Abstract
Botrytis cinerea is a necrotrophic pathogen that causes grey mold in many plant species, including crops and model plants of angiosperms. B. cinerea also infects and colonizes the bryophyte Physcomitrium patens (previously Physcomitrella patens), which perceives the pathogen and activates defense mechanisms. However, these defenses are not sufficient to stop fungal invasion, leading finally to plant decay. To gain more insights into B. cinerea infection and virulence strategies displayed during moss colonization, we performed genome wide transcriptional profiling of B. cinerea during different infection stages. We show that, in total, 1015 B. cinerea genes were differentially expressed in moss tissues. Expression patterns of upregulated genes and gene ontology enrichment analysis revealed that infection of P. patens tissues by B. cinerea depends on reactive oxygen species generation and detoxification, transporter activities, plant cell wall degradation and modification, toxin production and probable plant defense evasion by effector proteins. Moreover, a comparison with available RNAseq data during angiosperm infection, including Arabidopsis thaliana, Solanum lycopersicum and Lactuca sativa, suggests that B. cinerea has virulence and infection functions used in all hosts, while others are more specific to P. patens or angiosperms.
Keywords: Botrytis cinerea, transcriptome, P. patens, angiosperms, ROS, cell wall degrading enzymes, transporters, virulence, secretome
1. Introduction
Botrytis cinerea is a necrotrophic fungus that causes grey mold and infects more than 1000 plant species, including model plants and many crops such as tomato, lettuce and berries [1,2,3]. It is found worldwide causing disease in fruits, flowers and leaves, leading to pre- and post-harvest crop losses [4,5,6]. B. cinerea is considered the second most important plant-pathogenic fungus, based on scientific and economic significance [2], and therefore it is an extensively studied plant pathogen. This fungus mainly enters the plant via direct penetration or through natural openings or wounds [7]. This ascomycete is a necrotrophic pathogen with a broad host range that kills the plant cells and colonizes the dead tissues to acquire nutrients [7]. B. cinerea stimulates reactive oxygen species (ROS) production by the plant and exploits the host programmed cell death (PCD) machinery to cause infection [8,9]. The infection strategies are well described and they rely on several virulence factors, including toxins and plant cell wall degrading enzymes (PCWDEs), necrosis-secreted proteins, transporter proteins and enzymes that protect the fungus from oxidative stress [10]. After landing on the plant surface, spores germinate and appresoria-mediated penetration involves the formation of a penetration peg needed to enter into the host cells [11]. CWDEs such as pectinases, including poly-galacturonases (PGs) and pectate and pectin lyases, as well as cellulases, xylanases, cutinases, lipases, and proteases, are produced to breach the plant surface, allowing plant tissue colonization and release of carbohydrates for consumption [9]. The high capacity for plant cell wall degradation and modification of glycoconjugates and polysaccharides by B. cinerea was demonstrated by the presence in its genome of 367 genes encoding putative CAZymes (Carbohydrate-Active enZymes) [12]. Most CWDEs and cutinases are encoded by multigenic families, and mutants in these genes do not always affect fungal virulence, indicating that some of these enzymes may have partly redundant functions [9,13,14,15,16]. Host cell death is facilitated by the action of fungal phytotoxins, including the two main toxins: botrydial and botcinic acid [17,18]. B. cinerea produces oxalic acid leading to acidification of the plant tissue [19], and mutants unable to produce this metabolite do not colonize the host tissues [20].
Plant cells recognize B. cinerea rapidly and mount a defense response activating the production of antimicrobial compounds and antimicrobial Pathogenesis-Related proteins (PRs), increasing the number of hormones such as salicylic acid (SA), jasmonic acid (JA), ethylene (ET) and brassinosteroids (BR), and other genes with different roles in defense [21]. In turn, B. cinerea suppresses plant defenses in early stages by producing small RNAs and effector proteins, which enables the fungus to establish inside the host tissues and accumulate biomass prior to the necrotrophic phase [22]. In addition to angiosperms, B. cinerea also infects and colonizes mosses in nature (Polytrichum juniperinum; [23]), and under axenic laboratory conditions (Physcomitrium patens (previously Physcomitrella patens); [24]). Mosses, liverworts and hornworts are bryophytes and, due to their key phylogenetic position as members of the sister lineage of vascular plants, they represent ideal organisms to perform evolutionary studies on defense mechanisms activated in plants in response to biotic stress. Fossil records suggest the existence of pathogenic fungi interacting with plants 400 million years ago [25]. These microorganisms might have imposed a selective pressure on early land plants, leading to the development of plant defense mechanisms to respond to microbial infection. Similar as in angiosperms, B. cinerea colonization leads to host cell death and necrosis of P. patens tissues [24,26]. After landing on the moss surface, spores germinate, germ tubes elongate and appressoria penetrate the plant cell cytoplasm directly through the plant leaf cell walls, or by invasion of the intercellular spaces between leaf cells leading to cytoplasm infection [27]. In response to infection, P. patens activates a defense response that includes the production of ROS, SA and the precursor of JA cis-oxophytodienoic acid (OPDA), reinforcement of the cell walls, and induction of genes encoding PRs and enzymes involved in secondary metabolism [24,26]. However, the activation of these defense mechanisms is not sufficient to stop B. cinerea growth and colonization, leading to plant decay. Recently, we have shown that P. patens responds to this fungal pathogen with transcriptional reprogramming of 3072 plant genes, which encode proteins involved in pathogen perception, signaling, transcription, hormonal signaling, secondary metabolic pathways and proteins with diverse role in defense against biotic stress [28]. To gain more insights into B. cinerea infection and virulence strategies displayed during moss colonization, we focused on the transcriptional response of B. cinerea genes during the infection process of P. patens. The results demonstrate that fungal genes encoding enzymes involved in ROS production and detoxification, CAZymes, and other virulence factors such as transporters, toxins and cell death inducing factors, were upregulated in P. patens tissues. Moreover, comparative analysis of B. cinerea-upregulated genes during P. patens and three angiosperms infections suggests that some genes involved in the infection process and virulence are commonly upregulated in all plant hosts, while other genes with these functions are more specific to P. patens or angiosperms.
2. Materials and Methods
2.1. B. cinerea, P. patens Inoculation and Microscopy
A B. cinerea strain isolated from lemon plants [24] was cultured on potato dextrose agar (PDA) at 22 °C, with a photoperiod of 16-h light/8-h dark. To inoculate P. patens, a spore suspension of B. cinerea was prepared from a 12-day-old culture. For this, P. patens Gransden wild type colonies were cultivated axenically on cellophane overlaid solid BCDAT medium (1.6 g L−1 Hoagland’s, 1 mM MgSO4, 1.8 mM KH2PO4 pH 6.5, 10 mM KNO3, 45 μM FeSO4, 1 mM CaCl2, 5 mM ammonium tartrate, and 10 g L−1 agar) as described by Ashton and Cove [29]. Moss colonies were grown at 22 °C under standard long-day conditions (16-h light/8-h dark regime under 60–80 μmol m2 s−1 white light). After 3 weeks of growth, colonies were sprayed with a 2 × 105 spores/mL suspension of B. cinerea and each colony received approximately 5000 spores. Tissues were inoculated 5 h after the start of photoperiod. Three time points were analyzed; 4 h post inoculation (hpi), 8 hpi and 24 hpi. B. cinerea mycelium grown on PDA plates for 12 days was used as a control. Three independent biological replicates, consisting of 3 plates with 16 moss colonies each, at each infection time point, and mycelium grown on PDA plates were harvested for RNA extraction, immediately frozen in liquid nitrogen, and stored at −80˚C.
B. cinerea tissues were stained with 0.1% solophenyl flavine 7GFE 500 according to [26]. Fluorescence microscopy was performed with an Olympus BX61 microscope (Shinjuku-ku, Japan). Photographs were taken at 4 hpi, 8 hpi and 24 hpi.
2.2. RNA Extraction, RNA Sequencing, Data Processing and qPCR Analysis of P. patens-Infected Tissues
RNA was extracted using the RNeasy Plant Mini Kit, including a RNase-Free DNase I digestion in column (Qiagen, Germany). RNA quality control, library preparation, and sequencing were performed at Macrogen Inc. (Seoul, Korea). RNA integrity was checked before library preparation using an Agilent Technologies 2100 Bioanalyzer (Agilent Technologies). Libraries for each biological replicate were prepared for paired-end sequencing by TruSeq Stranded Total RNA LT Sample Prep Kit (Plant) with 1 μg input RNA, following the TruSeq Stranded Total RNA Sample Prep Guide, Part # 15,031,048 Rev. E. Sequencing was performed on Illumina platform (Illumina, CA, USA) by Macrogen Inc. (Seoul, Korea) to generate paired-end 101 bp reads, obtaining 41 to 64 M reads per sample with Q20 > 98.43%. RNA-seq processing steps were done through Galaxy platform (https://usegalaxy.org/). Raw reads quality was revised by FastQC software ver. 0.11.2 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and then preprocessed for both quality and adapter trimmings using Trimmomatic Version 0.38.0 software [30]. Additionally, to the default options, the following parameters were adjusted: adapter sequence TruSeq3 (paired-ended, for MiSeq and HiSeq), always keep both reads of PE, and SLIDINGWINDOW: 4:15 HEADCROP:12 MINLEN:50. Trimmed reads were mapped to the reference genome of B. cinerea isolate B05.10 (ASM14353v4) (http://fungi.ensembl.org/Botrytis_cinerea/Info/Index) using Hisat2 software [31]. The BAM files were obtained with Samtools View software ver. 1.9 and then sorted by name with Samtools Sort software ver. 2.0.3 [32], for further analysis. All raw RNA-Seq read data are deposited in the National Center for Biotechnology Information (NCBI) Short Read Archive (http://www.ncbi.nlm.nih.gov/sra/) under the BioProject accession code PRJNA647932.
Reads were counted using FeatureCounts software ver. 1.6.4 [33]. Additionally to default options, parameters were adjusted for: count fragments instead of reads, allow read to map to multiple features, and use reference sequence file GCA_000143535.4_ASM14353v4_genomic. Cluster analysis of replicates from each time point and control samples was performed by Principal Component Analysis (PCA) using pcaExplorer 2.16.0 software [34]. Differential expression analyses were performed using EdgeR software ver. 3.24.1 [35], with p-value adjusted threshold 0.05, p-value adjusted method of Benjamini and Hochberg [36] and Minimum log2 Fold Change 2. Counts were normalized to counts per million (cpm) with the TMM method and low expressed genes filtered for count values ≥ 3 in all samples. In this study, a false discovery rate (FDR) ≤ 0.05 was used to determine significant differentially expressed genes (DEGs) between B. cinerea grown on P. patens and B. cinerea grown on PDA (control), and expression values were represented by log2 ratio.
Expression level of 16 selected B. cinerea genes related to pathogenesis was analyzed to validate RNAseq results via quantitative reverse transcription PCR (RT-qPCR). cDNA was generated from 1 μg of RNA using RevertAid Reverse transcriptase (Thermo Scientific) and oligo (dT) according to the manufacturer’s protocol. RT-qPCR was performed using the QuantiNova Probe SYBR Green PCR Kit (Qiagen, Germany), according to manufacturer’s instructions, in an Applied Biosystems QuantStudio 3 thermocycler. Relative expression of each gene was normalized to the quantity of constitutively expressed BctubB, using the 2−ΔΔCt method [37]. Gene expression of B. cinerea grown on plant was expressed relative to B. cinerea grown on PDA, with its expression level set to one. Each data point is the mean value of three biological replicates. The significance for quantitative gene expression analysis was determined with Student’s t-test using GraphPad Prism software ver. 8.0.2. p-values < 0.05 were considered statistically significant. Primer pairs used for qPCR analyses are provided in Supplementary Table S1. All primer combinations showed amplification efficiencies greater than 95%.
2.3. RNA-seq Data from B. cinerea Grown on A. thaliana, S. lycopersicum, and L. sativa and Comparison with B. cinerea Grown on P. patens
RNA-seq Illumina sequence reads from B. cinerea grown in Arabidopsis thaliana [38], Lactuca sativa (lettuce) [39], and Solanum lycopersicum (tomato) [40], were obtained from NCBI. These plant hosts were chosen since A. thaliana represents a model plant for angiosperms, and S. lycopersicum and L. sativa are two important crops affected by B. cinerea. In all cases data correspond to plant leaves inoculated with a spore suspension of B. cinerea strain B05.10, using 1 × 105 spores/mL for A. thaliana (2000 spores per leaf), 2 × 105 spores/mL for S. lycopersicum (6000 spores per leaf) and 5 × 105 spores/mL for L. sativa (10.000 spores per leaf). The closest available time points after inoculation compared to our B. cinerea transcriptomes in P. patens were considered; A. thaliana 12 and 24 hpi, S. lycopersicum 16 and 23 hpi, and L. sativa 24 and 48 hpi. Hence, in this study the corresponding RNA-seq were analyzed: A. thaliana at 12 hpi (SRR3383472, SRR3383521, SRR3383545) and 24 hpi (SRR3383805, SRR3383815, SRR3383816), L. sativa at 24 hpi (SRA059059, split and extract data using barcode 24hpib1 CCGTCC, 24hpib2 GTCCGC and 24hpib3 GTGGCC) and 48 hpi (SRA059059, split and extract data using barcode: 48hpib1 CTTGTA, 48hpib2 TTAGGC and 48hpib3 GATCAG), S. lycopersicum at 16 hpi (SRR12676683, SRR12676682, SRR12676681) and 23 hpi (SRR12676681, SRR12676679, SRR12676678). RNA-seq processing steps were done through Galaxy platform (https://usegalaxy.org/) using the same pipeline as described before for P. patens RNAseq processing; setting TruSeq3 for single-ended in Trimmomatic 0.38.0 in accordance to Illumina single-end sequencing data available for A. thaliana, L. sativa and S. lycopersicum. Differential expression analyses were performed using EdgeR software ver. 3.24.1, using the same parameters as for P. patens analysis (p-value adjusted threshold 0.05, p-value adjusted by Benjamini and Hochberg method and Minimum log2 Fold Change 2). Counts were normalized to counts per million (cpm) with TMM method and low expressed genes filtered for count values ≥ 3 in all samples; a FDR ≤ 0.05 was used to determine significant differentially expressed genes (DEGs) between B. cinerea grown on A. thaliana, L. sativa, S. lycopersicum and B. cinerea grown on PDA (control), and expression values were represented by log2 ratio.
