Abstract
The systematic knowledge on the genus Hedysarum L. (Fabaceae: Hedysareae) is still incomplete. The species from the section Hedysarum are valuable forage and medicinal resources. For eight Hedysarum species, we constructed the integrated schematic map of their distribution within Eurasia based on currently available scattered data. For the first time, we performed cytogenomic characterization of twenty accessions covering eight species for evaluating genomic diversity and relationships within the section Hedysarum. Based on the intra- and interspecific variability of chromosomes bearing 45S and 5S rDNA clusters, four main karyotype groups were detected in the studied accessions: (1) H. arcticum, H. austrosibiricum, H. flavescens, H. hedysaroides, and H. theinum (one chromosome pair with 45S rDNA and one pair bearing 5S rDNA); (2) H. alpinum and one accession of H. hedysaroides (one chromosome pair with 45S rDNA and two pairs bearing 5S rDNA); (3) H. caucasicum (one chromosome pair with 45S rDNA and one chromosome pair bearing 5S rDNA and 45S rDNA); (4) H. neglectum (two pairs with 45S rDNA and one pair bearing 5S rDNA). The species-specific chromosomal markers detected in karyotypes of H. alpinum, H. caucasicum, and H. neglectum can be useful in taxonomic studies of this section.
Keywords: genus Hedysarum, species distribution areas, chromosome variability, minor 45S rDNA sites
1. Introduction
The genus Hedysarum L. (Fabaceae: Hedysareae) comprises about 200 species of annual or perennial herbs distributed in Asia, Europe, North Africa, and North America which adapt to diverse habitats including temperate forests, stepped, polar and high-mountain regions [1,2,3].
The systematic knowledge on the genus Hedysarum is still incomplete, with uncertainty in generic delimitation. Fairly recently, the most prevalent Fedtschenko’s classification [1], which divided Hedysarum into seven sections according to their habits and morphology of stems and loments, was revised by Choi and Ohashi [4]. Currently, based on comprehensive morphological studies, the section Gamotion [5] is renamed into the sect. Hedysarum [4]. However, within the sections of the genus, the presence of morphologically intermediate forms and also significant intraspecific morphological diversity complicate discrimination between closely related species [2,6]. Moreover, only scattered data on the species occurrence in Eurasia are currently available, and spatial overlap in species distributions is still uncertain.
There are some differences in carpological and anatomical features of fruits between the species from the sect. Hedysarum (=syn. Gamotion) and other sections of the genus Hedysarum [7]. Also, the medicinal species from this section contain different bioactive compounds in leaves and roots, which are responsible for their immunomodulatory, antioxidant, anti-tumor and anti-diabetic effects [8,9,10]. In particular, several species, H. neglectum Ledeb., H. alpinum L., H. theinum Krasnob., and H. flavescens Rgl. et Schmalh., are rich in xanthone magniferine and oligomeric catechins, which makes them valuable raw material for the production of multifunctional biologically active additives, and thus offers the opportunity to develop new effective herbal remedies, in particular, a multifunctional antiviral drug ‘Alpisarin’ [10,11,12,13,14,15]. Some species from the section Hedysarum, such as H. flavescense, H. neglectum and H. alpinum, are valuable forage resources in their distribution areas [16,17,18].
At the same time, natural resources of Hedysarum species are insufficient for ever-growing needs. In view of this, H. alpinum and H. theinum are already being cultivated and/or introduced into the cell culture by means of biotechnological techniques [19,20]. However, such approaches need comprehensive comparative studies of genetic traits in valuable species and their closest relatives. In the genus Hedysarum, karyological studies were mostly performed by simple monochrome staining of chromosomes [21,22,23] or C-banding [24]. As reported earlier, the species belonging to the sect. Hedysarum have the basic chromosome number x = 7 unlike the species from other sections having x = 8 [4,21,25]. FISH-based chromosomal localization of rRNA genes was carried out in five Algerian species [26,27]. Molecular cytogenetic analysis of other Hedysarum species has not been conducted, and intra- or interspecific variability in chromosome localization of rRNA genes still remains uncertain.
Currently, taxonomic and phylogenetic relationships within the genus Hedysarum are being studied intensively with the use of various molecular phylogenetic approaches [28,29,30,31,32,33,34,35]. Recent extensive phylogenetic studies of Hedysareae based on nuclear (ITS, ETS, PGDH, SQD1, TRPT) and plastid (psbA-trnH, trnC-petN, trnL-trnF, trnS-trnG, petN-psbM) DNA sequencing did not support the monophyly of the genus Hedysarum [33,34,35]. Duan et al. [33], using nrDNA ITS and three plastid regions (matK, trnL–F, trnH–psbA), recognized three clades within the genus Hedysarum corresponding to the sections Hedysarum, Multicaulia, and Stracheya. Later, however, the incongruent position of the Hedysarum s.s. clade between the nuclear and plastid trees was revealed that could be related to a chloroplast capture hypothesis via introgression [34]. The sect. Stracheya clade was resolved as sister to the sect. Hedysarum clade in both nuclear and plastid trees, which supports merging Stracheya into Hedysarum [34]. These findings are consistent with the differentiation of the genus Hedysarum into three sections, with the species grouped according to their basic chromosome numbers x = 7 (the section Hedysarum) or x = 8 (the sections Multicaulia and Stracheya) [4,21,25]. At the same time, within the sect. Hedysarum, comparative analyses of nuclear and plastid genome sequences have been performed only in few species, and genomes of a number closely related medicinal species distributed in Eurasia still remain unstudied [33,34,35], and comprehensive investigation of cytogenetic peculiarities is essential for clarifying their taxonomy and phylogenetic relationships.
In order to characterize intra- and interspecific genomic diversity and also evaluate relationships within the genus Hedysarum sect. Hedysarum, we performed molecular cytogenetic characterization of twenty Eurasian accessions covering eight closely related species. Additionally, spatial overlap in species distributions within Eurasia was analyzed and the integrated schematic map of the species areas was constructed.
2. Results
2.1. Distribution Areas of the Studied Hedysarum Species
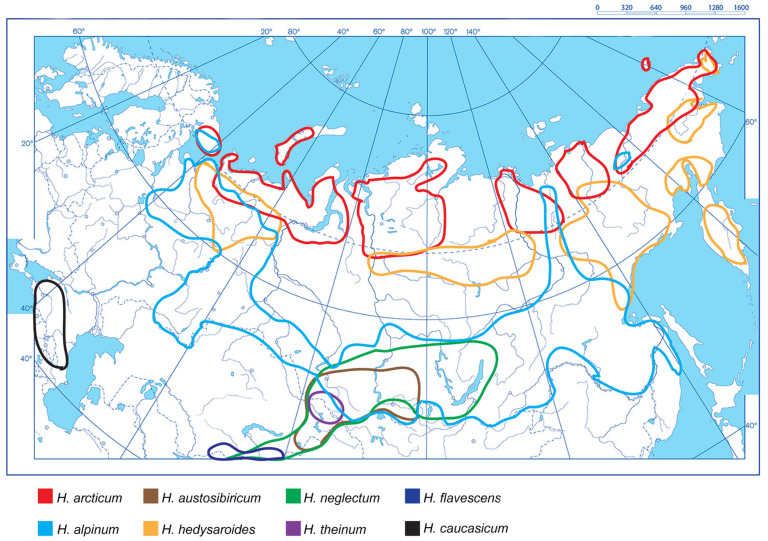
For H. alpinum, H. hedysaroides (L.) Schinz et Thell., H. arcticum B. Fedtsch., H. austrosibiricum B. Fedtsch., H. neglectum, H. theinum, H. flavescens, and H. caucasicum M. Bieb., we constructed an integrated schematic map of their distribution in the northern, central and eastern parts of Eurasia based on the analysis of the currently available data [6,16,17,18,36,37,38,39,40] (Figure 1).
