Abstract
Background: Early recurrence of Plasmodium vivax is a challenge for malaria control in the field, particularly because this species is associated with lower parasitemia, which hinders diagnosis and monitoring through blood smear testing. Early recurrences, defined as the persistence of parasites in the peripheral blood despite adequate drug dosages, may arise from resistance to chloroquine. The objective of the study was to estimate early recurrence of P. vivax in the Brazilian Amazon by using a highly-sensitive detection method, in this case, PCR. Methods: An ultra-sensitive qPCR that targeted mitochondrial DNA was used to compare a standard qPCR that targeted 18S rDNA to detect early recurrence of P. vivax in very low densities in samples from patients treated with chloroquine. Results: Out of a total of 312 cases, 29 samples (9.3%) were characterized as recurrences, from which 3.2% (10/312) were only detected through ultra-sensitive qPCR testing. Conclusions: Studies that report the detection of P. vivax early recurrences using light microscopy may severely underestimate their true incidence.
Keywords: malaria diagnosis, submicroscopic infection, chloroquine resistance, mitochondrial DNA
1. Introduction
Plasmodium vivax is estimated to be responsible for 7.5 million clinical cases of malaria worldwide in 2018, and 75% of this total occurs in the region of the Americas [1]. Chloroquine (CQ) in combination with primaquine (PQ) is the first-line treatment for P. vivax malaria in most endemic countries, but CQ efficacy is now under threat from the emergence and spread of CQ resistant species of P. vivax, the occurrence of which was first reported in Papua New Guinea in 1989 [2]. In areas where high-grade CQ-resistant P. vivax is prevalent, CQ-resistance is an important contributing factor to severe P. vivax malaria [3]. P. vivax resistance to CQ has been reported in the Amazon region in Brazil, with prevalence ranging from 4.4% to 11.5% [4,5,6,7].
In the absence of effective anti-hypnozoite treatment with 8-aminoquinoline, monitoring of CQ-resistant P. vivax malaria represents a special challenge due to the ability of a new infection to relapse from dormant liver stages over weeks to years. CQ-resistance is defined as the persistence or reappearance of parasites at plasma levels of CQ and its metabolite, desethylchloroquine (DCQ), above 100 ng/dL within the first 28 days (D28) after standard therapy with CQ [2,5,8]. Recurrent parasitemia that occurs after D28 is generally considered a relapse or new infection [8]. Higher CQ efficacy in preventing late recurrence compared with drugs with shorter half-lives, such as artemether–lumefantrine, has been observed in areas of low prevalence of CQ-resistance [9].
The usual methods for measuring CQ efficacy are prone to underestimating the true early recurrence rates, which is a fundamental barrier for adequate control and longer-term strategies targeting regional elimination. The aim of this study was to accurately estimate early recurrence of P. vivax in the Brazilian Amazon using a highly sensitive qPCR method.
2. Results
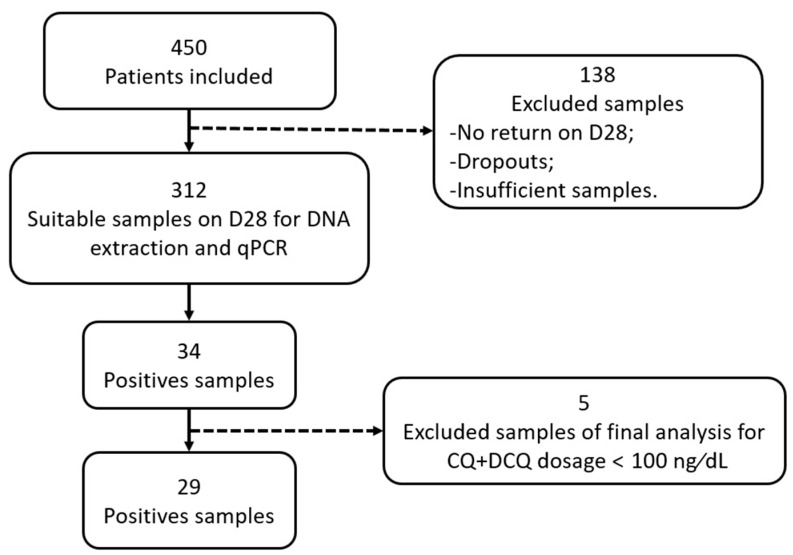
During the period of the study, 450 patients treated with CQ were enrolled. However, only 312 samples were available on D28 for DNA extraction and qPCR testing. Thirty-four (10.9%) samples were positive by Pv-mtCOX1 qPCR, although 5 samples were excluded from the final analysis because CQ/DCQ concentrations were less than 100 ng/dL (Figure 1).
