Abstract
Candida auris is an emerging multi-drug resistant pathogen with high mortality rate; nosocomial infections have been reported worldwide, causing a major challenge for clinicians and microbiological laboratories. The study aims to describe new cases of C. auris and detect drug resistance-associated mutations of C. auris by the sequencing of ERG11 and FKS1 genes. A total of six specimens were collected from blood, urine, ear swab, and groin screening samples. Isolates were incubated for 48 h on Sabouraud Dextrose agar (SDA) at 42 °C, then confirmed by MALDI-TOF MS. Furthermore, antifungal susceptibility testing was performed using the Vitek 2 system to detect Minimum Inhibitory Concentrations (MICs) of six antifungals. Sequences of 18S rRNA gene and ITS regions from isolates and phylogenetic analysis were performed. Gene sequencing was analysed to detect drug resistance-associated mutations by FKS1 and ERG11 genes sequencing. All C. auris isolates were confirmed by MALDI-TOF MS, and evolutionary analyses using sequences of 18S rRNA gene and ITS region. Antifungal susceptibility testing showed that all isolates were resistant to fluconazole. Sequencing of ERG11 and FKS1 genes from the isolates revealed the presence of two (F132Y and K143R) drug resistance-associated mutations in ERG11, however, FKS1 gene was devoid of mutations. The study sheds light on a public health threat of an emerging pathogen, and the hospital implemented strict contact screening and infection control precautions to prevent C. auris infection. Finally, there is a critical need to monitor the antifungal resistance in different geographical areas and implementation of efficient guidelines for treatment.
Keywords: Candida auris, nosocomial infections, emerging pathogen, resistance genes, Saudi Arabia
1. Introduction
Candida auris is an emerging pathogen that had been reported in the past decade as a rising threat and a challenging nosocomial infection [1]. C. auris tends to transmit rapidly from person to person, and persist on medical devices and on the surfaces of hospital areas [2,3].
It was first described in Japan from a culture of external ear canal in 2009, and within the last decade, C. auris was frequently isolated from the bloodstream, urinary, and respiratory tract. It has been reported from several countries in Africa, Asia, Europe, America, and the Middle East, with significant fatality rate [4].
The detection of C. auris is still a challenging dilemma for clinical laboratories because it is closely related to other Candida species such as, C. haemulonii, C. duobushaemulonii, and C. lusitaniae [5,6]. C. auris grows well on Sabouraud and chromogenic agar at 37 °C and 42 °C, and there are different efficient methods to identify C. auris, such as molecular techniques and MALDI-TOF MS [7,8].
Azoles, echinocandins, and amphotericin B are the three main classes of antifungal agents, the increasing fluconazole resistance could be explained by mutations in the ERG11 gene encoding lanosterol 14-alpha-demethylase, which has an essential role in the ergosterol synthesis pathway [9]. In contrast, echinocandin resistance remains quite low in most Candida species except Candida glabrata, and it is linked to mutations of FKS, which is the gene encoding the catalytic subunits of the enzyme β-1,3-D-glucan synthase that target the drug [10]. Antifungal sensitivity of C. auris can be evaluated by using either microdilution or disk diffusion test. However, identification of ERG11 and FKS1 genes mutations would be an advanced screening method to detect potentially resistant strains [11].
The first case from Saudi Arabia were reported in 2018, followed by other cases of C. auris infection from different cities [12,13,14,15,16]. The aim of this study is to describe new cases of C. auris infection and to detect antifungal resistance genes, namely, ERG11 and FKS1, in isolates of this global emerging pathogen.
2. Materials and Methods
2.1. Samples Collection and Microbial Identification
A total of six specimens were collected from five patients who were admitted to a tertiary care hospital in Al Khobar, Saudi Arabia. Specimens were collected from sterile and non-sterile sites including blood, urine, ear swab, and groin screening samples during the period from November 2018 to April 2019. Clinical microbiology testing was performed according to standard operative procedures in the hospital using different culture media. All clinical isolates were incubated for 48 h on Sabouraud Dextrose agar at 42 °C, then confirmed by MALDI-TOF MS, and the identification procedure was performed according to the manufacturer’s protocol and guidelines for yeast identification [17].
2.2. Antifungal Susceptibility Testing
In vitro AFST was performed using the Vitek 2 system (AST-YS08 card: bioMérieux, Hazelwood, MO, USA). Minimum Inhibitory Concentrations (MIC) of 6 antifungals were tested as per the manufacturer’s instructions; amphotericin B, fluconazole, voriconazole, caspofungin, micafungin, and Flucytosine. In each test, two reference strains, Candida parapsilosis ATCC 22019 and Candida krusei ATCC 6258, were included as control strains. The interpretation of the results was based on the following MIC breakpoints for C. auris published by the CDC: Amphotericin B, 2 µg/mL; fluconazole, 32 µg/mL; voriconazole, not available; caspofungin, 2 µg/mL; and micafungin, 4 µg/mL; and Flucytosine ≤ 1 μg/mL.
