Abstract
Familial hypercholesterolemia (FH) is a common autosomal codominant disorder, characterized by elevated low-density lipoprotein cholesterol levels causing premature atherosclerotic cardiovascular disease. About 2900 variants of LDLR, APOB, and PCSK9 genes potentially associated with FH have been described earlier. Nevertheless, the genetics of FH in a Russian population is poorly understood. The aim of this study is to present data on the spectrum of LDLR, APOB, and PCSK9 gene variants in a cohort of 595 index Russian patients with FH, as well as an additional systematic analysis of the literature for the period of 1995–2020 on LDLR, APOB and PCSK9 gene variants described in Russian patients with FH. We used targeted and whole genome sequencing to search for variants. Accordingly, when combining our novel data and the data of a systematic literature review, we described 224 variants: 187 variants in LDLR, 14 variants in APOB, and 23 variants in PCSK9. A significant proportion of variants, 81 of 224 (36.1%), were not described earlier in FH patients in other populations and may be specific for Russia. Thus, this study significantly supplements knowledge about the spectrum of variants causing FH in Russia and may contribute to a wider implementation of genetic diagnostics in FH patients in Russia.
Keywords: familial hypercholesterolemia, Russian, whole genome sequencing, LDLR, APOB, PCSK9
1. Introduction
Familial hypercholesterolemia (FH) is a common autosomal codominant disorder, characterized by elevated low-density lipoprotein (LDL) cholesterol levels causing premature atherosclerotic cardiovascular disease [1]. In two meta-analyses of 2020, similar results were obtained on the prevalence of heterozygous FH (HeFH) in the general population: one in 311 and one in 313, respectively [2,3]. The prevalence of homozygous FH (HoFH) is one in 300,000 [4]. Mutations in one of the three genes (low-density lipoprotein receptor gene (LDLR), apolipoprotein B gene (APOB) and proprotein convertase subtilisin/kexin type 9 gene (PCSK9)) cause both HeFH and HoFH, and these genes account for the vast majority of genetically confirmed cases of FH [1]. For LDLRAP1, LIPA, ABCG5 and ABCG8 genes, two mutant alleles act recessively, producing a severe phenotype consistent with HoFH, but only single families have been described [1]. About 2900 variants in the LDLR, APOB and PCSK9 genes potentially associated with FH have been described by the members of the ClinGen FH Variant Curation Expert Panel from 13 different countries [5]. Nevertheless, the genetics of FH in a Russian population is still poorly understood, with only about 60 variants of LDLR and APOB genes described in single publications [6,7,8,9,10]. The aim of this study is to present data on the spectrum of the LDLR, APOB and PCSK9 gene variants in a cohort of 595 index Russian patients with FH, and to perform an additional systematic analysis of the literature for the period of 1995–2020 on LDLR, APOB and PCSK9 gene variants described in Russian FH patients.
2. Materials and Methods
2.1. Clinical Description of the Patients
The study included index patients (n = 595) with clinically and genetically confirmed diagnosis of HeFH or HoFH examined by researchers at the National Medical Research Center for Therapy and Preventive Medicine (Moscow, Russia) and the National Medical Research Center for Cardiology (Moscow, Russia). HeFH was determined using the Dutch Lipid Clinical Network Criteria (DLCN) including the results of genetic testing [11]. This diagnosis was established when the DLCN score was six points or more. The diagnosis of HoFH was determined using the guidance of the European Atherosclerosis Society [4]. Blood for genetic analysis was stored in the Biobank of the National Medical Research Center for Therapy and Preventive Medicine (Moscow, Russia). Targeted sequencing and Sanger sequencing were performed at the National Medical Research Center for Therapy and Preventive Medicine (Moscow, Russia). Whole genome sequencing was performed at the Center for Strategic Planning of the Federal Medical Biological Agency (Moscow, Russia). This study was performed in accordance with the Declaration of Helsinki and was approved by the Committee on the Ethics issues in clinical cardiology of the National Medical Research Center for Cardiology (Moscow, Russia) and by the Institutional Review Boards of the National Research Center for Therapy and Preventive Medicine (Moscow, Russia) with written informed consent obtained from each participant and/or their legal representative, as appropriate.
2.2. Systematic Review
We performed a systematic review of all relevant peer-reviewed published articles involving patients with FH from Russia. The search strategy was designed to cover all articles published in English using three literature databases (Scopus, Web of Science and PubMed) from 1995 to July 2020. The search terms were: (“Familial hypercholesterolemia” OR “LDLR” OR “APOB” OR “PCSK9”) and (“Russia” OR “Russian”). The eligible articles were screened for both the titles and abstracts.
2.3. Molecular Genetic Analysis
2.3.1. Target Sequencing
DNA was isolated using the QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany). DNA concentration was assessed with a Qubit 4.0 fluorimeter (Thermo Fisher Scientific, Waltham, MA, USA). Target sequencing was performed with two platforms: Ion S5 (Thermo Fisher Scientific, Waltham, MA, USA) and Nextseq550 (Illumina, San Diego, CA, USA). For sequencing on Ion S5, DNA libraries were prepared on an Ion Chef System (Thermo Fisher Scientific, Waltham, MA, USA) using a custom panel designed automatically by Ion AmpliSeq Designer software v7.4.2 (Thermo Fisher Scientific, Waltham, MA, USA). The panel flanked exonic and adjacent intronic sequences of 25 genes (UTR + CDS + 100 bp padding). VCF files were generated from BAM files on a Torrent Server (Thermo Fisher Scientific, Waltham, MA, USA) with default parameters. VCF files were annotated using Ion Reporter (Thermo Fisher Scientific, Waltham, MA, USA) with Annotate Variants analysis tool. For Nextseq 550, the library preparation was performed using the SeqCap EZ Prime Choice Library kit (Roche, Basel, Switzerland). Two Roche panels were used, consisting of 24 (CDS + 25 bp padding) and 244 (CDS + 25 bp padding) genes. All three panels included the LDLR, APOB and PCSK9 genes. All stages of sequencing were carried out according to the manufacturers’ protocols. Reads were aligned to the reference genome (GRCh37). Sequencing analysis resulted in fastq files. Data processing was performed with BWA, Picard, bcftools, GATK3 and generally followed the GATK best practices for variant calling. We applied standard GATK hard filters for single nucleotide substitutions (MQ, QD, FS, SOR, MQRankSum, QUAL, ReadPosRankSum) and for short insertions and deletions (QD, FS, QUAL, ReadPosRankSum). Single nucleotide variants and short indels were annotated with ANNOVAR.
2.3.2. Whole Genome Sequencing and Bioinformatic Analysis
DNA was extracted from whole blood sample using QIAamp® DNA Mini Kit (Qiagen, Hilden, Germany). A WGS library was prepared using Nextera DNA Flex kit (Illumina, San Diego, CA, USA) according to manufacturer instructions. Paired-end sequencing (150 bp) was performed to mean sequencing coverage of 30× or more. Reads were aligned to the reference genome (GRCh38) and small variants were called using Dragen Bio-IT platform (Illumina, San Diego, CA, USA) and joint-called with GLnexus [12].
Structural variant (SV) calling was performed with smoove software [13]. Annotation was performed using an Ensembl Variant Effect Predictor (VEP) [14]. All variants were visually inspected in an Integrative Genomics Viewer (IGV) [15] and breakpoint regions were investigated with PCR and Sanger sequencing. Mobile elements (ME) SVA, LINE1 and Alu were called using MELT software [16] and annotated with VEP [14]. Images were prepared using the R programming language. For Figure 1 a trackViewer package was used [17].
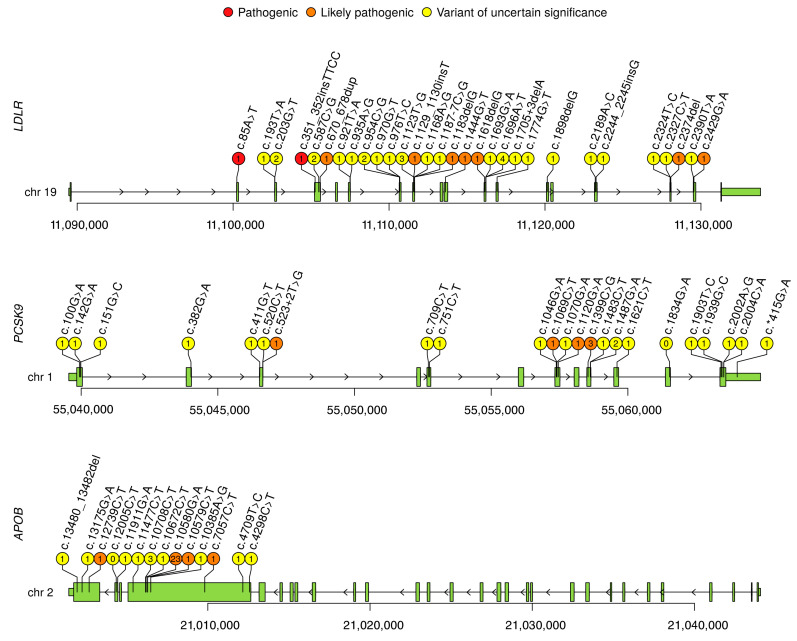
Figure 1.