To compare DEGs from P. patens and the three studied angiosperms, hierarchical clustering analysis of expressed genes were performed on log2 Fold-Change expression values using the “hclust” tool from R package “stats” ver. 3.6.0. To visualize the obtained data, heatmap plots were performed using the “heatmap.2” tool from R package “gplots” ver. 3.1.0.
2.4. Functional Clasification and Comparison with B. cinerea Secretomes
To test for GO terms enrichment in different sets of DEGs, GO and functional annotations were assigned using G: profiler (http://biit.cs.ut.ee/gprofiler/); which use data retrieved from Ensembl database and fungi, plants or metazoa specific versions of Ensembl Genomes [41]. Only annotated genes were used and GO terms with a FDR ≤ 0.05 were considered for our analysis.
B. cinerea DEGs obtained in the different plant species (P. patens and angiosperms) were compared with previously published B. cinerea secretomes [42,43]. Candidate effectors were searched for among these genes using EffectorP ver 2.0 (http://effectorp.csiro.au/).
3. Results
3.1. B. cinerea Differentially Expressed Genes during P. patens Infection
P. patens consists of juvenile proto-nemal filaments that further develop into leafy gametophores with rhizoids [44]. Proto-nemal filaments and leaves were infected by B. cinerea, as can be visualized by solophenyl flavine staining (Figure S1). B. cinerea infection process starts with the germination of spores at the plant surface at 4 h post inoculation (hpi), and continues with germ tubes elongation and formation of terminal swollen structures called appressoria at 8 hpi. Finally, hyphae proliferate and protonemal and leaf cells are invaded at 24 hpi. Accordingly, B. cinerea starts to enhance biomass production at 8 hpi, reaching high levels at 24 hpi [26]. Symptoms development were visible at 24 hpi with typical browning and maceration of the tissue (Figure S1).
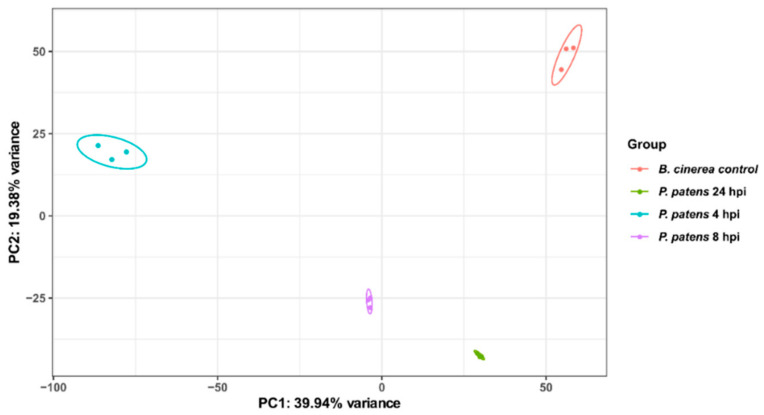
In order to identify B. cinerea molecular players involved in virulence and P. patens colonization, we performed transcriptional profiling of these three stages. Samples of B. cinerea infecting P. patens tissues (4, 8 and 24 hpi) and B. cinerea grown on PDA medium (control) generated a total of 312,737,769 clean reads after removing adapter sequences and low-quality reads (Table S2). Considering all samples, 0.12% to 92.86% of the reads in the libraries mapped successfully to the genome of B. cinerea (nuclear and mitochondria). Reads mapped uniquely to B. cinerea nuclear genome were considered for further analyses; 98,929 at 4 hpi, 796,752 at 8 hpi, 4,261,033 at 24 hpi and 53,254,525 for control B. cinerea samples. Biological variability within replicates was analyzed by principal component analysis (PCA). As shown in Figure 1, the first principal component (PC1) accounted for 40% of the total variation and separates the three time points (4, 8 and 24 hpi), and the control B. cinerea samples. Variability between biological replicates was very low as indicated.
Figure 1.
Analysis of B. cinerea RNA-Seq data during P. patens infection by principal component analysis (PCA). Variation among the three biological replicates per sample (P. patens 4 hpi, P. patens 8 hpi, P. patens 24 hpi and B. cinerea control), is shown. Colored dots denote each biological replicate.
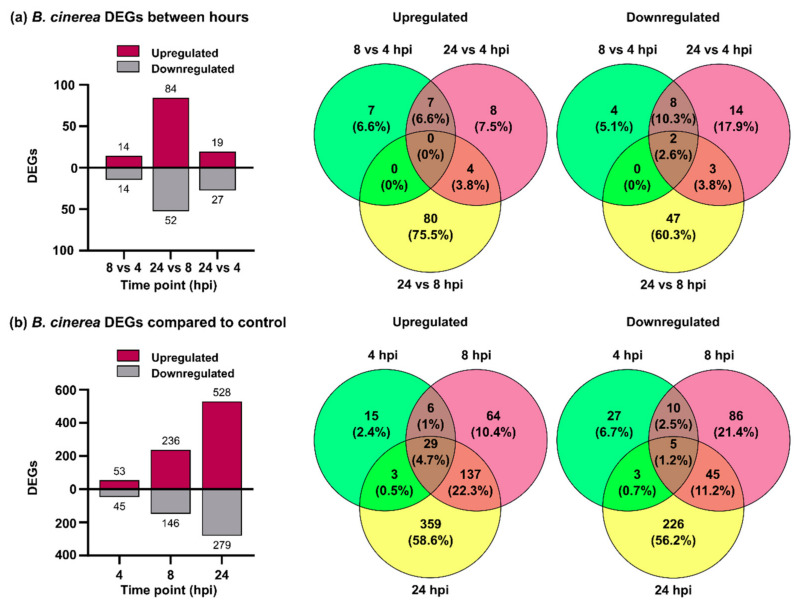
Differential expression was first analyzed between B. cinerea genes expressed in planta during the different infection time points. In total, 184 B. cinerea genes were differentially expressed, 106 were upregulated and 78 were downregulated (Figure 2a; Table S3). In the 8 versus 4 hpi comparison, 14 differentially expressed genes (DEGs) were upregulated and 14 DEGs were downregulated. The number of DEGs increased when 24 hpi was compared with 8 hpi; 84 and 52 DEGs were upregulated and downregulated, respectively. Finally, the comparison of 24 versus 4 hpi showed 19 upregulated and 27 downregulated DEGs. Given the low number of B. cinerea DEGs, we compared each time point of infection with PDA grown mycelium in order to obtain more information on the infection process. In total, 1015 B. cinerea DEGs were obtained and among them 613 were upregulated and 402 downregulated (Figure 2b; Table S3).
Figure 2.
Differentially expressed B. cinerea genes during P. patens infection. (a) Number of differentially expressed genes (DEGs) and Venn diagrams from B. cinerea comparisons at 8 vs. 4 hpi, 24 vs. 8 hpi and 24 vs. 4 hpi. (b) Number of DEGs and Venn diagrams from B. cinerea at 4, 8 and 24 hpi compared with potato dextrose agar (PDA) samples. Log2 FC ≥ 2.0 or ≤ −2.0 and false discovery rate (FDR) ≤ 0.05 were considered for DEGs identification. In Venn diagrams, the overlap of expressed fungal genes can be observed.
Based on this finding, we decided to consider the 1015 DEGs for further analyses. B. cinerea DEGs increased over time and more upregulated genes than downregulated genes were observed at the three time points. The number of upregulated B. cinerea genes increased from 53 at 4 hpi, to 236 at 8 hpi, and finally to 528 at 24 hpi. Among downregulated genes, 45, 146 and 279 were identified at 4, 8 and 24 hpi, respectively. A high proportion of upregulated genes present at 4 hpi were also identified at 8 hpi and 24 hpi (71% and 78%), and similarly 70% of upregulated genes at 8 hpi were also upregulated at 24 hpi.
3.2. B. cinerea Genes Encoding Secreted Cell Wall Degrading Enzymes and Other Virulence Factors Are Induced during P. patens Infection
Gene ontology (GO) term enrichment analysis and manual inspection of the identified DEGs were performed to identify the biological processes (BP), molecular function (MF) and cellular compartment (CC) mostly affected in B. cinerea during moss infection (Figure S2; Table S4). Enriched downregulated processes include terms related to ribonucleotide and nucleotide binding at 4 hpi, transporter activities at 4 and 8 hpi, and ion binding activities at 8 and 24 hpi. In addition, downregulated genes were also enriched in GO terms such as tetrapyrrole and heme binding, oxidoreduction processes and oxidoreductase activities, and FAD binding and monooxygenase activity at 24 hpi. For upregulated genes, GO terms enrichment at 4 hpi were related to organic acid and oxoacid metabolic processes, amino acids metabolic and biosynthetic processes, oxidoreduction processes such as nicotinamide adenine dinucleotide (NAD) binding, oxidoreductase activities acting on different compounds, and transporter activity. At 8 hpi, the most significantly enriched GO terms for upregulated genes were those involved in degradation of various cell wall components, including hydrolase activities hydrolyzing O-glycosyl compounds or acting on glycosyl bonds, PG activity, catalytic activity and carbohydrate metabolic process. All these GO terms were also enriched for upregulated genes at 24 hpi, and other GO terms included carbohydrate binding, hydrolase activity, polysaccharide binding, cellulose binding, alpha-L-arabinofuranosidase activity, cutinase activity, pectate lyase activity, carbon-oxygen lyase activity acting on polysaccharides, and arabinose metabolic process. In addition, intrinsic components of membrane were present at all time points, while the extracellular region was a common GO term for upregulated genes at 8 and 24 hpi, which is consistent with secretion of hydrolytic enzymes to degrade the plant cell wall. These results show that GO enrichment analysis also provided insights into temporal characteristics of fungal pathogenesis.