2.3. DNA Extraction and Sequencing of 18S rRNA Gene
Total genomic DNA from isolates, CA1, CA3, CA4, CA5, CA7, and CA8 were extracted as per the manufacturer’s instruction using Qiagen’s Yeast/Bact. Kit (Gentra Puregene Yeast/Bact. Kit, Qiagen, Hilden, Germany). The 18S rRNA gene (1655 bp) of all the strains was amplified using 18SrRNAF (5′-GCTTAATTTGACTCAACACGGGA-3′) and 18SrRNAR (5′-AGCTATCAATCTGTCAATCCTGTC-3′) primers, (MoleQule-On, Auckland, New Zealand) at annealing temperature at 61.8 °C using absolute master mix (MoleQule-On, Auckland, New Zealand) in T-Professional thermocycler (Biometra, Göttingen, Germany) for 35 cycles.
The PCR amplicons were purified using QIAquick PCR Purification Kit (Qiagen, Germany) after visualized the product using 2% agarose gel. The purified amplicons were sequenced using 3500 genetic analysers (Applied Biosystems, Forster City, CA, USA) with the forward and reverse primers used for the amplification using Big Dye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Forster City, CA, USA).
2.4. Molecular Identification
The 18S rRNA gene sequence from isolates CA1, CA3, CA4, CA5, CA7, and CA8 were aligned and analysed using Basic Local Alignment Search Tool, PHYMYCO-DB, and FungiDB [18,19]. An evolutionary relationship of samples was constructed using the Maximum Likelihood method based on the Tamura-Nei model in MEGA7 software package with bootstrap consensus tree from 500 replicates [20,21,22].
2.5. Sequencing of ITS Region
Primers were designed for the amplification of the internal transcribed spacer 1 (ITS1), 5.8S ribosomal RNA, and internal transcribed spacer 2 (ITS2); forward primer: ITSbF 5′-AGGAATTCCTAGTAAGCGCAAGT-3′ and reverse primer: ITSbR 5′-ATTTACCACCCACTTAGAGCT-3′ (Integrated DNA technologies, Coralville, IA, United States). ITS region was amplified at 57.5 °C annealing temperature using absolute master mix (MoleQule-On, Auckland, New Zealand) in T-Professional thermocycler (Biometra, Göttingen, Germany) for 35 cycles. Amplicons were visualized using 2% agarose gel and documented. The amplified ITS region amplicons (840 bp) were purified and sequenced using 3500 genetic analysers (Applied Biosystems, Forster City, CA, USA) with the forward primers (ITSbF) using Big Dye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Forster City, CA, USA). The sequences were analysed using BLASTn and UNITE advanced analyses.
2.6. Sequencing of Resistance-Associated Mutations
Primers for the FKS1 and ERG11 genes were designed and used for the study. FKS1 gene was amplified using forward (FKS1aF 5′ATGTCTTACGATAACAATCACAACTAC-3′), and reverse primers (FKS1aR 5′-AGTAAGATTCGGCCAACTTAGCAG-3′) (MoleQule-On, Auckland, New Zealand) with the 1900 bp amplicon. ERG11 gene was amplified using the forward (ERG11aF 5′- ATGGCCTTGAAGGACTGCATCGT-3′) and reverse primers (ERG11aR 5′-TTAGTAAACACAAGTCTCTCTTTTCTCCCA-3′) (MoleQule-On, Auckland, New Zealand) with the 1575 bp amplicon. FKS1 and ERG11 genes were amplified at 59.4 °C and 62.2 °C annealing temperature, respectively, using absolute master mix (MoleQule-On, New Zealand) in T-Professional thermocycler (Biometra, Göttingen, Germany) for 35 cycles. Amplicons were visualized using 2% agarose gel and documented. The amplified FKS1 and ERG11 gene amplicons were purified and sequenced using 3500 genetic analysers (Applied Biosystems, Forster City, CA, USA) with the forward and reverse primers (FKS1aF & FKS1aR for FKS1; ERG11aF and ERG11aR for ERG11) separately using Big Dye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Forster City, CA, USA). The sequences were analysed using Mutation Surveyor with GenBank: KY410388.1 and GenBank: NW_021640162.1 as reference sequences for ERG11 and FKS1, respectively.