Variants in LDLR, PSCK9, and APOB genes, specific for the Russian population. For the LDLR gene, due to the large quantity, only 30 novel variants found in this study are shown (with the exception of four large structural variants presented in Figure 2). Number of index patient is indicated in the circle (0 is for variants found in other studies), color indicates clinical interpretation: red, orange and yellow for pathogenic (P), likely pathogenic (LP) and variant of uncertain significance (VUS), respectively. Coordinates are given in hg38 assembly.
2.3.3. Clinical Interpretation
The following canonical transcripts were used in this work: NM_000527.5 (LDLR), NM_000384.3 (APOB), and NM_174936.4 (PCSK9). For clinical interpretation, short genetic variants with overall frequencies for European (non-Finnish) in the gnomAD database of <0.5%, or missing in the gnomAD, were selected. SV-only variants with frequencies of <0.5% for European (non-Finnish) were left for evaluation. No ME insertions were found for LDLR, APOB or PSCK9. Evaluation of the pathogenicity of the variants was carried out in accordance with the recommendations of the American College of Medical Genetics and Genomics (ACMG) with modifications [18]. The following types of variants are reported in the article: pathogenic (P), likely pathogenic (LP) and variant of unknown significance (VUS). All variants were analyzed for their presence in the databases (LOVD, ClinVar and HGMD) [5,19].
2.3.4. Sanger Sequencing
The validation of NGS results was done by Sanger sequencing. PCRs were performed in 20 μL of a mixture containing 0.2 mM of each nucleotide, 1× PCR buffer, 20 ng of the DNA, 10 ng of each primer, 2.5 U of DNA polymerase. Amplification was performed on a GeneAmp PCR System 9700 thermocycler (Thermo Fisher Scientific, Waltham, MA, USA) with the following parameters: 95 °C—300 s; 30 cycles: 95 °C—30 s, 62 °C—30 s, 72 °C—30 s; 72 °C—600 s. Before the Sanger reaction, the obtained amplicons were purified using ExoSAP-IT (Affymetrix, Santa Clara, CA, USA) according to the manufacturer’s protocol. The nucleotide sequence of PCR products was determined using the ABI PRISM® BigDye™ Terminator reagent kit v. 3.1 followed by analysis of the reaction products on an automated DNA sequencer Applied Biosystem 3500 DNA Analyzer (Thermo Fisher Scientific, Waltham, MA, USA).
3. Results
3.1. Systematic Literature Review
The search strategy described above yielded 665 citations; 474 remained after duplicate removal. After the analysis of the abstracts referring to genetic testing or LDLR, APOB and PCSK9 variants in FH patients, 27 articles were selected, of which 25 contained data on the LDLR, APOB, and PCSK9 variants, including three of previously published articles by our group [6,7,8,9,10,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39]. These articles describe 91 causal variants of LDLR gene, one variant of APOB, and one variant of PCSK9 (Figure 1, Table A1, Table A2 and Table A3 in Appendix A).
3.2. Genetic Test Results
In our study we performed genetic testing of 595 unrelated patients with FH, of which six patients demonstrated the phenotype of HoFH and the rest had clinical features of HeFH. Target sequencing was performed for 401 patients and whole genome sequencing was performed for 405 patients (both methods were performed for 211 patients). In 405 WGS patients we called SNPs, short indels, long SVs and ME insertions. We identified 122 different potentially causative variants in LDLR, 13 variants in APOB, and 21 variants in PCSK9 in 294 unrelated patients (Figure 1 and Figure 2, Table A1, Table A2 and Table A3). No potentially causative variants were found in 301 of 595 patients (50.6%). Out of these 294 patients, one patient was a true homozygote, four compound heterozygotes with two LDLR variants on different chromosomes (in trans), one compound heterozygote with two LDLR variants on the same chromosome (in cis), two compound heterozygotes with two LDLR variants of unknown mutual arrangement of alleles, six double heterozygotes (harboring two variants in two different genes) and one patient with three variants in three genes (Table A4), the rest were simple heterozygotes. A total of 34 variants in LDLR, six variants in APOB and six variants in PCSK9, were found in this study for the first time. Most of these variants were unique but some LDLR variants occurred in several unrelated patients: p.Cys68Phe, p.Pro196Arg, p.Cys318Trp, p.Tyr375Asp and p.Ile566Phe. Of 35 variants previously described in the literature [6,7,8,9,10,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39] only for the Russian population, six variants were also found in this study. Most of these variants were also unique, except for variant LDLR-p.Cys160Gly, that was found in six unrelated patients. Of all variants (the percentage of all identified potentially causative alleles (310 alleles found in this study)) the most common were: LDLR-p.Gly592Glu—9.4%, LDLR-p.Leu401His—9%, APOB-p.Arg3527Gln—7.4%, LDLR-p.Cys329Tyr—2.6%, LDLR-p.Cys160Gly—1.9%. Most of the variants described above were SNPs and short indels. Only five large SVs were found in this study and all of them in LDLR gene (Figure 2). Four novel deletions were found and a tandem duplication previously described in a patient of Czech origin (ClinVar ID: 251140). No ME insertions were found in any of the studied genes.
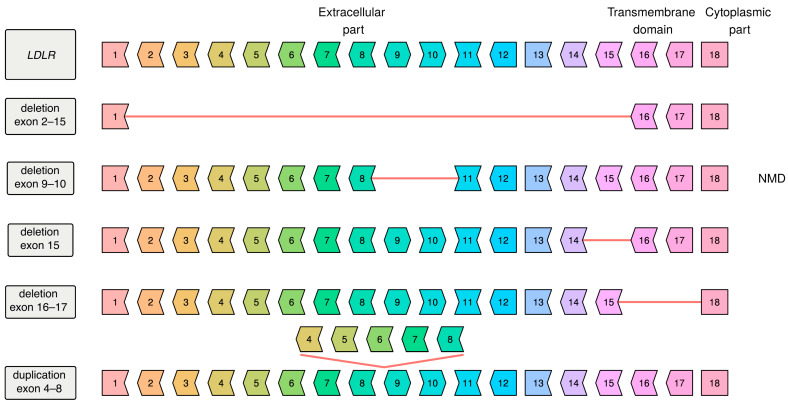
Figure 2.
Exonic structure of the native LDLR gene and its large structural variants found in this study. Exon border shape (flat and right or left pointing) shows the phase of the reading frame (+0; +1; +2); if borders don’t match, a frame shift occurs (deletions exon 9–10 and 16–17). NMD marks a variant that likely leads to the nonsense-mediated decay.
3.3. Description of All Variants in Russia
In total, when combining our data (156 LDLR, APOB and PCSK9 variants) and the data of the systematic review (91 LDLR, APOB and PCSK9 variants), we described 224 variants: 187 LDLR variants, 14 APOB variants, and 23 PCSK9 variants (Table A1, Table A2 and Table A3). A significant proportion of variants—36.1% (67 LDLR variants, six APOB variants and eight PCSK9 variants)— was not described in FH patients in other populations and may be specific for Russia.
In accordance with the criteria of pathogenicity, 38 LDLR variants were classified as pathogenic (P), 53 as likely pathogenic (LP) and 95 as variant of unknown significance (VUS). In the APOB gene there were four LP and 10 VUS, and in the PCSK9 gene four LP and 19 VUS (Table 1).
Table 1.
Variants, found in this study.
| Gene | Total (P/LP/VUS) | Possibly Unique including Novel for the Russian Population and Described Earlier (P/LP/VUS) | Novel (P/LP/VUS) | Described in Other World Populations |
|---|---|---|---|---|
| LDLR | 187 (38/53/95) * | 67 (11/19/37) | 34 (3/10/21) | 120 (27/34/58) |
| APOB | 14 (0/4/10) | 6 (0/1/5) | 6(0/1/5) | 8 (0/3/5) |
| PCSK9 | 23 (0/4/19) | 8(0/1/7) | 6 (0/1/5) | 5 (0/3/12) |
Novel: variants found in this study for the first time. Possibly unique for the Russian population: variants found in this study and previously described only for the Russian population. (*)—for one variant it was impossible to determine the category of pathogenicity. However, it was earlier described in the literature as pathogenic.
