Abstract
Members of the archaeal order Methanomassiliicoccales are methanogens mainly associated with animal digestive tracts. However, environmental members remain poorly characterized as no representatives not associated with a host have been cultivated so far. In this study, metabarcoding screening combined with quantitative PCR analyses on a collection of diverse non-host-associated environmental samples revealed that Methanomassiliicoccales were very scarce in most terrestrial and aquatic ecosystems. Relative abundance of Methanomassiliicoccales and substrates/products of methanogenesis were monitored during incubation of environmental slurries. A sediment slurry enriched in Methanomassiliicoccales was obtained from a freshwater sample. It allowed the reconstruction of a high-quality metagenome-assembled genome (MAG) corresponding to a new candidate species, for which we propose the name of Candidatus ‘Methanomassiliicoccus armoricus MXMAG1’. Comparison of the annotated genome of MXMAG1 with the published genomes and MAGs from Methanomassiliicoccales belonging to the 2 known clades (‘free-living’/non-host-associated environmental clade and ‘host-associated’/digestive clade) allowed us to explore the putative physiological traits of Candidatus ‘M. armoricus MXMAG1’. As expected, Ca. ‘Methanomassiliicoccus armoricus MXMAG1’ had the genetic potential to produce methane by reduction of methyl compounds and dihydrogen oxidation. This MAG encodes for several putative physiological and stress response adaptations, including biosynthesis of trehalose (osmotic and temperature regulations), agmatine production (pH regulation), and arsenic detoxication, by reduction and excretion of arsenite, a mechanism that was only present in the ‘free-living’ clade. An analysis of co-occurrence networks carried out on environmental samples and slurries also showed that Methanomassiliicoccales detected in terrestrial and aquatic ecosystems were strongly associated with acetate and dihydrogen producing bacteria commonly found in digestive habitats and which have been reported to form syntrophic relationships with methanogens.
Keywords: Methanomassiliicoccales, cultivation, methyl-compounds, environmental cluster, networks
1. Introduction
Methanogenic Archaea are responsible for production of biogenic methane (CH4) on Earth, reported to release one-half of the annual output of this strong greenhouse gas into the atmosphere [1]. Methanogens inhabit a large variety of anoxic terrestrial and aquatic environments, as well as various animal gastrointestinal tracts (GITs) [2]. In these ecosystems, methanogenesis occurs under reduced conditions, in areas where substrates are produced from fermentation (H2, CO2, acetate, or methylated compounds) or geological processes (H2, CO2, CO) [3,4,5]. Over the last decade, our knowledge of methanogenesis pathways and the taxa carrying out this process has changed considerably. For example, methoxylated aromatics and short-chain alkanes have been described as potential methanogenesis substrates for newly-described methanogenic lineages [6,7,8]. Furthermore, genome-centric metagenomics studies and cultivation efforts led to the discovery of several new taxa of methanogens phylogenetically distant from the euryarchaeal phylum [9,10,11,12]. Most of these taxa were predicted to perform a methyl-dependent hydrogenotrophic methanogenesis, challenging the paradigm of an ancestral euryarchaeal methanogenesis based on the consumption of H2/CO2 [8,12,13,14].
The recently described order Methanomassiliicoccales has been reported to perform a methyl-dependent hydrogenotrophic methanogenesis (reducing methyl-compounds with H2 as electron donor), that involves two types of heterodisulfide reductases, HdrB and HdrD to conserve energy [15,16,17]. In addition, Methanomassiliicoccales possess rather rare metabolic properties because they require the 22nd amino acid, pyrrolysine, encoded by an UAG (amber) codon [16].
Based on 16S rRNA and mcrA (methyl co-enzyme M subunit A) genes, this order encompasses at least two family-level lineages, respectively, called Methanomassiliicoccaceae and Candidatus ‘Methanomethylophilaceae’ [18,19,20], frequently associated with the two specific habitats (host-associated habitats versus non-host associated ones), in which they are found and probably reflecting their ecological adaptation [21,22,23,24].
The first cluster, referred to as the ‘host-associated’ clade, corresponding roughly to Ca. ‘Methanomethylophilaceae’, is the one that has been the most studied [24,25]. It comprises representatives originating exclusively from animal GITs (e.g., mammals, reptiles, and insects), suggesting an evolution specialized towards digestive tract microbiomes [23,24]. This clade has representatives grown in enrichment cultures, derived from animal and human GITs or kitchen wastes: Ca. ‘Methanomethylophilus alvus’, Ca. ‘Methanoplasma termitum’, and Ca. ‘Methanogranum caenicola’, as well as strains RumEn M2 and ISO4-H5 [20,21,22,23,26,27]. It has notably been studied in human GITs, appears to be increasing with age of the person, and was predominantly associated with elderly subjects being in good health condition [9,24,28,29,30].
The second cluster of Methanomassiliicoccales, referred in the literature to as the ‘free-living’ clade, corresponding roughly to Methanomassiliicoccaceae, is much less documented. It comprises mainly members detected by molecular methods in terrestrial and aquatic ecosystems, and known in their vast majority only through 16S rRNA or mcrA gene sequences. Sequences affiliated with this ‘free-living’ clade have been detected in various common and extreme habitats, such as seafloor sediments, peatland soils, terrestrial hot springs, or mangroves [23,31,32,33]. To date, in addition to fragmentary environmental DNA sequences detected through molecular PCR-based methods, our knowledge of this ‘free-living’ clade is limited to few metagenome-assembled genomes (MAGs) and three cultured strains sometimes predominantly active in these environments [15,16,23,34]. So far, the few cultured strains from this clade are paradoxically derived from human GIT, i.e., Methanomassiliicoccus luminyensis (M. luminyensis, the only strain of Methanomassiliicoccales isolated in pure culture) and Ca. ‘Methanomassiliicoccus intestinalis’ or from ruminant GITs, i.e., strain RumEn M1 [15,16,23]. However, no Methanomassiliicoccales belonging to the ‘free-living’ clade has been isolated or enriched from an aquatic or terrestrial environment.
The goal of this study was to culture representatives of the ‘free-living’ clade of Methanomassiliicoccales from a natural environment, to obtain information on their biology and make assumptions about possible adaptations to terrestrial and aquatic ecosystems, and from a more general point of view, on a type of methanogenesis that seems to be widespread within the recently discovered methanogenic lineages. Methanomassiliicoccales abundance was determined for 22 samples from a variety of natural environments worldwide, with various physico-chemical characteristics, out of which, six samples were selected to perform enrichment in slurries. A metagenomic approach was applied and led to reconstruct a high-quality MAG of a Methanomassiliicoccales strain from the ‘free-living’ clade coming from a natural setting, providing matter for a phylogenetic and functional characterization.
2. Materials and Methods
2.1. Site Description
Eighty-six samples were collected from reported reduced areas of various ecosystems (marine sediments, deep-sea hydrothermal vents, marine pockmarks, submarine mud volcano, deep-sea hypersaline anoxic brine, peatland soils, lakes, hot springs, river sediments) during several sampling expeditions from 40 worldwide locations and stored in the laboratory for further investigations. Location and detailed information on sampling sites, depths, and sampling expeditions are given in Table S1. After a first molecular screening (as described in Text S1) performed to detect Methanomassiliicoccales-containing samples by PCR targeting 16S rRNA gene sequences of Methanomassiliicoccales and then confirmation of their presence by metabarcoding, 22 positive samples were selected for further investigations (Table 1).
Table 1.
Origin and main characteristics of the environmental samples selected after a pre-screening molecular stage targeting Methanomassiliicoccales. Environmental parameters were measured in situ during cruises/expeditions. –, not determined.
| Sample Acronym | Sample Type | Geographical Origin | Water Depth (m) | Depth Below Subsurface/Subseafloor (cm) | pH | Temperature (°C) |
|---|---|---|---|---|---|---|
| COMRA | Deep-sea sediments | South-West Indian ocean | 2267 | 0–20 | 7.5 | – |
| DOUR | Coastal sediments | France (Bay of Morlaix, France) | 0 | 5–10 | 7.25 | 8 |
| HM1 | Mud from a mud volcano | Barents sea, Norway | 1285 | 0–1 | – | – |
| HM2 | Mud from a mud volcano | Barents sea, Norway | 1285 | 1–6 | – | – |
| HM3 | Mud from a mud volcano | Barents sea, Norway | 1285 | 6–11 | – | – |
| HM4 | Mud from a mud volcano | Barents sea, Norway | 1282 | 1–6 | – | – |
| KERG | Water from a geothermal hot spring | Kerguelen Island, Indian Ocean (French Southern and Antarctic lands) | – | – | 7.0 | 50 |
| KRY150 | Deep-Sea Hypersaline Anoxic Basin | Mediterranean sea (Kryos basin) | ~3338 | – | 7.5 | 15 |
| KRY238 | Deep-Sea Hypersaline Anoxic Basin | Mediterranean sea (Kryos basin) | ~3338 | – | 6.5 | 15 |
| MOUG1 | Peatland soil | Peatland in France (Commana, France) | – | 0–10 | 6.4 | 4 |
| MOUG2 | Peatland soil | Peatland in France (Commana, France) | – | – | 4.78 | 2 |
| MOUG3 | Peatland soil | Peatland in France (Commana, France) | – | 0–10 | 5.22 | 6 |
| MOUG4 | Peatland soil | Peatland in France (Commana, France) | – | 0–10 | 4.75 | 2.3 |
| MOUG5 | Freshwater sediments | Peatland in France (Commana, France) | 0.2 | 5–15 | 6.7 | 8 |
| MOZ1 | Sediments from a pockmarck area | Mozambique Channel (Madagascar shore) | 762 | 2–4 | 8.0 | 8 |
| MOZ2 | Sediments from a pockmarck area | Mozambique Channel (Madagascar shore) | 762 | 4–6 | 8.0 | 8 |
| MOZ3 | Sediments from a pockmarck area | Mozambique Channel (Madagascar shore) | 762 | 6–11 | 8.0 | 8 |
| PAV60 | Anoxic water from a meromictic lake | Pavin lake (France) | 60 | – | 5.3 | 4.2 |
| PAV70 | Anoxic water from a meromictic lake | Pavin lake (France) | 70 | – | 5.3 | 4.75 |
| PAV80 | Anoxic water from a meromictic lake | Pavin lake (France) | 80 | – | 5.3 | 5 |
| PENF | Freshwater sediments | Tributary of the French river Penfeld (Brest, France) | 0.15 | 0–5 | 6.0 | 8 |
| XIA | Water from a geothermal hot spring | Hot spring in China (Xiamen Botanical Garden) | – | 0 | 6.3 | 82 |
The 22 samples used for an in-depth study were the following ones: (i) five samples were collected in a peatland area (Brittany, France), including four soil samples (MOUG1–MOUG4) and one sediment sample from a freshwater stream (MOUG5); (ii) four samples were collected from the active center of the deep-sea methane-emitting Håkon Mosby mud volcano, in the Barents Sea (Arctic Ocean) (HM1–HM4); (iii) three anoxic samples were collected in the water column of the meromictic crater Lake Pavin (Auvergne, France) at 60, 70, and 80 m water depth (PAV60-PAV70-PAV80); (iv) one geothermal hot spring sediment sample was collected from the Xiada reservoir of the Xiamen botanical garden (China) (XIA); (v) one geothermal spring sediment sample was taken from a hot spring at the Kerguelen islands (Kerguelen archipelago, France, Indian Ocean) (KERG); (vi) two samples were originating from the athalassic (MgCl2-rich) deep-sea hypersaline anoxic brine (DHAB) Kryos, in the Mediterranean Sea, respectively, collected from layers at 150 g L−1 salts (mainly MgCl2) (KRY150) and 238 g L−1 (mainly MgCl2) (KRY238); (vii) three sediments samples were collected in a pockmark field in the Mozambique Channel (off the Madagascar island) (MOZ1–MOZ3); (viii) one superficial marine sediment sample was collected from a marine bay influenced by freshwater inputs (DOUR) (English Channel, France); (ix) one deep-sea sediment sample was collected from the South West Indian Ocean (COMRA); and (x), finally, a freshwater sediment sample was taken from a stream (PENF) (Brittany, France). All the investigated samples were described and conditioned during each cruise and field expedition before being stored at 4 °C under anoxic conditions for culture and at −80 °C for DNA extraction. More detailed descriptions of these samples are given in Text S2 and Table S1.
2.2. DNA Extraction
Due to the various nature and composition of the environmental matrices, a standardized DNA extraction protocol, combining mechanical lysis by bead-beating and chemical lysis, was applied in triplicate to each sample onto 0.5 g of environmental matrix. One negative control was included and contained 0.5 mL of DNA-free sterile water. The mechanical lysis step was followed by a chemical lysis, based on the use of detergents (SDS and sarkozyl) and proteinase K, and was followed by a phenol/chloroform/isoamyl alcohol extraction of nucleic acids. The detailed procedure is described in Text S3. Elution of total DNA extracts was performed in 100 µL EB buffer (10 mM Tris-Cl, pH 8.5). Nucleic acid solution quality was determined using the NanoDrop™ 8000 (Thermo Scientific, Waltham, MA, USA) spectrophotometer. Double-strand DNA concentration was measured using the kit Quantifluor™ dsDNA system (Promega, Madison, WI, USA), following the manufacturer’s instructions.
2.3. Metabarcoding Sequencing and Sequence Analysis
The hypervariable partial SSU V4 region of the community 16S rRNA genes were analyzed to characterize the prokaryotic diversity in bulk samples and in substrate-amended slurries (DOUR, MOUG2, and PENF, respectively). Amplifications were carried out using the primer pair 515F-Y (5′-GTG YCA GCM GCC GCG GTA A-3′) [35] and 806R (5′-GCA CTA CNV GGG TWT CTA AT-3′) [36], with barcode on the forward primer, following the indexing and PCR conditions described elsewhere [37]; see https://www.earthmicrobiome.org/protocols-and-standards/16s/. The amplicons were then sequenced by the company Molecular Research-MrDNA (Shallowater, TX, USA), using the Illumina MiSeq (2 × 300 bp, paired-end reads; amplicon size: ~390 bp) technology, as described elsewhere [37]; see https://www.earthmicrobiome.org/protocols-and-standards/16s/. Raw reads were processed using the FROGS (Find, Rapidly, Operational taxonomic units (OTUs) with Galaxy Solution) pipeline [38] implemented on Galaxy (v. r3.0-1.4; Toulouse platform (see https://www.genotoul.fr/)) to generate FASTQ file, perform read quality controls, merge paired-end reads, remove unique singletons, and perform clustering by aggregation. Operational taxonomic units (OTUs) were generated from representative sequences (253 ± 20% bp) clustered as described elsewhere [38]. Singletons and potential chimeras were then removed to discriminate against sequencing artefacts. This resulted in 2,074,891 paired-end reads, forming, respectively, 14,096 and 11,843 OTUs, for bulk samples and cultures, respectively. Taxonomic affiliations of OTU representative sequences were done against the release 138 of the SILVA SSU database (www.arb-silva.de) and then double-checked against the RDP Classifier (v. 2.12). The taxonomic position of all Methanomassiliicoccales-related sequences was further analyzed by integrating the representative sequences into a reference dendrogram, using the distance-based phylogeny reconstruction algorithm BioNJ, as detailed in Text S4. The tree was visualized with iToL (v. 5.7; https://itol.embl.de/) (Figure 2).
The 16S rRNA gene amplicons dataset was analyzed using the phyloseq package (v. 1.28.0 R) [39] on program R studio (v. 1.3.5001). The OTUs abundance, assigned taxonomy, and sample contextual data were integrated into a phyloseq object before removing potential reagent contaminants using the decontam package (v. 1.4.0) [40]. Then, the metagenomSeq package (v. 1.26.3) [41] was used to normalize the dataset of the different samples by cumulative sum scaling (CSS), a median-quantile normalization which corrects differences in library sizes. Finally, all statistical analyses were performed with the phyloseq package. Extraction of 16S rRNA gene sequences representative of the OTUs and phylogenetic analyses are detailed in Text S4. A Bray-Curtis index was calculated to assess the dissimilarity between total microbial communities in environmental samples by observing ecological distances.
2.4. Quantitative Polymerase Chain Reaction
The abundance of 16S rRNA genes from Methanomassiliicoccales in natural samples and substrate-amended slurries was estimated using quantitative PCR (qPCR) with the primers AS05 Fw (5′-GGG GTA GGG GTA AAA TCC TG-3′) (this study)—AS2 Rv modified (5′-AAC AAC TTC TCT CCG GCA CT-3′) [28]. Quantifications were performed in triplicate with 5 ng of DNA template (5 μL of 1 ng μL−1 DNA template) (+ negative controls). Amplification reactions were carried out in a StepOnePlus™ Real-Time PCR system (Applied Biosystems, Foster City, CA, USA), in a final volume of 25 μL using the kit PerfeCTa® SYBR®Green SuperMix (Quanta Biosciences, Gaithersburg, MD, USA, supplier: VWR), 200 nM primers targeting Methanomassiliicoccales partial 16S rRNA gene sequences, and 5 μL of DNA template. For 16S rRNA gene copies quantification, qPCR conditions were as follows: 1 initial denaturation step at 95 °C for 10 min, followed by 40 cycles of denaturation at 95 °C for 15 s, then annealing and extension at 58 °C for 60 s. qPCR specificity was checked based on melting curve analysis, PCR efficiency (90–110%), r² of standard curves (>0.99), Ct (≤30), electrophoretic migration of amplicons, and sequencing checking of the PCR products. Standard curves were obtained from 10−1 to 108 copies µL−1 of plasmids containing a full 16S rRNA gene insert of Methanomassiliicoccus luminyensis (DSM 25720) (cloned in the vector pCR™2.1-TOPO™ of the TOPO™TA cloning® kit (Invitrogen)). The r2 of standard curves obtained by qPCR were equal to or more than 0.992 and PCR efficiencies between 97 and 105%. The qPCR results were expressed in terms of copy gene numbers per gram of wet weight sample or per ml. For the environmental samples, they were considered as exploitable and biologically relevant when reaching at least 1 × 103 gene copies per ml or gram of wet weight of sample.