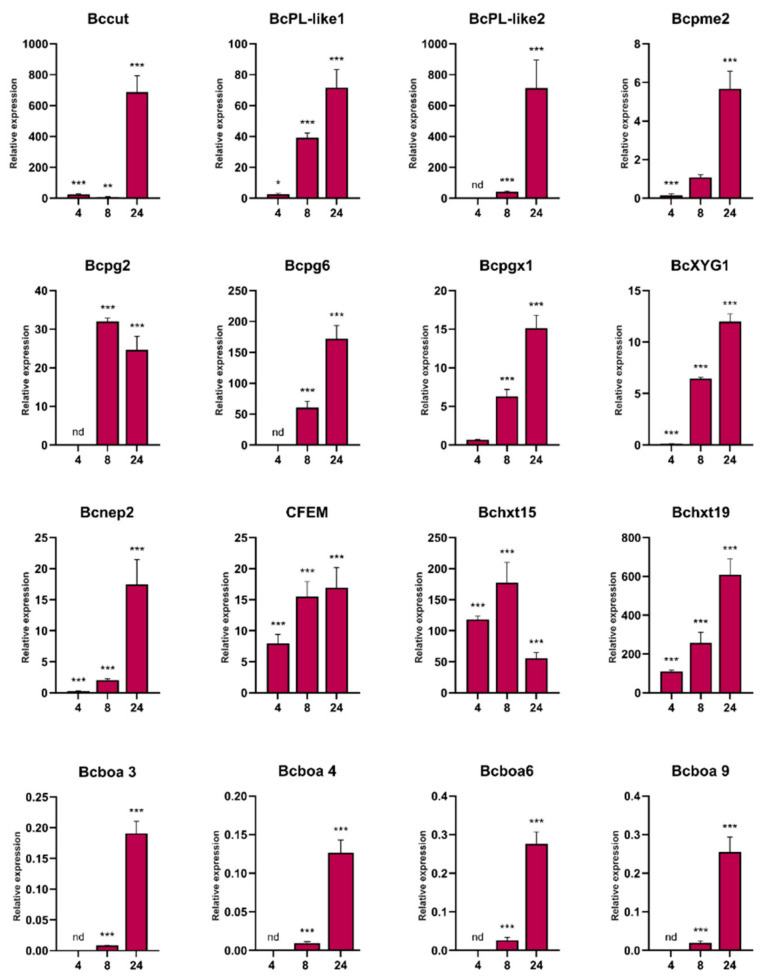
We further focused on upregulated genes since they could encode proteins involved in B. cinerea pathogenesis. In total, 29 DEGs were commonly upregulated at all time points, including genes encoding several major facilitator superfamily (MFS) transporters, two sugar transporters (Bchxt19 and Bchxt15), a nitrate reductase (BcniaD), a nitrite reductase (BcniiA), several types of hydrogenases, a common fungal extracellular membrane (CFEM) domain-containing protein (Bcin09g02270), a chitinase, and several hypothetical proteins. In addition, genes involved in the catabolic pathway of D-galacturonic acid [45], which is the major component of pectin polysaccharides [46], were upregulated during the infection process, including those encoding a 2-keto-3-deoxy-L-galactonate aldolase (Bclga1), galactonate dehydratase (Bclgd1) and galacturonate reductase (Bcgar2). To validate the expression profiles of the RNA-Seq data, some of these genes and others related to pathogenesis were selected for analysis using RT-qPCR. The results obtained by the two techniques showed a strong correlation (R2 = 0.9882) (Figure 3; Table S5). Among the common 137 upregulated genes present at 8 and 24 hpi, virulence components such as cutinases and secreted proteins including CAZymes involved in pectin (mainly homogalacturonan), hemicellulose (mainly xyloglucan) and cellulose degradation were identified, as well as Bcnep1 and Bcnep2 (Necrosis- and Ethylene-inducing Proteins), glutathione peroxidase and glutathione S-transferase (GST) (Table S6). At 8 hpi upregulated genes encode PGs (Bcpg2, Bcpg4, Bcpg6, Bcpgx1 and a second exo-PG), pectate lyases, pectin lyases, beta-glucosidases, a xyloglucan endo-β-glucanase (BcXYG1), an endo-beta-1,4-glucanase, a xylanase, cellobiose dehydrogenases, a cello-bio-hydrolase, and arabino-furanosidases (Table S6). At 24 hpi, the number of upregulated genes encoding enzymes for homogalacturonan degradation increased to seven PGs (Bcpg2, Bcpg4, Bcpg5, Bcpg6, Bcpgx1 and two other exo-PGs), four pectate lyases, four pectin lyases and a pectin-esterase. Genes encoding enzymes that target xyloglucan were upregulated at 24 hpi, included β-glucoside gluco-hydrolase, three beta-glucosidases and BcXYG1 (Table S6). In addition, genes encoding enzymes for xylan degradation were also upregulated at 24 hpi, including Bcxyn11A, Bcxyn11C and three other xylanases. Genes encoding secreted enzymes acting on rhamnogalacturonans backbone, xylan backbone, mannans, cellulose, and side-chains were also upregulated at 24 hpi. These DEGs included glycoside hydrolases, rhamnogalacturonases, beta-glucosidase, mannosidases, glucanases and cello-bio-hydrolase, arabino-furanosidase and beta-galactosidases, among others. In addition, several peptidases were upregulated at 8 and 24 hpi. Taken together, these results support the important role played by these fungal enzymes during P. patens infection.
Figure 3.
Validation of B. cinerea differentially expressed genes during P. patens infection by quantity reverse transcription RT-qPCR at 4, 8 and 24 hpi. The expression levels of B. cinerea genes infecting P. patens at the indicated time points are relative to the level of expression of B. cinerea grown in PDA. Genes are represented by the corresponding abbreviation as indicated in Table S1. BctubB was used as the reference gene. Results are reported as means ± standard deviation (SD) of three samples for each treatment. Asterisks indicate a statistically significant difference between B. cinerea grown in P. patens and in PDA (Students t-test, * p < 0.5, ** p <0.01; *** p < 0.005). Abbreviation: nd: no data.
Surprisingly, genes encoding enzymes involved in botcinic acid synthesis were downregulated at 8 and 24 hpi, indicating higher expression levels in fungi growing on PDA medium. However, when DEGs expressed in planta were compared, several genes involved in botcinic acid production were upregulated at 24 hpi compared to 8 hpi, including Bcboa3, Bcboa4, Bcboa5, Bcboa6, Bcboa7 and Bcboa9 (Table S3; Figure 3). Other genes that increased in planta during the progression of infection (24 compared to 8 hpi), included those encoding cutinases, PGs, a pectin lyase, Bcnep2, and MSF transporters, among others (Table S3).
3.3. Genes Encoding B. cinerea Virulence Factors and Candidate Effectors Are Upregulated during Moss Infection
In addition to CWDEs genes, other genes encoding virulence factors and known effectors were present among the upregulated B. cinerea genes during P. patens infection. These included the cell death inducing factors Bcnep1 and Bcnep2, the necrosis-inducing xylanase Xyn11A and xyloglucanase BcXYG1, as well as the necrosis inducing Bcpg2. Several genes encoding CFEM domain-containing proteins were also upregulated during moss infection, including CFEM1, which is important for pathogenesis (Table S6). Genes encoding two secreted CalciNeurine-Dependant (CND1 and CND3) genes with putative virulence functions, peptidases and proteinases were also upregulated in moss tissues, including secreted aspartic proteinase (Bcap9), peptidases and a metalloproteinase (Deuterolysin; Bcmp1). We searched for effectors among the upregulated B. cinerea DEGs encoding secreted fungal proteins, using EffectorP (Table S6). Eight candidate secreted effectors were identified, including Pectin lyase (Bcin04g00470), BcNEP1, the small secreted BcSSP2A, two ribonucleases (Bcin07g03720 and Bcin05g04720), and three hypothetical proteins. One of these hypothetical protein-encoding genes, Bcin04g03920, showed very high expression levels at 8 and 24 hpi. These results indicate the involvement of virulence factors during P. patens infection, and suggest that candidate secreted effectors may interfere with plant defenses.
3.4. Comparison of B. cinerea DEGs during the Interaction with P. patens and Three Different Angiosperms
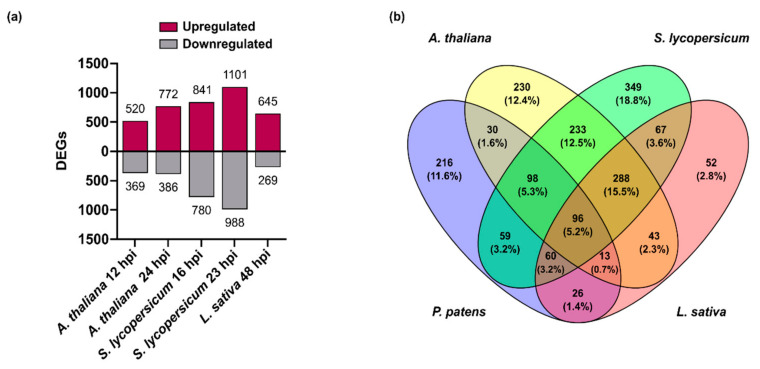
The expression pattern of B. cinerea genes encoding virulence factors and other proteins involved in the infection process of P. patens was compared with gene expression pattern during angiosperms infection, using available RNAseq data that were reanalyzed in this work. A summary of the number of reads mapped to the B. cinerea genome from each biological replicates in the different plant hosts is provided in Table S2. The percentage of reads that uniquely mapped to the B. cinerea genome relative to the total number of reads, in each sample, were in general similar between species, except for P. patens at 4–8 hpi and L. sativa at 24 hpi (lower percentage of reads), and S. lycopersicum at 23 hpi (higher percentage of reads). The number of B. cinerea genes targeted by these reads and the number of genes that passed FDR ≤ 0.05, for each plant species are shown in Table S7. Based on the low number of reads and DEGs in L. sativa 24 hpi, this time point was discarded for further analyses. PCA showed that PC1 accounted for 38% of the total variation and segregated P. patens from angiosperms (Figure S3). In addition, PC1 also allows the differentiation between early time point in angiosperms (A. thaliana 12 hpi and S. lycopersicum 16 hpi) and later time points (A. thaliana 24 hpi, S. lycopersicum 23 hpi and L. sativa 48 hpi). This result suggests that the transcriptome state of B. cinerea during early infection of the different angiosperms is comparable, and a similar scenario occurs within the late time points. The number of B. cinerea DEGs with |log2 FC| ≥ 2 in A. thaliana were 520 upregulated and 369 downregulated genes at 12 hpi, and 772 upregulated and 386 downregulated genes at 24 hpi. S. lycopersicum has 841 upregulated and 780 downregulated DEGs at 16 hpi and 1101 upregulated and 988 downregulated genes at 23 hpi, while L. sativa has 645 upregulated and 269 downregulated DEGs at 48 hpi (Figure 4a; Tables S8–S10).
Figure 4.
Differentially expressed B. cinerea genes during P. patens and angiosperms infection. (a) Number of differentially expressed genes (DEGs) from B. cinerea grown in different plant species at the indicated time points. (b) Venn diagram of B. cinerea upregulated DEGs in different plant species, showing overlap of upregulated fungal genes. P. patens: B. cinerea DEGs at 8 and 24 hpi; A. thaliana: B. cinerea DEGs at 12 and 24 hpi; S. lycopersicum: B. cinerea DEGs at 16 and 23 hpi; and L. sativa: B. cinerea DEGs at 48 hpi. For the Venn diagram, DEGs had log2 FC ≥ 2, FDR ≤ 0.05, and were expressed differentially in at least one time point for each plant species condition.
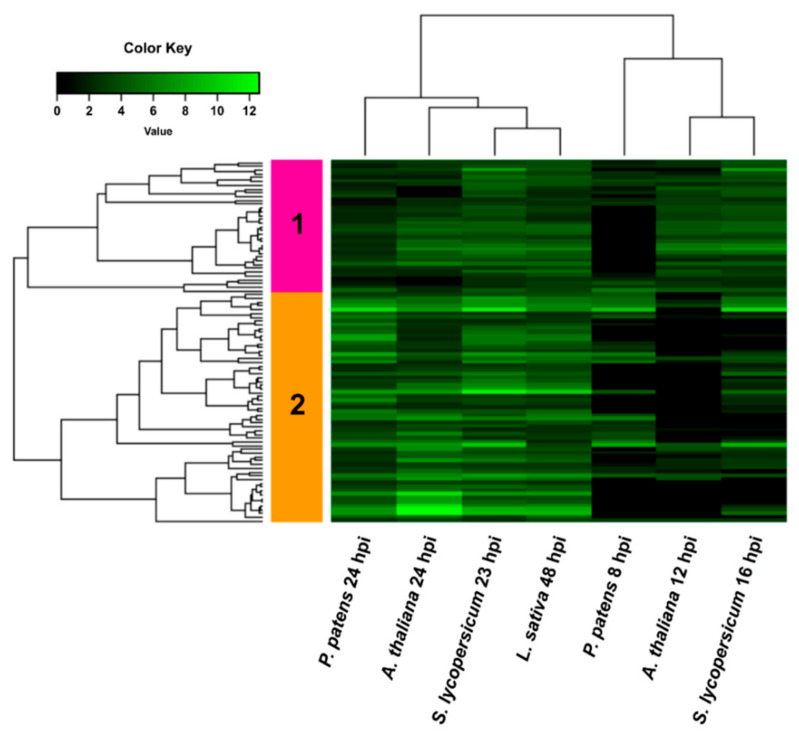
We first centered our analysis in the upregulated B. cinerea DEGs and show that 96 DEGs were commonly upregulated in all hosts, while 216 and 288 DEGs were only upregulated in P. patens and in the three angiosperms, respectively (Figure 4b). The 96 common upregulated DEGs showed an expression profile that separated samples into two groups by hierarchical clustering, one comprising the early time points and a second that included the late time points of infection in the different hosts (Figure 5; Table S11).
Figure 5.
Heatmap of hierarchical clustering of common upregulated B. cinerea DEGs during infection of P. patens and different angiosperms. DEGs correspond to the 96 common upregulated genes of B. cinerea grown in all plant species. Selected DEGs had log2 FC ≥ 2, FDR ≤ 0.05, and were expressed differentially in at least one time point for each plant species. For samples where no significant difference with control was observed (FDR > 0.05) or no data was available, the log2 FC was assigned as cero. See Table S11 for complete information.