4. Discussion
This study was based on the largest number of participants of any genetic FH study in Russia to date. Including collected literature data, this study reported 224 variants found in the Russian population, either novel or reported before, with 81 variants described only in Russian FH patients. These data on the spectrum of the LDLR, APOB and PCSK9 variants can be useful for clinical interpretation when carrying out a genetic diagnosis of FH in Russia. It also improved knowledge about the genetics of FH in general. Thus, according the results of this study, Russia is ranked fourth among countries with the largest number of variants described in FH patients, after the United Kingdom, the Netherlands and Italy [18]. In our study, we did not carry out a functional analysis of the identified variants and used ACMG recommendations to assess their pathogenicity. About half of the variants described here were assigned a category of uncertain significance and, possibly, in the future with the advent of new data, their causality may be revised. It would also be desirable to assess the clinical significance of the combined effect of two or more variants identified in patients with HeFH (Table A4).
The WGS-based SV analysis was performed for 405 patients for whom no relevant variants were found by targeted sequencing. The fact that no large SVs were found either in PCSK9 or in APOB may be explained by their gain-of-function pathogenicity model. Taking into account the literature data, nine large rearrangements in LDLR were described for the Russian patients earlier and their proportion of the total number of unique variants (n = 187) of the LDLR gene was 4.8%, which is slightly less than the share of large LDLR rearrangements in the ClinVar database (6.1%) [5]. The presence of large deletions, encompassing exonic LDLR regions, suggests that multiplex ligation-dependent probe amplification could be a useful method in genetic confirmation of FH.
5. Conclusions
This study significantly supplements knowledge about the spectrum of variants causing FH in Russia and may contribute to a wider implementation of genetic diagnostics in Russian FH patients.
Acknowledgments
The authors are grateful to the patients and their family for their continuous contributions and support of our research.
Appendix A
Table A1.
List of the LDLR variants described in Russian patients.
| Number of Index Patients 1 | Variant Data 2 | Exon | DNA Change | Protein Change | dbSNP ID | gnomAD MAF (v. 2.1.1) | ACMG Interpretation | ClinVar Interpretation | ClinVar ID | References |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 1i–15i | c.68-366_2312-791del | P | ||||||
| 0 | 6 | 2 | p.Cys27Trp | rs2228671 | VUS | P/LP | 226304 | [30] | ||
| 1 | 1 | 2 | c.85A > T | p.Arg29Ter | rs879254401 | P | P | 251011 | ||
| 0 | 6 | 2 | c.97C > T | p.Gln33Ter | rs121908024 | 0.000007963 | P | P | 3683 | [6,20,30] |
| 0 | 6 | 3 | c.191_313del | p.Leu64_Pro105delinsSer | P | [9] | ||||
| 0 | 5 | 3 | c.193_202delTCTGTCACCTinsGGACTTCA | p.Ser65Glyfs * 64 | LP | [8,10,25,27,29] | ||||
| 1 | 1 | 3 | c.193T > A | p.Ser65Thr | VUS | |||||
| 0 | 5 | 3 | c.195dupT | p.Val66Cysfs * 64 | rs879254435 | P | P | 251075 | [8,10,25,29] | |
| 2 | 4 | 3 | c.200C > T | p.Thr67Ile | rs1337448484 | 0.00001060 | VUS | VUS | 629411 | |
| 2 | 1 | 3 | c.203G > T | p.Cys68Phe | VUS | |||||
| 2 | 2 | 3 | c.230dup | p.Arg78ProfsTer55 | rs879254440 | P | P | 251083 | [30,37,39] | |
| 1 | 4 | 3 | c.241C > T | p.Arg81Cys | rs730882078 | 0.000007953 | VUS | P/LP/VUS | 183083 | |
| 0 | 6 | 3 | c.245G > C | p.Cys82Ser | VUS | VUS | 431509 | [10] | ||
| 1 | 4 | 3 | c.246C > A | p.Cys82Ter | rs875989891 | P | P | 226309 | ||
| 0 | 6 | 3 | c.285C > A | p.Cys95Ter | rs139400379 | P | P | 251115 | [20,21,30] | |
| 0 | 6 | 3i | c.313 + 1G > A | rs112029328 | 0.00002784 | P | P/LP | 3736 | [6,20] | |
| 0 | 5 | 3i | c.313 + 2T > G | LP | [10] | |||||
| 1 | 3 | 4–8 | c.317-1185dup | p.Pro106_Val395dup | LP | [9] | ||||
| 2 | 4 | 4 | c.326G > A | p.Cys109Tyr | rs121908042 | 0.000003996 | LP | P/LP | 226319 | |
| 2 | 4 | 4 | c.343C > T | p.Arg115Cys | rs774723292 | 0.00002792 | LP | P/LP/VUS | 251162 | |
| 1 | 4 | 4 | c.347G > A | p.Cys116Tyr | LP | |||||
| 0 | 6 | 4 | c.347_349delGCC | p.Cys116_His117delinsTyr | rs879254483 | LP | LP | 251164 | [20,26,30] | |
| 1 | 1 | 4 | c.351_352insTTCC | p.Asp118PhefsTer13 | P | [7] | ||||
| 1 | 5 | 4 | c.355_356insTTCC | p.Gly119ValfsTer12 | P | [9] | ||||
| 1 | 4 | 4 | c.420G > C | p.Glu140Asp | rs879254520 | LP | P/LP | 251216 | ||
| 0 | 5 | 4 | c.444T > G | p.Cys148Trp | rs879254528 | LP | LP | 251228 | [20,24,30] | |
| 0 | 6 | 4 | c.451G > C | p.Ala151Pro | rs763233960 | 0.00001195 | LP | VUS | 251234 | [20,28,30] |
| 6 | 2 | 4 | c.478T > G | p.Cys160Gly | rs879254540 | LP | LP | 251248 | [20,24,29,30] | |
| 0 | 6 | 4 | c.499T > C | p.Cys167Arg | rs879254547 | LP | P/LP | 251255 | [20,28] | |
| 1 | 4 | 4 | c.502G > C | p.Asp168His | rs200727689 | LP | P/LP | 251258 | ||
| 1 | 4 | 4 | c.519C > G | p.Cys173Trp | rs769318035 | 0.000007958 | LP | P/LP | 251277 | |
| 0 | 6 | 4 | c.530C > T | p.Ser177Leu | rs121908026 | 0.00001592 | LP | P/LP | 3686 | [9] |
| 1 | 3 | 4 | c.534T > G | p.Asp178Glu | rs879254566 | LP | P/LP | 251287 | [36] | |
| 0 | 6 | 4 | c.542C > T | p.Pro181Leu | rs557344672 | 0.000007958 | LP | LP | 431512 | [9] |
| 2 | 4 | 4 | c.551G > A | p.Cys184Tyr | rs121908039 | 0.00009554 | LP | P/LP | 3739 | |
| 2 | 1 | 4 | c.587C > G | p.Pro196Arg | VUS | |||||
| 0 | 6 | 4 | c.618T > G | p.Ser206Arg | rs879254595 | LP | P/LP | 251325 | [8,10,25,29] | |
| 5 | 3 | 4 | c.622G > A | p.Glu208Lys | rs879254597 | LP | P/LP | 251328 | [32] | |
| 0 | 6 | 4 | c.626G > A | p.Cys209Tyr | rs879254600 | LP | P/LP | 251332 | [20,28,30] | |
| 5 | 3 | 4 | c.654_656delTGG | p.Gly219del | rs121908027 | P | P/LP | 226329 | [6,20,23,29,30,31] | |