Figure 1.
Flowchart of processed samples.
2.1. Pv-mtCOX1 qPCR, Pv 18S rRNA qPCR and Thick Blood Smear Agreement Results
Clinical and laboratory characteristics of patients included in this study are presented in Table 1. Of the 312 samples, 29 (9.3%) were positive for Pv-mtCOX1 qPCR, 14 (4.5%) after analysis using Pv 18S rRNA qPCR and only 5 (1.6%) were positive via thick blood smear (TBS). Pv-mtCOX1 qPCR was consistent with the positivity of the samples from the other two tests.
Table 1.
Demographic and clinical data of individuals involved in this study.
| Parameter | Value |
|---|---|
| Sample size | 307 |
| Mean age, years (SD) | 39.4 (14.3) |
| No. of males (%) | 215 (70) |
| Parasite geometric mean D0 per mm3 (SD) | 3781.6 (4109.2) |
| No. with presence of gametocytes D0 under microscopy (%) | 259 (84.4) |
| Mean hemoglobin in D0, g/dL (SD) a | 13.4 (1.7) |
| Mean hemoglobin in D28, g/dL (SD) a | 13.6 (1.1) |
| No. with anemia (%) a,b | 57 (27.5) |
a Only 207 samples had data on hemoglobin. b According to World Health Organization (WHO) criteria (2001) [10].
Among the 29 samples that were positive by Pv-mtCOX1 qPCR on D28, 11 (37.9%) of these patients were positive under TBS microscopy between day 30 and day 38 of follow-up, while 4 (13.8%) were positive on day 42 and 2 (1.4%) after 95 days. Agreement between positive diagnoses between the Pv 18S rRNA qPCR test and TBS microscopy was seen for only 37.5% of the samples, with a kappa coefficient of 0.51. Between Pv-mtCOX1 qPCR and TBS, co-positivity was 17.2%, with a kappa value of 0.27. When the two molecular tests were compared, co-positivity was 51.7%, with a kappa value of 0.63.
2.2. Parasitological Quantifications
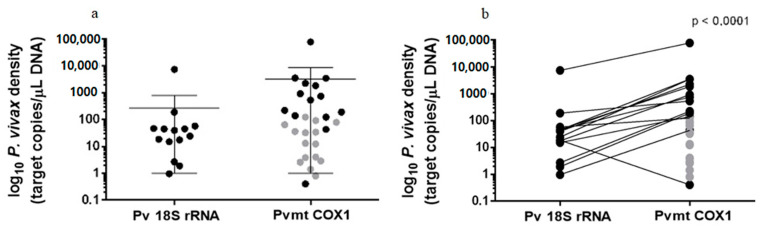
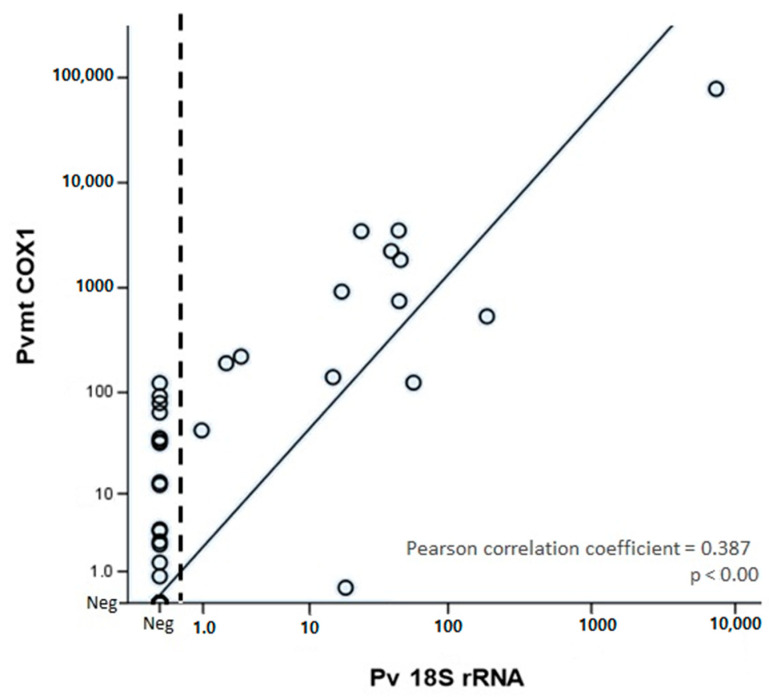
Regarding quantifications, the average obtained for the TBS was 1710.6 parasites/µL blood, and averages using Pv 18S rRNA qPCR were 561.9 DNA copies/μL and 3188.9 DNA copies/μL using Pv-mtCOX1 qPCR. The mean quantification obtained after using Pv-mtCOX1 qPCR was significantly higher than when using Pv 18S rRNA qPCR (Figure 2a,b) (p < 0.00010). Correlation showed that quantifications of the two qPCRs tend to increase together (Pearson correlation coefficient = 0.387, p < 0.00, Figure 3).
