Abstract
Head and neck squamous carcinoma (HNSCC) constitutes the sixth most prevalent cancer worldwide. The molecular pathogenesis of HNSCC includes disorders in cell cycle, intercellular signaling, proliferation, squamous cell differentiation and apoptosis. In addition to the genetic mutations, changes in HNSCC are also characterized by the accumulation of epigenetic alterations such as DNA methylation, histone modifications, non-coding RNA activity and RNA methylation. In fact, some of them may promote cancer formation and progression by controlling the gene expression machinery, hence, they could be used as biomarkers in the clinical surveillance of HNSCC or as targets for therapeutic strategies. In this review, we focus on the current knowledge regarding epigenetic modifications observed in HNSCC and its predictive value for cancer development.
Keywords: head and neck cancer, squamous cell carcinoma, epigenetics, DNA methylation, histone modification, non-coding RNA activity, RNA methylation, biomarkers
1. Introduction
Head and neck squamous cell carcinoma (HNSCC) is a common heterogeneous malignant cancer type originating from the squamous cells, located in the mucous membrane of the oral cavity, oropharynx, paranasal sinuses, nasal cavity, nasopharynx, larynx and hypopharynx [1]. The main prognostic features of HNSCC progression are the location, tumor size and the presence of distant metastases [2]. The estimated number of HNSCC accounts for more than 650,000 cases and 330,000 deaths annually [3]. Main and widely investigated contributors to the development of head and neck cancers are tobacco smoking, alcohol consumption as well as viral factors such as human papillomavirus (HPV) and Epstein–Barr virus infections [4,5,6,7]. The treatment of an HNSCC patient involves surgical eradication, radiotherapy (RT) and chemotherapy (CT). Moreover, the approved targeted drug is cetuximab, a monoclonal antibody targeting epidermal growth factor receptor (EGFR) for both HPV(+) and HPV(–) subtypes [8]. The treatment method depends on the type and stage of cancer, possible side effects and the patient’s overall health. Unfortunately, cetuximab and other therapies have a limited efficacy due to molecular and histological diversity of HNSCC [9].
Molecular pathogenesis of HNSCC is a complex process with a high rate of genetic heterogeneity. It is possible to distinguish alterations in the tumor suppressor pathways p53, p16INK4a and retinoblastoma (RB) which affects DNA damage response, apoptosis, cell cycle and genomic stability [10,11,12,13]. Additionally, overexpression of EGFR correlates with poor prognosis and metastatic potential of cancer cells [14]. Moreover, disorders in signaling pathways associated with Ras-mitogen activated protein kinase (Ras-MAPK) lead to disturbances in gene expression level involved in the cell proliferation, differentiation, apoptosis, angiogenesis and cell motility. Similarly, a higher rate of cancer recurrence and metastases is associated with mutations in NOTCH1-4 genes in HNSCC [15]. Moreover, PI3K-Akt/mTOR constitutes a frequently disturbed pathway in HNSCC and simultaneously is a cascade responsible for phosphorylation and activation of many proteins [16]. The activation of STAT3 pathway, crucial in many cancer types including HNSCC, leads to a malignant transformation of cells and protect them from recognition and degradation by cytotoxic T lymphocytes [17]. Furthermore, overexpression of hepatocyte growth factor receptor (MET) is correlated with cisplatin and EGFR-targeted therapies resistance as well as with poor prognosis for HNSCC patients [18]. Additionally, a nuclear transcription factor-κB (NF-κB) modulating the expression of genes involved in inflammation, immunity, proliferation, and apoptosis is constitutively activated in HNSCC, and affects the therapeutic resistance [19]. All these signaling cascades shape complex cellular conditions which ultimately affect squamous epithelial proliferation and differentiation, cell survival and metastatic phenotype.
It is crucial to bear in mind the fact that, carcinogenesis of HNSCC is driven not only by the accumulation of genetic alterations, but also by the changes in the epigenetic landscape. Epigenetic modifications found in HNSCC include DNA methylation, histone modification, non-coding RNA activity, as well as RNA methylation [20,21]. Since these modifications regulate the expression of target genes (tumor suppressor genes (TSGs) and oncogenes), they have become a focus of attention in cancer studies, also in terms of personalized therapy strategies. They may be involved in the pathology of the disease; therefore, they are considered candidates for diagnostic biomarkers and prognostic features of cancer. In this review, we discuss the current literature associated with the impact of epigenetic modification on the progression of head and neck squamous cell carcinoma.
2. The HNSCC Epigenetic Landscape and Its Clinical Implications
2.1. DNA Methylation
DNA methylation is one of the best investigated DNA modifications which modulates the expression of genes without affecting their nucleotide sequence. DNA methylation is a process of covalent conversion of a hydrogen atom into a methyl group at the fifth carbon of the pyrimidine ring of cytosine (5-methylcytosine, 5-mC). In fact, this modification constitutes an essential epigenetic marker recognized by specific proteins.
In mammals, 5-mC is highly accumulated in the DNA regions rich in CpG dinucleotides (so-called CpG islands) where 70–80% of cytosines are methylated [22]. About 60% of CpG islands are located in the gene promoter regions [23]. The presence of DNA methylation in promoters causes transcriptional repression by preventing the binding of transcription factors and by influencing interactions between enhancers and promoters [24]. Furthermore, 5-mCs are also found in repetitive sequences, gene bodies and intergenic regions. 9i [25,26]. DNA methylation is also found in non-CpG sites and it includes methylation at cytosine followed by adenine (CpA), thymine, (CpT) or another cytosine (CpC) [27]. On the other hand, non-CpG methylation is tissue-specific and functions as a transcriptional repressor by blocking transcription factors binding sites [28].
The enzymes responsible for DNA methylation belong to the DNA methyltransferase (DNMT) family: DNMT1, DNMT2, DNMT3A, DNMT3B and DNMT3L [29]. In fact, DNMT1 is responsible for maintaining methylation patterns after replication [30], whereas DNMT2 is a methyltransferase homologue which mainly methylates aspartic acid cytosine 38 in the tRNA anti-codon loop [31,32]. Recent reports have indicated that DNMT2 can also methylate other RNA molecules and short DNA segments in vitro [33,34]. Moreover, DNMT3A and DNMT3B are responsible for de novo DNA methylation and are particularly crucial in the embryonic development during determining methylation pattern. Last but not least, DNMT3L lacks catalytic activity, but supports DNMT3A/B in the binding of the methyl donor group S-adenosylmethionine (SAM) and regulates their multimerization and nuclear localization [35].
DNA methylation is a reversible modification which may occur as a passive or active mechanism. Passive DNA demethylation occurs by inhibition or lack of DNMTs activity, during DNA replication [35]. In contrast, active DNA demethylation is mediated by specific enzymes from the TET (Tet methylcytosine dioxygenase) family, regardless of DNA replication [36]. TET oxidize 5-methylcytosine to 5-hydroxymethylcytosine (5-hmC), 5-formylcytosine (5-fC) or 5-carboxylcytosine (5caC) with an altered preference. Furthermore, at least four mechanisms of demethylation to cytosine have been proposed [37]. The first mechanism suggests a replication-dependent passive dilution of 5-mC, whereas the second one includes an active replication-independent demethylation based on 5-mC removing to cytosine by thymine-DNA glycosylase (TDG) in the base excision repair (BER) mechanism. Another process is based on enzymatic 5-caC decarboxylation to cytosine and the last one is associated with the activation-induced deaminase/apolipoprotein B mRNA-editing enzyme complex (AID/APOBEC), which can deaminate 5-hmC to 5-hmU [38,39].
One of the cancer hallmarks is a global DNA hypomethylation and specific local hypermethylation of CpG islands [38]. Hypomethylation in cancerous tissues appears predominantly on multiple repeats elements (e.g., SAT2) and retrotransposons (e.g., LINE-1 and ALU) sequences leading to genomic instability and activation of oncogenes. Local hypermethylation of DNA is usually associated with CpG islands in promoters of tumor suppressor genes where their expression is downregulated [39]. In HNSCC patients, numerous aberrantly methylated genes have been identified. The altered methylation patterns of the selected genes have been correlated with HNSCC formation and progression, based on clinical data, and proposed as a potential biomarker of the disease progression with specific diagnostic significance (see Table 1). Unfortunately, there is a lack of research concerning its diagnostic potential in in vitro and/or in vivo assays.
Table 1.