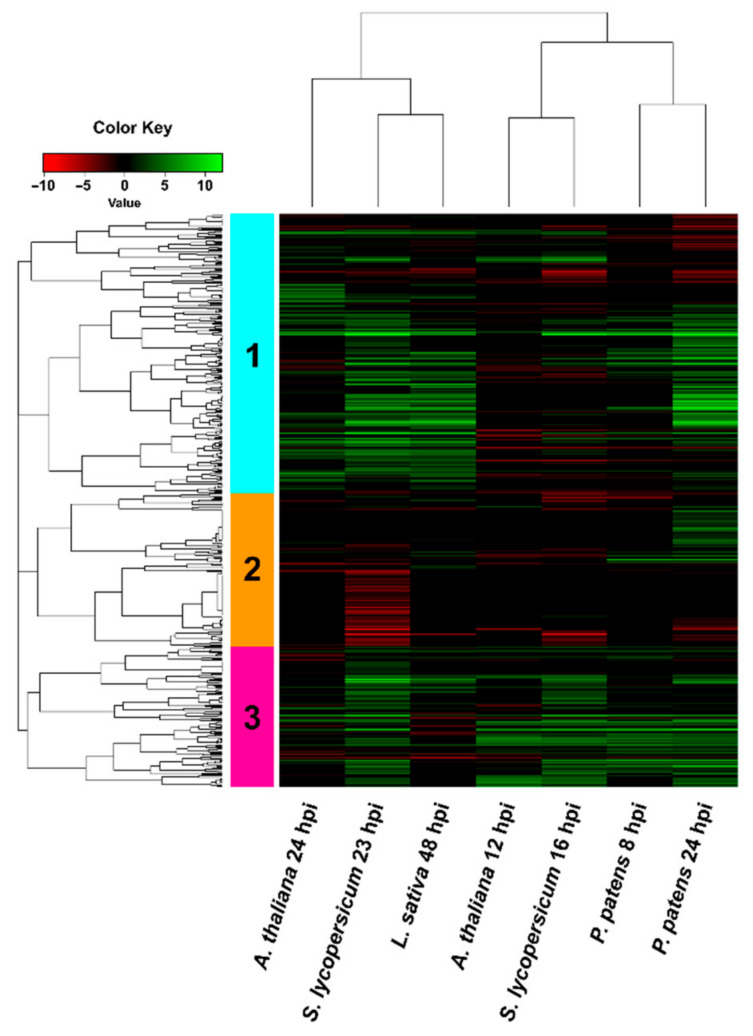
In addition, two gene clusters with different expression patterns were identified; cluster 1 included DEGs that were equally upregulated at early and late time points, while cluster 2 showed a higher number of upregulated DEGs during the late time points of infection compared with early time points. Common upregulated B. cinerea genes in all plant species encoded proteins with oxidoreductase activity, virulence factors (NEP2 and CFEM containing protein), transferases, transporters and hypothetical proteins, distributed in both clusters. Cluster 2 included several genes encoding secreted CAZymes such as Bcpg2, Bcpg6, pectin lyases, pectate lyase, rhamnogalacturonan acetyl-esterase, α-L-rhamnosidase, α-galactosidase, β-galactosidase, α-N-arabino-furanosidase, and Bcxyn11A. Based on this result, we looked into more detail to the expression pattern of genes encoding predicted secreted proteins of B. cinerea during P. patens and angiosperm infection (Figure 6; Table S12). Hierarchical clustering grouped the expression pattern of secretome-encoding genes into a group that included the late time points of angiosperms, and a second group including both time points of P. patens and the early time points of angiosperms. In addition, clustering identified genes with three expression patterns, cluster 1 has the highest number of upregulated B. cinerea DEGs. We focused in this cluster and observed that B. cinerea DEGs were detected in some but not all hosts. Expression patterns of fungal DEGs were similar during infection of S. lycopersicum 23 hpi and L. sativa 48 hpi, while expression profiles of fungal genes in angiosperms and P. patens differ at late infection stages.
Figure 6.
Hierarchical clustering (heat map) showing genes encoding secreted B. cinerea proteins during infection of P. patens and different angiosperms. Only DEGs that passed FDR ≤ 0.05 were considered in the analysis. For samples where no significant difference with control was observed (FDR > 0.05) or no data was available, the log2 FC was assigned as cero. See Table S12 for complete information.
Cluster 1 also included DEGs that are commonly upregulated in all plant species such as the previously mentioned Bcpg2, Bcpg6, pectate lyase, pectin lyase and other CAZymes, which were among the highest upregulated DEGs in all plant species. In addition, several CAZymes genes were only upregulated at late time points in all plant species except A. thaliana, including genes encoding endoglucanases, endo-β-mannosidase, rhamnogalacturonase, α-L-arabino-furanosidase, a cellobiohydrolase, a glycoside hydrolase and a xylanase. Finally, a small group of upregulated DEGs that encode putative secreted carboxylic ester hydrolase and hypothetical proteins were present at late time points in angiosperms and not in P. patens. Inversely, a group of upregulated DEGs present in P. patens at 24 hpi were not differentially expressed in angiosperms, including genes encoding β-glucoside gluco-hydrolase, endo-1,4-beta-xylanase (BcXyn11C), carboxylic ester hydrolase, peroxidase, and an exo-PG, among others.
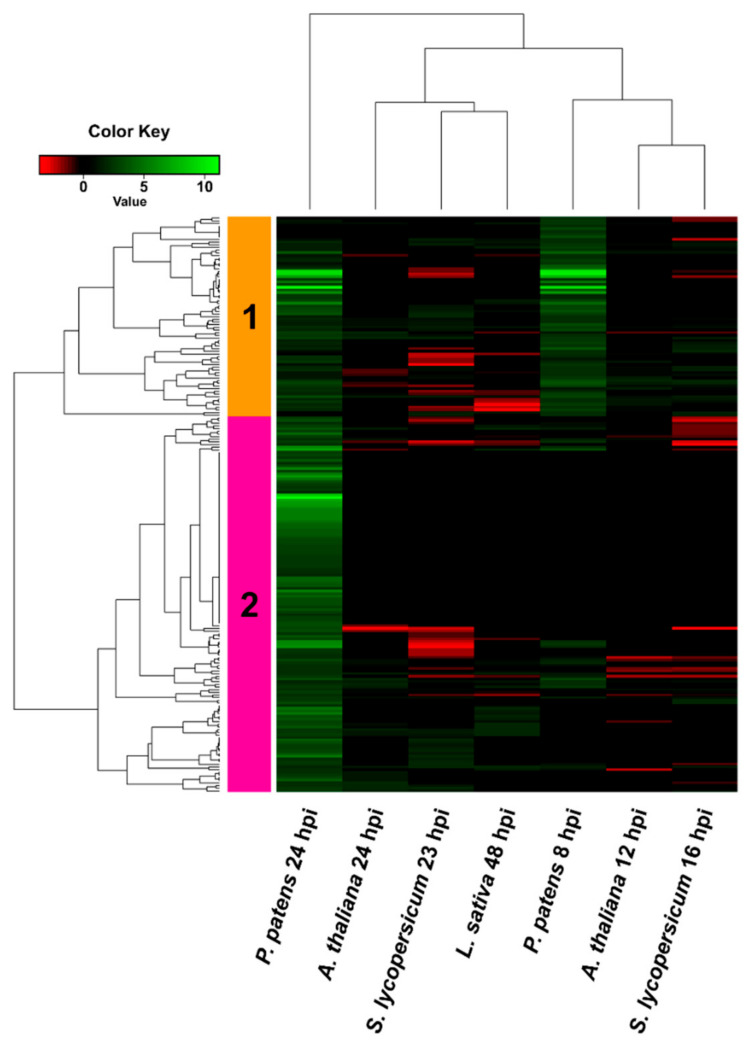
We further analyzed in more detail the 216 B. cinerea DEGs that were only upregulated in P. patens and showed very low or no expression in angiosperms. These DEGs were distributed in two clusters (Figure 7, Table S13): cluster 1 contained upregulated B. cinerea DEGs at 8 hpi and 24 hpi, while cluster 2 was mainly composed of upregulated DEGs at 24 hpi.
Figure 7.
Hierarchical clustering (heat map) showing B. cinerea genes only upregulated in P. patens. DEGs correspond to the 216 genes that were only upregulated when growing in P. patens. Selected DEGs had log2 FC ≥ 2, FDR ≤ 0.05, and were expressed differentially in at least one time point for each plant species. For samples where no significant difference with control was observed (FDR > 0.05) or no data was available, the log2 FC was assigned as cero. See Table S13 for complete information.
In cluster 1, four upregulated DEGs showed very high expression levels at 8 and 24 hpi, and encoded a D-malate dehydrogenase, a glyceraldehyde-3-phosphate dehydrogenase (GAPDH), a D-glycerate 3-kinase and a MFS transporter. Interestingly, 22 additional genes encoding putative fungal MSF transporters were upregulated in P. patens and in almost all cases expression was not detected or was repressed in angiosperms. Among other upregulated B. cinerea genes that were only upregulated in P. patens we found carboxylic ester hydrolases, cellobiose dehydrogenase, beta-glucosidases, a peroxidase, a polyketide synthase (BcPks8), and a high number of hypothetical proteins. Additionally, six Zn(2)-C6 transcription factors, including the BcGaaR GalA regulator involved in D-galacturonic acid utilization by B. cinerea [47], were upregulated during P. patens infection and not in angiosperms. Interestingly, Bcaba2 encoding a Cytochrome P450 monooxygenase, which is an ABA biosynthesis cluster protein, was only upregulated at 24 hpi in P. patens tissues.
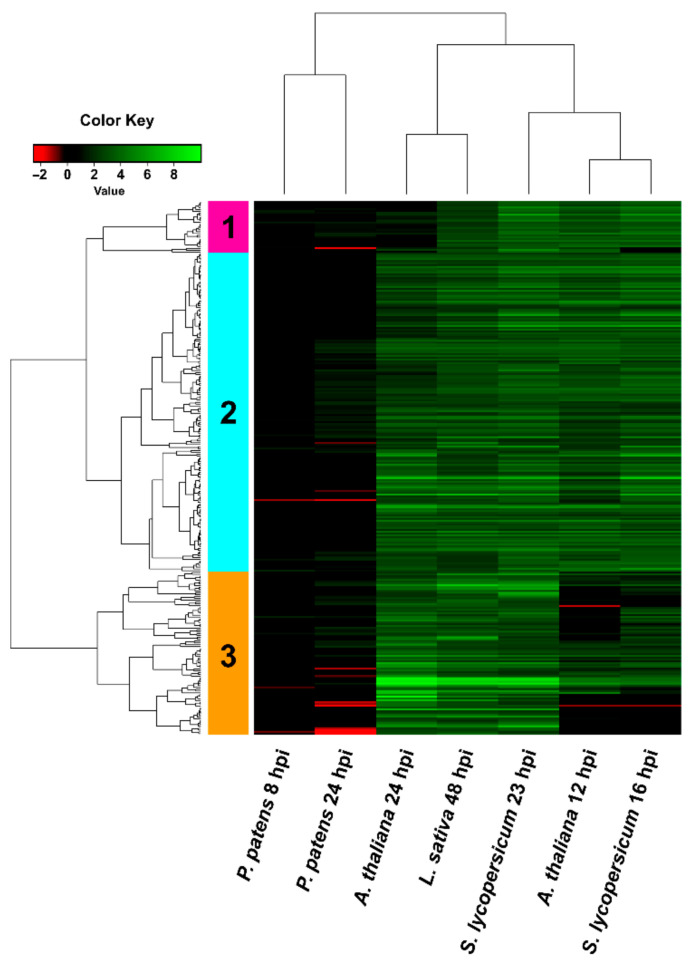
Among the 288 B. cinerea DEGs that were only upregulated in angiosperms (Figure 4b), we focused on those that were not detected or showed very low expression levels in P. patens (Figure 8; Table S14). Interestingly, among them we detected most genes of the botrydial gene cluster at least at one time point, including Bcbot1, Bcbot3, Bcbot4 and Bcbot5. In addition, Bcbot2 was highly upregulated in all angiosperms, while in P. patens an induction below log2 FC 2 was observed at 24 hpi. The major positive regulator of botrydial synthesis, the transcription factor Bcbot6 (Bcin12g06420), and a dehydrogenase (Bcbot7; Bcin12g06430), were only upregulated in the three angiosperms. Other fungal upregulated DEGs that were only detected in angiosperms included genes encoding glutaredoxins, GSTs, superoxide dis-mutases (SOD), other oxidoreductase related proteins, carboxyl ester hydrolases, transcription factors including Nmra and bZIP, and hypothetical proteins, among others. Taken together, these results reveal that several B. cinerea genes that play important roles in host infection have different expression patterns in the plant species analyzed.
Figure 8.
Hierarchical clustering (heat map) showing B. cinerea DEGs only upregulated in angiosperms. DEGs correspond to the 288 genes that were upregulated in all angiosperms and not in P. patens. Selected DEGs had log2 FC ≥ 2, FDR ≤ 0.05, and were expressed differentially in at least one time point for each plant species. For samples where no significant difference with control was observed (FDR > 0.05) or no data was available, the log2 FC was assigned as cero. See Table S14 for complete information.
Finally, we looked into B. cinerea DEGs that were only downregulated in P. patens or in angiosperms (Figure S4, Tables S15 and S16). In total 111 and 93 fungal DEGs were only downregulated in P. patens or angiosperms, respectively. From those, only two genes that were downregulated in angiosperms showed increased expression in P. patens that was close to Log2 FC=2 (Bcin06g02530: DUF2235 domain-containing protein and Bcin05g05110: hypothetical protein). In contrast, several genes that were downregulated in P. patens were upregulated in all angiosperms, including Bcbot4, and two genes encoding a hydrolase EHN domain-containing protein and a PKS. Other DEGs that were downregulated in P. patens and showed increased expression in some of the angiosperms included those encoding the aspartic proteinase Bccap1, two ABC transporters, an acyl transferase and several hypothetical proteins. Finally, Bcboa3-5 were downregulated in P. patens and showed no expression in most angiosperms. Taken together, our results show that common and differential expression patterns of genes with roles in virulence and infection exist between P. patens and angiosperms.
4. Discussion
Transcriptional profiling during the B. cinerea infection process has been performed in several angiosperms [38,39,40,48,49,50,51]. However, no information related to the molecular mechanisms used by B. cinerea during moss invasion is available. Mosses interact with a variety of fungal pathogens in nature and under controlled conditions in the laboratory [52,53]. To get insights into B. cinerea infection and virulence strategies developed during moss colonization, we performed a global expression profiling of different infection stages. Expression patterns of upregulated genes and GO enrichment analysis indicated that infection of moss tissues by B. cinerea depends on ROS generation and detoxification, transporter activities, plant cell wall degradation and modification, toxin production and probable plant defense evasion by effector proteins. Moreover, a comparison with available RNAseq data during angiosperm infection, including A. thaliana, S. lycopersicum and L. sativa, suggests that B. cinerea has probably virulence and infection functions that are used in all hosts, while others are more specific to P. patens or angiosperms.