2.5. Culture-Based Incubation Experiments
2.5.1. Medium Composition for Culture-Based Experiments with Environmental Slurries
Enrichment slurries targeting hydrogenotrophic methyl-dependent methanogens were performed with environmental samples either (i) having the highest Methanomassiliicoccales 16S rRNA copy number per gram of wet weight or ml quantified (ii) or corresponding to ecosystems with extreme and unusual physical-chemical parameters (KRY150—MgCl2-rich brine subjected to more than 33 MPa pressure; KERG—geothermal hot spring). The medium composition was designed based upon (i) previously published recipes used by culture collections (DSMZ medium 141) (Text S5) to grow or enriched Methanomassiliicoccales, (ii) on published enrichment media [15,17,42,43], and (iii) on metabolic information derived from metagenome-assembled genomes (MAGs) of Methanomassiliicoccales available in public databases [27]. Most of these recipes were based on substrate-rich composition, notably comprising a high concentration of methanol and/or methyl-compounds to promote Methanomassiliicoccales growth.
Basal medium contained, for 1 L of distilled water: 1 g NH4Cl, 1 g Na-acetate, 0.1 g peptone from casein (Merck, Germany), 0.5 g KH2PO4, 20 mL fatty-acid solution, 5 mL porcine hemin solution, 1 mL tungstate-selenite solution, 1 mL trace elements solution SL10 [44], 5 mL K3 vitamin solution, NaHCO3 (10% m/v), and resazurin (0.0001%) (see Text S5 for more details concerning the preparation of these solutions).
For culture-based incubation experiments performed with anoxic freshwater (PAV60), peat (MOUG2), geothermal hot spring water (KERG), and river sediments (PENF), 5 g NaCl, 0.4 g MgSO4·7H2O, and 0.05 g CaCl2·2H2O were initially added to the basal medium. Since the MOUG2 sample had relatively high acetate concentrations (>4 mM), no Na-acetate was added to the medium. For culture-based incubation experiments performed with marine samples (DOUR, KRY150), 20 g NaCl, 3.45 g MgSO4·7H2O, and 0.14 g CaCl2·2H2O were added to the basal medium. As Kryos DHAB is concentrated in magnesium [45], the magnesium concentration was adjusted to 150 g L−1 with MgCl2 for the slurry KRY150. The pH of the media were adjusted to 5.5 or 7.5 before boiling. After boiling, 0.5 g L−1 of cysteine HCl.H2O was added to the degassed medium under a N2/CO2 (80:20, v/v; 0.3 bar) flow, and the medium was then dispensed into 500 mL Schott flasks (135 mL of medium per flask). The flasks were closed with butyl rubber stoppers, and filled with a N2/CO2 (80:20, v/v; 0.3 bar) atmosphere before autoclaving. Vitamins, bicarbonates, trace elements, and reducing agent Na2S.9H2O (10% w/v) were then added to the medium before addition of substrates and cofactors from anoxic sterile stocks solutions: 40 mM trimethylamine (TMA), 40 mM methanol, 10 µM coenzyme M, and a gas phase of H2/CO2 (80:20, v/v; 1 bar). Since all published Methanomassiliicoccales MAGs seem to not contain the gene set required for coenzyme M production, 10 µM of coenzyme M were added to the media to promote their growth [27]. Media were then inoculated with 10% (w/v or v/v) original samples and incubated in the dark, without shaking, at 30 °C (DOUR, PAV60, PENF, MOUG2), 37 °C (KRY150), and 60 °C (KERG), respectively. When depleted, methanogenesis substrates were renewed by the addition of 40 mM TMA, 40 mM MeOH, and a renewal of the gas phase with H2/CO2 (80:20, v/v; 1 bar) to maintain methanogenesis activity. Uninoculated media were used as negative controls.
2.5.2. Monitoring of Culture-Based Incubation Experiments
The evolution of microbial diversity within substrate-amended slurries has been monitored by carrying out metabarcoding (as described in Section 2.3) and by quantifying Methanomassiliicoccales (as described in Section 2.4), on a weekly basis, on three technical replicates. Although the metabarcoding and qPCR analyses were made from the same DNA extracts, amplifications were performed with different polymerases and different primers (see Section 2.3 and Section 2.4). At the same time as these molecular analyses, an analytical monitoring of metabolic substrates and products, and, in particular, of those of hydrogenotrophic methyl-dependent methanogenesis, was carried out as described below.
2.5.3. Quantifications of Substrates and Metabolic Products
Headspace gas CH4, H2, N2, and CO2 were measured using a modified INFICON/Micro GC FUSION Gas Analyzer (INFICON, Basel, Switzerland) fitted with a pressure gauge and two conductivity detectors, and using argon as a carrier gas, as detailed in Text S6.
Samples for cation analyses were analyzed using a Dionex ICS-900 Ion Chromatography System (Dionex, Camberley, UK) coupled with a CERS 500 4 mm suppressor and a DS5 conductivity detector (40 °C) and fitted with an RFC-10 Reagent-Free Controller™, an ASDV autosampler, and an IonPac CS16 column maintained at 60 °C in an UltiMate™ 3000 Thermostated Column Compartment (Thermo Scientific, Waltham, MA, USA). Anions concentrations were quantified by anion chromatography as described elsewhere [46]. The detailed protocol is given in Text S6. Analysis of methanol concentrations was carried out using an Agilent 6890N Gas Chromatograph (GC) (Agilent Technologies, Santa Clara, CA, USA) coupled with a Flame Ionization Detector (FID) instrument as described in Text S6.
2.6. MAG Sequencing and Annotation
Metagenomic sequencing was carried out with the DNA extracted after 8 weeks of incubation of the slurry PENF (corresponding to the T8 sample of the culture-based experiment). This sample was in fact the one with the highest qPCR estimation in Methanomassiliicoccales. The aim was to reconstruct a MAG of Methanomassiliicoccales in order to learn more about the genetic, physiological and adaptive potential of the ‘free-living’ Methanomassiliicoccales cluster. A comparison of this MAG with genomes of ‘host-associated’ representatives was also intended to identify potential adaptations to the environment. Short read DNA sequencing was performed by Fasteris SA (Plan-les Ouates, Switzerland), using the NovaSeq 6000 technology (2 × 100 bp, 100,000,000 paired-end reads; Protocol genomic Nano, 250 bp insert size section). Libraries constructions and quality controls were performed by the sequencing facility and verified with FastQC (v. 0.11.8—https://www.bioinformatics.babraham.ac.uk/projects/fastqc/).
MAGs were reconstructed using the Let-it-bin pipeline (https://github.com/QuentinLetourneur/Let-it-bin). Reads were first trimmed with AlienTrimmer [47]. Redundant reads were then removed using khmer [48]. Filtered reads were assembled with Megahit [49] and metaSpades [50]. Binning was performed with MaxBin2 [51], MetaBAT [52], and MetaBAT2 [52]. MAGs assemblies’ statistics were obtained with Quast (v. 5.0.2; https://github.com/ablab/quast). MAG completeness and potential contamination were controlled with CheckM (v. 1.1.2—https://ecogenomics.github.io/CheckM/) [53]. Methanomassiliicoccales MAGs were identified by phylogenetic approaches described below. Two Methanomassiliicoccales MAGs obtained from metaSpades and Megahit assemblies contained contigs with identical sequences but interrupted at different loci. In order to improve the contig length, the contigs from these two MAGs were assembled with cap3 [54], leading to MXMAG1 MAG. A second MAG, MXMAG2, was obtained with metaSpades. Both were analyzed and annotated with the fast annotation software Prokka (v. 1.14.6—https://github.com/tseemann/prokka), Dfast (v. 1.2.5—https://github.com/nigyta/dfast_core), the online version of the RAST software (v. 2.0—http://rast.theseed.org/FIG/rast.cgi), and the MicroScope Microbial Genome Annotation and Analysis Platform (MaGe) (https://mage.genoscope.cns.fr/microscope/home/index.php), using KEGG and BioCyc databases with default parameters and databases for all of the five software/pipelines [55,56,57,58]. Functional annotations of predicted CDSs were further compared with NCBI (v. 2.10.0+), and UniProtKB database (release 2020_07).
2.7. Phylogenetic Position of the MAGs and Comparative Genomics
The archaeal MAGs obtained from the T8 sample metagenome were first placed in the archaeal phylogeny, including all major lineages of archaea to identify Methanomassiliicoccales representatives. The two Methanomassiliicoccales MAGs were then placed in a tree, including 17 other Methanomassiliicoccales genomes and 14 of the closest non-Methanomassiliicoccales genomes. Both phylogenies were based a concatenation of 40 conserved phylogenetic markers [8] were constructed in IQ-TREE with Maximum Likelihood (LG + F + G4) [59]. Average Nucleotide Identity (ANI) scores were calculated using the ANI calculator tool provided by the EzBioCloud web server (https://www.ezbiocloud.net/tools/ani) [60] and using the JSpeciesWS (http://jspecies.ribohost.com/jspeciesws/) [61], between the high-quality MAG and its closest relative organisms Methanomassiliicoccus luminyensis B10T (Accession number NZ_CAJE00000000.1) and Candidatus ‘Methanomassiliicoccus intestinalis’ Issoire-Mx1 (Accession number: NC_021353.1). DNA-DNA hybridization (DDH) estimate values were calculated using the genome-to-genome distance calculator (GGDC, v. 2.1, formula 2) [62] between the high-quality MAG and two closest relative organisms cited above.
Representative Methanomassiliicoccales genomes from the ‘host-associated’ and ‘free-living’ clades were compared to reconstructed MAGs using the MaGE platform’s Pan-genome Analysis tool (https://mage.genoscope.cns.fr/microscope/home/index.php), based on the clustering algorithm SiLiX (http://lbbe.univ-lyon1.fr/-SiLiX-.html) which clustered genomic CDSs by 50% or/and 80% amino-acid identity and 80% amino-acid alignment coverage, with permissive parameters. Resulting CDSs were blasted against the UniprotKB database, and hypothetical protein CDSs were analyzed with the InterProScan webserver (https://www.ebi.ac.uk/interpro/) for functional predictions.
2.8. Co-Occurrence Network Analysis
Network analysis was used to describe co-occurrences/putative associations between taxa within the environmental samples and in the substrate-amended slurries, respectively. On the basis of the co-occurrences of taxa found in the different samples and cultures, a matrix of correlation was computed between all the previously assigned OTUs, defined as mentioned previously (non-parametric Spearman). In this network, each node represented an OTU, and the links between nodes represented the positive correlation between them. The strategy used to analyze its topology was the same as described elsewhere [63]. In order to catch the most relevant information embedded in these co-occurrences, we started to analyze the networks at their respective percolation thresholds, which are the values at which the overall organization of the networks is a minimal giant cluster bearing the minimum number of links required for an overall connectivity of the system [64]. The visualization and statistical analysis of the network at these thresholds was done with Gephi (v. 0.9.2) [65]. Notably, the hierarchical structure was explored through a modularity algorithm [66]. An in-depth bibliographical analysis of the metabolic characteristics of known representatives of the taxa co-occurring in this network was carried out to focus on potential physiological characteristics described for the cultured taxa of these groups and determine if a putative physiology could be inferred for a given group (Table S5). The connectivity of Methanomassiliicoccales in each of the modules was characterized according to the metabolic characteristics of each taxon belonging to the same module.
2.9. Availability of Data and Materials
The MAGs’ bins generated and analyzed in this study are available under the Whole Genome Shotgun project deposited at DDBJ/ENA/GenBank under the accession numbers JAEILR000000000 v.1 (MXMAG1) and JAEILS000000000 v.1 (MXMAG2). The 16S rRNA gene sequences k119_35148 and k119_91804 are, respectively, available under the accession numbers MW383280 and MW383279. The 16S rRNA gene sequence generated by metabarcoding are available under the accession numbers MW386093-MW386158 in DDBJ/ENA/GenBank databases.
3. Results and Discussion
3.1. Diversity and Abundance of Methanomassiliicoccales in a Wide Range of Habitats
3.1.1. Molecular Pre-Screening of Various Environmental Samples
In order to explore the distribution of Methanomassiliicoccales associated with terrestrial and aquatic ecosystems a first molecular screening was carried out on 86 anoxic samples collected worldwide. These samples comprised a wide diversity of habitats (marine sediments, deep-sea hydrothermal vents, marine pockmarks, submarine mud volcano, deep-sea hypersaline anoxic brine, peatland soils, lakes, hot springs, river sediments) displaying a large range of physico-chemical conditions (pH, temperature, salinity, pressure). Methanomassiliicoccales were detected using PCR-based metabarcode screening (with the GoTaq® G2 DNA polymerase) in 26% (n = 22) of all samples, including coastal and deep-sea marine sediments, submarine mud volcano samples, anoxic lake waters, freshwater sediments, deep-sea brine samples, hot spring sediments, and peatland soils (Figure 1). A Bray-Curtis index calculated between these 22 samples revealed that the microbial community compositions were mainly structured according to sample type and by sampling site (see network topology in Figure 1; Table 1). So far, some of the anoxic habitats where Methanomassiliicoccales have been detected were reported to contain fermentation products and methylated compounds [67,68,69,70,71]. The methylated compounds result from the anaerobic degradation of organic compounds, such as pectin (for methanol) or N-methylated compounds (e.g., choline, betaine or trimethylamine-N-oxide), which are particularly represented in marine habitats [72], where they mainly serve as osmoregulators in organisms. A large part of this work has focused on marine habitats where the terminal degradation of organic matter results from microbial sulfate-reduction or methanogenesis reactions [68].
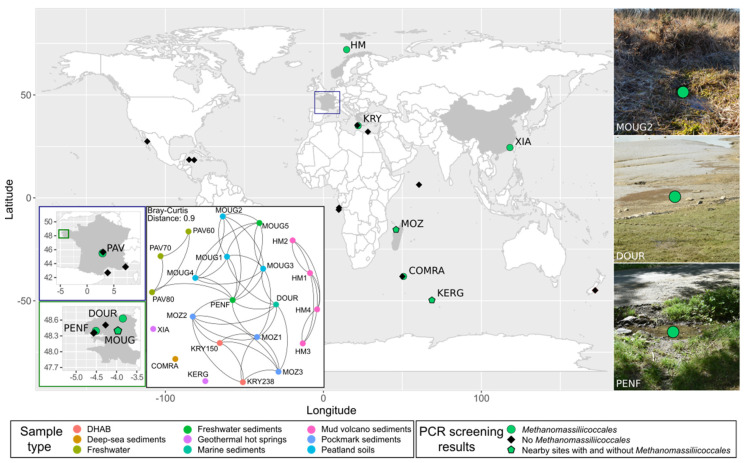
Figure 1.
Distribution of environmental samples screened for Methanomassiliicoccales. On the map, green dots indicate the presence of Operational taxonomic units (OTUs) assigned to Methanomassiliicoccales in the sample; black diamonds indicate samples in which no Methanomassiliicoccales was detected by PCR with primers targeting Methanomassiliicoccales. Green and black pentagons show the position of nearby sites characterized by the presence of Methanomassiliicoccales for some, and their absence for others. The Network based on Bray-Curtis index shows the dissimilarity in the composition of total microbial communities between samples; pictures on the right represent, respectively, the MOUG2, DOUD, and PENF sampling sites.
This work highlighted the presence of Methanomassiliicoccales in new geographical areas (Kerguelen archipelago in the Indian Ocean and the Brittany region in France, respectively), in environments where their presence had not yet been reported (submarine mud volcanos) and in habitats sometimes characterized by extreme conditions, such as high pressures, high salinities, high temperatures, or low pH (Table 1). As previously reported, the presence of Methanomassiliicoccales was detected in various ecosystems with various physical-chemical parameters, such as (i) in terrestrial hot springs where temperatures were between 50 and 82 °C (KERG, XIA) [31,73,74], (ii) in a large number of samples where temperatures were more moderate (between 8 and 30 °C), as in peatland soils, where the pH was slightly acidic (MOUG2, MOUG4) [23,75], (iii) in submarine mud volcanoes (HM1-4) and subseafloor sediments (COMRA) located at significant depths (between 1200 and 3338 m) [32,76], (iv) in non-saline (PAV60, PAV70, PAV80) [22,77] (v) to hypersaline (KRY150, KRY238) [45] environments, and (vi), finally, in a habitat subject to anthropogenic pollution and freshwater inputs (DOUR) [78]. These results reinforce the hypothesis that Methanomassiliicoccales might have a physiological diversity and potentially encompass mesophilic, thermophilic, halophilic, neutrophilic, acidophilic, and maybe even piezophilic taxa, as long as the amplified sequences were derived from active cells, and not from free DNA or dormant cells.