3. Results
3.1. Culture Identification and Antifungal Susceptibility Testing
All specimens were collected from Saudi male patients with diabetic and different comorbidity diseases. Clinical samples were isolated on culture media, then confirmed by MALDI-TOF MS analysis.
Patients presented with signs of infection including hematuria, sepsis, decompensated heart failure secondary to hospital acquired pneumonia, and ear discharge. Meropenem was prescribed before and after isolation of C. auris in the first two patients in Table 1. The fourth patient received tazocin, cefazoline, and ceftazidime before and after isolation. Other antimicrobial agents including tri-sulfa, and levofloxacin were administrated before isolation C auris. Then, after isolation, patients received colistin, and tigecycline. Antifungal drugs such as: Amphotericin B, caspofungin, and anidulafungin were prescribed for patients with invasive infections. With respect to the prognosis, patients were critically ill and have already passed away due to serious co-morbidity.
Table 1.
MICs (Minimum Inhibitory Concentrations) of tested antifungal agents for six Candida auris isolated from five patients with comorbidity diseases.
| Antifungal Agent | Isolates | |||||
|---|---|---|---|---|---|---|
| CA1 | CA3 | CA4 | CA5 | CA7 | CA8 | |
| Sex | Male | Male | Male | Male | Male | |
| Age | 62 | 85 | 26 | 77 | 68 | |
| Medical condition | DM, HTN, CAD, right MCA stroke | DM, HTN, COPD, CVA | Brain stem haemorrhage | DM, HTN, chronic anaemia, ischaemic stroke, subarachnoid haemorrhage | DM, HTN, post cardiac valve replacement | |
| Specimen | Urine | Urine | Groin | Blood | Urine | Ear swab |
| Amphotericin B | 0.5 | 8 | 0.5 | 8 | 8 | 0.5 |
| Fluconazole | 16 | 32 | 32 | 32 | 32 | 8 |
| Voriconazole | 0.5 | 0.5 | 0.5 | R–1 | 1 | ≥8 |
| Caspofungin | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 |
| Micafungin | ≤0.06 | ≤0.06 | ≤0.06 | ≤0.06 | ≤0.06 | ≤0.06 |
| Flucytosine | ≤1 | S–≤ 1 | S–≤ 1 | S–≤ 1 | S–≤ 1 | S–≤ 1 |
DM: Diabetes, HTN: Hypertension, CAD: Chronic artery disease, MCA: Middle cerebral artery, COPD: Chronic obstructive pulmonary disease, CVA: Cerebrovascular stroke.
Interpretation of antifungal susceptibility results showed that C. auris isolates were resistant to fluconazole with MICs ranged from 8 to 32 μg/mL. However, voriconazole with MIC, 0.5 μg/mL showed 3 (50%) resistant isolates, and the others (50%) were intermediate. Furthermore, 50% of C. auris isolates were resistant to Amphotericin B with MICs ranged from 8 to 32 μg/mL. Finally, caspofungin, micafungin, and flucytosine showed excellent activity with the tested isolates. Unfortunately, three patients died almost one month after detection of C. auris (Table 1).
3.2. ITS and 18S rRNA Sequencing
The sequences of internal transcribed spacer 1 (ITS1), 5.8S ribosomal RNA, and internal transcribed spacer 2 (ITS2) from all the samples were analysed using BLASTn and UNITE advanced analyses, and the results confirmed that all the strains are C. auris, as identified through 18S rRNA gene. Sequences of ITS region were submitted to GenBank, accession number: CA1: MW039128.1; CA3: MW039130.1; CA4: MW039131.1; CA5: MW039132.1; CA7: MW039134.1; and CA8: MW039135.1.