Hypermethylated gene with diagnostic value identified in head and neck squamous carcinoma (HNSCC).
| Gene | Tissue | Type of Study | Diagnostic Significance | References |
|---|---|---|---|---|
| ZNF14, ZNF160, ZNF420 | Tumor and saliva | Meta-analysis confirmed in patient samples | HNSCC detection and surveillance | [40] |
| hTERT | Blood leukocytes | Patient study | HNSCC detection | [41] |
| FAM135B | Tumor | Meta-analysis | Overall survival of HNSCC patients | [42] |
| CDKN2A | Tumor and saliva | Meta-analysis | HNSCC progression and metastases | [43] |
| ATM | Tumor | Patients study | HNSCC detection in early age and early tumor stage | [44] |
| MGMT | Tumor | Meta-analysis | Risk of HNSCC | [45] |
| DAPK | Tumor | Patients study | HNSCC HPV(–) detection in early stage | [46] |
| RASSF1A, MLH1, MGMT | Tumor | Patients and in vitro study | HNSCC and high proliferative potential of tumor cells detection | [47] |
| CTLA4 | Tumor | Patients study | HNSCC detection and surveillance | [48] |
| APC | Tumor | Patients study | Lower number of metastatic lymph nodes | [49] |
| CCNA1, TIMP3 | Tumor | Patients study | Risk of second primary carcinomas | [50] |
| ZIC4 | Tumor | Patients study | Risk of lymph node involvement | [51] |
| PROM1 | Tumor | Meta-analysis | HNSCC detection in early stage and invasion potential | [52] |
In addition, the genome-wide DNA methylation assays also possess significant predictive and diagnostic value. According to the literature, whole-genome analysis of DNA methylation has been carried out in the peripheral blood of HNSCC patients in which differently methylated CpG sites have been identified in comparison to controls [53,54,55,56]. This non-invasive approach allows to identify global specifically methylated (hypo- or hypermethylated) regions of the DNA, particularly within promoters of genes. Moreover, the array-based DNA methylation profiling of HNSCC allows to distinguish tumors in terms of environmental factors and contributes to a personalized therapy [57]. Additionally, there are differences in DNA methylation profiles between HPV(+) and HPV(–) HNSCC as shown on the whole-genome sequencing data. The HPV(+) tumors tend to be more globally methylated than HPV(–) [58]. Nevertheless, the novel promising non-invasive prognostic tool for HPV(+) are biomarkers, such as circulating tumor DNA (ctDNA) from blood. In fact, the DNA methylation of CALML5, DNAJC5G and LY6D genes found in ctDNA from HNSCC patients had high predictive value in early diagnosis [59].
2.2. Histone Modifications
Histone proteins undergo many different post-translational modifications such as acetylation, methylation, phosphorylation, ubiquitination or sumoylation which leads to global epigenetic alterations in cancer cells [60]. However, the most described mechanisms with prognostic potential for HNSCC development and progression include histone acetylation and methylation [61].
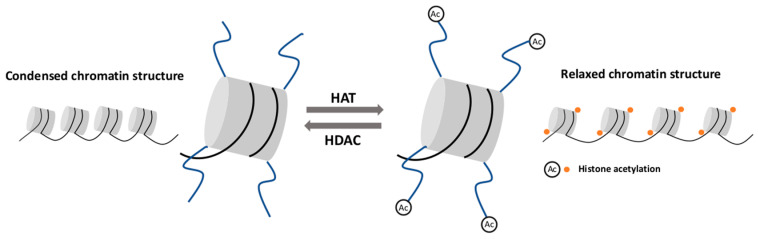
Histone acetylation is an important mechanism affecting the chromatin structure and regulating gene expression [62,63]. Histone acetyltransferase (HAT) is the enzyme responsible for attaching the acetyl group to a specific lysine residue, mostly on H3 and H4 histone [64]. Histone acetylation neutralizes the positive charge of lysine residues and relaxes the chromatin structure. This process is correlated with the recruitment of transcription coactivators and an increased transcription elongation performed by RNA polymerase II [62]. In principle, histone deacetylase (HDAC) is responsible for the removal of acetyl groups, restoring a positive charge to lysine residues and consequently, leading to chromatin condensation. This configuration limits the availability of DNA for transcription factors and results in transcriptional inhibition [65] (Figure 1).
Figure 1.
Histone acetylation and deacetylation. Histone acetyltransferase (HAT) attaches the acetyl group to the histone tail and leads to chromatin structure relaxation. Histone deacetylase (HDAC) removes the acetyl group and causes chromatin condensation.
One of the characteristic factors of the solid and metastatic tumor as HNSCC is hypoxic microenvironment [66]. In response to hypoxia, H3K2 is acetylated and activates the epithelial mesenchymal transition (EMT) correlated genes, including GLI1 and SMO genes, thus increasing the metastatic potential of the tumor. In fact, these genes may be considered as hypoxia-induced EMT biomarkers of HNSCC [67]. Furthermore, in oral squamous carcinoma (OSCC), acetylation of H3K27 increased the expression of long non-coding RNA (lncRNA) PLAC2, which induced Wnt/β-catenin signaling cascade and affected tumor growth and metastases. Hence, overexpression of PLAC2 may be a prognostic biomarker of metastases in OSCC [68]. Moreover, poor prognosis of the HNSCC patients may stem from chemoresistance. The overexpression of NF-κB protein complex leads both to histone deacetylation and to cisplatin resistance by means of reducing BRCA1 nuclear translocation in HNSCC. Therefore, NF-κB protein complex would constitute as chemoresistance biomarker for HNSCC [69]. Additionally, in vitro HNSCC cells assay shows global histone H3 hypoacetylation as compared to normal oral keratinocytes. Moreover, inhibition of HDAC leads to decreased number of cancer stem cells (CSC) and reduces the clonogenic sphere formation [70]. Interestingly, HDAC inhibitors also possess the ability to inactivate other genes such as ARF1 which affects the EGFR degradation and the inhibition of HNSCC cells invasion [71].
Histone methylation in a lysine (Lys or K) or arginine (Arg or R) residue constitutes another posttranslational modification which plays a vital role in gene regulation. These modifications can be recognized by multiple positive and negative regulators activating or repressing gene transcription [72]. According to the literature, lysine residues in histone can be mono-, di-, or tri-methylated. Di- and tri-methylation at H3K4, H3K36 and H3K79 are typically gene-activating, whereas H3K9 and H3K27 methylations are generally gene-repressive [73]. Moreover, H3K4me3 marks promoters, as well as H3K36 and H3K79 methylation occurs primarily over gene bodies [72,74]. Histone methyltransferases (HMT) includes histone lysine methyltransferases (HKMT) and protein/histone arginine methyltransferases (PRMT) [75]. Similarly, to other epigenetic modifications, histone methylation is also a reversible process. However, lysine-specific demethylases (KDMs) action is better understood, whereas arginine demethylation performed by PADI4 and JMJD6 demethylases is considerably less clear [76]. Alterations in histone methylation process have been observed in several cancers, such as gastric carcinoma [77], breast [78] or colon cancer [79], as well as hepatocellular carcinoma [80]. In the case of OSCC, the histone methylation of H3K4 is significantly different in comparison to normal tissues [81]. Furthermore, aberrant methylation of H3K9 carried out by G9a has been observed in HNSCC cells, and may be involved in the lymph node-related metastases and TGF-β-induced EMT [82]. Therefore, histone methylation profiles may be considered as biomarkers of HNSCC detection and metastases. In addition, an elevated level of histone methylation mark at H3K27me3 in HPV(+) HNSCC may, in turn, increase the tumorigenic potential and constitute a HNSCC diagnostic biomarker [83]. Moreover, H3K27me3 regulates the homeobox gene transcription in OSCC and plays a role in neoplastic phenotype of oral keratinocytes [84].
2.3. Non-Coding RNA Activity
Non-coding RNA (ncRNA) can be divided into small (less than 200 nucleotides) and large ncRNA. Small ncRNAs include small nuclear RNA (snoRNA), PIWI-interacting RNA (piRNA), small interfering RNA (siRNA) and microRNA (miRNA). The action of ncRNA is based on the transcriptional and post-transcriptional gene silencing by the specific pairing of bases with target sequences [85]. In this review, we mostly focus on the role of miRNA and lncRNA in HNSCC progression.
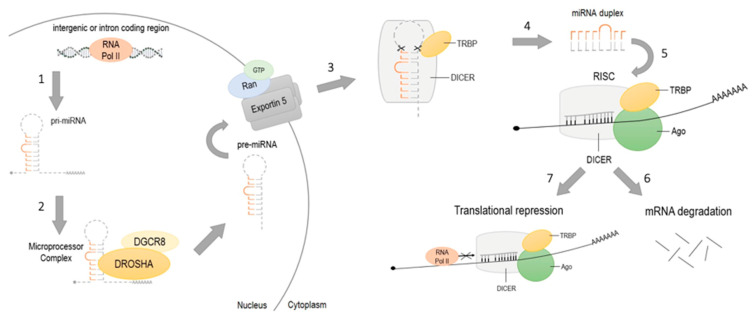
MicroRNAs are endogenous small non-coding RNAs regulating the expression of mRNA by interacting with the 3′ untranslated region (3′UTR) of target genes [86]. miRNAs may act as tumor suppressors or as oncogenes (oncomiRs), and play a crucial role in angiogenesis, cell proliferation and apoptosis [87]. Besides, there are several miRNAs influencing gene instability, immune evasion, tumor metastases and chemo- and radioresistance in tumorigenesis [88]. Maturation of miRNA consists of several stages (Figure 2). Transcription of miRNA from intergenic or intron coding region is typically performed by RNA polymerase II [89]. The transcription results in the 5’ capped and 3’ polyadenylated primary transcript (pri-miRNA) which forms hairpin structures. Nuclear protein DGCR8 recognizes pri-miRNA and targets it for Drosha, RNase III-driven cleavage. In fact, about 85 nucleotides long, released hairpin structure, are precursors to miRNA (pre-miRNA). The Ran/GTP/Exportin 5 complex transports pre-miRNA from the nucleus to the cytoplasm where pre-miRNA is processed by RNase III enzyme Dicer and TAR RNA binding protein (TRBA) to double-stranded, miRNA of about 20–22 nucleotides in length [90]. Single-stranded mature miRNA attaches to RNA-induced silencing complex (RISC) and guides RISC to the target mRNA. There are two ways of miRNA gene repression. Firstly, miRNA hybridizes to 3′UTR of the target genes, recruits RISC complex and leads to slitting and degradation of target mRNA. Secondly, miRNA can act as a blocker by connecting to the mRNA and inhibiting its translation [91].
Figure 2.