ROS production is important for B. cinerea virulence during plant invasion and increased levels of ROS in plant cells are beneficial to the pathogen, leading to accelerated colonization of host tissue [8]. We have previously observed that during P. patens-B. cinerea interaction, ROS are produced in P. patens cells after hyphal contact, probably resembling an oxidative burst response, and also in hyphal tips in contact with moss cells [26]. Similarly, in angiosperms, generation of H2O2 in the tip of the penetration pegs that breach the cuticle has been described [54]. ROS might assist in host invasion by providing substrates for oxidases that can modify the cuticle [9]. Moreover, endogenous ROS play crucial roles in B. cinerea conidial germination and host penetration [55,56]. Here, we show that oxidoreductase activities are enriched in upregulated fungal genes during early P. patens infection (4 hpi), including genes encoding BcniaD and BcniiA, that are further upregulated during the entire time course of infection analyzed. Similarly, BcniaD and BcniiA are both upregulated during A. thaliana, S. lycopersicum and L. sativa infection, but only at late time points. The relevance of this finding needs further investigation, since the production of nitric oxide (NO) by B. cinerea probably promotes fungal colonization of the plant tissue, although nitrate reductase is not the main system responsible for the production of NO in this pathogen [57]. Oxidoreductase processes continued during P. patens infection at 8 and 24 hpi, and genes encoding laccase, quinone reductase, GMC oxidoreductase, peroxidases, glutathione peroxidase (Bcgpx3), GST, and different types of oxidoreductases were upregulated. Laccase, quinone reductase and GMC oxidoreductase can be a source of ROS production needed for B. cinerea infection, while peroxidases, glutathione peroxidases as well as other detoxification enzymes may act to control ROS levels [58]. In addition, Bcin12g02910 encoding a cellobiose dehydrogenase, that is secreted at early time points of infection and could be responsible for ROS generation by B. cinerea [59], was upregulated during P. patens infection. Similarly, during B. cinerea colonization of A. thaliana, S. lycopersicum and L. sativa, a high number of ROS related genes were upregulated, including peroxidases, GSTs, catalases, SODs and different oxidoreductases. Consistently, oxidoreductase activities in the secretome of several Botrytis species, including B. cinerea, represented 10% of the total secretome, highlighting the importance of these activities during fungal infection [48].
ROS production contribute to host cell death, favoring invasion and further colonization of B. cinerea in the dead host tissue [60]. This pathogen facilitates host cell death by producing toxins, including botrydial and botcinic acid [17,18]. We only observed increased expression of fungal genes involved in botcinic acid production, including Bcboa3-7 and Bcboa9, during P. patens infection when we compared 24 hpi with 8 hpi, suggesting the involvement of this toxin in moss infection. Interestingly, genes encoding the biosynthesis enzymes of botrydial [61], were upregulated in A. thaliana, S. lycopersicum and L. sativa, while in P. patens only Bcbot2 encoding the sesquiterpene cyclase showed a slight induction. Other genes of the botrydial cluster, including Bcbot1, Bcbot3 and Bcbot4 that encode cytochrome P450 proteins, and Bcbot5 encoding a putative acetyl transferase, were not detected or were repressed. In addition, the major positive regulator of botrydial synthesis Bcbot6 that encode a putative Zn(II)2Cys6 transcription factor, and the dehydrogenase Bcbot7 that might be involved in the conversion of botrydial to dihydrobotrydial [62,63], were only upregulated in angiosperms and not in P. patens. The fact that botrydial related genes were not upregulated during moss infection was not expected. However, it was previously shown that mutations in botrydial biosynthetic genes and the Bcbot6 regulator did not alter the development and virulence of B. cinerea, which is in accordance with a redundant role of botcinic acid and botrydial in virulence of this fungal pathogen [64,65]. Other genes encoding secreted fungal proteins with necrosis-inducing activity such as NEP1 and NEP2 [66], were upregulated in P. patens and most analyzed angiosperms. An HR-like response with similar features as in angiosperms has been observed in B. cinerea-infected P. patens tissues [26], and these proteins may contribute to induce PCD in moss for fungi benefit. In addition, upregulation of several hydrolase-encoding genes during P. patens infection, such as Bcpg2, Xyn11A and xyloglucanase BcXYG1, with known cell death inducing activities in angiosperms [43,67,68,69], may contribute to moss cell death and facilitate fungal infection. These three hydrolases are able to induce a necrotic response independent of their catalytic activity, and Bcpg2 and Xyn11A are commonly upregulated genes in all analyzed plant hosts.
As expected for a pathogen with a necrotrophic lifestyle, we show that a large number of B. cinerea genes upregulated during P. patens and angiosperms infection, encode enzymes with hydrolytic activities. Consistently, hydrolase activity was reported as the most common molecular function of the B. cinerea secretome and other Botrytis species (approximately 25% of the total secretome) [48]. The production of extracellular proteins is shown to be regulated at the transcriptional level, suggesting a fine-tuning of B. cinerea secretome according to conditions that contribute to accomplish successful infection [70]. A high proportion of the upregulated fungal genes in P. patens and angiosperms encode CAZymes, including cutinases and PCWDEs with a role in host tissue breakdown and pathogenesis. Many of these enzymes were also detected in different studies of B. cinerea secretomes [59,70,71,72,73]. These extracellular enzymes are potential virulence factors that could play important roles in facilitating hyphal growth by softening the host tissues and converting complex plant material available for consumption [74]. Our expression profiles showed that the highest proportion of CAZymes genes were upregulated at late time points of infection, which is consistent with higher maceration rates and cell death of plant tissues, which are needed for effective colonization.
The initial interaction between plants and B. cinerea occurs at the plant cuticle, and upregulation of three cutinase-encoding genes in moss tissues suggests that as in other plants [9,15,16,75], these enzymes play a role in breaching P. patens cuticle for host penetration. Penetration pores were detected on the surface of P. patens cell walls and in intercellular spaces that are colonized by infection hyphae that subsequently invaded leaf cells [27]. Cutinases may be involved in gametophore colonization since fungal penetration pegs penetrate the tissues directly through leaf cell walls [53], and protonema cells have no cuticle [76]. In addition, some B. cinerea cutinases show high homology to acetylxylanesterases [77], and could be involved in degradation of xylans in moss tissues. Moreover, the expression pattern of cutinases varied among angiosperms infection and while BccutB was only upregulated in S. lycopersicum, other cutinases were expressed in more than one plant. Consistently, previous studies have shown that the expression levels of these enzymes depended on the host tissues [77]. Our results also revealed the presence of common upregulated genes encoding secreted CAZymes in all host plants, as well as CAZymes genes that were detected in some but not all plants hosts. Bcpg2, Bcpg6, two pectin lyases, a pectate lyase, rhamnogalacturonan acetyl-esterase, α-L-rhamnosidase, α-galactosidase, β-galactosidase, α-N-arabino-furanosidase, and Bcxyn11A were upregulated in all plant host. In accordance, PGs, pectin lyases, pectate lyases and Bcxyn11A are essential for virulence and fungal colonization into host tissues [69,75,77,78]. P. patens infection showed the highest number of upregulated genes encoding pectin and pectate lyases (8 genes), followed by S. lycopersicum infection (6 genes), and finally A. thaliana and L. sativa infection (4 genes). Moreover, several upregulated DEGs were present in P. patens but were not detected in angiosperms, including genes encoding β-glucoside gluco-hydrolase, BcXyn11C, a GPI anchored protein; poly(β-D-mannuronate) lyase, an exo-PG, a glycoside hydrolase and a cellobiose dehydrogenase, among others. In addition, several B. cinerea DEGs appeared to be preferentially expressed in one or two specific angiosperms. Remarkably, induced expression of secreted CAZymes could be related to cell wall composition in different plant species [77]. The cell walls of mosses and angiosperms are mainly composed of the same classes of polysaccharides with the exception of xylogalacturonan, which has not been detected in P. patens, and some differences exist inside chain composition and structure [79,80]. HG, β-1,4-galactan, α-1,5-arabinan, desterified pectin and RG-I have been identified in P. patens and some moss species may contain an RG-II-like polysaccharide [80]. In P. patens, the main upregulated B. cinerea DEGs were related to pectin homogalacturonan and XyG backbone-degrading enzymes. Furthermore, upregulation of Bclga1, Bclgd1 and Bcgar2 involved in the catabolic pathway of the final product released from pectin degradation, D-galacturonic acid, suggest that pectin depolymerizing and utilization of D-galacturonic acid is important as a carbon nutrient for B. cinerea, allowing efficient colonization of P. patens tissues. BcGaaR encoding a Zn2Cys6 transcription factor involved in D-galacturonic acid utilization [47], was also upregulated in moss tissues. B. cinerea mutants in Bclga1, Bclgd1 and Bcgar2, have affected virulence on N. benthamiana and A. thaliana leaves, but not on S. lycopersicum, indicating host-specific virulence [81]. Interestingly, this difference was correlated with the amount of D-galacturonic acid present in these hosts since S. lycopersicum has lower D-galacturonic acid content than A. thaliana and N. benthamiana leaves. At 24 hpi, upregulated DEGs encoded enzymes involved in HG, XyG backbone, RG-I backbone, xylan backbone, mannans, cellulose, and side-chains/adducts degradation and modification. Upregulation of Bcpme2 in P. patens and most angiosperms may facilitate the action of PGs by demethylating pectin to pectate. B. cinerea cellulases and hemi-cellulases (xylanases)-encoding genes were also upregulated during P. patens and angiosperms colonization. BcAra1 encoding an endo-arabinanase that carries out the breakdown of arabinan [82], was upregulated at late time points during P. patens, A. thaliana and S. lycopersicum infections. Interestingly, BcAra1 plays an important role during infection of A. thaliana where mutants provoke smaller lesions, while in N. benthamiana lesion size did not differ compared to the wild type strain, indicating that Bcara1 plays a host-dependent role in the virulence of B. cinerea [82]. Expression levels of BcAra1 in N. benthamiana were much lower than in A. thaliana indicating regulation at the transcriptional level that serves as a determinant of disease progression. Taken together, our results are consistent with host-dependent expression of genes related to cell-wall degradation and modification by B. cinerea.
Two of the highest expressed B. cinerea genes during P. patens infection encoded a malate dehydrogenase and GAPDH. Interestingly, malate dehydrogenase catalyzes the reversible conversion of oxalacetate and malate, and oxalacetate is a precursor of the pathogenicity factor oxalic acid [83]. Oxalic acid facilitates B. cinerea infection through different mechanisms, including enhancement of PGs activity to promote cell wall degradation, suppression of the plant oxidative burst, induction of HR, inhibition of plant–protection enzymes, and alteration of the cellular redox status in the plant [12]. Consistently, malate dehydrogenase has been identified in the secretome of different B. cinerea strains [84]. GAPDH functions in the glycolytic cycle and it may serves as a virulence factor as has been proposed for several pathogenic fungi [85]. In addition, other virulence genes were upregulated during P. patens infection, including BcCFEM1, which has a putative GPI modification site and is required for virulence and tolerance to osmotic and cell wall stress [43]. Other genes encoding CFEM domain-containing proteins were upregulated in the different hosts, which is consistent with their potential role in pathogenicity [86,87]. Upregulated genes encoding secreted proteins with putative virulence functions during P. patens and angiosperm infection included different CND proteins. The calcium/calcineurin signaling pathway controls the botrydial gene cluster, and calcineurin phosphatase has an essential role in virulence and was shown to be involved in B. cinerea appressorium formation [10,88]. Secreted proteinases and peptidases have been reported as virulence factors involved in host-tissue invasion and pathogenicity, and different types of fungal proteases were upregulated in P. patens and angiosperms, including aspartic proteinases [89]. Consistently, a high number of proteases were found in different B. cinerea secretomes [42,48,59]. Several studies have demonstrated the role of secreted effector genes in the establishment of B. cinerea infection via suppression of plant defense [42]. We identified several candidate effectors genes that were upregulated during P. patens infection and could interfere with moss defenses. They include known effectors such as BcNEP1 and BcSSP2 [90], two ribonucleases and three hypothetical proteins, including Bcin04g03920, which is highly expressed in P. patens, A. thaliana and S. lycopersicum. Ribonucleases are known effectors in other necrotrophic pathogens, such as Blumeria graminis, where they bind to host ribosomes and inhibit the action of plant ribosome-inactivating proteins that are known components of plant immune responses that lead to host cell death [91].
