Abstract
This paper aimed to investigate the molecular epidemiological features of the cfr gene in E. coli isolates in a typical swine farm during 2014–2017. A total of 617 E. coli isolates were screened for the cfr gene using PCR amplification. A susceptibility test, pulsed-field gel electrophoresis (PFGE), S1-PFGE, southern blotting hybridization, and the genetic context of the cfr gene were all used for analyzing all cfr-positive E. coli isolates. A conjugation experiment was conducted with the broth mating method using E. coli C600 as the recipient strain and 45 mcr-1-cfr-bearing E. coli isolates as the donor strain. Plasmids pHNEP124 and pHNEP129 were revealed by Illumina Miseq 2500. Eighty-five (13.7%) E. coli isolates were positive for the cfr gene and the prevalence of the cfr gene had significantly increased from 1.6% in 2014 to 29.1% in 2017. The Pulsed-Field Gel Electrophoresis (PFGE) analysis indicated that the spread of the cfr gene among E. coli isolates was mainly due to horizontal transfer. In addition, the cfr gene was primarily located on the plasmids between 28.8-kb to 60-kb in size, and the cfr gene was flanked by two copies of IS26 with the same orientation. Sequence analysis suggested that the plasmids pHNEP124 and pHNEP129 co-harboring the cfr and mcr-1 genes belonged to the plasmids IncP plasmid and IncX4 plasmid, respectively. In conclusion, this is the first study to report the high prevalence of the cfr gene among E. coli isolates and the first report of the complete genome sequence of IncP and IncX4 plasmids carrying the mcr-1 and cfr genes. The occurrence and dissemination of the cfr/mcr-1-carrying plasmids among E. coli isolates need further surveillance.
Keywords: multi-resistant cfr gene, E. coli, IS26-mediated, co-transfer with mcr-1, swine farm
1. Introduction
The multi-resistant cfr gene, encoded rRNA methyltransferase, confers cross-resistance to five chemically unrelated classes of antimicrobial agents, including phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A (called PhLOPSA phenotype) [1]. Moreover, it also decreases susceptibility to the 16-membered macrolides spiramycin and josamycin [2]. Since the first identified cfr gene in plasmid pSCFS1 from a Staphylococcus sciuri isolate, the transferable cfr gene has been detected in both Gram-positive and Gram-negative bacteria such as Staphylococcus, Enterococcus, Bacillus, Macrococcus, Jeotgalicoccus, Streptococcus, Proteus, Escherichia coli, and Morganella morganii [3,4,5]. Mobile gene elements such as insertion sequences (ISs) and plasmids can acquire antimicrobial resistance genes and play a vital role in the dissemination of cfr in both Gram-negative and Gram-positive genera [4,6].
Multidrug-resistant (MDR) Escherichia coli has become a worrisome issue that poses a threat to public health and it is also considered to be a major reservoir of antibiotic resistance genes that may be responsible for the treatment failure events in human clinical and veterinary medicine [7]. To date, a total of 24 cfr-positive E. coli isolates have been reported in food-producing animals from various sources in seven provinces of China and it is mainly located on various plasmid replicon types such as IncX4, IncF43:A-:B-, and F14:A-:B- [8]. Mobile colistin resistance gene mcr-1 has experienced global dissemination and has spread to over 40 countries or regions covering five continents since the first identification in 2015 [9,10]. Colistin-resistant bacteria has raised serious concern and increased risk to human and animal health because of colistin, which is used as a last-resort drug for treating MDR Gram-negative bacteria infections [11]. Moreover, there are no reports that the cfr gene co-exists with the mcr-1 gene among E. coli isolates. A previous study of our lab had reported that the cfr gene was been detected in E. coli with IncF43:A-:B- plasmid pHNEP28 and S. sciuri in a commercial swine farm in 2013 [12]. Thus, in the present study, we continued monitoring the prevalence of the cfr gene in E. coli isolates from this swine farm during 2014–2017. Interestingly, we found that the prevalence of cfr gene had rapidly increased in E. coli isolates, and we identified, for the first time, the cfr and mcr-1 genes coexisting in various plasmids such as IncP plasmid and IncX4 plasmid.
2. Results
2.1. The Prevalence of the cfr Gene and Detection of Other Resistance Genes
As shown in Table 1, a total of 85 (13.7%) E. coli isolates were positive for the cfr gene, and the prevalence of the cfr gene among 617 E. coli isolates had significantly increased from 2014 to 2017 (1.6% in 2014, 8.6% in 2015, 19.9% in 2016, and 29.1% in 2017). These strains were isolated from environmental samples including soil (n = 1) and sewage (n = 1), and from anal swab samples of different growth stages of pigs consisting of nursery pigs (n = 29), fattening pigs (n = 52), sows (n = 1), and boars (n = 1), through different stages of growth. Compared with the detectable rate of the cfr gene among E. coli isolates from different growth stages, the fattening pigs had the highest detectable rate of the cfr gene (25.2%), followed by nursery pigs (22.5%), boars (7.7%), and sows (0.6%). No cfr-carrying E. coli isolates were detected from suckling piglets (Table 1). Moreover, the encoding florfenicol efflux pump gene floR was detected in all of the cfr-positive E. coli isolates, while fexA, fexB, and optrA were not. It is worth noting that 45 (52.9%) cfr-positive E. coli isolates were positive for the mcr-1 gene (Figure 2). Compared with the previous reports, which found 24 cfr-positive E. coli isolates, this study observed that the mcr-1 gene coexisted in cfr-positive E. coli isolates with the high detection rate in this study.
Table 1.
Prevalence of the cfr gene in E. coli isolates from various sources, as well as information on drug use in the swine farm during the period of 2014–2017.
| Collected Date | Suckling Piglet (N) | Nursery Pig (N) | Fattening Pig (N) | Sow (N) | Boar (N) | Environment (N) | Total | History of Drug Use |
|---|---|---|---|---|---|---|---|---|
| Jun. 2014 | 33 | 35 (3) | 64 | 52 | 0 | 0 | 184 (3, 1.6%) | gentamycin, amoxicillin |
| Jul. 2015 | 28 | 39 (3) | 46 (10) | 38 | 0 | 0 | 151 (13, 8.6%) | florfenicol |
| Jul. 2016 | 8 | 32 (9) | 45 (18) | 51 | 4 (1) | 1 | 141 (28, 19.9%) | florfenicol, enrofloxacin, mequindox, kanamycin |
| Apr. 2017 | 14 | 23 (14) | 51 (24) | 36 (1) | 9 | 8 (2) | 141 (41, 29.1%) | gentamycin, amoxicillin, kanamycin |
| Total | 83 (0) | 129 (29, 22.5%) | 206 (52, 25.2%) | 177 (1, 0.6%) | 13 (1, 7.7%) | 9 (2, 22.2%) | 617 (85, 13.7%) | - |
Note: the number and detection rate of the cfr-positive E. coli isolates are listed in brackets.