Figure 1.
An integrated schematic map showing distribution of H. alpinum, H. arcticum, H. austrosibiricum, H. caucasicum, H. flavescens, H. hedysaroides, H. neglectum and H. theinum within the northern, central and eastern parts of Eurasia. The species names and correspondent colours of the lines indicating the boundaries of the species occurrence are specified under the maps.
In these regions, H. alpinum occurs in Kola Peninsula and North-Eastern Europe and also in Siberia, Kamchatka and Far East regions. H. arcticum is widespread in the Arctic and Sub-Arctic regions of the Eurasian continent from Arctic Scandinavia and Kola Peninsula to Chukotka Peninsula [6,16,36]. H. hedysaroides is distributed in the North-Eastern Europe, Central Siberia and the Far East regions [38].
H. austrosibiricum is an endemic species which normally occurs in the highlands of Southern Siberia (the Altai Mountains, Kuznetskii Alatau and the eastern edge of the Sayan Mountains) [37]. H. theinum is a highland alpine endemic species which grows in the Western Altai Mountains and also in the mountain ranges of Western Mongolia at altitudes of 1300–2000 m [36]. H. neglectum is distributed in the Altai Mountains, Kuznetskii Alatau, Western and Eastern Sayan Mountains and South-western parts of Tuva at lower altitudes (500–1500 m) compared to H. theinum [36]. H. flavescens occupies a narrow ecological niche growing on the limited small areas within the mountain ranges of the Western Tian Shan, Pamir-Alay Mountains [17,18]. This species normally occurs at altitudes of 2500–3100 m and less frequently, at medium altitudes (1000–2500 m) [17,18]. H. caucasicum is a highland species distributed in the Caucasus, Republic of Adygea and Krasnodar region at altitudes of 1500–3000 m [16,39,40].
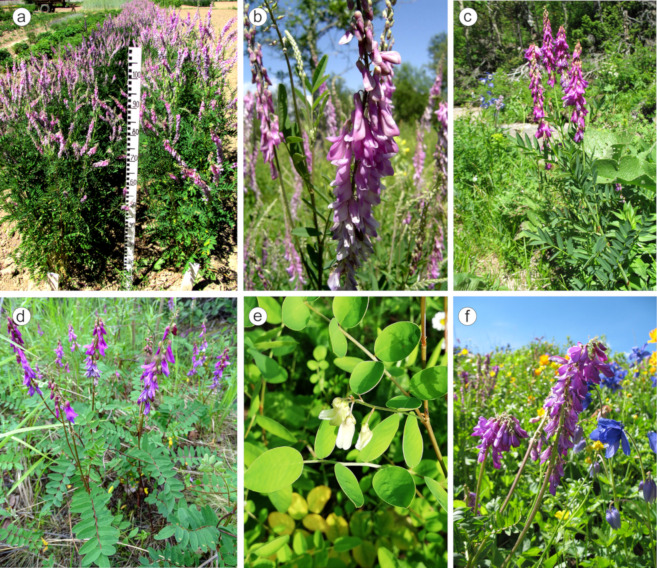
The constructed schematic map demonstrates ecological diversity in the species occurrences and also indicates the regions where several Hedysarum species grow together. The areas of H. alpinum, H. hedysaroides, and H. arcticum overlap partly, and the ranges of H. austrosibiricum, H. neglectum, H. theinum and H. alpinum overlap almost totally. Moreover, H. flavescens and H. neglectum have similar geographical areas within the mountain ranges of the Pamir and Altai Mountains. The distribution area of H. caucasicum confines to the Caucasus region and has no areas overlapping with the other studied species (Figure 1). In Figure 2, several studied Hedysarum species growing on a trial plot or in their natural habit are presented.
Figure 2.
H. alpinum plants growing on the trial plot (AIMAP, Moscow) (a) and wild populations of H. alpinum (the Zabaikalye region) (b), H. theinum (the Altai Mountains) (c), H. hedysaroides (the Magadan region) (d), H. flavescens (AIMAP, Moscow) (e) and H. neglectum (the Altai region) (f). The photos were taken by S.I. Romashkina (a,e) and I.Yu. Selyutina (b–d,f).
2.2. Chromosomal Structural Variations in the Studied Species
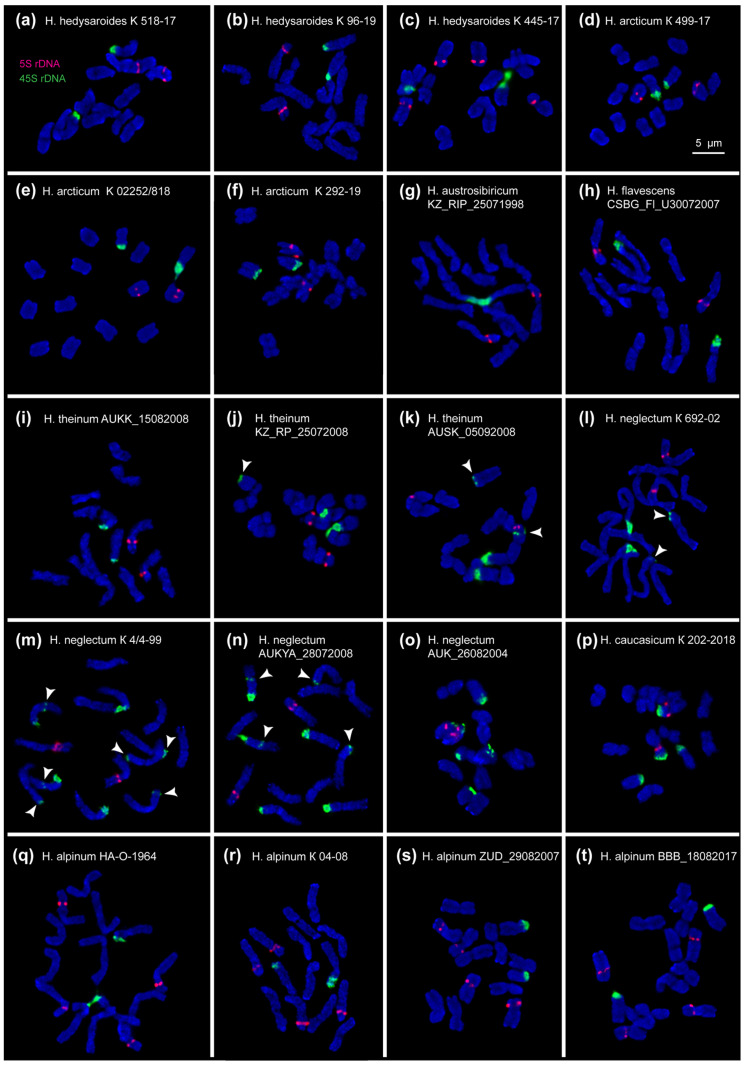
FISH mapping of 45S and 5S rDNA clusters on chromosomes of the studied Hedysarum species was performed for the first time (Figure 3).
Figure 3.