Maturation of miRNA. (1) Transcription of pri-miRNA from intergenic or intron coding regions by RNA polymerase II; (2) recognition of pri-miRNA by DGCR8 and Drosha-cleavage; resulting hairpin- structured pre-miRNA realizing; (3) pre-miRNA transportation from the nucleus to the cytoplasm by Ran/GTP/Exportin 5 complex; (4) pre-miRNA cleavage by Dicer and TAR RNA binding protein (TRBA) to 20–22 nucleotides miRNA; (5) single-stranded miRNA incorporation with Ago and RISC complex connection to target mRNA; gene silencing by (6) translational repression or by (7) mRNA degradation.
A high-throughput meta-analysis of miRNAs expression shows a long list of miRNAs associated with a poor prognosis, lower survival and metastases in HNSCCs [92]. The dysregulated expression patterns of selected miRNAs were correlated with the clinical stage, lymph node metastases and patient survival, indicating their effectiveness as molecular biomarkers for the HNSCC prognosis [93]. Moreover, the RNA interference mechanism, comprising the action of miRNA and siRNA, has become a valuable research tool for a more comprehensive understanding of the mechanisms regarding HNSCC pathogenesis [94]. Table 2 summarizes the miRNA involved in HNSCC progression.
Table 2.
Examples of miRNAs involved in the development of head and neck squamous carcinoma (HNSCC).
| Process | microRNA | Diagnostic Significance | References |
|---|---|---|---|
| (Up- or Downregulated) | |||
| Apoptosis | miR137 | downregulated | [95] |
| miR34 | upregulated | [96] | |
| miR17-92 | upregulated | [97] | |
| Gene instability | miR210 | upregulated | [98] |
| miR29 | downregulated | [99] | |
| Immune evasion | miR21 | upregulated | [100] |
| miR210 | downregulated | [101] | |
| Inflammation | miR26 | downregulated | [102] |
| miR218 | downregulated | [103] | |
| Metabolism | miR26 | downregulated | [102] |
| miR125b | downregulated | [104] | |
| Metastases | miR26 | upregulated | [105] |
| miR125b | upregulated | [105] | |
| miR139 | downregulated | [106] | |
| let-7d | upregulated | [107] | |
| miR200b | upregulated | [108] | |
| miR218 | downregulated | [109] | |
| miR96 | upregulated | [109] | |
| miR29 | downregulated | [99] | |
| miR200 | downregulated | [101] | |
| Proliferation | miR21 | upregulated | [100] |
| miR29 | downregulated | [99] | |
| miR139 | downregulated | [106] | |
| miR155 | upregulated | [110] | |
| Resistance to the radiotherapy and chemotherapy | miR210 | downregulated | [101] |
| miR31 | upregulated | [111] | |
| miR125b | downregulated | [104,109] | |
| miR96 | upregulated | [110] | |
| let-7d | downregulated | [107] | |
| miR205 | upregulated | [107] | |
| miR96 | upregulated | [109] |
Following, long non-coding RNAs consist of more than 200 nucleotides and lack protein-coding potential. They are involved in gene expression regulation at both the transcriptional and translational levels, and participate in tumorigenesis and tumor metastases [112,113]. Therefore, lncRNAs expression are promising biomarkers of cancer detection and expansion [114]. LncRNAs are found to play an important role also in HNSCC development. LncRNA ADAMTS9-AS2 expression is significantly upregulated in tongue squamous cell carcinoma (TSCC) of patients with lymph node metastases and follows poor prognostic criteria for advanced disease. The ADAMTS9-AS2 knockdown experiments in TSCC cell lines reduced the cell migration and invasion together with an inhibition of cell growth presented in vitro and in vivo models [115]. Additionally, high expression of lncRNA LINC00460 has been found in HNSCC patients and positively correlated with lymph metastases, pathological differentiation and tumor size [116]. On the other hand, in the case of laryngeal squamous cell cancer (LSCC), high expression of lncRNA MIR31HG is associated with HIF1A and p21 action which leads to an increased cancer cells proliferation [117]. Moreover, lncRNA may act as a tumor suppressor and inhibit tumor growth, e.g., overexpression of lncMX1-215 inhibits H3K27 acetylase resulting in a decreased proliferation of HNSCC cells and a reduced metastatic capacity in vitro and in vivo [118]. Furthermore, overexpression of MYOSLID lncRNA is correlated with upregulation of EMT-related markers, which points to the MYOSLID as a promising controlling biomarker of metastases in HNSCC [119]. Interestingly, Zhang et al. developed a multi-RNA-based model consisting of specific lncRNA, miRNA and mRNA with expression levels correlating with clinicopathological features of HNSCC and predicting survival risk of HNSCC [120]. To summarize, lncRNAs as well as microRNAs expression level has a potential to effectively predict the prognosis and tumorigenesis of HNSCC.
2.4. RNA Methylation
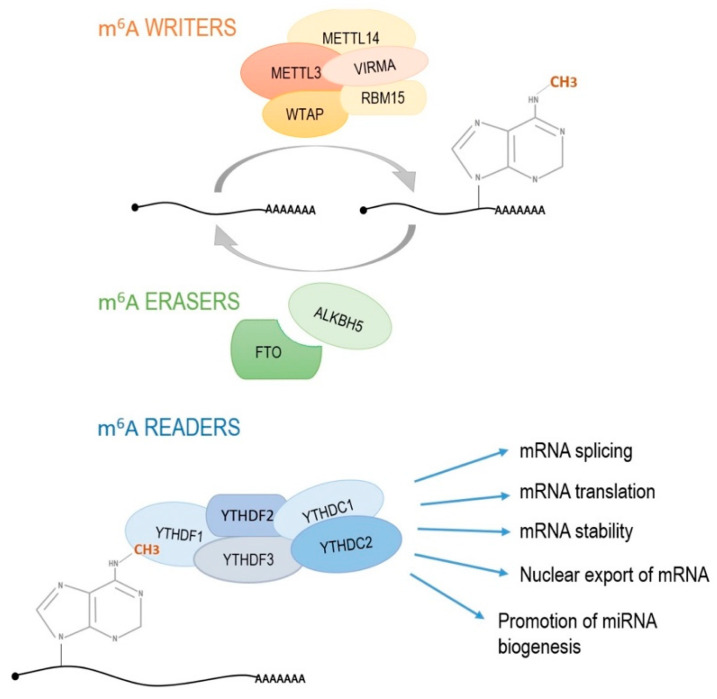
Methylation of adenosine at nitrogen-6 position (m6A) in RNA has recently received great attention from cancer researchers. In fact, the m6A has been considered as the most prevalent, dynamic and conserved internal transcriptional modification among more than 100 different chemical modifications of RNA [121,122]. Moreover, m6A is typically enriched near STOP codon and 3′UTR region containing 5′-RRACH-3′ sequence in which A3 becomes N6-methylated [123,124]. Reports suggest that this modification has been involved in all stages of RNA processing, including nuclear export, translation modulation to RNA degradation and initiation of miRNA biogenesis [125]. Additionally, m6A RNA methylation affects tumor initiation and progression by various mechanisms [126]. RNA methylation related effects are the result of the cooperation of multiprotein complexes known as “writers”, “erasers” and “readers” (Figure 3). The m6A methylase complex “writers” consist of:
-
(1)
main catalytic core enzyme which states methyltransferase like 3 (METTL3),
-
(2)
methyltransferase like 14 (METTL14) which structurally positions mRNA for methylation,
-
(3)
WT1-associated protein (WTAP) regulating the recruitment of methyltransferase complex to mRNA targets,
-
(4)
RNA-binding motif protein 15 (RBM15) which is responsible for moving the complex towards the appropriate m6A sites and the last “writer” protein,
-
(5)
Vir like m6A methyltransferase associated (VIRMA) with uncharacterized molecular function.
Figure 3.
RNA methylation process. Multiprotein complex writers are responsible for introducing the methyl group on adenine in position 6. Erasers remove the methyl group while readers recognize the presence of m6A methylation and induce processes, such as mRNA splicing, mRNA translation, mRNA stability, nuclear export of mRNA and miRNA biogenesis. METTL3, methyltransferase like 3; METTL14, methyltransferase like 14; VIRMA, Vir like m6A methyltransferase associated; WTAP, WT1-associated protein; RBM15, RNA-binding motif protein 15; FTO, fat mass and obesity-associated protein; ALKBH5, alkB homolog 5; YTHDF1-3 and YTHDC1,2, proteins with YT521-B homology (YTH) domain.
The “erasers” complex consists of demethylases FTO (fat mass and obesity-associated protein) and ALKBH5 (alkB homolog 5) which removes the methyl group. The “readers” complex which recognize the presence of the methyl group include YTHDF1-3, YTHDC1 and YTHDC2. These proteins possess YT521-B homology (YTH) domain and participate in the translation, stabilization, splicing and nuclear export of mRNA [127]. YTHDF1 recognizes m6A-modified mRNA and increases the translation efficiency. YTHDF2 recruits the CCR4-NOT deadenylase complex to destabilize and further decay target mRNAs. YTHDC1 is the nuclear m6A reader, involved in exon selection during gene splicing. In contrast, YTHDC2 is a putative RNA helicase which cooperates with the meiosis-specific coiled-coil domain-containing protein (MEIOC) and regulates mRNA level during meiosis [128].