2.2. Antibiotic Susceptibility Testing
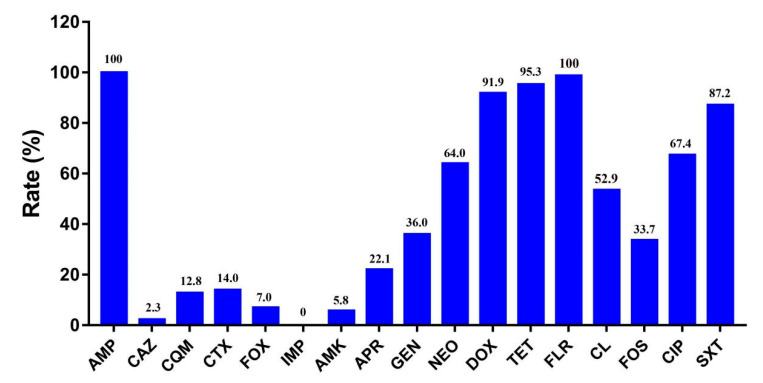
As shown in Figure 1, susceptibility testing showed that all cfr-positive E. coli isolates were highly resistant to ampicillin (100%), florfenicol (100%), tetracycline (95.3%), and doxycycline (91.9%), followed by sulfamethoxazole/trimethoprim (87.2%), ciprofloxacin (67.4%), neomycin (64.0%), colistin (52.9%), gentamycin (36.0%), and fosfomycin (33.7%). These isolates showed a lower resistance to apramycin (22.1%), cefotaxime (14.0%), cefquinome (12.8%), cefoxitin (7.0%), amikacin (5.8%), and ceftazidime (2.3%). All cfr-positive E. coli isolates were susceptible to imipenem. Interestingly, the prevalence of the mcr-1 gene among cfr-positive E. coli had decreased sharply from 86.4% before 2017 to 17.1% in 2017, which implies that the results may be a consequence of the ban on colistin as a feed additive for animals in China.
Figure 1.
Resistance rate of 85 cfr-positive E. coli isolates. AMP—ampicillin; CAZ—ceftazidime; CQM—cefquinome; CTX—cefotaxime; FOX—cefoxitin, IMP—imipenem; AMK—amikacin; APR— apramycin; GEN—gentamycin; NEO—neomycin; DOX—doxycycline; TET—tetracycline; FLR—florfenicol; CL—colistin; FOS—fosfomycin; CIP—ciprofloxacin; SXT—sulfamethoxazole/trimethoprim.
2.3. Pulsed-Field Gel Electrophoresis (PFGE) Patterns, Location, and Genetic Context of the cfr Gene
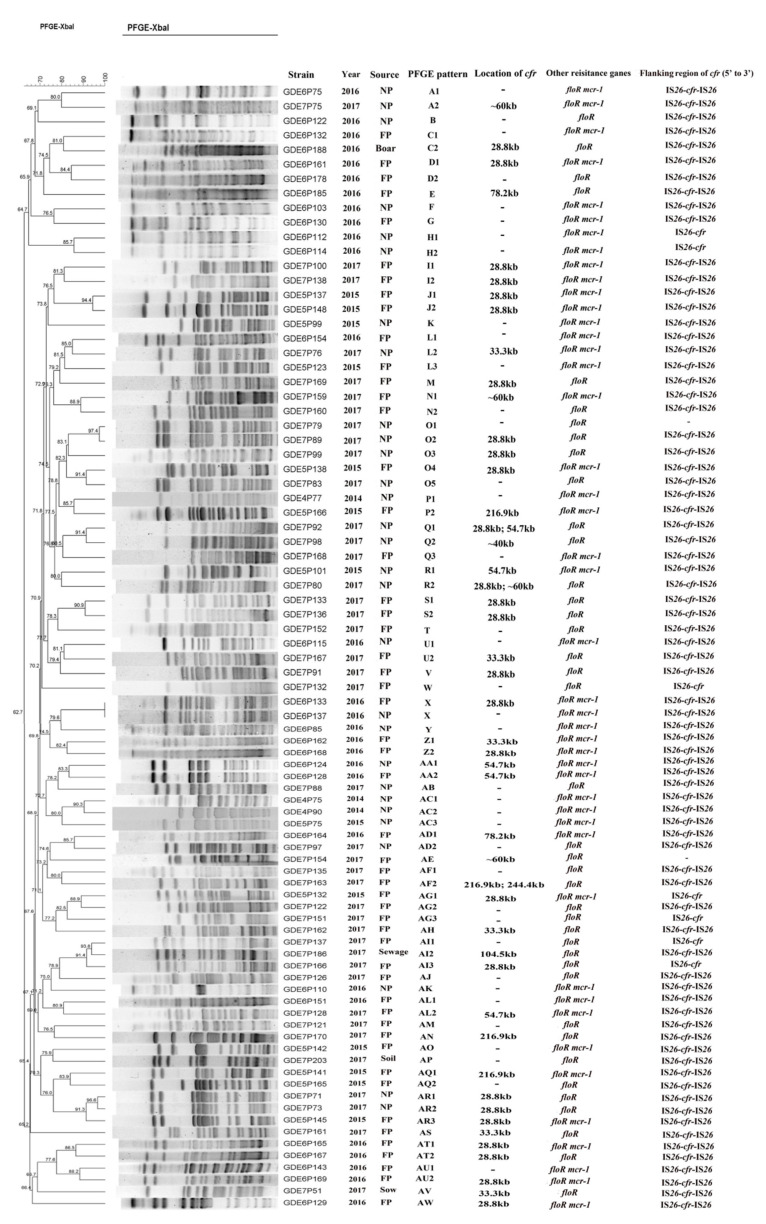
The PFGE analysis revealed that 85 cfr-positive E. coli isolates were grouped into 49 PFGE patterns, designated A to AW (Figure 2). The PFGE analysis of all cfr-bearing E. coli isolates showed that most of these isolates were genetically divergent and epidemiologically unrelated. This result suggests that the spread of the cfr gene among E. coli was mainly due to horizontal transfer. The S1-PFGE and Southern blot hybridization analyses confirmed that the cfr gene of 46 E. coli isolates was successfully located on plasmids ranging from 28.8 kb to 244.4 kb in size (Figure 2). Two cfr-bearing plasmids coexisted in three isolates (GDE7P80, GDE7P92, and GDE7P163). Moreover, the cfr gene was mainly distributed on the plasmids in size between 28.8 kb and 60 kb (Figure 2). In addition, the genetic structure of the cfr gene was flanked by two copies of IS26 with the same orientation in 76 E. coli isolates and one copy of IS26 was found to be located upstream of the cfr gene in seven E. coli isolates (Figure 2).