Plants perceive pathogens by detecting pathogen-associated molecular patterns (PAMPs) through pattern recognition receptors [92]. Several secreted enzymes of B. cinerea are recognized by plant leucine-rich repeat receptors, including PGs [93], and BcXYG1 [42], leading to activation of plant defense. Interestingly, P. patens induces rapidly the expression levels of a high number of genes encoding these type of receptors during B. cinerea infection [28]. However, further research is needed to understand if some of these receptors recognize PGs and other fungal PAMPs in moss tissues. Moreover, we observed that two genes encoding putative plant pectin methyl-esterase inhibitor were upregulated in P. patens after B. cinerea colonization [28]. These inhibitors could be involved in making the wall more resistant to degradation by fungal enzymes by increasing methylated pectin in the cell wall [94]. In response to plant defense activation, B. cinerea has developed different strategies to interfere with plant immunity, including secretion of hydrolases to detoxify antifungal secondary metabolites produced by the host. B. cinerea is capable of degrading stilbene phytoalexins [95] from grapevines, α- tomatine [96] from S. lycopersicum, and metabolize scopoletin from tobacco [97]. Moreover, cruciferous phytoalexins were detoxified by B. cinerea via either oxidative degradation or hydrolysis [98]. In addition, upregulation of genes encoding peptidases and proteases during P. patens and angiosperms infection, leads to amino acids release for fungal growth, degradation of plant cell wall proteins and defense proteins [59]. Plant pathogens, have transporters, such as ATP-binding cassette (ABC) and MFS transporters, involved in the excretion of plant compounds that are toxic for the pathogen and are produced by the host as a defensive strategy [99]. These transporters contribute to virulence and are especially useful for pathogens with a broad host range [99], including B. cinerea. Here, we show that during P. patens infection, B. cinerea induces the expression of a high number of genes encoding putative MFS transporters, some of which are very highly induced (Bcin07g06720 and Bcin09g05570). Interestingly, most of these MSF-transporter encoding genes were only upregulated during P. patens infection and not in angiosperms. The large number of MSF transporters gene members in the B. cinerea genome, 286 and 282 genes in strain T4 and strain B05.10, respectively, highlight their importance in counteracting the effect of antimicrobial compounds [100]. Consistently, a MFS and an ABC transporter contribute to virulence in B. cinerea and increase tolerance to glucosinolates and camalexin respectively, which are produced by A. thaliana as defensive compounds [100,101]. Moreover, Bcmfs1 is involved in tolerance to antifungal compounds such as camptothecin and cercosporin [102]. Our results suggest that MFS transporters could play an important role in P. patens–B. cinerea interaction, although further studies are needed to reveal whether these transporters are involved in detoxification of moss specific metabolites. Until now, only few compounds with antifungal activity have been identified in P. patens [103], and due to their requirement in improved defenses to biotic challengers during plant colonization, these metabolites are of particular interest.
5. Conclusions
Our transcriptional profiling of B. cinerea-infected plants demonstrated that common molecular mechanisms were involved in fungal virulence and the infection process in P. patens and angiosperms. However, differences in expression patterns of fungal CAZymes genes and putative MFS transporter genes between hosts revealed that some infection functions are specific to P. patens or angiosperms infections. A deeper understanding of the plant cell wall targets, as well as the identity of metabolites with antifungal activities of the encoded proteins by P. patens, will certainly increase our knowledge of bryophyte-fungal interactions. In addition, the use of B. cinerea mutants will contribute to uncover the molecular pathogenic mechanisms and regulatory network used by this pathogen during moss infection and their involvement during coevolution of pathogens with land plants.
Acknowledgments
The authors thank Ricardo Larraya for technical assistance.
Supplementary Materials
The following are available online at https://www.mdpi.com/2309-608X/7/1/11/s1, Figure S1: B. cinerea infection of P. patens tissues. Infection progress during time is shown; (a) spore germination at 4 hpi, (b) germ tubes elongation at 8 hpi, (c) proliferation of mycelium at 24 hpi, (d) appresorium formation at 8 hpi, (e) hyphae colonizing a leaf cell, and (f) hyphae colonizing a protonemal cell. Symptom development at 8 hpi (h), 24 hpi (i) and 24 h water-treated moss colonies (g) are shown. The arrow in (d) indicate an appresorium. The scale bar in a-f represents 10 μm and 0.5 cm in g-i. Figure S2: Enriched gene ontology (GO) terms of B. cinerea DEGs. The top 15 enrichment GO terms were chosen according to their statistical significance (−log10 FDR). GO enrichment are shown for upregulated and downregulated B. cinerea DEGs at 4, 8 and 24 hpi. See Table S5 for complete information. Figure S3: Analysis of B. cinerea RNA-Seq data during P. patens, A. thaliana, S. lycopersicum and L. sativa infection by principal component analysis (PCA). Colored dots denote each biological replicate. Figure S4: B. cinerea downregulated DEGs during P. patens and angiosperm infection. (a) Venn diagram of B. cinerea downregulated DEGs in different plant species, showing overlap of fungal repressed genes. P. patens: sum of B. cinerea DEGs at 8 and 24 hpi; A. thaliana: sum of B. cinerea DEGs at 12 and 24 hpi; S. lycopersicum: sum of B. cinerea DEGs at 16 and 23 hpi; and L. sativa: B. cinerea DEGs at 48 hpi. For Venn diagram, DEGs had log2 FC ≤ −2, FDR ≤ 0.05, and were expressed differentially in at least one time point for each plant species condition. (b) Hierarchical clustering (heat map) showing B. cinerea genes only downregulated in P. patens. Fungal DEGs corresponds to the 111 genes that were only downregulated in P. patens. (c) Hierarchical clustering showing B. cinerea genes only downregulated in angiosperms. Fungal DEGs corresponds to the 93 genes that were only downregulated in angiosperms. Table S1: List of qPCR primers used in this study. Table S2: Summary of mapped reads of B. cinerea in the RNA-Seq libraries. 1–3 indicate the three biological replicates of different plants species at the indicated time points. Table S3: List of B. cinerea differentially expressed genes (DEGs) during P. patens infection at 4, 8 and 24 hpi. Table S4: Enriched gene ontology (GO) terms (over-representation) for biological processes (BP), molecular function (MF) and cell compartment (CC) at 4, 8 and 24 hpi. Table S5: Validation of RNA-Seq data by qPCR assay: correlation of log2 FC values for 16 genes obtained by RNA-Seq and qPCR. Gene name and identification, log2 fold change from RNA-seq and RT-qPCR analysis and description are provided. Table S6: List of B. cinerea upregulated DEGs encoding secreted proteins during infection of P. patens at 4, 8 and 24 hpi. Table S7: Summary of numbers of B. cinerea analyzed genes. Table S8: List of B. cinerea differentially expressed genes (DEGs) during A. thaliana infection at 12 and 24 hpi. Table S9: List of B. cinerea differentially expressed genes (DEGs) during S. lycopersicum infection at 16 and 23 hpi. Table S10: List of B. cinerea differentially expressed genes (DEGs) during L. sativa infection at 48 hpi. Table S11: List of common upregulated genes in all plants species. B. cinerea DEGs were included when log2 FC was ≥ 2 and FDR ≤ 0.05 in at least one time point of each plant species. For samples where no significant difference with control was observed (FDR > 0.05) or no data was available, the log2 FC was assigned as cero. Table S12: List of genes encoding putative secreted B. cinerea proteins in P. patens, A. thaliana, L. lycopersicum and L. sativa at the indicated time points of infection. DEGs were considered when FDR was ≤0.05. For samples where no significant difference with control was observed (FDR > 0.05) or no data was available, the log2 FC was assigned as cero. Table S13: List of 96 B. cinerea DEGs that were only upregulated in P. patens. DEGs were considered when log2 FC was ≥ 2 and FDR ≤ 0.05 in at least one time point of each plant species. For samples where no significant difference with control was observed (FDR > 0.05) or no data was available, the log2 FC was assigned as cero. Table S14: List of 288 B. cinerea DEGs that were only upregulated in all angiosperms and not in P. patens. DEGs were considered when log2 FC was ≥ 2 and FDR ≤ 0.05 in at least one time point of each plant species. For samples where no significant difference with control was observed (FDR > 0.05) or no data was available, the log2 FC was assigned as cero. Table S15: List of 111 B. cinerea DEGs that were only downregulated in P. patens. DEGs were considered when log2 FC was ≤ −2 and FDR ≤ 0.05 in at least one time point of P. patens. For samples where no significant difference with control was observed (FDR > 0.05) or no data was available, the log2 FC was assigned as cero. Table S16: List of 93 B. cinerea DEGs that were only downregulated in all angiosperms and not in P. patens. DEGs were considered when log2 FC was ≤ −2 and FDR ≤ 0.05 in at least one time point of each plant species. For samples where no significant difference with control was observed (FDR > 0.05) or no data was available, the log2 FC was assigned as cero.
Author Contributions
Conceived and designed the experiments: I.P.D.L., A.A., G.R.; Performed the experiments: G.R. and L.V.; Analyzed and interpreted the data: I.P.D.L., A.A., G.R. and L.V.; Wrote the article: I.P.D.L.; participated in the drafting of this work A.A., G.R., L.V. and R.A.B.-G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by “Fondo Conjunto” Uruguay-México (AUCI-AMEXCID), “Agencia Nacional de Investigación e Innovación (ANII) (graduate fellowships)” Uruguay, “Programa de Desarrollo de las Ciencias Básicas (PEDECIBA)” Uruguay, and “Programa para Grupo de I + D Comisión Sectorial de Investigación Científica, Universidad de la República”, Uruguay.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Elad Y., Pertot I., Cotes Prado A.M., Stewart A. Plant hosts of Botrytis spp. In: Fillinger S., Elad Y., editors. Botrytis—The Fungus, the Pathogen and Its Management in Agricultural Systems. Springer; Berlin, Germany: 2015. pp. 413–486. [Google Scholar]
- 2.Dean R., van Kan J.A.L., Pretorius Z.A., Hammond-Kosack K.E., Di Pietro A., Spanu P.D., Rudd J.J., Dickman M., Kahmann R., Ellis J. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012;13:414–430. doi: 10.1111/j.1364-3703.2011.00783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williamson B., Tudzynski B., Tudzynski P., Van Kan J.A.L. Botrytis cinerea: The cause of grey mould disease. Mol. Plant Pathol. 2007;8:561–580. doi: 10.1111/j.1364-3703.2007.00417.x. [DOI] [PubMed] [Google Scholar]
- 4.Nicot P.C., Baille A. Aerial Plant Surface Microbiology. Springer; Boston, MA, USA: 2007. Integrated Control of Botrytis cinerea on Greenhouse Tomatoes; pp. 169–189. [Google Scholar]
- 5.Elad Y., Williamson B., Tudzynski P., Delen N. Botrytis: Biology, Pathology and Control. Springer Science and Business Media LLC; Dordrecht, The Netherlands: 2007. Botrytis spp. and Diseases They Cause in Agricultural Systems—An Introduction; pp. 1–8. [Google Scholar]
- 6.Fillinger S., Elad Y. Botrytis—The Fungus, the Pathogen and Its Management in Agricultural Systems. Springer; New York, NY, USA: 2016. [Google Scholar]
- 7.Holz G., Coertze S., Williamson B. The Ecology of Botrytis on Plant Surfaces. In: Elad Y., Williamson B., Tudzynski P., Delen N., editors. Botrytis: Biology, Pathology and Control. Springer; Dordrecht, The Netherlands: 2007. pp. 9–27. [Google Scholar]
- 8.Govrin E.M., Levine A. The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea. Curr. Biol. 2000;10:751–757. doi: 10.1016/S0960-9822(00)00560-1. [DOI] [PubMed] [Google Scholar]
- 9.Van Kan J.A. Licensed to kill: The lifestyle of a necrotrophic plant pathogen. Trends Plant Sci. 2006;11:247–253. doi: 10.1016/j.tplants.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 10.Choquer M., Fournier E., Kunz C., Levis C., Pradier J.-M., Simon A., Viaud M. Botrytis cinerea virulence factors: New insights into a necrotrophic and polyphageous pathogen. FEMS Microbiol. Lett. 2007;277:1–10. doi: 10.1111/j.1574-6968.2007.00930.x. [DOI] [PubMed] [Google Scholar]
