterozygous at rs4841132 (22-24). Causal inference analysis (25) was used with rs4841132 designated as the locus and PPP1R3B expression as the outcome/trait.
Functional Analysis in HuH-7 Cell Lines
The HuH-7 human hepatocarcinoma cell line (26) was used for functional analyses. The Alt-R CRISPR-Cas9 System (Integrated DNA Technologies) was used to produce a 105-bp deletion of exon 2 in LOC157273 including the variant rs4841132. In a separate set of experiments, the same system was used to produce a homozygous frameshift by the insertion of a T in the codon for amino acid 10 in the PPP1R3B gene. Guide RNA oligonucleotide sequences are shown in Supplementary Table 8 (4). Briefly, cells were seeded into 6-well plates with complete DMEM medium with 10% fetal bovine serum (FBS) plus 100 IU/mL penicillin and 100 µg/mL streptomycin at a density of 3 × 105 cells per well. After 24 hours, cells were transfected with either PPP1R3B or rs4841132 complexes (ie, gRNA+tracrRNA and Cas9 nuclease) using Lipofectamine RNAi Max transfection reagent (Thermo Fisher Scientific). DMEM medium was replaced 48 hours posttransfection and cells cultured for 2 days. Cells were single-cell cloned by limiting dilution. After 10 days, 50 µL 1× TrypLE enzyme solution was added to each well, transferred into 6 wells each, and cultured for 3 days in DMEM. DNA was isolated, PCR was performed around the putative insertion/deletion, and editing confirmed with Sanger sequencing.
For overexpression experiments, an overexpression plasmid for LOC157273 was made by transferring the LOC157273 cDNA into the pCMV6-Entry vector using the Gibson cloning kit with Q5 High-Fidelity DNA Polymerase (New England Biolabs). The cDNA insert was verified by EcoRI digest and bidirectionally sequenced using tiled primers with > 2× coverage. An overexpression plasmid for PPP1R3B was made by transferring the PPP1R3B cDNA cloned from MGH clone plasmid 50661 into the HinDIII site of pCMV6-Entry vector using the Gibson cloning kit with Q5 High-Fidelity DNA Polymerase (New England Biolabs). The cDNA insert was verified by EcoRI digest and bidirectionally sequenced using tiled primers with > 2× coverage.
For knockdown experiments, HuH-7 cells were seeded at a density of 2 × 105 cells per well in 6-well plates in C-DMEM medium. After 24 hours’ culture, 10 nM (final concentration) of LOC157273 antisense oligonucleotide (ASO) scramble 2 and 3 or a mixture of LOC157273 ASO1, ASO2, ASO3, ASO4, and ASO5 was transfected into cells using FUGENE-HD transfection reagent (Promega, WI). Cells were cultured for an additional 24 hours and then assayed. Antisense oligonucleotide sequences are shown in Supplementary Table 8 (4).
RT-quantitative PCR and Glycogen Assays for 30 nM Insulin and 200 µM BSA-bound Oleic Acid-treated LOC157273 105-bp Deletion in HuH-7 Cells
For LOC157273 105-bp deletion cell line studies, cells including mock, LOC157273 105-bp homozygous deletion, and LOC157273 105-bp heterozygous deletion cell lines were seeded in 12-well plates (5 × 105 cells/well). Cells were maintained at 37°C in 5% CO2 in low glucose (5.5 mM) DMEM medium with 10% FBS and 100 U/mL penicillin-streptomycin. After 40 hours of incubation, upon reaching confluence, cells were washed twice with PBS and treatment media was added. Treatment media consisted of serum-free DMEM, 12.5 mM glucose with 100 U/mL penicillin-streptomycin for 4 hours, and then cells were treated with either 30 nM of insulin or 200 µM of BSA-bound oleic acid with 12.5 mM glucose DMEM no-serum medium for 24 hours. In the control group, the same volume of treatment DMEM medium was added into each well. After 24 hours of treatment with either insulin or oleic acid, HuH-7 cells were washed and harvested. Cell lysate was collected with ddH2O (100 µL/well) as a glycogen sample or lysis with RIPA buffer (200 µL/well) for a protein sample and RNA samples were isolated from these cells by using TRIzol reagent. RNA samples were used to measured LOC157273 and PPP1R3B mRNA levels using RT-quantitative PCR (RT-qPCR). The superscript VILO reverse transcriptase kit (Life Technologies) was used to synthesize the first strand cDNA. RT-qPCR was performed using TaqMan Gene Expression assays and ELF1 probes (Life Technologies) were used as controls. Total cellular glycogen was quantified using the Glycogen Assay Kit (Sigma-Aldrich). Values were normalized by total cellular protein amount. Experiments were done in 6 replicates and differences were evaluated using a paired Student t test. Difference between experimental and control groups was considered significant at P < 0.05.
RT-qPCR and Glycogen Assays for 30 nM Insulin and 200 µM BSA-bound Oleic Acid-treated LOC157273 Overexpression in HuH-7 Cells
For LOC157273 overexpression studies, cells were transfected with the pCMV6-Entry vector expressing LOC157273 cDNA or a control empty pCMV6-Entry vector using FuGENE transfection reagent (Thermo Scientific). Posttransfection (48 hours), cells stably expressing these genes were selected with G-418 for 72 hours. Medium was changed and fresh G-418 was added every 2 days. After 3 days, cells stably overexpressing the LOC157273 were collected and the total cellular RNA extracted using TRIzol reagent. Overexpression was measured and confirmed by RT-qPCR assay. Stable LOC157273 overexpressing sublines were cultured in high glucose (25 mM) DMEM with 10% FBS and G-418 (10 µg/mL). After 24 hours, the medium was replaced with DMEM with 10% delipidated FBS and G-418 for another 24 hours. Then 30 nM insulin or 200 µM of BSA-bound oleic acid was added to culture medium. For the control group, the same volume of treatment DMEM medium was added into each well. After an additional 24 hours of incubation with insulin or oleic acid, cells were collected and cell lysis with RIPA buffer (200 µL/well) for protein sample and RNA samples were isolated from these cells by using TRIzol reagent. Cellular glycogen was extracted and quantified using the Glycogen Assay and Serum Triglyceride Determination kits (Sigma-Aldrich). Values were normalized by total cellular protein amount. LOC157273 and PPP1R3B expression levels were measured using RT-qPCR assay (Thermo Scientific, HS01934960). Experiments were done in 6 replicates and differences were evaluated using a paired t test. Difference between experimental group and control group was considered significant at P < 0.05.
RT-qPCR and Glycogen Assays for 30 nM Insulin and 200 µM BSA-bound Oleic Acid-treated LOC157273-ASO Knock Down in HuH-7 Cells
For knockdown experiments, HuH-7 cells were seeded at a density of 1.5 × 105 cells/well in 12-well plates in high glucose C-DMEM medium. After 24 hours, 20 nM (final concentration) of scrambled DNA 2 and 3 or a mixture of LOC157273 ASOs were transfected into cells using FUGENE-HD transfection reagent (Promega, WI). Cells were transfected for 24 hours and then C-DMEM medium was replaced with high glucose (25 mM) DMEM medium with 10% delipidated FBS for 24 hours. Then, 30 nM insulin or 200 µM of BSA-bound oleic acid was added to culture medium for another 24 hours. For the control group, the same volume of treatment DMEM medium was added to each well. After incubation with insulin or oleic acid, cells were collected and cells were lysed with RIPA buffer (200 µL/well) for protein sample and RNA samples were isolated using TRIzol reagent. Cellular glycogen and triglyceride were extracted and quantified using the Glycogen Assay and Serum Triglyceride Determination kits (Sigma-Aldrich). Values were normalized by total cellular protein amount. LOC157273 and PPP1R3B expression levels were measured using RT-qPCR (Thermo Scientific). Experiments were done in 6 replicates and differences were evaluated using a Student paired t test. Differences between experimental group and control group was considered significant at P < 0.05.