Figure 2.
Copy numbers of each target per sample. (a) Each dot represents one sample; gray indicates samples detected only by Pv-mtCOX1 qPCR. (b) Lines linking the same samples in both targets show the different quantifications between them.
Figure 3.
Correlation of log10 template copy numbers detected by Pv-mtCOX1 and Pv 18S rDNA qPCR.
2.3. Risk Factors
Univariate and multivariate (logistic regression) analyses and odds ratio were performed to determine certain potential risk factors, including being of the male gender, presence of gametocytes at microscopy on D0, asexual parasitemia in more than 5000 parasites and anemia on D0. Only the presence of gametocytes was found to be an independent risk factor, with an odds ratio (OR) of 3.98 in univariate analyses and 3.64 in multivariate analyses (p < 0.05 in both analyses, Table 2).
Table 2.
Univariate and multivariate (logistic regression) analyses of factors associated in D0 to early recurrence in patients with uncomplicated Plasmodium vivax malaria under supervised treatment with chloroquine plus primaquine.
| Factor | Univariate Analysis | Multivariate Analysis | ||||||
|---|---|---|---|---|---|---|---|---|
| Odds Ratio | 95% Confidence Interval | p-Value | Adjusted Odds Ratio | 95% Confidence Interval | p-Value | |||
| Lower | Upper | Lower | Upper | |||||
| Male gender | 0.72 | 0.30 | 1.76 | 0.471 | 0.91 | 0.29 | 2.84 | 0.872 |
| Presence of gametocytes under microscopy | 3.98 | 1.51 | 9.54 | 0.001 | 3.64 | 1.10 | 12.08 | 0.035 |
| Asexual parasites > 5000/mm3 | 2.14 | 0.72 | 6.37 | 0.161 | 1.03 | 0.26 | 4.03 | 0.968 |
| Anemia D0 a,b | 1.65 | 0.42 | 5.76 | 0.497 | 1.51 | 0.40 | 5.73 | 0.541 |
Bold type indicates significance. a Only 207 samples had data on hemoglobin. b According to WHO criteria (2001) [10].