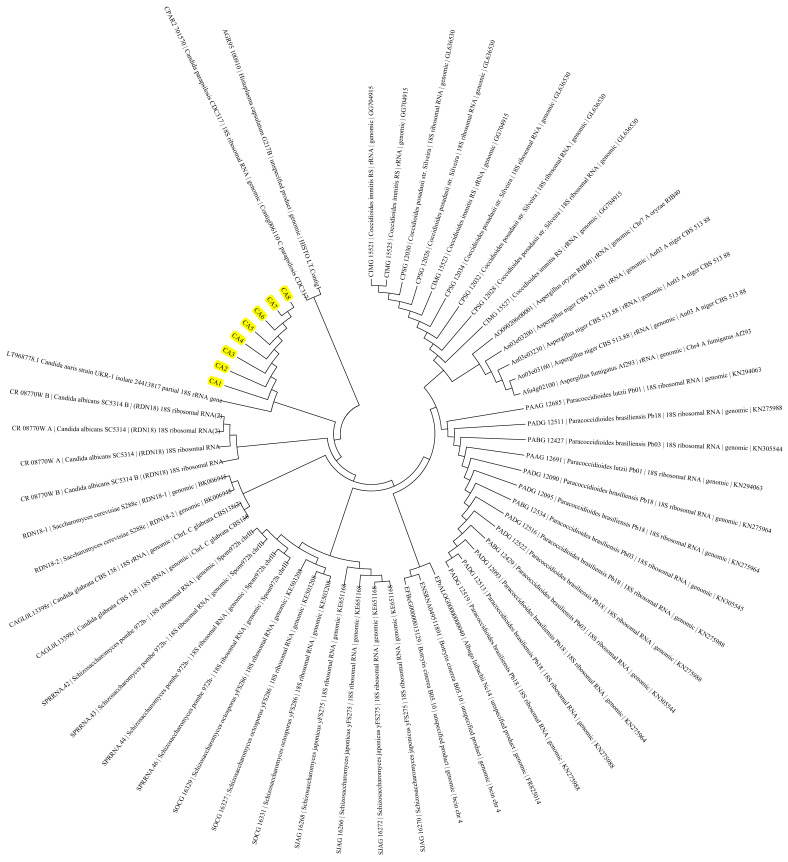
The evolutionary history was inferred by using the Maximum Likelihood method based on the Tamura-Nei model [22]. The bootstrap consensus tree inferred from 500 replicates is taken to represent the evolutionary history of the taxa analysed [20]. Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach, and then selecting the topology with superior log likelihood value. The analysis involved 58 nucleotide sequences. Codon positions included were 1st + 2nd + 3rd + Noncoding. All positions containing gaps and missing data were eliminated. Evolutionary analyses were conducted in MEGA7 [21]. There were an overall of 333 positions in the final dataset, and the tree with the highest log likelihood (−1643.31) was demonstrated (Figure 1). Evolutionary analysis of the standard sequences of C. auris and the six isolates indicated no genetic divergence among the six isolates on 18S rRNA gene and ITS region. However, whole genome comparisons are needed to confirm complete sequence divergences.
Figure 1.
Molecular Phylogenetic analysis of the 6 isolates (CA1, CA3, CA4, CA5, CA7, and CA8) of Candida auris by Maximum Likelihood method. Six isolates of Candida auris and internal positive controls are highlighted yellow.
3.3. Resistance Genes Mutations
All samples were successfully amplified for both ERG11 (1575 bp) and FKS1 (1900 bp) genes. Sequences of 18S rRNA gene were submitted to GenBank, accession number: CA1: MN658527.1; CA3: MN658529.1; CA4: MN658530.1; CA5: MN658531.1; CA7: MN658533.1; and CA8: MN658534.1.
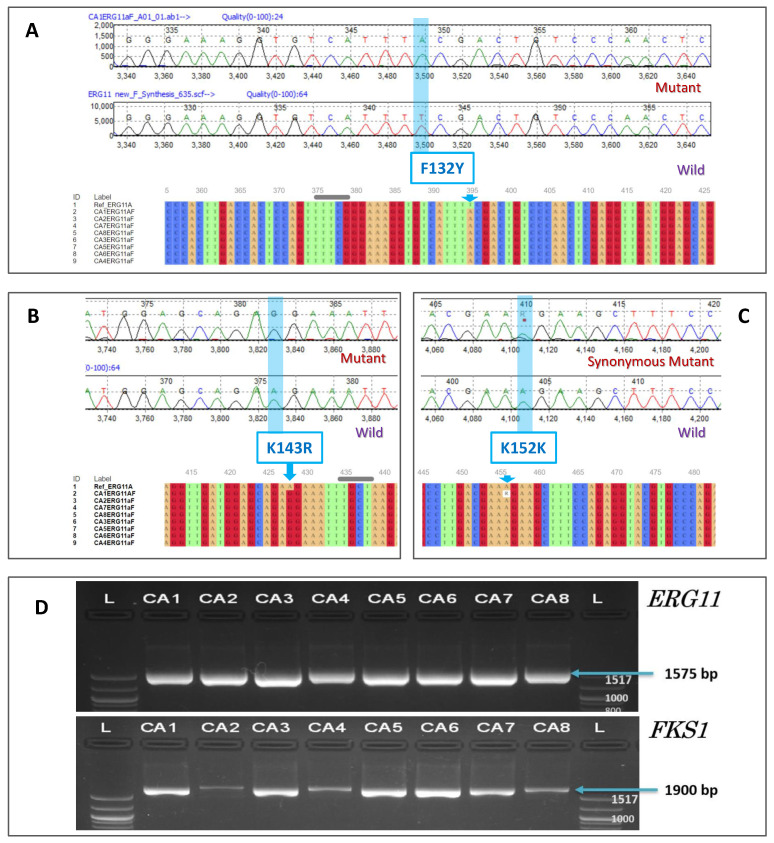
The amplicons were sequenced, and the sequences were analysed for the identification of the drug resistance-associated mutations in the ERG11 and FKS1 genes from the 6 isolates (CA1, CA3, CA4, CA5, CA7, and CA8) of Candida auris. The analysis revealed the presence of two (F132Y and K143R) drug resistance-associated mutations in ERG11 gene. Furthermore, a synonymous heterozygous K152K mutation was observed in a strain. However, no mutations were detected in the FKS1 genes (Table 2; Figure 2).