Figure 2.
Characterization of 85 cfr-positive E. coli isolates in this study. NP—nursery pig; FP—fattening pig. “–” indicates none detected.
2.4. Co-Transfer of the mcr-1 and cfr Genes
A conjugative experiment demonstrated that the plasmids carrying the mcr-1 and cfr genes of 30 E. coli were successfully transferred to E. coli C600 using Luria–Bertani (LB) agar plates containing streptomycin (3000 mg/L) and colistin (2 mg/L). Susceptibility testing indicated that all transconjugants were resistant to colistin, followed by ampicillin (n = 16), sulfamethoxazole/trimethoprim (n = 3), florfenicol (n = 3), apramycin (n = 2), and doxycycline (n = 2). One transconjugant exhibited resistance to fosfomycin and cefotaxime, and one transconjugant exhibited resistance to gentamycin (Table 2). Notably, apart from strains GDE6P133J and GDE6P151J carried in the floR gene, the rest of the cfr-positive transconjugants showed that the minimum inhibitory concentration (MIC) values of florfenicol and colistin were improved by 0.5–4-fold and 16–32-fold compared with the recipients, respectively. The PCR-based replicon typing (PBRT) analysis indicated that IncX4 plasmid (n = 19) was the most prevalent incompatible (Inc) plasmid type out of the transferable plasmids co-harboring the mcr-1 and cfr genes. Other plasmid types such as IncP, IncI2, and IncHI2 were also been detected in transconjugants. In accordance with the characterization of the cfr-positive transconjugants, 12 transformants were obtained and positive for mcr-1 and cfr genes (Table S1). Susceptibility testing indicated that all cfr-positive transformants were susceptible to florfenicol and the MIC values of florfenicol were improved 1–2 folds compared with E. coli DH5α. It is noteworthy that all cfr-positive transconjugants or transformants except for strains GDE6P133J and GDE6P151J failed to mediated resistance to florfenicol.
Table 2.
Characterization of 30 transconjugants carrying the mcr-1 and cfr genes.
| Strains | MIC Values | Replicon Type | Resistance Phenotype a | Resistance Genes | |
|---|---|---|---|---|---|
| Florfenicol | Colistin | ||||
| GDE5P101J | 2 | 4 | IncI2 | CL | cfr mcr-1 |
| GDE5P123J | 4 | 4 | IncI2 | AMP CL | cfr mcr-1 |
| GDE5P132J | 2 | 4 | IncX4 | CL | cfr mcr-1 |
| GDE5P137J | 4 | 8 | IncX4 | AMP APR CL | cfr mcr-1 |
| GDE5P138J | 4 | 4 | IncX4 | CL | cfr mcr-1 |
| GDE5P141J | 4 | 8 | IncHI2 | CL | cfr mcr-1 |
| GDE5P142J | 8 | 4 | IncX4 | CL | cfr mcr-1 |
| GDE5P145J | 2 | 8 | IncX4 | CL | cfr mcr-1 |
| GDE5P148J | 8 | 4 | - | AMP CL | cfr mcr-1 |
| GDE5P165J | 8 | 4 | IncX4 | CL | cfr mcr-1 |
| GDE6P75J | 8 | 4 | IncHI2 | AMP CTX CL FOS | cfr mcr-1 |
| GDE6P85J | 4 | 4 | IncX4 | CL | cfr mcr-1 |
| GDE6P103J | 4 | 4 | - | AMP CL | cfr mcr-1 |
| GDE6P110J | 8 | 2 | IncX4 | CL | cfr mcr-1 |
| GDE6P115J | 2 | 4 | - | AMP CL | cfr mcr-1 |
| GDE6P124J | 8 | 4 | IncP b | AMP CL | cfr mcr-1 |
| GDE6P128J | 8 | 4 | IncP | AMP CL | cfr mcr-1 |
| GDE6P129J | 4 | 4 | IncX4 | CL | cfr mcr-1 |
| GDE6P130J | 4 | 4 | IncX4 | AMP CL | cfr mcr-1 |
| GDE6P133J | >128 | 4 | IncX4 | AMP FLR CL | cfr mcr-1 floR |
| GDE6P143J | 4 | 4 | IncX4 | CL | cfr mcr-1 |
| GDE6P151J | >128 | 4 | IncX4 | AMP GEN FLR CL | cfr mcr-1 floR |
| GDE6P164J | 4 | 4 | IncX4 | CL | cfr mcr-1 |
| GDE6P165J | 4 | 4 | IncX4 | CL | cfr mcr-1 |
| GDE6P168J | 4 | 4 | IncX4 | AMP CL | cfr mcr-1 |
| GDE6P169J | 4 | 4 | IncX4 | AMP CL | cfr mcr-1 |
| GDE7P75J | 16 | 4 | IncX4 | CL SXT | cfr mcr-1 |
| GDE7P100J | 8 | 4 | IncHI2 IncX4 | AMP APR DOX CL SXT | cfr mcr-1 |
| GDE7P128J | 8 | 8 | IncI2 | AMP CL SXT | cfr mcr-1 |
| GDE7P166J | 16 | 2 | IncX4 | AMP DOX CL | cfr mcr-1 |
| coli C600 | 4 | 0.125 | - c | - | - |
| ATCC 25922 | 2 | 0.5 | - | - | - |
Note: (a) AMP—ampicillin; CTX—cefotaxime; APR—apramycin; GEN—gentamycin; FLR—florfenicol; DOX—doxycycline; CL—colistin; FOS—fosfomycin; SXT—sulfamethoxazole/trimethoprim. (b) The replicon type of IncP was detected using the primer list in Supplemental Table S1. (c) “–” indicates none detected.