RT-qPCR and Glycogen Assays for PPP1R3B Overexpression in HuH-7 Cells
For PPP1R3B overexpression studies, cells were transfected with the pCMV6-Entry vector expressing PPP1R3B3 cDNA or a control empty pCMV6-Entry vector using FuGENE transfection reagent (Thermo Scientific). Posttransfection (48 hours), cells stably expressing these genes were selected with G-418 for 72 hours. Medium was changed and fresh G-418 was added every 2 days. After 3 days, cells stably overexpressing the PPP1R3B3 were collected and the total cellular RNA extracted using TRIzol reagent. Overexpression was measured and confirmed by qRT-PCR assay. Stable overexpressing sublines were cultured in high glucose (4.5 mg/mL) DMEM with 10% FBS and G-418 (10 µg/mL). After 24 hours, the medium was replaced with DMEM with 10% delipidated FBS and G-418. Cells were processed and total protein and glycogen measured as described previously. Experiments were done in triplicate and differences were evaluated using a paired Student t test. Difference between experimental and control groups was considered significant at P < 0.05.
RT-qPCR and Glycogen Assays for PPP1R3B Knock Down in HuH-7 Cells
For PPP1R3B knock-down studies, PPP1R3B was knocked down using CRISPR-Cas9 to create a homozygous premature stop codon with nonsense-mediated decay. Cells including mock and homozygous PPP1R3B deletion were included. Cell lysate was collected with ddH2O (100 µL/well) as a glycogen sample or lysis with RIPA buffer (200 µL/well) for a protein sample and RNA samples were isolated using TRIzol reagent. PPP1R3B mRNA level was measured using RT-qPCR. Cells were processed and total protein and glycogen measured as described previously. Experiments were done in triplicate and differences were evaluated using a paired Student t test. Difference between experimental and control groups was considered significant at P < 0.05.
LOC157273 RNA In Situ Hybridization
HuH-7 cells were grown on poly-D-lysine coated glass coverslips in 6-well plates in high glucose (4.5 mg/mL) C-DMEM medium. After 24 hours, cells were washed with PBS, fixed in 4% (w/v) paraformaldehyde (Fisher Scientific) in PBS for 15 minutes, then LOC157273 RNA in situ hybridization was done with cells on the coverslips using RNAscope 2.5 Chromogenic Assay (Duplex) from Advanced Cell Diagnostics according to the manufacturer’s protocol. Cells were also probed with a baculoviral clone including the PPP1R3B and LOC157273 genomic region to visualize the locus.
Phenome-Wide Association Study (PheWAS)
We assessed the effect of rs4841132 on metabolic traits from previously published GWAS (19, 27-36).
Results
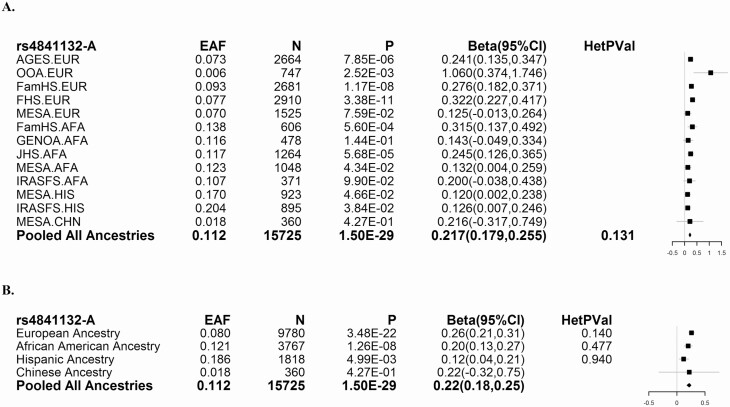
To determine whether variants near PPP1R3B associate with liver attenuation across ancestries, we examined genotype data for 16 234 participants from the Genetics of Obesity-associated Liver Disease (GOLD) Consortium (Supplementary Tables 1-5 (4)). We found that among variants on the exome chip, rs4841132-A had the strongest association with increased liver attenuation (Fig. 1). Two coding variants in PPP1R3B did not associate with liver attenuation (Supplementary Table 9 (4)). rs4841132-A (P = 0.32), unlike rs738409 (PNPLA3, P = 5 × 10-22) and rs58542926 (TM6SF2, P = 1.66 × 10-17) does not associate with hepatic steatosis as measured using magnetic resonance imaging proton density fat fraction in the UKBB (Supplementary Table 10 (4)). Rs4841132-A (P = 0.43), unlike rs738409 (PNPLA3, P = 1.9 × 10-24) and rs58542926 (TM6SF2, P = 1.5 × 10-5) does not associate with NAFLD in Michigan Genomics Initiative (MGI) confirming that rs4841132-A does not promote fatty liver disease (Supplementary Table 11 (4)).
Figure 1.
Association of rs4841132 with liver attenuation across studies and ancestries. (A) Effect of index variants on liver attenuation by study (European [EUR]; African American [AFA]; Hispanic [HIS]; Chinese [CHN]) and (B) by ancestry. Effect allele frequency (EAF) is listed with the corresponding P value (P), effect size on the inverse-normally transformed scale (Beta), and 95% confidence interval in effect size (95% CI) for association with liver attenuation adjusted for age, age2, sex, alcoholic drinks per week, and population substructure. Values greater than 0 indicate an increase in the amount of liver attenuation. For the pooled analyses, heterogeneity P values are also reported. Effect sizes and confidence intervals are summarized in the Forest plots at right. The solid vertical line represents beta = 0.
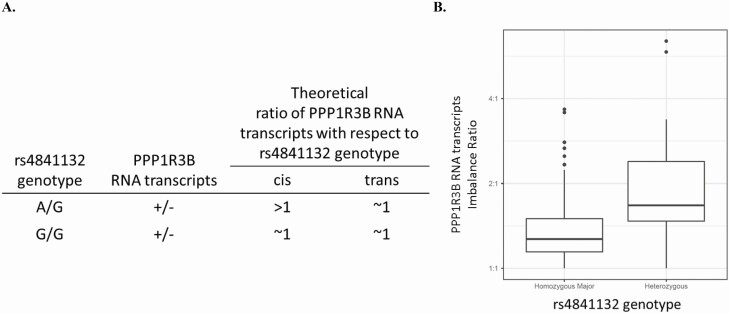
We then examined the effects of rs4841132 on expression of PPP1R3B in liver tissue from the MGH bariatric (18) and STARNET cardiac surgery (19) datasets. rs4841132-A was significantly associated with increased expression of PPP1R3B in liver in both datasets (Table 1, Supplementary Table 11A (4)). We found that there was allelic expression imbalance in STARNET at PPP1R3B (rs330915) for rs4841132 heterozygotes versus homozygotes (P < 8.94 × 10-5; Fig. 2, Supplementary Table 11B (4)), suggesting that the effect of rs4841132-A on PPP1R3B expression is in cis (ie, affects expression of the PPP1R3B allele on the same chromosome). rs4841132-A decreased LOC157273 RNA levels in both datasets (Table 1, Supplementary Table 12A (4)). Causal inference analysis (25) in STARNET did not support that rs4841132-A increased PPP1R3B expression was due to suppression of LOC157273 (or vice versa, P ≈ 1).
Table 1.