Variations in RNA methylation process contribute to tumor growth, progression, invasion and migration of cancer cells in acute myeloid leukemia [129], glioblastoma [130], lung cancer [131], breast cancer [132], liver cancer [133], bladder cancer [134] or pancreatic cancer [135]. In terms of head and neck cancers, disorders in establishing and reading of RNA methylation have been demonstrated in the case of nasopharyngeal carcinoma (NPC) and OSCC. On the basis of the TGCA HNSCC dataset, Zhao et al. demonstrated the significant differential expression of m6A RNA methylation regulators between tumor and normal samples [136]. More specifically, Zhang et al. identified an increased level of m6A RNA methylation in the ZNF750 gene coding sequence and correlated those changes with ZNF750 lower expression in NPC. The ZNF750 overexpression experiments show cell growth inhibition in NPC in vitro and in vivo models, and indicate the importance of m6A RNA methylation in gene expression regulation [21]. Expression of m6A machinery elements has also been found to be altered in squamous cell carcinoma. In OSCC patients, METTL3 gene is significantly upregulated in cancerous tissue samples compared to healthy counterparts and these changes correlated with the poor prognosis. The overexpression of METTL3 promoted proliferation, invasion and migration of OSCC cells in vitro, whereas the METTL3 knockdown inhibited the tumor growth in vivo [137]. In addition, m6A demethylase ALKBH5 is directly upregulated by DDX3, RNA helicase, which plays an important role in cell proliferation, invasion, and metastases in several kinds of neoplasms. This regulation leads to a decreased m6A methylation in FOXM1 and NANOG nascent transcript which contribute to chemoresistance in OSCC [138]. Thus, ALKBH5 has been suggested as a potential target for novel anticancer therapies, due to a direct correlation of its expression with primary HNSCC tumor size [139]. Moreover, the m6A modification of lncRNA LNCAROD, mediated by METTL3 and METTL14, enhanced its stability in the HNSCC cells. In in vitro experiments LNCAROD silencing inhibits cell proliferation, mobility, and tumorigenicity, whereas overexpression of LNCAROD in vivo demonstrated opposite results [140]. Considering the crucial role of m6A RNA methylation in cell metabolism and unquestionable effects of the disturbances in this process concerning carcinogenesis, RNA methylation as well as RNA methylation-related mechanisms definitely will be discussed in more detail and considered as a candidate for novel, promising HNSCC biomarkers and therapy goals.
3. Conclusions
Currently, epigenetic modifications gain more interest in the HNSCC carcinogenesis. Some of them promote cancer formation and progression by controlling the expression machinery. Consequently, the detailed characteristics of the epigenetic changes in HNSCC will ultimately deliver novel, critical prognostic and predictive factors, thus providing the necessary information regarding the treatment and anti-cancer therapies. Moreover, the detailed epigenome-wide profiling may improve both the diagnosis of cancer patients and a target personalized therapy. Although presently, with limited data regarding the mechanism and prognostic value for HNSCC, the role of RNA methylation in carcinogenesis is also worth emphasizing, particularly in terms of a better understanding of the molecular basis of HNSCC and new therapy strategies.
Abbreviations
| 3′UTR | 3′ untranslated region |
| 5-caC | 5-carboxylcytosine |
| 5-fC | 5-formylcytosine |
| 5-hMC | 5-hydroxymethylcytosine |
| 5-mC | 5-methylcytosine |
| AID/APOBEC | Activation- induced deaminase/apoplipoprotein B |
| ALKBH5 | AlkB homolog 5 |
| APC | Adenomatous polyposis coli |
| ATM | Ataxia-telangiectasia-mutated |
| BER | Base excision repair |
| BRCA1 | Breast cancer type I |
| CALML5 | Calmodulin like 5 |
| CCNA1 | Cyclin-A1 |
| CDKN2A | Cyclin Dependent Kinase Inhibitor 2A |
| CSC | Cancer steam cell |
| CT | Chemotherapy |
| ctDNA | Circulating DNA |
| CTLA4 | Cytotoxic T-Lymphocyte Associated Protein 4 |
| DAPK | Death-associated protein kinase |
| DNMT | DNA methyltrasnferase |
| EGFR | Epidermal growth factor receptor |
| EMT | Epithelial-mesenchymal transition |
| FAM135B | Family with sequence similarity 135 member B |
| FTO | FTO Alpha-ketoglutarate dependent dioxygenase |
| GLI1 | GLI1 Family Zinc Finger 1 |
| HAT | Histone acetyltransferase |
| HDAC | Histone deacetylase |
| HKMT | Histone lysine methyltransferase |
| HMT | Histone methyltransferase |
| HNSCC | Head and neck squamous cell carcinoma |
| HPV | Human papilloma virus |
| hTERT | Human telomerase reverse transcriptase |
| JMJD6 | Jumonji domain containing 6 |
| KDM | Lysine specific demethylase |
| LINE-1 | Long interspersed nuclear element 1 |
| LSCC | Laryngeal squamous cell carcinoma |
| LY6D | Lymphocyte antigen 6 family member D |
| m6A | N6-methyladenosine |
| MEIOC | Meiosis specific with coiled-coli domain |
| METTL | Methyltransferase like |
| MGMT | O-6-methylguanine-DNA methyltransferase |
| miRNA | microRNA |
| MLH1 | MutL homolog 1 |
| ncRNA | Non-coding RNA |
| NF-κB | Nuclear transcription factor-κB |
| NPC | Nasopharyngeal carcinoma |
| OSCC | Oral squamous cell carcinoma |
| PADI4 | Peptidyl arginine deiminase 4 |
| piRNA | PIWI-interacting RNA |
| Pl3K/Akt | Phosphatidylinositol 3-kinase/threonine protein kinase B |
| PRMT | Histone arginine methyltransferase |
| PROM1 | Prominin 1 |
| RASSF1 | Ras association domain family member 1 |
| RB | Retinoblastoma |
| RBM15 | RNA-binding motif protein 15 |
| RISC | RNA-induced silencing complex |
| RT | Radiotherapy |
| SAM | S-adenosylomethionine |
| SAT2 | Spermidine/spermine N1-acetyltransferase family member 2 |
| siRNA | Small interfering RNA |
| SMO | smoothened |
| snoRNA | Small nuclear RNA |
| STAT3 | Signal transducer and activator of transcription 3 |
| TDG | Thymine-DNA glycosylase |
| TET | Tet-methylcytosine dioxygenase |
| TGF-β | Transforming growth factor β |
| TIMP3 | TIMP metallopeptidase inhibitor 3 |
| TRBA | TAR RNA binding protein |
| TSCC | Tongue squamous cell carcinoma |
| TSG | Tumor suppressor gene |
| WTAP | WT1-associated protein |
| VIRMA | Vir like m6A methyltransferase associated |
| YTH | YT521-B homology domain |
| ZIC4 | Zic family member 4 |
| ZNF | Zinc-finger protein |
Funding
This work was funded by The Greater Poland Cancer Centre, grant number 31/08/2020/PRB/WCO/008 and grant number 29/01/2018/PRB/WCO/003.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Marur S., Forastiere A.A. Head and neck squamous cell carcinoma: Update on epidemiology, diagnosis, and treatment. Mayo Clin. Proc. 2016;91:386–396. doi: 10.1016/j.mayocp.2015.12.017. [DOI] [PubMed] [Google Scholar]
- 2.Sessions D.G., Spector G.J., Lenox J., Haughey B., Chao C., Marks J. Analysis of treatment results for oral tongue cancer. Laryngoscope. 2002;112:616–625. doi: 10.1097/00005537-200204000-00005. [DOI] [PubMed] [Google Scholar]
- 3.Bray F., Ferlay J., Soerjomataram I. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 4.Franceschi S., Talamini R., Barra S., Barón A.E., Negri E., Bidoli E., Serraino D., La Vecchia C. Smoking and drinking in relation to cancers of the oral cavity, pharynx, larynx, and esophagus in northern Italy. Cancer Res. 1990;50:6502–6507. [PubMed] [Google Scholar]
- 5.Argiris A., Karamouzis M.V., Raben D., Ferris R.L. Head and neck cancer. Lancet. 2008;371:1695–1709. doi: 10.1016/S0140-6736(08)60728-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gillison M.L., Koch W.M., Capone R.B., Spafford M., Westra W.H., Wu L., Zahurak M.L., Daniel R.W., Viglione M., Symer D.E., et al. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J. Natl. Cancer Inst. 2000;92:709–720. doi: 10.1093/jnci/92.9.709. [DOI] [PubMed] [Google Scholar]
- 7.Sisk E.A., Bradford C.R., Carey T.E., Paulino A., Robertson E. Epstein-Barr virus detected in a head and neck squamous cell carcinoma cell line derived from an immunocompromised patient. Arch. Otolaryngol. Head Neck Surg. 2003;129:1115–1124. doi: 10.1001/archotol.129.10.1115. [DOI] [PubMed] [Google Scholar]
- 8.Moskovitz J., Moy J., Ferris R.L. Immunotherapy for Head and Neck Squamous Cell Carcinoma. Curr. Oncol. Rep. 2018;20:22. doi: 10.1007/s11912-018-0654-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alsahafi E., Begg K., Amelio I., Raulf N., Lucarelli P., Sauter T., Tavassoli M. Clinical update on head and neck cancer: Molecular biology and ongoing challenges. Cell Death Dis. 2019;10:540. doi: 10.1038/s41419-019-1769-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gleich L.L., Li Y.Q., Biddinger P.W., Gartside P.S., Stambrook P.J., Pavelic Z.P., Gluckman J.L. The loss of heterozygosity in retinoblastoma and p53 suppressor genes as a prognostic indicator for head and neck cancer. Laryngoscope. 1996;106:1378–1381. doi: 10.1097/00005537-199611000-00013. [DOI] [PubMed] [Google Scholar]