Table 2.
List of resistance-associated mutations in ERG11 and FKS1 genes in the C. auris isolates.
| Gene | Mutation | CA1 | CA3 | CA4 | CA5 | CA7 | CA8 |
|---|---|---|---|---|---|---|---|
| ERG11 | F132Y | + | + | + | + | + | + |
| ERG11 | K143R | + | + | + | + | + | + |
| ERG11 | K152K | + | - | - | - | - | - |
| FKS1 | - | - | - | - | - | - | - |
Reference sequence used for ERG11 and FKS1 are GenBank: KY410388.1 and GenBank: NW_021640162.1, respectively. -: No substitutions of nucleotides observed. +: Substitutions of nucleotides observed.
Figure 2.
Resistance associated mutations in ERG11 gene. Reference sequence used for analysis: GenBank: KY410388.1. (A): Electropherogram of F132Y mutant. Bottom: Multiple sequence alignment of region with T > A. (B): Electropherogram of K143R mutant. Bottom: Multiple sequence alignment of region with A > G. (C): Electropherogram of K152K mutant. Bottom: Multiple sequence alignment of region with A > G (AAA > AAG). (D): The ERG11 (Lanosterol 14-alpha demethylase) and FKS1 (1,3-beta-glucan synthase component) gene PCR amplification from Candida auris. L: 100 bp ladder; CA1, CA3, CA4, CA5, CA7, and CA8: Isolates of Candida auris. CA2 (GenBank: MK910118.1) and CA6 (GenBank: MK910117.1): Internal positive controls of Candida auris.
4. Discussion
Candida auris is the first fungal considered as a public health threat, as it can spread easily among patients in hospitals and may cause serious diseases [23]. High risk factors associated with C. auris infections include using broad spectrum antibiotics, catheters, ICU admission [6,24], and diabetic patients, and most infections arise two to seven weeks after admission. C. auris infection is correlated with higher resistance to fluconazole and moderately low resistance to echinocandins, therefore echinocandin is recommended as an empirical therapy before antifungal susceptibility testing of collected strains [4].
According to a study that reported the first hospital outbreak of C. auris in a European hospital, most of clinical manifestations of C. auris included colonization followed by candidemia, wound infections, urinary catheter infections, and others [25].
Guidelines of the CDC and recent publications stated that invasive infections may develop after colonization, and prophylactic antifungal treatment should be considered in case of a patient colonized with C. auris subsequently deteriorates [26,27,28]. In the present study, these six isolates were the only cases during the time period, 90% of the patients were diabetics and using broad spectrum antibiotics, with an age range from 62 to 85, except one patient, who was 26 years old. Caspofungin was prescribed and the hospital instituted strict contact screening and infection control precautions according to CDC guidelines [4,29].
In Saudi Arabia, the first two cases of C. auris were detected one to three months after the patients’ admission to hospitals, and echinocandins had been used for successful treatment [12,13]. Four more cases were reported, where 90% of the isolates were misidentified as C. haemulonii, and in the same year, one case was confirmed via MALDI-TOF MS [14,15]. Recently, a study reported a high mortality rate of seven patients with C. auris infection [16].
Present antifungal susceptibility results concur with a study in Kuwait, which reported multidrug resistance and high mortality rate among cases of C. auris infections [30]. Additionally, two studies in Oman reported of C. auris occurrence at different hospitals, isolates were highly resistant to fluconazole and the onset of infection after hospitalization ranged from one to two months [31,32].
Sequencing results showed that drug resistance-associated mutations F132Y and K143R were identified in all isolates. These findings concur with a study in 2018, which concluded that F132Y and K143R mutations could be initial markers for azole resistance in South Asian and South American clade [33].