2.5. Plasmid Analysis of pHNEP124
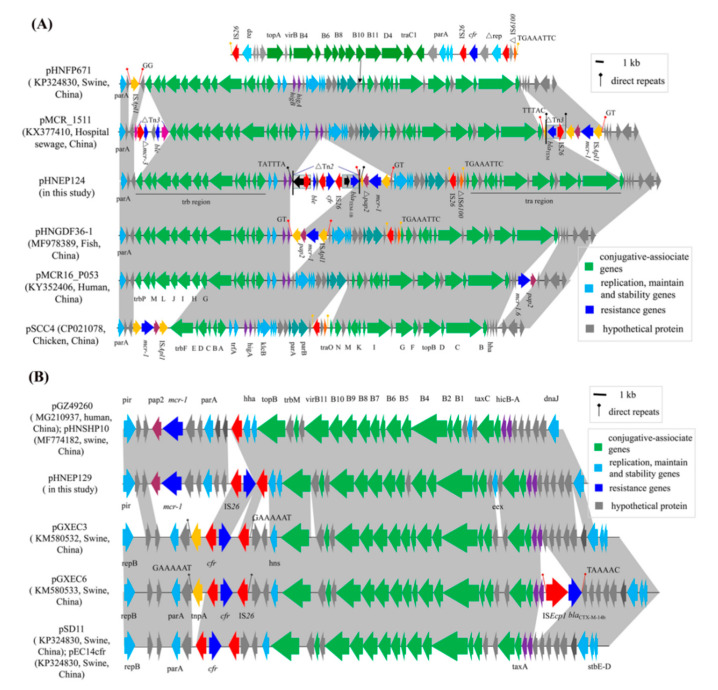
The sequence analysis revealed that plasmid pHNEP124 is 60430 base pairs (bp) in size with an average GC content of 47.01% and belonged to plasmid pMCR_1511-like IncP type plasmid, which consisted of a typical plasmid backbone and two mosaic variant regions. The backbone of plasmid pHNEP124 is comprised of the trfA encoding plasmid replication initiation protein, two par modules for plasmid partitioning, a toxin-antitoxin higA-B system, and a “KlcA-kleE” region encoding a host-lethal protein for plasmid maintenance and stability, as well as the two conjugative regions tra (~17.3-kb) and trb (~12.7-kb) for plasmid horizontal transfer. BLASTn analysis indicated that the backbone of the plasmid pHNEP124 shared a high identity (>99%) with those mcr-1-carrying IncP type plasmids found in Enterobacteriaceae, such as E. coli plasmid pHNGDF36-1 (Genbank accession no. MF978389), Klebsiella pneumoniae pMCR_1511 (Genbank accession no. KX377410), Salmonella enterica serovar Typhimurium pMCR16_P053 (Genbank accession no. KY352406), and Citrobacter braakii pSCC4 (Genbank accession no. CP021078), which are obtained from diverse sources such as fish products, hospital sewage, chickens, and humans. Moreover, it also showed high homology (>99%) to IncP plasmid pHNFP671 (Genbank accession no. KP324830) which carries the cfr gene and was isolated from swine feces in Guangdong Province, China. A comparative analysis of the plasmid pHNEP124 and other IncP plasmids indicated that IncP type plasmids have a conserved backbone structure, except plasmid pSCC4, which is missing an ~8.4 kb conjugative region (Figure 3A). In addition, plasmid pHNFP671, with ~35.5-kb insertion, contained a conjugative region and a cfr-carrying module (Figure 3A).
Figure 3.
Liner comparison of the cfr-carrying plasmids. Arrows indicate the positions of the genes and the direction. Regions with >99% homology are shaded in gray. ∆ indicates a truncated gene or mobile element. (A) Comparative analysis of plasmid pHNEP124 and other IncP-type plasmids. (B) Comparative analysis of plasmid pHNEP129 and other IncX4-type plasmids.
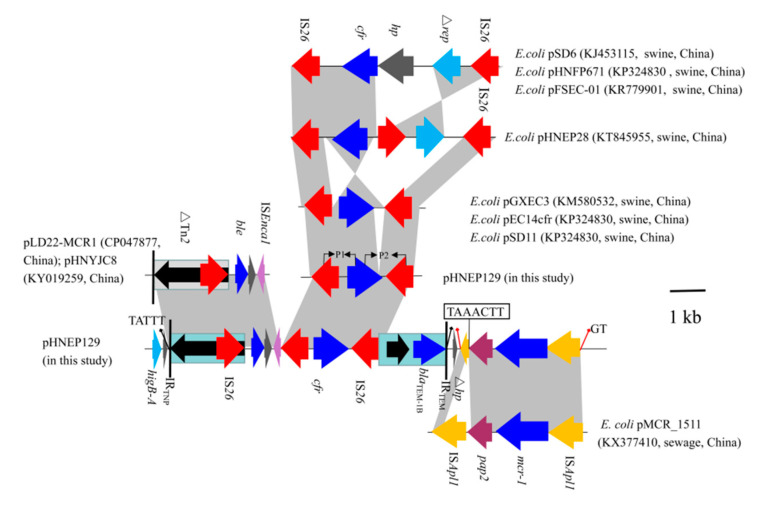
Although the IncP plasmid possessed a conserved backbone region, the variant regions were distinct. In plasmid pHNEP124, the first mosaic variant regions (~12.5-kb) contained two modules that carried the multi-resistance gene cfr, colistin resistance gene mcr-1, β-lactams resistance gene blaTEM-1B, and putative bleomycin resistance gene ble. The cfr-bearing modules (∆Tn2-IS26-ble-orf-∆ISEnca1-IS26-cfr-IS26-∆Tn2) was inserted in an open reading frame (orf) located downstream of higA and flanked by 5-bp (TATTT) direct repeats (DRs). In addition, the 3680 bp genetic structure (∆Tn2-IS26-ble-orf-∆ISEnca1) shared a 99% identity to the corresponding region in the E. coli IncHI2 plasmid pLD22-MCR1 (CP047877) and pHNYJC8 (KY019259), which were recovered from deer feces and chicken meat in China, respectively (Figure 4). The mcr-1-carrying module (ISApl1-mcr-1-∆pap2-∆ISApl1) was inserted downstream of the cfr-bearing modules and produced 2-bp (GT) DRs, which is in agreement with the mcr-1-carrying plasmid pHNGDF36-1 and pMCR_1511. Notably, the pap2 gene missed the stop codon and has a 7-bp (TAAACTT) homology sequence with delta ISApl1 that may be caused by recombination (Figure 4). Another variant region (IS26-hp-∆IS6100) was inserted into an orf gene and is flanked by 8-bp (TGAAATTC) DRs, which were also found in the same locus of plasmid pHNGDF36-1 and pSCC4, respectively.