2.4. Symptomatic Recurrences
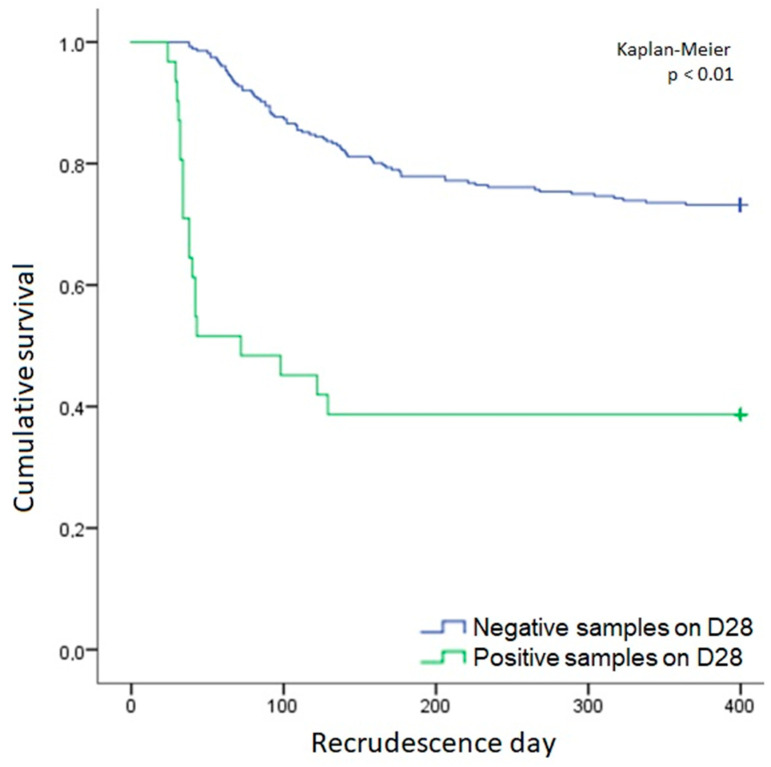
Information obtained via the SIVEP-Malaria platform (the official Brazilian Malaria Epidemiological Surveillance Information System) indicated that 30.1% (94/312) of individuals were confirmed to have symptomatic recurrences after the study follow-up on D28. Furthermore, 17 out of 29 individuals were positive on D28 according to testing using Pv-mtCOX1 qPCR (Supplementary file Table S2). The Kaplan–Meier method showed that the group that was positive on D28 when tested with Pv-mtCOX1 qPCR had recurrences earlier than the control group, with p < 0.01 (Figure 4). When analyzing clearance time by TBS soon after treatment initiation, no difference was observed between these groups (p > 0.05).
Figure 4.
Survival function calculated by the Kaplan–Meier method in relation to the day of patient’s recrudescence after the study follow-up period, showing the curves of the negative and positive samples for Pv-mtCOX1 qPCR on day 28.
3. Discussion
In this study, we compared different diagnostic methods for detecting early recurrence. Pv-mtCOX1 qPCR increased our ability to detect early recurrence. The quantification was superior and the samples with lower copy number were not detected through Pv 18S rRNA qPCR. These data confirm the high sensibility of the mitochondrial target gene, as shown previously in other studies [11,12,13,14,15] that reveal that it is present in at least 20 copies per cell [16].
These techniques are thousands of times more sensitive than rapid diagnostic tests (RDTs) and microscopy, and tens-to-hundreds of times more sensitive than standard PCR [17,18]. Gruenberg et al. [13] demonstrated the first target for use in an ultra-sensitive qPCR screening for P. vivax, namely qPCR Pv-mtCOX1.
As noted in this study, other studies have also found that PvMtCOX1 was more sensitive in the detection of P. vivax [11,14]. Hofman et al. [12] used Pv-mtCOX1 qPCR and obtained 62% positivity, while rRNA 18S only revealed 52% positivity in asymptomatic individuals from Papua New Guinea. Gruenberg et al. [14] observed that the use of Pv-mtCOX1 led to a significant increase in positivity in the range of 5.1%, 6.4% and 11.5% in Thailand, Brazil and Papua New Guinea, respectively.
Moreover, yearly detection using Pv-mtCOX1 qPCR could predict the recurrence of symptoms in these patients, well before the patient is symptomatic and seeks health care. Sexual stages appear early in the course of vivax infection and in parallel with asexual parasitemia [19], therefore, asymptomatic infections are able to transmit to mosquitos due the presence of gametocytes even when there is low parasitemia [20,21]. Possibly these patients have been transmitting to mosquitos until the parasitemia grows sufficiently to be diagnosed by TBS and then be treated again. Due to the presence of asymptomatic carriers of Plasmodium being more frequent and probably serving as reservoirs for mosquitoes, the application of molecular techniques for diagnosis has been of fundamental importance [22].
In contrast to our findings, Lin et al. [23] did not find the same association between microscopic gametocyte carriage and parasite drug resistance, and that gametocytemia was more common in those reporting antimalarial use within the past year.
However, this study had some limitations. It was not possible to differentiate between sexual and asexual forms of P. vivax, though we did perform clinical monitoring of the participants for a period of one year and, as a result, it was shown that malaria returned. In most cases, participants were positive using Pv-mtCOX1 and the recurrence time was shorter. Moreover, studies in Thailand and Indonesia demonstrated that without treatment P. vivax gametocytes are reported to persist in the peripheral circulation for a maximum of three days, and that the relationship between asexual and sexual stage parasitemia does not differ substantially between initial and recurrent infections [24]. Sexual stages appear early in the course of vivax malaria infection and increase jointly with the asexual parasitemia [19]. Another limitation was that data regarding recurrence episodes were accessed on the SIVEP-Malaria platform (Malaria Epidemiological Surveillance System), which is susceptible to underreporting issues.