Metabarcode sequencing targeting variable regions of 16S rRNAs gene sequences from Bacteria and Archaea and qPCR targeting Methanomassiliicoccales were then carried out to confirm these results and determine the diversity and abundance of Methanomassiliicoccales in these samples.
3.1.2. Diversity and Abundance of Methanomassiliicoccales
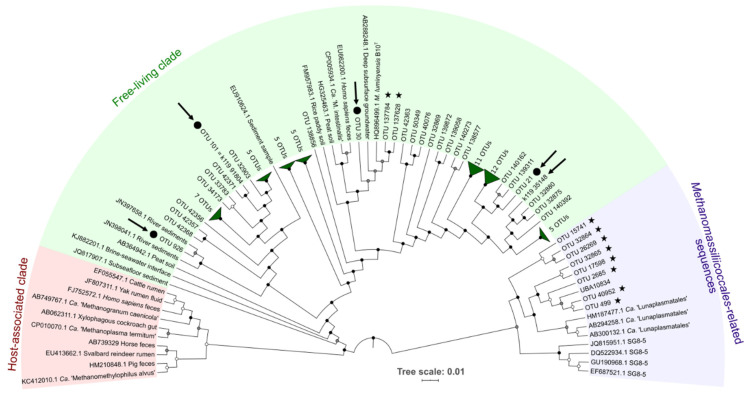
Metabarcoding analyses confirmed the presence of Methanomassiliicoccales in the 22 samples where the presence of Methanomassiliicoccales was initially detected. The diversity of Methanomassiliicoccales in these samples was low. Indeed, out of the 14,809 prokaryotic OTUs identified by processing the dataset, only 6 OTUs were confirmed to belong to Methanomassiliicoccales based on the automatic assignment made by SILVA 138 database and by finer phylogenetic analyses performed with a set of Methanomassiliicoccales sequences and closely related lineages (Tables S2 and S3). These 6 OTUs (OTUs 21, 30, 101, 926, 137,628, and 137,784) fell into the ‘free-living’ cluster of the Methanomassiliicoccales (Figure 2), in the Methanomassiliicoccaceae family. Among them, OTU30 was tightly related to M. luminyensis B10T 16S rRNA sequence (99.6% sequence similarity) and was present in all of the 22 selected samples, supporting the hypothesis that M. luminyensis might be inferred to a wide range of environmental conditions (Table 1) [24,79]. Eight other OTUs of the dataset were found to belong to very closely related lineages of Methanomassiliicoccales referenced as Ca. ‘Lunaplasmatales’ and UBA10834 [80].
Figure 2.
Dendrogram showing the relationships between 16S rRNA gene sequences (253 bp) detected in environmental samples, in substrate-amended slurry (PENF slurry) and inferred to metagenome-assembled genomes (MAGs), among Methanomassiliicoccales [22]. The 85 OTUs of Methanomassiliicoccales detected in this study are shown on this dendrogram. OTUs detected only in environmental samples are indicated by a black star. OTUs without a symbol were detected in the culture-based experiment performed with the sediment PENF. OTUs indicated by a circle were detected both in bulk samples and in the culture-based experiment. The 16S rRNA gene sequences k119_35148 and k119_91804 were extracted from the metagenome sequences of the culture-based experiment PENF, after 8 weeks of incubation. Sequences discussed in the text are indicated by black arrows. The Methanomassiliicoccales-related sequences encompassed representative 16S rRNA gene sequences of Ca. ‘Lunaplasmatales’, UBA10834 and SG8-5 [80]. This reconstruction was performed on partial 16S rRNA gene sequences (253 bp), using the BIONJ method [81], with the modifications of Jukes and Cantor and using 1000 bootstrap replicates. Black-filled dots indicate nodes with bootstrap supports <50%, grey-filled dots 50–75%, and white-filled dots show support values between 75 and 100%. The top-right insert indicates the relative abundance of the main Methanomassiliicoccales-affiliated OTUs in the substrate-amended slurry PENF, after 8 weeks of incubation.
The total abundance of Methanomassiliicoccales within samples was determined by quantitative PCR (Table S4). For samples where quantification has been possible, abundance of Methanomassiliicoccales was generally very low, ranging from 1.62 × 103 16S rRNA gene copies per gram of wet matrix in MOZ1 samples, to 7.5 × 104 16S rRNA gene copies per gram of wet weight soil in the peatland soil sample MOUG2. Many samples did not contain Methanomassiliicoccales abundances falling within the range set in this study as being biologically relevant for crude environmental samples (≥1 × 103 gene copies per ml or gram of wet weight of sample). These results were congruent with the literature, which indicate that these methanogenic archaea are very scarce in the environment [14,23]. However five samples contained non-negligible effectives of Methanomassiliicoccales: (i) soil samples from the Mougau peatland (MOUG1 and MOUG2) with respective abundances of 7.5 × 104 and 6.2 × 104 16S rRNA gene copies of Methanomassiliicoccales per gram of wet weight (and 34.4% and 1.7% of total known methanogen’s sequences in the metabarcoding dataset, respectively); (ii) sediments from the stream tributary to the Penfeld river (PENF; 5.3 × 104 16S rRNA gene copies per gram of wet weight, 69.8% of total methanogen’s sequences); (iii) a sample from the Pavin lake collected at 60 m depth (PAV60; 1.3 × 104 16S rRNA gene copies per ml; 60.0% of total methanogen’s sequences), and iv) a sediment sample from the Dourduff-en-Mer marine Bay (DOUR, 6.5 × 103 16S rRNA gene copies per gram of wet weight, 38.6% of total methanogen’s sequences) and a hot spring water sample from the Kerguelen Island (KERG, 6.4 × 103 16S rRNA gene copies per gram of wet weight, 79.5% of total methanogen’s sequences).
3.2. Enrichment Cultures Targeting Methanomassiliicoccales
Six samples were selected for enrichment cultures of Methanomassiliicoccales. Sample selection was based on: (i) qPCR abundance values of Methanomassiliicoccales exceeding 1 × 103 16S rRNA gene copy numbers per ml or gram of wet weight sample (DOUR, MOUG2, PAV60, PENF), and/or (ii) corresponding to ecosystems different from those in the gastrointestinal tract from which the Methanomassiliicoccales strains were isolated to date (KRY150—MgCl2-rich brine subjected to more than 33 MPa pressure; KERG—hot spring). To reach this objective, a medium was developed in this work to promote growth of Methanomassiliicoccales. This medium contained methyl compounds (MeOH and TMA, 40 mM as described by Reference [15,43]) and dihydrogen in order to promote methyl-dependent hydrogenotrophic methanogenesis.
Of the 6 substrate-amended slurries incubated and monitored using geochemical and molecular approaches for 10 weeks, 3 did not produce methane de novo (PAV, KERG, KRY150). Two others led to methane and ammonium production but without Methanomassiliicoccales enrichment (DOUR and MOUG2) (Figure S1). No methane production or cell growth was detected in the uninoculated control culture. The only culture that showed production of methane associated with an increase in the copy number of Methanomassiliicoccales 16S rRNA sequences over time was the PENF freshwater sediment sample (Figure 3).
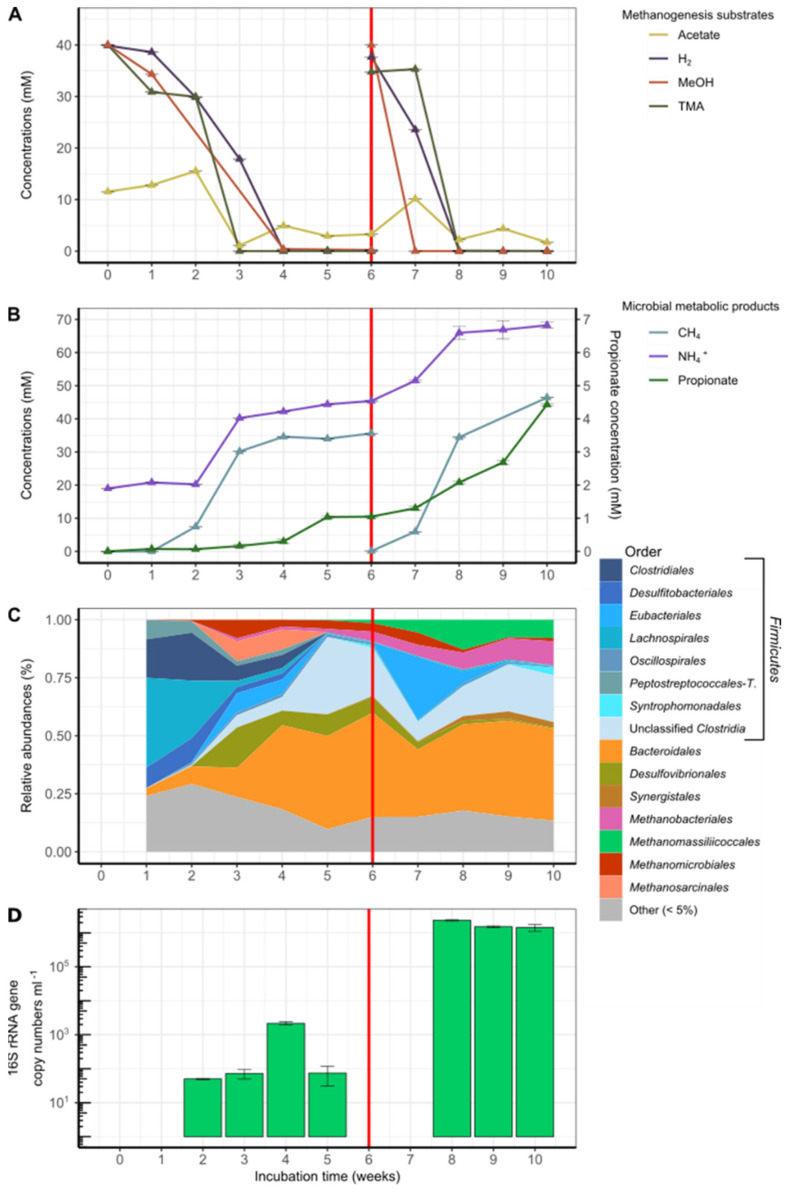
Figure 3.
(A) Consumption kinetics of methanogenesis substrates during the weeks of incubation of the substrate-amended slurry performed with the PENF sample; (B) Consumption kinetics of some metabolic products (methane, ammonia, propionate) during the weeks of incubation of the substrate-amended slurry performed with the PENF sample. Concentrations of propionate are indicated on the right vertical axis of the figure and other microbial products (CH4 and NH4+) are indicate on the left vertical axis. The vertical red line indicates a novel addition of trimethylamine (TMA), methanol and H2/CO2 at T6 to stimulate methanogenesis; (C) Trends in microbial diversity revealed by metabarcoding; (D) Methanomassiliicoccales abundances determined by qPCR (logarithmic scale). Methanomassiliicoccales could not be quantified by qPCR in 4 samples (T0, T1, T6, and T7). Average copy numbers are given on the chart bars. Renewal of methanogenesis substrates (H2, CO2, MeOH, TMA) after six weeks of incubation is represented by the vertical red lines. Peptostreptococcales-T.: Peptostreptococcales-Tissierellales.
After 6 weeks of incubation, the added substrates were consumed. Hence, slurries were reamended with MeOH, TMA, and H2 in order to overcome potential growth limitation of Methanomassiliicoccales. A constant increase in methane concentrations followed the consumption of these substrates in the first phase of the culture, as well as after the renewal of the substrates and the gas phase, suggesting that methanogenesis was occurring in this culture (Figure 3A,B). It is noteworthy that small amounts of dimethylamine (DMA) and monomethylamine (MMA) were detected one week after the beginning of the incubation (T1) (0.31 and 0.08 mM, respectively) and also one week after the new addition of TMA (T7) (0.16 and 0.10 mM, respectively). These products are intermediates in the degradation of TMA and probably reflect the progressive degradation of this substrate [72]. Additionally, a small amount (0.01 mM) of choline, a quaternary amine known to be degradable to TMA, ethanol, and acetate, by some fermentative microorganisms [68], was also detected at T6. A steady increase in propionate concentration over time also suggests that several fermentative microorganisms were active in this culture. In addition, variations in acetate concentrations were also observed throughout the incubation period, increasing from 11.53 to 15.53 mM between T0 and T2 before stabilizing between 1.08 and 4.94 mM between T3 and T10, interspersed with a sharp increase at T7 (10.14 mM), one week after the new addition of substrates. These variations reflect high microbial activity throughout the incubation period, since this compound can be a product and also a substrate of microbial metabolism (e.g., acetogenesis). Considering that the production of methane (respectively, 35.61 and 46.42 mol L−1 at T6 and T10) was higher than that of the initial available dihydrogen (respectively, 31.74 and 37.66 mol L−1 at T0 and T6), the stoichiometry of the methyl-dependent hydrogenotrophic methanogenesis was not conserved, confirming that several metabolisms were at work in this culture. In accordance, the metabarcoding analysis performed over time on this culture showed a large fraction of 16S rRNA gene sequences of methanogenic lineages (Methanobacteriales, Methanomassiliicoccales, Methanomicrobiales, and Methanosarcinales) and of taxa mainly reported in animal GITs [82,83] (Firmicutes (Clostridiales, Desulfitobacteriales, Eubacteriales, Lachnospirales, Peptostreptococcales-Tissierellales, and unclassified Clostridia), Bacteroidales, Synergistales, and taxa in lower relative abundance (<5%)), indicating that it was a complex microbial community, even after 10 weeks of incubation. Among the Methanomassiliicoccales-affiliated OTUs, sequences belonging to the OTUs 21, 30, 101, and 926 were detected throughout the incubation period and were predominant in the dataset.
In terms of overall diversity, the species richness slightly decreased over incubation time (from T1 to T10: Shannon index, 3.11 to 2.61; Simpson index, 0.88 to 0.82) (Figure S2). It is noteworthy that the majority of lineages in which relative abundance increases over time (Bacteroidales, Clostridiales, Desulfovibrionales, and Synergistales), as well as those of Methanomassiliicoccales, were similar to cohorts associated with Methanomassiliicoccales previously described that had been inoculated under mesophilic conditions with bovine rumen fluid, kitchen wastes, or mixtures of Gracilaria sp. algae and marine sediments [20,23,84]. In this culture, Desulfovibrionales were dominated by sequences affiliated to the genus Desulfovibrio, a genus within which certain species are known to produce TMA, ethanol, and acetate from choline fermentation [85]. A predominance of Bacteroidetes, Firmicutes, and Methanomassiliicoccales was also reported for an environmental sample, in this case, a birch forest soil in winter [86]. Apart from methanogenic Archaea, other Archaea represented a marginal fraction of the total prokaryotic diversity, accounting for 46.2% and 2.3% of total read sequences at T1 and T2, respectively, to less than 0.1% of total sequences after the second week of incubation (Figure S3A,B).
A decrease in the number of sequences affiliated with the genera Methanosarcina (mainly composed of representative strains using H2/CO2, or R-CH3 or acetate) and Methanomicrobiales (using H2/CO2/formate) was observed over time, while the number and relative fraction of sequences affiliated with the genus Methanobacterium (Methanobacteriales; mainly growing on H2/CO2) and the order Methanomassiliicoccales (using H2 + R-CH3) increased (Figure 3C). Between T1 and T5, the proportion of Methanomassiliicoccales sequences within total methanogenic archaea decreased from 29.2% to 4.6%. However, their relative abundance among methanogens increased after 6th week of incubation (16.4%) to reach a maximum value at T8 (63.1%) (Figure S3C). From the 8th week of incubation (T8), the relative fraction of Methanomassiliicoccales decreased slightly and that of Methanobacterium spp. (Methanobacteriales) increased slightly, to both stabilize at about 50%. Interestingly, some Methanobacterium representatives are able to use ammonium as the only source of nitrogen [87]. At 8 weeks incubation time (T8), 85 Methanomassiliicoccales OTUs were detected in the incubated slurry and were all affiliated to the ‘free-living’ environmental clade, forming various clusters of sequences within this clade (Figure 2), and representing, 63.1% of methanogens-affiliated sequences, 63.1% of all archaeal sequences (Figure S3D) and 14.9% of total sequences (Figure S3E). Interestingly, 22 OTUs, including OTU101 and OTU926, formed a cluster not related to the genus Methanomassiliicoccus. With the exception of 4 OTUs (OTUs 21, 30, 101, and 926, respectively), the majority of these sequences detected in the culture-based incubation experiment have not been detected by metabarcoding on the environmental sample, suggesting that these taxa were extremely scarce in situ.