Figure 4.
Comparison of the genetic context of cfr gene among E. coli strains. Arrows indicate the positions of the genes and the direction. Regions with >99% homology are shaded in gray. ∆ indicates a truncated gene or mobile element.
2.6. Plasmid Analysis of pHNEP129
The sequence analysis revealed that plasmid pHNEP129 is 35336 bp and belongs to the IncX4 plasmid, which is comprised of a typical IncX4 plasmid backbone including the encoding replication initiation protein gene pir, plasmid partitioning protein gene parA, encoding DNA relaxase gene taxC, a toxin-antitoxin system hicA-B, conjugative transfer protein gene trbM, and the virB gene. BLASTn analysis showed that plasmid pHNEP129 shares a >99% identify with 96% coverage to other mcr-1-carrying IncX4 plasmids such as plasmid pMCR1-NY (CP019908, E. coli, human) from the United States, plasmid pmcr1_IncX4 (KU761327, Klebsiella pneumoniae, human), plasmid pGZ49260 (MG210937, E. coli, human), and plasmid pHNSHP10 (MF774182, E. coli, swine) isolated from China (Figure 3B). Unlike those mcr-1-carrying IncX4 plasmids, plasmid pHNEP129 showed a 97–99% identity with 74% coverage to other cfr-bearing IncX4 plasmids such as E.coli plasmids pSD11 (KM212169), pGXEC3 (KM580532), pGXEC6 (KM580533), and pEC14cfr (KY865319) recovered from swine farms in different geographic locations of China (Guangdong, Guangxi and Liaoning province; Figure 3B).
A comparative analysis of the plasmid pHNEP129 and other IncX4 plasmids carried the mcr-1 or cfr genes, the backbone region of plasmid pHNEP129, except for the cfr-carrying module, was almost identical to the mcr-1-carrying plasmids such as plasmid pGZ49260 and pHNSHP10. However, the backbone region of the plasmid pHNEP129, except for the conjugative transfer region, showed a lower identity to the cfr-carrying plasmids such as pSD11, pGXEC3s and pEC14cfr (Figure 3B). In addition, the cfr-carrying module contained a cfr gene and two copies of IS26 in the same orientation located downstream of hns in plasmid pHNEP129, which was consistent with plasmid pSD11, pGXEC3, and pEC14cfr (Figure 3B). However, the cfr-carrying module (tnpA-IS26-cfr-IS26) of pGXEC3 and pGXEC6 was inserted in the IncX4 plasmid backbone with 7-bp (TAAAAAC) DRs, but no DRs were found in plasmid pHNEP129. This result indicates that the cfr-carrying module (IS26-cfr-IS26) is likely to be directly inserted into the mcr-1-positive IncX4 plasmid rather than evolving from cfr-positive plasmids such as pSD11 and pGXEC3.
The cfr-carrying module (2875-bp) showed a >99% identity to the cfr-positive plasmid pGXEC3 and pGXEC6. It is interesting to note that the cfr-carrying module has a 43 bp insertion upstream of the cfr gene and a 355 bp deletion downstream of the cfr gene compared with plasmid pGXEC3 and pSD11 (Figure 4).
3. Discussion
Food-producing animals, considered to be a “reservoir” of resistant genes, play a vital role in the dissemination of important resistance genes, such as the mcr-1 gene. The multiresistant cfr gene can confer a broad range of antibiotics resistance (exhibiting PhLOPSA phenotype) in Gram-positive bacteria, but it only mediated resistance to phenicols such as florfenicol in Gram-negative bacteria. In this study, the prevalence (13.7%) of the cfr gene in E. coli isolates was significantly higher than the previous reported 0.08–1.6% in different provinces of China [8,13,14], which suggested that the cfr gene may rapidly disseminate in a specific swine farm at a small scale but not in large areas of China. The cfr gene was detected in different sources, which suggests that the cfr gene had a high prevalence of circulation in a typical swine farm. To the best of our knowledge, this is the first report on the rapid increase of the cfr gene in E. coli and the first detection from environmental samples such as soil and sewage in a typical swine farm of Guangdong Province, China.
Compared with the detectable rate of the cfr gene in E. coli isolates from different growth stages, the higher prevalence of the cfr gene in E. coli isolates from nursery pigs and fattening pigs may be explained by the long-term use or overuse of antibacterial drugs such as florfenicol, amoxicillin, and gentamycin. Although florfenicol has not been used since 2017 in this swine farm, the detectable rate of the cfr gene was obviously increased in E. coli isolates, which may because the effects of florfenicol are difficult to eliminate in the short term. A similar result was observed in the spread of the mcr-1 gene in a swine farm in Shanghai, China [15]. Interestingly, the prevalence of the mcr-1 gene among cfr-positive E. coli had decreased sharply from 86.4% before 2017 to 17.1% in 2017, which may be due to the ban of colistin as a feed additive for animals in China in 2016 [16], which is in accordance with the prevalence of the mcr-1 gene in E. coli isolates from a swine farm in Guangdong Province, China [17].
The PFGE analysis of all cfr-bearing E. coli isolates showed that most of these isolates were epidemiologically unrelated. This result suggests that the spread of the cfr gene was mainly due to horizontal transfer, which is in agreement with the previous reports [8,12,14]. Previous studies have suggested that the cfr gene is mainly located on the plasmid of 23 E. coli isolates and on the chromosomal DNA of isolate FSEC-02, and that the IS26 element plays a vital role in the dissemination of the cfr gene [4,18]. The cfr-carrying module (IS26-cfr-IS26) was the most prevalent genetic surrounding in E. coli isolates except for one cfr-carrying module composed of a cfr gene and two copies of IS256, which was identified in the IncA/C plasmid pSCEC2 from porcine E. coli isolates [19]. In addition, two copies of IS26 in the same orientation have been described to form minicircles comprising the cfr gene and one copy of the IS26 element, which may accelerate the transfer of cfr gene by IS26-mediated recombination [20], but we failed to detect the minicircles in the current study (data not provided). Furthermore, compared with the previous reports about the genetic surrounding of the cfr gene, the cfr-carrying module in this study was distinct from the cfr-carrying module of plasmid pHNEP28 found in 2013 in this swine farm. Based on this, we assumed that the cfr-carrying module has contributed to the dissemination among E. coli isolates through the missing delta rep gene. Thus, the results provide compelling evidence that plasmids and insertion sequences such as IS26 are closely related to the spread and diffusion of the cfr gene in this swine farm.
