Abstract
Within the German national monitoring of zoonotic agents, antimicrobial resistance determination also targets carbapenemase-producing (CP) Escherichia coli by selective isolation from food and livestock. In this monitoring in 2019, the CP E. coli 19-AB01133 was recovered from pork shoulder. The isolate was assigned to the phylogenetic group B1 and exhibited the multi-locus sequence-type ST5869. Molecular investigations, including whole genome sequencing, of 19-AB01133 revealed that the isolate carried the resistance genes blaVIM-1, blaSHV-5 and blaCMY-13 on a self-transmissible IncA/C2 plasmid. The plasmid was closely related to the previously described VIM-1-encoding plasmid S15FP06257_p from E. coli of pork origin in Belgium. Our results indicate an occasional spread of the blaVIM-1 gene in Enterobacteriaceae of the European pig population. Moreover, the blaVIM-1 located on an IncA/C2 plasmid supports the presumption of a new, probably human source of carbapenemase-producing Enterobacteriaceae (CPE) entering the livestock and food chain sector.
Keywords: carbapenem, metallo-β-lactamase, IncA/C2, WGS, blaVIM-1
1. Introduction
Carbapenems are last resort antimicrobial agents against several infections caused by multidrug-resistant bacteria in human medicine. The World Health Organization defines them as “Highest Priority Critically Important Antimicrobials”. Nevertheless, the reports of carbapenem resistant bacteria are not limited to human medicine [1]. The repeated detection of carbapenemase-producing (CP) Enterobacteriaceae (CPE) in the German food-production chain is of considerable concern [1,2]. In general, CPE have been isolated sporadically from non-human sources (i.e., livestock, pets, wildlife) although carbapenems are not approved for veterinary application [2]. One of the main mechanisms of carbapenem resistance is the production of degrading enzymes, so called carbapenemases, which can hydrolyze almost all available β-lactams, including carbapenems [3]. The corresponding genes are often located on mobile genetic elements, in particular plasmids, which often carry additional antimicrobial, biocide, or heavy metal resistance genes [4]. These plasmids can be horizontally spread to bacteria under the selective pressure also imposed by non-β-lactam agents [5,6]. Since 2011, CPE isolates, which harbored VIM-1 encoding IncHI2 plasmids, were detected in livestock and food [7,8,9,10,11,12]. The first report described Salmonella Infantis and E. coli isolates detected in 2011 from fattening pigs in Germany [10]. They could persist in the animal host and closely related isolates could be detected in fattening pigs and minced meat in the following years [7,8]. Another VIM-1 report described the detection of this gene on an IncY plasmids in E. coli originated from seafood samples in Germany [13]. Besides the increasing reports of VIM-1 producing Enterobacteriaceae in the German food chain, the variants of carbapenemase genes increased as well. In 2019 within the annual resistance monitoring in the food chain in Germany, three CP E. coli were isolated. These include an OXA-48 producing E. coli [14] and a GES-5 producing E. coli [15], both isolated from pig feces, and another VIM-1 producing E. coli from a pork sample. This blaVIM-1 harboring E. coli isolate 19-AB01133 and its comprehensive molecular characteristics are described in this study. It shows great similarities to an E. coli isolate described by Garcia-Graells et al. (2020) [16] that was isolated from a pork sample in Belgium. Both isolates harbored almost identical IncA/C2 plasmids, which encode the blaVIM-1 gene. Furthermore, the genome data hint to a specific human entry source.
2. Materials and Methods
2.1. Isolate Origin and Antimicrobial Susceptibility Testing
The E. coli isolate 19-AB01133 was obtained in 2019 in the framework of the annual resistance monitoring in the food chain in Germany. The selective isolation of the CP E. coli was performed at a Federal State Laboratory, following Commission Implementing Decision (CID) 2013/652/EC and protocols provided by the European Union Reference Laboratory for Antimicrobial Resistance (EURL-AR) (https://www.eurl-ar.eu/protocols.aspx). The E. coli isolate 19-AB01133 was recovered from a shoulder meat sample of a pig raised in Germany. The pig was fattened and slaughtered in Brandenburg. The isolate was sent to the German Federal Institute for Risk Assessment for further confirmation and phenotypic and genotypic characterization. Antimicrobial susceptibility testing according to the EN ISO20776-1:2006 was conducted by broth microdilution using commercial plates (Thermo Fisher Scientific, Schwerte, Germany) with antimicrobial substances and concentrations defined by CID 2013/652/EC. The minimum inhibitory concentration (MIC) values were interpreted based on epidemiological cut-off values defined by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) (www.eucast.org) and fixed in CID 2013/652/EU.
