Abstract
Human immunodeficiency virus (HIV) causes acquired immune deficiency syndrome (AIDS) and enters the host cell via CD4 and either CC-chemokine receptor 5 (CCR) or CXC-chemokine receptor 4 (CXCR4). HIV is directly recognized by toll-like receptor 4 (TLR4) and affects downstream immune-related signal pathways. In addition, stimulated TLR4 inhibits HIV-1 invasion, and the rs4986790 single nucleotide polymorphism (SNP) (D299G) of the TLR4 gene contributes to the risk of HIV-1 infection in an Indian population. To evaluate whether the rs4986790 SNP of the TLR4 gene is related to vulnerability to HIV-1 infection, we collected genetic information from HIV-1 patients in previous studies and performed an association analysis with a matched control population obtained from the 1000 Genomes Project. In addition, to strengthen the results of association analysis, we performed a meta-analysis. We identified a strong association between the rs4986791 SNP and susceptibility to HIV infection in HIV-infected patients in previous studies and a matched control population obtained from the 1000 Genomes Project. In addition, we found that the G allele of the rs4986791 SNP in the TLR4 gene is strongly related to susceptibility to HIV infection in three Caucasian populations (odd ratio = 2.29, 95% confidence interval: 1.72–3.07, p = 1.438 × 10−7) and all four populations (odd ratio = 2.22, 95% confidence interval: 1.74–2.84, p = 2 × 10−10) in a meta-analysis. To the best our knowledge, this was the first meta-analysis on the association between the rs4986791 SNP of the TLR4 gene and susceptibility to HIV infection.
Keywords: HIV, susceptibility, TLR4, rs4986790, SNP, D299G, meta-analysis
1. Introduction
Human immunodeficiency virus (HIV) is a retrovirus that harbors positive sense single strand RNA as its viral genome. The viral genome is translated to viral structural proteins (Gag, Env and Pol), essential regulatory elements (Tat and Rev) and accessory regulatory proteins (Nef, Vpr, Vif and Vpu) [1,2]. It has been postulated that HIV originated from nonhuman primates and spread to humans through certain body fluids, including blood, semen, vaginal or rectal fluids and breast milk during the 1900s. HIV is internalized by host cells using host receptor proteins, including CD4 and either CC-chemokine receptor 5 (CCR) or CXC-chemokine receptor 4 (CXCR4). Thus, HIV mainly targets CD4+ T cells and can lead acquired immune deficiency syndrome (AIDS) by disarming the host immune system [3,4].
To prevent external infection, several pattern recognition receptors (PRRs) including toll-like receptors (TLRs), RIG-I-like receptors (RLRs) and NOD-like receptors (NLRs), recognize a pathogen-related pattern of external invaders and activate the host immune system through downstream regulators, including nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB), mitogen-activated protein kinase (MAPK) and interferon type I [5,6,7,8,9]. Among TLRs, a previous study reported that TLR4 located on the cell surface was upregulated in response to HIV-1 infection in monocyte-derived macrophage (MDM) and peripheral blood mononuclear cell (PBMC) [10]. In addition, the Tat protein of HIV-1 directly binds to TLR4 and activates tumor necrosis factor-α (TNF-α) and interleukin-10 (IL-10) [11]. These studies suggest an association between TLR4 and HIV infection. Remarkably, TLR4 stimulation protects HIV infection from CD4+ T cells in vitro [12]. In addition, a functional variation of TLR4, namely, the rs4986790 single nucleotide polymorphism (SNP) (D299G), contributes to risk of HIV-1 infection in an Indian population [13].
To validate whether the rs4986790 SNP of the TLR4 gene is associated with susceptibility to HIV-1 infection, we collected three studies that contain genetic information on ethnic backgrounds and allele frequencies of rs4986790 SNP of the TLR4 gene from HIV-infected patients [14,15,16]. The matched Caucasian control populations, including Iberian populations from Spain, Tuscans from Italy and northern and western Europeans from Utah, were obtained from the 1000 Genomes Project and used for an association analysis [17]. Then, we performed a meta-analysis by collecting data from eligible studies to evaluate the association between rs4986790 SNP of the TLR4 gene and susceptibility to HIV-1 infection.