The cfr gene is mainly found on various Inc type plasmids, including IncX4, IncA/C, IncF43:A-:B-, and IncF14:A-:B-, as well as untyped plasmids in E. coli isolates from food-producing animals in China [8]. Plasmids such as IncX4, IncI2, IncP, and IncHI2, which were considered to be the important vectors of the mcr-1 gene, play an important role in the global spread of the mcr-1 gene [10]. In the present study, cfr and mcr-1 were first detected in the existence in multiple plasmids such as IncX4, IncI2, and IncP. Notably, the IncP plasmid is a broad-host-range type of plasmid, which had been detected in various sources such as fish products, pig feces, hospital sewage, and humans in China [21,22,23]. It is considered an important carrier of the mcr-1 and mcr-3 genes, with the potential to mediate the spread of mcr-1 or mcr-3 genes from Enterobacteriaceae to other Gram-negative bacteria, such as Pseudomonas aeruginosa and Aeromonas spp. A previous study reported that mcr-1-bearing IncP plasmids have high conjugative frequencies and low fitness costs in hosts, which may facilitate the dissemination of mcr-1 among various hosts [23]. The IncP plasmid carried multiple resistance genes such as blaTEM-1B, ble, cfr, and mcr-1 genes compared with other IncP plasmids, but this needs to be investigated further in animal husbandry in order to prevent the dissemination of antibiotic-resistant genes. In addition, the cfr gene in plasmids pGXEC3 and pSD11 exhibited resistance to florfenicol but most of the plasmids in this study did not. Although the phenomenon of a “silent” cfr gene has been described in Enterococcus faecium isolates from swine and the patient, this has not been observed in E. coli before [24,25], and the mechanism of a “silent” cfr in E. coli was unknown in this study and needs further research in the future.
4. Materials and Methods
4.1. Detection of Multi-Resistant cfr Gene and Other Resistance Genes
A total of 861 samples including 846 anal swabs samples and 14 environmental samples, were collected from the swine farm from May 2014 to February 2017 and the information of the sample collection is listed in Table S2. All of the samples were cultured using LB broth and were incubated with 200 rpm/min at 37 °C for 14 h. The E. coli isolate was screened and purified using MacConkey agar plates without antibiotic selection pressure. Then, a non-duplicate colony was selected and identified using Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry (MALDI-TOF-MS) and 16S rRNA sequencing [26]. In this swine farm, all of the pigs were divided into five growth stages according to the age of the pigs including suckling piglets, nursery pigs, fattening pigs, sows, and boars. Common drugs were investigated and are listed in Table 1. The bacteria DNA was prepared through a boiling method and were used for the PCR amplification and sequencing analysis to detect the cfr gene using previously designed primers [27]. Considering that the cfr gene is likely to co-transfer with other florfenicol resistance genes and colistin-resistant genes in Gram-negative bacteria, the cfr-positive E. coli isolates were investigated further using PCR for other florfenicol genes and the mcr-1 gene, and the primers are listed in Supplemental Table S2.
4.2. Antibiotic Susceptibility Testing
Minimum inhibitory concentration (MIC) values of all cfr-positive E. coli isolates were found using an agar dilution method of 16 different drugs, including ampicillin, cefotaxime, cefquinome, ceftazidime, cefoxitin, imipenem, gentamycin, amikacin, apramycin, neomycin, doxycycline, tetracycline, florfenicol, fosfomycin, ciprofloxacin, and sulfamethoxazole/trimethoprim. The MIC values of colistin were determined via the broth microdilution method. MIC values were interpreted according to the document M100 and VET01-S4 of the Clinical and Laboratory Standards Institute [28,29]. Colistin (>2 mg/L) and florfenicol (>16 mg/L) were interpreted according to the clinical breakpoints or epidemiological cut-off values of the European Committee on Antimicrobial Susceptibility Testing (http://mic.eucast.org/Eucast2/). ATCC 25922 served as the control.
4.3. Pulsed-Field Gel Electrophoresis (PFGE) and Flanking Regions of the cfr Gene
The PFGE analysis of all cfr-positive strains with XbaI-digested genomic DNA was performed using the CHEF-MAPPER System (Bio-Rad Laboratories, Hercules, CA, USA) as described previously [30]. The PFGE patterns were analyzed using BioNumerics software (Applied Maths, Sint-Martens-Latem, Belgium) with a cut-off value at 80% of the similarity values in order to indicate different PFGE patterns according to a previous report [14]. The flanking regions of the cfr gene in the E. coli isolates were determined by PCR mapping and the sequences of the primers are listed in Supplemental Table S3.
4.4. S1-PFGE and Southern Blotting Hybridization
S1 nuclease pulsed-field gel electrophoresis combined with Southern blotting hybridization with the cfr probe was conducted to determine the location of the cfr gene in E. coli, as well as the size of the cfr-carrying plasmid according to previous protocols [12,15].
4.5. Conjugation Experiment and Transformation Assay
The conjugation experiment was conducted with the broth mating method using streptomycin-resistant E. coli C600 as the recipient strain. The 45 E. coli isolates carrying the mcr-1 and cfr genes were used as the donor strains. After 4 h of the culture of the donor strains and E. coli, C600 were mixed (ratio of 1:4) in Luria-Bertani (LB) broth, and then put into incubation for 4 h at 37 °C. The transconjugants were selected on LB agar plates supplemented with streptomycin (3000 mg/L) and florfenicol (10 mg/L) or colistin (2 mg/L). To obtain the transferable plasmids co-harboring the mcr-1 and cfr genes, the cfr/mcr-1-positive plasmids of the transconjugants were transformed into the E. coli DH5α (Takara) through electroporation. Transformants were selected on LB agar plates containing 10 mg/L florfenicol or 2 mg/L colistin. The antimicrobial susceptibility of the transconjugants or transformants was determined by either the agar dilution method or the broth microdilution method, and the presence of cfr, mcr-1, and floR genes in the transconjugants or transformants were identified by PCR.