| 1 | 3 | 4 | c.658_663delCCCGAC | p.Pro220_Asp221del | rs1555803409 | LP | P | 440589 | [9] | |
| 0 | 5 | 4–6 | Del 5 kb incl. ex. 4–6 | LP | [38] | |||||
| 3 | 4 | 4 | c.666C > A | p.Cys222Ter | rs756613387 | 0.000004005 | P | P | 251364 | |
| 1 | 4 | 4 | c.672_686delCAAATCTGACGAGGA | p.Asp224_Glu228del | rs1555803439 | LP | LP | 441189 | [7] | |
| 0 | 5 | 4 | c.670_671insG | p.Asp224GlyfsTer4 | rs879254629 | P | P | 251372 | [6,20,30] | |
| 1 | 1 | 4 | c.670_678dup | p.Asp224_Ser226dup | LP | [7] | ||||
| 3 | 4 | 4 | c.682G > A | p.Glu228Lys | rs121908029 | 0.00001614 | LP | P/LP | 3691 | [30,37,39] |
| 0 | 6 | 4 | c.682G > T | p.Glu228Ter | rs121908029 | 0.00001074 | P | P/LP | 226333 | [6,20,30] |
| 1 | 4 | 4 | c.693C > A | p.Cys231Ter | rs121908035 | P | P/LP | 3730 | ||
| 0 | 5 | 5 | p.Glu240Ter * | LP | [30] | |||||
| 0 | 6 | 5 | p.Glu240Lys * | VUS | P/LP/VUS | 200920 | [30] | |||
| 1 | 4 | 5 | c.768C > A | p.Asp256Glu | rs879254671 | VUS | LP | 438322 | ||
| 0 | 6 | 5 | p.Cys261Phe * | VUS | LP | 3740 | [30] | |||
| 0 | 6 | 5 | c.796G > A | p.Asp266Asn | rs875989907 | 0.00001193 | LP | P/LP | 226334 | [34,36] |
| 3 | 4 | 5 | c.798T > A | p.Asp266Glu | rs139043155 | 0.00003535 | VUS | P/LP/VUS | 161287 | |
| 2 | 3 | 5 | c.810C > A | p.Cys270Ter | rs773328511 | P | P | 251465 | [6,20,30] | |
| 1 | 4 | 6 | c.825_826delCT | p.Cys276ArgfsTer24 | rs879254691 | P | P | 251478 | ||
| 2 | 4 | 6 | c.829G > A | p.Glu277Lys | rs148698650 | 0.0005056 | VUS/LB | P/VUS/LB/B | 183097 | |
| 0 | 6 | 6 | p.Glu288Lys | VUS | P/LP/VUS | 161268 | [30] | |||
| 2 | 4 | 6 | c.905G > T | p.Cys302Phe | rs879254715 | LP | P | 430768 | ||
| 0 | 6 | 6 | c.922G > A | p.Glu308Lys | rs879254721 | VUS | LP | 251528 | [9] | |
| 0 | 6 | 6 | c.925_931delCCCATCA | p.Pro309LysfsTer59 | rs387906304 | P | P | 3729 | [6,8,10,20,22,25,29,30] | |
| 1 | 1 | 6 | c.921T > A | p.Asp307Glu | VUS | |||||
| 1 | 1 | 6 | c.935A > G | p.Glu312Gly | rs1380197577 | 0.000003984 | VUS | |||
| 0 | 5 | 6 | c.939_940 + 3delCGGTG | p.Cys313AspfsTer17 | rs879254727 | P | P | 251536 | [6,20,30] | |
| 3 | 3 | 6i | c.940 + 3_940 + 6del | VUS | P/VUS | 869390 | [9] | |||
| 0 | 5 | 5i_6i | c.817 + 303_940 + 943del | p.Val273_Cys313del | VUS | [32] | ||||
| 1 | 4 | 6i | c.941-3C > G | VUS | ||||||
| 1 | 4 | 6i | c.941-2A > G | rs112366278 | P | P/LP | 251554 | |||
| 1 | 4 | 7 | c.949G > A | p.Glu317Lys | rs746834464 | 0.00005311 | VUS | P/LP | 251567 | |
| 2 | 1 | 7 | c.954C > G | p.Cys318Trp | VUS | |||||
| 1 | 1 | 7 | c.970G > T | p.Gly324Cys | VUS | |||||
| 1 | 4 | 7 | c.974G > A | p.Cys325Tyr | rs879254746 | VUS | P/LP | 251580 | ||
| 1 | 1 | 7 | c.976T > C | p.Ser326Pro | VUS | |||||
| 8 | 3 | 7 | c.986G > A | p.Cys329Tyr | rs761954844 | 0.00002479 | VUS | P/LP/LB | 226344 | [6,9,20,30,36] |
| 0 | 6 | 7 | c.1009G > A | p.Glu337Lys | rs539080792 | 0.0000935 | VUS | VUS | 523729 | [34,36] |
| 0 | 6 | 7 | rs755757866 | NA (G > A)\0.000007967 (G > T) | LP (G > A)\NA (G > T) | 251600 (G > A)\NA (G > T) | [34,36] | |||
| 1 | 4 | 7 | c.1027G > A | p.Gly343Ser | rs730882096 | 0.00002832 | VUS | P/LP/VUS | 183106 | |
| 1 | 4 | 7 | c.1048C > T | p.Arg350Ter | rs769737896 | 0.000007977 | P | P | 226342 | |
| 1 | 3 | 7 | c.1054T > C | p.Cys352Arg | rs879254769 | VUS | LP | 251618 | [9,34,36] | |
| 3 | 4 | 7i | c.1061-8T > C | rs72658861 | 0.005498 | VUS/LB | VUS/PB/B | 36451 | ||
| 3 | 1 | 8 | c.1123T > G | p.Tyr375Asp | VUS | |||||
| 1 | 1 | 8 | c.1129_1130insT | p.Cys377LeufsTer1 | LP | |||||
| 1 | 4 | 8 | c.1162del | p.His388ThrfsTer25 | P | P | 226348 | |||
| 1 | 1 | 8 | c.1168A > G | p.Lys390Glu | VUS | |||||
| 1 | 1 | 8 | c.1183delG | p.V395fs | LP | |||||
| 1 | 4 | 8i | c.1186 + 1G > T | rs730880131 | P | P | 180403 | |||
| 1 | 1 | 8i-10i | c.1186 + 568_1586 + 1067del | LP | ||||||
| 3 | 4 | 8i | c.1187-10G > A | rs765696008 | 0.00002798 | VUS | P/LP | 226349 | ||
| 1 | 1 | 8i | c.1187-7C > G | VUS | ||||||
| 28 | 3 | 9 | c.1202T > A | p.Leu401His | rs121908038 | VUS | P/LP | 3735 | [6,20,34,36] | |
| 2 | 4 | 9 | c.1217G > A | p.Arg406Gln | rs552422789 | 0.00001593 | VUS | P/LP/VUS | 228798 | |
| 5 | 3 | 9 | c.1222G > A | p.Glu408Lys | rs137943601 | 0.000007965 | LP | P/LP | 36453 | [10] |
| 0 | 5 | 9 | p.Arg410Gly * | VUS | [30] | |||||
| 0 | 5 | 9 | p.Met412Val * | VUS | [30] | |||||
| 4 | 3 | 9 | c.1246C > T | p.Arg416Trp | rs570942190 | 0.00002389 | LP | P/LP | 183110 | [9,30,34,36] |
| 1 | 3 | 9 | c.1252G > T | p.Glu418Ter | rs869320651 | P | P | 251755 | [20,26,30] | |
| 0 | 5 | 9 | p.Glu418Gly * | VUS | [30] | |||||
| 0 | 6 | 9 | c.1277T > C | p.Leu426Pro | rs879254851 | VUS | P/LB | 251763 | [25] | |
| 1 | 4 | 9 | c.1285G > A | p.Val429Met | rs28942078 | 0.00001194 | LP | P/LP | 3694 | |
| 0 | 5 | 9 | c.1291_1331del41 | p.Ala431Ter | rs879254854 | LP | [6,20,30] | |||
| 1 | 4 | 9 | c.1292C > T | p.Ala431Val | VUS | |||||
| 0 | 6 | 9 | p.Leu432Arg * | VUS | [30] | |||||
| 0 | 5 | 9 | p.Asp433Glu * | rs778309692 | 0.000003980 | VUS | [30] | |||
| 0 | 6 | 9 | p.Asp433His * | VUS | [30] | |||||
| 0 | 6 | 9 | p.Asp433Tyr * | VUS | [30] | |||||
| 0 | 6 | 9 | c.1302delG | p.Glu435MetfsTer15 | P | [6,20,30] | ||||
| 1 | 4 | 9 | c.1322T > A | p.Ile441Asn | rs879254862 | VUS | LP | 251782 | ||
| 2 | 2 | 9 | c.1327T > C | p.Trp443Arg | rs773566855 | 0.000003980 | LP | [9,10,30,33] | ||
| 0 | 6 | 9 | c.1328G > A | p.Trp443Ter | rs879254866 | P | P | 251789 | [6,20] | |
| 0 | 6 | 9 | c.1340C > G | p.Ser447Cys | rs879254870 | VUS | LP | 251797 | [8,10,25] | |
| 0 | 6 | 9i | c.1358 + 1G > A | rs775924858 | P | P/LP | 251802 | [6,20] | ||
| 1 | 1 | 10 | c.1444G > T | p.Asp482Tyr | rs139624145 | LP | LP | 251845 | [30] | |
| 1 | 2 | 10 | c.1465T > A | p.Tyr489Asn | VUS | [9] | ||||
| 1 | 4 | 10 | c.1471A > G | p.Thr491Ala | VUS | |||||
| 1 | 4 | 10 | c.1474G > A | p.Asp492Asn | rs373646964 | 0.00002386 | VUS | P/LP/VUS | 161285 | |
| 1 | 4 | 10 | c.1502C > T | p.Ala501Val | rs755667663 | 0.000007954 | LP | P/LP | 251874 | |