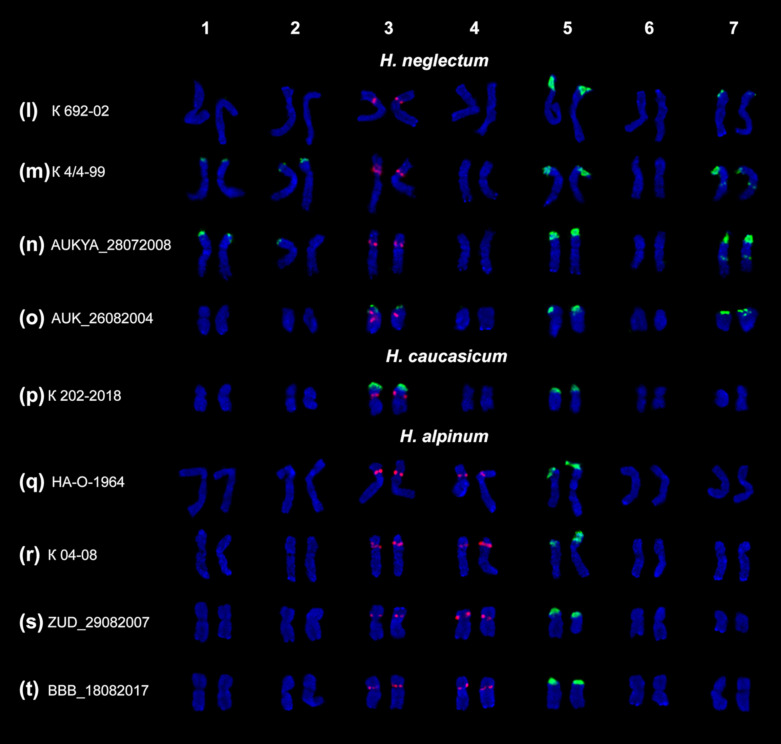
Metaphase plates of the studied accessions of H. hedysaroides (a–c), H. arcticum (d–f), H. austrosibiricum (g), H. flavescens (h), H. theinum (i–k), H. neglectum (l–o), H. caucasicum (p) and H. alpinum (q–t) after FISH with 45S rDNA (green) and 5S rDNA (red). DAPI chromosome staining—blue. Arrowheads point to polymorphic sites of 45S rDNA. Bar—5 μm.
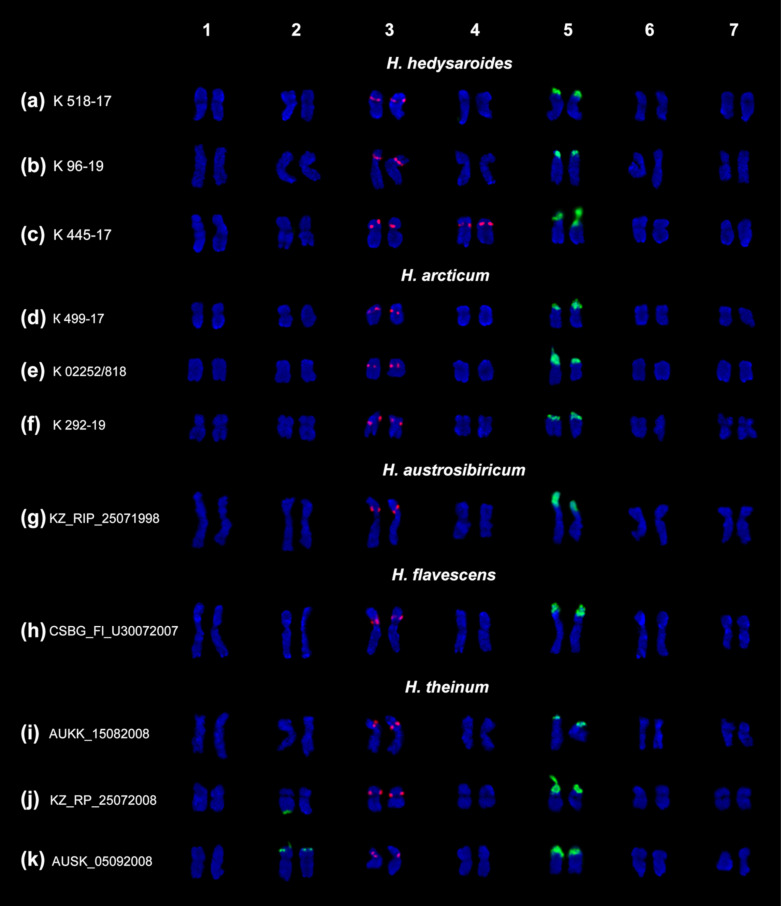
Based on chromosomal morphology, DAPI-banding patterns and distribution of 45S and 5S rDNA clusters, the species karyograms were constructed (Figure 4 and Figure 5).
Figure 4.
Karyotypes of the studied accessions of H. hedysaroides (a–c), H. arcticum (d–f), H. austrosibiricum (g), H. flavescens (h) and H. theinum (i–k). Karyograms of the metaphase plates shown in Figure 3 after FISH with 45S rDNA (green) and 5S rDNA (red). DAPI chromosome staining—blue.
Figure 5.
Karyotypes of the studied accessions of H. neglectum (l–o) and H. caucasicum (p) and H. alpinum (q–t). Karyograms of the metaphase plates shown in Figure 3 after FISH with 45S rDNA (green) and 5S rDNA (red). DAPI chromosome staining—blue.
All studied accessions have diploid karyotypes with 2n = 14 chromosomes. In H. flavescens and H. caucasicum, the chromosome numbers were determined for the first time. The examined karyotypes involved metacentric and submetacentric chromosomes (Figure 4h and Figure 5p).
In karyotypes of H. hedysaroides, H. arcticum, H. austrosibiricum, H. theinum and H. flavescens we observed one similar pair of major clusters of each 45S and 5S rDNA (Figure 4). In H. theinum, we also detected polymorphic minor 45S rDNA sites but only in chromosome pair 2 (in heteromorphic or homomorphic combinations) (Figure 4j,k).
Karyotypes of the examined accessions of H. neglectum and H. caucasicum included two pairs of satellite chromosomes (Figure 5l–p). The second major 45S rDNA cluster was localized in the short arms of chromosome pairs 3 (in H. caucasicum) and 7 (in H. neglectum). Also, in karyotypes of H. neglectum, polymorphic minor 45S rDNA sites were revealed in chromosome pairs 1, 2, 3 or 7 (Figure 5m–o).
In karyotypes of H. alpinum (Figure 5q–t) and one accession of H. hedysaroides (K 445-17) (Figure 4c), we observed two chromosome pars bearing major 5S rDNA clusters, without considerable changes in morphology of these chromosomes.
However, the chromosomal localization of rDNA clusters observed in the studied species, was not uniform, and polymorphic variants (in different combinations or independently) were revealed among individuals of the same accession as well as among different accessions.
Variant NOR-I was observed in H. austrosibiricum and H. flavescence. A satellite chromosome pair (5) was presented as a medium-sized submetacentric chromosome with a large satellite localized in the short arms. A 45S rDNA cluster was localized on the satellite and adjacent chromosome region (Figure 4g,h).
NOR-II was revealed in H. hedysaroides, H. arcticum and H. theinum. A satellite chromosome pair (5) was presented as a small-sized submetacentric chromosome with a 45S rDNA cluster localized in the distal region, and a small satellite was sometimes dispersed into a cloud (Figure 4a–f,i–k).
In karyotypes of H. alpinum (Figure 5q–t) and one accession of H. hedysaroides (K 445-17) (Figure 4c), we observed both variants (NOR-I and NOR-II) in homomorphic and/or heteromorphic combinations.