4. Material and Methods
4.1. Ethics Statement, Study Site and Selection of Patients
The study was approved by the Ethics Review Board at Fundação de Medicina Tropical Doutor Heitor Vieira Dourado (FMT-HVD) (approval number: 1074306/2015). All participants signed an informed consent form. The study was performed at Fundação de Medicina Tropical Doutor Heitor Vieira Dourado (FMT-HVD), Manaus, AM and included patients from 2011 to 2017.
The study included patients with P. vivax malaria of both genders, aged 6 months to 60 years, weight greater than 5 kg, with blood parasite density from 250 to 100,000 parasites/mL and axillary temperature ≥37.5 °C or history of fever in the last 48 h. Use of antimalarials in the previous 30 days, impossibility of being followed up for 42 days and clinical complications were considered exclusion criteria. Patients received treatment with 25 mg/kg of CQ phosphate over a period of 3 days (10 mg/kg on day 0 and 7.5 mg/kg on days 1 and 2) either associated or not with PQ 0.5 mg/kg/day for the following 7 days [25]. Patients were evaluated on days 0, 1, 2, 3, 7, 14, 28 and 42, as well as at any time during the follow-up period if they felt ill.
4.2. P. vivax Malaria Diagnosis by Thick Smears
Thick blood smear (TBS) was performed using the Walker technique and performed by experienced microscopists [26]. Parasite densities were calculated by counting the number of parasites per 200 leukocytes, and the number of parasites/µL per patient was also determined.
4.3. Detection of Plasmodium spp. By qPCR
DNA extraction was performed using the PureLink® Genomic DNA kit (Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer’s specifications. All DNA samples were subjected to Pv-mtCOX1 qPCR to detect Plasmodium vivax by targeting a conserved region of the cytochrome oxidase 1 gene [15]. For quantification of Pv-mtCOX1 copy numbers, in each experiment these were quantified using a standard curve generated from three dilutions of plasmids containing the respective targeted region in triplicate (102, 104 and 106 copies/μL). DNA was amplified using a 7500 fast Real-Time PCR System® (Thermo Fisher Scientific), with the use of the primers and Taqman fluorescence probes described in Supplementary file Table S1. Cycling parameters for PCR were an initial denaturation step at 95 °C for 10 min, 45 cycles of 15 s at 95 °C and 1 min at 60 °C. Each DNA samples was assayed in triplicate. All biology molecular procedures were performed at FMT-HVD.
P. vivax early recurrence was defined if qPCR was positive until D28. With CQ/DCQ plasma concentrations higher than 100 ng/dL, these patients were likely to have CQ-resistant parasites. In addition, individual data were collected from the Brazilian Malaria Epidemiological Surveillance System (SIVEP-Malaria) to verify their recurrence after the study follow-up.
4.4. CQ and DCQ Level
CQ and DCQ plasmatic levels were determined only in case of parasitological failure. Three aliquots of 100 µL of whole blood samples were spotted on filter paper for analysis by high performance liquid chromatography (HPLC), as previous described [26,27]. The sum of CQ and DCQ plasmatic levels was used to classify parasites as being an early recurrence based on the 100 ng/mL cutoff level [28,29,30].
4.5. Statistical Analyses
Co-positivity and co-negativity between the tests and the agreement evaluation were calculated by the Kappa coefficient using Pv-mtCOX1 qPCR as the standard method. Univariate and multivariate analyses were performed to identify factors associated to early recurrence. The difference between the averages of parasite density was estimated by the non-parametric Mann–Whitney test, and all tests considered a 95% confidence interval. Analyses and graphs were generated by the software Statistical Package for the Social Sciences (IBM_SPSS) v.21 and GraphPad Prism 7 Software. Statistical analyses were performed at FMT-HVD.
5. Conclusions
We conclude that Pv-mtCOX1 qPCR is an ultrasensitive instrument for the diagnosis of early recurrence samples when compared to the other methods tested. The results obtained show the existence of sub-notification when tests are done using TBS and highlight the need for development and implementation of more sensitive tests, though not only for diagnosis but also for follow-up of patients with vivax malaria in order to favor more assertive decisions aimed at the elimination of the disease.