| 0 | 6 | 10 | c.1532T > C | p.Leu511Ser | rs879254932 | VUS | LP | 251887 | [8,25,33] | |
| 1 | 4 | 10 | c.1561G > A | p.Ala521Thr | rs879254940 | VUS | VUS | 251898 | ||
| 1 | 4 | 10 | c.1577C > A | p.Pro526His | rs879254944 | VUS | VUS | 496019 | ||
| 1 | 4 | 10i | c.1586 + 5G > A | rs781362878 | 0.00003189 | VUS | LP/VUS | 251909 | ||
| 1 | 1 | 11 | c.1618delG | p.Ala540ProfsTer8 | LP | |||||
| 1 | 3 | 11 | c.1633G > A | p.Gly545Arg | rs879254965 | LP | P/LP | 251942 | [9] | |
| 0 | 6 | 11 | p.Gly549Asp * | rs28941776 | 0.00002386 | VUS | P/LP | 3698 | [30] | |
| 0 | 5 | 11 | c.1655_1672del | LP | [34,36] | |||||
| 1 | 4 | 11 | c.1672G > T | p.Glu558Ter | rs879254980 | P | P | 251964 | ||
| 0 | 5 | 11 | p.Glu558Lys * | VUS | [30] | |||||
| 0 | 5 | 11 | c.1686_1693delGCCCAATGinsT | p.Trp562CysfsTer5 | rs879254984 | P | P | 251968 | [8,25,33] | |
| 1 | 1 | 11 | c.1693G > A | p.Gly565Ser | rs1344561983 | 0.000003978 | VUS | |||
| 4 | 1 | 11 | c.1696A > T | p.Ile566Phe | VUS | |||||
| 1 | 1 | 11 | c.1705 + 3delA | VUS | ||||||
| 0 | 6 | 11 | p.Leu568Va l * | VUS | [30] | |||||
| 1 | 4 | 12 | c.1706-10G > A | rs17248882 | 0.002220 | VUS/LB | VUS/LB/B | 226368 | ||
| 1 | 4 | 12 | c.1708_1710delCTC | p.Leu571del | rs772492150 | 0.000007953 | VUS | |||
| 1 | 4 | 12 | c.1729T > C | p.Trp577Arg | rs879255000 | LP | P/LP | 252001 | ||
| 0 | 5 | 12 | c.1741A > C | p.Lys581Gln | VUS | [9] | ||||
| 2 | 4 | 12 | c.1747C > T | p.His583Tyr | rs730882109 | 0.0001025 | VUS | P/LP | 200921 | |
| 1 | 4 | 12 | c.1750T > C | p.Ser584Pro | rs879255010 | VUS | LP/VUS | 252015 | ||
| 2 | 2 | 12 | c.1756T > C | p.Ser586Pro | VUS | [9] | ||||
| 1 | 1 | 12 | c.1774G > T | p.Gly592Trp | VUS | |||||
| 29 | 3 | 12 | c.1775G > A | p.Gly592Glu | rs137929307 | 0.00005656 | LP | P/LP | 161271 | [9,20,21,30] |
| 2 | 4 | 12 | c.1784G > A | p.Arg595Gln | rs201102492 | 0.00003889 | VUS | P/LP/VUS | 183126 | |
| 0 | 5 | 12 | p.Leu605Val * | VUS | [30] | |||||
| 0 | 5 | 12 | p.Leu605Arg * | VUS | [30] | |||||
| 0 | 5 | 12 | p.Ala612Gly * | VUS | [30] | |||||
| 1 | 2 | 12i | c.1846-3T > G | VUS | [9] | |||||
| 0 | 5 | 13 | c.1855–1856insA | p.Phe619TyrfsTer26 | rs879255053 | P | P | 252082 | [6,20,30] | |
| 0 | 6 | 13 | c.1859G > C | p.Trp620Ser | VUS | [33] | ||||
| 1 | 3 | 13 | c.1864G > A | p.Asp622Asn | rs879255059 | LP | LP | 252092 | [6,20,30] | |
| 1 | 1 | 13 | c.1898delG | p.Arg633fs | VUS | |||||
| 2 | 4 | 13 | c.1898G > A | p.Arg633His | rs754536745 | 0.00002121 | VUS | P/LP/VUS | 226380 | |
| 0 | 5 | 13 | c.1936C > A | p.Leu646Ile | rs779940524 | 0.000003977 | VUS | LP | 252118 | [8,10,25] |
| 1 | 4 | 13 | c.1945C > T | p.Pro649Ser | rs879255080 | VUS | LP | 252121 | ||
| 3 | 4 | 13 | c.1955T > C | p.Met652Thr | rs875989936 | 0.000003977 | VUS | LP/VUS | 226382 | |
| 1 | 4 | 13 | c.1966C > A | p.His656Asn | rs762815611 | LP | B/LP/VUS | 252136 | ||
| 1 | 4 | 14 | c.1998G > A | p.Trp666Ter | rs752935814 | P | P | 252161 | ||
| 0 | 5 | 14 | c.1999T > A | p.Cys667Ser | rs150021927 | VUS | LP | 252162 | [6,20,30] | |
| 1 | 4 | 14 | c.2001_2002delTG | p.Cys667_Glu668delinsTer | rs1600743301 | LP | P | 630543 | ||
| 1 | 4 | 14 | c.2043C > A | p.Cys681Ter | rs121908031 | 0.000007959 | P | P/LP | 3699 | |
| 3 | 4 | 14 | c.2089G > C | p.Ala697Pro | rs776217028 | VUS | LP | 252213 | ||
| 0 | 6 | 14i–16i | c.2141-966_2390-330del | p.Glu714_Ile796del | LP | [9] | ||||
| 1 | 1 | 14i-15i | c.2141-799_2311 + 689del | LP | ||||||
| 1 | 1 | 15 | c.2189A > C | p.Lys730Thr | VUS | |||||
| 0 | 5 | 15 | c.2191delG | p.Val731SerfsTer6 | rs879255161 | P | P | 252253 | [8,25,29,33] | |
| 0 | 6 | 15 | c.2215C > T | p.Gln739Ter | rs370018159 | P | P/LP | 252258 | [9] | |
| 1 | 4 | 15 | c.2230C > T | p.Arg744Ter | rs200793488 | 0.000003979 | LP | P | 430795 | |
| 0 | 6 | 15 | c.2231G > A | p.Arg744Gln | rs137853963 | 0.0008030 | VUS | LP/VUS/LB/B | 68104 | [20,21] |
| 1 | 1 | 15 | c.2244_2245insG | p.Thr749AspfsTer33 | VUS | |||||
| 1 | 1 | 15i-17i | c.2312-2107_2547 + 620del | LP | ||||||
| 1 | 1 | 16 | c.2324T > C | p.Val775Ala | rs780300776 | 0.00002121 | VUS | LB/VUS | 440691 | |
| 0 | 5 | 16 | c.2326G > T | p.Ala776Ser | VUS | [32] | ||||
| 1 | 1 | 16 | c.2327C > T | p.Ala776Val | VUS | |||||
| 1 | 4 | 16 | c.2347A > C | p.Lys783Gln | rs769318961 | 0.000007954 | VUS | |||
| 1 | 1 | 16 | c.2374del | p.Ile792LeufsTer137 | LP | |||||
| 3 | 3 | 16 | c.2389G > A | p.Val797Met | rs750518671 | 0.000007957 | VUS | P/LP/VUS | 226393 | [6,20,30] |
| 1 | 4 | 16 | c.2389G > C | p.Val797Leu | rs750518671 | VUS | VUS | 565983 | ||
| 1 | 4 | 16 | c.2389 + 2T > G | rs879255188 | P | LP | 252302 | |||
| 4 | 3 | 16i | c.2389 + 5G > C | rs879255191 | VUS | VUS | 661713 | [9] | ||
| 2 | 4 | 16 | c.2389 + 5G > A | rs879255191 | VUS | P/LB | 252306 | |||
| 1 | 1 | 17 | c.2390T > A | p.Val797Glu | VUS | |||||
| 2 | 4 | 17 | c.2416_2417insG | p.Val806fs | P | P/LP | 252330 | |||
| 0 | 6 | 17 | c.2416dupG | p.Val806GlyfsTer11 | rs773618064 | P | P/LP | 252330 | [9] | |
| 1 | 1 | 17 | c.2429G > A | p.Trp810Ter | LP | |||||
| 1 | 4 | 17 | c.2448G > C | p.Lys816Asn | rs1399689294 | 0.00003186 | VUS | LP/VUS | 440698 | |
| 1 | 4 | 17 | c.2473A > G | p.Asn825Asp | rs879255215 | VUS | LP | 252340 | ||
| 1 | 3 | 17 | c.2479G > A | p.Val827Ile | rs137853964 | 0.0009193 | VUS/LB | LP/VUS/LB/B | 36462 | [6,20,34,36] |
| 1 | 4 | 17 | c.2531G > A | p.Gly844Asp | rs121908037 | VUS | LP | 3734 |
1 Only for variants found in this study a number of index patients is given. Variants from systematic review are labelled with “0”. 2 1–described only in this study, 2–described in this study and in other studies in Russia, 3–described in this study, in other studies in Russia and other countries, 4–described in this study and other countries, 5–did not occur in this study, described in other studies in Russia, 6–did not occur in this study, described in other studies in Russia and other countries. * No data on coding sequence alteration in reference.