A multi-centre study in India showed that 41% of isolates were resistant to two classes of antifungal agents and numerous ERG11 mutations have been detected in C. auris isolates from several geographic areas [34]. MALDI-TOF MS and gene sequencing are advanced and expensive techniques. Thus, there is a critical necessity to develop cost-effective techniques for the detection of this emerging pathogen in underdeveloped countries [35]. Rapid detection of ERG11 and FKS1 has the potential to overcome the deficiencies of existing Minimum Inhibitory Concentration (MIC) to detect azole and echinocandin resistant C. auris [11]. Mutations of ERG11 (F132Y and K143R) in C. auris were associated with to increased resistance to fluconazole [36].
The limitations of this study include the identification of the cause of death precisely, as those patients had comorbidity diseases, and only one isolate was obtained from a sterile site (bloodstream). Isolation of C. auris from groin and ear canal may merely reflect colonization rather than infection.
Three salient features of this study, descripting new cases of C. auris from different sites confirmed by MALDI TOF MS, ribosomal, ITS sequencing, and developing the phylogenetic analysis of the isolates, and sequence of ERG11 and FKS1 genes among C. auris isolates clearly confer drug resistant characteristics of the isolates.
Further studies are crucial to analyse Whole Genome Sequence (WGS) of C. auris isolates to discover the mechanism of resistance and potential effective treatment for this emerging pathogen.
Acknowledgments
The authors thank the Dean, Institute for Research and Medical Consultations (IRMC), Imam Abdulrahman Bin Faisal University, Dammam, Saudi Arabia for her continuous support encouragement. The authors thank Ranilo. M. Tumbaga, and Horace T. Pacifico for their support and assistance.
Abbreviations
The following abbreviations are used in this manuscript: C. auris: Candida auris; MALDI TOF MS: Matrix-Assisted Laser Desorption/Ionization-Time of Flight mass spectrometry; PCR: Polymerase Chain Reaction; IPP: Infection Prevention Program; CDC: Centers for Disease Control and prevention; ITS: Internal transcribed spacer; WGS: Whole Genome Sequencing.
Author Contributions
Conceptualization, R.A. and D.M.A.; data curation, N.M. and M.A.; formal analysis, R.A. and M.A.; investigation, N.M. and M.A.; methodology, R.A., D.M.A., N.M., B.A., and S.A.; resources, R.A., N.M., M.A., S.A., and J.F.B.; software, B.A., S.A., and J.F.B.; validation, B.A. and S.A.; writing—original draft, D.M.A. and J.F.B.; writing—review & editing, D.M.A., B.A., and J.F.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the department of Microbiology of the college of Medicine (Number 5 on 16/4/2020), Institutional Review Board of Imam Abdulrahman Bin Faisal University (IRB-2020-547-Med).
Informed Consent Statement
The clinical specimens including blood were obtained from hospitalized patients after obtaining verbal consent only as part of routine patient care and diagnostic work-up for the isolation and antifungal susceptibility testing of fungal pathogens. All patient’s personal identifiers were kept confidential and the results are reported in this manuscript on deidentified samples without revealing patient identity.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to privacy.
Conflicts of Interest
The authors declare no conflict of interests.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kordalewska M., Perlin D.S. Identification of drug resistant candida auris. Front. Microbiol. 2019;10 doi: 10.3389/fmicb.2019.01918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eyre D.W., Sheppard A.E., Madder H., Moir I., Moroney R., Quan T.P., Griffiths D., George S., Butcher L., Morgan M., et al. A candida auris outbreak and its control in an intensive care setting. N. Engl. J. Med. 2018;379:1322–1331. doi: 10.1056/NEJMoa1714373. [DOI] [PubMed] [Google Scholar]
- 3.Vallabhaneni S., Kallen A., Tsay S., Chow N., Welsh R., Kerins J., Kemble S.K., Pacilli M., Black S.R., Landon E., et al. Investigation of the First Seven Reported Cases of Candida auris, a Globally Emerging Invasive, Multidrug-Resistant Fungus-United States, May 2013-August 2016. Am. J. Transplant. 2017;17:296–299. doi: 10.1111/ajt.14121. [DOI] [PubMed] [Google Scholar]