2. Materials and Methods
2.1. Literature Search
A literature search was conducted in PubMed to identify studies reporting the rs4986790 SNP of the TLR4 gene. The following searching terms were used: “TLR4,” “SNP” and “HIV” combined with “polymorphism” or “susceptibility” (the last search update was performed on 18 July 2020). Irrelevant reports were excluded after the initial screening of titles and abstracts. Eligible studies should comply with the following inclusion criteria: (1) investigating the association between rs4986790 and HIV-1; (2) a cohort or case–control study; (3) genetic information of rs4986790 of HIV-1 infected patients (4) with full text; (5) published in English. Exclusion criteria were as follows: (1) animal studies; (2) case reports or reviews; (3) containing insufficient genotype data.
2.2. Association Analysis
We collected three studies that contain information on ethnic background and allele frequencies of the rs4986790 SNP of the TLR4 gene in HIV-infected patients [12,13,14]. Matched Caucasian control populations, including Iberian populations from Spain, Tuscans from Italy and northern and western Europeans from Utah, were obtained from the 1000 Genome Project [17]. Differences in allele frequencies between HIV-infected patients and control populations were analyzed using SAS version 9.4 (SAS Institute Inc., Cary, NC, USA). Statistical significance was determined by p-values obtained using the χ2 test.
2.3. Meta-Analysis
The strength of the association between the rs4986790 SNP of the TLR4 gene and susceptibility to HIV infection was evaluated by calculating crude additive odd ratios and 95% confident intervals for each study. The pooled odd ratios were calculated based on an additive genetic model (A allele vs. G allele). Heterogeneity was based on p-value and I2 value. A fixed effect model was used to calculate the pooled odd ratios. Publication bias was examined using Begg’s funnel plot and Egger’s weighted regression methods. All statistical analyses were conducted using the meta package of R program (https://www.r-project.org/).
3. Results
3.1. Strong Association between the rs4986790 SNP (D299G) of the TLR4 Gene and Susceptibility to HIV Infection in Three Caucasian Populations
We searched 20 research articles following searching terms: “TLR4,” “SNP” and “HIV” combined with “polymorphism” or “susceptibility” (the last search update was performed on 18 July 2020) on PubMed. After excluding duplicate articles, a total of four relevant studies were extracted from the databases based on our inclusion and exclusion criteria.
To identify an association between the rs4986790 SNP (D299G) and susceptibility to HIV infection, we performed an association analysis between HIV patients for whom information on ethnic background and allele frequencies of rs4986790 SNP of the TLR4 gene were reported in previous studies [14,15,16] and matched Caucasian control populations, including Iberian populations in Spain, Tuscans from Italy and northern and western Europeans from Utah, obtained from the 1000 Genomes Project. Notably, allele frequencies of the rs4986790 SNP of the TLR4 gene exhibited a strong association (p < 0.05) with susceptibility to HIV infection in all tested groups (Table 1).
Table 1.
Comparison of allele frequencies of rs4986790 (D299G) in the toll-like receptor 4 (TLR4) gene between human immunodeficiency virus (HIV)-infected patients in previous studies and matched control populations obtained from the 1000 Genomes Project.
| Population | Country | Year of Publication |