Table A2.
List of the PSCK9 variants described in Russian patients.
| Number of Index Patients 1 | Exon | DNA Change | Protein Change | dbSNP ID | gnomAD MAF (v. 2.1.1) | ACMG Interpretation | ClinVar Interpretation | ClinVar ID | References |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | c.100G > A | p.Glu34Lys | rs371030381 | 0.00001626 | VUCS | VUS | 536202 | |
| 1 | 1 | c.142G > A | p.Glu48Lys | rs1278890129 | 0.00002190 | VUCS | P/VUS | 440707 | |
| 1 | 1 | c.151G > C | p.Gly51Arg | VUCS | |||||
| 1 | 2 | c.382G > A | p.Gly128Ser | rs766314770 | 0.00003978 | VUCS | |||
| 1 | 3 | c.411G > T | p.Leu137Phe | VUCS | |||||
| 1 | 3 | c.520C > T | p.Pro174Ser | rs533273863 | 0.00007089 | VUCS | VUS | 496561 | |
| 1 | 3 | c.523 + 2T > G | LP | ||||||
| 1 | 5 | c.709C > T | p.Arg237Trp | rs148195424 | 0.0006952 | VUCS | VUS/LB | 265933 | |
| 1 | 5 | c.751C > T | p.Arg251Cys | rs778900671 | 0.00002009 | VUCS | |||
| 1 | 7 | c.1046G > A | p.Gly349Glu | VUCS | |||||
| 1 | 7 | c.1069C > T | p.Arg357Cys | rs148562777 | 0.0001450 | LP | VUS | 575758 | |
| 1 | 7 | c.1070G > A | p.Arg357His | rs370507566 | 0.00003978 | VUCS | VUS | 403288 | |
| 1 | 7 | c.1120G > A | p.Asp374Asn | rs137852912 | 0.00007079 | LP | |||
| 3 | 9 | c.1399C > G | p.Pro467Ala | rs772677312 | 0.00002829 | LP | LP/VUS | 265944 | |
| 1 | 9 | c.1483C > T | p.Arg495Trp | rs758999339 | 0.00001607 | VUCS | |||
| 2 | 9 | c.1487G > A | p.Arg496Gln | rs139669564 | 0.0002363 | VUCS | VUS/LB | 438338 | |
| 1 | 10 | c.1621C > T | p.Pro541Ser | rs369996097 | 0.00001056 | VUCS | |||
| 0 | 11 | c.1834G > A | p.Glu612Lys | VUCS | [32] | ||||
| 1 | 12 | c.1903T > C | p.Cys635Arg | VUCS | |||||
| 1 | 12 | c.1939G > C | p.Ala647Pro | VUCS | |||||
| 1 | 12 | c.2002A > G | p.Ser668Gly | rs775077080 | 0.00004790 | VUCS | VUS | 297707 | |
| 1 | 12 | c.2004C > A | p.Ser668Arg | rs762298323 | 0.00002397 | VUCS | VUS | 403291 | |
| 1 | c.*415G > A | VUCS | [32] |
1 Only for variants found in this study a number of index patients is given. Variants from systematic review are labelled with “0”.
Table A3.
List of the APOB variants described in Russian patients.
| Number of Index Patients 1 | Exon | DNA Change | Protein Change | dbSNP ID | gnomAD MAF (v. 2.1.1) | ACMG Interpretation | ClinVar Interpretation | Clinvar ID | References |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 26 | c.4298C > T | p.Ser1433Leu | rs200708197 | 7.583 × 105 | VUCS | VUS | 630306 | |
| 1 | 26 | c.4709T > C | p.Leu1570Ser | VUCS | |||||
| 1 | 26 | c.7057C > T | p.Gln2353Ter | LP | |||||
| 1 | 26 | c.10385A > G | p.Tyr3462Cys | VUCS | |||||
| 1 | 26 | c.10579C > T | p.Arg3527Trp | rs144467873 | 0.0001595 | LP | P/LP | 40223 | |
| 23 | 26 | c.10580G > A | p.Arg3527Gln | rs5742904 | 0.0002942 | LP | P/LP | 17890 | [9,35,36] |
| 1 | 26 | c.10672C > T | p.Arg3558Cys | VUCS | |||||
| 3 | 26 | c.10708C > T | p.His3570Tyr | rs201736972 | VUCS | ||||
| 1 | 26 | c.11477C > T | p.Thr3826Met | rs61744153 | 0.001592 | VUCS | LP/VUS/LB/B | 237735 | |
| 1 | 28 | c.11911G > A | p.Glu3971Lys | VUCS | |||||
| 0 | 28 | c.12005C > T | p.Ala4002Val | rs369364335 | 1.195 × 105 | VUCS | VUS | 898076 | [32] |
| 1 | 29 | c.12739C > T | p.Gln4247Ter | rs907126709 | LP | VUS | 544074 | ||
| 1 | 29 | c.13175G > A | p.Ser4392Asn | VUCS | |||||
| 1 | 29 | c.13480_13482del | p.Gln4494del | rs756545438 | VUCS | LP/VUS | 265896 |
1 Only for variants found in this study a number of index patients is given. Variants from systematic review are labelled with “0”.
Table A4.
Patients with multiple variants.