2.2. Phenotypic and Genotypic Characterization
Species confirmation of the isolate was conducted by MALDI-TOF Microflex LT/SH (Bruker Daltonics, Bremen, Germany) according to the manufacturer’s recommendations. Therefore, α-Cyano-4-hydroxycinnamic acid (HCCA, Bruker, MA, USA) was used as the matrix. Molecular determination of the carbapenem-resistance genes and initial phylogenetic typing were performed as previously described [14]. The E. coli 19-AB01133 was subjected to S1-nuclease pulsed-field gel electrophoresis (S1-PFGE) [17] (https://www.cdc.gov/pulsenet/pathogens/protocols.html) followed by Southern blot hybridization [18] against a digoxigenin-labelled blaVIM-1 probe while using DIG EasyHyb and DIG Wash and Block Buffer Set (Roche Diagnostics, Mannheim, Germany) according to the manufacturer’s recommendations. The transmissibility of the plasmid was investigated by in vitro filter-mating studies using E. coli K12 J53 as the recipient. Moreover, the plasmid pEC19-AB01133 was extracted by using the CosMCPrep Plasmid Purification Kit (Beckman Coulter, Krefeld, Germany) according to the manufacturer’s recommendations. The plasmids were transformed into highly competent E. coli DH10B (ElectroMAXTM DH10BTM Cells; InvitrogenTM, Thermo Fisher Scientific, Schwerte, Germany) by electroporation using disposable cuvettes with 1 mm gap and, 1.8 kV (E = 18 kV/cm) in a Bio-Rad MicroPulser (Bio-Rad Laboratories, Feldkirchen, Germany). Potential transconjugants and transformants were analyzed for their plasmid content and resistance phenotype. The E. coli isolate 19-AB01133 was subjected to short-read (MiSeq, Illumina, CA, USA) and long-read whole genome sequencing (MinIon; Oxford Nanopore, Oxford, UK) followed by a hybrid assembly of the obtained sequences using Unicycler v0.4.4 under default parameters [19]. Based on this assembly, the multi-locus sequence-type (MLST) as well as resistance and virulence genes were determined using online tools that were provided by the Danish Technical University (http://www.genomicepidemiology.org). The annotation was carried out by RAST2 provided by PATRIC (www.patricbrc.org). The assembly of the plasmid pEC19-AB01133 was deposited in GenBank (NCBI) under the accession number MT682138.
2.3. Farm Investigation
Three months after detecting the isolate 19-AB01133, the farm and the corresponding slaughterhouse were re-investigated. Therefore, composite fecal samples (n = 8) were taken from all pens. Moreover, ten environmental samples were taken in the farm. These samples include cobwebs, dust, water trough, wet residues and sock swabs. In the slaughterhouse, additional 17 samples were taken, including minced and sausage meat, fat, a smear of work surfaces and drains, curing water, hand washing water and more. These samples were analyzed according to the EURL method combined with a second enrichment step in lysogeny broth (LB) supplemented with 1 mg/L cefotaxime (CTX) and in LB supplemented with 0.125 mg/L meropenem (MEM). Afterwards, 10 µL of all enrichments (BPW, LB + CTX and LB + MEM) were streaked out on self-made selective agar (McConkey agar (McC) supplemented with 0.125 mg/L MEM and McC supplemented with 0.125 mg/L MEM and 1 mg/L CTX) and on chromID® CARBA SMART (bioMérieux, Nürtingen, Germany). Plates were incubated for 16–18 h at 37 ± 2 °C. Up to ten colonies with different morphologies were picked and further analyzed by real-time PCR to confirm the presence or absence of blaGES, blaKPC, blaNDM, blaOXA-48 and blaVIM [14] and by MALDI-ToF MS for species confirmation of presumptive CPE.