The total abundance of Methanomassiliicoccales increased over the incubation period, from 5.00 × 101 at T2, to 2.35 × 106 16S rRNA gene copies of Methanomassiliicoccales mL−1 at 8 weeks of incubation (T8), before decreasing slightly to 1.43 × 106 copies at T10 (Figure 3D). Unfortunately, no qPCR quantification of Methanomassiliicoccales could be obtained at T1, T6, and T7. Nevertheless, this nearly 5-log increase in the abundance of Methanomassiliicoccales demonstrated that there was enrichment (p-value < 0.0001, in ANOVA and Student’s t-test) and, therefore, growth of the Methanomassiliicoccales during incubation, on the medium developed in this study with the freshwater sediment sample PENF. This result is reinforced by the fact the number of Methanomassiliicoccales sequences also increased gradually and concomitantly over the incubation period, until time T8.
The highest levels of Methanomassiliicoccales abundance results were obtained after 8 weeks of incubation, both in terms of copy number of the 16S rRNA gene sequences quantified by qPCR and among the total methanogenic community detected by metabarcoding (63.1% of the total methanogenic sequences). At that time, OTU21 was the predominant OTU of Methanomassiliicoccales, representing 86.5% of the total sequences of Methanomassiliicoccales, followed by OTU101 (13.4%) and then by the sequences of the other Methanomassiliicoccales OTUs (<1% in total) (Table S2). This result guided us to retain the T8 point for a metagenomic analysis.
3.3. Methanomassiliicoccales Genomic Features
Two Methanomassiliicoccales-affiliated MAGs were reconstructed from the T8 point of the substrate-amended slurry made from the PENF sample: one of high-quality (MXMAG1) and the other of low-quality (MXMAG2).
3.3.1. Taxonomic Position and General Features of the MAGs
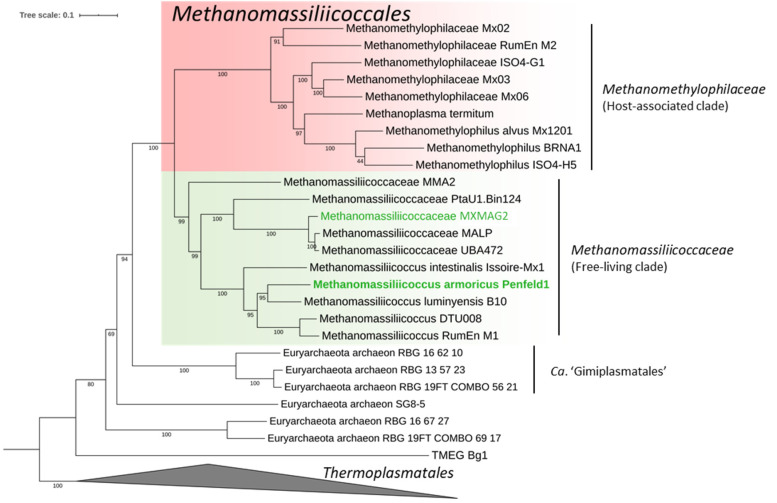
Following genome-wide taxonomic analysis (with IQ-TREE) based on concatenated protein reference trees, both MAGs were robustly placed in the order Methanomassiliicoccales (Figure 4). ANI scores for the MXMAG1 MAG calculated against M. luminyensis B10T and Ca. ‘Methanomassiliicoccus intestinalis’ Issoire-Mx1 had OrthoANIu values of 74.73% and 66.54%, respectively. These values are much lower than the threshold criterion for prokaryotic species delineation proposed to be 95–96% [88], suggesting that this MAG corresponds to a different Methanomassiliicoccales representative than those previously described.
Figure 4.
Maximum Likelihood phylogenomic tree (LG + F + G4) showing the position of the two Methanomassiliicoccales MAGs obtained in this study (in green) with respect to closely related MAGs and taxa. This phylogeny is based on a concatenation of 40 conserved phylogenetic markers (10,111 positions). The scale bar represents the average number of substitutions per site, and numbers indicate bootstrap replicates.
The digital DNA-DNA hybridization (DDH) estimate values between MXMAG1 MAG and M. luminyensis B10T was 19.60%, and the one with Ca. ‘Methanomassiliicoccus intestinalis’ Issoire-Mx1 was 21.40%, both of which were values well below the standard criterion (70%) for the delineation of a prokaryotic species [89], confirming that the MXMAG1 MAG belongs to a different genomic species than M. luminyensis and Ca. ‘Methanomassiliicoccus intestinalis’. Thus, we propose that MXMAG1 represents the type material of a novel Methanomassiliicoccus species, for which we propose the name Candidatus ‘Methanomassiliicoccus armoricus MXMAG1’ (ar.mo’ri.cus L. pl. fem. n. Armoricae, part of Gaul between the Seine and the Loire, including Brittany, where the sample used for the slurry were taken from; N.L. masc. adj. armoricus MXMAG1, pertaining to Brittany).
Apart from the 16S and 23S rRNA genes which were absent, the sequence of the MAG called Ca. ‘Methanomassiliicoccus armoricus MXMAG1’ met all of the criteria described elsewhere [90] to be considered a high-quality draft. This MAG consisted of 37 contigs with an overall size of 2,230,712 bp and a G+C content of 62.13 mol%. This size was intermediate between the genome of closest species, M. luminyensis and Ca. ‘M. intestinalis’, that are 2.6 Mbp and 1.9 Mbp, respectively [79]. CheckM estimated the genome to be 94.4% complete based on the presence of single-copy marker genes (9 markers were missing) and without contamination. Annotation with MaGe of Ca. ‘Methanomassiliicoccus armoricus MXMAG1’ resulted in the prediction of 2448 genomic objects, among which 2407 were protein-coding sequences. This reconstructed genome was relatively streamlined, with coding sequences covering approximately 90.87% of the entire genome. However, slightly different results were obtained with other annotation software: 2271 CDSs were found with RAST (745/1526 were not integrated to subsystem categories), 2261 CDSs with Prokka, and 2243 CDSs with Dfast.
Sequence of MXMAG2 consisted of 203 contigs with an overall size of 1,040,932 bp and a G + C content of 57.78 mol%. CheckM estimated the genome to be 48.2% complete based on the presence of single-copy marker genes (82 markers were missing) and with 2.1% contamination (4 markers were duplicated). Annotation with MaGe of MXMAG2 resulted in the prediction of 1305 genomic objects, among which 1281 were protein-coding sequences. MXMAG2 had a relatively streamlined genome with coding sequences covering approximately 91.34% of the entire genome. However, slightly different results were obtained with other annotation software: 1149 CDSs with Prokka and 954 CDSs with Dfast.
As for other Methanomassiliicoccales MAGs published previously, both MAGs did not contain one full complete 5S-16S-23S rRNA genes operon [79]. One and two 5S rRNA genes were found for MXMAG2 and Ca. ‘Methanomassiliicoccus armoricus MXMAG1’MAGs, respectively. A partial 542 bp 23S rRNA gene was found in MXMAG2. We detected 24 and 38 tRNA for MXMAG2 and Ca. ‘Methanomassiliicoccus armoricus MXMAG1’ MAGs, respectively, according to ARAGORN RNA finder (http://130.235.244.92/ARAGORN/) associated to 18 standard amino acids for Ca. ‘Methanomassiliicoccus armoricus MXMAG1’. Cysteine and tryptophan tRNA appeared to be missing, although the CDSs of the cysteine-tRNA and tryptophan-tRNA ligases were found.
The two Methanomassiliicoccales MAGs reconstructed did not contain any 16S rRNA sequences, probably due to the high degree of conservation of this gene and the closeness between 16S rRNA sequences of related organisms which make their binning challenging [91]. However, two 16S rRNA sequences referenced as k119_35148 and k119_91804 (1448 and 1397 bp, respectively) were independently reconstructed from the metagenomes and assigned to Methanomassiliicoccales (Table S3). Thus, it seems reasonable to assume that they correspond to the same 2 Methanomassiliicoccales genomes reflected by the MAGs. The similarity of the 16S rRNA gene sequence with the sequences of the closest cultured relatives, calculated using the “16-based ID” tool provided by the EzBioCloud web server (https://www.ezbiocloud.net/) showed that the sequence k119_35148 (probably the one corresponding to Ca. ‘Methanomassiliicoccus armoricus MXMAG1’) was quite closely related to M. luminyensis (98.23% pairwise similarity) and Ca. ‘M. intestinalis’ (97.61%), indicating the enrichment of an undescribed representative of Methanomassiliicoccus. This sequence, even if it diverges at a nucleotide base (probably due to a sequencing error in the 16S rRNA gene sequence extracted from the metagenome), most likely corresponds to that of OTU21 present in the metabarcode dataset throughout the PENF cultivation period, and also present in natural samples KERG, MOUG3, MOZ1–3, PAV60, and PENF. Since the level of similarity of the 16S rRNA gene sequence between sequence k119_35148 and those of its closest relatives was below the threshold value (98.7–99%) currently recommended for testing the genomic uniqueness of a new species, this value alone was sufficient to conclude that Archaea forming a new genomic species within the genus Methanomassiliicoccus have been enriched in this culture [92,93]. These results were congruent with the ANI and digital DDH scores calculated on Ca. ‘Methanomassiliicoccus armoricus MXMAG1’, reinforcing the idea that this sequence corresponds well to the Ca. ‘M. armoricus MXMAG1’. The nearly complete sequence of the 16S rRNA gene bearing the reference k119_91804 (probably the one corresponding to MXMAG2), covered completely identically the representative sequence OTU101 (Figure 2), which was present in the environmental samples KERG, MOZ1–3, PAV70, PAV80, and PENF. This sequence was more distant from the sequences of the species and genera of Methanomassiliicoccales described to date, with a pairwise similarity ranging from 88.96 to 91.84%, respectively, to the closest relatives Ca. ‘Methanogranum caenicola’ and M. luminyensis, respectively. Although these sequences were incomplete, these 16S rRNA gene sequence similarity scores are far below the standard threshold level (<94.5%) generally accepted for the delineation of a new genus [93]. Thus, an archaeon belonging to an undescribed genus of Methanomassiliicoccales, which would be widely distributed in natural ecosystems, has likely been enriched in this culture-based incubation experiment.
3.3.2. Putative Metabolic Pathways
An extensive genomic characterization has been performed on the high-quality MXMAG1 Ca. ‘Methanomassiliicoccus armoricus MXMAG1’ to explore the pathways of methanogenesis, the biology, and the possible adaptations of the ‘free-living’ clade Methanomassiliicoccales Archaea from which this MAG originates, which appear to be widespread in natural ecosystems (Figure 5).
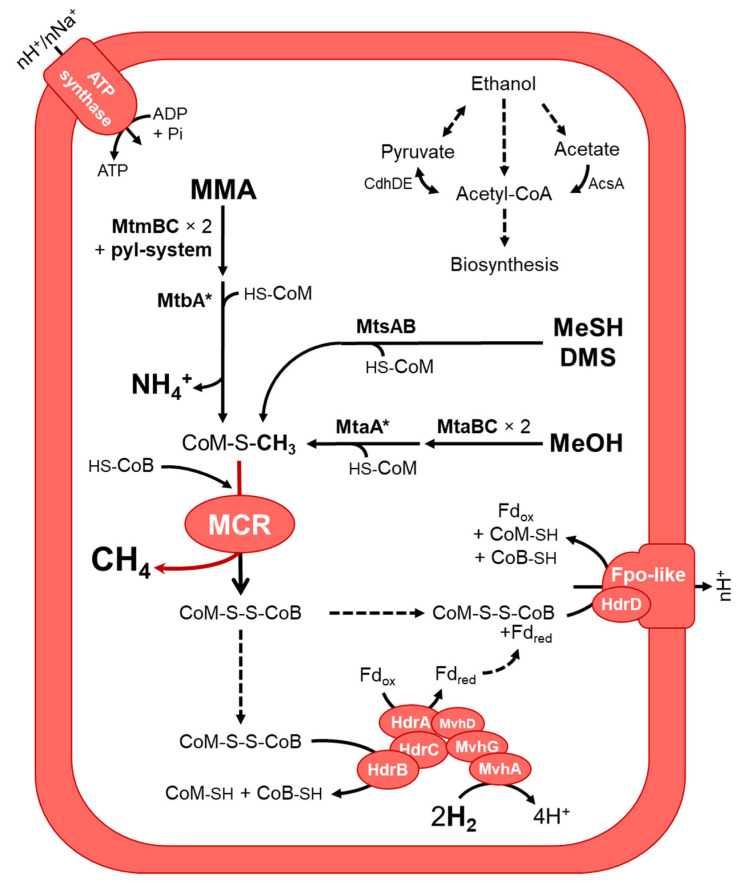
Figure 5.
Predicted metabolic pathways for methanogenesis, carbon assimilation, and energy conservation in Ca. ‘Methanomassiliicoccus armoricus MXMAG1’, belonging to the ‘free-living clade’ of Methanomassiliicoccales. Genes encoding the MtmBC and MtaBC enzymes were found in two copies in the MAG. The “*” symbol indicates the presence of enzyme homologous to the MtaA/MtbA which are found in other Methanomassiliicoccales for MeOH and MMA incorporation, as described elsewhere [22]. Legend: CdhDE, Acetyl-CoA decarbonylase/synthase complex subunits; DMS, dimethylsulfide; MeOH, methanol; MeSH, methanethiol, MMA, monomethylamine.
Consistent with previous studies conducted on Methanomassiliicoccales, the methyl-dependent hydrogenotrophic methanogenesis pathway was predicted in Ca. ‘Methanomassiliicoccus armoricus MXMAG1’ MAG. Hence, the metabolism of the Archaea, from which this MAG was derived, was also probably similar to the other described Methanomassiliicoccales [16,22,23,26]. Methyltransferases for the production of methane from methanol (MtaA/MtaBC), monomethylamine (MtbA/MtmBC), dimethylsufide and methanethiol (MtsAB) and HdrABC-MvhAGD electron-bifurcating complex (including an (NiFe)-hydrogenase) have been identified, suggesting that Ca. ‘Methanomassiliicoccus armoricus MXMAG1’ would be capable of reducing such methyl-compounds using H2 as electron donor. As for Ca. ‘Methanoplasma termitum’ [26], the genes encoding the methyltransferases involved in the use of di- and trimethylamine (mtbB and mttB, respectively) are absent from the sequence of Ca. ‘M. armoricus MXMAG1’. However, several non-binned Methanomassiliicoccales-affiliated mttB sequences have been found in the metagenome (Figure S4), suggesting that some Methanomassiliicoccales in this culture, and possibly even this one, were capable of reducing methyl-groups of TMA into methane. As in Methanomassiliicoccales from digestive environments, the presence of a complete pyl system (pylBCD/pylS), involved in the biosynthesis of the 22nd amino acid pyrrolysine, was found, and the presence of the amber codon was found in methyltransferase genes, as in other Methanomassiliicoccales methylamine methyltransferases [16,94]. A complete methyl-coenzyme M reductase (MCR) complex (mcrABG) gene set was present in the MAG. Of the N5-methyltetrahydromethanopterin: coenzyme M methyltransferase (MTR) complex, only mtrH gene was present, and none of the genes of the methyl branch of the H4MPT-type Wood-Ljungdahl pathway. Altogether, the presence of the identified genes in MXMAG1 strongly suggests that Ca. ‘M. armoricus’ produced energy from a methyl-dependent methanogenesis.
As described in all known Methanomassiliicoccales, Ca. ‘Methanomassiliicoccus armoricus MXMAG1’ harbored two heterodisulfide reductases systems (the methyl viologen-dependent hydrogenase/heterodisulfide reductase (MVR/HDR) complex (MvhADG/HdrABC) and the HdrD-liked bonded to the Fpo-like complex) involved in the regeneration of the heterodisulfide complex (CoM-S-S-CoB) and the energy-conserving [23,26,95]. Ca. ‘M. armoricus MXMAG1’ encoded also an ADP-forming acetyl-CoA synthetase (AscA) [13,23,26], suggesting that acetate could be used as a carbon source. The MAG sequence also encodes the enzymes that allow the synthesis of acetyl-CoA from pyruvate (a pyruvate synthase complex; a pyruvate dehydrogenase complex), and a complete ethanol degradation pathway to acetyl-CoA, suggesting that pyruvate and ethanol may also serve as a carbon source for cellular biosynthesis.
Fourteen out of the 22 proteinogenic amino acid biosynthesis pathways were predicted as complete in Ca. ‘Methanomassiliicoccus armoricus MXMAG1’ MAG: Ala, Arg, Asn, Asp, Cys, Glu, Gly, His, Ile, Lys (biosynthesis VI), Pyl, Thr, Trp, and Val. The presence of numerous genes coding for cobalamin (vitamin B12) ATP-binding transporters involved in cobalamin import (BtuD) suggests that this genomic strain was capable to import this vitamin. Its de novo synthesis pathway is not present on the 94.4% complete reconstructed MAG. Several complete central metabolic pathways were also identified in the MAG, including: (i) gluconeogenesis/glycolysis, (ii) anaplerotic CO2 fixation into oxaloacetate, (iii) the non-oxidative branch of the pentose phosphate pathway, and (iv) a fatty acid activation pathway. The presence of these complete paths, as well as those mentioned above, does not necessary imply that all of them were used.