Authors | Cases | Controls | p-Value | Reference | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Allele Frequencies, n (%) | Total, n | Allele Frequencies, n (%) | Total, n | ||||||||
| A | G | A | G | ||||||||
| Caucasian | USA | 2009 | Pine | 374 (93.03) | 28 (6.97) | 201 | 600 (95.85) | 26 (4.15) | 313 | 0.0486 | 1000 Genomes Project, [14] |
| Caucasian | Spain | 2010 | Pulido | 822 (87.82) | 114 (12.18) | 468 | 600 (95.85) | 26 (4.15) | 313 | 5.2687 × 10−8 | 1000 Genomes Project, [15] |
| Caucasian | Greece | 2010 | Papadopoulos | 369 (92.71) | 29 (7.29) | 199 | 600 (95.85) | 26 (4.15) | 313 | 0.0302 | 1000 Genomes Project, [16] |
| Asian | India | 2018 | Vidyant | 277 (86.56) | 43 (13.44) | 160 | 502 (92.96) | 38 (7.04) | 270 | 0.0019 | [13] |
3.2. Strong Association between the rs4986790 SNP of the TLR4 Gene and Susceptibility to HIV Infection Based on a Meta-Analysis
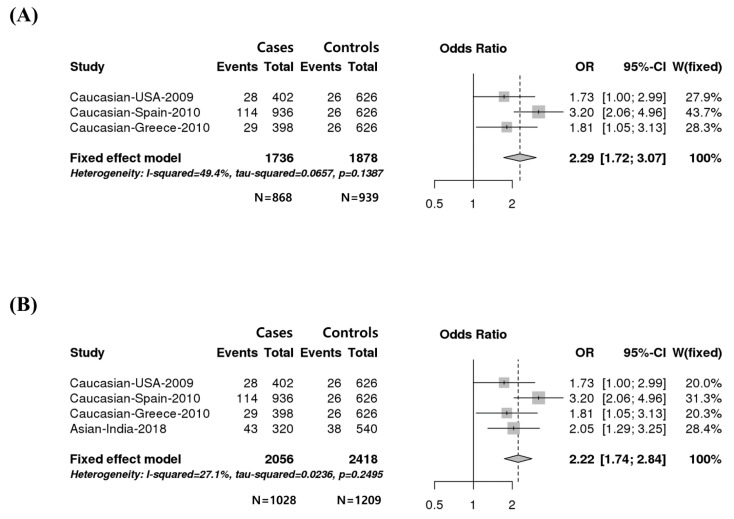
A total of four studies that reported the association between the rs4986790 SNP and susceptibility to HIV infection were identified from the literature and included in this meta-analysis (Figure 1). In total, 1028 HIV-infected patients and 1209 controls were included in the meta-analysis (Figure 1B). Detailed information on the eligible studies is presented in Table 1.
Figure 1.
(A) Forest plot of the association between the rs4986790 single nucleotide polymorphism (SNP) of the TLR4 gene and susceptibility to human immunodeficiency virus (HIV) infection in a Caucasian population. (B) Forest plot of the association between the rs4986790 SNP and susceptibility to HIV infection in four populations. Forest plots of odd ratios (ORs) were calculated using an additive model (A allele vs. G allele). The number of each column of “Cases” and “Controls” indicates number of alleles. “N” indicates the number of people.
Heterogeneity among collected studies was tested using the p-value and I2 value (Table 2). Given that heterogeneity was not found in these studies, we used a fixed effect model for the meta-analysis. In addition, given that the genetic information from previous studies was commonly available as allele frequencies, we used an additive model (A allele vs. G allele) for a meta-analysis. Our data revealed an association with the risk of HIV infection in Caucasian populations (odd ratio = 2.29, 95% confidence interval: 1.72–3.07, p = 1.438 × 10−7) and all four populations (odd ratio = 2.22, 95% confidence interval: 1.74–2.84, p = 2 × 10−10) (Figure 1).
Table 2.
Meta-analysis of the association between the rs4986790 SNP of the TLR4 gene and susceptibility to HIV infection.
| Populations | Adjust p-Value | Heterogeneity | Egger’s Test | |
|---|---|---|---|---|
| p-Value | I2 Value | |||
| Caucasian | 1.438 × 10−7 | 0.1387 | 49.4% | 0.0208 |
| Total | 2 × 10−10 | 0.2495 | 27.1% | 0.1598 |
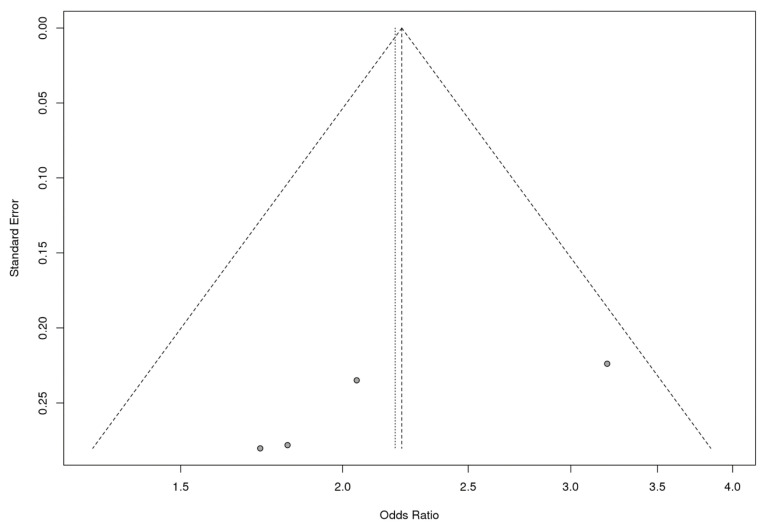
To examine potential publication bias, Begg’s and Egger’s tests were performed. The shape of the funnel plots revealed no evidence of obvious asymmetry (Figure 2). However, Egger’s test indicated the possibility of publication bias in a meta-analysis of the Caucasian subgroup (p = 0.0208) (Table 2). Sensitivity analyses were conducted to evaluate the impact of single studies on the pooled results by omitting individual studies in turn. In the Caucasian subgroup and the total group, after excluding one study (Caucasian-Spain-2010), the remaining studies showed similar results (the relative results are not provided in the text).