4.6. PBRT and Plasmid Analysis
PCR-based replicon typing (PBRT) was performed on all cfr/mcr-1-positive transconjugants/transformants using the primers as described previously [31,32]. Plasmids pHNEP124 and pHNEP129 bearing the cfr and mcr-1 genes from transformants GDE6P124T and GDE6P129T were extracted and purified using a Qiagen plasmid midi kit (Qiagen, Hilden, Germany), and were subjected to sequencing by Illumina Miseq 2500 (Illumina, San Diego, CA, USA). The sequence reads were assembled into contigs using SOAP denovo version 2.04, and the gaps between the contigs were linked by PCR and sequencing. The complete sequences of plasmids pHNEP124 and pHNEP129 were analyzed and annotated by IS finder (https://www-is.biotoul.fr/), BLASTn (https://blast.ncbi.nlm.nih.gov/Blast.cgi), ResFinder (https://cge.cbs.dtu.dk//services/ResFinder/), RAST [33], and Vector NTI program (Invitrogen, San Diego, CA, USA).
4.7. Nucleotide Sequence Accession Number
The nucleotide sequences of plasmids pHNEP124 and pHNEP129 were deposited in the GenBank database under the accession numbers MT667260 and MT667261, respectively.
5. Conclusions
This is the first study to report the high prevalence of the cfr gene among E. coli isolates in a typical swine farm from 2014–2017 and the first identification of the complete genome sequence of IncP and IncX4 plasmids co-harboring the mcr-1 and cfr genes. Florfenicol was extensively used for preventing and treating an infectious disease, which might have facilitated the spread of the cfr gene in this swine farm. Plasmids such as IncX4 and insertion sequence IS26 were responsible for the dissemination of the cfr gene in this study. The occurrence and dissemination of plasmids carrying the cfr and mcr-1 genes in E. coli isolates need further surveillance.
Acknowledgments
We thank the help of Jing Wang for the cfr-positive plasmid analysis.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-0817/10/1/33/s1, Table S1. Characterization of cfr-carrying transformants, Table S2: information of sample collection in the swine farm during the period of 2014–2017, Table S3: Primers used for PCR and PCR mapping in this study.
Author Contributions
Conceptualization, Z.Z., J.-H.L. and Z.M.; methodology, Z.M., J.L., L.C., X.L.; software, Z.M., W.X.; validation, Z.M., and J.L.; formal analysis, Z.Z., W.X. and Z.M.; investigation, Z.M., L.C., and X.L.; resources, J.-H.L. and Z.Z.; data curation, Z.M., J.-H.L. and Z.Z.; writing—original draft preparation, Z.M., Z.Z.; writing—review and editing, Z.M., J.L., L.C., W.X., J.-H.L. and Z.Z.; visualization, Z.M., X.L.; supervision, J.-H.L. and Z.Z.; project administration, Z.Z.; funding acquisition, Z.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Key R&D Program of China (Grant Number 2018YFD0500300) and the National Natural Science Foundation of China (Grant Number 31672608).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Long K.S., Poehlsgaard J., Kehrenberg C., Schwarz S., Vester B. The Cfr rRNA methyltransferase confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics. Antimiocrob. Agents Chemother. 2006;50:2500–2505. doi: 10.1128/AAC.00131-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith L.K., Mankin A.S. Transcriptional and translational control of the mlr operon, which confers resistance to seven classes of protein synthesis inhibitors. Antimicrob. Agents Chemother. 2008;52:1703–1712. doi: 10.1128/AAC.01583-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kehrenberg C., Schwarz S., Jacobsen L., Hansen L.H., Vester B.A. New mechanism for chloramphenicol, florfenicol and clindamycin resistance: Methylation of 23S ribosomal RNA at A2503. Mol. Microbiol. 2005;57:1064–1073. doi: 10.1111/j.1365-2958.2005.04754.x. [DOI] [PubMed] [Google Scholar]
- 4.Shen J., Wang Y., Schwarz S. Presence and dissemination of the multiresistance gene cfr in Gram-positive and Gram-negative bacteria. J. Antimicrob. Chemother. 2013;68:1697–1706. doi: 10.1093/jac/dkt092. [DOI] [PubMed] [Google Scholar]
- 5.Chen Y., Lei C., Zuo L., Kong L., Kang Z., Zeng J., Zhang X., Wang H. A novel cfr-carrying Tn7 transposon derivative characterized in Morganella morganii of swine origin in China. J. Antimicrob. Chemother. 2018;74:603–606. doi: 10.1093/jac/dky494. [DOI] [PubMed] [Google Scholar]
- 6.Partridge S.R., Kwong S.M., Firth N., Jensen S.O. Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 2018;31:e00088-17. doi: 10.1128/CMR.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poirel L., Madec J.-Y., Lupo A., Schink A.-K., Kieffer N., Nordmann P., Schwarz S. Antimicrobial resistance in Escherichia coli. Microbiol. Spectrum. 2018;6:ARBA-0026-2017. doi: 10.1128/microbiolspec.ARBA-0026-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X., Zhu Y., Hua X., Chen F., Wang C., Zhang Y., Liu S., Zhang W. F14:A-:B- and IncX4 Inc group cfr-positive plasmids circulating in Escherichia coli of animal origin in Northeast China. Vet. Microbiol. 2018;217:53–57. doi: 10.1016/j.vetmic.2018.02.003. [DOI] [PubMed] [Google Scholar]
- 9.Liu Y.Y., Wang Y., Walsh T.R., Yi L.X., Zhang R., Spencer J., Doi Y., Tian G., Dong B., Huang X., et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016;16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 10.Sun J., Zhang H., Liu Y.H., Feng Y. Towards Understanding MCR-like Colistin Resistance. Trends. Microbiol. 2018;26:794–808. doi: 10.1016/j.tim.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 11.Baron S., Hadjadj L., Rolain J.M., Olaitan A.O. Molecular mechanisms of polymyxin resistance: Knowns and unknowns. Int. J. Antimicrob. Agents. 2016;48:583–591. doi: 10.1016/j.ijantimicag.2016.06.023. [DOI] [PubMed] [Google Scholar]