The poor quality of the MXMAG2 MAG reconstruction allowed only partial metabolic pathways to be predicted. This MAG had genes encoding a nearly complete MCR complex (the mcrB subunit was missing) and several genes encoding subunits of a Fpo-like complex (fpoMN) and an MVR/HDR complex (mvhAB and hdrBC, respectively). The presence of the mtbC gene coding for the dimethyltransferase suggests that DMA could be used for methanogenesis. The presence of the Methanomassiliicoccales mtaA/mtbA gene encoding for a methyltransferase involved in methanol and methylamines incorporation was also in line with this assumption [22]. Unlike the high-quality MAG, the MXMAG2 encoded a choline trimethylamine-lyase activating enzyme (cutD). This enzyme may be involved in the production of TMA from choline, possibly allowing the utilization of choline as substrate for methanogenesis. As described in Ca. ‘Methanomassiliicoccus armoricus MXMAG1’, this MAG encoded the ADP-forming acetyl-CoA synthetase, as well as the pyruvate dehydrogenase complex.
3.3.3. Putative Physiological and Stress Response Functions
Several putative adaptive mechanisms known to be involved in microbial stress responses have been identified in Ca. ‘Methanomassiliicoccus armoricus MXMAG1’ MAG. Among them, complete biosynthesis and degradation pathways of trehalose were predicted, as well as 8 genes coding for a sub-unit (malK) of a trehalose/maltose importer. This could suggest that trehalose might be used to cope with external factors of the environment, such as temperatures, dehydration, or osmotic/ oxidative stresses, or might be used as a storage compound, as described in other prokaryotes [96,97]. However, functional analyses will be useful to confirm this hypothesis.
As described in many microbial lineages including some methanogens, Ca. ‘M. armoricus MXMAG1’ MAG encoded a pyruvoyl-dependent arginine decarboxylase [98,99]. The presence of the gene encoding this enzyme suggests that Ca. ‘M. armoricus MXMAG1’ might regulate intracellular pH by converting a L-arginine to agmatine and CO2 under acidic conditions [100]. In addition, this MAG encodes enzymes involved in (i) the degradation of agmatine to putrescine via an agmatinase; (ii) possessed a complete pathway for putrescine biosynthesis; and (iii) possessed numerous copies of the gene encoding the pH-dependent bidirectional putrescine-ornithine transporter PotE [100,101]. Putrescine is a common polyamine that could promote the growth of most living cells. Although present in most methanogenic archaea, the function of putrescine remains unclear [102,103]. The presence of complete pathways involved in putrescine incorporation and biosynthesis suggests that this compound might play a role in Ca. ‘M. armoricus MXMAG1’ physiology.
Several genes involved in arsenic metabolism were also found in this MAG. Arsenic is a harmful toxic pollutant at low concentrations that is widely distributed in natural environments (i.e., groundwater, sediments, soils), with the potential to inhibit methanogenesis depending on arsenic speciation and concentration [104,105]. Ca. ‘Methanomassiliicoccus armoricus MXMAG1’ genome contains a complete arsRDABC operon, similar to the one found in Escherichia coli [106]. The arsRDABC operon is involved in the arsenic detoxication and the active release of this compound outside the cell. The only difference with the canonical arsenic detoxification pathway is that the arsB gene is replaced in MXMAG1 by the acr3 gene, which codes for a protein with the same function as ArsB (arsenite efflux pump). It has been shown that various Bacteria and Archaea prevent arsenic toxicity by using this pathway which, ironically, convert arsenate to the more toxic arsenite [106,107,108]. It should be noted that some MAGs of the sister lineage of Methanomassiliicoccales named Candidatus ‘Gimiplasmatales’ have also been reported to encode genes of this operon [109]. Interestingly, a comparative genomic analysis conducted on 5 genomes/MAGs belonging to the ‘host-associated’ clade and on 20 genomes/MAGs belonging to the ‘free-living’ clade showed that the complete arsRDABC operon was only present in MXMAG1 and MXMAG2. Several genes of this operon were also present in the genome of M. luminyensis strain B10T and in the MAG referenced as RumEn M1. Genes coding for the arsenical pump-driving ATPase were not present in the genomes/MAGs from the host-associated clade analysed in this study. Furthermore, genes involved in arsenic methylation (e.g., arsM) by other methanogens were not found in Ca. ‘M. armoricus MXMAG1’ MAG nor in Methanomassiliicoccales genome comparisons [110]. Thus, reduction and excretion of arsenite could represents the only arsenic detoxification pathway available for Methanomassiliicoccales. It is noteworthy that the detoxication pathway of arsenate to arsenite was also present in MXMAG2. Finally, in opposition to M. luminyensis, Ca. ‘Methanomassiliicoccus armoricus MXMAG1’ and Ca. ‘Methanomassiliicoccus intestinalis’ do not harbor the hgcAB operon [24,111]. This operon encodes an enzyme involved in the methylation of inorganic mercury (Hg) to form the highly toxic methylmercury [112,113]. Among methanogens, this metabolic capacity has only been confirmed for M. luminyensis and 7 species from various origins distributed among the Methanomicrobia class [113]. Therefore, if confirmed by functional approaches on isolates, this would demonstrate that this ability would not be uniformly present within the genus Methanomassiliicoccus, but only in some species of the genus.
3.4. Co-Occurrence Network Analysis
To explore potential interactions between Methanomassiliicoccales and other microorganisms, co-occurrence network analyses of 16S rRNA genes sequences were performed. These analyses were carried out on the 22 environmental samples and on culture-based incubation experiments performed with high substrate concentrations (Figure 6).
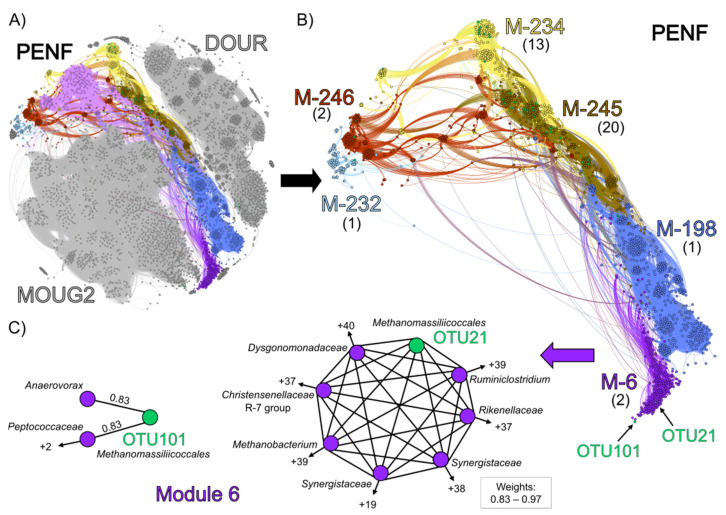
Figure 6.
Co-occurrence network reconstructed based on 16S rRNA gene-sequencing data from substrate-amended slurries (DOUR, PENF, and MOUG2, respectively), based on a Spearman rank’s correlation between OTUs and calculated with a percolation threshold of 0.79. (A) Clusters in slurries DOUR, PENF, and MOUG2, respectively. Modules in PENF network are colored to highlight them; (B) modules (M-6, module 6; M-198, module 198; M-232, module 232; M-234, module 234 and M-245, module 245, respectively) containing Methanomassiliicoccales in the cluster from the PENF culture-based incubation experiment. Modules consisting of a single Methanomassiliicoccales (modules 22 and 68, respectively) were excluded. The number of Methanomassiliicoccales in each module is shown in brackets; (C) non-random co-occurrences between Methanomassiliicoccales and other prokaryotes in module 6 showing co-occurrences of Methanomassiliicoccales (OTU21 and OTU101, respectively) with putative acetate- and H2-producing bacteria. Black arrows followed by a plus and a number indicate the number of taxa non-randomly co-occurring with other prokaryotes outside the groups represented. The weights of the co-occurrences are written above the link (OTU101) or in the insert below the cluster of the OTU21.
Four modules were identified, each of which comprised Methanomassiliicoccales sequences sharing strong non-random associations (weight >0.7) with other taxa: module 1104 (OTU21), module 117 (OTU101), module 0 (OTU30), and module 927 (OTU926) (Table S5). Module 117 included 119 other OTUs distributed in more than 20 order-level lineages. Among others, this module contained 8 Woesearchaeales, 7 Aminicenantales, 18 Omnitrophales, and 13 Marine Benthic Group D/Deep-sea Hydrothermal Vent Euryarchaeota Group-1 (MBGD/DHVEG-1), as well as 3 Methanofastidiosales (hydrogenotrophic methyl-dependent methanogenesis) and a Methanothermus (Methanobacteriales; hydrogenotrophic methanogenesis). Interestingly, Woesearchaeotales and Aminicentales present in this module have recently been proposed to maintain a nutritional relationship with various hydrogenotrophic methanogens, notably Methanobacteria, Methanofastidiosales, Methanomicrobia, and/or Methanomassiliicoccales [34,114,115,116]. When using a percolation threshold with a high-value of 0.83, OTU101 showed the highest non-random co-occurrence with two MBGD/DHVEG-1 sequences (weights: 0.85 and 0.87, respectively), and OTU30 showed significant non-random-co-occurrences with the poorly described bacterial phylum NKB15 and with a Microgenomatia (weights: 0.84 and 0.85, respectively) (Table S5). At this threshold of 0.83, no OTU was branched with OTU21 or OTU926. Thus, to observe their associated connectivity, a slightly lower threshold was used (0.7) (Figure S5) [117]. At this value, the Methanomassiliicoccales OTUs 21, 30, 101, and 926 showed significant non-random co-occurrences (weights: 0.70–0.87) with, respectively, 3, 5, 6, and 11 OTUs. Under these conditions, Anaerolineaceae, Anaerovorax, Aminicenantales, Geobacteraceae, MBGD/DHVEG-1, Sporomusaceae, and Woesearchaeales were the main lineages sharing strong connections with Methanomassiliicoccales. Interestingly, Anaerolineaceae, Anaerovorax, Geobacteraceae, and Sporomusaceae have also been reported to form syntrophic relationships with acetoclastic and hydrogenotrophic methanogens, notably Methanobacteriales, Methanofastidiosales, and Methanomassiliicoccales [118,119,120,121,122]. In contrast, if active, the Desulfatiglans and the Methanoregulaceae (Methanomicrobia) associated with OTU30 and OTU926, respectively, could represent hypothetical competitors for the acetate and/or H2 resources of Methanomassiliicoccales [123,124,125].
Similarly, a co-occurrence network analysis was also performed (percolation threshold: 0.79) on the substrate-amended slurries incubated over 10 weeks (DOUR, MOUG2 and PENF) to reveal the narrow potential interactions between Methanomassiliicoccales and other microbial taxa (Figure 6). Our aim was to assess whether the microbial composition of the cultured community could be likely correlated with the enrichment of Methanomassiliicoccales in the PENF slurry. The overall topology of the network highlighted 3 clusters of nodes that clearly corresponded to the different culture-based incubation experiments (Figure 6A): (i) the cluster of nodes over-represented in the Dourduff-en-Mer slurry sampled throughout the incubation, is located at the top of the network, (ii) the cluster of nodes over-represented in the Mougau samples is located at the bottom, and (iii) the cluster of nodes over-represented in the Penfeld samples is located in between. The modularity analysis showed that 40 of the 42 the Methanomassiliicoccales-affiliated OTUs were detected inside modules from the Penfeld Cluster. This is congruent with the absence of Methanomassiliicoccales detected by metabarcoding approach in the DOURD and MOUG2 incubated slurries (Figure S1). In terms of overall community composition (Figure 6B), modules 232, 234, 245, and 246 showed close taxonomic patterns dominated by Firmicutes and Bacteroidetes, taxa similar to those found in animal GITs from which most of the known Methanomassiliicoccales have been enriched or isolated so far [20,21,22,26]. These modules were predominantly composed of Bacteroidales (Dysgonomonadaceae and Rikenellaceae) and various Clostridiales (Christensenellaceae R-7 group, Gracilibacteraceae, Hungateiclostridiaceae, Oscillospiraceae, Ruminococcaceae) mainly reported to be able to produce acetate and/or H2 as fermentative end products and sometimes to form partnerships with hydrogen-consuming methanogens (i.e., Methanospirillum hungatei, Methanobrevibacter smithii) (Table S5) [82,126,127,128,129,130,131,132]. Such bacteria pattern could possibly be explained by the utilization of a medium rich in organic compounds which could be used as substrates for fermentation or acetogenesis. As we used a complex culture medium comprising various substrates for methanogenesis, several methanogenic Archaea mainly affiliated to Methanobacteriales, Methanomicrobiales, and/or Methanosarcinales were maintained throughout the incubation process and were also found in these modules. Despite strong similarities in microbial community composition with the four other modules (modules 232, 234, 245 and 246), module 6 contained a slightly higher taxonomic diversity among Firmicutes. This module contained the OTU21 and OTU101, which co-occurred strongly with Ca. ‘Methanomassiliicoccus armoricus MXMAG1’ and MXMAG2, respectively. Since these OTUs were predominant throughout the PENF culture-based incubation experiment and were also present in various environmental samples examined in this study, we focused on these two OTUs to determine whether Methanomassiliicoccales would significantly co-occur with given taxa (Figure 6C). OTU101 showed high non-random associations with an Anaerovoracaceae OTU (Peptostreptococcales-Tissierellales) (weight: 0.83) and a Peptococcaceae OTU (weight: 0.82). Several representatives from these lineages have been reported to ferment amino acids and to produce acetate or H2 and to carry out a functional or syntrophic relationships with hydrogenotrophic methanogens that scavenge hydrogen to keep fermentation of fatty acids and alcohols energetically favorable (e.g., Methanospirillum hungatei) (Table S5) [118,120,133,134]. OTU21 showed strong non-random co-occurrences with 43 taxa (weights: 0.79–0.95). The six strongest non-random associations (weights: >0.9) occurred with taxa known to produce acetate and dihydrogen by fermentation: A Dysgonomonadaceae (weight: 0.96), two Synergistaceae (weights: 0.95 and 0.93), a Ruminiclostridium (weight: 0.95), a Christensenellaceae R-7 group (weight: 0.92) and a Rikenellaceae (weight: 0.91). Members of the Synergistaceae family and the Ruminiclostridium genus have been reported to be able to produce short-chain fatty acids from several proteins and peptides, and acetate from various complex substrates (Table S5) [135,136]. A representative of the genus Methanobacterium, potentially a competitor for H2, was also strongly associated with OTU21 (weight: 0.91).
The results of these network analyses reinforce the hypothesis that Methanomassiliicoccales of the ‘free-living’ clade appear to display strong co-occurrences with acetate- and H2-producing bacteria, suggesting that they might maintain a metabolic interaction similar to that described between fermentative bacteria and other hydrogen-utilizing methanogens, by scavenging H2 and promoting acetate production [136,137,138] (Figure 7).
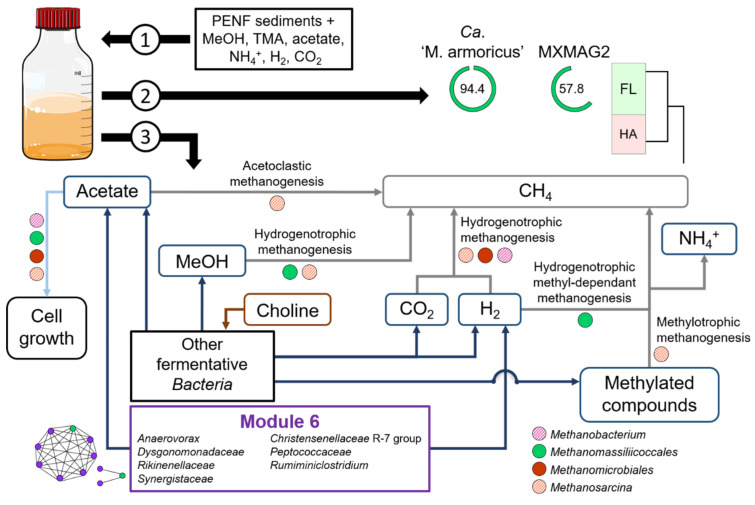
Figure 7.
Schematic diagram representing the substrates amended to the original slurry in the culture-based incubation experiment PENF (1) which led to the reconstruction of two Methanomassiliicoccales MAGs (Ca. ‘M. armoricus MXMAG1′ and MXMAG2) affiliated to the ‘free-living’ clade (2). Tentative reconstruction of the microbial metabolic interactions and substrate functions in this culture-based incubation experiment (3). The purple frame represents the module 6 of the co-occurrence network analysis, composed of reported fermentative Bacteria which maintained strong non-random co-occurrences with the dominant Methanomassiliicoccales OTUs sequences; FL: ‘Free-living’ clade; HA: ‘Host-associated’ clade; numbers in green rings indicate the completeness of the two Methanomassiliicoccales MAGs; dark blue arrows indicate fermentative products; grey arrows indicate the methanogenesis pathways probably occurring in this slurry; brown arrow indicates a fermentative substrate found in the enrichment slurry; light blue arrow indicates that acetate is potentially consume for methanogens cell growth. The colored circles indicate the dominant methanogenic taxa during the enrichment process and their known metabolic properties. The hatched color indicates that this property is present in only a part of the taxa of the lineage.