(A) Liver eQTL results for PPP1R3B and LOC157273 RNA; (B) PPP1R3B and LOC157273 gene expression changes with a 100 bp deletion eliminating rs4841132 region in human HuH7 cells
| (A)Cohort | Variant | Effect allele | Transcript | Fold-change | P value |
|---|---|---|---|---|---|
| Massachusetts General Hospital | |||||
| rs4841132 | A | PPP1R3B | 1.12 | 1.00E-22 | |
| rs4841132 | A | LOC157273 | 0.90 | 2.38E-14 | |
| Stockholm-Tartu Atherosclerosis Reverse Networks Engineering Task Study | |||||
| rs4841132 | A | PPP1R3B | 1.39 | 3.50E-05 | |
| rs4841132 | A | LOC157273 | 0.45 | 2.00E-25 | |
| (B) | |||||
| Gene | Fold-change | P value | |||
| PPP1R3B | 1.66 | < 0.01 | |||
| LOC157273 | 0.34 | < 0.01 |
Cohort: Massachusetts General Hospital, expression by array (17)) or STARNET (expression by RNA sequencing (18)). Effect allele, allele for which direction of effect is given; transcript, transcript for the gene product (standard symbol) in which change in expression noted; fold-change: fold-change in expression of transcript per effect allele in liver; P value: P value of association of the variant with gene probe intensity (Massachusetts General Hospital) or adjusted gene read counts (STARNET).
Figure 2.
Allele-specific expression shows that rs4841132-A increases PPP1R3B expression in cis. (A) Theoretical ratio of PPP1R3B RNA transcripts with respect to rs4841132 genotype and (B) PPP1R3B allelic balance is closer to 1 in rs4841132-G/G individuals than in rs4841132-A/G individuals suggesting a cis effect of rs4841132-A. Box plots: dark line indicates the median value, a box indicating the 25th and 75th percentiles, whiskers extending from the median for twice the interquartile range, and individual points representing measurements outside that interval.
The expression of PPP1R3B was examined in GTEx (37) and found to be expressed not only in liver but also in adipose tissue, muscle, and blood vessels (Supplementary Fig. 1A (38)). Even though rs4841132-A is an eQTL for PPP1R3B in liver (Table 1), rs4841132-A was not a statistically significant eQTL for PPP1R3B in subcutaneous or visceral fat in the MGH dataset (Supplementary Table 11A (4)). This was due to a lower effect of the variant on expression rather than to a difference in sample size. LOC157273 was expressed at very low levels in multiple tissues in GTEx (Supplementary Fig. 1B (38)).
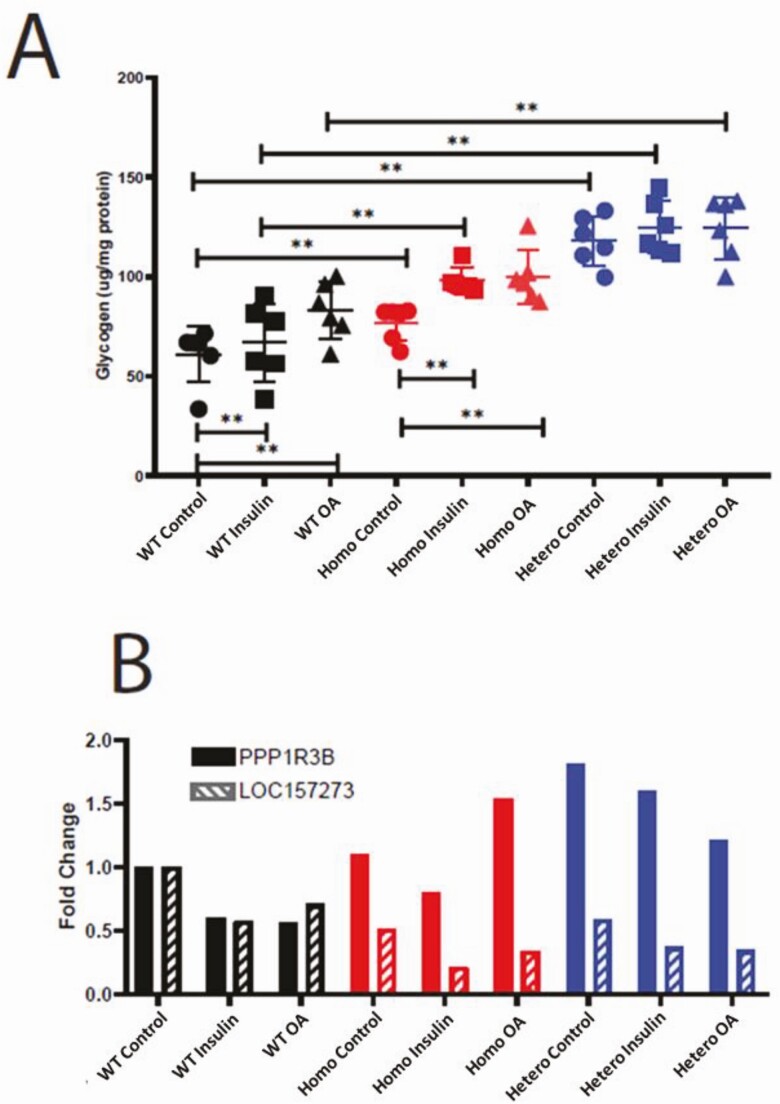
To test whether changes in the genomic region of rs4841132 affected PPP1R3B expression and glycogen levels, we used CRISPR-Cas9 to create 105 bp homozygous or heterozygous deletion of the area inclusive of this variant in HuH-7 cells. We measured glycogen in cells that had been glucose starved and then changed to medium glucose media with or without insulin or oleic acid. We found that insulin or oleic acid stimulation slightly increased glycogen levels in wild-type HuH-7 cells with a concomitant decrease in PPP1R3B and LOC157273 RNA. Cells with a homozygous or heterozygous 105 bp deletion of LOC157273 at rs4841132 had higher glycogen levels in control and stimulated conditions. Uniformly, cells with a homozygous or heterozygous 105bp deletion of LOC157273 at rs4841132 had increased PPP1R3B RNA and decreased LOC157273 RNA under control and stimulated conditions (Fig. 3, Table 1). This expression pattern matches what is seen in the human liver eQTL data from our 2 cohorts (Table 1) and shows that these engineered cell lines perfectly model rs4841132-A effects on LOC157273 and PPP1R3B RNA expression. The effect of insulin and oleic acid on PPP1R3B RNA was magnified in cells with a homozygous or heterozygous 105 bp deletion of LOC157273 at rs4841132 where PPP1R3B RNA increased by 1.34- and 2.76-fold with insulin and 2.68- and 2.16-fold with oleic acid versus control (Fig. 3B). These results suggest that there may be a gene by environment interaction effect at this locus. Although cells with a heterozygous 105 bp deletion of LOC157273, under some circumstances, had higher glycogen and PPP1R3B RNA than cells with a homozygous 105 bp deletion of LOC157273, this was not always the case (oleic acid stimulated cells had more PPP1R3B RNA in homozygous than heterozygous cells). That we see an effect on glycogen in cells with a homozygous and heterozygous 105 bp deletion of LOC157273 suggests that the effect is dominant.
Figure 3.
CRISPR/Cas9 105 bp deletion of rs4841132 region increases glycogen, PPP1R3B RNA, and decreases LOC157273 RNA in HuH-7 cells. (A) Fold-change in glycogen in μg/mg protein. (B) Quantitative PCR of PPP1R3B and LOC157273 normalized to wild-type (WT) control (black). Control, no stimulation of cells (circles); hetero: heterozygously deleted rs4841132 region (blue); homo, homozygously deleted rs4841132 region (red); insulin, cells treated with 30 μM insulin (squares); OA, cells treated with 200 μM oleic acid (triangles). *P < 0.05; **P < 0.01.