3. Results and Discussion
The phenotypic analysis of E. coli 19-AB01133 indicated a non-wildtype phenotype for the tested β-lactam antimicrobial agents, including penicillins (ampicillin MIC > 64 mg/L), cephalosporins (cefotaxime MIC 64 mg/L, ceftazidime MIC > 128 mg/L, cefepime MIC > 32 mg/L, cefoxitin MIC > 64 mg/L, ceftazidime MIC > 8 mg/L) and carbapenems (imipenem MIC 8 mg/L, ertapenem MIC > 2 mg/L and meropenem MIC 8 mg/L), and also for (fluoro)quinolones (ciprofloxacin MIC > 8 mg/L, nalidixic acid MIC > 128 mg/L) and aminoglycosides (gentamicin MIC > 32 mg/L) (Table 1). Further typing assigned the isolate to the phylogenetic group B1. This group is often associated with enhanced antimicrobial resistance but low virulence potential [20,21]. The virulence potential of the E. coli 19-AB01133 is composed of the genes iss, ifpA and gad, which code for an increased serum survival, for long polar fimbriae and for a glutamate decarboxylase, respectively. The S1-PFGE with subsequent Southern blot hybridization and in vitro filter-mating and transformation studies identified a single, conjugative ~190 kb plasmid carrying the blaVIM-1 gene.
Table 1.
Minimal inhibitory concentrations (MIC) of following antimicrobial agents (mg/L) to 19-AB01133 and its transkonjugant TK_19AB01133. MIC values classified as non-wildtype are colored in red. Ampicillin (AMP); Azithromycin (AZI); Cefepime (FEP); Chloramphenicol (CHL); Ciprofloxacin (CIP); Colistin (COL); Ertapenem (ERP); Cefotaxime (FOT); Cefoxitin (FOX); Gentamicin (GEN); Imipenem (IMI); Kanamycin (KAN); Meropenem (MERO); Nalidixic acid (NAL); Sulfamethoxazole (SMX); Ceftazidime (TAZ); Tetracycline (TET); Tigecycline (TGC); Trimethophrim (TMP).
| Isolate | AMP | AZI | CHL | CIP | COL | ERP | FEP | FOT | FOX | GEN | IMI | MERO | NAL | SMX | TAZ | TET | TGC | TMP |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 19-AB01133 | >64 | 16 | 16 | >8 | ≤1 | >2 | >32 | >64 | >64 | 32 | 8 | 8 | >128 | >1024 | >8 | 4 | ≤0.25 | >32 |
| TK_19-AB01133 | >64 | 8 | 8 | ≤0.015 | ≤1 | >2 | >32 | >64 | >64 | 4 | 8 | 4 | ≤4 | >1024 | >8 | 4 | ≤0.25 | >32 |
| K12 J53 | 4 | 8 | ≤8 | ≤0.015 | ≤1 | ≤0.015 | ≤0.06 | ≤0.25 | 4 | 2 | 0.25 | ≤0.03 | ≤4 | ≤8 | ≤0.5 | 4 | 0.5 | ≤0.25 |
The bioinformatic analysis revealed assigned E. coli 19-AB01133 to the sequence type ST5869, which has not been described before from livestock and food in Germany. The plasmid pEC19-AB01133 (190,205 bp) exhibited an IncA/C2 replicon type. The characteristics of the presented isolate 19-AB01133 and its plasmid pEC19-AB01133 are summarized in the Supplemental Material Table S1. The encoded blaVIM-1 gene is highly likely to be responsible for the resistance to the tested carbapenems (imipenem, ertapenem and meropenem) in the E. coli isolate 19-AB01133. To the best of our knowledge, blaVIM-1-carrying IncA/C2 plasmids were detected sporadically in human samples [22,23], but so far only once in livestock and food in Europe [16]. Recently, Garcia-Grealls and her colleagues (2020) described an almost identical VIM-1 encoding plasmid (99.99% identical over the entire plasmid nucleotide sequence, Acc. No. PRJNA564835) of E. coli (Acc. No. MN477204.1) isolated from minced pork in Belgium in 2015 [16]. The corresponding isolate was assigned to the same sequence type, which might indicate a clonal relationship. This observation leads to the question of repeated contamination of pigs and pork with this strain, or to a persistence of this strain in Central European pig production. The E. coli isolate 19-AB01133 further showed a close relationship to the E. coli isolate TZ 116, which is used by the EURL-AR as second (backup) strain for the validation of selective plates for the detection of CP E. coli (https://www.eurl-ar.eu/protocols.aspx). This backup strain was isolated from a human sample. According to personal communication, the strain was not used in the isolating laboratory at that time or in our lab and thus, a cross-contamination can be excluded.