Based on large-scale metagenome analysis, co-occurrence between the Methanomassiliicoccales genera Ca. “Methanomethylophilus” and Methanomassiliicoccus with various TMA-producing Bacteria in a GIT context was recently proposed, supporting the hypothesis of an interaction between these taxa and gut bacteria to earn TMA and H2 [26]. However, metabolic interactions, such as those suggested here as a hypothesis, have also recently been highlighted between Ca. ‘Methanomethylophilaceae’ and various acetogenic Bacteria in another GIT context [111]. Thus, these results reinforce the idea that Methanomassiliicoccales could develop relationships with acetogenic Bacteria, possibly to access acetate for cell growth in both GIT and environmental contexts [12,44]. Nevertheless, it cannot be excluded that another type of functional interaction would be at work between Methanomassiliicoccales and their associated microbial cohorts. One or more other metabolite(s) produced by associated microorganisms could stimulate the growth of Methanomassiliicoccales [24,26]. It also cannot be excluded that the interaction promoting the growth of Methanomassiliicoccales is simply functional/metabolic. Finally, a strong metabolic relationship, close to syntrophy, based on H2 moderation between Methanomassiliicoccales and various fermentative bacteria could explain why the isolation of Methanomassiliicoccales representatives in clonal culture, from natural environments, has not been successful so far.
4. Conclusions
Among the diverse marine and terrestrial ecosystem samples screened in this study, Methanomassiliicoccales never appeared to represent a significant fraction of the microbial diversity. Nevertheless, cultural approaches allowed us to obtain a slurry significantly enriched in Methanomassiliicoccales, from freshwater sediments. This enrichment allowed the reconstruction of a high-quality MAG (MXMAG1) corresponding to a new candidate genomic species named Candidatus ‘Methanomassiliicoccus armoricus MXMAG1’. This genomic species seems to be present in several ecosystems and is affiliated to the ‘free-living’ clade of Methanomassiliicoccales. Analysis of its genome showed that the archaea from which this genome is derived have probably an energy-producing metabolism similar to previously described Methanomassiliicoccales, based on the consumption of methylated compounds and hydrogen to produce methane. Furthermore, the comparison of this MAG with previously published genomes of the two functional clades of Methanomassiliicoccales revealed that representatives from the ‘free-living’ clade, to which MXMAG1 belongs, encoded for predicted various mechanisms involved in intracellular equilibrium to counteract certain environmental parameters. Finally, network analyses showed that Methanomassiliicoccales from the ‘free-living’ clade were strongly co-occurring with some putative acetate and dihydrogen-producing bacteria previously reported to form syntrophic relationships with methanogens in animal GITs.
Acknowledgments
The authors thank the co-chiefs, crew and shipboard scientific parties of the oceanographic expeditions that provide samples. We thank Jean-François Brugère for the discussions we had at the birth of this project. We are grateful to Antje Boetius, Marc Le Romancer, Sabrina Berlendis and Telesphore Sime-Ngando, who provided us samples from, respectively, the Håkon Mosby mud volcano, the Kerguelen archipelago, the Kryos Basin and the Pavin lake. We thank Valérie-Cueff-Gauchard and Guillaume Lannuzel for logistical and technical help with the qPCR. We thank Xavier Philippon and Ludovic Stephan for technical help with ionic chromatography and GC-FID analyses. LABGeM (CEA/Genoscope & CNRS UMR8030), the France Génomique and French Bioinformatics Institute national infrastructures (funded as part of Investissement d’Avenir program managed by Agence Nationale pour la Recherche, contracts ANR-10-INBS-09 and ANR-11-INBS-0013) are acknowledged for support within the MicroScope annotation platform.
Abbreviations
| ADP | Adenosine diphosphate |
| Ala | Alanine |
| ANI | Average Nucleotide Identity |
| ANOVA | Analysis of variance |
| Arg | Arginine |
| ASCII | American Standard Code for Information Interchange |
| ATP | Adenosine triphosphate |
| Asp | Asparagine |
| BIOM | Biological Observation Matrix |
| Bp | Base pair |
| BSA | Bovine Serum Albumin Acetylated |
| Ca. | Candidatus |
| CERS | Cation Electrolytically Regenerated Suppressor |
| CDS | Coding DNA sequences |
| CSS | Cumulative Sum Scaling |
| Cys | Cystine |
| DES | Deionized Elution Buffer |
| DDH | DNA-DNA Hybridization |
| Dfast | DDBJ Fast Annotation and Submission Tool |
| DHAB | Deep-sea Hypersaline Anoxic Brine |
| DHVEG-1 | Deep-sea Hydrothermal Vent Euryarchaeota Group-1 |
| DMA | Dimethylamine |
| DMS | Dimethylsulfide |
| DNA | Desoxyribonucleic Acid |
| DSMZ | Deutsche Sammlung von Mikroorganismen und Zellkulturen |
| EB | Elution Buffer |
| FID | Flame Ionization Detector |
| GC | Gas Chromatograph |
| GGDC | Genome to Genome Distance Calculator |
| GIT | Gastro-Intestinal Tract |
| Glu | Glutamine |
| Gly | Glycine |
| GTDB-Tk | Genome Taxonomy Database Toolkit |
| HDR | HeteroDisulfide Reductase |
| His | Histidine |
| Ile | Isoleucine |
| iTOL | interactive Tree Of Life |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| Lys | Lysine |
| MAG | Metagenome-Assembled Genome |
| MaGe | MicroScope Microbial Genome Annotation and Analysis Platform |
| MBGD | Marine Benthic Group D |
| MCR | Methyl-Coenzyme M Reductase |
| MeOH | Methanol |
| MeSH | Methanethiol |
| MetaBAT | Metagenome Binning with Abundance and Tetra-nucleotide frequencies |
| MMA | Monomethylamine |
| MUSCLE | Multiple Sequence Comparison by Log- Expectation |
| NCBI | National Center for Biotechnology Information |
| OTU | Operational Taxonomic Unit |
| PAMELA | Passive Margins Exploration Laboratories |
| PCI | Phenol Chloroform Isoamyl alcohol |
| PCR | Polymerase Chain Reaction |
| Pyl | Pyrrolysine |
| Prokka | rapid prokaryotic genome annotation |
| qPCR | Quantitative Polymerase Chain Reaction |
| Quast | Quality Assessment Tool |
| RAST | Rapid Annotations using Subsystems Technology |
| RDP | Ribosomal Database Project |
| rRNA | ribosomal Ribonucleic Acid |
| SDS | Sodium Dodecyl Sulfate |
| SSU | Small Subunit |
| TE | Tris-Ethylene diamine tetra acetic acid |
| Thr | Threonine |
| TMA | Trimethylamine |
| tRNA | transfer RNA |
| Trp | Tryptophane |
| UniProtKB | Universal Protein resource Knowledgebase |
| Val | Valine |
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/9/1/30/s1: Supplementary Material: Text S1. Pre-metabarcoding screening. Text S2. Origin and detailed description of environmental samples. Text S3. Total DNA extraction for metabarcoding screening. Text S4. Details of the barcoding and the phylogenetic analyses. Text S5. Media composition. Text S6. Quantification of substrates and metabolic products. Supplementary Tables and Figures: Figure S1. Trends in microbial diversity revealed by metabarcoding in DOUR (top) and MOUG2 (bottom) substrate-amended slurries over time. Figure S2. Evolution of the prokaryotic alpha-diversity during the incubation of the PENF sample, in the culture-based incubation experiment. Figure S3. Microbial diversity patterns revealed by metabarcoding (16S rRNA gene copy numbers) in the PENF substrate-amended slurry over time. Figure S4. Phylogenetic tree showing the relationships between the trimethylamine methyltransferases (mttB) sequences extracted from the metagenome (in red), and closest mttB sequences in public databases. Figure S5. Co-occurrence network reconstructed based on 16S rRNA gene sequencing data from bulk environmental samples, based on a Spearman rank’s correlation between OTUs and calculated with a percolation threshold of 0.70. Table S1. Origin and location of the 86 environmental samples screened as a first approach for the research of Methanomassiliicoccales. Table S2. Abundance of Methanomassiliiccoccales, other methanogens and total prokaryotic OTU 16S rRNA, representative sequences generated by metabarcoding sequencing in environmental samples and enrichment cultures PENF, DOUR, and MOUG2. Table S3. List of the Methanomassiliicoccales 16S rRNA gene sequences generated by metagenomics sequencing from the PENF enrichment culture (8th week of incubation), and 16S rRNA gene representative sequences of OTUs generated by metabarcoding sequencing. Table S4. Results of the quantification of Methanomassiliicoccales by qPCR in the different samples. Table S5. Taxonomic assignation of the taxa non-randomly associated with Methanomassiliicoccales OTUs and their reported metabolic features, within the PENF substrate-amended slurry.
Author Contributions
Conceptualization, M.C., K.A., G.B., L.T.; formal analysis, M.C., G.B., E.R., Y.M., M.A., A.S., K.A.; Writing—original draft preparation, M.C., K.A.; writing—review and editing, all co-authors; supervision, K.A., L.T.; funding acquisition, K.A. All authors have read and agreed to the published version of the manuscript.
Funding
The study was supported by grants from the French Ministry of Higher Education and Research, the Région Bretagne and the LabexMer (ANR-10-LABX-1) for MC. This work was supported by the program MERLIN Abyss, the “Laboratoire d’Excellence” LabexMER Axis 3 programs ENDIV and CULTIVENT (ANR-10-LABX-1), and the Sino-French IRP 1211 MicrobSea. Samples from the Mozambic Channel were provided in the frame of the PAMELA collaborative project (Ifremer/TOTAL SA).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kirschke S., Bousquet P., Ciais P., Saunois M., Canadell J.G., Dlugokencky E.J., Bergamaschi P., Bergmann D., Blake D.R., Bruhwiler L., et al. Three decades of global methane sources and sinks. Nat. Geosci. 2013;6:813–823. doi: 10.1038/ngeo1955. [DOI] [Google Scholar]
- 2.Chaban B., Ng S.Y.M., Jarrell K.F. Archaeal habitats—From the extreme to the ordinary. Can. J. Microbiol. 2006;52:73–116. doi: 10.1139/w05-147. [DOI] [PubMed] [Google Scholar]
- 3.Zinder S.H. Physiological Ecology of Methanogens. In: Ferry J.G., editor. Methanogenesis: Ecology, Physiology, Biochemistry & Genetics. Chapman & Hall; New York, NY, USA: 1993. [DOI] [Google Scholar]
- 4.Garcia J.-L., Patel B.K.C., Ollivier B. Taxonomic, Phylogenetic, and Ecological Diversity of Methanogenic Archaea. Anaerobe. 2000;6:205–226. doi: 10.1006/anae.2000.0345. [DOI] [PubMed] [Google Scholar]
- 5.Parkes R.J., Cragg B., Roussel E., Webster G., Weightman A., Sass H. A review of prokaryotic populations and processes in sub-seafloor sediments, including biosphere: Geosphere interactions. Marine Geol. 2014;352:409–425. doi: 10.1016/j.margeo.2014.02.009. [DOI] [Google Scholar]
- 6.Evans P.N., Parks D.H., Chadwick G.L., Robbins S.J., Orphan V.J., Golding S.D., Tyson G.W. Methane metabolism in the archaeal phylum Bathyarchaeota revealed by genome-centric metagenomics. Science. 2015;350:434–438. doi: 10.1126/science.aac7745. [DOI] [PubMed] [Google Scholar]
- 7.Mayumi D., Mochimaru H., Tamaki H., Yamamoto K., Yoshioka H., Suzuki Y., Kamagata Y., Sakata S. Methane production from coal by a single methanogen. Science. 2016;354:222–225. doi: 10.1126/science.aaf8821. [DOI] [PubMed] [Google Scholar]
- 8.Borrel G., Adam P.S., McKay L.J., Chen L.-X., Sierra-García I.N., Sieber C.M.K., Letourneur Q., Ghozlane A., Andersen G.L., Li W.-J., et al. Wide diversity of methane and short-chain alkane metabolisms in uncultured archaea. Nat. Microbiol. 2019;4:603–613. doi: 10.1038/s41564-019-0363-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borrel G., Brugère J.-F., Gribaldo S., Schmitz R.A., Moissl-Eichinger C. The host-associated archaeome. Nat. Rev. Microbiol. 2020;18:622–636. doi: 10.1038/s41579-020-0407-y. [DOI] [PubMed] [Google Scholar]
- 10.Nobu M.K., Narihiro T., Kuroda K., Mei R., Liu W.-T. Chasing the elusive Euryarchaeota class WSA2: Genomes reveal a uniquely fastidious methyl-reducing methanogen. ISME J. 2016;10:2478–2487. doi: 10.1038/ismej.2016.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vanwonterghem I., Evans P.N., Parks D.H., Jensen P.D., Woodcroft B.J., Hugenholtz P., Tyson G.W. Methylotrophic methanogenesis discovered in the archaeal phylum Verstraetearchaeota. Nat. Microbiol. 2016;1:16170. doi: 10.1038/nmicrobiol.2016.170. [DOI] [PubMed] [Google Scholar]
- 12.Adam P.S., Borrel G., Brochier-Armanet C., Gribaldo S. The growing tree of Archaea: New perspectives on their diversity, evolution and ecology. ISME J. 2017;11:2407–2425. doi: 10.1038/ismej.2017.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borrel G., Adam P.S., Gribaldo S. Methanogenesis and the Wood—Ljungdahl Pathway: An Ancient, Versatile, and Fragile Association. Genome Biol. Evol. 2016;8:1706–1711. doi: 10.1093/gbe/evw114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans P.N., Boyd J.A., Leu A.O., Woodcroft B.J., Parks D.H., Hugenholtz P., Tyson G.W. An evolving view of methane metabolism in the Archaea. Nat. Rev. Microbiol. 2019;17:219–232. doi: 10.1038/s41579-018-0136-7. [DOI] [PubMed] [Google Scholar]
- 15.Dridi B., Fardeau M.-L., Ollivier B., Raoult D., Drancourt M. Methanomassiliicoccus luminyensis gen. nov., sp. nov., a methanogenic archaeon isolated from human faeces. Int. J. Syst. Evol. Micr. 2012;62:1902–1907. doi: 10.1099/ijs.0.033712-0. [DOI] [PubMed] [Google Scholar]
- 16.Borrel G., Gaci N., Peyret P., O’Toole P.W., Gribaldo S., Brugère J.-F. Unique Characteristics of the Pyrrolysine System in the 7th Order of Methanogens: Implications for the Evolution of a Genetic Code Expansion Cassette. Archaea. 2014;2014:1–11. doi: 10.1155/2014/374146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kröninger L., Berger S., Welte C., Deppenmeier U. Evidence for the involvement of two heterodisulfide reductases in the energy-conserving system of Methanomassiliicoccus luminyensis. FEBS J. 2016;283:472–483. doi: 10.1111/febs.13594. [DOI] [PubMed] [Google Scholar]
- 18.Borrel G., Harris H.M.B., Tottey W., Mihajlovski A., Parisot N., Peyretaillade E., Peyret P., Gribaldo S., O’Toole P.W., Brugère J.-F. Genome Sequence of “Candidatus Methanomethylophilus alvus” Mx1201, a Methanogenic Archaeon from the Human Gut Belonging to a Seventh Order of Methanogens. J. Bacteriol. 2012;194:6944–6945. doi: 10.1128/JB.01867-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaci N., Borrel G., Tottey W., O’Toole P.W., Brugere J.-F. Archaea and the human gut: New beginning of an old story. World J. Gastroenterol. 2014;20:16062–16078. doi: 10.3748/wjg.v20.i43.16062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iino T., Tamaki H., Tamazawa S., Ueno Y., Ohkuma M., Suzuki K., Igarashi Y., Haruta S. Candidatus Methanogranum caenicola: A Novel Methanogen from the Anaerobic Digested Sludge, and Proposal of Methanomassiliicoccaceae fam. nov. and Methanomassiliicoccales ord. nov., for a Methanogenic Lineage of the Class Thermoplasmata. Microbes Environ. 2013;28 doi: 10.1264/jsme2.ME12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paul K., Nonoh J.O., Mikulski L., Brune A. “Methanoplasmatales,” Thermoplasmatales-Related Archaea in Termite Guts and Other Environments, Are the Seventh Order of Methanogens. Appl. Environ. Microbiol. 2012;78:8245–8253. doi: 10.1128/AEM.02193-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borrel G., O’Toole P.W., Harris H.M.B., Peyret P., Brugère J.-F., Gribaldo S. Phylogenomic Data Support a Seventh Order of Methylotrophic Methanogens and Provide Insights into the Evolution of Methanogenesis. Genome Biol. Evol. 2013;5:1769–1780. doi: 10.1093/gbe/evt128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Söllinger A., Schwab C., Weinmaier T., Loy A., Tveit A.T., Schleper C., Urich T. Phylogenetic and genomic analysis of Methanomassiliicoccales in wetlands and animal intestinal tracts reveals clade-specific habitat preferences. FEMS Microbiol. Ecol. 2016;92 doi: 10.1093/femsec/fiv149. [DOI] [PubMed] [Google Scholar]