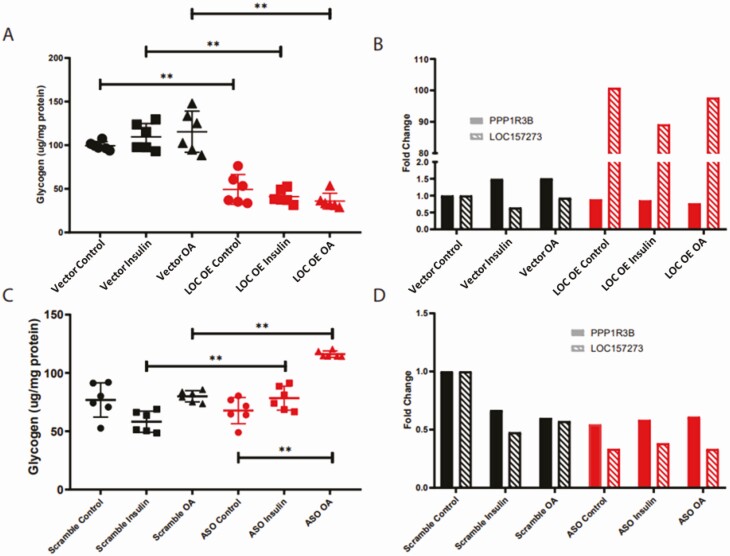
The variant rs4841132 is located in the second exon of a noncoding region of the genome denoted LOC157273. LOC157273 is not found in nonprimates and is found in only some primate species. We did not find homology of LOC157273 with any coding gene in the genome. LOC157273 appears to be widely expressed at low levels (Supplementary Fig. 1B (38)). To test whether increase or decrease in LOC157273 RNA would phenocopy the effects of rs4841132-A, we overexpressed LOC157273 in HuH-7 cells using a cytomegalovirus promoter or reduced its levels using ASOs, respectively. Increasing LOC157273 decreased glycogen levels irrespective of stimulation status (Fig. 4A). Despite LOC157273 being more than 90-fold higher than baseline, PPP1R3B RNA was only slightly reduced from baseline (Fig. 4B). When LOC157273 was decreased, there was a slight increase in glycogen after stimulation with insulin or oleic acid but not without stimulation (Fig. 4C). When LOC157273 RNA was reduced, PPP1R3B RNA was lower than baseline in both unstimulated and stimulated conditions (Fig. 4D). Therefore, even though overexpression or knockdown of LOC157273 RNA decreased and increased glycogen, respectively, its effects on PPP1R3B RNA did not match the pattern seen with rs4841132-A in human liver.
Figure 4.
LOC157273 overexpression and knockdown decreases and increases glycogen and has variable effects on PPP1R3B RNA levels in Huh-7 cells. Overexpression of vector only or vector with LOC157273. (A) Fold change in glycogen in μg/mg protein and (B) quantitative PCR of PPP1R3B and LOC157273 normalized to vector control. Antisense oligonucleotide knockdown of LOC157273. (C) Fold-change in glycogen in μg/mg protein and (D) quantitative PCR of PPP1R3B and LOC157273 normalized to scrambled control. ASO, cells exposed to LOC157273 antisense oligonucleotides; control, no stimulation of cells (circles); insulin, cells treated with 30 μM insulin (squares); LOC, cells expressing LOC157273; OA; cells treated with 200uM oleic acid (triangles); scrambled: cells exposed to scrambled oligonucleotide; vector, cells expressing vector. *P < 0.05; **P < 0.01.
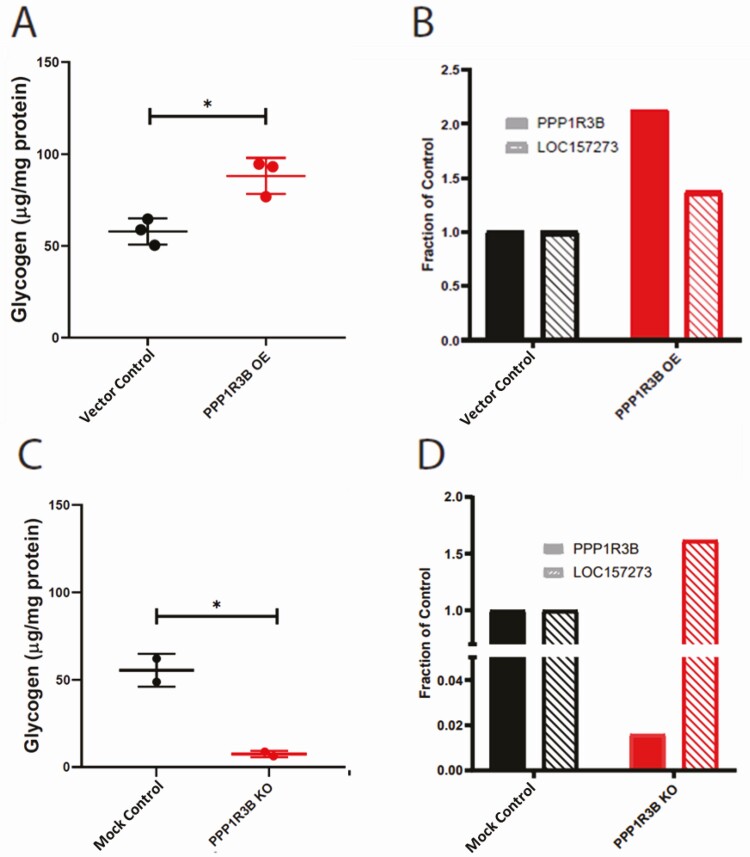
To determine whether increased or decreased expression of PPP1R3B RNA would phenocopy the effects of rs4841132-A, PPP1R3B was overexpressed using a cytomegalovirus promoter or knocked down using CRISPR-Cas9 to create a homozygous premature stop codon with nonsense mediated decay. We found that increasing PPP1R3B expression increased glycogen (Fig. 5A). Cells expressing the premature stop in PPP1R3B had decreased glycogen (Fig. 5C). In both increased and decreased PPP1R3B, expression of LOC157273 RNA increased (Fig. 5B, D), which does not mimic the pattern seen with rs4841132-A in human liver.
Figure 5.
Overexpression and knockout of PPP1R3B increases and decreases glycogen and has variable effects on LOC157273 RNA levels in HuH-7 cells. Overexpression of vector or vector with PPP1R3B. (A) Fold-change in glycogen in μg/mg protein and (B) QPCR of PPP1R3B and LOC157273 normalized to vector control. Knockout of PPP1R3B with CRISPR-Cas9 engineered insertion causing protein early termination and nonsense mediated decay. (C) Fold-change in glycogen in μg/mg protein and (D) quantitative PCR of PPP1R3B and LOC157273 normalized to HuH-7 control. Control, no stimulation of cells; KO, knock-out; OE, overexpression; vector, cells expressing vector. P value between experimental (red) and control (black) conditions are noted, with * representing P < 0.05.
Causal inference analysis in the STARNET dataset suggested that PPP1R3B expression is not dependent on LOC157273 expression and vice versa. Allelic imbalance analysis (Fig. 2) suggested that the effect of rs4841132-A on PPP1R3B RNA was in cis. To determine whether LOC157273 RNA was present on the DNA in cis to potentially produce an effect, we carried out fluorescence in situ hybridization for LOC157273 in HuH-7 cells. LOC157273 was found in 2 distinct loci in the nucleus but these were not at the genomic site where LOC157273 or PPP1R3B are transcribed (Supplementary Fig. 2 (38)). Taken together, these data suggest that the LOC157273 RNA is not acting in cis to affect PPP1R3B RNA levels. Altering levels of LOC157273 can affect glycogen levels but this is not through affecting PPP1R3B RNA levels and vice versa. These data demonstrate that perturbation of the DNA region at rs4841132 mimics the decrease in LOC157273 RNA, increase in PPP1R3B RNA and increase in glycogen seen with rs4841132-A, whereas altering the levels of PPP1R3B or LOC157273 RNA individually does not result in this pattern of effects.