Figure 2.
Begg’s funnel plot for the meta-analysis of the rs4986790 SNP of the TLR4 gene in four populations.
4. Discussion
TLR4 is located on the cell surface and senses bacterial lipopolysaccharide (LPS). Thus, previous studies have revealed the association between TLR4 and bacterial-related phenotypes, including tuberculosis, cirrhosis, ascites, scrub typhus and Crohn’s disease [18,19]. In this context, the rs4986790 SNP (D299G) of the TLR4 gene has also been investigated to explain the variation of disease-related phenotypes based on the risk allele of the rs4986790 SNP. In addition, differences in the local crystal structure of the TLR4-MD2-LPS complex were noted between TLR4 with the wild type allele and the G299 allele [20,21]. Since genetic variations confer functional alteration, these studies indicate that the rs4986790 SNP of the TLR4 gene is related to anti-bacterial functions of TLR4 [22,23,24]. Interestingly, TLR4/MD2 complex directly interacts with the Tat protein of HIV-1, and HIV-1 modulates the expression level of TLR4 and downstream immune responses, including the NF-κB pathway [25,26]. In addition, the rs4986790 SNP is associated with the risk of HIV-1 infection in an Indian population [13]. Since TLR4 plays a dual role in HIV-1 infection and anti-bacterial function, rs4986790 SNP, which is associated with structural and functional alteration of TLR4, showed a relationship with the susceptibility of HIV infection and bacterial-related phenotype, including tuberculosis. These studies indicate the possibility of the association between the rs4986790 SNP of the TLR4 gene and susceptibility to HIV infection. Thus, we performed an association analysis in Caucasian HIV-infected patients from previous studies. When considering the ethnic background of Caucasian HIV-infected patients, genetic information on control populations was not available in the previous Caucasian studies. Thus, we selected and matched control populations, including Iberian populations from Spain, Tuscans from Italy and northern and western Europeans from Utah from the 1000 Genomes Project. Remarkably, we identified a strong association between the rs4986790 SNP and susceptibility to HIV infection in all tested groups (Table 1). However, since these results were highly dependent on the frequency of the control Caucasian population from the 1000 Genome Project, further validation is highly desirable in other control Caucasian populations. Furthermore, a case-control study using direct sequencing from HIV-1 infected patients in Caucasians, North and South Americans and East Asians has not been performed thus far. Thus, further study to confirm an association between rs4986790 SNP and vulnerability of HIV-1 infection is needed. The number of each column of “Allele frequencies” indicates number of alleles. “Total, n” indicates the number of people. Bold text indicates statistically significant results (p < 0.05). The control population obtained from 1000 Genome Project contains Iberian populations from Spain (IBS), Tuscans from Italy (TSI) and northern and western Europeans from Utah (CEU). To strengthen the conclusions, we performed a meta-analysis. Notably, we identified strong associations in Caucasian and all four populations (Figure 1, Table 2). Although publication bias was found in the Caucasian population, both heterogeneity and publication biases were alleviated by omitting one study (Caucasian-Spain-2010). Given that association studies on the TLR4 gene in HIV-infected patients are rare, further investigations using a larger number and various ethnic groups, including Africans and East Asians, are highly desirable in the future. In addition, given that another SNP, namely, the rs4986791 SNP (T399I) of the TLR4 gene, is also associated with bacterial infection, further validation of the association of rs4986791 SNP in HIV-infected patients should be performed in future studies.
5. Conclusions
In conclusion, we identified a strong association between the rs4986791 SNP and susceptibility to HIV infection between HIV-infected patients in previous studies and a matched control population obtained from the 1000 Genomes Project. We also found that the G allele of the rs4986791 SNP of the TLR4 gene is potent risk factors of HIV infection in Caucasians and all four populations assessed in this meta-analysis. To the best of our knowledge, this was the first meta-analysis assessment of the association between the rs4986791 SNP of the TLR4 gene and susceptibility to HIV infection.