- 11.Gourgues M., Brunet-Simon A., Lebrun M.-H., Levis C. The tetraspanin BcPls1 is required for appressorium-mediated penetration of Botrytis cinerea into host plant leaves. Mol. Microbiol. 2003;51:619–629. doi: 10.1046/j.1365-2958.2003.03866.x. [DOI] [PubMed] [Google Scholar]
- 12.Amselem J., Cuomo C.A., Van Kan J.A.L., Viaud M., Benito E.P., Couloux A., Coutinho P.M., De Vries R.P., Dyer P.S., Fillinger S., et al. Genomic Analysis of the Necrotrophic Fungal Pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet. 2011;7:e1002230. doi: 10.1371/journal.pgen.1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Espino J., Brito N., Noda J., González C. Botrytis cinerea endo-ß-1,4-glucanase Cel5A is expressed during infection but is not required for pathogenesis. Physiol. Mol. Plant Pathol. 2005;66:213–221. doi: 10.1016/j.pmpp.2005.06.005. [DOI] [Google Scholar]
- 14.Kars I., McCalman M., Wagemakers L., Van Kan J.A.L. Functional analysis of Botrytis cinerea pectin methylesterase genes by PCR-based targeted mutagenesis: Bcpme1 and Bcpme2 are dispensable for virulence of strain B05.10. Mol. Plant Pathol. 2005;6:641–652. doi: 10.1111/j.1364-3703.2005.00312.x. [DOI] [PubMed] [Google Scholar]
- 15.Van Kan J.A.L., Klooster J.W.V., Wagemakers C.A.M., Dees D.C.T., Van Der Vlugt-Bergmans C.J.B. Cutinase A of Botrytis cinerea is Expressed, but not Essential, During Penetration of Gerbera and Tomato. Mol. Plant Microbe Interact. 1997;10:30–38. doi: 10.1094/MPMI.1997.10.1.30. [DOI] [PubMed] [Google Scholar]
- 16.Reis H., Pfiffi S., Hahn M. Molecular and functional characterization of a secreted lipase from Botrytis cinerea. Mol. Plant Pathol. 2005;6:257–267. doi: 10.1111/j.1364-3703.2005.00280.x. [DOI] [PubMed] [Google Scholar]
- 17.Colmenares A.J., Aleu J., Durán-Patrón R., Collado I.G.G., Hernández-Galán R. The Putative Role of Botrydial and Related Metabolites in the Infection Mechanism of Botrytis cinerea. J. Chem. Ecol. 2002;28:997–1005. doi: 10.1023/A:1015209817830. [DOI] [PubMed] [Google Scholar]
- 18.Van Kan J.A.L., Stassen J.H.M., Mosbach A., Van Der Lee T.A.J., Faino L., Farmer A.D., Papasotiriou D.G., Zhou S., Seidl M.F., Cottam E., et al. A gapless genome sequence of the fungus Botrytis cinerea. Mol. Plant Pathol. 2016;18:75–89. doi: 10.1111/mpp.12384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manteau S., Abouna S., Lambert B., Legendre L. Differential regulation by ambient pH of putative virulence factor secretion by the phytopathogenic fungus Botrytis cinerea. FEMS Microbiol. Ecol. 2003;43:359–366. doi: 10.1111/j.1574-6941.2003.tb01076.x. [DOI] [PubMed] [Google Scholar]
- 20.Kunz C., Vandelle E., Rolland S., Poinssot B., Bruel C., Cimerman A., Zotti C., Moreau E., Vedel R., Pugin A., et al. Characterization of a new, nonpathogenic mutant of Botrytis cinerea with impaired plant colonization capacity. New Phytol. 2006;170:537–550. doi: 10.1111/j.1469-8137.2006.01682.x. [DOI] [PubMed] [Google Scholar]
- 21.AbuQamar S.F., Moustafa K., Tran L.S. Mechanisms and strategies of plant defense against Botrytis cinerea. Crit. Rev. Biotechnol. 2017;37:262–274. doi: 10.1080/07388551.2016.1271767. [DOI] [PubMed] [Google Scholar]
- 22.Veloso J., Van Kan J.A. Many Shades of Grey in Botrytis–Host Plant Interactions. Trends Plant Sci. 2018;23:613–622. doi: 10.1016/j.tplants.2018.03.016. [DOI] [PubMed] [Google Scholar]
- 23.Racovitza A. Muséum National d’Histoire naturelle. Volume 10. Museum National D’histoire Naturelle; Paris, France: 1959. Etude systématique et bryologique des champignons bryophiles; p. 288. [Google Scholar]
- 24.Ponce De León I., Oliver J.P., Castro A., Gaggero C., Bentancor M., Vidal S. Erwinia carotovora elicitors and Botrytis cinerea activate defense responses in Physcomitrella patens. BMC Plant Biol. 2007;7:52. doi: 10.1186/1471-2229-7-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krings M., Taylor T.N., Hass H., Kerp H., Dotzler N., Hermsen E.J. Fungal endophytes in a 400-million-yr-old land plant: Infection pathways, spatial distribution, and host responses. New Phytol. 2007;174:648–657. doi: 10.1111/j.1469-8137.2007.02008.x. [DOI] [PubMed] [Google Scholar]
- 26.Ponce De León I., Schmelz E.A., Gaggero C., Castro A., Álvarez A., Montesano M. Physcomitrella patens activates reinforcement of the cell wall, programmed cell death and accumulation of evolutionary conserved defence signals, such as salicylic acid and 12-oxo-phytodienoic acid, but not jasmonic acid, upon Botrytis cinerea infection. Mol. Plant Pathol. 2012;13:960–974. doi: 10.1111/j.1364-3703.2012.00806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan H.Q., Zhang T.T., Lan S.C., Jiang S. Ultrastructural study on the interaction between Physcomitrella patens and Botrytis cinerea. Plant Pathol. 2017;67:42–50. doi: 10.1111/ppa.12720. [DOI] [Google Scholar]
- 28.Reboledo G., Agorio A., Vignale L., Batista-García R., Ponce de León I. Transcriptional profiling reveals conserved and species-specific plant defense responses during the interaction of Physcomitrium patens with Botrytis cinerea. Plant Mol. Biol. 2020 doi: 10.1007/s11103-021-01116-0. under review. [DOI] [PubMed] [Google Scholar]
- 29.Ashton N.W., Cove D.J. The isolation and preliminary characterisation of auxotrophic and analogue resistant mutants of the moss, Physcomitrella patens. Mol. Genet. Genom. 1977;154:87–95. doi: 10.1007/BF00265581. [DOI] [Google Scholar]
- 30.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim D., Langmead B., Salzberg S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liao Y., Smyth G.K., Shi W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2013;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 34.Martini F., Binder H. pcaExplorer: An R/Bioconductor package for interacting with RNA-seq principal components. BMC Bioinform. 2019;20:1–8. doi: 10.1186/s12859-019-2879-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benjamini Y., Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Stat. Methodol. 1995;57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 37.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 38.Coolen S., Proietti S., Hickman R., Olivas N.H.D., Huang P.-P., Van Verk M.C., Van Pelt J.A., Wittenberg A.H., De Vos M., Prins M., et al. Transcriptome dynamics of Arabidopsis during sequential biotic and abiotic stresses. Plant J. 2016;86:249–267. doi: 10.1111/tpj.13167. [DOI] [PubMed] [Google Scholar]
- 39.De Cremer K., Mathys J., Voß C., Froenicke L., Michelmore R., Cammue B.P.A., De Coninck B. RNAseq-based transcriptome analysis of Lactuca sativa infected by the fungal necrotroph Botrytis cinerea. Plant Cell Environ. 2013;36:1992–2007. doi: 10.1111/pce.12106. [DOI] [PubMed] [Google Scholar]
- 40.Srivastava D.A., Arya G.C., Pandaranayaka E.P.J., Manasherova E., Prusky D., Elad Y., Frenkel O., Harel A. Transcriptome Profiling Data of Botrytis cinerea Infection on Whole Plant Solanum lycopersicum. Mol. Plant Microbe Interact. 2020;33:1103–1107. doi: 10.1094/MPMI-05-20-0109-A. [DOI] [PubMed] [Google Scholar]
- 41.Raudvere U., Kolberg L., Kuzmin I., Arak T., Adler P., Peterson H., Vilo J. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update) Nucleic Acids Res. 2019;47:W191–W198. doi: 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heard S., Brown N.A., Hammond-Kosack K. An Interspecies Comparative Analysis of the Predicted Secretomes of the Necrotrophic Plant Pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS ONE. 2015;10:e0130534. doi: 10.1371/journal.pone.0130534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu W., Ronen M., Gur Y., Minz-Dub A., Masrati G., Ben-Tal N., Savidor A., Sharon I., Eizner E., Valerius O., et al. BcXYG1, a Secreted Xyloglucanase from Botrytis cinerea, Triggers Both Cell Death and Plant Immune Responses. Plant Physiol. 2017;175:438–456. doi: 10.1104/pp.17.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cove D.J., Knight C.D., Lamparter T. Mosses as model systems. Trends Plant Sci. 1997;2:99–105. doi: 10.1016/S1360-1385(96)10056-X. [DOI] [Google Scholar]
- 45.Zhang L., Thiewes H., Van Kan J.A. The d-galacturonic acid catabolic pathway in Botrytis cinerea. Fungal Genet. Biol. 2011;48:990–997. doi: 10.1016/j.fgb.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 46.Mohnen D. Pectin structure and biosynthesis. Curr. Opin. Plant Biol. 2008;11:266–277. doi: 10.1016/j.pbi.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 47.Zhang L., Lubbers R.J., Simon A., Stassen J.H., Ribera P.R.V., Viaud M., Van Kan J.A. A novel Zn2Cys6transcription factor BcGaaR regulates D-galacturonic acid utilization in Botrytis cinerea. Mol. Microbiol. 2016;100:247–262. doi: 10.1111/mmi.13314. [DOI] [PubMed] [Google Scholar]
- 48.Valero-Jiménez C.A., Veloso J., Staats M., Van Kan J.A. Comparative genomics of plant pathogenic Botrytis species with distinct host specificity. BMC Genom. 2019;20:1–12. doi: 10.1186/s12864-019-5580-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kong W., Chen N., Liu T., Zhu J., Wang J., He X., Jingqi W. Large-Scale Transcriptome Analysis of Cucumber and Botrytis cinerea during Infection. PLoS ONE. 2015;10:e0142221. doi: 10.1371/journal.pone.0142221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haile Z.M., Guzman E.G.N.-D., Moretto M., Sonego P., Engelen K., Zoli L., Moser C., Baraldi E. Transcriptome Profiles of Strawberry (Fragaria vesca) Fruit Interacting With Botrytis cinerea at Different Ripening Stages. Front. Plant Sci. 2019;10:1131. doi: 10.3389/fpls.2019.01131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haile Z.M., Malacarne G., Pilati S., Sonego P., Moretto M., Masuero D., Vrhovsek U., Engelen K., Baraldi E., Moser C. Dual Transcriptome and Metabolic Analysis of Vitis vinifera cv. Pinot Noir Berry and Botrytis cinerea During Quiescence and Egressed Infection. Front. Plant Sci. 2020;10:1704. doi: 10.3389/fpls.2019.01704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davey M.L., Currah R.S. Interactions between mosses (Bryophyta) and fungi. Can. J. Bot. 2006;84:1509–1519. doi: 10.1139/b06-120. [DOI] [Google Scholar]
- 53.Ponce De León I. The Moss Physcomitrella patens as a Model System to Study Interactions between Plants and Phytopathogenic Fungi and Oomycetes. J. Pathog. 2011;2011:1–6. doi: 10.4061/2011/719873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tenberge K.B., Beckedorf M., Hoppe B., Schouten A., Solf M., Driesch M.V.D. In Situ Localization of AOS in Host-Pathogen Interactions. Microsc. Microanal. 2002;8:250–251. doi: 10.1017/S1431927602100067. [DOI] [Google Scholar]
- 55.Zhang M., Sun C., Liu Y., Feng H., Chang H., Cao S., Li G., Yang S., Hou J., Zhu-Salzman K., et al. Transcriptome analysis and functional validation reveal a novel gene, BcCGF1, that enhances fungal virulence by promoting infection-related development and host penetration. Mol. Plant Pathol. 2020;21:834–853. doi: 10.1111/mpp.12934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hou J., Feng H., Chang H., Liu Y., Li G., Yang S., Sun C., Zhang M., Yuan Y., Sun J., et al. The H3K4 demethylase Jar1 orchestrates ROS production and expression of pathogenesis-related genes to facilitate Botrytis cinerea virulence. New Phytol. 2019;225:930–947. doi: 10.1111/nph.16200. [DOI] [PubMed] [Google Scholar]
- 57.Turrion-Gomez J.L., Benito E.P. Flux of nitric oxide between the necrotrophic pathogen Botrytis cinerea and the host plant. Mol. Plant Pathol. 2011;12:606–616. doi: 10.1111/j.1364-3703.2010.00695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Siegmund U., Viefhues A. Botrytis—The Fungus, the Pathogen and Its Management in Agricultural Systems. Springer; Cham, Germany: 2015. Reactive Oxygen Species in the Botrytis—Host Interaction; pp. 269–289. [Google Scholar]
- 59.Espino J.J., Gutiérrez-Sánchez G., Brito N., Shah P., Orlando R., González C. The Botrytis cinerea early secretome. Proteom. 2010;10:3020–3034. doi: 10.1002/pmic.201000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tiedemann A. Evidence for a primary role of active oxygen species in induction of host cell death during infection of bean leaves with Botrytis cinerea. Physiol. Mol. Plant Pathol. 1997;50:151–166. doi: 10.1006/pmpp.1996.0076. [DOI] [Google Scholar]