Apart from this control strain, some more blaVIM-1 harboring IncA/C2 plasmids of isolates from human origin showed a close relationship to the described plasmid pEC19-AB01133. For example, Drieux et al. (2012) [23] described a plasmid (Acc. No. JQ824049) from a Greek Providencia stuartii multiresistant strain. By comparing this plasmid with the plasmid pEC19-AB01133, a query coverage of 87% was observed. Further, plasmids from Enterobacteriaceae of clinical and environmental samples from Canada showed also high similarities [24] to the described plasmid pEC17-AB01133. The plasmid pIncAC-KP4898 (Acc. No. KY882285.1) and the plasmid pRIVM0001_VIM-1 (Acc. No. MH220284.1) had a query coverage of 70% and the plasmid pKPC_CAV1344 (Acc. No. CP011622.1) a query coverage of 81%. Taken together, the close relationship and the repeated detection of the strain in food chain samples hints to a spill-over of the strain from humans to food.
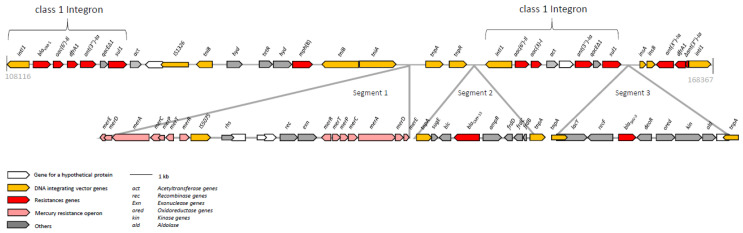
The plasmid pEC19-AB01133 harbored a variety of antimicrobial resistance genes, which are located within a ~60 kb multiresistance region (Figure 1). The blaVIM-1 gene is located on a class 1 integron. In addition, the displayed region exhibited three further interesting segments. The first segment comprised two mercury resistance operons (Figure 1), which confer narrow-spectrum resistance to inorganic mercury [25]. Two tnpA genes encompass the 5768 bp segment 2. This DNA-segment, originally identified in Citrobacter freundii (Acc. No. AY339625.2), encodes the small multidrug resistance efflux transporter SugE and the AmpC β-lactamase CMY-13 [26]. To date, the occurrence of blaCMY-13 has only sporadically been reported [26]. Another neighboring class 1 integron conferred resistance to aminoglycosides [aac(6′)-Il, aac(3)-l, ant(3”)-Ia] and to sulfonamides (sul1). Downstream of this class 1 integron, the third segment was integrated. This segment carried the ESBL β-lactamase gene blaSHV-5. The blaSHV-5 gene has been previously reported from isolates of E. coli, Klebsiella pneumoniae and Providencia stuartii from humans, livestock and wildlife [27], but not yet in animals or food in Germany.
Figure 1.
Schematic illustration of the multidrug-resistance region of plasmid pEC19-AB01133. The reading frames are displayed as arrows with the arrowhead showing the direction of transcription. The numbers refer to the whole plasmid sequence of pEC19-AB01133, which is deposited in the GenBank database under accession no. MT682138.