To determine the impact of rs4841132 on cardiometabolic disease, we examined the effect of rs4841132-A on outpatient measures and diagnoses from the MGI biobank (Fig. 6, Supplementary Table 11 (4)). The minor allele rs4841132-A (minor allele frequency [MAF] = 0.09) was associated with higher alanine aminotransferase (P = 4.22 × 10-4), a marker of hepatocellular damage, alkaline phosphatase (P = 2.46 × 10-5), a marker of liver infiltrative disease and cirrhosis (P = 0.012) while associating with decreased total serum bilirubin (P = 3.55 × 10-3). rs4841132-A was not associated with a diagnosis of NAFLD (P = 0.44) (Supplementary Table 10 (4)) consistent with our previous data (2).
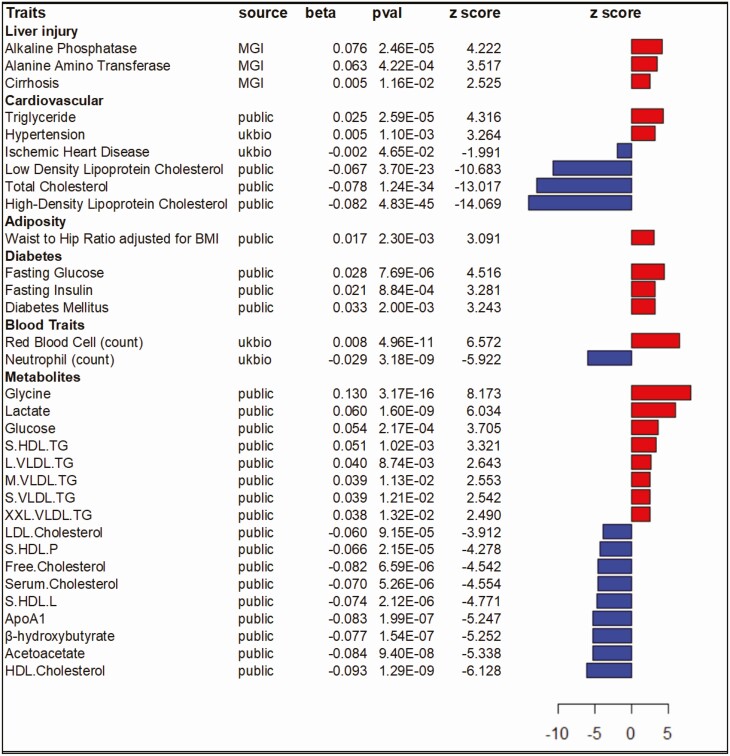
Figure 6.
Pleiotropic effects of rs4841132. Each row summarizes the effect of rs4841132-A on a trait using an additive model. Trait names (Traits) are grouped by system. Sources include: GWAS summary statistics (public), the Michigan Genomics Initiative (MGI), or the UK Biobank (UKBB). For each trait, effect sizes (beta) are in common units with a P value (pval) and corresponding z score from the largest analysis in source manuscripts. The z scores are graphically summarized to the right; a red bar indicates rs4841132-A associates with an increase, whereas a blue bar indicates a decrease. GWAS, genome-wide association study.
Examination of the effect of rs4841132-A in published GWAS of metabolic traits (Fig. 5, Supplementary Tables 13 (4)) (27-36) revealed rs4841132-A was significantly associated with increased serum triglyceride (P = 2.59 × 10-5) and lower serum high density lipoprotein cholesterol (HDL; P = 4.83 × 10-45), low density lipoprotein cholesterol (LDL; p = 3.70 × 10-23) and total serum cholesterol (P = 1.24 × 10-34). rs4841132-A was significantly associated with higher waist-to-hip ratio (P = 2.30 × 10-3) adjusted for body mass index, fasting blood glucose (P = 7.69 × 10-6), insulin (P = 8.84 × 10-4), serum lactate (P = 1.60 × 10-9), and increased prevalence of type 2 diabetes (P = 2.00 × 10-3). In the UKBB (Fig. 5, Supplementary Table 14 (4)), rs4841132-A was significantly associated with increased hypertension (P = 1.10 × 10-3) and red blood cell percent/count; but, associated with decreased neutrophil percent/count (P = 3.18 × 10-9) and decreased prevalence of ischemic heart disease (P = 0.04). In a large meta-analysis of nuclear magnetic resonance (NMR)-measured serum metabolites (39) (Fig. 5, Supplementary Table 13 (4)), rs4841132-A was associated (P < 0.011) with higher glycine, lactate, glucose, triglyceride levels on very-low-density lipoprotein (VLDL), and triglyceride levels on small HDL. rs4841132-A was associated with decreased HDL cholesterol particles, phospholipids, lipids, cholesterol esters, and free cholesterol on HDL. rs4841132-A was also associated with lower products of fatty acid oxidation (β-hydroxybutyrate, acetoacetate), APOA1, and free and total HDL, LDL, and total cholesterol. These define a metabolic signature of rs4841132-A and perhaps of increased liver glycogen status. Two coding variants in PPP1R3B did not associate with cardiometabolic traits similar to what is seen with rs4841132-A (Supplementary Tables 15-19 (4)).
Discussion
Using multiple population-based cohorts, we have shown that a common variant, rs4841132, near PPP1R3B, was associated with liver attenuation. Our query of published studies revealed rs4841132-A was also associated with increased obesity, hypertension, insulin resistance, and decreased serum HDL, total serum cholesterol, and ischemic heart disease (ie, promotion of metabolic disease), but not myocardial infarction (MI). The minor allele, rs4841132-A, was associated with liver damage but not with a diagnosis of NAFLD in MGI consistent with prior reports (40, 41). We also report that rs4841132-A does not associate with liver fat in UKBB. This lack of association of rs4841132-A with NAFLD/liver fat is consistent with our previous report that rs4240624, which is in high linkage disequilibrium (LD) with rs4841132, was not associated with histologic measures of NAFLD including steatosis, nonalcoholic steatohepatitis, and fibrosis (2, 3). These data suggest that rs4841132-A promotes liver disease through a mechanism separate from promoting NAFLD. Taken together, the presence of liver damage, increased serum lactate, and increased triglyceride levels with rs4841132-A parallels what is seen in individuals with liver glycogen storage diseases.
Metabolic syndrome consists of having 3 or more of the following: increased abdominal obesity (waist circumference), hypertension, serum triglyceride, insulin resistance, and low HDL (42). Traits associated with rs4841132-A include all 5 criteria. In addition, rs4841132-A was associated with end products of increased glycolysis (ie, increased lactate, serum triglyceride, and glycine). These metabolites can be used in place of glucose as an energy source by peripheral tissues and are often elevated with increased glycogen storage and glycogen storage diseases (43). rs4841132-A was associated with lower β-hydroxybutyrate and acetoacetate suggesting that beta-oxidation of fatty acids was reduced, further increasing levels of fatty acids and triglyceride. Excess liver triglyceride can be transported to other tissues via VLDL, which may explain why rs4841132-A was associated with increased serum extra-large VLDL particle concentrations, VLDL triglyceride content, and total serum triglyceride content. Interestingly, fasting glucose levels in individuals with rs4841132-A were slightly increased as were insulin levels which is not typical of most glycogen storage diseases where often hypoglycemia is seen (43). This suggests that rs4841132-A may lead to insulin resistance. The molecular mechanism(s) through which rs4841132-A promotes insulin resistance, hypertension, and decreased serum HDL are not clear. The strongest effect of rs4841132-A was on reducing HDL and, in particular, HDL particle concentrations. This suggests that some forms of low HDL reflect high glycogen stores and explains the well-known association between low physical activity and low HDL (44) that, from our results here, may represent high liver glycogen. Further, our findings suggest that a primary defect increasing glycogen storage (caused by rs4841132-A) can secondarily reduce HDL levels, suggesting that the known beneficial effects of exercise on metabolic syndrome traits may be mediated, in part, through a primary reduction in glycogen energy stores. Thus, our data suggest that interventions targeting HDL directly rather than the problem of excess energy stores and increased hepatic glycogen will likely not be effective in curbing development or treatment of metabolic syndrome and may help explain the failure of HDL increasing medications to improve metabolic syndrome (45).