- 61.Pinedo C., Wang C.-M., Pradier J.-M., Dalmais B., Choquer M., Le Pêcheur P., Morgant G., Collado I.G., Cane D.E., Viaud M. Sesquiterpene Synthase from the Botrydial Biosynthetic Gene Cluster of the Phytopathogen Botrytis cinerea. ACS Chem. Biol. 2008;3:791–801. doi: 10.1021/cb800225v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Siewers V., Viaud M., Jimenez-Teja D., Collado I.G., Gronover C.S., Pradier J.-M., Tudzynski B., Tudzynski P. Functional Analysis of the Cytochrome P450 Monooxygenase Gene bcbot1 of Botrytis cinerea Indicates That Botrydial Is a Strain-Specific Virulence Factor. Mol. Plant Microbe Interact. 2005;18:602–612. doi: 10.1094/MPMI-18-0602. [DOI] [PubMed] [Google Scholar]
- 63.Collado I.G.G., Viaud M. Botrytis—The Fungus, the Pathogen and Its Management in Agricultural Systems. Springer; Cham, Germany: 2015. Secondary Metabolism in Botrytis cinerea: Combining Genomic and Metabolomic Approaches; pp. 291–313. [Google Scholar]
- 64.Porquier A., Morgant G., Moraga J., Dalmais B., Luyten I., Simon A., Pradier J.-M., Amselem J., Collado I.G., Viaud M. The botrydial biosynthetic gene cluster of Botrytis cinerea displays a bipartite genomic structure and is positively regulated by the putative Zn(II)2Cys6 transcription factor BcBot6. Fungal Genet. Biol. 2016;96:33–46. doi: 10.1016/j.fgb.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 65.Dalmais B., Schumacher J., Moraga J., Le Pêcheur P., Tudzynski B., Collado I.G., Viaud M. The Botrytis cinerea phytotoxin botcinic acid requires two polyketide synthases for production and has a redundant role in virulence with botrydial. Mol. Plant Pathol. 2011;12:564–579. doi: 10.1111/j.1364-3703.2010.00692.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Arenas Y.C., Kalkman E.R., Schouten A., Dieho M., Vredenbregt P., Uwumukiza B., Oses-Ruiz M., Van Kan J.A. Functional analysis and mode of action of phytotoxic Nep1-like proteins of Botrytis cinerea. Physiol. Mol. Plant Pathol. 2010;74:376–386. doi: 10.1016/j.pmpp.2010.06.003. [DOI] [Google Scholar]
- 67.Noda J., Brito N., González C. The Botrytis cinerea xylanase Xyn11A contributes to virulence with its necrotizing activity, not with its catalytic activity. BMC Plant Biol. 2010;10:38. doi: 10.1186/1471-2229-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kars I., Krooshof G.H., Wagemakers L., Joosten R., Benen J.A.E., Van Kan J.A.L. Necrotizing activity of five Botrytis cinerea endopolygalacturonases produced in Pichia pastoris. Plant J. 2005;43:213–225. doi: 10.1111/j.1365-313X.2005.02436.x. [DOI] [PubMed] [Google Scholar]
- 69.Brito N., Espino J.J., González C. The Endo-β-1,4-Xylanase Xyn11A Is Required for Virulence in Botrytis cinerea. Mol. Plant Microbe Interact. 2006;19:25–32. doi: 10.1094/MPMI-19-0025. [DOI] [PubMed] [Google Scholar]
- 70.Li B., Wang W., Zong Y., Qin G., Tian S. Exploring Pathogenic Mechanisms of Botrytis cinerea Secretome under Different Ambient pH Based on Comparative Proteomic Analysis. J. Proteome Res. 2012;11:4249–4260. doi: 10.1021/pr300365f. [DOI] [PubMed] [Google Scholar]
- 71.Shah P., Atwood J.A., Orlando R., El Mubarek H., Podila G.K., Davis M.R. Comparative Proteomic Analysis of Botrytis cinerea Secretome. J. Proteome Res. 2009;8:1123–1130. doi: 10.1021/pr8003002. [DOI] [PubMed] [Google Scholar]
- 72.Fernández-Acero F.J., Colby T., Harzen A., Carbú M., Wieneke U., Cantoral J.M., Schmidt J. 2-DE proteomic approach to the Botrytis cinerea secretome induced with different carbon sources and plant-based elicitors. Proteomics. 2010;10:2270–2280. doi: 10.1002/pmic.200900408. [DOI] [PubMed] [Google Scholar]
- 73.Shah P., Powell A.L., Orlando R., Bergmann C., Gutierrez-Sanchez G. Proteomic Analysis of Ripening Tomato Fruit Infected by Botrytis cinerea. J. Proteome Res. 2012;11:2178–2192. doi: 10.1021/pr200965c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Staples R.C., Mayer A. Putative virulence factors of Botrytis cinerea acting as a wound pathogen. FEMS Microbiol. Lett. 1995;134:1–7. doi: 10.1111/j.1574-6968.1995.tb07905.x. [DOI] [Google Scholar]
- 75.Leroch M., Kleber A., Silva E., Coenen T., Koppenhöfer D., Shmaryahu A., Valenzuela P.D.T., Hahn M. Transcriptome Profiling of Botrytis cinerea Conidial Germination Reveals Upregulation of Infection-Related Genes during the Prepenetration Stage. Eukaryot. Cell. 2013;12:614–626. doi: 10.1128/EC.00295-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee S.B., Yang S.U., Pandey G., Kim M., Hyoung S., Choi D., Shin H.-Y., Suh M.C. Occurrence of land-plant-specific glycerol-3-phosphate acyltransferases is essential for cuticle formation and gametophore development in Physcomitrella patens. New Phytol. 2019;225:2468–2483. doi: 10.1111/nph.16311. [DOI] [PubMed] [Google Scholar]
- 77.Blanco-Ulate B., Morales-Cruz A., Amrine K.C.H., Labavitch J.M., Powell A.L.T., Cantu D. Genome-wide transcriptional profiling of Botrytis cinerea genes targeting plant cell walls during infections of different hosts. Front. Plant Sci. 2014;5:435. doi: 10.3389/fpls.2014.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Have A.T., Breuil W.O., Wubben J.P., Visserb J., Van Kan J.A. Botrytis cinerea Endopolygalacturonase Genes Are Differentially Expressed in Various Plant Tissues. Fungal Genet. Biol. 2001;33:97–105. doi: 10.1006/fgbi.2001.1269. [DOI] [PubMed] [Google Scholar]
- 79.Moller I., Sørensen I., Bernal A.J., Blaukopf C., Lee K., Øbro J., Pettolino F., Roberts A., Mikkelsen J.D., Knox J.P., et al. High-throughput mapping of cell-wall polymers within and between plants using novel microarrays. Plant J. 2007;50:1118–1128. doi: 10.1111/j.1365-313X.2007.03114.x. [DOI] [PubMed] [Google Scholar]
- 80.Roberts A.W., Roberts E.M., Haigler C.H. Moss cell walls: Structure and biosynthesis. Front. Plant Sci. 2012;3:166. doi: 10.3389/fpls.2012.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang L., van Kan J.A.L. Botrytis cinerea mutants deficient in D-galacturonic acid catabolism have a perturbed virulence on Nicotiana benthamiana and Arabidopsis; but not on tomato. Mol. Plant Pathol. 2013;14:19–29. doi: 10.1111/j.1364-3703.2012.00825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nafisi M., Stranne M., Zhang L., Van Kan J.A.L., Sakuragi Y. The Endo-Arabinanase BcAra1 Is a Novel Host-Specific Virulence Factor of the Necrotic Fungal Phytopathogen Botrytis cinerea. Mol. Plant Microbe Interact. 2014;27:781–792. doi: 10.1094/MPMI-02-14-0036-R. [DOI] [PubMed] [Google Scholar]
- 83.Lyon G.D., Goodman B.A., Williamson B. Botrytis: Biology, Pathology and Control. Springer; Dordrecht, The Netherlands: 2007. Botrytis cinerea Perturbs Redox Processes as an Attack Strategy in Plants; pp. 119–141. [Google Scholar]
- 84.González-Fernández R., Aloria K., Valero-Galván J., Redondo I., Arizmendi J.M., Jorrín-Novo J.V. Proteomic analysis of mycelium and secretome of different Botrytis cinerea wild-type strains. J. Proteom. 2014;97:195–221. doi: 10.1016/j.jprot.2013.06.022. [DOI] [PubMed] [Google Scholar]
- 85.Pandey V., Singh M., Pandey D., Kumar A. Integrated proteomics, genomics, metabolomics approaches reveal oxalic acid as pathogenicity factor in Tilletia indica inciting Karnal bunt disease of wheat. Sci. Rep. 2018;8:1–14. doi: 10.1038/s41598-018-26257-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Guyon K., Balagué C., Roby D., Raffaele S. Secretome analysis reveals effector candidates associated with broad host range necrotrophy in the fungal plant pathogen Sclerotinia sclerotiorum. BMC Genom. 2014;15:1–19. doi: 10.1186/1471-2164-15-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Seifbarghi S., Borhan M.H., Wei Y., Coutu C., Robinson S.J., Hegedus D.D. Changes in the Sclerotinia sclerotiorum transcriptome during infection of Brassica napus. BMC Genom. 2017;18:1–37. doi: 10.1186/s12864-017-3642-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Viaud M., Brunet-Simon A., Brygoo Y., Pradier J.-M., Levis C. Cyclophilin A and calcineurin functions investigated by gene inactivation, cyclosporin A inhibition and cDNA arrays approaches in the phytopathogenic fungus Botrytis cinerea. Mol. Microbiol. 2003;50:1451–1465. doi: 10.1046/j.1365-2958.2003.03798.x. [DOI] [PubMed] [Google Scholar]
- 89.Have A.T., Espino J.J., Dekkers E., Van Sluyter S.C., Brito N., Kay J., González C., Van Kan J.A. The Botrytis cinerea aspartic proteinase family. Fungal Genet. Biol. 2010;47:53–65. doi: 10.1016/j.fgb.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 90.Denton-Giles M., McCarthy H., Sehrish T., Dijkwel Y., Mesarich C.H., Bradshaw R.E., Cox M.P., Dijkwel P.P. Conservation and expansion of a necrosis-inducing small secreted protein family from host-variable phytopathogens of the Sclerotiniaceae. Mol. Plant Pathol. 2020;21:512–526. doi: 10.1111/mpp.12913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pennington H.G., Jones R., Kwon S., Bonciani G., Thieron H., Chandler T., Luong P., Morgan S.N., Przydacz M., Bozkurt T., et al. The fungal ribonuclease-like effector protein CSEP0064/BEC1054 represses plant immunity and interferes with degradation of host ribosomal RNA. PLoS Pathog. 2019;15:e1007620. doi: 10.1371/journal.ppat.1007620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Boller T., Felix G. A Renaissance of Elicitors: Perception of Microbe-Associated Molecular Patterns and Danger Signals by Pattern-Recognition Receptors. Annu. Rev. Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- 93.Zhang L., Kars I., Essenstam B., Liebrand T.W., Wagemakers L., Elberse J., Tagkalaki P., Tjoitang D., Ackerveken G.V.D., Van Kan J.A. Fungal Endopolygalacturonases Are Recognized as Microbe-Associated Molecular Patterns by the Arabidopsis Receptor-Like Protein RESPONSIVENESS TO BOTRYTIS POLYGALACTURONASES1. Plant Physiol. 2014;164:352–364. doi: 10.1104/pp.113.230698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lionetti V., Raiola A., Camardella L., Giovane A., Obel N., Pauly M., Favaron F., Cervone F., Bellincampi D. Overexpression of Pectin Methylesterase Inhibitors in Arabidopsis Restricts Fungal Infection by Botrytis cinerea. Plant Physiol. 2007;143:1871–1880. doi: 10.1104/pp.106.090803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sbaghi M., Jeandet P., Bessis R., Leroux P. Degradation of stilbene-type phytoalexins in relation to the pathogenicity of Botrytis cinerea to grapevines. Plant Pathol. 1996;45:139–144. doi: 10.1046/j.1365-3059.1996.d01-101.x. [DOI] [Google Scholar]
- 96.Quidde T., Osbourn A., Tudzynski P. Detoxification of α-tomatine by Botrytis cinerea. Physiol. Mol. Plant Pathol. 1998;52:151–165. doi: 10.1006/pmpp.1998.0142. [DOI] [Google Scholar]
- 97.El Oirdi M., Trapani A., Bouarab K. The nature of tobacco resistance against Botrytis cinerea depends on the infection structures of the pathogen. Environ. Microbiol. 2010;12:239–253. doi: 10.1111/j.1462-2920.2009.02063.x. [DOI] [PubMed] [Google Scholar]
- 98.Pedras M.S.C., Hossain S., Snitynsky R.B. Detoxification of cruciferous phytoalexins in Botrytis cinerea: Spontaneous dimerization of a camalexin metabolite. Phytochemistry. 2011;72:199–206. doi: 10.1016/j.phytochem.2010.11.018. [DOI] [PubMed] [Google Scholar]
- 99.VanEtten H., Temporini E., Wasmann C. Phytoalexin (and phytoanticipin) tolerance as a virulence trait: Why is it not required by all pathogens? Physiol. Mol. Plant Pathol. 2001;59:83–93. doi: 10.1006/pmpp.2001.0350. [DOI] [Google Scholar]
- 100.Vela-Corcía D., Srivastava D.A., Dafa-Berger A., Rotem N., Barda O., Levy M. MFS transporter from Botrytis cinerea provides tolerance to glucosinolate-breakdown products and is required for pathogenicity. Nat. Commun. 2019;10:1–11. doi: 10.1038/s41467-019-10860-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Stefanato F.L., Abou-Mansour E., Buchala A., Kretschmer M., Mosbach A., Hahn M., Bochet C.G., Métraux J.-P., Schoonbeek H.-J. The ABC transporter BcatrB from Botrytis cinerea exports camalexin and is a virulence factor on Arabidopsis thaliana. Plant J. 2009;58:499–510. doi: 10.1111/j.1365-313X.2009.03794.x. [DOI] [PubMed] [Google Scholar]
- 102.Hayashi K., Schoonbeek H.-J., De Waard M.A. Bcmfs1, a Novel Major Facilitator Superfamily Transporter from Botrytis cinerea, Provides Tolerance towards the Natural Toxic Compounds Camptothecin and Cercosporin and towards Fungicides. Appl. Environ. Microbiol. 2002;68:4996–5004. doi: 10.1128/AEM.68.10.4996-5004.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ponce De León I., Montesano M. Adaptation Mechanisms in the Evolution of Moss Defenses to Microbes. Front. Plant Sci. 2017;8:366. doi: 10.3389/fpls.2017.00366. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.