The farm and the corresponding slaughter facility were re-investigated three months later. Thereby, 35 samples from feces, farm and slaughter facility environment and different meat products were taken. No CPE were detected in these samples. This suggests that the strain was unable to persist on the farm and spread of the VIM-1 plasmid did not occur. Nevertheless, the detection of an isolate with such high similarities to the Belgian isolates might hint to the ability for it to persist within the pork production chain [15]. A re-introduction by human, i.e., by farmers or workers, is also possible as there was no obvious link between the two fattening pig farms.
The detection of multiresistance IncA/C2 plasmids identified in E. coli isolates from food chain is a threat to public health. Previous studies from Germany described the location of blaVIM-1 only on IncHI2 or IncY plasmids. However, for IncA/C2 plasmids, a high impact on the dissemination of antimicrobial resistance genes has been reported [20]. The detection of new plasmid types and the recurrence of carbapenemase-producing isolates in the German pork production chain emphasizes the importance of the CPE monitoring [4,8,9]. The occurrence of the blaVIM-1 located on an IncA/C2 plasmid in an E. coli isolate from pork suggests a new, probably human source of CPE entering the food chain.
Acknowledgments
We gratefully acknowledge the support of the regional laboratories and authorities in collecting the samples and providing the isolates in the framework of the monitoring. We also thank the laboratory team, especially Romy Fuhrmeister and Silvia Schmoger, for excellent technical assistance.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/9/1/29/s1. Table S1: General information about the isolate 19-AB01133 and its plasmid pEC19-AB01133.
Author Contributions
Data curation, N.P.; formal analysis, S.S.; investigation, N.P., J.A.H., M.G., B.-A.T. and A.I.; project administration, A.I.; visualization, N.P.; writing—original draft, N.P., J.A.H. and A.I.; writing—review and editing, J.A.H., M.G., A.K., B.-A.T., B.M., S.S., D.M. and A.I. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the German Federal Institute for Risk Assessment (BfR, 43-001 and 1322-648) and the European Joint Project (EJP) IMPART funded by the European Union’s Horizon 2020 research and innovation programme under Grant Agreement No 773830. The work of JAH was supported by the BMG project GÜCCI. The work of SS was supported by the German Federal Ministry of Education and Research (BMBF) under project number 01KI1727D as part of the Research Network Zoonotic Infectious Diseases.
Data Availability Statement
Data is contained within the article and supplementary material. Moreover, the plasmid sequence was deposited in GenBank (NCBI) under the accession number MT682138.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.WHO. Advisory Group on Integrated Surveillance of Antimicrobial Resistance (AGISAR) Critically Important Antimicrobials for Human Medicine: Ranking of Antimicrobial Agents for Risk Management of Antimicrobial Resistance due to Non-Human Use. 5th ed. World Health Organization; Geneva, Switzerland: 2017. [Google Scholar]
- 2.Köck R., Daniels-Haardt I., Becker K., Mellmann A., Friedrich A.W., Mevius D., Schwarz S., Jurke A. carbapenem-resistant enterobacteriaceae in wildlife, food-producing, and companion animals: A systematic review. Clin. Microbiol. Infect. 2018;24:1241–1250. doi: 10.1016/j.cmi.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 3.Salahuddin P., Kumar A., Kha A.U. Structure, function of serine and Metallo-β-lactamases and their Inhibitors. Curr. Protein. Pept. Sci. 2018;19:130–144. doi: 10.2174/0929866524666170724160623. [DOI] [PubMed] [Google Scholar]