Variant NOR-negl denotes chromosome pair 7 with a 45S rDNA cluster localized in the distal region and a small satellite, which was often dispersed and poorly stained with DAPI. NOR-negl was revealed in karyotypes of H. neglectum together with NOR-I (chromosome pair 5) (Figure 5l–o). Additionally, in H. neglectum accessions K 4/4-99 and AUKYA_28072008, we detected minor polymorphic sites of 45S rDNA localized in the median regions of the long arms of chromosome pair 7 (Figure 5m,n).NOR-cauc denotes a pair of chromosomes (3) with a small dispersed satellite bearing a 45S rDNA cluster (localized in the distal region) and also a 5S rDNA cluster (revealed in the pericentromeric region of the short arms). Both NOR-cauc and NOR-II (chromosome 5) variants were detected in karyotypes of H. caucasicum (Figure 5p).
In H. neglectum (accessions K 4/4-99 and AUKYA_28072008), minor sites 45S rDNA were revealed in the distal regions of the short arms of chromosome pairs 1 and 2 (in homomorphic and/or heteromorphic variants) (Figure 5m,n). In H. theinum, minor 45S rDNA clusters were revealed in the distal regions of the short (accession AUSK_05092008; homomorphic variant) (Figure 4k) and the long (accession KZ_RP_25072008; heteromorphic variant) (Figure 4j) arms of chromosome pair 2.
Besides, we revealed several variants of localization of 5S rDNA clusters.
A similar variant 5S-sim was observed in all examined accessions. A 5S rDNA cluster was localized in the proximal region of the short arms of chromosome pair 3 (Figure 4 and Figure 5). In two accessions of H. neglectum, K 692-02 (in three plants) and AUK_26082004 (in all studied plants), variant 5S-negl was detected. On chromosome pair 3, we observed a major 5S rDNA cluster localized similar to variant 5S-sim, and also a minor 45S rDNA site revealed in the distal region of the short arms (Figure 5o).
Besides, in karyotypes of all studied accessions of H. alpinum (Figure 5q–t) and one accession of H. hedysaroides (K 445-17) (Figure 4c), we revealed variant 5S-sim (chromosome 3) together with variant 5S-alp which denoted a 5S rDNA cluster localized in the proximal region of the short arms of chromosome pairs 4.
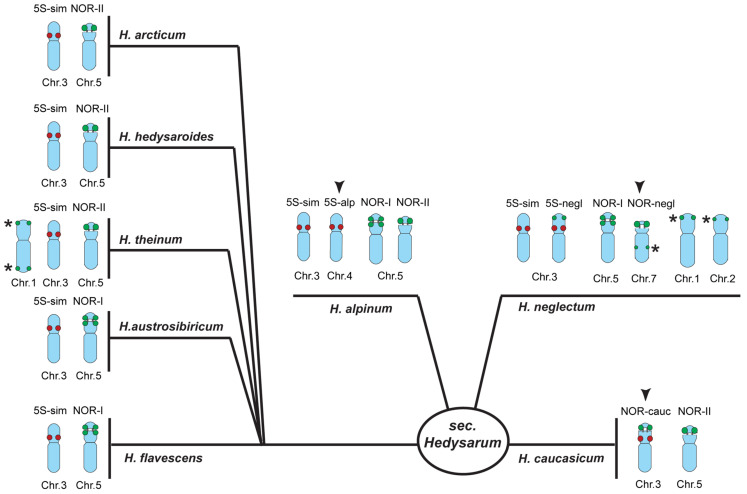
Based on chromosomal localization of 45S and 5S rDNA clusters, we divided all studied species from the sect. Hedysarum into four main karyotypic groups: (1) H. arcticum, H. austrosibiricum, H. flavescens, H. hedysaroides and H. theinum; (2) H. alpinum and one accession K 445-17 of H. hedysaroides; (3) H. caucasicum and (4) H. neglectum (Figure 6).
Figure 6.
Four main groups of karyotypes within the sect. Hedysarum based on chromosome localization of 45S rDNA and 5S rDNA clusters. Heads of arrows indicate the revealed species-specific chromosome markers. Asterisks denote polymorphic minor 45S rDNA sites.
3. Discussion
Previous phylogenetic data indicated non-monophyly of the genus Hedysarum in relation to Onobrychis, Greuteria, Eversmannia, Corethrodendron, Taverniera, and Ebenus. At the same time, both the cp and nr trees resolved the sect. Hedysarum as a monophyletic clade [33,35]. The species of the sect. Hedysarum have the basic chromosome number x = 7 in contrast to the other sections of the genus Hedysarum having the basic chromosome number x = 8 [4,21,25,36]. Karyotypes of the examined here accessions of H. alpinum, H. arcticum, H. austrosibiricum, H. caucasicum, H. flavescens, H. hedysaroides, H. neglectum, and H. theinum also comprise seven chromosome pairs. Moreover, we did not reveal any tetraploid karyotypes among the studied accessions though tetraploids were described earlier for these species [25,36,41]. In the present study, all examined species from the sect. Hedysarum were divided into four main groups according to the chromosome localization of 45S and 5S rDNA clusters: (1) H. arcticum, H. austrosibiricum, H. flavescens, H. hedysaroides and H. theinum; (2) H. alpinum and one accession K 445-17 of H. hedysaroides; (3) H. caucasicum and (4) H. neglectum. In karyotypes of all studied species we observed similar distribution of major 5S and 45S rDNA clusters on chromosome pairs 3 and 5, correspondingly that might suggest their close relationship. These similarities in distribution of major 45S and 5S rDNA clusters are consistent with the monophyletic origin of the sect. Hedysarum [33,35]. Besides, in karyotypes of the species from groups 2, 3, and 4, additional 5S and 45S rDNA clusters were revealed on other chromosome pairs, which can be related to chromosome rearrangements as well as intra- and interspecific hybridization events occurred during speciation.
The sect. Hedysarum is the widest-ranging section within the genus Hedysarum. Plant species belonging to this section are distributed in temperate to boreal regions of the Northern Hemisphere and occur in various habitats such as alpine and arctic meadows, stone grasslands, deserts and seashores [4,16]. The constructed here integrated schematic map allowed us to specify the species occurrence and analyze spatial overlap in species distributions within Eurasia. In the regions where populations of related species grow together, introgressive hybridization events can occur. Several new species having hybrid background were earlier detected within the Hedysarum sect. Multicaulia [42,43]. At the same time, according to the constructed map, H. flavescens and H. caucasicum have rather narrow distribution ranges. These species normally grow in high mountain areas [16,17,18,40]. Most probably, the both species are relics of former distribution areas, which were probably much wider, and now are isolated from the other members having large overlapping areas in boreal and arctic zones.
H. alpinum, H. hedysaroides, H. arcticum, and H. neglectum are the most common species of the sect. Hedysarum [16,36]. Among them, H. alpinum covers the widest ranges of habitats including Europe, Siberia, the Far East, China, the Korean Peninsula and northern parts of Mongolia [36]. H. theinum is an endemic species growing in the Altai Mountains [36,37]. The distribution area of this species overlaps (partly or almost totally) with the ranges of H. hedysaroides, H. arcticum, H. austrosibiricum, and H. neglectum. These species can occupy different ecological niches, in particular, H. theinum grows in the subalpine and, rarely, alpine meadows, whereas H. neglectum occurs in the forest zone [36,37,44]. Nonetheless, there are still problems with the correct plant identification, especially in the regions with multiple species occurrence. This can be related to hybridization and introgression events as well as morphological similarities observed in closely related species of this section [2,4,6]. The chromosome analysis with the use of cytogenomic markers could specify some closely related species of the sect. Hedysarum.