- 4.Osei Sekyere J. Candida auris: A systematic review and meta-analysis of current updates on an emerging multidrug-resistant pathogen. Microbiologyopen. 2018;7 doi: 10.1002/mbo3.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Girard V., Mailler S., Chetry M., Vidal C., Durand G., van Belkum A., Colombo A.L., Hagen F., Meis J.F., Chowdhary A. Identification and typing of the emerging pathogen Candida auris by matrix-assisted laser desorption ionisation time of flight mass spectrometry. Mycoses. 2016;59:535–538. doi: 10.1111/myc.12519. [DOI] [PubMed] [Google Scholar]
- 6.Chowdhary A., Sharma C., Meis J.F. Candida auris: A rapidly emerging cause of hospital-acquired multidrug-resistant fungal infections globally. PLoS Pathog. 2017;13:1–10. doi: 10.1371/journal.ppat.1006290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahmad A., Spencer J.E., Lockhart S.R., Singleton S., Petway D.J., Bagarozzi D.A., Herzegh O.T. A high-throughput and rapid method for accurate identification of emerging multidrug-resistant Candida auris. Mycoses. 2019;62:513–518. doi: 10.1111/myc.12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rossato L., Colombo A.L. Candida auris: What have we learned about its mechanisms of pathogenicity? Front. Microbiol. 2018;9:1–6. doi: 10.3389/fmicb.2018.03081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Cássia Orlandi Sardi J., Silva D.R., Soares Mendes-Giannini M.J., Rosalen P.L. Candida auris: Epidemiology, risk factors, virulence, resistance, and therapeutic options. Microb. Pathog. 2018;125:116–121. doi: 10.1016/j.micpath.2018.09.014. [DOI] [PubMed] [Google Scholar]
- 10.Perlin D.S. Echinocandin Resistance in Candida. Clin. Infect. Dis. 2015;61:S612–S617. doi: 10.1093/cid/civ791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hou X., Lee A., Jiménez-Ortigosa C., Kordalewska M., Perlin D.S., Zhao Y. Rapid Detection of ERG11-Associated Azole Resistance and FKS-Associated Echinocandin Resistance in Candida auris. Antimicrob. Agents Chemother. 2019;63:e01811-18. doi: 10.1128/AAC.01811-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abdalhamid B., Almaghrabi R., Althawadi S., Omrani A. First report of Candida auris infections from Saudi Arabia. J. Infect. Public Health. 2018;11:598–599. doi: 10.1016/j.jiph.2018.05.010. [DOI] [PubMed] [Google Scholar]
- 13.Elsawy A., Alquthami K., Alkhutani N., Marwan D., Abbas A. The second confirmed case of Candida auris from Saudi Arabia. J. Infect. Public Health. 2019;12:907–908. doi: 10.1016/j.jiph.2019.07.011. [DOI] [PubMed] [Google Scholar]
- 14.Abanamy H., Alsharif T., Solomon R., AlAlwan B., Al-Johani S. Reporting 4 Candida auris in National Guard Hospital, Riyadh/ Saudi Arabia- ClinicalKey. J. Infect. Public Health. 2019;12:139–140. doi: 10.1016/j.jiph.2018.10.107. [DOI] [Google Scholar]
- 15.Hudhaiah D., Elhadi N. Prevalence and Genotypes of Nosocomial Clostridium difficile Infections in the Eastern Province of the Kingdom of Saudi Arabia: A Multi-Centre Prospective Study. J. Clin. Diagn. Res. 2019;13:16–20. doi: 10.7860/JCDR/2019/38321.12694. [DOI] [Google Scholar]
- 16.Almaghrabi R.S., Albalawi R., Mutabagani M., Atienza E., Aljumaah S., Gade L., Forsberg K., Litvintseva A., Althawadi S. Molecular characterisation and clinical outcomes of Candida auris infection: Single-centre experience in Saudi Arabia. Mycoses. 2020;63:452–460. doi: 10.1111/myc.13065. [DOI] [PubMed] [Google Scholar]
- 17.Duyvejonck H., Cools P., Decruyenaere J., Roelens K., Noens L., Vermeulen S., Claeys G., Decat E., Van Mechelen E., Vaneechoutte M. Validation of High Resolution Melting Analysis (HRM) of the amplified ITS2 region for the detection and identification of yeasts from clinical samples: Comparison with culture and MALDI-TOF based identification. PLoS ONE. 2015;10:1–17. doi: 10.1371/journal.pone.0132149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahé S., Duhamel M., Le Calvez T., Guillot L., Sarbu L., Bretaudeau A., Collin O., Dufresne A., Kiers E.T., Vandenkoornhuyse P. PHYMYCO-DB: A Curated Database for Analyses of Fungal Diversity and Evolution. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0043117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Basenko E.Y., Pulman J.A., Shanmugasundram A., Harb O.S., Crouch K., Starns D., Warrenfeltz S., Aurrecoechea C., Stoeckert C.J., Kissinger J.C., et al. FungiDB: An integrated bioinformatic resource for fungi and oomycetes. J. Fungi. 