- 24.Borrel G., McCann A., Deane J., Neto M.C., Lynch D.B., Brugère J.-F., O’Toole P.W. Genomics and metagenomics of trimethylamine-utilizing Archaea in the human gut microbiome. ISME J. 2017;11:2059–2074. doi: 10.1038/ismej.2017.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De la Cuesta-Zuluaga J., Spector T.D., Youngblut N.D., Ley R.E. Genomic insights into adaptations of TMA-utilizing methanogens to diverse habitats including the human gut. bioRxiv. 2020 doi: 10.1101/2020.09.17.302828. Unpublished work. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lang K., Schuldes J., Klingl A., Poehlein A., Daniel R., Brune A. New Mode of Energy Metabolism in the Seventh Order of Methanogens as Revealed by Comparative Genome Analysis of “Candidatus Methanoplasma termitum”. Appl. Environ. Microbiol. 2015;81:1338–1352. doi: 10.1128/AEM.03389-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y., Leahy S.C., Jeyanathan J., Henderson G., Cox F., Altermann E., Kelly W.J., Lambie S.C., Janssen P.H., Rakonjac J., et al. The complete genome sequence of the methanogenic archaeon ISO4-H5 provides insights into the methylotrophic lifestyle of a ruminal representative of the Methanomassiliicoccales. Stand. Genomic Sci. 2016;11:59. doi: 10.1186/s40793-016-0183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mihajlovski A., Doré J., Levenez F., Alric M., Brugère J.-F. Molecular evaluation of the human gut methanogenic archaeal microbiota reveals an age-associated increase of the diversity. Environ. Microbiol. Rep. 2010;2:272–280. doi: 10.1111/j.1758-2229.2009.00116.x. [DOI] [PubMed] [Google Scholar]
- 29.Clemente J.C., Pehrsson E.C., Blaser M.J., Sandhu K., Gao Z., Wang B., Magris M., Hidalgo G., Contreras M., Noya-Alarcón Ó., et al. The microbiome of uncontacted Amerindians. Sci. Adv. 2015;1:e1500183. doi: 10.1126/sciadv.1500183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vanderhaeghen S., Lacroix C., Schwab C. Methanogen communities in stools of humans of different age and health status and co-occurrence with bacteria. FEMS Microbiol. Lett. 2015;362 doi: 10.1093/femsle/fnv092. [DOI] [PubMed] [Google Scholar]
- 31.De León K.B., Gerlach R., Peyton B.M., Fields M.W. Archaeal and bacterial communities in three alkaline hot springs in Heart Lake Geyser Basin, Yellowstone National Park. Front. Microbiol. 2013;4 doi: 10.3389/fmicb.2013.00330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou Z., Chen J., Cao H., Han P., Gu J.-D. Analysis of methane-producing and metabolizing archaeal and bacterial communities in sediments of the northern South China Sea and coastal Mai Po Nature Reserve revealed by PCR amplification of mcrA and pmoA genes. Front. Microbiol. 2015;5 doi: 10.3389/fmicb.2014.00789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li W., Guan W., Chen H., Liao B., Hu J., Peng C., Rui J., Tian J., Zhu D., He Y. Archaeal communities in the sediments of different mangrove stands at Dongzhaigang, China. J. Soils Sediments. 2016;16:1995–2004. doi: 10.1007/s11368-016-1427-0. [DOI] [Google Scholar]
- 34.Zhang C.-J., Pan J., Liu Y., Duan C.-H., Li M. Genomic and transcriptomic insights into methanogenesis potential of novel methanogens from mangrove sediments. Microbiome. 2020;8:94. doi: 10.1186/s40168-020-00876-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parada A.E., Needham D.M., Fuhrman J.A. Every base matters: Assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 2016;18:1403–1414. doi: 10.1111/1462-2920.13023. [DOI] [PubMed] [Google Scholar]
- 36.Apprill A., McNally S., Parsons R., Weber L. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 2015;75:129–137. doi: 10.3354/ame01753. [DOI] [Google Scholar]
- 37.Walters W., Hyde E.R., Berg-Lyons D., Ackermann G., Humphrey G., Parada A., Gilbert J.A., Jansson J.K., Caporaso J.G., Fuhrman J.A., et al. Improved Bacterial 16S rRNA Gene (V4 and V4-5) and Fungal Internal Transcribed Spacer Marker Gene Primers for Microbial Community Surveys. mSystems. 2016;1 doi: 10.1128/mSystems.00009-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Escudié F., Auer L., Bernard M., Mariadassou M., Cauquil L., Vidal K., Maman S., Hernandez-Raquet G., Combes S., Pascal G. FROGS: Find, Rapidly, OTUs with Galaxy Solution. Bioinformatics. 2018;34:1287–1294. doi: 10.1093/bioinformatics/btx791. [DOI] [PubMed] [Google Scholar]
- 39.McMurdie P.J., Holmes S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davis N.M., Proctor D.M., Holmes S.P., Relman D.A., Callahan B.J. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome. 2018;6:226. doi: 10.1186/s40168-018-0605-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paulson J.N., Pop M., Bravo H.C. metagenomeSeq: Statistical Analysis for Sparse High-Throughput Sequencing. [(accessed on 20 November 2020.)]; Available online: http://cbcb.umd.edu/software/metagenomeSeq.
- 42.Kemnitz D., Kolb S., Conrad R. Phenotypic characterization of Rice Cluster III archaea without prior isolation by applying quantitative polymerase chain reaction to an enrichment culture. Environ. Microbiol. 2005;7:553–565. doi: 10.1111/j.1462-2920.2005.00723.x. [DOI] [PubMed] [Google Scholar]
- 43.Kröninger L., Gottschling J., Deppenmeier U. Growth Characteristics of Methanomassiliicoccus luminyensis and expression of methyltransferase encoding genes. Archaea. 2017:2756573. doi: 10.1155/2017/2756573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Widdel F., Kohring G.-W., Mayer F. Studies on dissimilatory sulfate-reducing bacteria that decompose fatty acids. Arch. Microbiol. 1983;134:286–294. doi: 10.1007/BF00407804. [DOI] [PubMed] [Google Scholar]
- 45.Yakimov M.M., Cono V.L., Spada G.L., Bortoluzzi G., Messina E., Smedile F., Arcadi E., Borghini M., Ferrer M., Schmitt-Kopplin P., et al. Microbial community of the deep-sea brine Lake Kryos seawater—Brine interface is active below the chaotropicity limit of life as revealed by recovery of mRNA. Environ. Microbiol. 2015;17:364–382. doi: 10.1111/1462-2920.12587. [DOI] [PubMed] [Google Scholar]
- 46.Webster G., Rinna J., Roussel E.G., Fry J.C., Weightman A.J., Parkes R.J. Prokaryotic functional diversity in different biogeochemical depth zones in tidal sediments of the Severn Estuary, UK, revealed by stable-isotope probing. FEMS Microbiol. Ecol. 2010;72:179–197. doi: 10.1111/j.1574-6941.2010.00848.x. [DOI] [PubMed] [Google Scholar]
- 47.Criscuolo A., Brisse S. AlienTrimmer removes adapter oligonucleotides with high sensitivity in short-insert paired-end reads. Commentary on Turner (2014) Assessment of insert sizes and adapter content in FASTQ data from NexteraXT libraries. Front. Genet. 2014;5 doi: 10.3389/fgene.2014.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Crusoe M.R., Alameldin H.F., Awad S., Boucher E., Caldwell A., Cartwright R., Charbonneau A., Constantinides B., Edvenson G., Fay S., et al. The khmer software package: Enabling efficient nucleotide sequence analysis. F1000Research. 2015;4:900. doi: 10.12688/f1000research.6924.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li D., Luo R., Liu C.-M., Leung C.-M., Ting H.-F., Sadakane K., Yamashita H., Lam T.-W. MEGAHIT v1.0: A fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods. 2016;102:3–11. doi: 10.1016/j.ymeth.2016.02.020. [DOI] [PubMed] [Google Scholar]
- 50.Nurk S., Meleshko D., Korobeynikov A., Pevzner P.A. metaSPAdes: A new versatile metagenomic assembler. Genome Res. 2017;27:824–834. doi: 10.1101/gr.213959.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu Y.-W., Simmons B.A., Singer S.W. MaxBin 2.0: An automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics. 2016;32:605–607. doi: 10.1093/bioinformatics/btv638. [DOI] [PubMed] [Google Scholar]
- 52.Kang D.D., Froula J., Egan R., Wang Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ. 2015;3:e1165. doi: 10.7717/peerj.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parks D.H., Imelfort M., Skennerton C.T., Hugenholtz P., Tyson G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang X., Madan A. CAP3: A DNA Sequence Assembly Program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 56.Brettin T., Davis J.J., Disz T., Edwards R.A., Gerdes S., Olsen G.J., Olson R., Overbeek R., Parrello B., Pusch G.D., et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015;5:8365. doi: 10.1038/srep08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vallenet D., Calteau A., Cruveiller S., Gachet M., Lajus A., Josso A., Mercier J., Renaux A., Rollin J., Rouy Z., et al. MicroScope in 2017: An expanding and evolving integrated resource for community expertise of microbial genomes. Nucleic Acids Res. 2017;45:D517–D528. doi: 10.1093/nar/gkw1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tanizawa Y., Fujisawa T., Nakamura Y. DFAST: A flexible prokaryotic genome annotation pipeline for faster genome publication. Bioinformatics. 2018;34:1037–1039. doi: 10.1093/bioinformatics/btx713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Minh B.Q., Schmidt H.A., Chernomor O., Schrempf D., Woodhams M.D., von Haeseler A., Lanfear R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoon S.-H., Ha S.-M., Kwon S., Lim J., Kim Y., Seo H., Chun J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Micr. 2017;67:1613–1617. doi: 10.1099/ijsem.0.001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Richter M., Rosselló-Móra R., Oliver Glöckner F., Peplies J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics. 2016;32:929–931. doi: 10.1093/bioinformatics/btv681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meier-Kolthoff J.P., Auch A.F., Klenk H.-P., Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aires T., Moalic Y., Serrao E.A., Arnaud-Haond S. Hologenome theory supported by cooccurrence networks of species-specific bacterial communities in siphonous algae (Caulerpa) FEMS Microbiol. Ecol. 2015;91 doi: 10.1093/femsec/fiv067. [DOI] [PubMed] [Google Scholar]
- 64.Moalic Y., Desbruyères D., Duarte C.M., Rozenfeld A.F., Bachraty C., Arnaud-Haond S. Biogeography Revisited with Network Theory: Retracing the History of Hydrothermal Vent Communities. Syst. Biol. 2012;61:127. doi: 10.1093/sysbio/syr088. [DOI] [PubMed] [Google Scholar]
- 65.Bastian M., Heymann S., Jacomy M. Gephi: An Open Source Software for Exploring and Manipulating Networks; Proceedings of the 3rd International AAAI Conference on Weblogs and Social Media; San Jose, CA, USA. 17–20 May 2009; pp. 361–362. [Google Scholar]
- 66.Blondel V.D., Guillaume J.-L., Lambiotte R., Lefebvre E. Fast unfolding of communities in large networks. J. Stat. Mech. 2008;2008:P10008. doi: 10.1088/1742-5468/2008/10/P10008. [DOI] [Google Scholar]
- 67.Lovley D.R., Klug M.J. Methanogenesis from Methanol and Methylamines and Acetogenesis from Hydrogen and Carbon Dioxide in the Sediments of a Eutrophic Lake. Appl. Environ. Microbiol. 1983;45:1310–1315. doi: 10.1128/AEM.45.4.1310-1315.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.King G.M. Metabolism of Trimethylamine, Choline, and Glycine Betaine by Sulfate-Reducing and Methanogenic Bacteria in Marine Sediments. Appl. Environ. Microbiol. 1984;48:719–725. doi: 10.1128/AEM.48.4.719-725.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Oren A. Formation and breakdown of glycine betaine and trimethylamine in hypersaline environments. Antonie van Leeuwenhoek. 1990;58:291–298. doi: 10.1007/BF00399342. [DOI] [PubMed] [Google Scholar]
- 70.Lomans B.P., Smolders A., Intven L.M., Pol A., Op D., Drift C.V.D. Formation of dimethyl sulfide and methanethiol in anoxic freshwater sediments. Appl. Environ. Microbiol. 1997;63:4741–4747. doi: 10.1128/AEM.63.12.4741-4747.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hanna E., Keller J.K., Chang D., de Bruyn W., Zalman C. The potential importance of methylated substrates in methane production within three northern Minnesota peatlands. Soil Biol. Biochem. 2020;150:107957. doi: 10.1016/j.soilbio.2020.107957. [DOI] [Google Scholar]
- 72.Watkins A.J., Roussel E.G., Webster G., Parkes R.J., Sass H. Choline and N,N-Dimethylethanolamine as Direct Substrates for Methanogens. Appl. Environ. Microbiol. 2012;78:8298–8303. doi: 10.1128/AEM.01941-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Merkel A.Y., Podosokorskaya O.A., Chernyh N.A., Bonch-Osmolovskaya E.A. Occurrence, diversity, and abundance of methanogenic archaea in terrestrial hot springs of Kamchatka and Saõ Miguel Island. Microbiology. 2015;84:577–583. doi: 10.1134/S002626171504013X. [DOI] [Google Scholar]
- 74.Merkel A.Y., Podosokorskaya O.A., Sokolova T.G., Bonch-Osmolovskaya E.A. Diversity of methanogenic archaea from the 2012 terrestrial hot spring (Valley of Geysers, Kamchatka) Microbiology. 2016;85:342–349. doi: 10.1134/S0026261716030073. [DOI] [Google Scholar]
- 75.Grodnitskaya I.D., Trusova M.Y., Syrtsov S.N., Koroban N.V. Structure of microbial communities of peat soils in two bogs in Siberian tundra and forest zones. Microbiology. 2018;87:89–102. doi: 10.1134/S0026261718010083. [DOI] [Google Scholar]
- 76.Becker K.W., Elling F.J., Yoshinaga M.Y., Söllinger A., Urich T., Hinrichs K.-U. Unusual Butane- and Pentanetriol-Based Tetraether Lipids in Methanomassiliicoccus luminyensis, a Representative of the Seventh Order of Methanogens. Appl. Environ. Microbiol. 2016;82:4505–4516. doi: 10.1128/AEM.00772-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Biderre-Petit C., Jézéquel D., Dugat-Bony E., Lopes F., Kuever J., Borrel G., Viollier E., Fonty G., Peyret P. Identification of microbial communities involved in the methane cycle of a freshwater meromictic lake. FEMS Microbiol. Ecol. 2011;77:533–545. doi: 10.1111/j.1574-6941.2011.01134.x. [DOI] [PubMed] [Google Scholar]
- 78.Chen Y., Mi T., Liu Y., Li S., Zhen Y. Microbial Community Composition and Function in Sediments from the Pearl River Mouth Basin. J. Ocean Univ. China. 2020;19:941–953. doi: 10.1007/s11802-020-4225-7. [DOI] [Google Scholar]
- 79.Borrel G., Parisot N., Harris H.M., Peyretaillade E., Gaci N., Tottey W., Bardot O., Raymann K., Gribaldo S., Peyret P., et al. Comparative genomics highlights the unique biology of Methanomassiliicoccales, a Thermoplasmatales-related seventh order of methanogenic archaea that encodes pyrrolysine. BMC Genom. 2014;15:679. doi: 10.1186/1471-2164-15-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zinke L.A., Evans P.N., Schroeder A.L., Parks D.H., Varner R.K., Rich V.I., Tyson G.W., Emerson J.B. Evidence for non-methanogenic metabolisms in globally distributed archaeal clades basal to the Methanomassiliicoccales. Environ. Microbiol. 2020 doi: 10.1111/1462-2920.15316. [DOI] [PubMed] [Google Scholar]