The systematic positions of H. arcticum, H. hedysaroides, and H. austrosibiricum are still uncertain due to their high morphological similarities. In different reports, H. arcticum was described as an independent plant species [16,36] or a subspecies of H. hedysaroides [6,45]. Our results indicate that H. arcticum and H. hedysaroides have overlapping geographical areas. According to recent phylogenetic cp data, H. hedysaroides and H. arcticum are grouped in one clade [33,35].
H. austrosibiricum was earlier classified as a subspecies of H. hedysaroides [6], and currently, H. austrosibiricum is considered an independent species because of its narrow mountain range confined to Southern Siberia [36]. In the present study, in karyotypes of H. hedysaroides, we detected heteromorphic combinations of homologues chromosomes bearing 45S rDNA (NOR-I and NOR-II variants). At the same time, in karyotypes of H. arcticum and H. austrosibiricum, we revealed a single variant (different for each species) of chromosomes bearing 45S rDNA. These observations might suggest that they are actually independent species.
H. flavescens and also other yellow-flowering species of the sect. Hedysarum are considered as a primary mesophilic group distributed in highlands of the Pamir-Alay and Tien Shan Mountains. It is suggested that a part of these species could spread across Eurasia and create centers of secondary speciation, in particular, in Siberia [17,18]. The geographical distribution of H. flavescens mostly corresponds to the distribution areas (mountainous regions of Central Asia) of the species forming basal clades within the sect. Hedysarum. However, H. flavescens was not examined in the previous phylogenetic studies [33,34,35]. This suggests that Mountains of Central Asia could be the ancestral area for the species from this section. Currently, H. flavescens has a narrow distribution area in the Pamir-Alay and Tien Shan Mountains. However, this species belongs to the group of other species of the sect. Hedysarum (H. arcticum, H. austrosibiricum, H. hedysaroides, and H. theinum) having similar karyotypes and distributed in different regions of Eurasia. Therefore, it cannot be excluded that the genome of H. flavescens might be the closest to the ancestor genome.
H. theinum is known to be morphologically different from closely related H. neglectum and considered an independent species [44]. The constructed integrated schematic map of the species distribution illustrates that ranges of H. neglectum and H. theinum overlap. However, H. theinum is an endemic species grown in the highlands (1300–2000 m above sea level). H. neglectum occurs a wider range of environmental conditions (mostly, in the forest zone) compared to H. theinum [36,37]. These species also differ considerably from each other in chemical composition as well as amount of biologically active substances in their leaves and roots [9,44]. According to our results, H. theinum and H. neglectum have some differences in their karyotype structure. In H. neglectum, we detected two pairs of satellite chromosomes though only one pair was revealed in H. theinum, which might suggest that they were different species.
Moreover, in H. neglectum, we observed a number of polymorphic minor 45S rDNA sites localized on chromosome pairs 1, 2, 3, and 7, and the chromosome localization of these minor 45S rDNA loci was specific for accessions grown in different locations. In karyotypes of H. theinum, minor 45S rDNA loci were detected only on chromosome pairs 2. These minor loci were also polymorphic; their presence and chromosome localization varied depending on the growing area (e.g., Kazakhstan or the Altai Mountains). Our results are consistent with data on ISSR analyses which demonstrated increased intraspecific variability in H. theinum [30] and a higher level of interpopulation variability of the electrophoretic spectra of seed polypeptides in H. neglectum compared to H. theinum [31].
In the present study, the revealed variability in chromosome localization of minor rDNA clusters probably indicates that active processes of 45S rDNA redistribution still occur in the genomes of H. neglectum and H. theinum. High recombination rate and also increased activity of mobile elements facilitate changes in number of rDNA sites [46,47,48,49]. It was earlier assumed that the variations in number of 45S rDNA clusters observed in karyotypes of different species of Anacyclus could result from interspecific hybridization events occurring in the regions where ranges of these species overlapped or along the edge of their distribution areas [50]. Also, variations in the number of 45S rDNA loci were observed in karyotypes of plant species (Phaseolus vulgaris, Capsicum species, Brassica rapa) grown in different locations [51,52,53]. On the other hand, no variations in number and location of 45S rDNA clusters were earlier revealed among accessions of Hedysarum perrauderianum from different regions of Algeria [26].
In both H. caucasicum and H. neglectum, one of two pairs of satellite chromosomes was presented by a variant of chromosome 5. In karyotypes of H. neglectum, the other satellite chromosome (7) was detected only in this species and can be used as a species-specific chromosome marker. In karyotype of H. caucasicum, the other satellite chromosome pair (3) bearing both 5S and 45S rDNA clusters is also a unique variant which can be used as an additional chromosome marker for identification of this species. H. caucasicum is known to occupy a narrow isolated area [40], and this fact has apparently helped to maintain such a unique variant of a satellite chromosome pair in its karyotype.
As mention above, H. alpinum and H. neglectum have overlapping areas. However, these species differed in localization of 45S and 5S rDNA clusters. Our results on chromosome analysis are consistent with the previous phylogenetic data indicating that H. alpinum and H. neglectum belong to separate clades [33,35]. H. alpinum holds a special position among the studied species, as it is considered to be a conserved type for the genus Hedysarum [4]. In this species, all examined accessions had the similar karyotypes. Moreover, we did not reveal any differences in chromosomal localization of 45S and 5S rDNA clusters between the introduced and wild populations. At the same time, in karyotypes of H. alpinum, we detected two chromosome pairs bearing 5S rDNA clusters unlike the other species of this section having one chromosome pair with 5S rDNA. The exception was one accession of H. hedysaroides (K 445-17) which also had two chromosome pairs bearing 5S rDNA. It cannot be excluded, however, that this accession has been misidentified by a taxonomist, and in fact, it also belongs to H. alpinum. It is therefore possible that the presence of the second chromosome pair (4) bearing 5S rDNA in a karyotype can be used as additional species-specific character in taxonomic studies of the sect. Hedysarum.
4. Materials and Methods
4.1. Plant Material
In the present study, we examined twenty plant accessions covering eight Hedysarum species obtained from different seed sources (detailed in Table 1) including the collection of All-Russian Institute of Medicinal and Aromatic Plants (AIMAP), Moscow, RF. Wild plant accessions were collected and identified by Dr. I. Yu. Selyutina and Dr. N.A. Karnaukhova, the Central Siberian Botanical Garden (CSBG), SB RAS, RF and Dr. N.A. Suprun, Volgograd Regional Botanical Garden (VRBG), Volgograd, RF.
Table 1.