- 11.Peltonen J.K., Helppi H.M., Pääkkö P., Turpeenniemi-Hujanen T., Vähäkangas K.H. p53 in head and neck cancer: Functional consequences and environmental implications of TP53 mutations. Head Neck Oncol. 2010;2:36. doi: 10.1186/1758-3284-2-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomas S., Balan A., Balaram P. The expression of retinoblastoma tumor suppressor protein in oral cancers and precancers: A clinicopathological study. Dent. Res. J. 2015;12:307–314. doi: 10.4103/1735-3327.161427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan L.S., Fredrik P., Ker L., Yu F.G., Wang D.Y., Goh B.C., Loh K.S., Lim C.M. High-risk HPV genotypes and P16INK4a expression in a cohort of head and neck squamous cell carcinoma patients in Singapore. Oncotarget. 2016;7:86730–86739. doi: 10.18632/oncotarget.13502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bossi P., Resteghini C., Paielli N., Licitra L., Pilotti S., Perrone F. Prognostic and predictive value of EGFR in head and neck squamous cell carcinoma. Oncotarget. 2016;7:74362–74379. doi: 10.18632/oncotarget.11413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukusumi T., Califano J.A. The NOTCH Pathway in Head and Neck Squamous Cell Carcinoma. J. Dent. Res. 2018;97:645–653. doi: 10.1177/0022034518760297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marquard F.E., Jücker M. PI3K/AKT/mTOR signaling as a molecular target in head and neck cancer. Biochem. Pharmacol. 2020;172:113729. doi: 10.1016/j.bcp.2019.113729. [DOI] [PubMed] [Google Scholar]
- 17.Geiger J.L., Grandis J.R., Bauman J.E. The STAT3 pathway as a therapeutic target in head and neck cancer: Barriers and innovations. Oral Oncol. 2016;56:84–92. doi: 10.1016/j.oraloncology.2015.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cho Y.A., Kim E.K., Heo S.J., Cho B.C., Kim H.R., Chung J.M., Yoon S.O. Alteration status and prognostic value of MET in head and neck squamous cell carcinoma. J. Cancer. 2016;7:2197–2206. doi: 10.7150/jca.16686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allen C.T., Ricker J.L., Chen Z., Van Waes C. Role of activated nuclear factor-kappaB in the pathogenesis and therapy of squamous cell carcinoma of the head and neck. Head Neck. 2007;29:959–971. doi: 10.1002/hed.20615. [DOI] [PubMed] [Google Scholar]
- 20.Castilho R.M., Squarize C.H., Almeida L.O. Epigenetic Modifications and Head and Neck Cancer: Implications for Tumor Progression and Resistance to Therapy. Int. J. Mol. Sci. 2017;18:1506. doi: 10.3390/ijms18071506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang P., He Q., Lei Y., Li Y., Wen X., Hong M., Zhang J., Ren X., Wang Y., Yang X., et al. m6A-mediated ZNF750 repression facilitates nasopharyngeal carcinoma progression. Cell Death Dis. 2018;9:1169. doi: 10.1038/s41419-018-1224-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lorincz M.C., Dickerson D.R., Schmitt M., Groudine M. Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nat. Struct. Mol. Biol. 2004;11:1068–1075. doi: 10.1038/nsmb840. [DOI] [PubMed] [Google Scholar]
- 23.Antequera F. Structure, function and evolution of CpG island promoters. Cell Mol. Life Sci. 2003;60:1647–1658. doi: 10.1007/s00018-003-3088-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ha P.K., Califano J.A. Promoter methylation and inactivation of tumour-suppressor genes in oral squamous-cell carcinoma. Lancet Oncol. 2006;7:77–82. doi: 10.1016/S1470-2045(05)70540-4. [DOI] [PubMed] [Google Scholar]
- 25.Zheng Y., Joyce B.T., Liu L., Zhang Z., Kibbe W.A., Zhang W., Hou L. Prediction of genome-wide DNA methylation in repetitive elements. Nuc. Acids Res. 2017;45:8697–8711. doi: 10.1093/nar/gkx587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones P. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 27.Chodavarapu R.K., Feng S., Bernatavichute Y.V., Chen P.Y., Stroud H., Yu Y., Hetzel J.A., Kuo F., Kim J., Cokus S.J., et al. Relationship between nucleosome positioning and DNA methylation. Nature. 2010;466:388–392. doi: 10.1038/nature09147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo J.U., Su Y., Shin J.H., Shin J., Li H., Xie B., Zhong C., Hu S., Le T., Fan G., et al. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat. Neurosci. 2014;17:215–222. doi: 10.1038/nn.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng X., Blumenthal R.M. Mammalian DNA methyltransferases: A structural perspective. Structure. 2008;16:341–350. doi: 10.1016/j.str.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng J., Zhou Y., Campbell S.L., Le T., Li E., Sweatt D., Silva A.J., Fan G. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat. Neurosci. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tuorto F., Liebers R., Musch T., Schaefer M., Hofmann S., Kellner S., Frye M., Helm M., Stoecklin G., Lyko F. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat. Struct. Mol. Biol. 2012;19:900–905. doi: 10.1038/nsmb.2357. [DOI] [PubMed] [Google Scholar]
- 32.Jeltsch A., Ehrenhofer-Murray A., Jurkowski T.P., Lyko F., Reuter G., Ankri S., Nellen W., Schaefer M., Helm M. Mechanism and biological role of Dnmt2 in Nucleic Acid Methylation. RNA Biol. 2017;14:1108–1123. doi: 10.1080/15476286.2016.1191737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaiser S., Jurkowski T.P., Kellner S., Schneider D., Jeltsch A., Helm M. The RNA methyltransferase Dnmt2 methylates DNA in the structural context of a tRNA. RNA Biol. 2017;14:1241–1251. doi: 10.1080/15476286.2016.1236170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiao J., Liu Y., Wu F., Liu R., Xie Y., Yang Q., Li Y., Liu M., Li S., Tang H. miR-639 Expression Is Silenced by DNMT3A- Mediated Hypermethylation and Functions as a Tumor Suppressor in Liver Cancer Cells. Mol. Ther. 2019;28:1–12. doi: 10.1016/j.ymthe.2019.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Gowher H., Jeltsch A. Mammalian DNA methyltransferases: New discoveries and open questions. Biochem. Soc. Trans. 2018;46:1191–1202. doi: 10.1042/BST20170574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maiti A. Mechanism of Active DNA Demethylation: Recent Progress in Epigenetics. J. Biomol. Res. Ther. 2012;1:1–2. doi: 10.4172/2167-7956.1000e106. [DOI] [Google Scholar]
- 37.Rawłuszko-Wieczorek A.A., Siera A., Jagodziński P.P. TET proteins in cancer: Current ‘state of the art’. Crit. Rev. Oncol. Hematol. 2015;96:425–436. doi: 10.1016/j.critrevonc.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 38.Suzuki M.M., Bird A. DNA methylation landscapes: Provocative insights from epigenomics. Nat. Rev. Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 39.Robertson K. DNA methylation and human disease. Nat. Rev. Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 40.Gaykalova D.A., Vatapalli R., Wei Y., Tsai H.L., Wang H., Zhang C., Hennessey P.T., Guo T., Tan M., Li R., et al. Outlier Analysis Defines Zinc Finger Gene Family DNA Methylation in Tumors and Saliva of Head and Neck Cancer Patients. PLoS ONE. 2015;10 doi: 10.1371/journal.pone.0142148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sobecka A., Blaszczak W., Barczak W., Golusinski P., Rubis B., Masternak M.M., Suchorska W.M., Golusinski W. hTERT promoter methylation status in peripheral blood leukocytes as a molecular marker of head and neck cancer progression. J. Appl. Genet. 2018;59:453–461. doi: 10.1007/s13353-018-0458-1. [DOI] [PubMed] [Google Scholar]
- 42.Zhou C., Ye M., Ni S., Li Q., Ye D., Li J., Shen Z., Deng H. DNA methylation biomarkers for head and neck squamous cell carcinoma. Epigenetics. 2018;13:398–409. doi: 10.1080/15592294.2018.1465790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou C., Shen Z., Ye D., Li Q., Deng H., Liu H., Li J. The Association and Clinical Significance of CDKN2A Promoter Methylation in Head and Neck Squamous Cell Carcinoma: A Meta-Analysis. Cell Physiol. Biochem. 2018;50:868–882. doi: 10.1159/000494473. [DOI] [PubMed] [Google Scholar]
- 44.Ai L., Vo Q.N., Zuo C., Li L., Ling W., Suen J.Y., Hanna E., Brown K.D., Fan C.Y. Ataxia-telangiectasia-mutated (ATM) gene in head and neck squamous cell carcinoma: Promoter hypermethylation with clinical correlation in 100 cases. Cancer Epidemiol. Biomarkers Prev. 2004;13:150–156. doi: 10.1158/1055-9965.EPI-082-3. [DOI] [PubMed] [Google Scholar]