Metabolic syndrome is associated with increased risk of MI in epidemiological studies (46). It has been recently suggested that variants that increase serum triglyceride production from liver increase risk of MI (47). This adds to the more traditional view that increased serum LDL cholesterol promotes MI. However, rs4841132-A dissociates the serum triglyceride from serum cholesterol levels. In rs4841132-A individuals, reduced prevalence of MI tracked with reduced serum total LDL cholesterol and not with increased total serum triglyceride. This is particularly evident in the NMR metabolomics analyses where most rs4841132-A-associated VLDL fractions were enriched for triglyceride at the expense of cholesterol. This suggests that at least higher triglyceride levels in rs4841132-A individuals do not increase MI risk. Our results suggest that strategies to reduce serum LDL cholesterol are more likely to reduce risk of MI than strategies to reduce serum triglyceride in rs4841132 individuals. Although lower cholesterol synthesis can be seen with high bilirubin levels, here rs4841132-A associates with lower total serum cholesterol and lower serum bilirubin. The mechanism behind this remains to be determined.
rs4841132-A was associated with increased red blood cell count and decreased neutrophil count. Most glycogen storage diseases are associated with anemia (48). The mechanism to explain why rs4841132-A is associated with increased red blood cell percentage is not known. Neutropenia can be seen in glycogen storage disease type 1b, where the transporter for glucose-6-phosphate is affected and causes a problem with dephosphorylation of glucose-6-phosphate to glucose to cause fasting hypoglycemia (43). rs4841132-A was not associated with fasting hypoglycemia, so it is not clear whether the mechanism causing neutropenia is the same in rs4841132-A as in glycogen storage disease type 1b.
PPP1R3B encodes the glycogen-targeting subunit for protein phosphatase 1, which increases the activity of glycogen synthase and decreases the activity of glycogen phosphorylase, thereby increasing glycogen accumulation (49). We show that deleting the rs4841132 region increases PPP1R3B RNA, decreases LOC157273 RNA, and increases glycogen, perfectly phenocopying the effects of rs4841132-A. Although increasing PPP1R3B or decreasing LOC157273 RNA can increase glycogen, altering the expression of these genes separately does not invariably decrease LOC157273 RNA or increase PPP1R3B RNA, respectively, which is what is seen with rs4841132-A. This suggests that the deletion of the region and an effect of the DNA alteration in cis better mimics the biology of rs4841132-A which also affects expression of PPP1R3B in cis, than altering the RNA of these genes. However, because altering PPP1R3B and LOC157273 RNA levels can independently affect glycogen levels, we cannot rule out more complex mechanisms of action at this locus. These results corroborate the work of others showing that overexpressing PPP1R3B in human cell lines and in mice increases liver glycogen (49, 50). We show that the population attributable risk of rs4841132-A in increasing liver density (glycogen) across ancestries is large (odds of increased liver attenuation of 2.4) for a common variant and leads to identifiable human phenotypes and disease. This is somewhere in between a strong Mendelian effect and a negligibly small common effect and warrants further investigation for clinical intervention. Loss of PPP1R3B in mouse liver decreases liver glycogen (50, 51). We note that 2 missense variants in PPP1R3B (rs3748140, Gly48Glu, MAF = 0.03; and rs61756425, Ser41Arg, MAF = 0.02) did not associate with lower CT-measured liver attenuation in our meta-analysis, nor with any other disease or metabolic measure in MGI, UKBB, or the NMR serum metabolite meta-analysis. This may be due to low power because of low allele frequency, our inability to quantitate low glycogen using CT, or to these alleles not having strong effects on protein function. HuH-7 cells with a 1 bp insertion in both copies of PPP1R3B, resulting in a premature stop codon, significantly decreased (but did not eliminate) glycogen in our experiments. Why then has PPP1R3B not been reported as a glycogen storage disease gene type 0? One possibility is that complete loss of PPP1R3B may be lethal in humans. Alternatively, and consistent with our results, residual glycogen synthesis may still occur in the absence of PPP1R3B, which may prevent the subtle clinical manifestations of glycogen storage disease type 0.
These results represent the largest genetic analysis of liver attenuation, allowing us to identify mechanisms by which this common noncoding variant appears to promote glycogen storage disease. Adding to our understanding of human metabolic disease, we show that elevated hepatic glycogen is one cause of metabolic syndrome that does not invariably increase risk of MI. We also show that elevated hepatic glycogen in the population does lead to cirrhosis; however, this work is not without limitations. Some of the phenotypes examined are from hospital-based collections that may vary from the population norm. Further, phenotypes of any particular individual with rs4841132-A may vary from population averages based on other personal characteristics, including genetic background, environmental exposures, and behaviors. Likewise, we cannot rule out the contributory role of other variants in LD with rs4841132 but these would have to be within the 105 bp window deleted, which functionally explains the phenotype. Finally, we cannot rule out that LOC157273 RNA may have effects on liver or other human phenotypes.
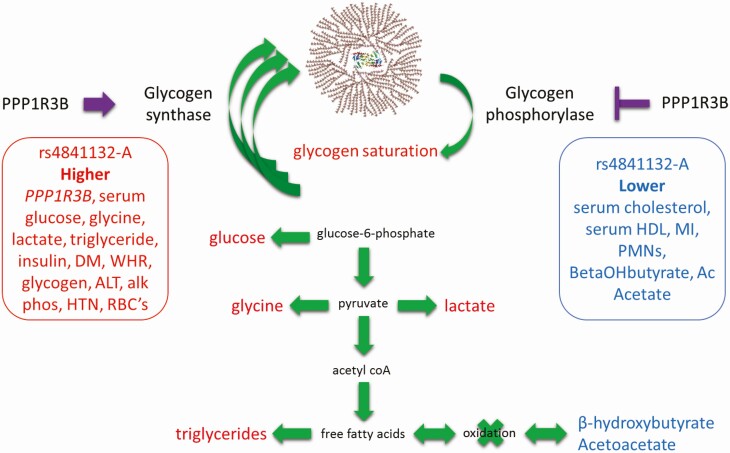
In summary, we show that deleting the region encompassing rs4841132-A increases PPP1R3B RNA, decreases LOC157273 RNA, and increases glycogen that phenocopies the pathology of rs4841132-A, which we propose causes pathology by increasing hepatic glycogen storage (Fig. 7). rs4841132-A promotes metabolic syndrome but protects against MI. These results are medically important in that they define the mechanism by which a noncoding variant causes human disease and suggest that metabolic syndrome may be a high glycogen state and that high serum triglycerides do not invariably lead to increased myocardial infarction.
Figure 7.
Physiological model of the metabolic effects of rs4841132-A. Purple arrow and inhibition sign note putative increased and inhibitory effects of increased PPP1R3B on the enzyme activity of glycogen synthase and glycogen phosphorylase, respectively. Green arrows represent possible net flow of metabolites to account for traits, diseases, and metabolite levels seen in the population. Traits, diseases, or metabolites that are increased or decreased in the population are noted in red and blue, respectively.
Acknowledgments
We are indebted to the participants for their willingness to participate in the study. This work was performed under the auspices of the Genetics of Obesity-Related Liver Disease Consortium. We thank Eric Boerwinkle, PhD, from the Human Genetics Center and Institute of Molecular Medicine and Division of Epidemiology, University of Texas Health Science Center, Houston, TX, USA, and Julie Cunningham, PhD, from the Department of Health Sciences Research, Mayo Clinic College of Medicine, Rochester, MN, USA, for their help with genotyping.