The list of the studied plant accessions.
| Species | Accession Number/Voucher | Origin/Seed Source |
|---|---|---|
| H. alpinum L. | HA-O-1964 | Omsk region, RF/germplasm collection of CSBG, 2009 |
| H. alpinum L. | K 04-08 | Unknown/germplasm collection of AIMAP, 2017 |
| H. alpinum L. | ZUD_29082007 | 51°7′ N; 112°11′ E; Zabaikalye region, RF/collected by Dr. N.A. Karnaukhova, 2007 |
| H. alpinum L. | BBB_18082017 | 53°58′ N; 113°35′ E; Bauntovskii region, Buryatia, RF/collected by Dr. I.Yu. Selyutina, 2017 |
| H. arcticum B. Fedtsch. | K 02252/818_24072016 | 67°25′ N; 65°11′ E; lake Shchuch’e, Komi Republic, RF/collected by Dr. N.A. Suprun, 2016 |
| H. arcticum B. Fedtsch. | K 499-17 | VRBG/germplasm collection of AIMAP, 2017 |
| H. arcticum B. Fedtsch. | K 292-19 | VRBG/germplasm collection of AIMAP, 2017 |
| H. austrosibiricum B. Fedtsch. | KZ_RIP_25071998 | 50°19′ N; 83°50′ E; Ivanovskiy Ridge, 2000 m above sea level, Kazakhstan/collected by Dr. I.Yu. Selyutina, 1998 |
| H. caucasicum M. Bieb. | K 202-2018 | Gothenburg Botanical Garden, Sweden/germplasm collection of AIMAP, 2018 |
| H. flavescens Rgl. et Schmalh. | CSBG_Fl_U30072007 | University of Uppsala Botanical Garden, Sweden, Uppsala/germplasm collection of CSBG, 2008 |
| H. hedysaroides (L.) Schinz et Thell. | K 518-17 | Botanical Garden of the University of Vienna, Austria/germplasm collection of AIMAP, 2017 |
| H. hedysaroides (L.) Schinz et Thell. | K 445-17 | University of Tartu Botanical Gardens, Estonia/germplasm collection of AIMAP, 2017 |
| H. hedysaroides (L.) Schinz et Thell. | K 96-19 | Botanical Garden of the University of Vienna, Austria/germplasm collection of AIMAP, 2017 |
| H. neglectum Ledeb. | K 4/4-99 | Kaira Yalbak Mountains, Altai region, RF/germplasm collection of AIMAP, 2017 |
| H. neglectum Ledeb. | K 692-02 | Altai region/germplasm collection of AIMAP, 2017 |
| H. neglectum Ledeb. | AUK_26082004 | 50°39′ N; 87°43′ E; Kubadru River, Altai region, RF/collected by Dr. I.Yu. Selyutina, 2004 |
| H. neglectum Ledeb. | AUKYA_28072008 | 50°52′ N; 85°15′ E; Yabogansky pass, Altai region, RF/collected by Dr. N.A. Karnaukhova, 2008 |
| H. theinum Krasnob. | AUKK_15082008 | 50°59′ N; 84°13′ E; Kumir river, Altai region, RF/collected by Dr. I.Yu. Selyutina, 2008 |
| H. theinum Krasnob. | KZ_RP_25072008 | 50°18′ N; 83°38′ E; Ridderskii region, Kazakhstan/collected by Dr. I.Yu. Selyutina, 2008 |
| H. theinum Krasnob. | AUSK_05092008 | 50°4′ N; 85°13′ E; Ust-Koksinsky region; Altai Mountains, RF/collected by Dr. N.A. Karnaukhova, 2008 |
4.2. Chromosome Spread Preparation
Plant seeds were scarified and put into hot (60–75 °C) water for 5–10 min. Then they were germinated in Petri dishes for 3–7 days at 22 °C. For cell cycle synchronization and accumulation of mitotic divisions, excised root tips (of 0.5 cm long) were incubated in ice water for 24 h. After this pre-treatment, the root tips were fixed in ethanol:acetic acid (3:1) for 3–24 h at room temperature. Before squashing, the roots were transferred into 1% acetocarmine solution in 45% acetic acid for 15 min. The cover slips were removed after freezing in liquid nitrogen. The slides were dehydrated in 96% ethanol and then air dried.
4.3. FISH Procedure
For a FISH procedure, following probes were used: pTa71 which comprised a 9 kb long DNA sequence of common wheat including 18S, 5.8S and 26S (45S) rDNA [54]; pTa794 which comprised a 420 bp long DNA sequence of wheat including 5S rDNA [55]. These DNA probes were labelled directly with fluorochromes SpectrumAqua or SpectrumRed by nick translation according to manufacturers’ protocols (Abbott Molecular, Wiesbaden, Germany). The FISH procedure was carried out as described previously [56]. After hybridization, the slides were washed respectively in 0.1 × SSC at 50 °C for 5 min, 2 × SSC at 37 °C for 10 min, 2 × SSC at RT for 5 min and 1 × PBS at RT for 3 min. Then the slides were dehydrated through a graded ethanol series, air dried and stained with 0.1 μg/mL DAPI (4′,6-diamidino-2-phenylindole) (Serva, Heidelberg, Germany) in Vectashield mounting medium (Vector laboratories, Peterborough, UK).
4.4. Chromosome Analysis
At least five plants of each accession and fifteen metaphase plates from each sample were analyzed. The slides were examined using an Olympus BX-61 epifluorescence microscope (Olympus, Tokyo, Japan). Images were captured with a monochrome charge-coupled device camera (Cool Snap, Roper Scientific, Inc., Sarasota, FL, USA). Then they were processed with Adobe Photoshop 10.0 software (Adobe, Birmingham, AL, USA).
5. Conclusions
For eight Hedysarum species, we constructed the integrated schematic map of their distribution within Eurasia based on currently available scattered data. For the first time, we performed cytogenomic characterization of twenty accessions covering eight closely related species from the sect. Hedysarum. Intra- and interspecific variability of chromosomes bearing 45S and 5S rDNA clusters was detected. Based on chromosome analysis, we revealed four main karyotype groups in the studied species: (1) H. arcticum, H. austrosibiricum, H. flavescens, H. hedysaroides, and H. theinum; (2) H. alpinum and one accession K 445-17 of H. hedysaroides; (3) H. caucasicum; and (4) H. neglectum. H. caucasicum, H. neglectum, additional cytogenomic markers were observed. The detected species-specific chromosomal markers can be useful in taxonomic studies of this section.
Acknowledgments
The authors acknowledge N.A. Karnaukhova (the Central Siberian Botanical Garden (CSBG), SB RAS, Novosibirsk, RF) and N.A. Suprun (Volgograd Regional Botanical Garden (VRBG), Volgograd, RF) for providing us valuable plant materials.
Author Contributions
Conceptualization, O.Y.Y. and O.V.M.; formal analysis, O.Y.Y., T.E.S., S.A.Z., and A.V.A.; investigation, O.Y.Y., T.E.S., I.Y.S., S.I.R., S.A.Z., and A.V.A.; methodology, O.Y.Y., T.E.S., S.A.Z., and O.V.M.; resources, I.Y.S. and S.I.R.; supervision, O.V.M.; visualization, O.Y.Y., A.V.A., I.Y.S., and S.I.R.; writing—original draft, O.Y.Y., I.Y.S., A.V.A., and O.V.M.; writing—review & editing, O.Y.Y., T.E.S., A.V.A., and O.V.M. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Russian Foundation for Basic Research (project No. 18-04-01091) and the Program of Fundamental Research of State Academies (project No. 01201363824).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data are contained within the article.