| Patient Numbers | Phenotype | Variant 1 (Gene/Variant/Zygosity) |
Variant 2 (Gene/Variant/Zygosity) |
Cis/Trans Position (Evidence) |
|---|---|---|---|---|
| 423 | HoFH | LDLR:p.Gly592Glu (het) | LDLR:p.Glu418Ter (het) | Trans (genetic test of relatives) |
| 474 | Severe HetFH | LDLR:p.Gly592Glu (het) | LDLR:p.Ala771Glufs*9 (het) | Trans (long read sequencing) |
| 166 | HoFH | LDLR:c.941-3C > G (het) | LDLR:p.Cys329Tyr (het) | Trans (genetic test of relatives) |
| 722 | HoFH | LDLR:c.940 + 3_940 + 6del (het) | LDLR:p.Arg416Trp (het) | Unknown |
| 668 | HoFH | LDLR:p.Cys329Tyr (het) | LDLR:p.Gly592Glu (het) | Trans (genetic test of relatives) |
| 675 | HoFH | LDLR:p.Trp577Arg (hom) | ||
| 355 | HoFH | LDLR:p.Ile441Asn (het) | LDLR:p.Ile792LeufsTer137 (het) | Unknown |
| 687 | HetFH | LDLR:p.Leu401His (het) | PCSK9:p.Arg357Cys (het) | |
| 211 | Severe HetFH | LDLR:p.Cys329Tyr (het) | LDLR:p.Gly592Glu (het) | Cis (genetic test of relatives) |
| 336 | HetFH | LDLR:p.Lys390Glu (het) | PCSK9:p.Glu34Lys (het) | |
| R-6 | HetFH | LDLR:p.Val429Met (het) | APOB:p.Glu3971Lys (het) | |
| R-35 | HetFH | LDLR:p.Gly592Glu (het) | APOB:p.Arg3527Gln (het) | |
| R-83 | HetFH | LDLR:p.Gly592Glu (het) | APOB:p.Ser4392Asn (het) + PCSK9:p.Gly51Arg (het) | |
| R-115 | HetFH | APOB:p.Arg3527Gln (het) | PCSK9:p.Pro174Ser (het) | |
| 969 | HetFH | APOB:p.Arg3527Gln (het) | APOB:p.Gln4494del (het) | Unknown |
Author Contributions
Conceptualization, A.M., A.K. and E.Z.; methodology, A.M., A.E., A.K., E.Z. and A.P.; software, E.Z., A.P., A.Z., V.R., A.B. (Anna Bukaeva) and S.M.; validation, A.K., E.S. (Evgeniia Sotnikova), M.D., O.K. and O.S.; investigation, A.M., A.E., A.K., E.Z., E.S. (Evgeniia Sotnikova), A.P., A.Z., P.M., T.R., A.B. (Anastasia Blokhina), M.D., Z.K., A.B. (Anna Bukaeva), A.L., V.M. (Valeriya Mikova), E.S. (Ekaterina Snigir), A.A. and D.K.; resources, M.P., S.M. and V.M. (Valentin Makarov); data curation, A.M., A.E., A.K., A.B. (Anna Bukaeva) and E.S. (Ekaterina Snigir); writing, A.M., A.K. and E.Z.; writing—review and editing, A.E., A.K., V.R. and A.B. (Anna Bukaeva); visualization, A.K. and A.B. (Anna Bukaeva); supervision, V.K., S.B. and O.D.; project administration, A.M.; funding acquisition, S.B., S.Y. and O.D. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by State assignment No AAAA-A18-118041790111-0. V.R. acknowledges support by the RFBR and DFG research project No 20-54-12008.
Institutional Review Board Statement
This study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the Ethics Committees in clinical cardiology of the National Medical Research Center for Cardiology (a statement on ethics approval No.144, 27 April 2009) and of the National Research Center for Therapy and Preventive Medicine (a statement on ethics approval №04-04/17, 6 June 2017).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Berberich A.J., Hegele R.A. The complex molecular genetics of familial hypercholesterolaemia. Nat. Rev. Cardiol. 2019;16:9–20. doi: 10.1038/s41569-018-0052-6. [DOI] [PubMed] [Google Scholar]
- 2.Hu P., Dharmayat K.I., Stevens C.A.T., Sharabiani M.T.A., Jones R.S., Watts G.F., Genest J., Ray K.K., Vallejo-Vaz A.J. Prevalence of familial hypercholesterolemia among the general population and patients with atherosclerotic cardiovascular disease. Circulation. 2020;141:1742–1759. doi: 10.1161/CIRCULATIONAHA.119.044795. [DOI] [PubMed] [Google Scholar]
- 3.Beheshti S.O., Madsen C.M., Varbo A., Nordestgaard B.G. Worldwide prevalence of familial hypercholesterolemia: Meta-analyses of 11 million subjects. J. Am. Coll. Cardiol. 2020;75:2553–2566. doi: 10.1016/j.jacc.2020.03.057. [DOI] [PubMed] [Google Scholar]
- 4.Cuchel M., Bruckert E., Ginsberg H.N., Raal F.J., Santos R.D., Hegele R.A., Kuivenhoven J.A., Nordestgaard B.G., Descamps O.S., Steinhagen-Thiessen E. Homozygous familial hypercholesterolaemia: New insights and guidance for clinicians to improve detection and clinical management. A position paper from the Consensus Panel on Familial Hypercholesterolaemia of the European Atherosclerosis Society. Eur. Heart J. 2014;35:2146–2157. doi: 10.1093/eurheartj/ehu274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iacocca M.A., Chora J.R., Carrié A., Freiberger T., Leigh S.E., Defesche J.C., Kurtz C.L., DiStefano M.T., Santos R.D., Humphries S.E., et al. ClinVar database of global familial hypercholesterolemia-associated DNA variants. Hum. Mutat. 2018;39:1631–1640. doi: 10.1002/humu.23634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zakharova F.M., Damgaard D., Mandelshtam M.Y., Golubkov V.I., Nissen P.H., Nilsen G.G., Stenderup A., Lipovetsky B.M., Konstantinov V.O., Denisenko A.D., et al. Familial hypercholesterolemia in St.-Petersburg: The known and novel mutations found in the low density lipoprotein receptor gene in Russia. BMC Med. Genet. 2005;6:6. doi: 10.1186/1471-2350-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meshkov A.N., Malyshev P.P., Kukharchuk V.V. Familial hypercholesterolemia in Russia: Genetic and phenotypic characteristics. Ter. Arkhiv. 2009;81:23–28. (In Russian) [PubMed] [Google Scholar]
- 8.Komarova T.Y., Korneva V.A., Kuznetsova T.Y., Golovina A.S., Vasilyev V.B., Mandelshtam M.Y. Familial hypercholesterolemia mutations in Petrozavodsk: No similarity to St. Petersburg mutation spectrum. BMC Med. Genet. 2013;14:128. doi: 10.1186/1471-2350-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Semenova A.E., Sergienko I.V., García-Giustiniani D., Monserrat L., Popova A.B., Nozadze D.N., Ezhov M.V. Verification of underlying genetic cause in a cohort of Russian patients with familial hypercholesterolemia using targeted next generation sequencing. J. Cardiovasc. Dev. Dis. 2020;7:16. doi: 10.3390/jcdd7020016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Korneva V.A., Kuznetsova T.Y., Bogoslovskaya T.Y., Polyakov D.S., Vasilyev V.B., Orlov A.V., Mandelshtam M.Y. Cholesterol levels in genetically determined familial hypercholesterolaemia in Russian Karelia. Cholesterol. 2017;2017:9375818. doi: 10.1155/2017/9375818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Civeira F. Guidelines for the diagnosis and management of heterozygous familial hypercholesterolemia. Atherosclerosis. 2004;173:55–68. doi: 10.1016/j.atherosclerosis.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 12.Lin M.F., Rodeh O., Penn J., Bai X., Reid J.G., Krasheninina O., Salerno W.J. GLnexus: Joint variant calling for large cohort sequencing. bioRxiv. 2018:343970. doi: 10.1101/343970. [DOI] [Google Scholar]
- 13.Smoove. [(accessed on 10 December 2020)]; Available online: https://github.com/brentp/smoove.