Acknowledgments
This work was supported by NRF (National Research Foundation of Korea) Grant funded by the Korean Government (NRF-2019-Fostering Core Leaders of the Future Basic Science Program/Global Ph.D. Fellowship Program).
Abbreviations
| HIV | Human immunodeficiency virus |
| SNP | Single nucleotide polymorphism |
| TLR4 | Toll-like receptor 4 |
| AIDS | Acquired immune deficiency syndrome |
| CCR | CC-chemokine receptor 5 |
| CXCR4 | CXC-chemokine receptor 4 |
| NF-κB | Nuclear factor kappa-light-chain-enhancer of activated B cells |
| TNF-α | Tumor necrosis factor-α |
Author Contributions
Y.-C.K. and B.-H.J. conceived and designed the experiments. Y.-C.K. and B.-H.J. analyzed the data. Y.-C.K. and B.-H.J. wrote the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Basic Science Program through the National Research Foundation (NRF) of Korea funded by the Ministry of Education, Science and Technology (2018R1D1A1B07048711). This research was supported by the Basic Science Research Program through the National Research Foundation (NRF) of Korea funded by the Ministry of Education (2017R1A6A1A03015876).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data generated or analysed during this study are available from the corresponding author on reasonable request.
Conflicts of Interest
The authors declare no conflict of interest, financial or otherwise.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Deeks S.G., Overbaugh J., Phillips A., Buchbinder S. HIV infection. Nat. Rev. Dis. Primers. 2015;1:15035. doi: 10.1038/nrdp.2015.35. [DOI] [PubMed] [Google Scholar]
- 2.Barre-Sinoussi F., Ross A.L., Delfraissy J.F. Past, present and future: 30 years of HIV research. Nat. Rev. Microbiol. 2013;11:877–883. doi: 10.1038/nrmicro3132. [DOI] [PubMed] [Google Scholar]
- 3.Moir S., Chun T.W., Fauci A.S. Pathogenic mechanisms of HIV disease. Annu Rev. Pathol. 2011;6:223–248. doi: 10.1146/annurev-pathol-011110-130254. [DOI] [PubMed] [Google Scholar]
- 4.Ghosn J., Taiwo B., Seedat S., Autran B., Katlama C. Hiv. Lancet. 2018;392:685–697. doi: 10.1016/S0140-6736(18)31311-4. [DOI] [PubMed] [Google Scholar]
- 5.Beutler B.A. TLRs and innate immunity. Blood. 2009;113:1399–1407. doi: 10.1182/blood-2008-07-019307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawasaki T., Kawai T. Toll-like receptor signaling pathways. Front. Immunol. 2014;5:461. doi: 10.3389/fimmu.2014.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vidya M.K., Kumar V.G., Sejian V., Bagath M., Krishnan G., Bhatta R. Toll-like receptors: Significance, ligands, signaling pathways, and functions in mammals. Int. Rev. Immunol. 2018;37:20–36. doi: 10.1080/08830185.2017.1380200. [DOI] [PubMed] [Google Scholar]
- 8.Brubaker S.W., Bonham K.S., Zanoni I., Kagan J.C. Innate immune pattern recognition: A cell biological perspective. Annu. Rev. Immunol. 2015;33:257–290. doi: 10.1146/annurev-immunol-032414-112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takeuchi O., Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 10.Hernandez J.C., Stevenson M., Latz E., Urcuqui-Inchima S. HIV type 1 infection up-regulates TLR2 and TLR4 expression and function in vivo and in vitro. AIDS Res. Hum. Retrovir. 2012;28:1313–1328. doi: 10.1089/aid.2011.0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ben Haij N., Leghmari K., Planes R., Thieblemont N., Bahraoui E. HIV-1 Tat protein binds to TLR4-MD2 and signals to induce TNF-α and IL-10. Retrovirology. 2013;10:123. doi: 10.1186/1742-4690-10-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cromarty R., Sigal A., Liebenberg L.J.P., McKinnon L.R., Abdool Karim S.S., Passmore J.S., Archary D. Diminished HIV Infection of Target CD4+ T Cells in a Toll-Like Receptor 4 Stimulated in vitro Model. Front. Immunol. 2019;10:1705. doi: 10.3389/fimmu.2019.01705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vidyant S., Chatterjee A., Dhole T.N. A single-nucleotide polymorphism in TLR4 is linked with the risk of HIV-1 infection. Br. J. Biomed. Sci. 2019;76:59–63. doi: 10.1080/09674845.2018.1559486. [DOI] [PubMed] [Google Scholar]