Conflicts of Interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Fedtschenko B.A. The genus Hedysarum. Trudy Imp. S. Petersburgsk Bot. Sada. 1902;19:185–342. [Google Scholar]
- 2.Roskov Y.R. Hedysarea. In: Yakovlev G.P., Sytin A.K., Roskov Y.R., editors. Legumes of Northern Eurasia: A Checklist. Royal Botanic Gardens; London, UK: 1996. pp. 379–426. [Google Scholar]
- 3.Choi B.-H., Ohashi H. Proposal to conserve the name Hedysarum (Leguminosae: Papilionoideae) with a conserved type. Taxon. 1998;47:877. doi: 10.2307/1224197. [DOI] [Google Scholar]
- 4.Choi B.-H., Ohashi H., Ohashi B.-H.C. Generic criteria and an infrageneric system for Hedysarum and related genera (Papilionoideae-Leguminosae) Taxon. 2003;52:567–576. doi: 10.2307/3647455. [DOI] [Google Scholar]
- 5.Basiner T.F. Enumeratio monographica specierum generis Hedysari. Bull. Phys. Math. Acad. Sci. (Petersb.) 1845;4:305–315. [Google Scholar]
- 6.Yurtsev B.A. Arctic flora of the USSR [Arkticheskaya flora SSSR] Volume 9 Nauka; Saint Petersburg, Russia: 1986. [Google Scholar]
- 7.Mironov Y.M. Pericarp anatomy of East European species of the genus Hedysarum L. (Papilionaceae): Sections Gamotion and Multicaulia. Bull. Mosc. Soc. Nat. Ser. Biol. 2000;105:50–53. [Google Scholar]
- 8.Choi B.H. Foliar Flavonoids of the genus Hedysarum and related genera (tribe Hedysareae—Leguminosae) Korean J. Plant Taxon. 1994;24:259–264. doi: 10.11110/kjpt.1994.24.4.259. [DOI] [Google Scholar]
- 9.Vysochina G.I., Kukushkina T.A., Karnaukhova N.A., Selyutina I.Y. Flavonoids of wild and introduced plants of several species of the Hedysarum L Genus. Chem. Sustain. Dev. 2011;19:327–333. [Google Scholar]
- 10.Dong Y.-M., Tang D., Zhang N., Li Y., Zhang C., Li L., Li M.-H. Phytochemicals and biological studies of plants in genus Hedysarum. Chem. Cent. J. 2013;7:124. doi: 10.1186/1752-153X-7-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rybachenko A.I., Krivut B.A., Georgievskii V.P. Fluorodensitometric determination of mangiferin and isomangiferin in Hedysarum flavescens and H. alpinum. Chem. Nat. Compd. 1976;12:395–396. doi: 10.1007/BF00564793. [DOI] [Google Scholar]
- 12.Komissarenko A.N., Nadezhina T.P., Komissarenko N.F. Flavonoids and xanthones of Hedysarum connatum and H. alpinum. Chem. Nat. Compd. 1994;30:521–522. doi: 10.1007/BF00630415. [DOI] [Google Scholar]
- 13.Liu Y., Zhang Q.-Y., Zhao Y.-Y., Wang B., Hai L.-Q., Ying Y.-P., Chen H.-B. Saponins from the roots of Hedysarum polybotrys. Biochem. Syst. Ecol. 2007;35:389–391. doi: 10.1016/j.bse.2006.12.008. [DOI] [Google Scholar]
- 14.Nechepurenko I.V., Polovinka M.P., Komarova N.I., Korchagina D.V., Salakhutdinov N.F., Nechepurenko S.B. Low-molecular-weight phenolic compounds from Hedysarum theinum roots. Chem. Nat. Compd. 2008;44:31–34. doi: 10.1007/s10600-008-0009-9. [DOI] [Google Scholar]
- 15.Zinner N.S., Vysochina G.I., Kukushkina T.A., Sviridova T.P. Biologically active substances Hedysarum alpinum and H. theinum (Fabaceae) introduced to Tomsk region. Tomsk State Univ. J. Biol. 2010;4:116–122. [Google Scholar]
- 16.Fedtschenko B.A. Hedysarum. In: Komarov V.L., Shishkin B.K., Bobrov E.G., editors. Flora URSS. Volume 13. Akad. Scient. URSS; Moscow, Russia: 1948. pp. 259–379. [Google Scholar]
- 17.Bajtenov M.S. Genus Hedysarum L. In: Pavlov N.V., editor. Flora of Kazahstan. Volume V. Izd-vo AN Kazah. SSR; Alma-Ata, Kazakhstan: 1961. pp. 418–442. [Google Scholar]
- 18.Bajtenov M.S. Flora of Kazakhstan. Generic Complex of Flora. Volume 2 Gylym; Almaty, Kazakhstan: 2001. [Google Scholar]
- 19.Sviridova T.P., Zinner N.S. Prospects of cultivation Hedysarum alpinum L. and H. theinum Krasnob. in conditions of Tomsk region. Tomsk State Univ. J. Biol. 2008;2:5–11. [Google Scholar]
- 20.Erst A.A., Zheleznichenko T.V., Novikova T.I., Dorogina O.V., Banaev E.V. Ecological and geographic variability of Hedysarum theinum and features of its propagation in vitro. Contemp. Probl. Ecol. 2014;7:67–71. doi: 10.1134/S1995425514010053. [DOI] [Google Scholar]
- 21.Arslan E., Ertugrul K., Tugay O., Dural H. Karyological studies of the genus Onobrychis Mill. and the related genera Hedysarum L. and Sartoria Boiss. and Heldr. (Fabaceae, Hedysareae) from Turkey. Caryologia. 2012;65:11–17. doi: 10.1080/00087114.2012.678079. [DOI] [Google Scholar]
- 22.Zvyagina N.S., Dorogina O.V., Catalan P. Genetic relatedness and taxonomy in closely related species of Hedysarum (Fabaceae) Biochem. Syst. Ecol. 2016;69:176–187. doi: 10.1016/j.bse.2016.10.001. [DOI] [Google Scholar]
- 23.Kumar P., Rana P.K., Singhal V.K., Singh H., Kholia B.S. Chromosome count, meiotic abnormalities and pollen sterility in Lahaul sweetvetch (Hedysarum astragaloides Benth. ex Baker, Fabaceae), an endemic and threatened species from India. Acta Bot. Croat. 2018;77:203–208. doi: 10.2478/botcro-2018-0020. [DOI] [Google Scholar]
- 24.Issolah R., Benhizia H., Khalfallah N. Karyotype variation within some natural populations of Sulla (Hedysarum coronarium L., Fabaceae) in Algeria. Genet. Resour. Crop Evol. 2006;53:1653. doi: 10.1007/s10722-005-5287-5. [DOI] [Google Scholar]
- 25.Tropicos.org Missouri Botanical Garden. IPCN Chromosome Reports. [(accessed on 21 November 2020)]; Available online: http://www.tropicos.org/Name/40022354.