Acknowledgments
We would like to thank the laboratory teams in the Laboratory of Toxicology of the Federal University of Pará, Brazil and all heath workers in the malaria outpatient hospital at Fundação de Medicina Tropical Heitor Vieira Dourado, Manaus, Brazil.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-0817/10/1/19/s1, Table S1: Oligonucleotide primers and probes used in qPCR assays; Table S2: Recurrence of patients that were positive when tested using Pv-mtCOX1 qPCR on D28 and reported on the SIVEP-Malaria database (TBS) until 180 days after follow-up study.
Author Contributions
G.C.M., L.R.A.B. and A.C.G.d.A. conceived and designed the study. L.R.A.B., E.L.d.S., A.C.G.d.A., Y.E.A.R.S. were responsible for the execution of the study. A.M.S. and J.L.F.V. were responsible for acquisition of data. L.R.A.B. and G.C.M. analyzed the data. L.R.A.B. and G.C.M. wrote the paper. A.C.G.d.A., A.M.S., M.d.G.C.A., Q.B., W.M.M. and M.V.G.d.L. carried out the critical revision of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES)—Finance Code 001 and Research Support Foundation of Amazonas (FAPEAM) through PPSUS, PROESTADO and PAPAC (Call No. 005/2019) projects. W.M.M. and M.V.G.L. are CNPq research fellows.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Ethics Review Board at Fundação de Medicina Tropical Doutor Heitor Vieira Dourado (FMT-HVD) (approval number: 1074306/2015).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author (GCM), upon reasonable request, due to ethical restrictions.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.World Malaria Report 2019. [(accessed on 18 September 2020)]; Available online: https://www.who.int/publications-detail-redirect/9789241565721.
- 2.Rieckmann K.H., Davis D.R., Hutton D.C. Plasmodium vivax resistance to chloroquine? Lancet. 1989;334:1183–1184. doi: 10.1016/S0140-6736(89)91792-3. [DOI] [PubMed] [Google Scholar]
- 3.Price R.N., Douglas N.M., Anstey N.M. New developments in Plasmodium vivax malaria: Severe disease and the rise of chloroquine resistance. Curr. Opin. Infect. Dis. 2009;22:430–435. doi: 10.1097/QCO.0b013e32832f14c1. [DOI] [PubMed] [Google Scholar]
- 4.Marques M.M., Costa M.R.F., Santana Filho F.S., Vieira J.L.F., Nascimento M.T.S., Brasil L.W., Nogueira F., Silveira H., Reyes-Lecca R.C., Monteiro W.M., et al. Plasmodium vivax Chloroquine Resistance and Anemia in the Western Brazilian Amazon. Antimicrob. Agents Chemother. 2014;58:342–347. doi: 10.1128/AAC.02279-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alecrim M.D., Alecrim W., Macêdo V. Plasmodium vivax resistance to chloroquine (R2) and mefloquine (R3) in Brazilian Amazon region. Rev. Soc. Bras. Med. Trop. 1999;32:67–68. doi: 10.1590/S0037-86821999000100013. [DOI] [PubMed] [Google Scholar]
- 6.de Santana Filho F.S., de Lima Arcanjo A.R., Chehuan Y.M., Costa M.R., Martinez-Espinosa F.E., Vieira J.L., Barbosa M.d.G.V., Alecrim W.D., Alecrim M.D. Chloroquine-Resistant Plasmodium vivax, Brazilian Amazon. Emerg. Infect. Dis. 2007;13:1125–1126. doi: 10.3201/eid1307.061386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siqueira A.M., Alencar A.C., Melo G.C., Magalhaes B.L., Machado K., Alencar Filho A.C., Kuehn A., Marques M.M., Manso M.C., Felger I., et al. Fixed-Dose Artesunate–Amodiaquine Combination vs Chloroquine for Treatment of Uncomplicated Blood Stage P. vivax Infection in the Brazilian Amazon: An Open-Label Randomized, Controlled Trial. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2017;64:166–174. doi: 10.1093/cid/ciw706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baird J.K., Leksana B., Masbar S., Fryauff D.J., Sutanihardja M.A., Suradi, Wignall F.S., Hoffman S.L. Diagnosis of Resistance to Chloroquine by Plasmodium vivax: Timing of Recurrence and Whole Blood Chloroquine Levels. Am. J. Trop. Med. Hyg. 1997;56:621–626. doi: 10.4269/ajtmh.1997.56.621. [DOI] [PubMed] [Google Scholar]