2018;4:39. doi: 10.3390/jof4010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Felsenstein J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution. 1985;39:783. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 21.Kumar S., Stecher G., Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamura K., Masatoshi N. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993;10 doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 23.Nett J.E. Candida auris: An emerging pathogen “incognito”? PLoS Pathog. 2019;15:6–11. doi: 10.1371/journal.ppat.1007638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bravo Ruiz G., Lorenz A. What do we know about the biology of the emerging fungal pathogen of humans Candida auris? Microbiol. Res. 2021;242:126621. doi: 10.1016/j.micres.2020.126621. [DOI] [PubMed] [Google Scholar]
- 25.Schelenz S., Hagen F., Rhodes J.L., Abdolrasouli A., Chowdhary A., Hall A., Ryan L., Shackleton J., Trimlett R., Meis J.F., et al. First hospital outbreak of the globally emerging Candida auris in a European hospital. Antimicrob. Resist. Infect. Control. 2016;5:1–7. doi: 10.1186/s13756-016-0132-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Das S., Tigga R., Rai G., Singh P.K., Datt S., Tyagi A., Singh N.P. Candida auris colonization in an immunocompetent patient: A new threat in medical ICU. Med. Mycol. Case Rep. 2018;21:54–56. doi: 10.1016/j.mmcr.2018.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cortegiani A., Misseri G., Fasciana T., Giammanco A., Giarratano A., Chowdhary A. Epidemiology, clinical characteristics, resistance, and treatment of infections by Candida auris. J. Intensive Care. 2018;6:69. doi: 10.1186/s40560-018-0342-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Healthcare Professionals FAQ | Candida auris | Fungal Diseases | CDC. [(accessed on 8 May 2020)]; Available online: https://www.cdc.gov/fungal/candida-auris/c-auris-health-qa.html.
- 29.Saudi CDC (Weqaya) Infection Prevention and Control. Recommendations of Candida auris. Ministry of Health; Riyadh, Saudi Arabia: 2020. [Google Scholar]
- 30.Khan Z., Ahmad S., Benwan K., Purohit P., Al-Obaid I., Bafna R., Emara M., Mokaddas E., Abdullah A.A., Al-Obaid K., et al. Invasive Candida auris infections in Kuwait hospitals: Epidemiology, antifungal treatment and outcome. Infection. 2018;46:641–650. doi: 10.1007/s15010-018-1164-y. [DOI] [PubMed] [Google Scholar]
- 31.Mohsin J., Hagen F., Al-Balushi Z.A.M., de Hoog G.S., Chowdhary A., Meis J.F., Al-Hatmi A.M.S. The first cases of Candida auris candidaemia in Oman. Mycoses. 2017;60:569–575. doi: 10.1111/myc.12647. [DOI] [PubMed] [Google Scholar]
- 32.Al-Siyabi T., Al Busaidi I., Balkhair A., Al-Muharrmi Z., Al-Salti M., Al’Adawi B. First report of Candida auris in Oman: Clinical and microbiological description of five candidemia cases. J. Infect. 2017;75:373–376. doi: 10.1016/j.jinf.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 33.Healey K.R., Kordalewska M., Ortigosa C.J., Singh A., Berrío I., Chowdhary A., Perlin D.S. Limited ERG11 mutations identified in isolates of candida auris directly contribute to reduced azole susceptibility. Antimicrob. Agents Chemother. 2018;62:1–4. doi: 10.1128/AAC.01427-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chowdhary A., Prakash A., Sharma C., Kordalewska M., Kumar A., Sarma S., Tarai B., Singh A., Upadhyaya G., Upadhyay S., et al. Multicentre study of antifungal susceptibility patterns among 350 Candida auris isolates (2009-17) in India: Role of the ERG11 and FKS1 genes in azole and echinocandin resistance. J. Antimicrob. Chemother. 2018;73:891–899. doi: 10.1093/jac/dkx480. [DOI] [PubMed] [Google Scholar]
- 35.Lone S.A., Ahmad A. Candida auris—the growing menace to global health. Mycoses. 2019;62:620–637. doi: 10.1111/myc.12904. [DOI] [PubMed] [Google Scholar]
- 36.Farrer R.A., Gade L., Chow N.A., Loparev V.N., Juieng P., Berkow E.L., Litvintseva A.P., Cuomo C.A. Genomic insights into multidrug-resistance, mating and virulence in Candida auris and related emerging species. Nat. Commun. 2018;9:1–13. doi: 10.1038/s41467-018-07779-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to privacy.