- 12.Liu X.-Q., Wang J., Li W., Zhao L.-Q., Lu Y., Liu J.-H., Zeng Z.-L. Distribution of cfr in Staphylococcus spp. and Escherichia coli Strains from Pig Farms in China and Characterization of a Novel cfr-Carrying F43:A-:B- Plasmid. Front. Microbiol. 2017;8:329. doi: 10.3389/fmicb.2017.00329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y., He T., Schwarz S., Zhou D., Shen Z., Wu C., Wang Y., Ma L., Zhang Q., Shen J. Detection of the staphylococcal multiresistance gene cfr in Escherichia coli of domestic-animal origin. J. Antimicrob. Chemother. 2012;67:1094–1098. doi: 10.1093/jac/dks020. [DOI] [PubMed] [Google Scholar]
- 14.Deng H., Sun J., Ma J., Li L., Fang L.-X., Zhang Q., Liu Y.-H., Liao X.-P. Identification of the multi-resistance gene cfr in Escherichia coli isolates of animal origin. PLoS ONE. 2014;9:e102378. doi: 10.1371/journal.pone.0102378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu R., Yi L.X., Yu L.F., Wang J., Liu Y., Chen X., Lv L., Yang J., Liu J.-H. Fitness Advantage of mcr-1-Bearing IncI2 and IncX4 Plasmids in Vitro. Front. Microbiol. 2018;9:331. doi: 10.3389/fmicb.2018.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walsh T.R., Wu Y. China bans colistin as a feed additive for animals. Lancet Infect. Dis. 2016;16:1102–1103. doi: 10.1016/S1473-3099(16)30329-2. [DOI] [PubMed] [Google Scholar]
- 17.Li W., Hou M., Liu C., Xiong W., Zeng Z. Dramatic decrease of the colistin resistance in E. coli from a typical farm following the restriction of use of colistin in China. Int. J. Antimicrob. Agents. 2019;53:707–708. doi: 10.1016/j.ijantimicag.2019.03.021. [DOI] [PubMed] [Google Scholar]
- 18.Zhang R., Sun B., Wang Y., Lei L., Schwarz S., Wu C. Characterization of a cfr-carrying plasmid from porcine Escherichia coli that closely resembles plasmid pEA3 from the plant pathogen Erwinia amylovora. Antimicrob. Agents Chemother. 2016;60:658–661. doi: 10.1128/AAC.02114-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang W.-J., Xu X.-R., Schwarz S., Wang X.-M., Dai L., Zheng H.-J., Liu S. Characterization of the IncA/C plasmid pSCEC2 from Escherichia coli of swine origin that harbours the multiresistance gene cfr. J. Antimicrob. Chemother. 2014;69:385–389. doi: 10.1093/jac/dkt355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun J., Deng H., Li L., Chen M.-Y., Fang L.X., Yang Q.-E., Liu Y.-H., Liao X.-P. Complete nucleotide sequence of cfr-carrying IncX4 plasmid pSD11 from Escherichia coli. Antimicrob. Agents Chemother. 2015;59:738–741. doi: 10.1128/AAC.04388-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao F., Feng Y., Lü X., McNally A., Zong Z. IncP Plasmid Carrying Colistin Resistance Gene mcr-1 in Klebsiella pneumoniae from Hospital Sewage. Antimicrob. Agents Chemother. 2017;61:e02229-16. doi: 10.1128/AAC.02229-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu L., Feng Y., Lü X., McNally A., Zong Z. New Variant of mcr-3 in an Extensively Drug-Resistant Escherichia coli Clinical Isolate Carrying mcr-1 and blaNDM-5. Antimicrob. Agents Chemother. 2017;61:e01757-17. doi: 10.1128/AAC.01757-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lv L., Cao Y., Yu P., Huang R., Wang J., Wen Q., Zhi C., Zhang Q., Liu J.-H. Detection of mcr-1 Gene among Escherichia coli Isolates from Farmed Fish and Characterization of mcr-1-Bearing IncP Plasmids. Antimicrob. Agents Chemother. 2018;62:e02378-17. doi: 10.1128/AAC.02378-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y., Wang Y., Schwarz S., Wang S., Chen L., Wu C., Shen J. Investigation of a multiresistance gene cfr that fails to mediate resistance to phenicols and oxazolidinones in Enterococcus faecalis. J. Antimicrob. Chemother. 2014;69:892–898. doi: 10.1093/jac/dkt459. [DOI] [PubMed] [Google Scholar]
- 25.Brenciani A., Morroni G., Vincenzi C., Manso E., Mingoia M., Giovanetti E., Varaldo P.E. Detection in Italy of two clinical Enterococcus faecium isolates carrying both the oxazolidinone and phenicol resistance gene optrA and a silent multiresistance gene cfr. J. Antimicrob. Chemother. 2016;71:1118–1189. doi: 10.1093/jac/dkv438. [DOI] [PubMed] [Google Scholar]
- 26.Kim T.W., Kim Y.H., Kim S.E., Lee J.H., Park C.S., Kim H.Y. Identification and distribution of bacillus species in doenjang by whole-cell protein patterns and 16S rRNA gene sequence analysis. J. Microbiol. Biotechnol. 2010;20:1210–1214. doi: 10.4014/jmb.1002.02008. [DOI] [PubMed] [Google Scholar]
- 27.Kehrenberg C., Schwarz S. Distribution of florfenicol resistance genes fexA and cfr among chloramphenicol-resistant Staphylococcus isolates. Antimicrob. Agents Chemother. 2006;50:1156–1163. doi: 10.1128/AAC.50.4.1156-1163.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.CLSI . Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2013. Approved Standard fourth Edition and Supplement. Documents VET01-A4 and VET01-S2. [Google Scholar]
- 29.CLSI . Performance Standards for Antimicrobial Susceptibility Testing. 30th ed. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2020. CLSI Supplement M100. [Google Scholar]
- 30.Barton B.M., Harding G.P., Zuccarelli A.J. A general method for detecting and sizing large plasmids. Anal. Biochem. 1995;226:235–240. doi: 10.1006/abio.1995.1220. [DOI] [PubMed] [Google Scholar]
- 31.Carattoli A., Bertini A., Villa L., Falbo V., Hopkins K.L., Threlfall E.J. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods. 2005;63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 32.Johnson T.J., Bielak E.M., Fortini D., Hansen L.H., Hasman H., Debroy C., Nolan L.K., Carattoli A. Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae. Plasmid. 2012;68:43–50. doi: 10.1016/j.plasmid.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 33.Aziz R.K., Bartels D., Best A.A., DeJongh M., Disz T., Edwards R.A., Formsma K., Gerdes S., Glass E.M., Kubal M., et al. The RAST server: Rapid annotations using subsystems technology. BMC Genom. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.