- 4.Bush K., Fisher J.F. Epidemiological expansion, structural studies, and clinical challenges of new β-lactamases from gram-negative bacteria. Annu. Rev. Microbiol. 2011;65:455–478. doi: 10.1146/annurev-micro-090110-102911. [DOI] [PubMed] [Google Scholar]
- 5.Partridge S.R., Kwong S.M., Firth N., Jensen S.O. Mobile genetic elements associated with antimicrobial resistance. Clin. Microbial. Rev. 2018;31:e00088-17. doi: 10.1128/CMR.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Potter R.F., D′Souza A.W., Dantas G. The rapid spread of carbapenem-resistant enterobacteriaceae. Drug Resist. Updat. 2016;29:30–46. doi: 10.1016/j.drup.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Irrgang A., Fischer J., Grobbel M., Schmoger S., Skladnikiewicz-Ziemer T., Thomas K., Hensel A., Tenhagen B.-A., Kasbohrer A., Käsbohrer A. Recurrent detection of VIM-1-producing Escherichia coli clone in German pig production. J. Antimicrob. Chemother. 2017;72:944–946. doi: 10.1093/jac/dkw479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borowiak M., Szabo I., Baumann B., Junker E., Hammerl J.A., Kaesbohrer A., Malorny B., Fischer J. VIM-1-producing Salmonella infantis isolated from swine and minced pork meat in Germany. J. Antimicrob. Chemother. 2017;72:2131–2133. doi: 10.1093/jac/dkx101. [DOI] [PubMed] [Google Scholar]
- 9.Fischer J., Rodriguez I., Schmoger S., Friese A., Roesler U., Helmuth R., Guerra B. Salmonella enterica subsp. enterica producing VIM-1 carbapenemase isolated from livestock farms. J. Antimicrob. Chemother. 2013;68:478–480. doi: 10.1093/jac/dks393. [DOI] [PubMed] [Google Scholar]
- 10.Fischer J., San Jose M., Roschanski N., Schmoger S., Baumann B., Irrgang A., Friese A., Roesler U., Helmuth R., Guerra B. Spread and persistence of VIM-1 carbapenemase-producing enterobacteriaceae in three German swine farms in 2011 and 2012. Vet. Microbiol. 2017;200:118–123. doi: 10.1016/j.vetmic.2016.04.026. [DOI] [PubMed] [Google Scholar]
- 11.Irrgang A., Tenhagen B.-A., Pauly N., Schmoger S., Kaesbohrer A., Hammerl J.A. Characterization of VIM-1-producing, E. coli isolated from a German fattening pig farm by an improved isolation procedure. Front. Microbiol. 2019;10:2256. doi: 10.3389/fmicb.2019.02256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pauly N., Hammerl J.A., Schwarz S., Grobbel M., Meemken D., Malorny B., Tenhagen B.-A., Käsbohrer A., Irrgang A. Co-occurrence of the blaVIM-1 and blaSHV-12 genes on an IncHI2 plasmid of an Escherichia coli isolate recovered from German livestock. J. Antimicrob. Chemother. 2020 doi: 10.1093/jac/dkaa436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roschanski N., Guenther S., Vu T.T.T., Fischer J., Semmler T., Huehn S., Alter T., Roesler U. VIM-1 carbapenemase-producing Escherichia coli isolated from retail seafood, Germany 2016. Eurosurveillance. 2017;22:17-00032. doi: 10.2807/1560-7917.ES.2017.22.43.17-00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Irrgang A., Pauly N., Tenhagen B.-A., Grobbel M., Kaesbohrer A., Hammerl A.J. A spill-over from public health? First detection of an OXA-48-producing Escherichia coli in a German pig farm. Microorganisms. 2020;8:855. doi: 10.3390/microorganisms8060855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Irrgang A., Tausch S.H., Pauly N., Grobbel M., Kaesbohrer A., Hammerl J.A. First detection of GES-5-producing escherichia coli from livestock-an increasing diversity of carbapenemases recognized from german pig production. Microorganisms. 2020;8:1593. doi: 10.3390/microorganisms8101593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia-Graells C., Berbers B., Verhaegen B., Vanneste K., Marchal K., Roosens N.H.C., Botteldoorn N., De Keersmaecker S.C.J. First detection of a plasmid located carbapenem resistant bla(VIM-1) gene in E. coli isolated from meat products at retail in Belgium in 2015. Int. J. Food Microbiol. 2020;324:108624. doi: 10.1016/j.ijfoodmicro.2020.108624. [DOI] [PubMed] [Google Scholar]