Financial Support: The AGES study has been funded by National Institutes of Health (NIH) contracts N01-AG-1-2100 and 271201200022C, the NIA Intramural Research Program, Hjartavernd (the Icelandic Heart Association), and the Althingi (the Icelandic Parliament). The study is approved by the Icelandic National Bioethics Committee, VSN: 00-063. The researchers are indebted to the participants for their willingness to participate in the study. The Framingham Heart Study is conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with Boston University (Contract No. N01-HC-25195 and HHSN268201500001I). Funding for SHARe Affymetrix genotyping was provided by NHLBI Contract N02-HL64278. SHARe Illumina genotyping was provided under an agreement between Illumina and Boston University. Funding for Affymetrix genotyping of the FHS Omni cohorts was provided by Intramural NHLBI funds from Andrew D. Johnson and Christopher J. O’Donnell. HRS is supported by the National Institute on Aging (NIA U01AG009740). The genotyping was funded separately by the National Institute on Aging (RC2 AG036495, RC4 AG039029). Genotyping was conducted by the NIH Center for Inherited Disease Research (CIDR) at Johns Hopkins University. Genotyping quality control and final preparation of the data were performed by the Genetics Coordinating Center at the University of Washington. MESA and the MESA SHARe projects are conducted and supported by the NHLBI in collaboration with MESA investigators. Support for MESA is provided by contracts HHSN268201500003I, N02-HL-64278, N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169, UL1-TR-000040, UL1-TR-001079, UL1-TR-001420, UL1-TR-001881, and DK063491. MESA Family is conducted and supported by the NHLBI in collaboration with MESA investigators. Support is provided by grants and contracts R01HL071051, R01HL071205, R01HL071250, R01HL071251, R01HL071258, R01HL071259, by the National Center for Research Resources, Grant UL1RR033176, and the National Center for Advancing Translational Sciences, Grant UL1TR001881. The WHI program is funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, and U.S. Department of Health and Human Services through contracts N01WH22110, 24152, 32100-2, 32105-6, 32108-9, 32111-13, 32115, 32118-32119, 32122, 42107-26, 42129-32, and 44221. This manuscript was not prepared in collaboration with investigators of the Women’s Health Initiative (WHI), has not been reviewed and/or approved by the WHI, and does not necessarily reflect the opinions of the WHI investigators or the NHLBI. The authors acknowledge the University of Michigan Medical School Central Biorepository for providing biospecimen processing, storage, management, and distribution services in support of the research reported in this publication. Funding for the Insulin Resistance Atherosclerosis Family Study was provided by the National Heart, Lung and Blood Institute grants 5R01HL060944, 5R01HL061019, 5R01HL060919, 5R01HL060894, and 5R01HL061210. The Family Heart Study was supported by grant R01-DK-089256 from National Institute of Diabetes and Digestive and Kidney Diseases and grant R01HL117078 from NHLBI. Support for the Genetic Epidemiology Network of Arteriopathy (GENOA) study was provided by the NHLBI (HL054457, HL054464, HL054481, HL087660, and HL085571) of the National Institutes of Health. E.K.S., Y.H.C., Y.C., S.K.H., L.C.S., and B.H. are supported by NIH grants RO1 DK106621, RO1 DK107904 and The University of Michigan Department of Internal Medicine.
Glossary
Abbreviations
- ASO
antisense oligonucleotide
- CT
computed tomography
- FBS
fetal bovine serum
- GOLD
Genetics of Obesity-associated Liver Disease
- GWAS
genome-wide association study
- HDL
high-density lipoprotein
- ICD
International Classification of Diseases
- IQS
imputation quality score
- LDL
low-density lipoprotein
- MAF
minor allele frequency
- MI
myocardial infarction
- MGH
Massachusetts General Hospital
- MGI
Michigan Genomics Initiative
- NAFLD
nonalcoholic fatty liver disease
- NMR
nuclear magnetic resonance
- RT-qPCR
RT-quantitative PCR
- UKBB
UK Biobank
- VLDL
very-low-density lipoprotein.
Additional Information
Disclosures: L.Y.A. is an employee of GSK. E.E.S. is chief executive officer of Sema4. J.O. consulted for Regeneron during the course of this work. The remaining authors have nothing to disclose.
Data Availability
Some or all data generated or analyzed during this study are included in this published article or in the data repositories listed in References. Genetic data for rs4841132 and phenotypic data from the GOLD Consortium is available from the individual cohort studies. The UKBB genomic and phenotypic data supporting this publication are publicly available from the Roslin Institute, University of Edinburgh (http://geneatlas.roslin.ed.ac.uk/). Restrictions apply to the availability of some or all data generated or analyzed during this study to preserve patient confidentiality or because they were used under license. The corresponding author will on request detail the restrictions and any conditions under which access to some data may be provided. The Michigan Genomics Initiative (MGI) genomic and phenotypic data are not publicly available due to restrictions on participant privacy. MGI data can be made available on reasonable request to the corresponding author with permission of the University of Michigan Institutional Review Board.
References
- 1. Ozen H. Glycogen storage diseases: new perspectives. World J Gastroenterol. 2007;13(18):2541-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Speliotes EK, Yerges-Armstrong LM, Wu J, et al. ; NASH CRN; GIANT Consortium; MAGIC Investigators; GOLD Consortium . Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 2011;7(3):e1001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gorden A, Yang R, Yerges-Armstrong LM, et al. ; GOLD Consortium . Genetic variation at NCAN locus is associated with inflammation and fibrosis in non-alcoholic fatty liver disease in morbid obesity. Hum Hered. 2013;75(1):34-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Supplementary Tables. 10.6084/m9.figshare.13061324. [DOI] [Google Scholar]
- 5. Harris TB, Launer LJ, Eiriksdottir G, et al. Age, Gene/Environment Susceptibility-Reykjavik Study: multidisciplinary applied phenomics. Am J Epidemiol. 2007;165(9): 1076-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Higgins M, Province M, Heiss G, et al. NHLBI Family Heart Study: objectives and design. Am J Epidemiol. 1996;143(12):1219-1228. [DOI] [PubMed] [Google Scholar]
- 7. Mahmood SS, Levy D, Vasan RS, Wang TJ. The Framingham Heart Study and the epidemiology of cardiovascular disease: a historical perspective. Lancet. 2014;383(9921):999-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Investigators TF. Multi-center genetic study of hypertension: the family blood pressure program (FBPP). Hypertension. 2002;39(1):3-9. [DOI] [PubMed] [Google Scholar]
- 9. Henkin L, Bergman RN, Bowden DW, et al. Genetic epidemiology of insulin resistance and visceral adiposity. The IRAS Family Study design and methods. Ann Epidemiol. 2003;13(4):211-217. [DOI] [PubMed] [Google Scholar]
- 10. Fuqua SR, Wyatt SB, Andrew ME, et al. Recruiting African-American research participation in the Jackson Heart Study: methods, response rates, and sample description. Ethn Dis. 2005;15(4 Suppl 6):S6-18. [PubMed] [Google Scholar]
- 11. Bild DE, Bluemke DA, Burke GL, et al. Multi-Ethnic Study of Atherosclerosis: objectives and design. Am J Epidemiol. 2002;156(9):871-881. [DOI] [PubMed] [Google Scholar]
- 12. Rampersaud E, Bielak LF, Parsa A, et al. The association of coronary artery calcification and carotid artery intima-media thickness with distinct, traditional coronary artery disease risk factors in asymptomatic adults. Am J Epidemiol. 2008;168(9):1016-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sorkin J, Post W, Pollin TI, O’Connell JR, Mitchell BD, Shuldiner AR. Exploring the genetics of longevity in the Old Order Amish. Mech Ageing Dev. 2005;126(2):347-350. [DOI] [PubMed] [Google Scholar]