- 9.Hwang J., Alemayehu B.H., Reithinger R., Tekleyohannes S.G., Takele T., Birhanu S.G., Demeke L., Hoos D., Melaku Z., Kassa M., et al. In Vivo Efficacy of Artemether-Lumefantrine and Chloroquine against Plasmodium vivax: A Randomized Open Label Trial in Central Ethiopia. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0063433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.WHO . Safe and Rational Clinical Use of Blood. World Health Organization; Geneva, Switzerland: 2018. [(accessed on 6 November 2018)]. Available online: http://www.who.int/bloodsafety/clinical_use/en/ [Google Scholar]
- 11.Cunha M.G., Medina T.S., Oliveira S.G., Marinho A.N., Póvoa M.M., Ribeiro-dos-Santos A.K.C. Development of a Polymerase Chain Reaction (PCR) method based on amplification of mitochondrial DNA to detect Plasmodium falciparum and Plasmodium vivax. Acta Trop. 2009;111:35–38. doi: 10.1016/j.actatropica.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 12.Hofmann N.E., Gruenberg M., Nate E., Ura A., Rodriguez-Rodriguez D., Salib M., Mueller I., Smith T.A., Laman M., Robinson L.J., et al. Assessment of ultra-sensitive malaria diagnosis versus standard molecular diagnostics for malaria elimination: An in-depth molecular community cross-sectional study. Lancet Infect. Dis. 2018 doi: 10.1016/S1473-3099(18)30411-0. [DOI] [PubMed] [Google Scholar]
- 13.Gruenberg M., Moniz C.A., Hofmann N.E., Wampfler R., Koepfli C., Mueller I., Monteiro W.M., Lacerda M., de Melo G.C., Kuehn A., et al. Plasmodium vivax molecular diagnostics in community surveys: Pitfalls and solutions. Malar. J. 2018;17 doi: 10.1186/s12936-018-2201-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Britton S., Cheng Q., Grigg M.J., Poole C.B., Pasay C., William T., Fornace K., Anstey N.M., Sutherland C.J., Drakeley C., et al. Sensitive Detection of Plasmodium vivax Using a High-Throughput, Colourimetric Loop Mediated Isothermal Amplification (HtLAMP) Platform: A Potential Novel Tool for Malaria Elimination. PLoS Negl. Trop. Dis. 2016;10:e0004443. doi: 10.1371/journal.pntd.0004443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gruenberg M., Moniz C.A., Hofmann N.E., Koepfli C., Robinson L.J., Nate E., Monteiro W.M., de Melo G.C., Kuehn A., Siqueira A.M., et al. Utility of ultra-sensitive qPCR to detect Plasmodium falciparum and Plasmodium vivax infections under different transmission intensities. Malar. J. 2020;19:319. doi: 10.1186/s12936-020-03374-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Preiser P.R., Wilson R.J., Moore P.W., McCready S., Hajibagheri M.A., Blight K.J., Strath M., Williamson D.H. Recombination Associated with Replication of Malarial Mitochondrial DNA. EMBO J. 1996;15:684–693. doi: 10.1002/j.1460-2075.1996.tb00401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adams M., Joshi S.N., Mbambo G., Mu A.Z., Roemmich S.M., Shrestha B., Strauss K.A., Johnson N.E., Oo K.Z., Hlaing T.M., et al. An ultrasensitive reverse transcription polymerase chain reaction assay to detect asymptomatic low-density Plasmodium falciparum and Plasmodium vivax infections in small volume blood samples. Malar. J. 2015;14:520. doi: 10.1186/s12936-015-1038-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imwong M., Hanchana S., Malleret B., Rénia L., Day N.P.J., Dondorp A., Nosten F., Snounou G., White N.J. High-Throughput Ultrasensitive Molecular Techniques for Quantifying Low-Density Malaria Parasitemias. J. Clin. Microbiol. 2014;52:3303–3309. doi: 10.1128/JCM.01057-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mckenzie F.E., Wongsrichanalai C., Magill A.J., Forney J.R., Permpanich B., Lucas C., Erhart L.M., O’Meara W.P., Smith D.L., Sirichaisinthop J., et al. Gametocytemia in Plasmodium vivax and Plasmodium Falciparum Infections. J. Parasitol. 