- 26.Benhizia H., Benhizia Y., Ghernoub L. Meiotic behaviour and karyotype features of endangered endemic fodder species Hedysarum perrauderianum (Fabaceae) in some populations from Algeria. Caryologia. 2013;66:195–204. doi: 10.1080/00087114.2013.821838. [DOI] [Google Scholar]
- 27.Benhizia H., Benhizia Y., Djeghar R., Siljak-Yakovlev S., Pustahija F., Khalfallah N. Cytogenetic characterization, nuclear genome size, and pollen morphology of some Hedysarum L. taxa (Fabaceae) from Algeria, with emphasis on the origin of H. perrauderianum Coss. & Durieu. Genet. Resour. Crop Evol. 2020 doi: 10.1007/s10722-020-01016-0. [DOI] [Google Scholar]
- 28.Marghali S., Panaud O., Lamy F., Ghariani S., Sarr A., Marrakchi M., Trifi-Farah N. Exploration of intra- and inter-population genetic diversity in Hedysarum coronarium L. Genet. Resour. Crop Evol. 2005;52:277–284. doi: 10.1007/s10722-005-5459-3. [DOI] [Google Scholar]
- 29.Bushman B.S., Larson S.R., Peel M., Pfrender M.E. Population structure and genetic diversity in North American Hedysarum boreale Nutt. Crop Sci. 2007;47:1281–1288. doi: 10.2135/cropsci2006.11.0702. [DOI] [Google Scholar]
- 30.Zvyagina N.S., Dorogina O.V. Genetic differentiation of Altai-Sayan endemic Hedysarum theinum Krasnob (Fabaceae) by inter-simple sequence repeat analysis. Russ. J. Genet. 2013;49:1030–1035. doi: 10.1134/S102279541310013X. [DOI] [PubMed] [Google Scholar]
- 31.Agafonova M., Agafonova O. Polymorphism of seed polypeptides in closely related species Hedysarum theinum Krasnob. and H. neglectum Ledeb. (Fabaceae) Turczaninowia. 2002;5:72–78. [Google Scholar]
- 32.Dorogina O.V., Agafonova M.A. Identification of closely related species Hedysarum theinum, H. neglectum and H. austrosibiricum (Fabaceae) by seed storage globulins. Bot. Zh. 2004;89:1637–1645. [Google Scholar]
- 33.Duan L., Wen J., Yang X., Liu P.L., Arslan E., Ertuğrul K., Chang Z.Y. Phylogeny of Hedysarum and tribe Hedysareae (Leguminosae: Papilionoideae) inferred from sequence data of ITS, matK, trnL-F and psbA-trnH. Taxon. 2015;64:49–64. doi: 10.12705/641.26. [DOI] [Google Scholar]
- 34.Liu P.L., Wen J., Duan L., Arslan E., Ertuğrul K., Chang Z.Y. Hedysarum L. (Fabaceae: Hedysareae) is not monophyletic—Evidence from phylogenetic analyses based on five nuclear and five plastid sequences. PLoS ONE. 2017;12:e0170596. doi: 10.1371/journal.pone.0170596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nafisi H., Kazempour-Osaloo S., Mozaffarian V., Schneeweiss G.M. Molecular phylogeny and divergence times of the genus Hedysarum (Fabaceae) with special reference to section Multicaulia in Southwest Asia. Plant Syst. Evol. 2019;305:1001–1017. doi: 10.1007/s00606-019-01620-3. [DOI] [Google Scholar]
- 36.Kurbatsky V.I. Genus Hedysarum L. In: Polozhii A., editor. Flora Sibiri (Flora of Siberia), Fabaceae (Leguminosae) Volume 9. Nauka; Novosibirsk, Russia: 1994. pp. 153–166. [Google Scholar]
- 37.Malyshev L.I. Manual of High Mountain Plants of the Southern Siberia. Nauka; Saint Petersburg, Russia: 1968. [Google Scholar]
- 38.Kharkevich S.S. Vascular Plants of the Soviet Far East. V.4. Nauka; Saint Petersburg, Russia: 1989. [Google Scholar]
- 39.Litvinskaya S.A., Murtazaliyev R.A. Flora of the North Caucasus: Atlas Continuant. Fiton XXI; Moscow, Russia: 2013. [Google Scholar]
- 40.Imachuyeva D.R., Serebryanaya F.K. The current state of study of plants of Hedysarum L. florae of the Caucasus. Pharm. Pharmacol. 2016;4:4–32. doi: 10.19163/2307-9266-2016-4-6-4-32. [DOI] [Google Scholar]
- 41.Kurbatsky V.I., Malahova L.A. Chromosome numbers for some species of Hedysarum L. from the territory of Siberia. Tomsk State Univ. 1992;89:3–5. [Google Scholar]
- 42.Kulikov P.V. Floristic findings in the Southern Ural (Chelyabinsk region) Bot. Zh. 1998;83:137–145. [Google Scholar]
- 43.Knyasev M.S. A new hybrid species of the genus Hedysarum (Fabaceae) from the East Europe. Bot. Zh. 2011;96:1122–1126. [Google Scholar]
- 44.Krasnoborov I.M., Azovtsev G.R., Orlov V.P. A new species of the genus Hedysarum L. (Fabaceae L.) from southern Siberia. Bot. Zh. 1985;70:968–973. [Google Scholar]
- 45.Tutin T.G., Heywood V.H., Burges N.A., Moore D.M., Valentine D.H., Walters S.M., Webb D.A. Flora Europaea, Leguminosae. Vol. 2 Cambridge University Press; Cambridge, UK: 1968. [Google Scholar]
- 46.Kobayashi T. A new role of the rDNA and nucleolus in the nucleus—rDNA instability maintains genome integrity. Bioessays. 2008;30:267–272. doi: 10.1002/bies.20723. [DOI] [PubMed] [Google Scholar]
- 47.Schubert I., Wobus U. In situ hybridization confirms jumping nucleolus organizing regions in Allium. Chromosoma. 1985;92:143–148. doi: 10.1007/BF00328466. [DOI] [Google Scholar]
- 48.Raskina O., Belyayev A., Nevo E. Quantum speciation in Aegilops: Molecular cytogenetic evidence from rDNA cluster variability in natural populations. Proc. Natl. Acad. Sci. USA. 2004;101:14818–14823. doi: 10.1073/pnas.0405817101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roa F., Guerra M. Distribution of 45S rDNA sites in chromosomes of plants: Structural and evolutionary implications. BMC Evol. Biol. 2012;12:225. doi: 10.1186/1471-2148-12-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rosato M., Álvarez I., Feliner G.N., Rosselló J.A. High and uneven levels of 45S rDNA site-number variation across wild populations of a diploid plant genus (Anacyclus, Asteraceae) PLoS ONE. 2017;12:e0187131. doi: 10.1371/journal.pone.0187131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pedrosa-Harand A., de Almeida C.C., Mosiolek M., Blair M.W., Schweizer D., Guerra M. Extensive ribosomal DNA amplification during Andean common bean (Phaseolus vulgaris L.) evolution. Theor. Appl. Genet. 2006;112:924–933. doi: 10.1007/s00122-005-0196-8. [DOI] [PubMed] [Google Scholar]
- 52.Scaldaferro M.A., da Cruz M.V., Cecchini N.M., Moscone E.A. FISH and AgNor mapping of the 45S and 5S rRNA genes in wild and cultivated species of Capsicum (Solananceae) Genome. 2016;59:95–113. doi: 10.1139/gen-2015-0099. [DOI] [PubMed] [Google Scholar]
- 53.Amosova A.V., Samatadze T.E., Mozgova G.V., Kipen V.N., Dubovskaya A.G., Artemyeva Y.O.Y., Zoshchuk S.A., Lemesh V.A., Muravenko O.V. Genomic markers associated with cold-hardiness in Brassica rapa L. Mol. Biol. 2020;54:541–552. doi: 10.1134/S0026893320040032. [DOI] [PubMed] [Google Scholar]
- 54.Gerlach W.L., Bedbrook J.R. Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Res. 1979;7:1869–1885. doi: 10.1093/nar/7.7.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gerlach W.L., Dyer T.A. Sequence organization of the repeating units in the nucleus of wheat which contain5S rRNA genes. Nucleic Acids Res. 1980;8:4851–4855. doi: 10.1093/nar/8.21.4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yurkevich O.Y., Samatadze T.E., Levinskikh M.A., Zoshchuk S.A., Signalova O.B., Surzhikov S.A., Sychev V.N., Amosova A.V., Muravenko O.V. Molecular cytogenetics of Pisum sativum L. grown under spaceflight-related stress. BioMed Res. Int. 2018;2018:1–10. doi: 10.1155/2018/4549294. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are contained within the article.