- 17.Guerra B., Junker E., Miko A., Helmuth R., Mendoza M.C. Characterization and localization of drug resistance determinants in multidrug-resistant, integron-carrying Salmonella enterica serotype typhimurium strains. Microb. Drug Resist. 2004;10:83–91. doi: 10.1089/1076629041310136. [DOI] [PubMed] [Google Scholar]
- 18.Rodriguez I., Barownick W., Helmuth R., Mendoza M.C., Rodicio M.R., Schroeter A., Guerra B. Extended-spectrum {beta}-lactamases and AmpC {beta}-lactamases in ceftiofur-resistant Salmonella enterica isolates from food and livestock obtained in Germany during 2003-07. J. Antimicrob. Chemother. 2009;64:301–309. doi: 10.1093/jac/dkp195. [DOI] [PubMed] [Google Scholar]
- 19.Hadziabdic S., Fischer J., Borowiak M., Malorny B., Juraschek K., Kaesbohrer A., Guerra B., Deneke C., Gonzalez-Zorn B., Szabo I. The blaNDM-1-carrying IncA/C2 plasmid underlies structural alterations and cointegrate formation in vivo. Antimicrob. Agents Chemother. 2019;63 doi: 10.1128/AAC.00380-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harmer C.J., Hall R.M. The A to Z of A/C plasmids. Plasmid. 2015;80:63–82. doi: 10.1016/j.plasmid.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 21.Johnson J.R., Kuskowski M.A., Owens K., Gajewski A., Winokur P.L. Phylogenetic origin and virulence genotype in relation to resistance to fluoroquinolones and/or extended-spectrum cephalosporins and cephamycins among Escherichia coli isolates from animals and humans. J. Infect. Dis. 2003;188:759–768. doi: 10.1086/377455. [DOI] [PubMed] [Google Scholar]
- 22.Antonelli A., D’Andrea M.M., Montagnani C., Bartolesi A.M., Di Pilato V., Fiorini P., Torricelli F., Galli L., Rossolini G.M. Newborn bacteraemia caused by an aeromonas caviae producing the VIM-1 and SHV-12 beta-lactamases, encoded by a transferable plasmid. J. Antimicrob. Chemother. 2016;71:272–274. doi: 10.1093/jac/dkv304. [DOI] [PubMed] [Google Scholar]
- 23.Drieux L., Decré D., Frangeul L., Arlet G., Jarlier V., Sougakoff W. Complete nucleotide sequence of the large conjugative pTC2 multireplicon plasmid encoding the VIM-1 metallo-β-lactamase. J. Antimicrob. Chemother. 2013;68:97–100. doi: 10.1093/jac/dks367. [DOI] [PubMed] [Google Scholar]
- 24.Kohler P., Tijet N., Kim H.C., Johnstone J., Edge T., Patel S.N., Seah C., Willey B., Coleman B., Green K., et al. The Toronto invasive bacterial diseases network. Dissemination of Verona integron-encoded Metallo-β-lactamase among clinical and environmental enterobacteriaceae isolates in Ontario, Canada. Sci. Rep. 2020;10:18580. doi: 10.1038/s41598-020-75247-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Osborn A.M., Bruce K.D., Strike P., Ritchie D.A. Distribution, diversity and evolution of the bacterial mercury resistance (mer) operon. FEMS Microbiol. Rev. 1997;19:239–262. doi: 10.1111/j.1574-6976.1997.tb00300.x. [DOI] [PubMed] [Google Scholar]
- 26.Miriagou V., Tzouvelekis L.S., Villa L., Lebessi E., Vatopoulos A.C., Carattoli A., Tzelepi E. CMY-13, a novel inducible cephalosporinase encoded by an Escherichia coli plasmid. Antimicrob. Agents Chemother. 2004;48:3172–3174. doi: 10.1128/AAC.48.8.3172-3174.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liakopoulos A., Mevius D., Ceccarelli D. A review of SHV extended-spectrum β-lactamases: Neglected yet ubiquitous. Front. Microbial. 2016;7:1374. doi: 10.3389/fmicb.2016.01374. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is contained within the article and supplementary material. Moreover, the plasmid sequence was deposited in GenBank (NCBI) under the accession number MT682138.