- 14.Pine S.O., McElrath M.J., Bochud P.Y. Polymorphisms in toll-like receptor 4 and toll-like receptor 9 influence viral load in a seroincident cohort of HIV-1-infected individuals. AIDS. 2009;23:2387–2395. doi: 10.1097/QAD.0b013e328330b489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pulido I., Leal M., Genebat M., Pacheco Y.M., Saez M.E., Soriano-Sarabia N. The TLR4 ASP299GLY polymorphism is a risk factor for active tuberculosis in Caucasian HIV-infected patients. Curr. HIV Res. 2010;8:253–258. doi: 10.2174/157016210791111052. [DOI] [PubMed] [Google Scholar]
- 16.Papadopoulos A.I., Ferwerda B., Antoniadou A., Sakka V., Galani L., Kavatha D., Panagopoulos P., Poulakou G., Kanellakopoulou K., van der Meer J.W., et al. Association of toll-like receptor 4 Asp299Gly and Thr399Ile polymorphisms with increased infection risk in patients with advanced HIV-1 infection. Clin. Infect. Dis. 2010;51:242–247. doi: 10.1086/653607. [DOI] [PubMed] [Google Scholar]
- 17.1000 Genomes Project Consortium. Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mukherjee S., Karmakar S., Babu S.P. TLR2 and TLR4 mediated host immune responses in major infectious diseases: A review. Braz. J. Infect. Dis. 2016;20:193–204. doi: 10.1016/j.bjid.2015.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janardhanan J., Joseph Martin S., Astrup E., Veeramanikandan R., Aukrust P., Abraham O.C., Varghese G.M. Single-nucleotide polymorphisms in Toll-like receptor (TLR)-2, TLR4 and heat shock protein 70 genes and susceptibility to scrub typhus. J. Hum. Genet. 2013;58:707–710. doi: 10.1038/jhg.2013.89. [DOI] [PubMed] [Google Scholar]
- 20.Ohto U., Yamakawa N., Akashi-Takamura S., Miyake K., Shimizu T. Structural analyses of human Toll-like receptor 4 polymorphisms D299G and T399I. J. Biol. Chem. 2012;287:40611–40617. doi: 10.1074/jbc.M112.404608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anwar M.A., Choi S. Structure-Activity Relationship in TLR4 Mutations: Atomistic Molecular Dynamics Simulations and Residue Interaction Network Analysis. Sci. Rep. 2017;7:43807. doi: 10.1038/srep43807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim Y.C., Won S.Y., Jeong B.H. Identification of Prion Disease-Related Somatic Mutations in the Prion Protein Gene (PRNP) in Cancer Patients. Cells. 2020;9:1480. doi: 10.3390/cells9061480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim Y.C., Jeong M.J., Jeong B.H. Strong association of regulatory single nucleotide polymorphisms (SNPs) of the IFITM3 gene with influenza H1N1 2009 pandemic virus infection. Cell Mol. Immunol. 2020;17:662–664. doi: 10.1038/s41423-019-0322-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Won S.Y., Kim Y.C., Jeong B.H. First Report of the Potential Bovine Spongiform Encephalopathy (BSE)-Related Somatic Mutation E211K of the Prion Protein Gene (PRNP) in Cattle. Int. J. Mol. Sci. 2020;21:4246. doi: 10.3390/ijms21124246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ben Haij N., Planes R., Leghmari K., Serrero M., Delobel P., Izopet J., BenMohamed L., Bahraoui E. HIV-1 Tat Protein Induces Production of Proinflammatory Cytokines by Human Dendritic Cells and Monocytes/Macrophages through Engagement of TLR4-MD2-CD14 Complex and Activation of NF-kappaB Pathway. PLoS ONE. 2015;10:e0129425. doi: 10.1371/journal.pone.0129425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bahraoui E., Serrero M., Planes R. HIV-1 Tat—TLR4/MD2 interaction drives the expression of IDO-1 in monocytes derived dendritic cells through NF-kappaB dependent pathway. Sci. Rep. 2020;10:8177. doi: 10.1038/s41598-020-64847-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analysed during this study are available from the corresponding author on reasonable request.