- 81.Gascuel O. BIONJ: An improved version of the NJ algorithm based on a simple model of sequence data. Mol. Biol. Evol. 1997;14:685–695. doi: 10.1093/oxfordjournals.molbev.a025808. [DOI] [PubMed] [Google Scholar]
- 82.Goodrich J.K., Waters J.L., Poole A.C., Sutter J.L., Koren O., Blekhman R., Beaumont M., Van Treuren W., Knight R., Bell J.T., et al. Human Genetics Shape the Gut Microbiome. Cell. 2014;159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xie F., Zhang L., Jin W., Meng Z., Cheng Y., Wang J., Zhu W. Methane Emission, Rumen Fermentation, and Microbial Community Response to a Nitrooxy Compound in Low-Quality Forage Fed Hu Sheep. Curr. Microbiol. 2019;76:435–441. doi: 10.1007/s00284-019-01644-5. [DOI] [PubMed] [Google Scholar]
- 84.Azizi A., Kim W., Lee J.H. Comparison of microbial communities during the anaerobic digestion of Gracilaria under mesophilic and thermophilic conditions. World J. Microbiol. Biotechnol. 2016;32:158. doi: 10.1007/s11274-016-2112-6. [DOI] [PubMed] [Google Scholar]
- 85.Baker F.D., Papiska H.R., Campbell L.L. Choline fermentation by Desulfovibrio desulfuricans. J. Bacteriol. 1962;84:973–978. doi: 10.1128/JB.84.5.973-978.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Du C., Xu C.-Y., Jian J.-S., He W.-X., Hou L., Geng Z.-C. Seasonal dynamics of bacterial communities in a Betula albosinensis forest. Eur. J. Soil Sci. 2018;69:666–674. doi: 10.1111/ejss.12568. [DOI] [Google Scholar]
- 87.Bhatnagar L., Jain M.K., Aubert J.-P., Zeikus J.G. Comparison of Assimilatory Organic Nitrogen, Sulfur, and Carbon Sources for Growth of Methanobacterium Species. Appl. Environ. Microbiol. 1984;48:785–790. doi: 10.1128/AEM.48.4.785-790.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Richter M., Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA. 2009;106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wayne L.G., Brenner D.J., Colwell R.R., Grimont P.A.D., Kandler O., Krichevsky M.I., Moore L.H., Moore W.E.C., Murray R.G.E., Stackebrandt E., et al. Report of the Ad Hoc Committee on Reconciliation of Approaches to Bacterial Systematics. Int. J. Syst. Evol. Microbiol. 1987;37:463–464. doi: 10.1099/00207713-37-4-463. [DOI] [Google Scholar]
- 90.Bowers R.M., Kyrpides N.C., Stepanauskas R., Harmon-Smith M., Doud D., Reddy T.B.K., Schulz F., Jarett J., Rivers A.R., Eloe-Fadrosh E.A., et al. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat. Biotechnol. 2017;35:725–731. doi: 10.1038/nbt.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Parks D.H., Rinke C., Chuvochina M., Chaumeil P.-A., Woodcroft B.J., Evans P.N., Hugenholtz P., Tyson G.W. Recovery of nearly 8000 metagenome-assembled genomes substantially expands the tree of life. Nat. Microbiol. 2017;2:1533–1542. doi: 10.1038/s41564-017-0012-7. [DOI] [PubMed] [Google Scholar]
- 92.Stackebrandt E., Ebers J. Taxonomic parameters revisited: Tarnished gold standards. Microbiol. Today. 2006;33:152–155. [Google Scholar]
- 93.Yarza P., Yilmaz P., Pruesse E., Glöckner F.O., Ludwig W., Schleifer K.-H., Whitman W.B., Euzéby J., Amann R., Rosselló-Móra R. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 2014;12:635–645. doi: 10.1038/nrmicro3330. [DOI] [PubMed] [Google Scholar]
- 94.Krzycki J.A. Function of genetically encoded pyrrolysine in corrinoid-dependent methylamine methyltransferases. Curr. Opin. Chem. Biol. 2004;8:484–491. doi: 10.1016/j.cbpa.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 95.Kröninger L., Steiniger F., Berger S., Kraus S., Welte C.U., Deppenmeier U. Energy conservation in the gut microbe Methanomassiliicoccus luminyensis is based on membrane-bound ferredoxin oxidation coupled to heterodisulfide reduction. FEBS J. 2019;286:3831–3843. doi: 10.1111/febs.14948. [DOI] [PubMed] [Google Scholar]
- 96.Nicolaus B., Gambacorta A., Basso A.L., Riccio R., De Rosa M., Grant W.D. Trehalose in Archaebacteria. Syst. Appl. Microbiol. 1988;10:215–217. doi: 10.1016/S0723-2020(88)80003-1. [DOI] [Google Scholar]
- 97.Elbein A.D., Pan Y.T., Pastuszak I., Carroll D. New insights on trehalose: A multifunctional molecule. Glycobiology. 2003;13:17R–27R. doi: 10.1093/glycob/cwg047. [DOI] [PubMed] [Google Scholar]
- 98.Graham D.E., Xu H., White R.H. Methanococcus jannaschii Uses a Pyruvoyl-Dependent Arginine Decarboxylase in Polyamine Biosynthesis. J. Biol. Chem. 2002;277:23500–23507. doi: 10.1074/jbc.M203467200. [DOI] [PubMed] [Google Scholar]
- 99.Tolbert W.D., Graham D.E., White R.H., Ealick S.E. Pyruvoyl-Dependent Arginine Decarboxylase from Methanococcus jannaschii: Crystal Structures of the Self-Cleaved and S53A Proenzyme Forms. Structure. 2003;11:285–294. doi: 10.1016/S0969-2126(03)00026-1. [DOI] [PubMed] [Google Scholar]
- 100.Richard H.T., Foster J.W. Acid resistance in Escherichia coli. Adv. Appl. Microbiol. 2003;52:167–186. doi: 10.1016/s0065-2164(03)01007-4. [DOI] [PubMed] [Google Scholar]
- 101.Sugiyama Y., Nakamura A., Matsumoto M., Kanbe A., Sakanaka M., Higashi K., Igarashi K., Katayama T., Suzuki H., Kurihara S. A Novel Putrescine Exporter SapBCDF of Escherichia coli. J. Biol. Chem. 2016;291:26343–26351. doi: 10.1074/jbc.M116.762450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Scherer P., Kneifel H. Distribution of polyamines in methanogenic bacteria. J. Bacteriol. 1983;154:1315–1322. doi: 10.1128/JB.154.3.1315-1322.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Michael A.J. Polyamine function in archaea and bacteria. J. Biol. Chem. 2018;293:18693–18701. doi: 10.1074/jbc.TM118.005670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sierra-Alvarez R., Cortinas I., Yenal U., Field J.A. Methanogenic Inhibition by Arsenic Compounds. Appl. Environ. Microbiol. 2004;70:5688–5691. doi: 10.1128/AEM.70.9.5688-5691.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jomova K., Jenisova Z., Feszterova M., Baros S., Liska J., Hudecova D., Rhodes C.J., Valko M. Arsenic: Toxicity, oxidative stress and human disease. J. Appl. Toxicol. 2011;31:95–107. doi: 10.1002/jat.1649. [DOI] [PubMed] [Google Scholar]
- 106.Chen J., Rosen B. Biosensors for Inorganic and Organic Arsenicals. Biosensors. 2014;4:494–512. doi: 10.3390/bios4040494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Field J.A., Sierra-Alvarez R., Cortinas I., Feijoo G., Moreira M.T., Kopplin M., Gandolfi A.J. Facile reduction of arsenate in methanogenic sludge. Biodegradation. 2004;15:185–196. doi: 10.1023/B:BIOD.0000026697.10029.b2. [DOI] [PubMed] [Google Scholar]
- 108.Liu Z., Rensing C., Rosen B.P. Encyclopedia of Inorganic and Bioinorganic Chemistry. John Wiley & Sons, Ltd.; Chichester, WS, UK: 2013. Toxicity: Resistance Pathways for Metalloids and Toxic Metals; pp. 1–13. [DOI] [Google Scholar]
- 109.Hu W., Pan J., Wang B., Guo J., Li M., Xu M. Metagenomic insights into the metabolism and evolution of a new Thermoplasmata order (Candidatus Gimiplasmatales) Environ. Microbiol. 2020 doi: 10.1111/1462-2920.15349. [DOI] [PubMed] [Google Scholar]
- 110.Bentley R., Chasteen T.G. Microbial Methylation of Metalloids: Arsenic, Antimony, and Bismuth. Microbiol. Mol. Biol. Rev. 2002;66:250–271. doi: 10.1128/MMBR.66.2.250-271.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Podar M., Gilmour C.C., Brandt C.C., Soren A., Brown S.D., Crable B.R., Palumbo A.V., Somenahally A.C., Elias D.A. Global prevalence and distribution of genes and microorganisms involved in mercury methylation. Sci. Adv. 2015;1:e1500675. doi: 10.1126/sciadv.1500675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Parks J.M., Johs A., Podar M., Bridou R., Hurt R.A., Smith S.D., Tomanicek S.J., Qian Y., Brown S.D., Brandt C.C., et al. The Genetic Basis for Bacterial Mercury Methylation. Science. 2013;339:1332–1335. doi: 10.1126/science.1230667. [DOI] [PubMed] [Google Scholar]
- 113.Ma M., Du H., Wang D. Mercury methylation by anaerobic microorganisms: A review. Crit. Rev. Environ. Sci. Technol. 2019;49:1893–1936. doi: 10.1080/10643389.2019.1594517. [DOI] [Google Scholar]
- 114.Gies E.A., Konwar K.M., Beatty J.T., Hallam S.J. Illuminating Microbial Dark Matter in Meromictic Sakinaw Lake. Appl. Environ. Microbiol. 2014;80:6807–6818. doi: 10.1128/AEM.01774-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kadnikov V.V., Mardanov A.V., Beletsky A.V., Karnachuk O.V., Ravin N.V. Genome of the candidate phylum Aminicenantes bacterium from a deep subsurface thermal aquifer revealed its fermentative saccharolytic lifestyle. Extremophiles. 2019;23:189–200. doi: 10.1007/s00792-018-01073-5. [DOI] [PubMed] [Google Scholar]
- 116.Liu X., Li M., Castelle C.J., Probst A.J., Zhou Z., Pan J., Liu Y., Banfield J.F., Gu J.-D. Insights into the ecology, evolution, and metabolism of the widespread Woesearchaeotal lineages. Microbiome. 2018;6:102. doi: 10.1186/s40168-018-0488-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Martínez-Álvaro M., Auffret M.D., Stewart R.D., Dewhurst R.J., Duthie C.-A., Rooke J.A., Wallace R.J., Shih B., Freeman T.C., Watson M., et al. Identification of Complex Rumen Microbiome Interaction Within Diverse Functional Niches as Mechanisms Affecting the Variation of Methane Emissions in Bovine. Front. Microbiol. 2020;11 doi: 10.3389/fmicb.2020.00659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Liu P., Klose M., Conrad R. Temperature effects on structure and function of the methanogenic microbial communities in two paddy soils and one desert soil. Soil Biol. Biochem. 2018;124:236–244. doi: 10.1016/j.soilbio.2018.06.024. [DOI] [Google Scholar]
- 119.Drake H.L., Gössner A.S. Bergey’s Manual of Systematics of Archaea and Bacteria. John Wiley & Sons, Inc.; Hoboken, NJ, USA: 2015. Sporomusa ; pp. 1–7. [DOI] [Google Scholar]
- 120.Schink B. Bergey’s Manual of Systematics of Archaea and Bacteria. John Wiley & Sons, Inc.; Hoboken, NJ, USA: 2015. Anaerovorax ; pp. 1–3. [DOI] [Google Scholar]
- 121.Zhang S., Chang J., Lin C., Pan Y., Cui K., Zhang X., Liang P., Huang X. Enhancement of methanogenesis via direct interspecies electron transfer between Geobacteraceae and Methanosaetaceae conducted by granular activated carbon. Bioresour. Technol. 2017;245:132–137. doi: 10.1016/j.biortech.2017.08.111. [DOI] [PubMed] [Google Scholar]
- 122.Snell-Castro R., Méndez-Acosta H.O., Arreola-Vargas J., González-Álvarez V., Pintado-González M., González-Morales M.T., Godon J.J. Active prokaryotic population dynamics exhibit high correlation to reactor performance during methane production from acid hydrolysates of Agave tequilana var. azul bagasse. J. Appl. Microbiol. 2019;126:1618–1630. doi: 10.1111/jam.14234. [DOI] [PubMed] [Google Scholar]
- 123.Sakai S., Ehara M., Tseng I.-C., Yamaguchi T., Bräuer S.L., Cadillo-Quiroz H., Zinder S.H., Imachi H. Methanolinea mesophila sp. nov., a hydrogenotrophic methanogen isolated from rice field soil, and proposal of the archaeal family Methanoregulaceae fam. nov. within the order Methanomicrobiales. Int. J. Syst. Evol. Microbiol. 2012;62:1389–1395. doi: 10.1099/ijs.0.035048-0. [DOI] [PubMed] [Google Scholar]
- 124.Suzuki D., Li Z., Cui X., Zhang C., Katayama A. Reclassification of Desulfobacterium anilini as Desulfatiglans anilini comb. nov. within Desulfatiglans gen. nov., and description of a 4-chlorophenol-degrading sulfate-reducing bacterium, Desulfatiglans parachlorophenolica sp. nov. Int. J. Syst. Evol. Microbiol. 2014;64:3081–3086. doi: 10.1099/ijs.0.064360-0. [DOI] [PubMed] [Google Scholar]
- 125.Zinder S., Bräuer S. Bergey’s Manual of Systematics of Archaea and Bacteria. John Wiley & Sons, Inc.; Hoboken, NJ, USA: 2016. Methanoregula ; pp. 1–8. [DOI] [Google Scholar]
- 126.Morotomi M., Nagai F., Watanabe Y. Description of Christensenella minuta gen. nov., sp. nov., isolated from human faeces, which forms a distinct branch in the order Clostridiales, and proposal of Christensenellaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2012;62:144–149. doi: 10.1099/ijs.0.026989-0. [DOI] [PubMed] [Google Scholar]
- 127.Graf J. The Family Rikenellaceae. In: Rosenberg E., DeLong E.F., Lory S., Stackebrandt E., Thompson F., editors. The Prokaryotes: Other Major Lineages of Bacteria and The Archaea. Springer; Berlin/Heidelberg, Germany: 2014. pp. 857–859. [DOI] [Google Scholar]
- 128.Lee Y.-J., Wiegel J. Bergey’s Manual of Systematics of Archaea and Bacteria. John Wiley & Sons, Inc.; Hoboken, NJ, USA: 2017. Gracilibacteraceae ; pp. 1–3. [DOI] [Google Scholar]
- 129.Shiratori-Takano H., Ueda K. Bergey’s Manual of Systematics of Archaea and Bacteria. John Wiley & Sons, Inc.; Hoboken, NJ, USA: 2017. Lutispora ; pp. 1–6. [DOI] [Google Scholar]
- 130.Murakami T., Segawa T., Takeuchi N., Sepúlveda G.B., Labarca P., Kohshima S., Hongoh Y. Metagenomic analyses highlight the symbiotic association between the glacier stonefly Andiperla willinki and its bacterial gut community. Environ. Microbiol. 2018;20:4170–4183. doi: 10.1111/1462-2920.14420. [DOI] [PubMed] [Google Scholar]
- 131.Lee J., Koo T., Yulisa A., Hwang S. Magnetite as an enhancer in methanogenic degradation of volatile fatty acids under ammonia-stressed condition. J. Environ. Manag. 2019;241:418–426. doi: 10.1016/j.jenvman.2019.04.038. [DOI] [PubMed] [Google Scholar]
- 132.Waters J.L., Ley R.E. The human gut bacteria Christensenellaceae are widespread, heritable, and associated with health. BMC Biol. 2019;17:83. doi: 10.1186/s12915-019-0699-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Müller N., Worm P., Schink B., Stams A.J.M., Plugge C.M. Syntrophic butyrate and propionate oxidation processes: From genomes to reaction mechanisms. Environ. Microbiol. Rep. 2010;2:489–499. doi: 10.1111/j.1758-2229.2010.00147.x. [DOI] [PubMed] [Google Scholar]
- 134.Chojnacka A., Szczęsny P., Błaszczyk M.K., Zielenkiewicz U., Detman A., Salamon A., Sikora A. Noteworthy Facts about a Methane-Producing Microbial Community Processing Acidic Effluent from Sugar Beet Molasses Fermentation. PLoS ONE. 2015;10:e0128008. doi: 10.1371/journal.pone.0128008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Vartoukian S.R., Palmer R.M., Wade W.G. The division “Synergistes”. Anaerobe. 2007;13:99–106. doi: 10.1016/j.anaerobe.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 136.Yutin N., Galperin M.Y. A genomic update on clostridial phylogeny: Gram-Negative spore formers and other misplaced clostridia. Environ. Microbiol. 2013;15:2631–2641. doi: 10.1111/1462-2920.12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.McInerney M.J., Struchtemeyer C.G., Sieber J., Mouttaki H., Stams A.J.M., Schink B., Rohlin L., Gunsalus R.P. Physiology, Ecology, phylogeny, and genomics of microorganisms capable of syntrophic metabolism. Ann. NY Acad. Sci. 2008;1125:58–72. doi: 10.1196/annals.1419.005. [DOI] [PubMed] [Google Scholar]
- 138.Sieber J.R., McInerney M.J., Gunsalus R.P. Genomic Insights into Syntrophy: The Paradigm for Anaerobic Metabolic Cooperation. Annu. Rev. Microbiol. 2012;66:429–452. doi: 10.1146/annurev-micro-090110-102844. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MAGs’ bins generated and analyzed in this study are available under the Whole Genome Shotgun project deposited at DDBJ/ENA/GenBank under the accession numbers JAEILR000000000 v.1 (MXMAG1) and JAEILS000000000 v.1 (MXMAG2). The 16S rRNA gene sequences k119_35148 and k119_91804 are, respectively, available under the accession numbers MW383280 and MW383279. The 16S rRNA gene sequence generated by metabarcoding are available under the accession numbers MW386093-MW386158 in DDBJ/ENA/GenBank databases.