- 45.Cai F., Xiao X., Niu X., Shi H., Zhong Y. Aberrant Methylation of MGMT Promoter in HNSCC: A Meta-Analysis. PLoS ONE. 2016;11 doi: 10.1371/journal.pone.0163534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reis R., Santos J., Abreu P.M., Dettogni R.S., Santos E., Stur E., Agostini L.P., Anders Q.S., Alves L., Valle I., et al. Hypermethylation status of DAPK, MGMT and RUNX3 in HPV negative oral and oropharyngeal squamous cell carcinoma. Genet. Mol. Biol. 2020;43:e20190334. doi: 10.1590/1678-4685-gmb-2019-0334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koutsimpelas D., Pongsapich W., Heinrich U., Mann S., Mann W.J., Brieger J. Promoter methylation of MGMT, MLH1 and RASSF1A tumor suppressor genes in head and neck squamous cell carcinoma: Pharmacological genome demethylation reduces proliferation of head and neck squamous carcinoma cells. Oncol. Rep. 2012;27:1135–1141. doi: 10.3892/or.2012.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Basu B., Chakraborty J., Chandra A., Katarkar A., Baldevbhai J., Dhar Chowdhury D., Ray J.G., Chaudhuri K., Chatterjee R. Genome-wide DNA methylation profile identified a unique set of differentially methylated immune genes in oral squamous cell carcinoma patients in India. Clin. Epigenetics. 2017;9:13. doi: 10.1186/s13148-017-0314-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim Y.T., Park J.Y., Jeon Y.K., Park S.J., Song J.Y., Kang C.H., Sung S.W., Kim J.H. Aberrant promoter CpG island hypermethylation of the adenomatosis polyposis coli gene can serve as a good prognostic factor by affecting lymph node metastasis in squamous cell carcinoma of the esophagus. Dis. Esophagus. 2009;22:143–150. doi: 10.1111/j.1442-2050.2008.00862.x. [DOI] [PubMed] [Google Scholar]
- 50.Rettori M.M., de Carvalho A.C., Longo A.L., de Oliveira C.Z., Kowalski L.P., Carvalho A.L., Vettore A.L. TIMP3 and CCNA1 hypermethylation in HNSCC is associated with an increased incidence of second primary tumors. J. Trans. Med. 2013;11:316. doi: 10.1186/1479-5876-11-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paluszczak J., Wiśniewska D., Kostrzewska-Poczekaj M., Kiwerska K., Grénman R., Mielcarek-Kuchta D., Jarmuż-Szymczak M. Prognostic significance of the methylation of Wnt pathway antagonists-CXXC4, DACT2, and the inhibitors of sonic hedgehog signaling-ZIC1, ZIC4, and HHIP in head and neck squamous cell carcinomas. Clin. Oral Investig. 2017;21:1777–1788. doi: 10.1007/s00784-016-1946-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hu Z., Liu H., Zhang X., Hong B., Wu Z., Li Q., Zhou C. Promoter hypermethylation of CD133/PROM1 is an independent poor prognosis factor for head and neck squamous cell carcinoma. Medicine. 2020;99:e19491. doi: 10.1097/MD.0000000000019491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Khongsti S., Lamare F.A., Shunyu N.B., Ghosh S., Maitra A., Ghosh S. Whole genome DNA methylation profiling of oral cancer in ethnic population of Meghalaya, North East India reveals novel genes. Genomics. 2018;110:112–123. doi: 10.1016/j.ygeno.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 54.Smith I.M., Mydlarz W.K., Mithani S.K., Califano J.A. DNA global hypomethylation in squamous cell head and neck cancer associated with smoking, alcohol consumption and stage. Int J. Cancer. 2007;121:1724–1728. doi: 10.1002/ijc.22889. [DOI] [PubMed] [Google Scholar]
- 55.Hsiung D.T., Marsit C.J., Houseman E.A., Eddy K., Furniss C.S., McClean M.D., Kelsey K.T. Global DNA methylation level in whole blood as a biomarker in head and neck squamous cell carcinoma. Cancer Epidemiol. Biomarkers Prev. 2007;16:108–114. doi: 10.1158/1055-9965.EPI-06-0636. [DOI] [PubMed] [Google Scholar]
- 56.Langevin S.M., Koestler D.C., Christensen B.C., Butler R.A., Wiencke J.K., Nelson H.H., Houseman E.A., Marsit C.J., Kelsey K.T. Peripheral blood DNA methylation profiles are indicative of head and neck squamous cell carcinoma: An epigenome-wide association study. Epigenetics. 2012;7:291–299. doi: 10.4161/epi.7.3.19134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zheng H., Momeni A., Cedoz P.L., Vogel H., Gevaert O. Whole slide images reflect DNA methylation patterns of human tumors. NPJ Genom. Med. 2020;5:11. doi: 10.1038/s41525-020-0120-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Colacino J.A., Dolinoy D.C., Duffy S.A., Sartor M.A., Chepeha D.B., Bradford C.R., McHugh J.B., Patel D.A., Virani S., Walline H.M., et al. Comprehensive analysis of DNA methylation in head and neck squamous cell carcinoma indicates differences by survival and clinicopathologic characteristics. PLoS ONE. 2013;8:e54742. doi: 10.1371/journal.pone.0054742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Misawa K., Imai A., Matsui H., Kanai A., Misawa Y., Mochizuki D., Mima M., Yamada S., Kurokawa T., Nakagawa T., et al. Identification of novel methylation markers in HPV-associated oropharyngeal cancer: Genome-wide discovery, tissue verification and validation testing in ctDNA. Oncogene. 2020;39:4741–4755. doi: 10.1038/s41388-020-1327-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Khan S.A., Reddy D., Gupta S. Global histone post-translational modifications and cancer: Biomarkers for diagnosis, prognosis and treatment. World J. Biol. Chem. 2015;6:333–345. doi: 10.4331/wjbc.v6.i4.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Le J.M., Squarize C.H., Castilho R.M. Histone modifications: Targeting head and neck cancer stem cells. World J. Stem Cells. 2014;6:511–525. doi: 10.4252/wjsc.v6.i5.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Verdone L., Caserta M., Di Mauro E. Role of histone acetylation in the control of gene expression. Biochem. Cell Biol. 2005;83:344–353. doi: 10.1139/o05-041. [DOI] [PubMed] [Google Scholar]
- 63.Annunziato A.T., Hansen J.C. Role of histone acetylation in the assembly and modulation of chromatin structures. Gene Expr. 2000;9:37–61. doi: 10.3727/000000001783992687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marmorstein R., Zhou M.M. Writers and readers of histone acetylation: Structure, mechanism, and inhibition. Cold Spring Harb. Perspect. Biol. 2014;6 doi: 10.1101/cshperspect.a018762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hanigan T.W., Danes J.M., Taha T.Y., Frasor J., Petukhov P.A. Histone deacetylase inhibitor-based chromatin precipitation for identification of targeted genomic loci. J. Biol. Methods. 2018;5 doi: 10.14440/jbm.2018.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bredell M.G., Ernst J., El-Kochairi I., Dahlem Y., Ikenberg K., Schumann D.M. Current relevance of hypoxia in head and neck cancer. Oncotarget. 2016;7:50781–50804. doi: 10.18632/oncotarget.9549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang J.Q., Yan F.Q., Wang L.H., Yin W.J., Chang T.Y., Liu J.P., Wu K.J. Identification of new hypoxia-regulated epithelial-mesenchymal transition marker genes labeled by H3K4 acetylation. Genes Chromosomes Cancer. 2020;59:73–83. doi: 10.1002/gcc.22802. [DOI] [PubMed] [Google Scholar]
- 68.Chen F., Qi S., Zhang X., Wu J., Yang X., Wang R. lncRNA PLAC2 activated by H3K27 acetylation promotes cell proliferation and invasion via the activation of Wnt/β‑catenin pathway in oral squamous cell carcinoma. Int. J. Oncol. 2019;54:1183–1194. doi: 10.3892/ijo.2019.4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Almeida L.O., Abrahao A.C., Rosselli-Murai L.K., Giudice F.S., Zagni C., Leopoldino A.M., Squarize C.H., Castilho R.M. NFκB mediates cisplatin resistance through histone modifications in head and neck squamous cell carcinoma (HNSCC) FEBS Open Bio. 2013;4:96–104. doi: 10.1016/j.fob.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Giudice F.S., Pinto D.S. Jr Nör, J.E.; Squarize, C.H.; Castilho, R.M. Inhibition of histone deacetylase impacts cancer stem cells and induces epithelial-mesenchyme transition of head and neck cancer. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0058672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.He L., Gao L., Shay C., Lang L., Lv F., Teng Y. Histone deacetylase inhibitors suppress aggressiveness of head and neck squamous cell carcinoma via histone acetylation-independent blockade of the EGFR-Arf1 axis. J. Exp. Clin. Cancer Res. 2019;38:84. doi: 10.1186/s13046-019-1080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jambhekar A., Dhall A., Shi Y. Roles and regulation of histone methylation in animal development. Nat. Rev. Mol. Cell Biol. 2019;20:625–641. doi: 10.1038/s41580-019-0151-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Peters A.H., Kubicek S., Mechtler K., O’Sullivan R.J., Derijck A.A., Perez-Burgos L., Kohlmaier A., Opravil S., Tachibana M., Shinkai Y., et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell. 2003;12:1577–1589. doi: 10.1016/S1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 74.Bannister A.J., Schneider R., Myers F.A., Thorne A.W., Crane-Robinson C., Kouzarides T. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J. Biol. Chem. 2005;280:17732–17736. doi: 10.1074/jbc.M500796200. [DOI] [PubMed] [Google Scholar]