2006;92:1281–1285. doi: 10.1645/GE-911R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martins-Campos K.M., Kuehn A., Almeida A., Duarte A.P.M., Sampaio V.S., Rodriguez Í.C., da Silva S.G.M., Ríos-Velásquez C.M., Lima J.B.P., Pimenta P.F.P., et al. Infection of Anopheles aquasalis from symptomatic and asymptomatic Plasmodium vivax infections in Manaus, western Brazilian Amazon. Parasit. Vectors. 2018;11 doi: 10.1186/s13071-018-2749-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Almeida A.C.G., Kuehn A., Castro A.J.M., Vitor-Silva S., Figueiredo E.F.G., Brasil L.W., Brito M.A.M., Sampaio V.S., Bassat Q., Felger I., et al. High proportions of asymptomatic and submicroscopic Plasmodium vivax infections in a peri-urban area of low transmission in the Brazilian Amazon. Parasit. Vectors. 2018;11 doi: 10.1186/s13071-018-2787-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phommasone K., van Leth F., Imwong M., Henriques G., Pongvongsa T., Adhikari B., Peto T.J., Promnarate C., Dhorda M., Sirithiranont P., et al. The use of ultrasensitive quantitative-PCR to assess the impact of primaquine on asymptomatic relapse of Plasmodium vivax infections: A randomized, controlled trial in Lao PDR. Malar. J. 2020;19:4. doi: 10.1186/s12936-019-3091-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin J.T., Patel J.C., Levitz L., Wojnarski M., Chaorattanakawee S., Gosi P., Buathong N., Chann S., Rekol H., Thay K., et al. Gametocyte Carriage, Antimalarial Use, and Drug Resistance in Cambodia, 2008–2014. Am. J. Trop. Med. Hyg. 2018;99:1145–1149. doi: 10.4269/ajtmh.18-0509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Douglas N.M., Simpson J.A., Phyo A.P., Siswantoro H., Hasugian A.R., Kenangalem E., Poespoprodjo J.R., Singhasivanon P., Anstey N.M., White N.J., et al. Gametocyte Dynamics and the Role of Drugs in Reducing the Transmission Potential of Plasmodium vivax. J. Infect. Dis. 2013;208:801–812. doi: 10.1093/infdis/jit261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ministério da Saúde, Secretaria de Vigilância em Saúde . Departamento de Imunização Guia Prático de Tratamento da Malária no Brasil. Ministério da Saúde, Secretaria de Vigilância em Saúde; Brasília, Brazil: 2020. [Google Scholar]
- 26.WHO . World Malaria Report 2017. WHO; Geneva, Switzerland: 2017. [Google Scholar]
- 27.Dua V.K., Kar P.K., Gupta N.C., Sharma V.P. Determination of chloroquine and desethylchloroquine in plasma and blood cells of Plasmodium vivax malaria cases using liquid chromatography. J. Pharm. Biomed. Anal. 1999;21:199–205. doi: 10.1016/S0731-7085(99)00097-7. [DOI] [PubMed] [Google Scholar]
- 28.Patchen L.C., Mount D.L., Schwartz I.K., Churchill F.C. Analysis of filter-paper-absorbed, finger-stick blood samples for chloroquine and its major metabolite using high-performance liquid chromatography with fluorescence detection. J. Chromatogr. 1983;278:81–89. doi: 10.1016/S0378-4347(00)84758-1. [DOI] [PubMed] [Google Scholar]
- 29.Price R.N., von Seidlein L., Valecha N., Nosten F., Baird J.K., White N.J. Global extent of chloroquine-resistant Plasmodium vivax: A systematic review and meta-analysis. Lancet Infect. Dis. 2014;14:982–991. doi: 10.1016/S1473-3099(14)70855-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.WHO . Methods for Surveillance of Antimalarial Drug Efficacy. World Health Organization; Geneva, Switzerland: 2009. [(accessed on 18 September 2020)]. Available online: https://www.who.int/docs/default-source/documents/publications/gmp/methods-for-surveillance-of-antimalarial-drug-efficacy.pdf?sfvrsn=29076702_2. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author (GCM), upon reasonable request, due to ethical restrictions.