- 14. Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Canela-Xandri O, Rawlik K, Tenesa A.. Data from: an atlas of genetic associations in UK Biobank. 2017. Deposited August 18, 2017. https://www.biorxiv.org/content/early/2017/08/18/176834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhou W, Zhao Z, Nielsen JB, et al. Scalable generalized linear mixed model for region-based association tests in large biobanks and cohorts. Nat Genet. 2020;52(6):634-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Maguire LH, Handelman SK, Du X, Chen Y, Pers TH, Speliotes EK. Genome-wide association analyses identify 39 new susceptibility loci for diverticular disease. Nat Genet. 2018;50(10):1359-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Greenawalt DM, Dobrin R, Chudin E, et al. A survey of the genetics of stomach, liver, and adipose gene expression from a morbidly obese cohort. Genome Res. 2011;21(7): 1008-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Franzén O, Ermel R, Cohain A, et al. Cardiometabolic risk loci share downstream cis- and trans-gene regulation across tissues and diseases. Science. 2016;353(6301):827-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yan H, Yuan W, Velculescu VE, Vogelstein B, Kinzler KW. Allelic variation in human gene expression. Science. 2002;297(5584):1143. [DOI] [PubMed] [Google Scholar]
- 23. Johnson AD, Zhang Y, Papp AC, et al. Polymorphisms affecting gene transcription and mRNA processing in pharmacogenetic candidate genes: detection through allelic expression imbalance in human target tissues. Pharmacogenet Genomics. 2008;18(9):781-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Smith RM, Webb A, Papp AC, et al. Whole transcriptome RNA-Seq allelic expression in human brain. BMC Genomics. 2013;14:571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Millstein J, Zhang B, Zhu J, Schadt EE. Disentangling molecular relationships with a causal inference test. BMC Genet. 2009;10:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang Y, Tong S, Tai AW, Hussain M, Lok AS. Hepatitis B virus core promoter mutations contribute to hepatocarcinogenesis by deregulating SKP2 and its target, p21. Gastroenterology. 2011;141(4):1412-1421, 1421.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Locke AE, Kahali B, Berndt SI, et al. ; LifeLines Cohort Study; ADIPOGen Consortium; AGEN-BMI Working Group; CARDIOGRAMplusC4D Consortium; CKDGen Consortium; GLGC; ICBP; MAGIC Investigators; MuTHER Consortium; MIGen Consortium; PAGE Consortium; ReproGen Consortium; GENIE Consortium; International Endogene Consortium . Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518(7538):197-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lu Y, Day FR, Gustafsson S, et al. New loci for body fat percentage reveal link between adiposity and cardiometabolic disease risk. Nat Commun. 2016;7:10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wood AR, Esko T, Yang J, et al. ; Electronic Medical Records and Genomics (eMEMERGEGE) Consortium; MIGen Consortium; PAGEGE Consortium; LifeLines Cohort Study . Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet. 2014;46(11):1173-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dupuis J, Langenberg C, Prokopenko I, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42(2):105-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Willer CJ, Schmidt EM, Sengupta S, et al. ; Global Lipids Genetics Consortium . Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45(11):1274-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Replication DIG, Meta-analysis C, Asian Genetic Epidemiology Network Type 2 Diabetes C, et al. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet. 2014;46(3):234-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nikpay M, Goel A, Won HH, et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015;47(10):1121-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ehret GB, Munroe PB, Rice KM, et al. ; International Consortium for Blood Pressure Genome-Wide Association Studies; CARDIoGRAM consortium; CKDGen Consortium; KidneyGen Consortium; EchoGen consortium; CHARGE-HF consortium . Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478(7367):103-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kilpelainen TO, Carli JF, Skowronski AA, et al. Genome-wide meta-analysis uncovers novel loci influencing circulating leptin levels. Nat Commun. 2016;7:10494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tin A, Balakrishnan P, Beaty TH, et al. GCKR and PPP1R3B identified as genome-wide significant loci for plasma lactate: the Atherosclerosis Risk in Communities (ARIC) study. Diabet Med. 2016;33(7):968-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Consortium GT, Laboratory DA, Coordinating Center -Analysis Working G, et al. Genetic effects on gene expression across human tissues. Nature. 2017;550(7675):204-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Supplementary Figures. 10.6084/m9.figshare.13061390. [DOI] [Google Scholar]
- 39. Kettunen J, Tukiainen T, Sarin AP, et al. Genome-wide association study identifies multiple loci influencing human serum metabolite levels. Nat Genet. 2012;44(3):269-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Romeo S, Kozlitina J, Xing C, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40(12):1461-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sookoian S, Castaño GO, Scian R, et al. Genetic variation in transmembrane 6 superfamily member 2 and the risk of nonalcoholic fatty liver disease and histological disease severity. Hepatology. 2015;61(2):515-525. [DOI] [PubMed] [Google Scholar]
- 42. National Cholesterol Education Program Expert Panel on Detection E, Treatment of High Blood Cholesterol in A. Third Report of the National Cholesterol Education Program (NCEP) expert panel on detection, evaluation, and treatment of high blood cholesterol in adults (Adult Treatment Panel III) final report. Circulation. 2002;106(25):3143-3421. [PubMed] [Google Scholar]
- 43. Wolfsdorf JI, Weinstein DA. Glycogen storage diseases. Rev Endocr Metab Disord. 2003;4(1):95-102. [DOI] [PubMed] [Google Scholar]
- 44. Nikkilä EA, Kuusi T, Myllynen P. High density lipoprotein and apolipoprotein A-i during physical inactivity. Demonstration at low levels in patients with spine fracture. Atherosclerosis. 1980;37(3):457-462. [DOI] [PubMed] [Google Scholar]
- 45. Keene D, Price C, Shun-Shin MJ, Francis DP. Effect on cardiovascular risk of high density lipoprotein targeted drug treatments niacin, fibrates, and CETP inhibitors: meta-analysis of randomised controlled trials including 117,411 patients. Bmj. 2014;349:g4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Grundy SM, Brewer HB Jr, Cleeman JI, Smith SC Jr, Lenfant C; American Heart Association; National Heart, Lung, and Blood Institute . Definition of metabolic syndrome: report of the National Heart, Lung, and Blood Institute/American Heart Association conference on scientific issues related to definition. Circulation. 2004;109(3):433-438. [DOI] [PubMed] [Google Scholar]
- 47. Liu DJ, Peloso GM, Yu H, et al. ; Charge Diabetes Working Group; EPIC-InterAct Consortium; EPIC-CVD Consortium; GOLD Consortium; VA Million Veteran Program . Exome-wide association study of plasma lipids in >300,000 individuals. Nat Genet. 2017;49(12):1758-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang DQ, Carreras CT, Fiske LM, et al. Characterization and pathogenesis of anemia in glycogen storage disease type Ia and Ib. Genet Med. 2012;14(9):795-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Agius L. Role of glycogen phosphorylase in liver glycogen metabolism. Mol Aspects Med. 2015;46:34-45. [DOI] [PubMed] [Google Scholar]
- 50. Mehta MB, Shewale SV, Sequeira RN, Millar JS, Hand NJ, Rader DJ. Hepatic protein phosphatase 1 regulatory subunit 3B (Ppp1r3b) promotes hepatic glycogen synthesis and thereby regulates fasting energy homeostasis. J Biol Chem. 2017;292(25):10444-10454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Stender S, Smagris E, Lauridsen BK, et al. Relationship between genetic variation at PPP1R3B and levels of liver glycogen and triglyceride. Hepatology. 2018;67(6):2182-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Some or all data generated or analyzed during this study are included in this published article or in the data repositories listed in References. Genetic data for rs4841132 and phenotypic data from the GOLD Consortium is available from the individual cohort studies. The UKBB genomic and phenotypic data supporting this publication are publicly available from the Roslin Institute, University of Edinburgh (http://geneatlas.roslin.ed.ac.uk/). Restrictions apply to the availability of some or all data generated or analyzed during this study to preserve patient confidentiality or because they were used under license. The corresponding author will on request detail the restrictions and any conditions under which access to some data may be provided. The Michigan Genomics Initiative (MGI) genomic and phenotypic data are not publicly available due to restrictions on participant privacy. MGI data can be made available on reasonable request to the corresponding author with permission of the University of Michigan Institutional Review Board.