- 75.Song Y., Wu F., Wu J. Targeting histone methylation for cancer therapy: Enzymes, inhibitors, biological activity and perspectives. J. Hematol. Oncol. 2016;9:49. doi: 10.1186/s13045-016-0279-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang J., Jing L., Li M., He L., Guo Z. Regulation of histone arginine methylation/demethylation by methylase and demethylase. Mol. Med. Rep. 2019;19:3963–3971. doi: 10.3892/mmr.2019.10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cai L., Ma X., Huang Y., Zou Y., Chen X. Aberrant histone methylation and the effect of Suv39H1 siRNA on gastric carcinoma. Oncol. Rep. 2014;31:2593–2600. doi: 10.3892/or.2014.3135. [DOI] [PubMed] [Google Scholar]
- 78.Zhang L., Deng L., Chen F., Yao Y., Wu B., We L., Mo Q., Song Y. Inhibition of histone H3K79 methylation selectively inhibits proliferation, self-renewal and metastatic potential of breast cancer. Oncotarget. 2014;5:10665–10677. doi: 10.18632/oncotarget.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kang K.A., Piao M.J., Ryu Y.S., Kang H.K., Young W. Interaction of DNA demethylase and histone methyltransferase upregulates Nrf2 in 5-fluorouracil-resistant colon cancer cells. Oncotarget. 2016;7:40594–40620. doi: 10.18632/oncotarget.9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mizuno Y., Maemura K., Tanaka Y., Hirata A., Futaki S., Hamamoto H., Taniguchi K., Hayashi M., Uchiyama K., Shibata M.A., et al. Expression of delta-like 3 is downregulated by aberrant DNA methylation and histone modification in hepatocellular carcinoma. Oncol. Rep. 2018;39:2209–2216. doi: 10.3892/or.2018.6293. [DOI] [PubMed] [Google Scholar]
- 81.Mancuso M., Matassa D.S., Conte M., Colella G., Rana G., Fucci L., Piscopo M. H3K4 histone methylation in oral squamous cell carcinoma. Acta Biochem. Pol. 2009;56:405–410. doi: 10.18388/abp.2009_2473. [DOI] [PubMed] [Google Scholar]
- 82.Liu S., Ye D., Guo W., Yu W., He Y., Hu J., Wang Y., Zhang L., Liao Y., Song H., et al. G9a is essential for EMT-mediated metastasis and maintenance of cancer stem cell-like characters in head and neck squamous cell carcinoma. Oncotarget. 2015;6:6887–6901. doi: 10.18632/oncotarget.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lindsay C.D., Kostiuk M.A., Harris J., O’Connell D.A., Seikaly H., Biron V.L. Efficacy of EZH2 inhibitory drugs in human papillomavirus-positive and human papillomavirus-negative oropharyngeal squamous cell carcinomas. Clin. Epigenetics. 2017;9:95. doi: 10.1186/s13148-017-0390-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Marcinkiewicz K.M., Gudas L.J. Altered histone mark deposition and DNA methylation at homeobox genes in human oral squamous cell carcinoma. J. Cell Physiol. 2014;229:1405–1416. doi: 10.1002/jcp.24577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang K.C., Chang H.Y. Molecular mechanisms of long non-coding RNAs. Mol. Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bartel D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 87.Hammond S.M. MicroRNAs as tumor suppressors. Nat. Genet. 2007;39:582–583. doi: 10.1038/ng0507-582. [DOI] [PubMed] [Google Scholar]
- 88.Hanahan D., Weinberg R.A. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 89.Lee Y., Kim M., Han J., Yeom K.H., Lee S., Baek S.H., Kim V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bohnsack M.T., Czaplinski K., Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cannell I.G., Kong Y.W., Bushell M. How do microRNAs regulate gene expression? Biochem. Soc. Trans. 2008;36:1224–1231. doi: 10.1042/BST0361224. [DOI] [PubMed] [Google Scholar]
- 92.Schneider A., Victoria B., Lopez Y.N., Suchorska W., Barczak W., Sobecka A., Golusinski W., Masternak M.M., Golusinski P. Tissue and serum microRNA profile of oral squamous cell carcinoma patients. Sci. Rep. 2018;8:675. doi: 10.1038/s41598-017-18945-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lubov J., Maschietto M., Ibrahim I., Mlynarek A., Hier M., Kowalski L.P., Alaoui-Jamali M.A., da Silva S.D. Meta-analysis of microRNAs expression in head and neck cancer: Uncovering association with outcome and mechanisms. Oncotarget. 2017;8:55511–55524. doi: 10.18632/oncotarget.19224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sobecka A., Barczak W., Suchorska W.M. RNA interference in head and neck oncology. Oncol. Lett. 2016;12:3035–3040. doi: 10.3892/ol.2016.5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Langevin S.M., Stone R.A., Bunker C.H., Lyons-Weiler M.A., LaFramboise W.A., Kelly L., Seethala R.R., Grandis J.R., Sobol R.W., Taioli E. MicroRNA-137 promoter methylation is associated with poorer overall survival in patients with squamous cell carcinoma of the head and neck. Cancer. 2011;1:1454–1462. doi: 10.1002/cncr.25689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Metheetrairut C., Chotigavanich C., Amornpichetkul K., Keskool P., Ongard S., Metheetrairut C. Expression levels of miR-34-family microRNAs are associated with TP53 mutation status in head and neck squamous cell carcinoma. Eur. Arch. Otorhinolaryngol. 2019;276:521–533. doi: 10.1007/s00405-018-5223-x. [DOI] [PubMed] [Google Scholar]
- 97.Liu F., Zhang F., Li X., Liu Q., Liu W., Song P., Qiu Z., Dong Y., Xiang H. Prognostic role of miR-17-92 family in human cancers: Evaluation of multiple prognostic outcomes. Oncotarget. 2017;8:69125–69138. doi: 10.18632/oncotarget.19096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gee H.E., Camps C., Buffa F.M., Patiar S., Winter S.C., Betts G., Homer J., Corbridge R., Cox G., West C.M.L., et al. hsa-mir-210 Is a Marker of Tumor Hypoxia and a Prognostic Factor in Head and Neck Cancer. Cancer. 2010;116:2148–2158. doi: 10.1002/cncr.25009. [DOI] [PubMed] [Google Scholar]
- 99.Koshizuka K., Kikkawa N., Hanazawa T., Yamada Y., Okato A., Arai T., Katada K., Okamoto Y., Seki N. Inhibition of integrin β1-mediated oncogenic signalling by the antitumor microRNA-29 family in head and neck squamous cell carcinoma. Oncotarget. 2017;11:3663–3676. doi: 10.18632/oncotarget.23194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Arantes L.M., Laus A.C., Melendez M.E., de Carvalho A.C., Sorroche B.P., De Marchi P.R., Evangelista A.F., Scapulatempo-Neto C., de Souza Viana L., Carvalho A.L. MiR-21 as prognostic biomarker in head and neck squamous cell carcinoma patients undergoing an organ preservation protocol. Oncotarget. 2017;7:9911–9921. doi: 10.18632/oncotarget.14253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Arunkumar G., Deva Magendhra Rao A.K., Manikandan M., Prasanna Srinivasa Rao H., Subbiah S., Ilangovan R., Murugan A.K., Munirajan A.K. Dysregulation of miR-200 family microRNAs and epithelial-mesenchymal transition markers in oral squamous cell carcinoma. Oncol. Lett. 2018;15:649–657. doi: 10.3892/ol.2017.7296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen C.Y., Chang J.T., Ho Y.F., Shyu A.B. MiR-26 down-regulates TNF-α/NF-κB signalling and IL-6 expression by silencing HMGA1 and MALT1. Nucleic. Acids Res. 2016;44:3772–3787. doi: 10.1093/nar/gkw205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kinoshita T., Hanazawa T., Nohata N., Kikkawa N., Enokida H., Yoshino H., Yamasaki T., Hidaka H., Nakagawa M., Okamoto Y., et al. Tumor suppressive microRNA-218 inhibits cancer cell migration and invasion through targeting laminin-332 in head and neck squamous cell carcinoma. Oncotarget. 2012;3:1386–1400. doi: 10.18632/oncotarget.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chen Y.F., Wei Y.Y., Yang C.C., Liu C.J., Yeh L.Y., Chou C.H., Chang K.W., Lin S.C. miR-125b suppresses oral oncogenicity by targeting the anti-oxidative gene PRXL2A. Redox Biol. 2019;22:101140. doi: 10.1016/j.redox.2019.101140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.González-Arriagada W.A., Olivero P., Rodríguez B., Lozano-Burgos C., de Oliveira C.E., Coletta R.D. Clinicopathological significance of miR-26, miR-107, miR-125b, and miR-203 in head and neck carcinomas. Oral Dis. 2018;24:930–939. doi: 10.1111/odi.12872. [DOI] [PubMed] [Google Scholar]
- 106.Sannigrahi M.K., Sharma R., Singh V., Panda N.K., Rattan V., Khullar M. Role of Host miRNA Hsa-miR-139-3p in HPV-16-Induced Carcinomas. Clin. Cancer Res. 2017;23:3884–3895. doi: 10.1158/1078-0432.CCR-16-2936. [DOI] [PubMed] [Google Scholar]
- 107.Kolenda T., Guglas K., Teresiak A., Bliźniak R., Lamperska K. Low let-7d and high miR-205 expression levels positively influence HNSCC patient outcome. J. Biomed. Sci. 2019;26:17. doi: 10.1186/s12929-019-0511-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hess A.K., Müer A., Mairinger F.D., Weichert W., Stenzinger A., Hummel M., Budach V., Tinhofer I. MiR-200b and miR-155 as predictive biomarkers for the efficacy of chemoradiation in locally advanced head and neck squamous cell carcinoma. Eur. J. Cancer. 2017;77:3–12. doi: 10.1016/j.ejca.2017.02.018. [DOI] [PubMed] [Google Scholar]