- 14.McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R.S., Thormann A., Flicek P., Cunningham F. The Ensembl variant effect predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gardner E.J., Lam V.K., Harris D.N., Chuang N.T., Scott E.C., Pittard W.S., Mills R.E., 1000 Genomes Project Consortium. Devine S.E. The Mobile Element Locator Tool (MELT): Population-scale mobile element discovery and biology. Genome Res. 2017;27:1916–1929. doi: 10.1101/gr.218032.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ou J., Zhu L.J. trackViewer: A Bioconductor package for interactive and integrative visualization of multi-omics data. Nat. Methods. 2019;16:453–454. doi: 10.1038/s41592-019-0430-y. [DOI] [PubMed] [Google Scholar]
- 18.Chora J.R., Medeiros A.M., Alves A.C., Bourbon M. Analysis of publicly available LDLR, APOB, and PCSK9 variants associated with familial hypercholesterolemia: Application of ACMG guidelines and implications for familial hypercholesterolemia diagnosis. Genet. Med. 2018;20:591–598. doi: 10.1038/gim.2017.151. [DOI] [PubMed] [Google Scholar]
- 19.Leigh S., Futema M., Whittall R., Taylor-Beadling A., Williams M., den Dunnen J.T., Humphries S.E. The UCL low-density lipoprotein receptor gene variant database: Pathogenicity update. J. Med. Genet. 2017;54:217–223. doi: 10.1136/jmedgenet-2016-104054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zakharova F.M., Tatishcheva Y.A., Golubkov V.I., Lipovetsky B.M., Konstantinov V.O., Denisenko A.D., Faergeman O., Vasilyev V.B., Mandelshtam M.Y. Familial hypercholesterolemia in St. Petersburg: Diversity of mutations argues against a strong founder effect. Russ. J. Genet. 2007;43:1046–1052. doi: 10.1134/S1022795407090116. [DOI] [PubMed] [Google Scholar]
- 21.Zakharova F.M., Golubkov V.I., Mandelshtam M.J., Lipovetskii B.M., Gaitskhoki V.S. Identification of novel missense mutation G571E, novel silent mutation H229H, nonsense mutation C74X, and four single nucleotide polymorphisms in the Low-Density Lipoprotein Receptor gene in patients with familial hypercholesterolemia from St. Petersburg. Russ. J. Bioorganic Chem. 2001;27:349–351. doi: 10.1023/A:1012300632671. [DOI] [PubMed] [Google Scholar]
- 22.Komarova T.Y., Golovina A.S., Grudinina N.A., Zakharova F.M., Korneva V.A., Lipovetskii B.M., Serebrenitskaya M.P., Konstantinov V.O., Vasilyev V.B., Mandelshtam M.Y. “Finnish” mutations in LDL Receptor gene: A rare cause of familial hypercholesterolemia in St. Petersburg and Petrozavodsk. Bull. Exp. Biol. Med. 2013;155:380–383. doi: 10.1007/s10517-013-2159-4. [DOI] [PubMed] [Google Scholar]
- 23.Durst R., Colombo R., Shpitzen S., Avi L.B., Friedlander Y., Wexler R., Raal F.J., Marais D.A., Defesche J.C., Mandelshtam M.Y., et al. Recent origin and spread of a common Lithuanian mutation, G197del LDLR, causing familial hypercholesterolemia: Positive selection is not always necessary to account for disease incidence among Ashkenazi Jews. Am. J. Hum. Genet. 2001;68:1172–1188. doi: 10.1086/320123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chakir K., Skobeleva N.A., Shevtsov S.P., Konstantinov V.O., Denisenko A.D., Schwartz E.I. Two Novel Slavic Point Mutations in the Low-Density Lipoprotein Receptor Gene in Patients with Familial Hypercholesterolemia from St. Petersburg, Russia. Mol. Genet. Metab. 1998;63:31–34. doi: 10.1006/mgme.1997.2614. [DOI] [PubMed] [Google Scholar]
- 25.Korneva V.A., Bogoslovskaia T.I., Kuznetsova T.I., Mandel’shtam M.I., Vasil’ev V.B. Familial hypercholesterolemia due to a new mutation in the low density lipoprotein receptor gene. Klin. Med. 2014;92:49–53. (In Russian) [PubMed] [Google Scholar]
- 26.Chakir K., Ju M.M., Shevtsov S.P., Golubkov V.I., Skobeleva N.A., Shur Y.A., Zakharova F.M., Lipovetskyi B.M., Konstantinov V.O., Denisenko A.D., et al. Two Novel Low-Density Lipoprotein Receptor Gene Mutations (E397X and 347delGCC) in St. Petersburg Familial Hypercholesterolemia. Mol. Genet. Metab. 1998;65:311–314. doi: 10.1006/mgme.1998.2762. [DOI] [PubMed] [Google Scholar]
- 27.Korneva V.A., Kuznetsova T.I., Komarova T.I., Golovina A.S., Mandel’shtam M.I., Konstantinov V.O., Vasil’ev V.B. A case of familial hypercholesterolemia caused by a novel mutation p. FsS65:D129X of human low density lipoprotein receptor gene. Kardiologiia. 2013;53:50–54. (In Russian) [PubMed] [Google Scholar]
- 28.Tatishcheva Y.A., Mandelshtam M.Y., Golubkov V.I., Lipovetsky B.M., Gaitskhoki V.S. Four New Mutations and Two Polymorphic Variants of the Low-Density Lipoprotein Receptor Gene in Familial Hypercholesterolemia Patients from St. Petersburg. Russ. J. Genet. 2001;37:1082–1087. doi: 10.1023/A:1011973817437. [DOI] [Google Scholar]
- 29.Komarova T.Y., Golovina A.S., Grudinina N.A., Zakharova F.M., Korneva V.A., Lipovetsky B.M., Serebrenitskaya M.P., Konstantinov V.O., Vasilyev V.B., Mandelshtam M.Y. New mutations in low-density lipoprotein receptor gene in familial hypercholesterolemia patients from Petrozavodsk. Russ. J. Genet. 2013;49:673–676. doi: 10.1134/S1022795413040066. [DOI] [PubMed] [Google Scholar]
- 30.Voevoda M.I., Kulikov I.V., Shakhtshneider E.V., Maksimov V.N., Pilipenko I.V., Tereschenko I.P., Kobzev V.F., Romaschenko A.G., Nikitin Y.P. The spectrum of mutations in the low-density lipoprotein receptor gene in the Russian population. Russ. J. Genet. 2008;44:1191–1194. doi: 10.1134/S1022795408100074. [DOI] [PubMed] [Google Scholar]
- 31.Mandelshtam M., Chakir K., Shevtsov S., Golubkov V., Skobeleva N., Lipovetsky B., Konstantinov V., Denisenko A., Gaitskhoki V., Schwartz E. Prevalence of Lithuanian mutation among St. Petersburg Jews with familial hypercholesterolemia. Hum. Mutat. 1998;12:255–258. doi: 10.1002/(SICI)1098-1004(1998)12:4<255::AID-HUMU6>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 32.Averkova A.O., Brazhnik V.A., Speshilov G.I., Rogozhina A.A., Koroleva O.S., Zubova E.A., Galyavich A.S., Tereshenko S.N., Boyeva O.I., Zateyshchikov D.A. Targeted sequencing in patients with clinically diagnosed hereditary lipid metabolism disorder and acute coronary syndrome. Bull. RSMU. 2018;5:80–85. doi: 10.24075/brsmu.2018.061. [DOI] [Google Scholar]
- 33.Korneva V.A., Kuznetsova T.Y., Murtazina R.Z., Didio A.V., Bogoslovskaya T.Y., Mandelshtam M.Y., Vasilyev V.B. The Familial Hypercholesterolemia caused by a novel human Low Density Lipoprotein Receptor Gene mutation c.1327 T>C (p.W433R) Kardiologiia. 2017;57:12–16. (In Russian) [PubMed] [Google Scholar]
- 34.Shakhtshneider E., Orlov P., Ivanoshchuk D., Makarenkova K., Ragino Y., Voevoda M. Analysis of the LDLR gene variability in patients with Familial Hypercholesterolemia in Russia using targeted high throughput resequencing. Atherosclerosis. 2017;263:e227. doi: 10.1016/j.atherosclerosis.2017.06.740. [DOI] [Google Scholar]
- 35.Pogoda T., Metelskaya V., Perova N., Limborska S. Detection of the apoB-3500 mutation in a Russian family with coronary heart disease. Hum. Hered. 1998;48:291–292. doi: 10.1159/000022819. [DOI] [PubMed] [Google Scholar]
- 36.Shakhtshneider E., Ivanoshchuk D., Orlov P., Timoshchenko O., Voevoda M. Analysis of the LDLR, APOB, PCSK9 and LDLRAP1 genes variability in patients with Familial Hypercholesterolemia in West Siberia using targeted high throughput resequencing. Atherosclerosis. 2019;287:e285. doi: 10.1016/j.atherosclerosis.2019.06.883. [DOI] [Google Scholar]
- 37.Meshkov A.N., Stambol’skiĭ D.V., Krapivner S.R., Bochkov V.N., Kukharchuk V.V., Malyshev P.P. Low density lipoprotein receptor gene mutations in patients with clinical diagnosis of familial hypercholesterolemia. Kardiologiia. 2004;44:58–61. (In Russian) [PubMed] [Google Scholar]
- 38.Mandelshtam M.J., Lipovetskyi B.M., Schwartzman A.L., Gaitskhoki V.S. A novel deletion in the low-density lipoprotein receptor gene in a patient with familial hypercholesterolemia from Petersburg. Hum. Mutat. 1993;2:256–260. doi: 10.1002/humu.1380020404. [DOI] [PubMed] [Google Scholar]
- 39.Meshkov A.N., Stambol’skiĭ D.V., Nikitina L.A., Abdullaev S.M., Bochkov V.N., Tkachuk V.A., Kukharchuk V.V., Malyshev P.P. Genetic factors of risk of ischemic heart disease development in patients with familial hypercholesterolemia. Kardiologiia. 2005;45:10–14. (In Russian) [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.