- 109.Vahabi M., Pulito C., Sacconi A., Donzelli S., D’Andrea M., Manciocco V., Pellini R., Paci P., Sanguineti G., Strigari L., et al. miR-96-5p targets PTEN expression affecting radio-chemosensitivity of HNSCC cells. J. Exp. Clin. Cancer Res. 2019;29:141. doi: 10.1186/s13046-019-1119-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rather M.I., Nagashri M.N., Swamy S.S., Gopinath K.S., Kumar A. Oncogenic microRNA-155 down-regulates tumor suppressor CDC73 and promotes oral squamous cell carcinoma cell proliferation: Implications for cancer therapeutics. J. Biol. Chem. 2013;288:608–618. doi: 10.1074/jbc.M112.425736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang L.L., Li H.X., Yang Y.Y., Su Y.L., Lian J.S., Li T., Xu J., Wang X.N., Jin N., Liu X.F. MiR-31 is a potential biomarker for diagnosis of head and neck squamous cell carcinoma. Int. J. Clin. Exp. Pathol. 2018;1:4339–4345. [PMC free article] [PubMed] [Google Scholar]
- 112.Schmitt A.M., Chang H.Y. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jiang M.C., Ni J.J., Cui W.Y., Wang B.Y., Zhuo W. Emerging roles of lncRNA in cancer and therapeutic opportunities. Am. J. Cancer Res. 2019;9:1354–1366. [PMC free article] [PubMed] [Google Scholar]
- 114.Yang B., Shen J., Xu L., Chen Y., Che X., Qu X., Liu Y., Teng Y., Li Z. Genome-Wide Identification of a Novel Eight-lncRNA Signature to Improve Prognostic Prediction in Head and Neck Squamous Cell Carcinoma. Front. Oncol. 2019;18:898. doi: 10.3389/fonc.2019.00898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Li Y., Wan Q., Wang W., Mai L., Sha L., Mashrah M., Lin Z., Pan C. LncRNA ADAMTS9-AS2 promotes tongue squamous cell carcinoma proliferation, migration and EMT via the miR-600/EZH2 axis. Biomed. Pharmacother. 2019;112 doi: 10.1016/j.biopha.2019.108719. [DOI] [PubMed] [Google Scholar]
- 116.Jiang Y., Cao W., Wu K., Qin X., Wang X., Li Y., Binbin Y., Zhang Z., Wang X., Yan H., et al. LncRNA LINC00460 promotes EMT in head and neck squamous cell carcinoma by facilitating peroxiredoxin-1 into the nucleus. J. Exp. Clin. Cancer Res. 2019;38:365. doi: 10.1186/s13046-019-1364-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang R., Ma Z., Feng L., Yang Y., Tan C., Shi Q., Lian M., He S., Ma H., Fang J. LncRNA MIR31HG targets HIF1A and P21 to facilitate head and neck cancer cell proliferation and tumorigenesis by promoting cell-cycle progression. Mol. Cancer. 2018;17:162. doi: 10.1186/s12943-018-0916-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ma H., Chang H., Yang W., Lu Y., Hu J., Jin S. A novel IFNα-induced long non-coding RNA negatively regulates immunosuppression by interrupting H3K27 acetylation in head and neck squamous cell carcinoma. Mol. Cancer. 2020;19:4. doi: 10.1186/s12943-019-1123-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Xiong H.G., Li H., Xiao Y., Yang Q.C., Yang L.L., Chen L., Bu L.L., Zhang W.F., Zhang J.L., Sun Z.J. Long noncoding RNA MYOSLID promotes invasion and metastasis by modulating the partial epithelial-mesenchymal transition program in head and neck squamous cell carcinoma. J. Exp. Clin. Cancer Res. 2019;38:278. doi: 10.1186/s13046-019-1254-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang C., Cao W., Wang J., Liu J., Liu J., Wu H., Li S., Zhang C. A prognostic long non-coding RNA-associated competing endogenous RNA network in head and neck squamous cell carcinoma. Peer J. 2020;8:e9701. doi: 10.7717/peerj.9701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Song J., Yi C. Chemical Modifications to RNA: A New Layer of Gene Expression Regulation. ACS Chem. Biol. 2017;12:316–325. doi: 10.1021/acschembio.6b00960. [DOI] [PubMed] [Google Scholar]
- 122.Mongan N.P., Emes R.D., Archer N. Detection and analysis of RNA methylation. F1000Res. 2019;8 doi: 10.12688/f1000research.17956.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ke S., Alemu E.A., Mertens C., Gantman E.C., Fak J.J., Mele A., Haripal B., Zucker-Scharff I., Moore M.J., Park C.Y., et al. A majority of m6A residues are in the last exons, allowing the potential for 3’ UTR regulation. Genes Dev. 2015;1:2037–2053. doi: 10.1101/gad.269415.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., Cesarkes K., Jacob-Hirsch J., Amariglio N., Kupiec M., et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 125.Yue Y., Liu J., He C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 2015;29:1343–1355. doi: 10.1101/gad.262766.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chen X.Y., Zhang J., Zhu J.S. The role of m6A RNA methylation in human cancer. Mol. Cancer. 2019;18:103. doi: 10.1186/s12943-019-1033-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lesbirel S., Wilson S.A. The m6A‑methylase complex and mRNA export. Biochim. Biophys. Acta Gene Regul. Mech. 2019;1862:319–328. doi: 10.1016/j.bbagrm.2018.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Liao S., Sun H., Xu C. YTH Domain: A Family of N 6 -methyladenosine (m6A) Readers. Geno. Prot. Bioin. 2018;16:99–107. doi: 10.1016/j.gpb.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Paris J., Morgan M., Campos J., Spencer G.J., Shmakova A., Ivanova I., Mapperley C., Lawson H., Wotherspoon D.A., Sepulveda C., et al. Targeting the RNA m6A Reader YTHDF2 Selectively Compromises Cancer Stem Cells in Acute Myeloid Leukemia. Cell Stem Cell. 2019;3:137–148. doi: 10.1016/j.stem.2019.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cui Q., Shi H., Ye P., Li L., Qu Q., Sun G., Sun G., Lu Z., Huang Y., Yang C.G., et al. m6A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells. Cell Rep. 2017;14:2622–2634. doi: 10.1016/j.celrep.2017.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Liu J., Ren D., Du Z., Wang H., Zhang H., Jin Y. m6A demethylase FTO facilitates tumor progression in lung squamous cell carcinoma by regulating MZF1 expression. Biochem. Biophys. Res. Commun. 2018;502:456–464. doi: 10.1016/j.bbrc.2018.05.175. [DOI] [PubMed] [Google Scholar]
- 132.Wu L., Wu D., Ning J., Liu W., Zhang D. Changes of N6-methyladenosine modulators promote breast cancer progression. BMC Cancer. 2019;19:326. doi: 10.1186/s12885-019-5538-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chen M., Wei L., Law C.T., Tsang F.H., Shen J., Cheng C.L., Tsang L.H., Ho D.W., Chiu D.K., Lee J.M., et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent post-transcriptional silencing of SOCS2. Hepatology. 2018;67:2254–2270. doi: 10.1002/hep.29683. [DOI] [PubMed] [Google Scholar]
- 134.Han J., Wang J.Z., Yang X., Yu H., Zhou R., Lu H.C., Yuan W.B., Lu J.C., Zhou Z.J., Lu Q., et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol. Cancer. 2019;22:110. doi: 10.1186/s12943-019-1036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Taketo K., Konno M., Asai A., Koseki J., Toratani M., Satoh T., Doki Y., Mori M., Ishii H., Ogawa K. The epitranscriptome m6A writer METTL3 promotes chemo- and radioresistance in pancreatic cancer cells. Int. J. Oncol. 2018;52:621–629. doi: 10.3892/ijo.2017.4219. [DOI] [PubMed] [Google Scholar]
- 136.Zhao X., Cui L. Development and validation of a m6A RNA methylation regulators-based signature for predicting the prognosis of head and neck squamous cell carcinoma. Am. J. Cancer Res. 2019;9:2156–2169. [PMC free article] [PubMed] [Google Scholar]
- 137.Zhao W., Cui Y., Liu J., Ma X., Qi X., Wang Y., Liu Z., Ma S., Liu J., Wu J. METTL3 facilitates oral squamous cell carcinoma tumorigenesis by enhancing c-myc syability via YTHDF1-mediated m6A modification. Mol. Ther. Nucleic Acids. 2020;20:1–12. doi: 10.1016/j.omtn.2020.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Shriwas O., Priyadarshini M., Samal S.K., Rath R., Panda S., Das Majumdar S.K., Muduly D.K., Botlagunta M., Dash R. DDX3 modulates cisplatin resistance in OSCC through ALKBH5-mediated m6A-demethylation of FOXM1 and NANOG. Apoptosis. 2020;25:233–246. doi: 10.1007/s10495-020-01591-8. [DOI] [PubMed] [Google Scholar]
- 139.Pilžys T., Marcinkowski M., Kukwa W., Garbicz D., Dylewska M., Ferenc K., Mieczkowski A., Kukwa A., Migacz E., Wołosz D., et al. ALKBH overexpression in head and neck cancer: Potential target for novel anticancer therapy. Sci. Rep. 2019;9:13249. doi: 10.1038/s41598-019-49550-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ban Y., Tan P., Cai J., Li J., Hu M., Zhou Y., Mei Y., Tan Y., Li X., Zeng Z., et al. LNCAROD is stabilized by m6A methylation and promotes cancer progression via forming a ternary complex with HSPA1A and YBX1 in head and neck squamous cell carcinoma. Mol. Oncol. 2020;14:1282–1296. doi: 10.1002/1878-0261.12676. [DOI] [PMC free article] [PubMed] [Google Scholar]